Abstract
To cause infection, a virus enters a host cell, replicates, and assembles, with the resulting new viral progeny typically released into the extracellular environment to initiate a new infection round. Virus entry, replication, and assembly are dynamic and coordinated processes that require precise interactions with host components, often within and surrounding a defined subcellular compartment. Accumulating evidence pinpoints the endoplasmic reticulum (ER) as a crucial organelle supporting viral entry, replication, and assembly. This review focuses on the molecular mechanism by which different viruses co-opt the ER to accomplish these crucial infection steps. Certain bacterial toxins also hijack the ER for entry. An interdisciplinary approach, using rigorous biochemical and cell biological assays coupled with advanced microscopy strategies, will push to the next level our understanding of the virus-ER interaction during infection.
An ideal conduit for certain viruses to enter the cytosol is the ER-to-cytosol retro-translocation machinery in the ER. Furthermore, budding at the ER membrane plays a role during viral replication and assembly.
To trigger infection, a virus binds to receptors on a host cell’s plasma membrane. This interaction induces virus internalization, and initiates a complex journey of the viral particle into the host’s interior that leads to either nonproductive or productive infection (Mercer et al. 2010). In nonproductive infection, the virus may be targeted to and trapped in organelles unsupportive of viral membrane fusion or penetration, events which normally enable the viral nucleic acid access to the host cytosol or nucleus. Alternatively, the virus could be transported to a degradative intracellular compartment in which it is destroyed. In contrast, for productive infection, a viral particle must avoid these nonproductive routes and traffic along a pathway that allows it to reach the appropriate replication and assembly site. Successful infection is usually completed when the newly assembled particle is released into the extracellular milieu, in which it can promote another infection round. Thus, the ability to co-opt a host cell entry pathway leading to efficient replication and assembly ultimately dictates the fate of an incoming virus.
For proper entry, replication, and assembly, viruses often rely on the complex membranous network surrounding and residing within the host cell, such as the plasma, endolysosomal, and endoplasmic reticulum (ER) membranes. Selecting the suitable membrane system requires several considerations. To support entry, the membranous system must possess triggers capable of inducing the necessary conformational changes that facilitate viral membrane fusion or penetration (Inoue et al. 2011). Examples of cellular triggers include receptors, low pH, proteases, chaperones, and reductases. Additionally, because viral replication and assembly often occur in the context of virus-induced membranous structures derived from host membranes, the membranous network of choice should accommodate these remodeling reactions (Miller and Krijnse-Locker 2008). Moreover, as a virus commonly manipulates the host immune system to sustain infection, a membrane’s ability to provide the virus with such an opportunity would offer tremendous advantages during the infection course (Takeuchi and Akira 2009).
A wealth of data implicates the endoplasmic reticulum (ER), one of the most elaborate membranous networks in a cell (Shibata et al. 2009), as the organelle many viruses exploit during infection. This review focuses on how viruses co-opt the ER to enter, replicate, and assemble in the target cell. We will also draw parallels from the mechanisms by which bacterial toxins use the ER for entry. Together, these insights should unveil clues regarding why many viruses select the ER during infection.
THE ER
Structurally, the ER is a continuous membranous system consisting of the nuclear envelope, and peripheral sheets and tubules emanating from it (Voeltz et al. 2002). Recent studies suggest the membrane sheets correspond to the rough ER whereas the tubules represent the smooth ER (Voeltz et al. 2006; Shibata et al. 2010). Functionally, the rough ER is responsible for translating secretory and transmembrane proteins, whereas the smooth ER possesses specialized roles including lipid and glycogen metabolism (reviewed in Hopkins 1978). Once a newly synthesized protein is translated and folded properly in the ER lumen, it exits the ER to reach the Golgi apparatus via membrane budding mediated by the COPII complex. By contrast, should a protein misfold in the ER, an endogenous ER quality control system called ER-associated degradation (ERAD) alleviates the build-up of misfolded ER proteins (Brodsky and Skach 2011; Smith et al. 2011). To do so, a network of ER factors recognizes and retro-translocates the misfolded protein to the cytosol. On reaching the cytosol, another cohort of cytosolic factors engage and ubiquitinate the substrate, targeting it to the proteasome for degradation.
Strikingly, some of ER’s general properties are beneficial to viruses. For example, as the ER-to-cytosol retro-translocation machinery is an inherent apparatus in the ER, it represents an ideal conduit for certain viruses and bacterial toxins to enter the cytosol.
Additionally, ER membrane’s ability to undergo constant budding reactions plays a crucial role during viral replication and assembly when viruses deform and rearrange the ER membrane to generate ER-derived structures used to support these processes.
POLYOMAVIRUS CO-OPTS THE ER DURING ENTRY
A decisive virus entry step necessitates the incoming viral particle to breach a host cell’s membrane barrier (Fig. 1). This barrier could be the plasma (Fig. 1, pathway 1), endolysosome (Fig. 1, pathway 2), or ER (Fig. 1, pathway 3) membrane. Although many viruses such as influenza virus, HIV, human poliovirus (PV), and adenovirus cross the plasma or endolysosome membrane (Fig. 1, pathways 1 and 2), only polyomavirus (Py) family members penetrate the ER membrane during entry (Fig. 1, pathway 3).
Figure 1.
Virus entry across different cellular membranes. To enter cells, viruses penetrate the plasma membrane (PM) (pathway 1), endolysosome membrane (pathway 2), or ER membrane (pathway 3) to reach the cytosol. Whereas many viruses including influenza virus, HIV, and PV breach the plasma or endolysosome membranes, only Py transports across the ER membrane during cell entry.
As Py lacks a surrounding lipid bilayer that defines enveloped viruses, it is classified as a nonenveloped virus. Prominent Py family members include the murine Py (mPy), simian virus 40 (SV40), and the human BK (BKV), JC (JCV), and Merkel Cell (MCPy) polyomaviruses. Although mPy and SV40 can induce tumors in experimental animals (Gross 1953; Sweet and Hilleman 1960), human polyomavirus’s role as human cancer-causing agents is less certain. Perhaps the strongest link is observed in MCPy, in which the virus is positively correlated with incidences of Merkel cell carcinoma, a rare but aggressive skin cancer of neuroendocrine origin (Feng et al. 2008). Regardless of their role in cancer, human polyomaviruses are well-established causative agents for other human diseases, including hemorrhagic cystitis and nephropathy (by BKV), and the fatal demyelinating disease progressive multifocal leukoencephalopathy (by JCV) (Jiang et al. 2009a).
Structurally, Py particles are composed of a layer of the outer coat protein VP1. This single protein, arranged as 72 pentamers, forms the shell surrounding the viral genome (Liddington et al. 1991; Stehle et al. 1994). Each VP1 pentamer engages the internal protein VP2 or VP3 via hydrophobic interactions (Chen et al. 1998). Additionally, VP1 binds directly to its DNA genome harbored within the viral particle (Carbone et al. 2004). To stabilize the virus structure, each VP1 within a pentamer extends its carboxy-terminal arm to an adjacent pentamer. Calcium ions bound to the virus further strengthen this interpentamer interaction (Stehle et al. 1996). Finally, a network of VP1 disulfide bonds reinforces its overall architecture (Stehle et al. 1996).
