Abstract
Much of the mammalian skeleton is composed of bones that originate from cartilage templates through endochondral ossification. Elucidating the mechanisms that control endochondral bone development is critical for understanding human skeletal diseases, injury response, and aging. Mouse genetic studies in the past 15 years have provided unprecedented insights about molecules regulating chondrocyte formation, chondrocyte maturation, and osteoblast differentiation, all key processes of endochondral bone development. These include the roles of the secreted proteins IHH, PTHrP, BMPs, WNTs, and FGFs, their receptors, and transcription factors such as SOX9, RUNX2, and OSX, in regulating chondrocyte and osteoblast biology. This review aims to integrate the known functions of extracellular signals and transcription factors that regulate development of the endochondral skeleton.
Endochondral ossification produces bone via a cartilage intermediate. Key extracellular signals (e.g., bone morphogenetic proteins), their receptors, and transcription factors (e.g., SOX9) are essential for this process.
1. INTRODUCTION
The osseous tissues in mammals are formed via two distinct processes during embryogenesis. Intramembranous bone formation produces many of the craniofacial bones directly from mesenchymal condensations. In contrast, endochondral ossification, the principal process responsible for forming much of the mammalian skeleton, generates bone via a cartilage intermediate. The transition from cartilage to bone is tightly coupled with chondrocyte, osteoblast, and vascular differentiation. This article focuses on the current understanding of the molecular basis for several key processes of endochondral ossification, including formation of the mesenchymal condensation, chondrocyte differentiation and maturation, and osteoblast development.
2. MESENCHYMAL CONDENSATION
Endochondral bone development begins with the condensation of mesenchymal cells of either neural crest in the craniofacial region (e.g., middle ear bones and temporal bones) or mesoderm elsewhere in the body (Fig. 1A). Formation of a mesenchymal condensation is a prerequisite for subsequent chondrogenic differentiation (Thorogood and Hinchliffe 1975). In the limb bud the condensation forms mainly through active congregation of cells without changes in cell proliferation. These condensations can be visualized by the dense packing of cells, the high affinity to the lectin peanut agglutinin, and the transient up-regulation of versican, tenascin, syndecan, N-CAM, and N-cadherin (Hall and Miyake 2000).
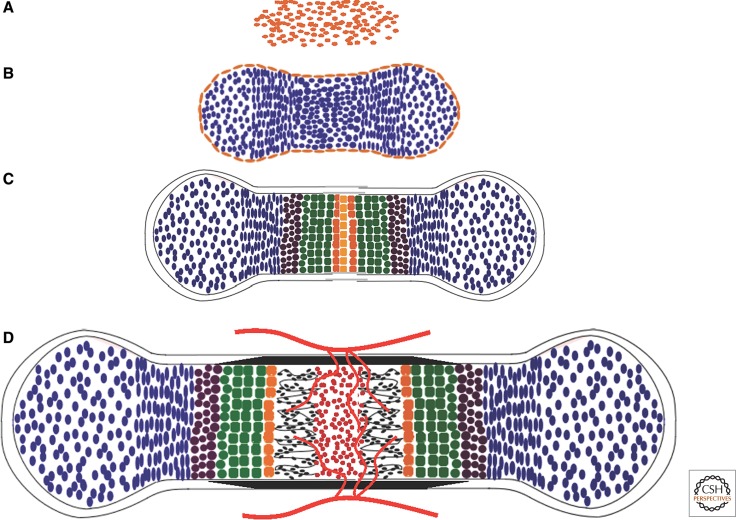
Figure 1.
Endochondral bone development. (A) Mesenchymal condensation. (B) Chondrocyte differentiation. Cells centrally located within the mesenchymal condensation differentiate to chondrocytes (blue), whereas the peripheral cells form the perichondrium (gold). (C) Chondrocyte maturation. After the initial proliferation, chondrocytes at the center of the cartilage primordium undergo progressive maturation through prehypertrophy (brown), hypertrophy (green), and terminal hypertrophy (orange). (D) Cartilage vascularization and bone collar formation. Following terminal hypertrophy of chondrocytes, blood vessels (red lines) from the surrounding tissue invade the center of the hypertrophic zone, concurrent with formation of the bone collar (black) from the surrounding perichondrium. Vascular invasion leads to resorption of cartilage matrix, formation of the marrow (red), and deposition of bone (black) within the marrow cavity.
The molecular drivers for mesenchymal condensation are not well understood. Based on expression analyses as well as studies with neutralizing antibodies, N-CAM and N-cadherin, which mediate Ca2+-dependent and -independent cell–cell adhesion, respectively, are implicated in mediating the cell-cell adhesion during condensation (DeLise et al. 2000). However, N-CAM-deficient mice developed to adulthood with no known defect in chondrogenesis (Cremer et al. 1994), and N-cadherin-deficient embryos died prematurely (at embryonic day 10 [E10]), precluding analyses of mesenchymal condensation (Radice et al. 1997). Hoxa13 and Hoxd13 have been shown genetically to control mesenchymal condensation in the autopod (Box 1) of the mouse (Fromental-Ramain et al. 1996; Stadler et al. 2001). Hoxa13- and Hoxd13-deficient embryos display defects in cell–cell adhesion mediated through Eph–ephrin signaling molecules that are involved in self-sorting of undifferentiated mesenchyme into skeletal precursors (Wada et al. 1998; Lu et al. 2008). Specifically, in the Hoxa13–/– autopod, the mesenchyme expressed a reduced level of EphA7 that normally demarcates the digit primordia, whereas ephrin A3, a high-affinity ligand for the EphA7 receptor normally marking the periphery of presumptive digits, was expressed diffusively in the autopod mesenchyme in the mutant embryo (Stadler et al. 2001). Importantly, the expression of other cell surface and proadhesion molecules including E-cadherin, N-CAM, EphA2, EphA4, ephrin A1, and ephrin A2 was not altered in the Hoxa13–/– embryos, indicating a specific relationship between Hoxa13 and Eph7A in the distal limb mesenchyme. Future research is needed to determine whether other Hox genes similarly control the condensations for the other limb elements and the axial skeleton.
BOX 1. AUTOPOD, ZEUGOPOD, AND STYLOPOD OF THE LIMB.
The vertebrate limb proximal–distal axis is subdivided into three morphogenetic domains, from distal to proximal, the autopod, the zeugopod, and the stylopd. Autopod, the most distal skeletal element, gives rise to the carpal bones (wrist), metacarpals, and phalangeal bones (digits) in the forelimb and the tarsal bones (ankle), metatarsals, and phalangeal bones (toes) in the hindlimb. Zeugopod, the middle skeletal element, gives rise to the radius and the ulna in the forelimb and the tibia and the fibula in the hindlimb. Stylopod, the most proximal element, gives rise to the humerus in the forelimb and the femur in the hindlimb.
Bone morphogenetic proteins (BMPs), members of the transforming growth factor-β (TGF-β) superfamily, are essential for the formation of chondrogenic mesenchymal condensations. BMPs transduce signals by binding to complexes of type I and II serine/threonine kinase receptors. Ligand binding induces phosphorylation of the receptors, which in turn phosphorylates and activates receptor SMADs (R-SMADs) 1, 5, and 8. The activated R-SMADs then complex with SMAD4 to enter the nucleus, eventually regulating gene expression (Feng and Derynck 2005; Massague et al. 2005). Additionally, BMPs can activate p38 and function through SMAD-independent mechanisms (Pogue and Lyons 2006). Studies with micromass cultures of limb bud mesenchyme have indicated that BMP signaling is required for the formation of condensations, although the relative contribution of SMAD-dependent versus -independent mechanisms is not clear (Pizette and Niswander 2000). Indeed, simultaneous deletion of Bmp2 and Bmp4 in prechondrogenic limb mesenchyme (with Prx1-Cre)3 led to the loss of zeugopod elements and to defective joint articulations (Bandyopadhyay et al. 2006). Conversely, genetic deletion of Noggin, a secreted antagonist of BMPs, resulted in enlarged cartilage elements and the failure to form joints (Brunet et al. 1998). Similarly, in humans, mutations in NOGGIN cause two autosomal dominant disorders: proximal symphalangism and multiple synostosis syndrome, both characterized by multiple joint fusions (Gong et al. 1999). The fact that in the mouse study not all cartilage elements were equally affected by the loss of Bmp2 and Bmp4 suggests that different skeletal elements may require different levels or specificity of BMP signaling.
The role of BMP signaling in chondrogenic mesenchymal condensation has been further dissected. Live imaging of limb mesenchymal cells undergoing successive phases of chondrogenesis in vitro revealed that BMP signaling was required for the coalescence of smaller aggregates into a tight cluster with a distinct outer boundary, a prerequisite step for chondrogenic differentiation (Barna and Niswander 2007). This event occurred independently of SOX9 (a critical factor for chondrocyte differentiation; see below), because Sox9-null cells can coalesce, even though they subsequently segregate from the condensations and adopt a distinctive “fibroblastoid” morphology. This finding appears to be at odds with that of conditional inactivation of SOX9 in limb mesenchyme (with Prx1-Cre), which led to the absence of mesenchymal condensations (Akiyama et al. 2002). However, it is likely that SOX9 is dispensable for the initial formation, but necessary for maintaining the condensation. Alternatively, the lack of mesenchymal condensations could be secondary to the failure in chondrocyte differentiation, which is known to require SOX9 (see below). Nonetheless, the molecular basis for this early SOX9-independent requirement of BMP signaling is not yet understood.
Fibroblast growth factor (FGF) signaling functions in the limb mesenchyme before the formation of condensations. FGFs are a large family of proteins (22 members in humans or mice) that carry out diverse biological functions in vertebrates (Itoh and Ornitz 2008). Most FGFs function by binding to cell surface tyrosine kinase FGF receptors (FGFR1–FGFR4 in humans and mice), leading to phosphorylation and activation of multiple signaling modules, including mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K), signal transducer and activator of transcription 1 (STAT1), and protein kinase C (PKC) (Eswarakumar et al. 2005; Turner and Grose 2010). Mutations in FGFRs are responsible for a large range of congenital skeletal disorders in humans (Ornitz and Marie 2002; Su et al. 2008). FGFRs 1 and 2 are expressed throughout limb bud mesenchyme before formation of condensations. Although conditional inactivation of Fgfr2 in mesenchymal precursor cells (with Dermo1-Cre)4 does not cause a major effect on cartilage (Yu et al. 2003), inactivation of both FGFRs 1 and 2 in the limb bud mesenchyme (with Prx1-Cre) results in smaller skeletal elements. Analysis of precondensation limb buds shows that FGF signaling provides an essential cell survival signal for the mesenchyme (Yu and Ornitz 2008).