To initiate infection, Py binds to glycolipid receptors called gangliosides on the plasma membrane (Smith et al. 2003; Tsai et al. 2003; Low et al. 2006). After internalization, the virus is transferred first to the endolysosome (Eash et al. 2004; Querbes et al. 2006; Qian et al. 2009; Engel et al. 2011) and then the ER (Tsai et al. 2003; Gilbert and Benjamin 2004; Qian et al. 2009). Py transport to the ER, a phenomenon documented over 20 years ago by Helenius and coworkers using electron microscopy (EM) (Kartenbeck et al. 1989), is unique as most extracellular ligands do not reach the ER after endocytosis. On reaching the ER, Py penetrates the ER membrane to gain access to the cytosol and then the nucleus, in which ensuing transcription and replication of the viral genome lead to lytic infection or cell transformation. Insights into how Py crosses the ER membrane to gain entry into the cytosol are emerging. Conceptually, Py’s entry into the cytosol via penetration of the ER membrane can be divided into three steps. First, the virus undergoes host-triggered conformational changes in the ER, priming it for membrane penetration. Second, the structurally altered virus transports across the ER membrane. Third, the virus is released into the cytosol.
In the first membrane penetration step, ER factors perturb the major forces stabilizing Py’s structure to generate a membrane transport-competent intermediate. One of the major forces is the VP1 disulfide bond network. ER-resident redox-active protein disulfide isomerase (PDI) family members, including the canonical PDI, ERp57, and ERp72, can isomerize or reduce the virus disulfide bonds (Fig. 2A, step 1a) (Gilbert et al. 2006; Schelhaas et al. 2007; Walczak and Tsai 2011; Nelson et al. 2012). However, the precise combination of PDI proteins engaging a specific Py family member may differ because of subtle differences in the viral disulfide bond arrangements. For some Pys, a dimeric redox-inactive PDI protein called ERp29 untangles the VP1 carboxy-terminal arms (Magnuson et al. 2005; Rainey-Barger et al. 2007a,b; Nelson et al. 2012) exposing hydrophobic VP2 and VP3 to generate a hydrophobic viral particle (Fig. 2A, step 1b) (Magnuson et al. 2005; Rainey-Barger et al. 2007a; Geiger et al. 2011). The hydrophobic virus is likely maintained in a soluble state by binding to the Hsp70 chaperone BiP (Fig. 2A, step 1c) (Geiger et al. 2011; Goodwin et al. 2011) in a reaction regulated by the J-domain containing cochaperone ERdj3 (Goodwin et al. 2011). The J domain typically stimulates Hsp70’s ATPase activity to control Hsp70-substrate interaction (Kampinga and Craig 2010). No ER factors have been found to extract Py’s bound calcium ions. Despite the actions of the ER factors on Py, biochemical analyses have revealed that the virus remains as a large and intact particle in the ER (Inoue and Tsai 2011).
Figure 2.
Model of Py and CT transport across the ER membrane. (A) On reaching the ER, PDI family members (i.e., PDI, ERp72, and ERp57) disrupt disulfide bonds on Py’s surface (step 1a). ERp29 then extrudes the VP1 carboxy-terminal arm to expose VP2 and VP3 (step 1b), generating a hydrophobic virus. This particle is likely maintained in a soluble state by the BiP-ERdj3 chaperones (step 1c). The hydrophobic virus then integrates into the ER membrane (step 2). ERAD membrane components including Derlin-1, Derlin-2, Sel1L, BAP29/31, and RMA1 are postulated to mediate virus transport across the ER membrane. How Py is released into the cytosol is not well-understood (step 3), but may rely on the ER membrane J proteins (i.e., DNAJ B12, B14, and C18) stimulating binding between the virus and cytosolic Hsp70; reiterative cycles of this binding “pull” the virus into the cytosol. (B) CT arrives to the ER by binding to the host ganglioside GM1 receptor. In the ER, CT is transferred to the Derlin-1-Hrd1 membrane proteins (step 1) potentially mediated by BiP-J protein chaperones. An unidentified reductase reduces CTA to generate the CTA1 peptide (step 2). Next, PDI (bound to Derlin-1 and Hrd1), in its reduced state, unfolds CTA1 (step 3); ensuing PDI oxidation by Ero1 releases the unfolded toxin from PDI. The unfolded toxin presumably crosses the Hrd1 complex to reach the cytosol (step 4). Finally, an unidentified cytosolic factor extracts the toxin into the cytosol to complete the transport event (step 5).
In the second step, the hydrophobic virus integrates into the ER membrane to initiate membrane penetration (Fig. 2A, step 2) (Daniels et al. 2006; Rainey-Barger et al. 2007b; Geiger et al. 2011). At this juncture, ERAD membrane components including Derlin-1 (Schelhaas et al. 2007; Jiang et al. 2009b), Derlin-2 (Lilley et al. 2006), Sel1L (Schelhaas et al. 2007), RMA1 (Geiger et al. 2011), and BAP29/BAP31 (Geiger et al. 2011), have been proposed to facilitate Py’s exit to the cytosol. However, as none of them have been shown to interact with Py physically, their actions on the virus may be transient or indirect. Additionally, given the relatively large viral particle size (40–50 nm in diameter) in the ER (Inoue and Tsai 2011) compared to cellular proteins, it is unlikely to cross a protein-conducting channel of the ERAD machinery that accommodates smaller cellular proteins. Instead, we hypothesize that Py penetrates the ER lipid bilayer directly. Trapping Py on the ER membrane to ascertain its interacting partners under this condition should distinguish these possibilities. Py penetration across a lipid bilayer and not a protein channel will be consistent with the general mechanism by which nonenveloped viruses breach their limiting membranes (Tsai 2007). If the large Py particle transports across the lipid bilayer, we envision this process to deform the ER membrane physically. The advent of electron tomography (ET) capable of visualizing cellular structures in three dimension (3D) at high resolutions may reveal such ER membrane remodeling during virus penetration. Clearly, deciphering Py’s precise membrane transport mechanism will require more rigorous biochemical and imaging approaches.
In the final entry step, Py is released into the cytosol (Fig. 2A, step 3). How this step is accomplished is the least understood. The cytosolic ATPase p97, which normally extracts misfolded cellular proteins into the cytosol during ERAD (Ye et al. 2001), plays only a minor role in releasing Py into the cytosol (Geiger et al. 2011). Furthermore, although there is evidence implicating the proteasome at this stage (Schelhaas et al. 2007; Jiang et al. 2009b; Inoue and Tsai 2011), it is unlikely to play a direct role. If cytosol release and virus disassembly are coupled, the cytosolic chaperone Hsc70 may be the ideal candidate for catalyzing the release step as it disassembles Py in vitro (Chromy et al. 2006). Because all of these proposed cytosolic factors play many other cellular functions, whether they execute a direct role in catalyzing Py release into the cytosol remains inconclusive. What is needed is the establishment of an in vitro translocation assay in which defined cytosolic factors can be added to an ER membrane fraction preloaded with Py; these putative factors’ ability to stimulate virus release from the ER membrane would offer compelling evidence of their direct role in promoting cytosol release.
Interestingly, a recent report identified an ER membrane-bound DNA J protein family (i.e., DNAJ B12, B14, and C18) in facilitating Py ER-to-cytosol transport (Goodwin et al. 2011). As these Hsp70 co-chaperones’ J domains are expected to face the ER membrane’s cytosolic surface, they may stimulate Hsp70’s ATPase activity to promote Hsp70-Py interaction; reiterative cycles of this binding eventually “pull” the virus into the cytosol. This scenario is reminiscent of the manner by which BiP “pulls” substrates into the ER lumen in a reaction controlled by the J-domain containing Sec63 protein during posttranslational translocation (Matlack et al. 1999). Why so many members of this DNA J protein family promote Py’s release into the cytosol from the ER is unclear.