3. CHONDROCYTE DIFFERENTIATION
Following mesenchymal condensation, cells in the core of the condensations differentiate into chondrocytes that secrete a cartilage matrix rich in types II, IX, and XI collagen and specific proteoglycans such as aggrecan, whereas the expression of type I collagen is suppressed. Cells at the periphery of the condensation form the perichondrium, which continues to express type I collagen and demarcates the developing skeletal element from the surrounding mesenchyme (Fig. 1B) (Caplan and Pechak 1987).
3.1. Sox Proteins
SOX9, a transcription factor of the SRY-related high mobility group box family of proteins, is the earliest known nuclear factor that is required for chondrogenesis. The importance of SOX9 in chondrogenesis is evident from the findings that heterozygous mutations within and around the Sox9 gene cause campomelic dysplasia, a severe form of human chondrodysplasia (Foster et al. 1994; Wagner et al. 1994). Similarly, haploinsufficiency of SOX9 results in chondrodysplasia in the mouse, and complete loss of SOX9 in prechondrogenic limb mesenchyme abolishes chondrogenesis altogether (Bi et al. 1999, 2001; Akiyama et al. 2002; Kist et al. 2002). More recently, live imaging techniques applied to micromass cultures of limb mesenchyme indicated that SOX9 is dispensable for the initial formation of mesenchymal condensations but necessary for the subsequent steps toward chondrocyte differentiation (Barna and Niswander 2007). Although SOX9 is known to regulate expression of a number of genes that encode chondrocyte-specific matrix proteins such as collagen II and aggrecan (de Crombrugghe et al. 2000), it is not clear whether these genes mediate the role of SOX9 during the early steps of chondrogenesis.
In addition to its early role, SOX9 regulates subsequent chondrocyte differentiation in collaboration with SOX5 and SOX6 (Smits et al. 2001; Akiyama et al. 2002). At the molecular level, the three Sox proteins cooperatively bind and activate the genes for many cartilage-specific extracellular matrix components (Lefebvre et al. 1998; Han and Lefebvre 2008). Thus, SOX9, SOX5, and SOX6 constitute a trio of transcription factors essential for chondrocyte differentiation.
3.2. Extracellular Signals Regulating Chondrocyte Differentiation
In addition to its role in mesenchymal condensation, BMP signaling also stimulates chondrocyte differentiation after the condensation stage. Simultaneous deletion of the BMP type I receptors, BMPR1A and BMPR1B, in chondrogenic cells (with Col2a1-Cre)5 resulted in a severe form of general chondrodysplasia in which a majority of the skeletal elements formed through endochondral ossification were absent (Yoon et al. 2005). Moreover, simultaneous removal of SMAD1 and SMAD5 by the same approach similarly resulted in severe chondrodysplasia (Retting et al. 2009). Thus, BMP signaling through R-SMADs is indispensable for the formation of cartilage following the activation of the Col2a1 gene. Interestingly, deletion of SMAD4, the binding partner for SMAD1, 5, or 8, with a similar Col2a1-Cre line did not severely affect cartilage formation (Zhang et al. 2005). The discrepancy between the R-SMAD- and SMAD4-deficient mice raises the possibility that BMP–SMAD1/5/8 signaling may control cartilage development in a SMAD4-independent manner. Indeed, BMPs were shown to induce R-SMAD nuclear localization in SMAD4-null colon cancer cells (Liu et al. 1997), but whether this occurs during chondrocyte differentiation remains to be investigated.
In contrast to BMP, some WNT proteins inhibit chondrocyte differentiation. The mammalian WNT family includes 19 members, and WNT proteins control morphogenesis and tissue patterning in a wide variety of organs (Wodarz and Nusse 1998). Depending on the cell context, WNTs interact with several types of membrane receptors, including those of the FRIZZLED (FZ) family and the low-density lipoprotein receptor-related proteins LRP5 and LRP6, to activate a variety of intracellular signaling cascades. In the β-catenin-dependent pathway, binding of WNTs stabilizes β-catenin, which enters the nucleus to interact with the lymphoid enhancer factor/T-cell factor (LEF/TCF) family of transcription factors and activates the transcription of downstream target genes. β-Catenin-independent pathways include those mediated by PKC or the Rho family of small GTPases, the latter of which can also participate in WNT/β-catenin signaling (Wu et al. 2008). In chick embryonic limbs and limb bud micromass cultures, ectopic expression of WNT1 or WNT7A inhibited chondrocyte differentiation but did not affect formation of mesenchymal condensations (Rudnicki and Brown 1997; Stott et al. 1999). The antichondrogenic role of WNT proteins may be mediated through β-catenin, as overexpression of a stabilized form of β-catenin in mouse embryonic limb mesenchyme resulted in a near complete loss of all limb cartilage elements (Hill et al. 2005). Continued exposure to WNT/β-catenin signaling appears to redirect limb mesenchymal cells to the soft connective tissue, but this process can be prevented by FGF signaling (ten Berge et al. 2008). WNT/β-catenin signaling also has further inhibitory action at stages after Col2a1 expression has been activated. Conditional overexpression of a stabilized β-catenin by Col2a1-Cre can lead to severe achondrodysplasia (Akiyama et al. 2004).
Similar to WNT/β-catenin signaling, Notch signaling also suppresses chondrogenesis. NOTCH signaling mediates communication between neighboring cells to control cell fate decisions in all metazoans (Artavanis-Tsakonas et al. 1999; Chiba 2006). The mammalian genome encodes four NOTCH receptors (NOTCH1 to -4) and at least five ligands (Jagged1 and -2 and Delta-like 1, 3, and 4). In the canonical NOTCH pathway, binding of the ligands to the NOTCH receptors present on the neighboring cell surface triggers two successive intramembranous proteolytic cleavages of the receptors, with the second event mediated by the γ-secretase complex and resulting in the release of the Notch intracellular domain (NICD) (Schroeter et al. 1998; Kopan and Goate 2000; Kopan and Ilagan 2009). On its release from the plasma membrane, NICD translocates to the nucleus, where it interacts with a transcription factor of the CSL family (RBPJκ/CBF-1 in mammals) to activate transcription of target genes (Honjo 1996). Expression studies revealed that multiple NOTCH ligands and receptors are expressed in the prechondrogenic mesenchyme. Abolition of NOTCH signaling in the embryonic limb mesenchyme by inactivating either the catalytic subunits of γ-secretase or the transcriptional effector RBPJκ accelerated chondrocyte differentiation (Dong et al. 2010). Conversely, enforced expression of the constitutively active NICD in limb mesenchyme completely abolished chondrogenesis, whereas simultaneous removal of RBPJκ fully rescued the chondrogenic defect caused by NICD overexpression (Dong et al. 2010). Similarly, overexpression of NICD in chondrogenic precursors (with Col2a1-Cre) resulted in generalized chondrodysplasia (Mead and Yutzey 2009). Thus, NOTCH signaling may inhibit both mesenchymal condensation and subsequent chondrocyte differentiation.
Retinoid signaling via the retinoic acid receptors (RARs) also acts to inhibit early chondrocyte differentiation. In transgenic mice expressing a constitutively active RARα in limb mesenchyme from the Hoxb-6 promoter, mesenchymal cells participated in condensations but failed to differentiate into chondrocytes (Cash et al. 1997; Weston et al. 2000). Conversely, the addition of an RARα-selective antagonist to cultures of these cells was sufficient to stimulate chondrocyte differentiation. However, a more recent study with mice harboring deletions of different RARs established a predominant role for RARγ in mediating the inhibitory role of retinoic acid in chondrogenesis (Shimono et al. 2011). Mechanistically, RARγ agonists appear to suppress the response to BMP signaling in mesenchymal cells and to alter their differentiation potential.
4. GROWTH PLATE DEVELOPMENT
Following formation of the cartilage primordia, initially all chondrocytes undergo rapid proliferation that drives the linear growth of the skeletal elements. At a certain stage specific for each element (e.g., ∼E14.5 for mouse tibia), chondrocytes in the center undergo progressive maturation. These cells eventually exit the cell cycle and undergo hypertrophy associated with the secretion of type X collagen (Poole 1991). The terminal hypertrophic chondrocytes express additional molecular markers such as matrix metalloproteinase 13 (MMP13) and are generally believed to undergo apoptosis (Fig. 1C). These changes are accompanied by vascular invasion of the hypertrophic cartilage and by differentiation of the inner perichondrium cells into osteoblasts, which secrete bone matrix to form the bone collar (Fig. 1D) (Caplan and Pechak 1987). Mouse knockout studies have shown that MMP13 plays a critical role in cleaving the extracellular matrix proteins within the hypertrophic cartilage and thus facilitating vascular invasion (Inada et al. 2004; Stickens et al. 2004). Vascular invasion of the hypertrophic cartilage leads to further degradation of the mineralized matrix by MMP9 produced by a resorptive cell type (the so-called chondroclast) (Vu et al. 1998), whose formation may be stimulated by RANKL (receptor activator of NF-κB ligand) produced by hypertrophic chondrocytes (Xiong et al. 2011). Vascular invasion also brings with it osteoprogenitors that differentiate into osteoblasts, which establish the primary ossification center to generate the trabecular bone (Maes et al. 2010). The orderly maturation of chondrocytes in the embryonic growth plate produces zones of proliferation, hypertrophy, and bone formation, linearly progressing from the articular ends (epiphysis) to the midshaft (diaphysis) of the skeletal element. The proliferative zone can be further divided into regions of round versus columnar chondrocytes based on their distinct morphology (Fig. 2). Continued proliferation of the less mature chondrocytes at the epiphysis, followed by their hypertrophy and their eventual replacement by trabecular bone near the diaphysis, results in a distal displacement of the growth plate and longitudinal growth of each skeletal element (Long 2012b). The orderly progression of the growth plate is critical for proper development of the endochondral skeleton (Kronenberg 2003) and is subject to regulation by key extracellular signals and nuclear factors that are discussed below.
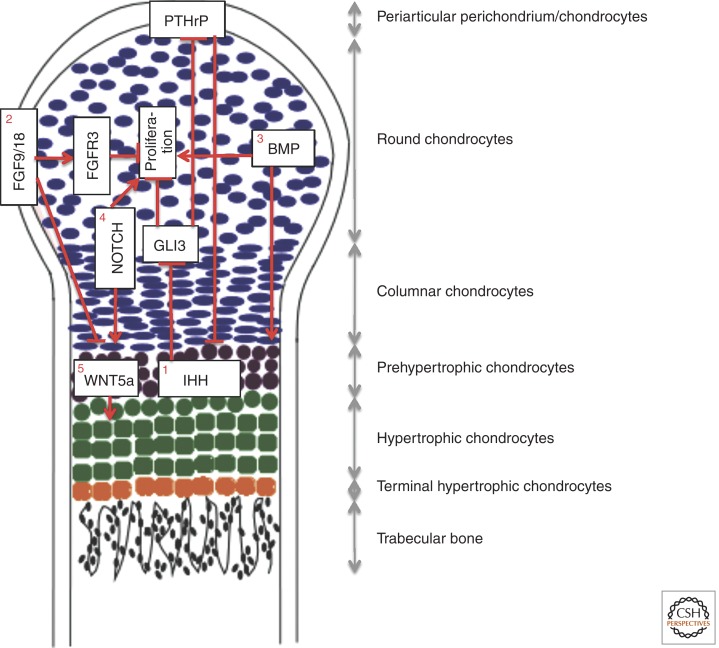
Figure 2.
Extracellular signals regulating growth plate development. Depicted is a longitudinal section through one of two growth plates of a mouse long bone during late embryogenesis (E15.5–E19). The growth plate at this stage is without a secondary ossification center and is organized into distinct domains as indicated. (1) IHH and PTHrP coordinate chondrocyte proliferation and maturation through a negative-feedback mechanism. IHH produced by pre- and early hypertrophic chondrocytes stimulates chondrocyte proliferation and PTHrP transcription through derepression of GLI3. PTHrP in turn suppresses chondrocyte maturation associated with IHH expression. Direct IHH signaling also regulates the formation of columnar chondrocytes from round chondrocytes (not depicted here). (2) FGF9/18 from the perichondrium suppresses chondrocyte proliferation and maturation. FGFR3 expressed in chondrocytes is a likely receptor for FGF9/18 to suppress proliferation in the growth plate late in embryonic development and during postnatal bone growth. FGF9/18 may use other yet-to-be-established mechanisms to suppress chondrocyte maturation in the early embryo. (3) BMPs expressed by both chondrocytes and perichondrial cells promote proliferation and maturation. (4) NOTCH signaling in chondrocytes promotes proliferation and maturation. (5) WNT5A expressed by prehypertrophic chondrocytes stimulates hypertrophy.
4.1. Extracellular Signals Regulating Growth Plate Development
4.1.1. PTHrP
PTHrP (parathyroid hormone-related peptide) is a paracrine factor expressed at highest levels by cells of the periarticular perichondrium and at a lower level by proliferating chondrocytes near the articular surface (Fig. 2) (Lee et al. 1995). The receptor for PTHrP (PTHR1) is expressed at low levels by proliferating chondrocytes but at high levels by the maturing chondrocytes before they become hypertrophic (Vortkamp et al. 1996; St-Jacques et al. 1999). Targeting inactivation of the gene encoding either PTHrP or its receptor in the mouse resulted in neonatal lethal short-limb dwarfism caused by premature hypertrophy of chondrocytes (Karaplis et al. 1994; Lanske et al. 1996), thus identifying a key role for PTH signaling in suppressing the onset of chondrocyte hypertrophy in the growth plate. Conversely, misexpression of PTHrP in all chondrocytes markedly delayed chondrocyte hypertrophy, resulting in a completely cartilaginous endochondral skeleton at birth (Weir et al. 1996). Similarly in humans, inactivating mutations in PTHR1 cause Blomstrand chondrodysplasia, characterized by advanced skeletal maturation with shortened long bones and increased bone density (Jobert et al. 1998; Karaplis et al. 1998; Karperien et al. 1999), whereas gain-of-function mutations in PTHR1 cause Jansen metaphyseal chondrodysplasia (Schipani et al. 1995, 1996), which was recapitulated in mice overexpressing such a mutant receptor (Schipani et al. 1997). Mechanistically, PTHrP appears to delay chondrocyte hypertrophy, mainly by activating cAMP-dependent signaling that both increases the activity of SOX9 (Huang et al. 2001; Guo et al. 2002), and regulates the HDAC4–MEF2C complex (see below).
4.1.2. IHH
The expression of PTHrP in the periarticular region is strictly dependent on Indian hedgehog (IHH), a member of the Hedgehog (HH) family that in mammals also includes Sonic hedgehog (SHH) and Desert hedgehog (DHH). The HH family of proteins plays fundamental roles in animal development and is conserved from flies to humans (Ingham and McMahon 2001; McMahon et al. 2003; Huangfu and Anderson 2006). The HH signal is transduced through Smoothened (SMO), a seven-pass transmembrane domain protein, and ultimately controls the processing and subcellular localization of the Gli transcription factors (GLI1 to -3) that regulate expression of downstream target genes. In the developing cartilage, IHH is primarily expressed by chondrocytes immediately before hypertrophy (prehypertrophic chondrocytes) and also by early hypertrophic chondrocytes. IHH signals to both the proliferative chondrocytes and the overlying perichondrial cells (Vortkamp et al. 1996; St-Jacques et al. 1999). Ihh–/– embryos exhibited a severe reduction in chondrocyte proliferation and premature hypertrophy of chondrocytes (St-Jacques et al. 1999; Long et al. 2001). Subsequent genetic manipulation of Smo in chondrocytes revealed that direct IHH input was required for chondrocyte proliferation (Long et al. 2001), but not for the regulation of chondrocyte hypertrophy. Instead, hypertrophy depended primarily on PTHrP, which was induced by IHH (St-Jacques et al. 1999; Karp et al. 2000; Long et al. 2001). Direct IHH signaling also regulates the formation of columnar chondrocytes from round chondrocytes within the proliferating zone (Kobayashi et al. 2005; Hilton et al. 2007). The control of PTHrP by IHH appears to be through direct HH signaling in the target cells, as localized removal of Smo (with Col2a1-CreERTM)6 led to a corresponding loss of PTHrP expression within the periarticular region (Hilton et al. 2007). Finally, the role of IHH in chondrocyte proliferation and PTHrP expression is mainly mediated through the derepression of GLI3 repressor function, as simultaneous removal of both GLI3 and IHH restored normal proliferation, PTHrP expression, and hypertrophy (Hilton et al. 2005; Koziel et al. 2005). Overall, these studies support a model in which IHH and PTHrP jointly regulate chondrocyte proliferation and maturation.
4.1.3. FGF
In addition to the IHH/PTHrP regulatory axis, FGF signaling also controls growth plate development. Conditional inactivation of Fgfr1 in chondrocytes (Col2a1-Cre) delayed maturation of hypertrophic chondrocytes (Jacob et al. 2006), whereas inactivation of Fgfr2 in skeletal mesenchymal precursor cells (Dermo1-Cre) resulted in skeletal dwarfism and decreased bone density, without causing major effects on cartilage (Yu et al. 2003). Fgfr3–/– mice have increased levels of chondrocyte proliferation (Deng et al. 1996; Eswarakumar and Schlessinger 2007) and an expanded hypertrophic zone that is most evident postnatally (Colvin et al. 1996; Deng et al. 1996). In contrast, overexpression of activated FGFR3 (achondroplasia or thanatophoric dysplasia mutation) in the growth plate of transgenic mice reduced chondrocyte proliferation and resulted in decreased numbers of cells in the prehypertrophic and hypertrophic zones (Naski et al. 1998; Chen et al. 2001; Iwata et al. 2001). Interestingly, in Fgfr3–/– mice, IHH expression and signaling in chondrocytes was increased relative to wild-type mice, whereas in the mice overexpressing activated FGFR3, IHH expression and signaling was inhibited (Naski et al. 1998; Chen et al. 1999; Li et al. 1999). In contrast to phenotypes observed in mice, IHH and PTHR1 expression were not altered in human achondroplasia or thanatophoric dysplasia growth plate tissue (Cormier et al. 2002). Thus, FGFR3 signaling may inhibit chondrocyte proliferation and hypertrophy in part through indirect modulation of the IHH/PTHrP axis, but additional mechanisms may also mediate the pathological consequences of FGFR3 activating mutations.
The mechanisms by which FGFR3 signaling regulates chondrocyte proliferation are not well defined. Early studies showed that activation of FGFR3 in growth plate chondrocytes leads to phosphorylation of STAT1 and inhibition of chondrocyte proliferation (Li et al. 1999). However, more recent studies have shown that activation of FGFR3 down-regulates AKT activity to cause decreased chondrocyte proliferation (Priore et al. 2006). Additionally, activation of MEK1 in chondrocytes leads to a chondrodysplasia phenotype accompanied by decreased chondrocyte differentiation (Murakami et al. 2004). Furthermore, MEK1 activation inhibited the skeletal overgrowth seen in mice lacking Fgfr3. Thus, FGFR3 may regulate chondrocyte proliferation through STAT1 and AKT, but differentiation through the MAPK pathway.
The physiologic FGF ligand(s) for FGFR3 in the growth plate remain to be identified. However, FGF9 and FGF18 are good candidates because both are expressed in the adjacent perichondrium (Liu et al. 2002; Ohbayashi et al. 2002; Hung et al. 2007), and FGF18 is also expressed at low levels by growth plate chondrocytes (Lazarus et al. 2007). In addition, both ligands are known to activate FGFR3 in vitro (Zhang et al. 2006). More importantly, studies of Fgf9–/– and Fgf18–/– mice and mice with an activating mutation of FGFR3 show that FGF9, FGF18, and FGFR3 can promote chondrocyte proliferation at early stages of embryonic development (E14.5–15.5) (Iwata et al. 2000; Liu et al. 2002; Hung et al. 2007). In the late embryonic growth plate, loss-of-function studies show that FGF18 and FGFR3 function to inhibit proliferation and accelerate maturation (Colvin et al. 1996; Deng et al. 1996; Liu et al. 2002). The observation that Fgf9–/– and Fgf18–/– mice begin to manifest accelerated chondrocyte maturation during embryogenesis when the Fgfr3–/– embryos are relatively normal suggests that FGF9 and FGF18 may use receptor(s) other than FGFR3 to regulate growth plate development at early embryonic stages.