ER AS THE ENTRY SITE FOR CHOLERA TOXIN
Similar to Py, certain bacterial toxins including cholera toxin (CT) and shiga toxin (ST) hijack the ERAD machinery to reach the cytosol to induce cytotoxicity (Teter and Holmes 2002; Lencer and Tsai 2003; Lord et al. 2005). As CT is this toxin group’s prototype, we briefly describe how it co-opts the ERAD machinery during entry. On reaching the ER from the cell surface, CT is transferred to a membrane complex composed of the Derlin-1 (Bernardi et al. 2008; Dixit et al. 2008) and Hrd1 E3 ubiquitin ligase (Bernardi et al. 2010) membrane proteins (Fig. 2B, step 1) postulated to form the ERAD translocation channel (Carvalho et al. 2010; Smith et al. 2011). How this complex captures CT is unknown. After reaching the Derlin-1-Hrd1 complex, the toxin’s CTA subunit is reduced to generate the toxic CTA1 fragment (Fig. 2B, step 2). PDI (bound to the Derlin-1-Hrd1 complex) then acts as a redox-dependent chaperone to unfold CTA1 (Fig. 2B, step 3) (Forster et al. 2009; Tsai et al. 2001). In its reduced state, PDI binds to and unfolds CTA1; PDI oxidation by Ero1 alters PDI’s conformation, enabling it to release the unfolded toxin (Tsai and Rapoport 2002; Moore et al. 2010). A recent high-resolution PDI X-ray structure confirms that it undergoes a redox-driven structural change (Wang et al. 2012). That CTA1 hijacks ER redox proteins for cytosol entry parallels Py’s use of these factors during its ER membrane transport.
Once CTA1 is released from PDI, it presumably transports across the Hrd1 complex (Fig. 2B, step 4) (Carvalho et al. 2010). The driving force propelling the toxin to the cytosol is unknown. One possibility is CTA1’s propensity to refold (Rodighiero et al. 2002). In this scenario, the toxin’s ability to refold rapidly on emerging the ER membrane’s cytosolic surface prevents it from backsliding. A cytosolic factor then extracts the folded toxin from the ER membrane, releasing it into the cytosol (Fig. 2B, step 5). Similar to Py, p97 plays a modest role in this step (Abujarour et al. 2005; Kothe et al. 2005). Thus, we postulate another cytosolic factor ejects the toxin into the cytosol. An obvious distinction between CTA1 and a typical ERAD substrate’s fate in the cytosol is that the toxin is not degraded by the proteasome. How the toxin evades this degradative machinery is unclear.
ER’S ROLE IN VIRUS REPLICATION AND ASSEMBLY
After entry into the cytosol, an incoming bacterial toxin exerts its cytotoxic effect. In contrast, an incoming viral particle after entry replicates and assembles to generate new progenies to complete its infection cycle. Viral replication and assembly are often supported by a host cell’s membranous network. These membranes function as scaffolds to recruit viral and host components necessary for replication and assembly. As examples, influenza virus (Compans and Dimmock 1969) and HIV (Ono and Freed 2001) assembly is supported by the plasma membrane (Fig. 3, site 1), Semliki Forest virus (Kujala et al. 2001) and Rubella virus (Magliano et al. 1998), RNA replication by the endolysosome (Fig. 3, site 2), Flock House virus (Miller et al. 2001), genome replication by the mitochondria (Fig. 3, site 3), and Herpes simplex virus (Mettenleiter et al. 2006) assembly by the inner nuclear membrane (Fig. 3, site 4).
Figure 3.
Viral replication and assembly in different membranous networks. Distinct host membranes support replication and assembly of different viruses. As examples, influenza virus and HIV assembly is supported by the plasma membrane (site 1), Semliki Forest virus and Rubella virus RNA replication by the endolysosome (site 2), Flock House virus genome replication by the mitochondria (site 3), Herpes simplex virus assembly by the inner nuclear membrane (site 4), and Hepatitis C virus, Dengue virus, vaccinia virus, and rotavirus replication and assembly by the ER membrane (site 5).
Remarkably, diverse viruses including RNA and DNA viruses belonging to both the enveloped and nonenveloped virus families rearrange the ER membrane to generate a vast array of ER-derived structures, with each structure postulated to facilitate viral replication and assembly (Fig. 3, site 5) (Stephens and Compans 1988; Miller and Krijnse-Locker 2008; den Boon and Ahlquist 2010). For instance, the enveloped hepatitis C virus (HCV) promotes formation of an ER-derived membranous matrix referred to as “membranous webs” important for viral replication and assembly (Egger et al. 2002), and the enveloped Dengue virus (DENV) generates so-called vesicle packets and convoluted membranes also presumed to facilitate genome replication and virus assembly (Welsch et al. 2009). For the enveloped vaccinia virus (VV), the only known DNA virus to replicate in the cytosol and not the nucleus, the ER membrane encloses discrete cytoplasmic foci in which active viral DNA replication is thought to take place (Tolonen et al. 2001). In the case of the nonenveloped rotavirus, the virus buds into the ER lumen to generate a transient enveloped intermediate, with removal of the viral envelope in the ER lumen required to complete virus assembly. We will not describe each case in detail as they are already covered in many excellent reviews (Miller and Krijnse-Locker 2008; den Boon and Ahlquist 2010). Instead, we focus here on how HCV induces ER-derived membranous webs to support its replication and assembly, and how rotavirus uses the ER for assembly. These examples illustrate ER’s remarkable versatility in facilitating different stages of the virus life cycle.
HCV, which infects approximately 170 million people worldwide, causes chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. This virus is a member of the Flavivirus family. Its positive-strand RNA genome encodes ten structural and nonstructural proteins (i.e., core, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (Reed and Rice 2000). The core proteins and the RNA genome constitute the nucleocapsid, and are surrounded by a host-derived envelope. Embedded in this envelope are the E1 and E2 glycoproteins that mediate viral attachment and entry. To infect cells, HCV binds to host receptors, becomes endocytosed, and undergoes pH-dependent membrane fusion in an endosomal compartment that enables the viral core proteins and genome access to the cytosol where genome replication ensues (Ploss and Evans 2012).
HCV replicates its genome specifically on the cytosolic surface of a virus-induced, ER-derived membranous system called the membranous web (Fig. 4A). Functionally, this web harbors the HCV replication complex that promotes replication with assistance from viral nonstructural proteins along with an increasing list of cellular factors (Wolk et al. 2008). Although the HCV membranous web is found juxtaposed to the rough ER (Egger et al. 2002), whether it is physically connected to or detached from the ER membrane network remains unclear. Web formation is thought to initiate when the ER membrane-bound viral nonstructural protein NS5A recruits and activates phosphatidylinositol 4-kinase-α (PI4KIIIα) (Fig. 4A) (Berger et al. 2009, 2011; Reiss et al. 2011), thereby increasing the local concentration of phosphatidylinositol-4-phosphate (PI4P). PI4P might itself be an important membrane constituent that supports membranous web structural integrity. Alternatively, PI4P may recruit downstream effector proteins such as oxysterol-binding protein 1 (OSBP1) to form a sterol-rich environment surrounding the membranous web essential for HCV replication (Amako et al. 2009). Recruitment of the host protein proline-serine-threonine phosphatase-interacting protein 2 (PSTPIP2) by the nonstructural proteins NS4B and NS5A to the membranous web also appears to mediate web formation (Chao et al. 2012). Interestingly, PSTPIP2 contains a F-BAR domain that has membrane-deforming activity (Frost et al. 2008). This property enables PSTPIP2 to induce membrane curvature on the ER membrane to promote web formation (Chao et al. 2012). In this context, NS4B itself can also cause web formation (Egger et al. 2002), suggesting that both viral and host components play key roles in sculpting the ER-derived membranous web.
Figure 4.