4.1.4. BMP
BMP signaling regulates multiple aspects of growth plate development (Pogue and Lyons 2006). Although mouse embryos completely lacking both Bmpr1a and Bmpr1b in chondrogenic cells (with Col2a1-Cre) (Bmpr1aCKO/Bmpr1b–/–) failed to form many cartilage elements, those carrying one remaining Bmpr1b allele (Bmpr1aCKO/Bmpr1b+/−) developed all elements but exhibited marked defects in chondrocyte proliferation, survival, and hypertrophy (Yoon et al. 2006). Similar defects were observed in the limbs of mice lacking Smad1 and Smad5 in chondrogenic cells (with Col2a1-Cre) (Retting et al. 2009). In embryos lacking either Bmpr1a/b or Smad1/5, IHH expression and signaling was reduced and may have contributed to the overall phenotype (Yoon et al. 2006; Retting et al. 2009). On the contrary, FGF signaling was increased in these mutant growth plates. Further supporting the antagonism between BMP and FGF signaling, the expression of Bmp4 was suppressed in both growth plate chondrocytes and in the surrounding perichondrium of mice overexpressing activated FGFR3 (Naski et al. 1998). Thus, proper balancing of BMP and FGF signaling may be important for normal development of the embryonic growth plate.
4.1.5. WNT
Members of the WNT family also regulate the proper progression of growth plate chondrocytes. WNT5A and WNT5B are expressed by chondrocytes during the transition from proliferative to hypertrophic zone in the mouse. Wnt5a–/– mice exhibited a marked delay in chondrocyte hypertrophy (Yang et al. 2003). Interestingly, overexpression of either WNT5A or WNT5B in chondrocytes also delayed hypertrophy in both mouse and chicken embryos (Hartmann and Tabin 2000; Yang et al. 2003). Thus, a proper level of WNT5A/B signaling is necessary for the normal transition of chondrocytes to hypertrophy.
4.1.6. NOTCH
NOTCH signaling modulates development of the growth plate chondrocytes. Deletion of both NOTCH1 and NOTCH2 or of RBPJκ in the limb mesenchyme (with Prx1-Cre) before chondrogenesis did not grossly affect the formation of cartilage anlagen but reduced chondrocyte proliferation and delayed both the onset and the terminal progression of chondrocyte hypertrophy in the mouse embryo (Hilton et al. 2008; Dong et al. 2010; Kohn et al. 2012). Conversely, forced expression of the intracellular domain of Notch1 (NICD) in differentiated chondrocytes (with tamoxifen induction of Col2-CreERT2)7 accelerated both the onset and terminal progression of hypertrophy in an RBPJκ-dependent manner (Kohn et al. 2012). Thus, proper Notch signaling within the growth plate is critical for normal proliferation and maturation of chondrocytes. Future studies are necessary to elucidate how NOTCH intersects with the other extracellular signals to ensure proper development of the growth plate (Fig. 2).
4.2. Nuclear Factors Regulating Growth Plate Development
In addition to the extracellular signals described above, a number of nuclear factors have also been shown to regulate chondrocyte hypertrophy. RUNX2, a runt-domain transcription factor, plays an important role in promoting chondrocyte hypertrophy. RUNX2 is initially expressed within the chondrogenic mesenchyme, subsequent to and dependent on SOX9 expression (Akiyama et al. 2002, 2005). Following the formation of cartilage anlagen, RUNX2 becomes more restricted to the perichondrial cells and osteoblasts, but it is reexpressed in the prehypertrophic and early hypertrophic chondrocytes. Several lines of evidence have supported RUNX2 as an important positive regulator of the hypertrophic program. First, Runx2–/– mice lack hypertrophic chondrocytes in multiple skeletal elements (Inada et al. 1999; Kim et al. 1999). Second, forced expression of RUNX2 in all chondrocytes leads to premature chondrocyte hypertrophy, and also rescues the defects in hypertrophy observed in Runx2–/– mice (Takeda et al. 2001; Ueta et al. 2001; Tu et al. 2012b), whereas expression of a dominant–negative RUNX2 mutant inhibits chondrocyte hypertrophy (Ueta et al. 2001). The role of RUNX2 in chondrocyte hypertrophy appears to be shared with RUNX3, as simultaneous deletion of both completely blocked hypertrophy in all endochondral skeletal elements (Yoshida et al. 2004). Finally, RUNX2 may also function in the perichondrium to regulate development of the growth plate through indirect mechanisms, as RUNX2 regulates the perichondrial expression of FGF18, which in turn signals to chondrocytes (Hinoi et al. 2006).
The direct effect of RUNX2 on chondrocyte hypertrophy appears to be regulated by HDAC4, a member of the class II histone deacetylases (Vega et al. 2004). Like RUNX2, HDAC4 is expressed in the prehypertrophic and early hypertrophic chondrocytes. Genetic deletion of HDAC4, like overexpression of RUNX2, greatly accelerated chondrocyte hypertrophy. Conversely, overexpression of HDAC4 in all chondrocytes inhibited hypertrophy, mimicking the effect of RUNX2 deletion. Biochemically, HDAC4 physically interacts with RUNX2 and reduces RUNX2 binding to DNA. Thus, HDAC4 prevents premature hypertrophy of chondrocytes in part by directly suppressing RUNX2 activity.
HDAC4 also suppresses chondrocyte hypertrophy by inhibiting MEF2C activity. MEF2C, a member of the myocyte enhancer factor-2 (MEF2) family, is expressed by prehypertrophic and early hypertrophic chondrocytes (Arnold et al. 2007). Conditional deletion or expression of a dominant–negative mutant of MEF2C in chondrocytes impairs hypertrophy, whereas an activated form induces precocious chondrocyte hypertrophy. MEF2C stimulates hypertrophy partly by increasing RUNX2 expression. Moreover, HDAC4 and MEF2C antagonize each other genetically, and HDAC4 functions as a corepressor of MEF2C to suppress chondrocyte hypertrophy.
The activity of HDAC4 in turn is regulated by other factors. PTHrP induces dephosphorylation of HDAC4 and promotes its nuclear translocation and repression of MEF2 transcriptional activity (Kozhemyakina et al. 2009). Conversely, salt-inducible kinase 3 (SIK3) is required for the exclusion of HDAC4 from the nuclei so that hypertrophy could occur, as Sik3–/– mice exhibited a marked delay in chondrocyte hypertrophy (Sasagawa et al. 2012). Thus, the HDAC4–MEF2C complex appears to be a central node of regulation for chondrocyte hypertrophy.
Additional regulators of chondrocyte hypertrophy have been identified. SOX9 has been proposed to both prevent cells from prematurely entering the prehypertrophic stage and to promote the subsequent hypertrophy (Dy et al. 2012). The transcription factors FoxA2 and FoxA3 are necessary for chondrocyte hypertrophy (Ionescu et al. 2012). SHOX2, a homeodomain-containing transcription factor, is necessary for RUNX2/RUNX3 expression and hypertrophy within the stylopod (see Box 1) (Cobb et al. 2006; Yu et al. 2007). Finally, Zfp521, a zinc finger-containing transcriptional coregulator, suppresses chondrocyte hypertrophy and partly mediates the function of PTHrP in this process (Correa et al. 2010; Seriwatanachai et al. 2011). Overall, chondrocyte hypertrophy is tightly regulated by a multitude of extracellular signals and nuclear factors. How the extracellular signals integrate with the nuclear factors represents an important area of future research.
5. OSTEOBLASTOGENESIS
Coupled with chondrocyte hypertrophy, osteoblast differentiation first occurs within the perichondrium and continues within the marrow cavity following vascularization of the hypertrophic cartilage. The differentiation of osteoblasts from their mesenchymal progenitors is regulated by specific extracellular signals and transcription factors as discussed below.
5.1. Extracellular Signals Regulating Osteoblast Differentiation
5.1.1. IHH Signaling
IHH critically regulates osteoblast differentiation during endochondral skeletal development. Ihh–/– mice completely lack osteoblasts within the endochondral skeleton, although intramembranous osteoblasts still form (St-Jacques et al. 1999). Moreover, IHH signaling is directly required for perichondrial cells to initiate osteoblast differentiation, as cells genetically deficient in SMO (an obligatory transducer of the HH signal) exhibit a cell-autonomous defect in osteoblast differentiation (Long et al. 2004). In the absence of IHH, the mesenchymal progenitors in the perichondrium fail to express RUNX2, which is indispensible for osteoblast differentiation. Interestingly, although the activation of RUNX2 expression by IHH in perichondrial cells can be achieved through removal of the GLI3 repressor, induction of Osterix (OSX or SP7), an essential transcription factor acting downstream from RUNX2, requires both the suppression of GLI3 and activation of GLI2 (Hilton et al. 2005; Joeng and Long 2009). In addition to osteoblast differentiation in the perichondrium, HH signaling is also necessary for trabecular bone formation, as indicated by lack of trabecular bone in mouse embryos lacking SMO in osteoprogenitor cells (Long et al. 2004). Thus, IHH, secreted by the prehypertrophic and early hypertrophic chondrocytes, controls the onset of osteoblast differentiation in the endochondral skeleton.
5.1.2. WNT Proteins
The WNT proteins have important roles in regulating osteoblast differentiation. Most notably, mice lacking β-catenin in mesenchymal progenitors fail to develop mature osteoblasts (Day et al. 2005; Hill et al. 2005; Hu et al. 2005; Rodda and McMahon 2006). Specifically, β-catenin is required for the progression from the RUNX2-positive to the OSX-positive stage, and from OSX-positive cells to mature osteoblasts (Hu et al. 2005; Rodda and McMahon 2006). Similarly, deletion of both LRP5 and LRP6 in mesenchymal progenitors results in loss of osteoblasts (Joeng et al. 2011). Interestingly, deletion of β-catenin in either mesenchymal progenitors or OSX-positive cells caused ectopic cartilage formation in place of bone, possibly owing to a fate switch for the bipotential progenitors (Day et al. 2005; Hill et al. 2005; Hilton et al. 2005; Rodda and McMahon 2006). Finally, β-catenin signaling appears to function downstream from IHH, as β-catenin deletion does not impair IHH signaling in the perichondrium (containing osteoblast progenitors), whereas IHH removal abolishes β-catenin signaling in that compartment (Hu et al. 2005; Mak et al. 2006). Thus, IHH and WNT signaling sequentially regulate osteoblast differentiation in the endochondral skeleton during embryogenesis.
WNT signaling has been further implicated in regulating bone accrual during postnatal life. In humans, loss-of-function mutations in the gene encoding the WNT coreceptor LRP5 cause osteoporosis–pseudoglioma syndrome (Gong et al. 2001), a form of juvenile-onset osteoporosis. Conversely, a different set of mutations in Lrp5 that render the coreceptor refractory to extracellular WNT inhibitors DKK1 and Sclerostin (encoded by Sost) cause high-bone-mass syndrome (Boyden et al. 2002; Little et al. 2002; Ai et al. 2005; Ellies et al. 2006; Semenov and He 2006). Further supporting the role of LRP5 signaling in bone formation, loss-of-function or loss-of-expression mutations in Sost, which are expected to enhance LRP5 signaling, result in the bone-thickening diseases known as sclerosteosis or van Buchem disease, respectively (Balemans et al. 2001, 2002; Brunkow et al. 2001; Staehling-Hampton et al. 2002). Furthermore, mice deficient in Lrp5 (Kato et al. 2002) or Wnt10b (Bennett et al. 2005) exhibit lower bone mass postnatally, whereas Lrp6 haploinsufficiency further decreases bone mass in Lrp5-null mice (Holmen et al. 2004). Conversely, mice expressing LRP5 with mutations that cause high bone mass in humans also exhibit a high-bone-mass phenotype (Yadav et al. 2008; Cui et al. 2011). Analyses of the LRP5 mutant mice revealed that LRP5 predominantly affects osteoblast number and function in postnatal animals, with no obvious effect on osteoclasts (Kato et al. 2002; Yadav et al. 2008; Cui et al. 2011). Thus, genetic evidence has shown the importance of LRP5 signaling in postnatal bone formation.
The mechanism through which LRP5 controls postnatal bone accrual is not well understood. Because deletion of β-catenin in mature osteoblasts or osteocytes did not produce a similar bone phenotype to Lrp5-deficient mice (i.e., increase in osteoblast number and activity with no obvious effect on osteoclasts) (Glass et al. 2005; Holmen et al. 2005; Kramer et al. 2010), LRP5 could function independently of β-catenin, at an earlier stage within the osteoblast lineage, or extrinsic to skeletal tissue. The potential stage specificity of LRP5 function was addressed by two different groups but with conflicting results. Whereas one group reported that deletion of Lrp5, either in mature osteoblasts (ColI-Cre)8 (Yadav et al. 2008) or in mesenchymal progenitors (Dermo1-Cre, and therefore all cells of the osteoblast lineage) (Yadav et al. 2010), did not produce any bone phenotype, a second group showed that deletion of Lrp5 predominantly in osteocytes (differentiated from osteoblasts; Dmp1-Cre)9 resulted in less bone (Cui et al. 2011). The different findings support competing models. Whereas the first group argues that LRP5 functions in the duodenum to suppress circulating levels of serotonin, which in turn directly inhibits osteoblast proliferation (Yadav et al. 2008), the second group believes that LRP5 functions directly in the osteoblast lineage (Cui et al. 2011). Thus, the precise mechanism for LRP5 regulation of osteoblasts remains to be further elucidated.
β-Catenin-independent WNT signaling also has been implicated in regulating osteoblast differentiation. WNT-induced G-protein-coupled phosphatidylinositol signaling and PKCδ activation have been shown to promote osteoblast differentiation by stimulating the progression from RUNX2- to OSX-positive cells through an unknown mechanism (Tu et al. 2007). In addition, WNT5A is believed to promote osteoblast differentiation by inducing suppressive histone methylation at the promoters of peroxisome proliferator-activated receptor-γ (PPARγ) target genes, thereby inhibiting the adipogenic program (Takada et al. 2007). On the other hand, WNT10B inhibits adipogenesis by suppressing the expression of the adipogenic transcription factors PPARγ and C/EBPα (CCAAT/enhancer-binding protein-α) (Ross et al. 2000). Exactly how inhibition of adipogenesis by either WNT5A or WNT10B leads to osteoblast differentiation is not clear. Finally, deletion of Frizzled9 decreased bone mass in part through down-regulation of ISG15, a ubiquitinlike protein modifier, without impairing β-catenin signaling (Albers et al. 2011). Thus, WNT signaling regulates osteoblast differentiation through multiple mechanisms.
5.1.3. NOTCH Signaling
Mouse genetic studies suggest that NOTCH signaling suppresses osteoblast differentiation. Removal of either presenilin 1 (PS1), PS2, NOTCH1, NOTCH2, or RBPJκ in the embryonic limb mesenchyme markedly enhanced osteoblast formation, resulting in high bone mass in adolescent mice (Hilton et al. 2008; Tu et al. 2012a). The high-bone-mass phenotype in NOTCH-deficient mice coincided with decreased numbers of bone marrow mesenchymal progenitors, supporting a model in which NOTCH signaling normally suppresses osteoblast differentiation from progenitor cells. The suppression seems to occur at a stage before OSX activation, as deletion of RBPJκ in OSX-positive cells did not cause a similar phenotype (Tu et al. 2012a). Mechanistically, NOTCH signaling inhibits osteoblast differentiation partly by inducing HEY1 and HEYL, which suppress osteoblast differentiation, both through decreasing RUNX2 transcriptional activity (Hilton et al. 2008), and by reducing the expression of NFATC1, a positive regulator of osteoblast differentiation (Tu et al. 2012a).
Consistent with the negative role of NOTCH signaling in osteoblast differentiation, NOTCH1 haploinsufficiency in humans causes ectopic ossification in the aortic valves (Garg et al. 2005; Mohamed et al. 2006), whereas gain-of-function mutations in NOTCH2 are responsible for Hadju–Cheney syndrome, a disorder of severe and progressive bone loss (Isidor et al. 2011; Simpson et al. 2011). Similarly, transgenic mice globally overexpressing HEY1 show progressive osteopenia (Salie et al. 2010). Interestingly, whereas NOTCH activation suppressed osteoblast differentiation from early precursors (Zanotti et al. 2008), at a later stage it caused pathological overproduction of immature osteoblasts, similar to what occurs in certain human osteosarcomas (Engin et al. 2008, 2009; Tao et al. 2010). These studies highlight the stage-specific functions of constitutive NOTCH activation in the osteoblast lineage.
5.1.4. BMP Signaling
Genetic studies have uncovered important roles for BMP2 and BMP4 in promoting osteoblast differentiation. Knockout experiments showed that a critical threshold level of BMP2 and BMP4 signaling is required for the formation of mature osteoblasts by controlling the transition from RUNX2- to OSX-positive cells (Bandyopadhyay et al. 2006). Interestingly, mice lacking only BMP2 in the limb mesenchyme form bone during embryogenesis but display a clear defect in postnatal bone mineral density, resulting in frequent fractures that fail to heal (Tsuji et al. 2006). Thus, BMP2 and BMP4 appear to redundantly control osteoblast differentiation during embryogenesis, with BMP2 having a unique role in postnatal bone formation.
BMP3 may counteract the activities of BMP2 and BMP4 to maintain a proper bone mass in vivo, as Bmp3-null mice had more trabecular bone than wild-type mice (Daluiski et al. 2001). Recent studies have shown that BMP3 engages the BMP type II receptor Acvr2b to inhibit signaling by BMP2 or BMP4 (Kokabu et al. 2012). In the postnatal skeleton, BMP3 is mainly produced by osteoblasts and osteocytes and it in turn reduces differentiation from the earlier-stage osteoblast lineage cells (Kokabu et al. 2012). Thus, BMP3 may function in a negative-feedback mechanism to ensure the production of a proper number of osteoblasts.
The importance of BMP signaling in bone has also been supported by studies of the BMP receptor, BMPR1A. Deletion of BMPR1A in preosteoblasts and osteoblasts, either in utero or postnatally, unexpectedly increased bone mass as a result of reduced bone resorption, even though bone formation was also decreased (Kamiya et al. 2008a,b, 2010). However, the precise role of BMPR1A signaling during osteoblast differentiation has not yet been elucidated, nor is it clear whether BMPR1A signals predominantly through the SMAD-dependent pathway. Nonetheless, BMPR1A appears to function through SMAD4 to promote the function of mature osteoblasts, as deletion of either molecule in mature osteoblasts decreased osteoblast function (Mishina et al. 2004; Tan et al. 2007).
5.1.5. FGF Signaling
FGF signaling has been implicated in osteoblast differentiation. A number of Fgfs are expressed in the perichondrium, where osteoblast differentiation occurs during endochondral bone development. These include Fgf1, -2, -6, -7, -8, -9, -17, and -18 (Colvin et al. 1999; Xu et al. 1999; Liu et al. 2002; Hung et al. 2007; Lazarus et al. 2007), although they are not all expressed uniformly in all skeletal elements. Importantly, Fgf18–/– embryos exhibited defects in the formation of mature osteoblasts despite normal RUNX2 expression (Ohbayashi et al. 2002; Liu et al. 2007).