ER-derived membranous web as the HCV replication and assembly site (A) HCV replication and assembly occur in the context of a virus-induced, ER-derived membrane matrix called the membranous web. It remains unclear whether the membranous web is connected to or detached from the ER membrane network. To initiate web formation, the viral nonstructural protein NS5A recruits and activates PI4KIIIα, generating PI4P. PI4P may be crucial to maintain web structural integrity, or it could recruit OSBP1 to create a sterol-rich environment that supports HCV RNA replication. Membrane remodeling proteins such as the viral nonstructural protein NS4B and host protein PSTPIP2 sculpt the ER membrane to promote web formation. (B) The membranous web is also thought to be the HCV assembly site. Lipid droplet (LD), situated proximal to the replication complex on the web surface, captures core proteins. Core proteins are released from LD, delivered to the assembly site on the membranous web, and encapsulate the RNA genome, generating the nucleocapsid (step 1). This reaction is mediated by P7 and the nonstructural proteins NS2, NS3, and NS4A. Nucleocapsid formation is thought to take place within an invagination on the membranous web distinct from where replication occurs. Once the nucleocapsid is formed, it buds into the lumen of the membranous web to form the enveloped virus with concomitant incorporation of the E1 and E2 glycoproteins (step 2). The HCV particle is then secreted to the extracellular milieu.
In addition to supporting replication, the membranous web is also postulated to be the virus assembly site. Within this web, lipid droplets (LDs), a storage organelle for neutral lipids in cells (Guo et al. 2009), execute a key viral assembly role (Miyanari et al. 2007). Acting as a scaffold, LD captures HCV core proteins on its surface (Fig. 4B). The LD-core protein interaction is crucial for production of infectious virus (Boulant et al. 2007; Shavinskaya et al. 2007). As LD is located proximal to the replication complex positioned on the surface of the membranous web, core proteins released from LD encapsulate the newly synthesized RNA genome, forming the nucleocapsid (Fig. 4B, step 1). This process likely occurs within an invagination on the membranous web distinct from where replication takes place. The viral P7 and nonstructural proteins NS2, NS3, and NS4A have been implicated in the release of core proteins from the LD (Boson et al. 2011; Counihan et al. 2011). Once the nucleocapsid is formed, it buds into the lumen of the membranous web, generating an enveloped viral particle with E1 and E2 glycoproteins displayed on the surface (Fig. 4B, step 2). If the membranous web remains connected to the ER membrane network, the newly assembled viral particle can be secreted from the cell via the classic secretory pathway. Should the web be disconnected from the ER, the virus-containing vesicle can reconnect with the secretory pathway by fusing with elements of the secretory pathway such as the Golgi complex.
Replication and assembly of DENV, a flavivirus that causes dengue fever, show interesting parallels to and differences from HCV. For instance, akin to HCV’s NS4B, DENV’s nonstructural protein NS4A rearranges the ER membrane (Miller et al. 2007), leading to formation of vesicle packets and convoluted membranes (Welsch et al. 2009). Additional viral and host components (Khadka et al. 2011) are likely recruited to the ER membrane to promote this reaction. DENV also employs LD for particle formation (Samsa et al. 2009), similar to HCV assembly. However, in contrast to the less obvious physical connection between the HCV-induced membranous web and ER, high-resolution ET analyses show that the DENV-induced ER-derived structures are clearly interconnected in one continuous ER membrane network (Welsch et al. 2009). Perhaps a similar approach using ET will better clarify the physical relationship between HCV’s membranous web and the ER membrane network.
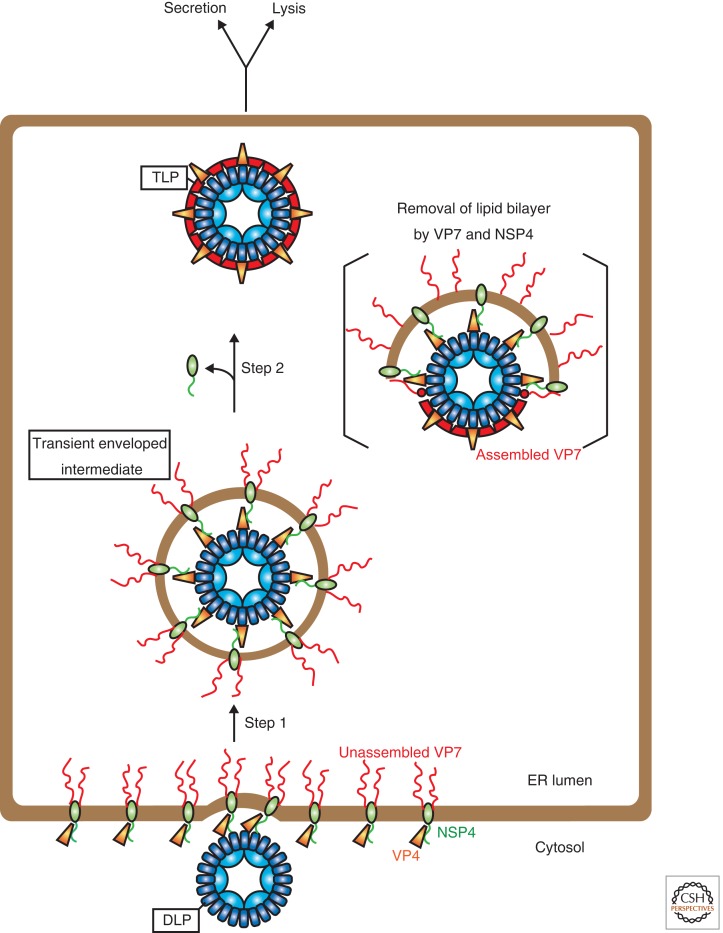
The ER also aids in rotavirus assembly, particularly during the late stage of this process. This virus belongs to the reovirus family and is an important causative agent for childhood diarrhea. Structurally, the capsid is an icosahedral particle with a diameter of approximately 100 nm; its double-stranded RNA genome is enclosed within the particle (Trask et al. 2012). The capsid consists of three layers: the inner most layer (called core shell), the middle layer, and the outer layer. The core shell is formed by 120 copies of VP2 dimers arranged as an icosahedron, with the replication complex and the genome encapsulated (McClain et al. 2010). The middle layer is formed by up to 260 VP6 trimers that interact with VP2 dimers to stabilize the core shell (Mathieu et al. 2001). The outer layer is composed of VP7 and VP4: VP7 constitutes a large part of the outer layer and makes extensive contact with the middle layer, whereas VP4 protrudes radially from the particle as spike-like structures (Settembre et al. 2011). A viral intermediate containing the core shell and middle layer is called a double-layer particle (DLP) and a mature infectious particle harboring all three layers is referred to as a triple-layer particle (TLP). DLP-to-TLP morphogenesis is intimately associated with the ER.
To form TLP, DLP must gain access into the ER lumen to acquire the outer layer (Fig. 5). Once formed, TLP exits the host cell via lysis (Musalem and Espejo 1985) or secretion (Jourdan et al. 1997). The unusual DLP-to-TLP conversion is composed of two steps. The first step requires DLP budding into the ER lumen, generating a transient enveloped intermediate (Fig. 5, step 1). The viral nonstructural membrane protein NSP4 executes a key role in this step. As a tetramer on the ER membrane (Bowman et al. 2000), NSP4 recruits DLP and VP4 to the ER membrane’s cytosolic surface (Petrie et al. 1984). The energy source required to deform the ER membrane to bud DLP into the ER lumen is not clear, although multivalent interactions between NSP4 and DLP may trigger this reaction (Taylor et al. 1993). VP4 is dispensable for budding as its knockdown generates TLP (Dector et al. 2002), albeit spike-less as the resulting TLP lacks VP4. That the ER membrane supports DLP budding into the ER lumen reflects this membrane’s propensity for promoting budding reactions, even though this reaction proceeds in the opposite direction to conventional ER budding.
Figure 5.