Studies of FGF receptors have revealed their diverse roles in the regulation of osteoblasts. In particular, FGFR1 signaling at an early developmental stage promotes osteoblast differentiation without affecting RUNX2 expression, but FGFR1 signaling in mature osteoblasts inhibits their mineralization activity (Jacob et al. 2006). In contrast, FGFR2 promotes the proliferation of preosteoblasts and the anabolic function of mature osteoblasts (Yu et al. 2003), partly through up-regulation of RUNX2 expression (Eswarakumar et al. 2002, 2004). Finally, mice lacking FGFR3 showed an increase in osteoblast number but a decrease in osteoid mineralization (Valverde-Franco et al. 2004). Thus, FGF signaling through different receptors has diverse roles in regulating preosteoblast proliferation, osteoblast differentiation, as well as the activity of mature osteoblasts, but the precise stages at which FGFs regulate proliferation and differentiation, potential redundancy between FGFRs, and the intracellular signaling cascades responsible for each function remain to be elucidated. In summary, all major developmental signaling pathways regulate specific aspects of osteoblast differentiation and/or function (Fig. 3).
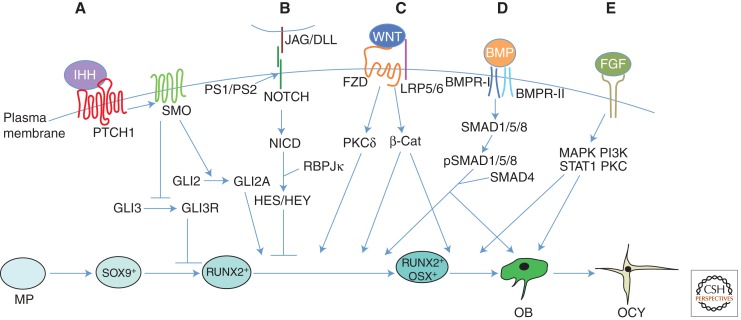
Figure 3.
Extracellular signals regulating osteoblast differentiation. Model is based on studies of the mouse limb skeleton. Osteoblasts differentiate from mesenchymal progenitors (MP) through distinct developmental stages marked by expression of key transcription factors including SOX9, RUNX2, and OSX. Mature osteoblasts (OB) can further differentiate to osteocytes (OCY) or bone lining cells (not depicted) or undergo apoptosis (not depicted). (A) Indian hedgehog (IHH) signaling is required for osteoblast differentiation during endochondral bone development. IHH binding to the receptor Patched homolog 1 (PTCH1) activates signaling through Smoothened (SMO), thereby inhibiting the generation of the proteolytically cleaved GLI3 repressor (GLI3R) and promoting the generation of the full-length GLI2 activator (GLI2A). Whereas derepression of GLI3R is sufficient to generate RUNX2+ cells, both derepression of GLI3R and activation of GLI2A are necessary for progression to the RUNX2+OSX+ stage. (B) NOTCH signaling inhibits osteoblast differentiation. Following binding to their ligands, Jagged (JAG) or Delta-like (DLL), Notch receptors are proteolytically cleaved by the γ-secretase complex, leading to release of the Notch intracellular domain (NICD) from the plasma membrane. NICD interacts with RBPJκ and together they activate downstream target genes, including HES (Hairy and Enhancer of Split) and HEY (HES-related with YRPW motif) family transcription factors, ultimately leading to inhibition of osteoblast differentiation, seemingly at a stage before OSX activation. (C) WNT signaling promotes osteoblast differentiation. During β-catenin-dependent WNT signaling, β-catenin is stabilized following binding of WNT to its receptors Frizzled (FZD) and lipoprotein receptor-related protein 5 (LRP5) or LRP6, leading to the transcription of β-catenin target genes and ultimately stimulating progression from the RUNX2+ stage to the RUNX2+OSX+ stage, and from RUNX2+OSX+ cells to mature osteoblasts. WNT can also signal independently of LRP5/6 and β-catenin through protein kinase Cδ (PKCδ), promoting progression to the RUNX2+OSX+ stage through an unknown mechanism. (D) Bone morphogenetic protein (BMP) signaling stimulates osteoblast differentiation and function. Binding of BMP2 or BMP4 to their receptors results in phosphorylation of SMAD1, SMAD5, or SMAD8. These can then form a complex with their partner, SMAD4, and enter the nucleus to regulate gene expression, ultimately promoting the transition to RUNX2+OSX+ cells and enhancing the function of mature osteoblasts; however, a direct role for SMAD signaling in osteoblast differentiation remains to be shown. (E) Fibroblast growth factor (FGF) signaling has diverse roles in osteoblast lineage cells. FGFs function by binding to cell surface Tyr kinase FGF receptors (FGFR1–FGFR4 in humans and mice), leading to the activation of multiple signaling modules. FGF signaling regulates preosteoblast proliferation and osteoblast differentiation, as well as the function of mature osteoblasts. However, the precise stages at which FGFs regulate proliferation and differentiation, and the intracellular signaling cascades responsible for each function, remain to be elucidated. BMPR, BMP receptor; MAPK, mitogen-activated protein kinase; PI3K, phosphoinositide 3-kinase; STAT1, signal transducer and activator of transcription 1.
5.2. Transcription Factors Controlling Osteoblast Differentiation
5.2.1. RUNX2 Is Essential for Osteoblast Differentiation
RUNX2 is indispensable for osteoblast differentiation during both endochondral and intramembranous ossification. Homozygous deletion of RUNX2 or its nuclear targeting signal in mice resulted in a complete lack of osteoblasts (Komori et al. 1997; Otto et al. 1997), whereas haploinsufficiency of RUNX2 in either mice or humans causes cleidocranial dysplasia (Lee et al. 1997; Mundlos et al. 1997; Choi et al. 2001). Interestingly, in the absence of RUNX2, the perichondrium (normally containing osteogenic progenitors) becomes hypoplastic (Komori et al. 1997; Otto et al. 1997), indicating that RUNX2 may be necessary for the production and/or maintenance of the progenitors. In addition to osteoblast differentiation, RUNX2 is necessary for the proper function of mature osteoblasts (Ducy et al. 1999).
5.2.2. OSX Is Required Downstream from RUNX2
OSX, a transcription factor containing three C2H2-type zinc fingers, is necessary for osteoblast differentiation. Deletion of OSX results in a complete absence of osteoblasts in the mouse embryo, even though expression of RUNX2 is relatively normal (Nakashima et al. 2002). These results, together with the observation that Osx expression is abolished in Runx2–/– mice (Nakashima et al. 2002), indicate that OSX functions downstream from RUNX2 as another indispensable regulator of osteoblast differentiation. However, there are important differences between the phenotypes of Runx2–/– and Osx–/– mice. Whereas RUNX2 deletion leads to a hypoplastic perichondrium, loss of OSX causes ectopic cartilage formation beneath a thickened perichondrium at the midshaft of long bones (where a bone collar normally forms), possibly because of a fate switch of progenitors from osteoblasts to chondrocytes (Nakashima et al. 2002). Beyond its role in embryonic osteoblast differentiation, OSX is also critical for postnatal osteoblast and osteocyte formation and function (Zhou et al. 2010).
5.2.3. ATF4 Regulates Osteoblast Differentiation
ATF4, a member of the bZIP family of transcription factors, is believed to play important roles at later stages of osteoblast differentiation. Misregulation of ATF4 activity has been linked with the skeletal abnormalities seen in human patients with Coffin–Lowry syndrome and neurofibromatosis type I (Yang et al. 2004; Elefteriou et al. 2006). Mechanistically, ATF4 directly regulates the expression of osteocalcin and RANKL in osteoblasts (Yang et al. 2004; Elefteriou et al. 2005) and also promotes amino acid import into osteoblasts to support robust protein synthesis (Yang et al. 2004). Interestingly, members of the forkhead box O (FOXO) family of transcription factors may interact with ATF4 to promote amino acid import in osteoblasts (Ambrogini et al. 2010; Rached et al. 2010). Conversely, the leucine-zipper-containing nuclear protein FIAT seems to antagonize ATF4 activity (Yu et al. 2005). More recently, ATF4 was shown to act in chondrocytes to promote osteoblast differentiation in the mouse embryo, possibly through stimulation of IHH (Wang et al. 2012). Thus, ATF4 may regulate osteoblast differentiation during embryogenesis in part through an indirect mechanism.
5.2.4. Other Transcription Factors
Numerous nuclear factors have been shown to synergize with RUNX2 to promote osteoblast differentiation (Karsenty 2009; Long 2012a). These include MAF, TAZ, SATB2, RB, GLI2, DLX5, MSX2, and BAPX1, which function in different ways, including stimulating Runx2 expression, enhancing RUNX2 activity, or acting as coactivators (Tribioli and Lufkin 1999; Satokata et al. 2000; Thomas et al. 2001; Robledo et al. 2002; Cui et al. 2003; Hong et al. 2005; Lee et al. 2005; Dobreva et al. 2006; Shimoyama et al. 2007; Calo et al. 2010; Nishikawa et al. 2010). Conversely, some other nuclear factors (TWIST1, HAND2, ZFP521, STAT1, Schnurri 3, GLI3, HOXA2, and the HES/HEY proteins) have been found to suppress RUNX2 levels or activity (Kanzler et al. 1998; Kim et al. 2003; Bialek et al. 2004; Garg et al. 2005; Jones et al. 2006; Hilton et al. 2008; Ohba et al. 2008; Funato et al. 2009; Hesse et al. 2010). These factors also use a range of mechanisms, including blocking RUNX2 DNA binding, nuclear translocation, and protein expression.
Several transcription factors participate in the regulation of osteoblast differentiation by modulating OSX (Long 2012a). These include the tumor suppressor p53, which decreases OSX levels through an unknown mechanism (Wang et al. 2006), and NFATC1, a calcium-sensitive transcription factor that stimulates osteoblast differentiation by enhancing OSX transcriptional activity (Koga et al. 2005; Winslow et al. 2006).
Members of the AP1 transcription factor family have been shown to regulate osteoblast differentiation or function, but their relationship with RUNX2, OSX, or ATF4 is not yet clear. In particular, loss of the AP1 protein FRA1 in the mouse embryo led to a low-bone-mass phenotype owing to reduced production of bone matrix proteins (Eferl et al. 2004), whereas transgenic overexpression of FRA1 resulted in high bone mass resulting from increased osteoblast differentiation (Jochum et al. 2000). Similarly, transgenic overexpression of ΔFosB, a naturally occurring isoform of the AP1 protein FOSB, increased bone mass through cell-autonomous stimulation of osteoblast differentiation and function (Sabatakos et al. 2000; Kveiborg et al. 2004). Conversely, genetic deletion of the AP1 factor JUNB reduced osteoblast differentiation in a cell-autonomous manner (Kenner et al. 2004).
6. CLOSING REMARKS
Genetic studies in the mouse in the past 15 years have greatly enhanced our understanding about the molecular mechanisms governing endochondral development. In particular, the roles of several major developmental pathways and key transcription factors in chondrocyte differentiation, hypertrophy, and osteoblast differentiation have been elucidated. For clarity, we have presented the regulation of chondrocytes and osteoblasts separately. In reality, development of the two cell lineages is tightly coupled during normal development. For instance, IHH is expressed by chondrocytes but regulates both chondrocyte maturation and osteoblast differentiation. A major challenge for future research is to understand how the various extracellular signals and nuclear factors integrate to execute specific developmental, homeostatic, and reparative functions. In addition, vascularization of the hypertrophic cartilage is of critical importance for proper development of the endochondral skeleton. Although a number of secreted factors including vascular endothelial growth factor A (VEGFA), IHH, and FGFs have been implicated (Gerber et al. 1999; Zelzer et al. 2002; Hung et al. 2007; Liu et al. 2007; Joeng and Long 2009), exactly how cartilage vascularization is regulated in a spatially and temporally specific manner remains an important topic for future research.
ACKNOWLEDGMENTS
Work in the authors’ laboratories is supported by U.S. National Institutes of Health grants AR055923, DK065789, and AR060456 (F.L.) and HD049808 (D.M.O.). The authors apologize for the omission of many references owing to space limitations.
Prx1-Cre targets early limb bud mesenchyme in the mouse at E9.5, and a subset of craniofacial mesenchyme (Logan et al. 2002).
Dermo1(Twist2)-Cre is expressed in limb and other mesenchyme beginning at E10.5 (Yu et al. 2003).
Col2a1-Cre targets the differentiating chondrogenic cells (Ovchinnikov et al. 2000).
Col2a1-CreERTM targets mainly chondrocytes upon administration of tamoxifen at proper times during embryogenesis (Hilton et al. 2007).
Col2-CreERT2 targets chondrocytes upon injection of tamoxifen (Chen et al. 2007).
ColI-Cre driven by the 2.3-kb promoter of mouse Colα1(I) targets mature osteoblasts (Dacquin et al. 2002).
Dmp1-Cre mainly targets osteocytes (Lu et al. 2007).
Editors: Patrick P.L. Tam, W. James Nelson, and Janet Rossant
Additional Perspectives on Mammalian Development available at www.cshperspectives.org
REFERENCES
- Ai M, Holmen SL, Van Hul W, Williams BO, Warman ML 2005. Reduced affinity to and inhibition by DKK1 form a common mechanism by which high bone mass-associated missense mutations in LRP5 affect canonical Wnt signaling. Mol Cell Biol 25: 4946–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B 2002. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev 16: 2813–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama H, Lyons JP, Mori-Akiyama Y, Yang X, Zhang R, Zhang Z, Deng JM, Taketo MM, Nakamura T, Behringer RR, et al. 2004. Interactions between Sox9 and β-catenin control chondrocyte differentiation. Genes Dev 18: 1072–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama H, Kim JE, Nakashima K, Balmes G, Iwai N, Deng JM, Zhang Z, Martin JF, Behringer RR, Nakamura T, et al. 2005. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc Natl Acad Sci 102: 14665–14670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers J, Schulze J, Beil FT, Gebauer M, Baranowsky A, Keller J, Marshall RP, Wintges K, Friedrich FW, Priemel M, et al. 2011. Control of bone formation by the serpentine receptor Frizzled-9. J Cell Biol 192: 1057–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrogini E, Almeida M, Martin-Millan M, Paik JH, Depinho RA, Han L, Goellner J, Weinstein RS, Jilka RL, O’Brien CA, et al. 2010. FoxO-mediated defense against oxidative stress in osteoblasts is indispensable for skeletal homeostasis in mice. Cell Metab 11: 136–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold MA, Kim Y, Czubryt MP, Phan D, McAnally J, Qi X, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN 2007. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell 12: 377–389 [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ 1999. Notch signaling: Cell fate control and signal integration in development. Science 284: 770–776 [DOI] [PubMed] [Google Scholar]
- Balemans W, Ebeling M, Patel N, Van Hul E, Olson P, Dioszegi M, Lacza C, Wuyts W, Van Den Ende J, Willems P, et al. 2001. Increased bone density in sclerosteosis is due to the deficiency of a novel secreted protein (SOST). Hum Mol Genet 10: 537–543 [DOI] [PubMed] [Google Scholar]
- Balemans W, Patel N, Ebeling M, Van Hul E, Wuyts W, Lacza C, Dioszegi M, Dikkers FG, Hildering P, Willems PJ, et al. 2002. Identification of a 52 kb deletion downstream of the SOST gene in patients with van Buchem disease. J Med Genet 39: 91–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay A, Tsuji K, Cox K, Harfe BD, Rosen V, Tabin CJ 2006. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet 2: e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barna M, Niswander L 2007. Visualization of cartilage formation: Insight into cellular properties of skeletal progenitors and chondrodysplasia syndromes. Dev Cell 12: 931–941 [DOI] [PubMed] [Google Scholar]
- Bennett CN, Longo KA, Wright WS, Suva LJ, Lane TF, Hankenson KD, MacDougald OA 2005. Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc Natl Acad Sci 102: 3324–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B 1999. Sox9 is required for cartilage formation. Nat Genet 22: 85–89 [DOI] [PubMed] [Google Scholar]
- Bi W, Huang W, Whitworth DJ, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B 2001. Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc Natl Acad Sci 98: 6698–6703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, Wu H, Yu K, Ornitz DM, Olson EN, et al. 2004. A twist code determines the onset of osteoblast differentiation. Dev Cell 6: 423–435 [DOI] [PubMed] [Google Scholar]
- Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, Wu D, Insogna K, Lifton RP 2002. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med 346: 1513–1521 [DOI] [PubMed] [Google Scholar]
- Brunet LJ, McMahon JA, McMahon AP, Harland RM 1998. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science 280: 1455–1457 [DOI] [PubMed] [Google Scholar]
- Brunkow ME, Gardner JC, Van Ness J, Paeper BW, Kovacevich BR, Proll S, Skonier JE, Zhao L, Sabo PJ, Fu Y, et al. 2001. Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot-containing protein. Am J Hum Genet 68: 577–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Quintero-Estades JA, Danielian PS, Nedelcu S, Berman SD, Lees JA 2010. Rb regulates fate choice and lineage commitment in vivo. Nature 466: 1110–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan AI, Pechak DG 1987. The cellular and molecular embryology of bone formation. In Bone and mineral research (ed. Peck WA), Vol. 5, pp. 117–183 Elsevier, New York [Google Scholar]
- Cash DE, Bock CB, Schughart K, Linney E, Underhill TM 1997. Retinoic acid receptor α function in vertebrate limb skeletogenesis: A modulator of chondrogenesis. J Cell Biol 136: 445–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Adar R, Yang X, Monsonego EO, Li C, Hauschka PV, Yayon A, Deng CX 1999. Gly369Cys mutation in mouse FGFR3 causes achondroplasia by affecting both chondrogenesis and osteogenesis. J Clin Invest 104: 1517–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li C, Qiao W, Xu X, Deng C 2001. A Ser365 → Cys mutation of fibroblast growth factor receptor 3 in mouse downregulates Ihh/PTHrP signals and causes severe achondroplasia. Hum Mol Genet 10: 457–465 [DOI] [PubMed] [Google Scholar]
- Chen M, Lichtler AC, Sheu TJ, Xie C, Zhang X, O’Keefe RJ, Chen D 2007. Generation of a transgenic mouse model with chondrocyte-specific and tamoxifen-inducible expression of Cre recombinase. Genesis 45: 44–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S 2006. Notch signaling in stem cell systems. Stem Cells 24: 2437–2447 [DOI] [PubMed] [Google Scholar]
- Choi JY, Pratap J, Javed A, Zaidi SK, Xing L, Balint E, Dalamangas S, Boyce B, van Wijnen AJ, Lian JB, et al. 2001. Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc Natl Acad Sci 98: 8650–8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb J, Dierich A, Huss-Garcia Y, Duboule D 2006. A mouse model for human short-stature syndromes identifies Shox2 as an upstream regulator of Runx2 during long-bone development. Proc Natl Acad Sci 103: 4511–4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM 1996. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat Genet 12: 390–397 [DOI] [PubMed] [Google Scholar]
- Colvin JS, Feldman B, Nadeau JH, Goldfarb M, Ornitz DM 1999. Genomic organization and embryonic expression of the mouse fibroblast growth factor 9 gene. Dev Dyn 216: 72–88 [DOI] [PubMed] [Google Scholar]
- Cormier S, Delezoide AL, Benoist-Lasselin C, Legeai-Mallet L, Bonaventure J, Silve C 2002. Parathyroid hormone receptor type 1/Indian hedgehog expression is preserved in the growth plate of human fetuses affected with fibroblast growth factor receptor type 3 activating mutations. Am J Pathol 161: 1325–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa D, Hesse E, Seriwatanachai D, Kiviranta R, Saito H, Yamana K, Neff L, Atfi A, Coillard L, Sitara D, et al. 2010. Zfp521 is a target gene and key effector of parathyroid hormone-related peptide signaling in growth plate chondrocytes. Dev Cell 19: 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer H, Lange R, Christoph A, Plomann M, Vopper G, Roes J, Brown R, Baldwin S, Kraemer P, Scheff S, et al. 1994. Inactivation of the N-CAM gene in mice results in size reduction of the olfactory bulb and deficits in spatial learning. Nature 367: 455–459 [DOI] [PubMed] [Google Scholar]