ER as the final rotavirus assembly site. Infectious rotavirus particle formation requires DLP-to-TLP conversion. This process requires two steps that occur in or surrounding the ER. The first step is DLP budding into the ER lumen, generating a viral enveloped intermediate (step 1). This step is initiated when NSP4 recruits DLP and VP4 to the ER membrane. ER membrane deformation by the DLP-NSP4-VP4 complex generates a transient enveloped intermediate. Because unassembled VP7 is tethered on the ER membrane’s lumenal surface prior to budding, formation of the transient enveloped intermediate “captures” VP7 that remains topologically facing the ER lumen. The second step necessitates removal of the transient membrane layer (step 2), a process likely mediated by VP7 and NSP4. This step is accompanied by VP7 conformational change that enables it to dock and assemble on DLP. Additionally, NSP4 is released from DLP during this process. Mature TLP finally exits the host cell via lysis or secretion.
How does VP7 assemble on DLP’s surface after budding? Using its signal sequence, VP7 translocates into the ER lumen where the signal sequence is cleaved. However, this cleaved unassembled VP7 remains tethered to the ER membrane via noncovalent interactions with its signal peptide (Stirzaker and Both 1989). VP7 also interacts with NSP4’s amino terminus on the ER membrane (Maass and Atkinson 1990). Thus the emerging picture of the transient enveloped intermediate is an ER-derived lipid bilayer encasing a VP4-bound DLP with unassembled VP7 decorated on the outer membrane surface (Fig. 5, transient enveloped intermediate).
In the second maturation step, the transient membrane layer is removed, which enables VP7 to dock and assemble on the VP4-bound DLP, forming infectious TLP; NSP4 release from DLP is coordinated with these events (Fig. 5, step 2). Although the mechanics of this process remains unclear, VP7 is thought to play a crucial role as VP7 knockdown fails to block viral budding but causes enveloped intermediate accumulation in the ER lumen (Lopez et al. 2005; Cuadras et al. 2006). How might VP7 promote envelope shedding? In addition to serving as a structural protein, VP7 possesses membrane lytic activity (Charpilienne et al. 1997). There is evidence that unassembled VP7 tethered on ER membrane’s lumenal side exists in a different conformation when compared to its native assembled structure on TLP’s surface (Kabcenell et al. 1988). Thus, VP7 likely undergoes conformational rearrangements after budding, exposing its membrane-lytic portion to disrupt envelope integrity. Although NSP4 also has membrane lytic activity (Browne et al. 2000), its function in removing the transient envelope is difficult to assess as NSP4 knockdown attenuates DLP assembly severely (Lopez et al. 2005). Topologically, VP7 but not NSP4 of the transient enveloped intermediate is exposed to the ER lumen. Thus, should ER factors trigger membrane disruption, VP7 would be the likely candidate to engage the ER proteins. Importantly, knockdown of ER-resident chaperones including BiP and PDI perturb proper rotavirus ER morphogenesis (Maruri-Avidal et al. 2008), further supporting ER’s role during rotavirus DLP-to-TLP assembly.
Clarifying how the transient enveloped intermediate’s membrane is removed to facilitate TLP formation is unquestionably the most crucial step in illuminating rotavirus assembly. What ER factors impart conformational changes on VP7 to render it membrane penetration-competent? What is the nature of these VP7 structural changes? Does the envelope simply “dissolve” to release DLP or does DLP cross the envelope leaving an intact membrane behind, potentially containing NSP4? And finally, how does VP7 dock and assemble on DLP as NSP4 is coordinately removed from DLP? Perhaps a reconstitution system in which ER proteins are added to various rotavirus ER intermediates to drive the next assembly step will provide a better understanding of this dynamic process. Moreover, high-resolution cryo-EM and ET approaches could be useful in revealing a 3D view of the ER-localized enveloped intermediate.
ER’S FUNCTION DURING VIRAL IMMUNE EVASION
In addition to supporting entry, replication, and assembly, the ER also affords viruses an opportunity to manipulate the host immune system to sustain the infection course. Prominent examples include proteasomal destruction of MHC class I molecule triggered by the human cytomegalovirus (HCMV)-encoded transmembrane proteins US2 and US11 (Wiertz et al. 1996; Machold et al. 1997) in a pathway that requires the ERAD machinery. By degrading MHC class I molecules, HCMV effectively prevents infected cells from properly presenting viral antigens on their surface. Similarly, the murine γ herpesvirus encodes a membrane-bound E3 ubiquitin ligase called mK3 that promotes MHC class I degradation via the ERAD pathway (Wang et al. 2006). Another salient example is observed in HIV, where its Vpu protein also coopts the ERAD machinery to down-regulate the host entry receptor CD4 (Willey et al. 1992; Magadan et al. 2010). CD4 down-regulation leads to a series of events including disruption of T cell activation (Lanzavecchia et al. 1988), ultimately contributing to robust HIV infection. As there are many reviews on how viruses use the ER to thwart the host immune system (Loureiro and Ploegh 2006; Lindwasser et al. 2007; Hansen and Bouvier 2009; Jackson et al. 2011), this topic will not be covered extensively in this article. Nonetheless, it unveils another aspect of the ER function that viruses hijack to maintain infection.
CONCLUDING REMARKS
A virus navigates through the host cell’s environment to chart the most effective infection strategy. This strategy not only requires it to evade pathways that trap or degrade itself, but more importantly, delivers it to an appropriate intracellular destination where it can replicate and assemble. Increasing evidence shows host membranes play crucial roles in supporting virus infection. There is no better example than the ER organelle, where many viruses have evolved to co-opt ER’s lumenal and membrane contents to achieve proper entry, replication, or assembly.
Although only Py family members use the ER for entry, many viruses including HCV, DENV, VV, and rotavirus, use the ER membrane to support their replication and assembly. In the case of Py entry, elegant studies have identified ER factors that prime the virus for ER membrane penetration. However, elucidating the specific ER membrane machinery and cytosolic factors that complete the transport reaction remains a challenge. This viral entry field should look to the field of ER membrane translocation where transport of a defined cellular substrate across the ER membrane can be reconstituted using purified translocation components (Brundage et al. 1990; Akimaru et al. 1991; Gorlich and Rapoport 1993). Such a feat can be accomplished for Py when more rigorous biochemical strategies are applied.
Powerful imaging approaches, in particular cryo-ET, have revealed formation of virus-induced ER-derived structures considered critical for viral replication and assembly. In many instances, co-localization of these structures with the replicated genome and assembling intermediates implicate these structures as virus replication and assembly sites. Although plausible, convincingly showing these structures support replication and assembly requires the imaging findings to be reinforced by in vitro and cell-based studies. For example, can the virus-induced ER-derived structures be isolated and shown to be active in promoting a specific replication or assembly step? In cells, will blocking formation of the ER-derived structures prevent virus replication or assembly? A more comprehensive approach combining classical biochemical and cell biological methods coupled with state-of-the-art imaging techniques is needed to establish firmly a causal relationship between these ER-derived structures and their roles in viral infection.
Historically, clarifying the nature of virus–host cell interactions has illuminated fundamental cellular processes. In this review, we observe how ER’s potential functions can be unveiled through its interaction with viruses. Specifically, ER-resident factors not only interact with endogenous substrates but also viral pathogens to trigger their entry and assembly. Additionally, the ER membrane that normally accommodates budding reactions can undergo virus-induced structural rearrangements to generate ER-derived structures postulated to be important for replication and assembly. Although not discussed, the abundant ER membrane surface can also serve to recruit different viral components to promote efficient genome replication. Such revelations regarding ER’s functional capacity will only expand as we continue to explore its role in facilitating pathogen infection.
ACKNOWLEDGMENTS
The authors thank Dr. Christiane Wobus (Michigan) and Christopher Walczak (Michigan) for critical review of this manuscript. We apologize for not citing the many excellent works of our colleagues because of space constraints. B.T. is a Burroughs Wellcome Fund Investigator in Pathogenesis of Infectious Disease and is supported by the NIH.
Footnotes
Editors: Susan Ferro-Novick, Tom A. Rapoport, and Randy Schekman
Additional Perspectives on The Endoplasmic Reticulum available at www.cshperspectives.org
REFERENCES
- Abujarour RJ, Dalal S, Hanson PI, Draper RK 2005. p97 Is in a complex with cholera toxin and influences the transport of cholera toxin and related toxins to the cytoplasm. J Biol Chem 280: 15865–15871 [DOI] [PubMed] [Google Scholar]
- Akimaru J, Matsuyama S, Tokuda H, Mizushima S 1991. Reconstitution of a protein translocation system containing purified SecY, SecE, and SecA from Escherichia coli. Proc Natl Acad Sci 88: 6545–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amako Y, Sarkeshik A, Hotta H, Yates J 3rd, Siddiqui A 2009. Role of oxysterol binding protein in hepatitis C virus infection. J Virol 83: 9237–9246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G 2009. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III α in hepatitis C virus replication. Proc Natl Acad Sci 106: 7577–7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KL, Kelly SM, Jordan TX, Tartell MA, Randall G 2011. Hepatitis C virus stimulates the phosphatidylinositol 4-kinase III α-dependent phosphatidylinositol 4-phosphate production that is essential for its replication. J Virol 85: 8870–8883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi KM, Forster ML, Lencer WI, Tsai B 2008. Derlin-1 facilitates the retro-translocation of cholera toxin. Mol Biol Cell 19: 877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi KM, Williams JM, Kikkert M, van Voorden S, Wiertz EJ, Ye Y, Tsai B 2010. The E3 ubiquitin ligases Hrd1 and gp78 bind to and promote cholera toxin retro-translocation. Mol Biol Cell 21: 140–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boson B, Granio O, Bartenschlager R, Cosset FL 2011. A concerted action of hepatitis C virus p7 and nonstructural protein 2 regulates core localization at the endoplasmic reticulum and virus assembly. PLoS Pathog 7: e1002144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulant S, Targett-Adams P, McLauchlan J 2007. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J Gen Virol 88: 2204–2213 [DOI] [PubMed] [Google Scholar]
- Bowman GD, Nodelman IM, Levy O, Lin SL, Tian P, Zamb TJ, Udem SA, Venkataraghavan B, Schutt CE 2000. Crystal structure of the oligomerization domain of NSP4 from rotavirus reveals a core metal-binding site. J Mol Biol 304: 861–871 [DOI] [PubMed] [Google Scholar]
- Brodsky JL, Skach WR 2011. Protein folding and quality control in the endoplasmic reticulum: Recent lessons from yeast and mammalian cell systems. Curr Opin Cell Biol 23: 464–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne EP, Bellamy AR, Taylor JA 2000. Membrane-destabilizing activity of rotavirus NSP4 is mediated by a membrane-proximal amphipathic domain. J Gen Virol 81: 1955–1959 [DOI] [PubMed] [Google Scholar]
- Brundage L, Hendrick JP, Schiebel E, Driessen AJ, Wickner W 1990. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell 62: 649–657 [DOI] [PubMed] [Google Scholar]
- Carbone M, Ascione G, Chichiarelli S, Garcia MI, Eufemi M, Amati P 2004. Chromosome-protein interactions in polyomavirus virions. J Virol 78: 513–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Stanley AM, Rapoport TA 2010. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell 143: 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao TC, Su WC, Huang JY, Chen YC, Jeng KS, Wang HD, Lai MM 2012. Proline-serine-threonine phosphatase-interacting protein 2 (PSTPIP2), a host membrane-deforming protein, is critical for membranous web formation in hepatitis C virus replication. J Virol 86: 1739–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpilienne A, Abad MJ, Michelangeli F, Alvarado F, Vasseur M, Cohen J, Ruiz MC 1997. Solubilized and cleaved VP7, the outer glycoprotein of rotavirus, induces permeabilization of cell membrane vesicles. J Gen Virol 78: 1367–1371 [DOI] [PubMed] [Google Scholar]
- Chen XS, Stehle T, Harrison SC 1998. Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry. EMBO J 17: 3233–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chromy LR, Oltman A, Estes PA, Garcea RL 2006. Chaperone-mediated in vitro disassembly of polyoma- and papillomaviruses. J Virol 80: 5086–5091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compans RW, Dimmock NJ 1969. An electron microscopic study of single-cycle infection of chick embryo fibroblasts by influenza virus. Virology 39: 499–515 [DOI] [PubMed] [Google Scholar]
- Counihan NA, Rawlinson SM, Lindenbach BD 2011. Trafficking of hepatitis C virus core protein during virus particle assembly. PLoS Pathog 7: e1002302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadras MA, Bordier BB, Zambrano JL, Ludert JE, Greenberg HB 2006. Dissecting rotavirus particle-raft interaction with small interfering RNAs: Insights into rotavirus transit through the secretory pathway. J Virol 80: 3935–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels R, Rusan NM, Wadsworth P, Hebert DN 2006. SV40 VP2 and VP3 insertion into ER membranes is controlled by the capsid protein VP1: Implications for DNA translocation out of the ER. Mol Cell 24: 955–966 [DOI] [PubMed] [Google Scholar]
- Dector MA, Romero P, Lopez S, Arias CF 2002. Rotavirus gene silencing by small interfering RNAs. EMBO Rep 3: 1175–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon JA, Ahlquist P 2010. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol 64: 241–256 [DOI] [PubMed] [Google Scholar]
- Dixit G, Mikoryak C, Hayslett T, Bhat A, Draper RK 2008. Cholera toxin up-regulates endoplasmic reticulum proteins that correlate with sensitivity to the toxin. Exp Biol Med (Maywood) 233: 163–175 [DOI] [PubMed] [Google Scholar]
- Eash S, Querbes W, Atwood WJ 2004. Infection of vero cells by BK virus is dependent on caveolae. J Virol 78: 11583–11590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger D, Bienz K 2002. Recombination of poliovirus RNA proceeds in mixed replication complexes originating from distinct replication start sites. J Virol 76: 10960–10971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol 76: 5974–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel S, Heger T, Mancini R, Herzog F, Kartenbeck J, Hayer A, Helenius A 2011. Role of endosomes in simian virus 40 entry and infection. J Virol 85: 4198–4211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, Moore PS 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319: 1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster ML, Mahn JJ, Tsai B 2009. Generating an unfoldase from thioredoxin-like domains. J Biol Chem 284: 13045–13056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De Camilli P, Unger VM 2008. Structural basis of membrane invagination by F-BAR domains. Cell 132: 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger R, Andritschke D, Friebe S, Herzog F, Luisoni S, Heger T, Helenius A 2011. BAP31 and BiP are essential for dislocation of SV40 from the endoplasmic reticulum to the cytosol. Nat Cell Biol 13: 1305–1314 [DOI] [PubMed] [Google Scholar]
- Gilbert J, Benjamin T 2004. Uptake pathway of polyomavirus via ganglioside GD1a. J Virol 78: 12259–12267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J, Ou W, Silver J, Benjamin T 2006. Downregulation of protein disulfide isomerase inhibits infection by the mouse polyomavirus. J Virol 80: 10868–10870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin EC, Lipovsky A, Inoue T, Magaldi TG, Edwards AP, Van Goor KE, Paton AW, Paton JC, Atwood WJ, Tsai B, et al. 2011. BiP and multiple DNAJ molecular chaperones in the endoplasmic reticulum are required for efficient simian virus 40 infection. mBio 2: e00101–00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Rapoport TA 1993. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell 75: 615–630 [DOI] [PubMed] [Google Scholar]
- Gross L 1953. A filterable agent, recovered from Ak leukemic extracts, causing salivary gland carcinomas in C3H mice. Proc Soc Exp Biol Med 83: 414–421 [DOI] [PubMed] [Google Scholar]
- Guo Y, Cordes KR, Farese RV Jr, Walther TC 2009. Lipid droplets at a glance. J Cell Sci 122: 749–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TH, Bouvier M 2009. MHC class I antigen presentation: Learning from viral evasion strategies. Nat Rev Immunol 9: 503–513 [DOI] [PubMed] [Google Scholar]
- Hopkins CR 1978. Structure and function of cells: A text for students in medicine and science. Saunders, London/Philadelphia [Google Scholar]
- Inoue T, Tsai B 2011. A large and intact viral particle penetrates the endoplasmic reticulum membrane to reach the cytosol. PLoS Pathog 7: e1002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Moore P, Tsai B 2011. How viruses and toxins disassemble to enter host cells. Annu Rev Microbiol 65: 287–305 [DOI] [PubMed] [Google Scholar]
- Jackson SE, Mason GM, Wills MR 2011. Human cytomegalovirus immunity and immune evasion. Virus Res 157: 151–160 [DOI] [PubMed] [Google Scholar]
- Jiang M, Abend JR, Johnson SF, Imperiale MJ 2009a. The role of polyomaviruses in human disease. Virology 384: 266–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Abend JR, Tsai B, Imperiale MJ 2009b. Early events during BK virus entry and disassembly. J Virol 83: 1350–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdan N, Maurice M, Delautier D, Quero AM, Servin AL, Trugnan G 1997. Rotavirus is released from the apical surface of cultured human intestinal cells through nonconventional vesicular transport that bypasses the Golgi apparatus. J Virol 71: 8268–8278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabcenell AK, Poruchynsky MS, Bellamy AR, Greenberg HB, Atkinson PH 1988. Two forms of VP7 are involved in assembly of SA11 rotavirus in endoplasmic reticulum. J Virol 62: 2929–2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 11: 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartenbeck J, Stukenbrok H, Helenius A 1989. Endocytosis of simian virus 40 into the endoplasmic reticulum. J Cell Biol 109: 2721–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadka S, Vangeloff AD, Zhang C, Siddavatam P, Heaton NS, Wang L, Sengupta R, Sahasrabudhe S, Randall G, Gribskov M, et al. 2011. A physical interaction network of dengue virus and human proteins. Mol Cell Proteomics 10: M111.012187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe M, Ye Y, Wagner JS, De Luca HE, Kern E, Rapoport TA, Lencer WI 2005. Role of p97 AAA-ATPase in the retrotranslocation of the cholera toxin A1 chain, a non-ubiquitinated substrate. J Biol Chem 280: 28127–28132 [DOI] [PubMed] [Google Scholar]
- Kujala P, Ikaheimonen A, Ehsani N, Vihinen H, Auvinen P, Kaariainen L 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J Virol 75: 3873–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzavecchia A, Roosnek E, Gregory T, Berman P, Abrignani S 1988. T cells can present antigens such as HIV gp120 targeted to their own surface molecules. Nature 334: 530–532 [DOI] [PubMed] [Google Scholar]
- Lencer WI, Tsai B 2003. The intracellular voyage of cholera toxin: Going retro. Trends Biochem Sci 28: 639–645 [DOI] [PubMed] [Google Scholar]
- Liddington RC, Yan Y, Moulai J, Sahli R, Benjamin TL, Harrison SC 1991. Structure of simian virus 40 at 3.8-A resolution. Nature 354: 278–284 [DOI] [PubMed] [Google Scholar]
- Lilley BN, Gilbert JM, Ploegh HL, Benjamin TL 2006. Murine polyomavirus requires the endoplasmic reticulum protein Derlin-2 to initiate infection. J Virol 80: 8739–8744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindwasser OW, Chaudhuri R, Bonifacino JS 2007. Mechanisms of CD4 downregulation by the Nef and Vpu proteins of primate immunodeficiency viruses. Curr Molec Med 7: 171–184 [DOI] [PubMed] [Google Scholar]
- Lopez T, Camacho M, Zayas M, Najera R, Sanchez R, Arias CF, Lopez S 2005. Silencing the morphogenesis of rotavirus. J Virol 79: 184–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord JM, Roberts LM, Lencer WI 2005. Entry of protein toxins into mammalian cells by crossing the endoplasmic reticulum membrane: Co-opting basic mechanisms of endoplasmic reticulum-associated degradation. Curr Top Microbiol Immunol 300: 149–168 [DOI] [PubMed] [Google Scholar]
- Loureiro J, Ploegh HL 2006. Antigen presentation and the ubiquitin-proteasome system in host-pathogen interactions. Adv Immunol 92: 225–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low JA, Magnuson B, Tsai B, Imperiale MJ 2006. Identification of gangliosides GD1b and GT1b as receptors for BK virus. J Virol 80: 1361–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass DR, Atkinson PH 1990. Rotavirus proteins VP7, NS28, and VP4 form oligomeric structures. J Virol 64: 2632–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machold RP, Wiertz EJ, Jones TR, Ploegh HL 1997. The HCMV gene products US11 and US2 differ in their ability to attack allelic forms of murine major histocompatibility complex (MHC) class I heavy chains. J Exp Med 185: 363–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magadan JG, Perez-Victoria FJ, Sougrat R, Ye Y, Strebel K, Bonifacino JS 2010. Multilayered mechanism of CD4 downregulation by HIV-1 Vpu involving distinct ER retention and ERAD targeting steps. PLoS Pathog 6: e1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magliano D, Marshall JA, Bowden DS, Vardaxis N, Meanger J, Lee JY 1998. Rubella virus replication complexes are virus-modified lysosomes. Virology 240: 57–63 [DOI] [PubMed] [Google Scholar]
- Magnuson B, Rainey EK, Benjamin T, Baryshev M, Mkrtchian S, Tsai B 2005. ERp29 triggers a conformational change in polyomavirus to stimulate membrane binding. Mol Cell 20: 289–300 [DOI] [PubMed] [Google Scholar]
- Maruri-Avidal L, Lopez S, Arias CF 2008. Endoplasmic reticulum chaperones are involved in the morphogenesis of rotavirus infectious particles. J Virol 82: 5368–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu M, Petitpas I, Navaza J, Lepault J, Kohli E, Pothier P, Prasad BV, Cohen J, Rey FA 2001. Atomic structure of the major capsid protein of rotavirus: Implications for the architecture of the virion. EMBO J 20: 1485–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlack KE, Misselwitz B, Plath K, Rapoport TA 1999. BiP acts as a molecular ratchet during posttranslational transport of prepro-α factor across the ER membrane. Cell 97: 553–564 [DOI] [PubMed] [Google Scholar]
- McClain B, Settembre E, Temple BR, Bellamy AR, Harrison SC 2010. X-ray crystal structure of the rotavirus inner capsid particle at 3.8 A resolution. J Mol Biol 397: 587–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J, Schelhaas M, Helenius A 2010. Virus entry by endocytosis. Annu Rev Biochem 79: 803–833 [DOI] [PubMed] [Google Scholar]
- Mettenleiter TC, Klupp BG, Granzow H 2006. Herpesvirus assembly: A tale of two membranes. Curr Opin Microbiol 9: 423–429 [DOI] [PubMed] [Google Scholar]
- Miller S, Krijnse-Locker J 2008. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol 6: 363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DJ, Schwartz MD, Ahlquist P 2001. Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J Virol 75: 11664–11676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Kastner S, Krijnse-Locker J, Buhler S, Bartenschlager R 2007. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J Biol Chem 282: 8873–8882 [DOI] [PubMed] [Google Scholar]
- Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol 9: 1089–1097 [DOI] [PubMed] [Google Scholar]
- Moore P, Bernardi KM, Tsai B 2010. The Ero1α-PDI redox cycle regulates retro-translocation of cholera toxin. Mol Biol Cell 21: 1305–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musalem C, Espejo RT 1985. Release of progeny virus from cells infected with simian rotavirus SA11. J Gen Virol 66: 2715–2724 [DOI] [PubMed] [Google Scholar]
- Nelson CD, Derdowski A, Maginnis MS, O’Hara BA, Atwood WJ 2012. The VP1 subunit of JC polyomavirus recapitulates early events in viral trafficking and is a novel tool to study polyomavirus entry. Virology 428: 30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Freed EO 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc Natl Acad Sci 98: 13925–13930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie BL, Greenberg HB, Graham DY, Estes MK 1984. Ultrastructural localization of rotavirus antigens using colloidal gold. Virus Res 1: 133–152 [DOI] [PubMed] [Google Scholar]
- Ploss A, Evans MJ 2012. Hepatitis C virus host cell entry. Curr Opin Virol 2: 14–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian M, Cai D, Verhey KJ, Tsai B 2009. A lipid receptor sorts polyomavirus from the endolysosome to the endoplasmic reticulum to cause infection. PLoS Pathog 5: e1000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querbes W, O’Hara BA, Williams G, Atwood WJ 2006. Invasion of host cells by JC virus identifies a novel role for caveolae in endosomal sorting of noncaveolar ligands. J Virol 80: 9402–9413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey-Barger EK, Magnuson B, Tsai B 2007a. A chaperone-activated nonenveloped virus perforates the physiologically relevant endoplasmic reticulum membrane. J Virol 81: 12996–13004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey-Barger EK, Mkrtchian S, Tsai B 2007b. Dimerization of ERp29, a PDI-like protein, is essential for its diverse functions. Mol Biol Cell 18: 1253–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed KE, Rice CM 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr Top Microbiol Immunol 242: 55–84 [DOI] [PubMed] [Google Scholar]
- Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS, et al. 2011. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9: 32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodighiero C, Tsai B, Rapoport TA, Lencer WI 2002. Role of ubiquitination in retro-translocation of cholera toxin and escape of cytosolic degradation. EMBO Rep 3: 1222–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsa MM, Mondotte JA, Iglesias NG, Assuncao-Miranda I, Barbosa-Lima G, Da Poian AT, Bozza PT, Gamarnik AV 2009. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog 5: e1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelhaas M, Malmstrom J, Pelkmans L, Haugstetter J, Ellgaard L, Grunewald K, Helenius A 2007. Simian virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell 131: 516–529 [DOI] [PubMed] [Google Scholar]
- Settembre EC, Chen JZ, Dormitzer PR, Grigorieff N, Harrison SC 2011. Atomic model of an infectious rotavirus particle. EMBO J 30: 408–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shavinskaya A, Boulant S, Penin F, McLauchlan J, Bartenschlager R 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J Biol Chem 282: 37158–37169 [DOI] [PubMed] [Google Scholar]
- Shibata Y, Hu J, Kozlov MM, Rapoport TA 2009. Mechanisms shaping the membranes of cellular organelles. Annu Rev Cell Dev Biol 25: 329–354 [DOI] [PubMed] [Google Scholar]
- Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA 2010. Mechanisms determining the morphology of the peripheral ER. Cell 143: 774–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AE, Lilie H, Helenius A 2003. Ganglioside-dependent cell attachment and endocytosis of murine polyomavirus-like particles. FEBS Lett 555: 199–203 [DOI] [PubMed] [Google Scholar]
- Smith MH, Ploegh HL, Weissman JS 2011. Road to ruin: Targeting proteins for degradation in the endoplasmic reticulum. Science 334: 1086–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehle T, Yan Y, Benjamin TL, Harrison SC 1994. Structure of murine polyomavirus complexed with an oligosaccharide receptor fragment. Nature 369: 160–163 [DOI] [PubMed] [Google Scholar]
- Stehle T, Gamblin SJ, Yan Y, Harrison SC 1996. The structure of simian virus 40 refined at 3.1 A resolution. Structure 4: 165–182 [DOI] [PubMed] [Google Scholar]
- Stephens EB, Compans RW 1988. Assembly of animal viruses at cellular membranes. Annu Rev Microbiol 42: 489–516 [DOI] [PubMed] [Google Scholar]
- Stirzaker SC, Both GW 1989. The signal peptide of the rotavirus glycoprotein VP7 is essential for its retention in the ER as an integral membrane protein. Cell 56: 741–747 [DOI] [PubMed] [Google Scholar]
- Sweet BH, Hilleman MR 1960. The vacuolating virus, S.V. 40. Proc Soc Exp Biol Med 105: 420–427 [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S 2009. Innate immunity to virus infection. Immunol Rev 227: 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JA, O’Brien JA, Lord VJ, Meyer JC, Bellamy AR 1993. The RER-localized rotavirus intracellular receptor: A truncated purified soluble form is multivalent and binds virus particles. Virology 194: 807–814 [DOI] [PubMed] [Google Scholar]
- Teter K, Holmes RK 2002. Inhibition of endoplasmic reticulum-associated degradation in CHO cells resistant to cholera toxin, Pseudomonas aeruginosa exotoxin A, and ricin. Infect Immun 70: 6172–6179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolonen N, Doglio L, Schleich S, Krijnse Locker J 2001. Vaccinia virus DNA replication occurs in endoplasmic reticulum-enclosed cytoplasmic mini-nuclei. Mol Biol Cell 12: 2031–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask SD, McDonald SM, Patton JT 2012. Structural insights into the coupling of virion assembly and rotavirus replication. Nat Rev Microbiol 10: 165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B 2007. Penetration of nonenveloped viruses into the cytoplasm. Annu Rev Cell Dev Biol 23: 23–43 [DOI] [PubMed] [Google Scholar]
- Tsai B, Rapoport TA 2002. Unfolded cholera toxin is transferred to the ER membrane and released from protein disulfide isomerase upon oxidation by Ero1. J Cell Biol 159: 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B, Rodighiero C, Lencer WI, Rapoport TA 2001. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell 104: 937–948 [DOI] [PubMed] [Google Scholar]
- Tsai B, Gilbert JM, Stehle T, Lencer W, Benjamin TL, Rapoport TA 2003. Gangliosides are receptors for murine polyoma virus and SV40. EMBO J 22: 4346–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Rolls MM, Rapoport TA 2002. Structural organization of the endoplasmic reticulum. EMBO Rep 3: 944–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124: 573–586 [DOI] [PubMed] [Google Scholar]
- Walczak CP, Tsai B 2011. A PDI family network acts distinctly and coordinately with ERp29 to facilitate polyomavirus infection. J Virol 85: 2386–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ye Y, Lencer W, Hansen TH 2006. The viral E3 ubiquitin ligase mK3 uses the Derlin/p97 endoplasmic reticulum-associated degradation pathway to mediate down-regulation of major histocompatibility complex class I proteins. J Biol Chem 281: 8636–8644 [DOI] [PubMed] [Google Scholar]
- Wang C, Yu J, Huo L, Wang L, Feng W, Wang CC 2012. Human protein-disulfide isomerase is a redox-regulated chaperone activated by oxidation of domain a′. J Biol Chem 287: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5: 365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiertz EJ, Jones TR, Sun L, Bogyo M, Geuze HJ, Ploegh HL 1996. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84: 769–779 [DOI] [PubMed] [Google Scholar]
- Willey RL, Maldarelli F, Martin MA, Strebel K 1992. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J Virol 66: 7193–7200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk B, Buchele B, Moradpour D, Rice CM 2008. A dynamic view of hepatitis C virus replication complexes. J Virol 82: 10519–10531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414: 652–656 [DOI] [PubMed] [Google Scholar]