- Cui CB, Cooper LF, Yang X, Karsenty G, Aukhil I 2003. Transcriptional coactivation of bone-specific transcription factor Cbfa1 by TAZ. Mol Cell Biol 23: 1004–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Niziolek PJ, Macdonald BT, Zylstra CR, Alenina N, Robinson DR, Zhong Z, Matthes S, Jacobsen CM, Conlon RA, et al. 2011. Lrp5 functions in bone to regulate bone mass. Nat Med 17: 684–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacquin R, Starbuck M, Schinke T, Karsenty G 2002. Mouse α1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast. Dev Dyn 224: 245–251 [DOI] [PubMed] [Google Scholar]
- Daluiski A, Engstrand T, Bahamonde ME, Gamer LW, Agius E, Stevenson SL, Cox K, Rosen V, Lyons KM 2001. Bone morphogenetic protein-3 is a negative regulator of bone density. Nat Genet 27: 84–88 [DOI] [PubMed] [Google Scholar]
- Day TF, Guo X, Garrett-Beal L, Yang Y 2005. Wnt/β-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell 8: 739–750 [DOI] [PubMed] [Google Scholar]
- de Crombrugghe B, Lefebvre V, Behringer RR, Bi W, Murakami S, Huang W 2000. Transcriptional mechanisms of chondrocyte differentiation. Matrix Biol 19: 389–394 [DOI] [PubMed] [Google Scholar]
- DeLise AM, Fischer L, Tuan RS 2000. Cellular interactions and signaling in cartilage development. Osteoarthritis Cartilage 8: 309–334 [DOI] [PubMed] [Google Scholar]
- Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P 1996. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell 84: 911–921 [DOI] [PubMed] [Google Scholar]
- Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Farinas I, Karsenty G, Grosschedl R 2006. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell 125: 971–986 [DOI] [PubMed] [Google Scholar]
- Dong Y, Jesse AM, Kohn A, Gunnell LM, Honjo T, Zuscik MJ, O’Keefe RJ, Hilton MJ 2010. RBPjκ-dependent Notch signaling regulates mesenchymal progenitor cell proliferation and differentiation during skeletal development. Development 137: 1461–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy P, Starbuck M, Priemel M, Shen J, Pinero G, Geoffroy V, Amling M, Karsenty G 1999. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev 13: 1025–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dy P, Wang W, Bhattaram P, Wang Q, Wang L, Ballock RT, Lefebvre V 2012. Sox9 directs hypertrophic maturation and blocks osteoblast differentiation of growth plate chondrocytes. Dev Cell 22: 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eferl R, Hoebertz A, Schilling AF, Rath M, Karreth F, Kenner L, Amling M, Wagner EF 2004. The Fos-related antigen Fra-1 is an activator of bone matrix formation. EMBO J 23: 2789–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elefteriou F, Ahn JD, Takeda S, Starbuck M, Yang X, Liu X, Kondo H, Richards WG, Bannon TW, Noda M, et al. 2005. Leptin regulation of bone resorption by the sympathetic nervous system and CART. Nature 434: 514–520 [DOI] [PubMed] [Google Scholar]
- Elefteriou F, Benson MD, Sowa H, Starbuck M, Liu X, Ron D, Parada LF, Karsenty G 2006. ATF4 mediation of NF1 functions in osteoblast reveals a nutritional basis for congenital skeletal dysplasiae. Cell Metab 4: 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellies DL, Viviano B, McCarthy J, Rey JP, Itasaki N, Saunders S, Krumlauf R 2006. Bone density ligand, Sclerostin, directly interacts with LRP5 but not LRP5G171V to modulate Wnt activity. J Bone Miner Res 21: 1738–1749 [DOI] [PubMed] [Google Scholar]
- Engin F, Yao Z, Yang T, Zhou G, Bertin T, Jiang MM, Chen Y, Wang L, Zheng H, Sutton RE, et al. 2008. Dimorphic effects of Notch signaling in bone homeostasis. Nat Med 14: 299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin F, Bertin T, Ma O, Jiang MM, Wang L, Sutton RE, Donehower LA, Lee B 2009. Notch signaling contributes to the pathogenesis of human osteosarcomas. Hum Mol Genet 18: 1464–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswarakumar VP, Schlessinger J 2007. Skeletal overgrowth is mediated by deficiency in a specific isoform of fibroblast growth factor receptor 3. Proc Natl Acad Sci 104: 3937–3942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswarakumar VP, Monsonego-Ornan E, Pines M, Antonopoulou I, Morriss-Kay GM, Lonai P 2002. The IIIc alternative of Fgfr2 is a positive regulator of bone formation. Development 129: 3783–3793 [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, Horowitz MC, Locklin R, Morriss-Kay GM, Lonai P 2004. A gain-of-function mutation of Fgfr2c demonstrates the roles of this receptor variant in osteogenesis. Proc Natl Acad Sci 101: 12555–12560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J 2005. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev 16: 139–149 [DOI] [PubMed] [Google Scholar]
- Feng XH, Derynck R 2005. Specificity and versatility in TGF-β signaling through Smads. Annu Rev Cell Dev Biol 21: 659–693 [DOI] [PubMed] [Google Scholar]
- Foster JW, Dominguez-Steglich MA, Guioli S, Kwok C, Weller PA, Stevanovic M, Weissenbach J, Mansour S, Young ID, Goodfellow PN, et al. 1994. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature 372: 525–530 [DOI] [PubMed] [Google Scholar]
- Fromental-Ramain C, Warot X, Messadecq N, LeMeur M, Dolle P, Chambon P 1996. Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development 122: 2997–3011 [DOI] [PubMed] [Google Scholar]
- Funato N, Chapman SL, McKee MD, Funato H, Morris JA, Shelton JM, Richardson JA, Yanagisawa H 2009. Hand2 controls osteoblast differentiation in the branchial arch by inhibiting DNA binding of Runx2. Development 136: 615–625 [DOI] [PubMed] [Google Scholar]
- Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D 2005. Mutations in NOTCH1 cause aortic valve disease. Nature 437: 270–274 [DOI] [PubMed] [Google Scholar]
- Gerber HP, Vu TH, Ryan AM, Kowalski J, Werb Z, Ferrara N 1999. VEGF couples hypertrophic cartilage remodeling, ossification and angiogenesis during endochondral bone formation. Nat Med 5: 623–628 [DOI] [PubMed] [Google Scholar]
- Glass DA, Bialek P 2nd, Ahn JD, Starbuck M, Patel MS, Clevers H, Taketo MM, Long F, McMahon AP, Lang RA, et al. 2005. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev Cell 8: 751–764 [DOI] [PubMed] [Google Scholar]
- Gong Y, Krakow D, Marcelino J, Wilkin D, Chitayat D, Babul-Hirji R, Hudgins L, Cremers CW, Cremers FP, Brunner HG, et al. 1999. Heterozygous mutations in the gene encoding noggin affect human joint morphogenesis. Nat Genet 21: 302–304 [DOI] [PubMed] [Google Scholar]
- Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D, et al. 2001. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell 107: 513–523 [DOI] [PubMed] [Google Scholar]
- Guo J, Chung UI, Kondo H, Bringhurst FR, Kronenberg HM 2002. The PTH/PTHrP receptor can delay chondrocyte hypertrophy in vivo without activating phospholipase C. Dev Cell 3: 183–194 [DOI] [PubMed] [Google Scholar]
- Hall BK, Miyake T 2000. All for one and one for all: Condensations and the initiation of skeletal development. Bioessays 22: 138–147 [DOI] [PubMed] [Google Scholar]
- Han Y, Lefebvre V 2008. L-Sox5 and Sox6 drive expression of the aggrecan gene in cartilage by securing binding of Sox9 to a far-upstream enhancer. Mol Cell Biol 28: 4999–5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C, Tabin CJ 2000. Dual roles of Wnt signaling during chondrogenesis in the chicken limb. Development 127: 3141–3159 [DOI] [PubMed] [Google Scholar]
- Hesse E, Saito H, Kiviranta R, Correa D, Yamana K, Neff L, Toben D, Duda G, Atfi A, Geoffroy V, et al. 2010. Zfp521 controls bone mass by HDAC3-dependent attenuation of Runx2 activity. J Cell Biol 191: 1271–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TP, Spater D, Taketo MM, Birchmeier W, Hartmann C 2005. Canonical Wnt/β-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell 8: 727–738 [DOI] [PubMed] [Google Scholar]
- Hilton MJ, Tu X, Cook J, Hu H, Long F 2005. Ihh controls cartilage development by antagonizing Gli3, but requires additional effectors to regulate osteoblast and vascular development. Development 132: 4339–4351 [DOI] [PubMed] [Google Scholar]
- Hilton MJ, Tu X, Long F 2007. Tamoxifen-inducible gene deletion reveals a distinct cell type associated with trabecular bone, and direct regulation of PTHrP expression and chondrocyte morphology by Ihh in growth region cartilage. Dev Biol 308: 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton MJ, Tu X, Wu X, Bai S, Zhao H, Kobayashi T, Kronenberg HM, Teitelbaum SL, Ross FP, Kopan R, et al. 2008. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med 14: 306–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoi E, Bialek P, Chen YT, Rached MT, Groner Y, Behringer RR, Ornitz DM, Karsenty G 2006. Runx2 inhibits chondrocyte proliferation and hypertrophy through its expression in the perichondrium. Genes Dev 20: 2937–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmen SL, Giambernardi TA, Zylstra CR, Buckner-Berghuis BD, Resau JH, Hess JF, Glatt V, Bouxsein ML, Ai M, Warman ML, et al. 2004. Decreased BMD and limb deformities in mice carrying mutations in both Lrp5 and Lrp6. J Bone Miner Res 19: 2033–2040 [DOI] [PubMed] [Google Scholar]
- Holmen SL, Zylstra CR, Mukherjee A, Sigler RE, Faugere MC, Bouxsein ML, Deng L, Clemens TL, Williams BO 2005. Essential role of beta-catenin in postnatal bone acquisition. J Biol Chem 280: 21162–21168 [DOI] [PubMed] [Google Scholar]
- Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, Mueller E, Benjamin T, Spiegelman BM, Sharp PA, et al. 2005. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science 309: 1074–1078 [DOI] [PubMed] [Google Scholar]
- Honjo T 1996. The shortest path from the surface to the nucleus: RBP-Jκ/Su(H) transcription factor. Genes Cells 1: 1–9 [DOI] [PubMed] [Google Scholar]
- Hu H, Hilton MJ, Tu X, Yu K, Ornitz DM, Long F 2005. Sequential roles of Hedgehog and Wnt signaling in osteoblast development. Development 132: 49–60 [DOI] [PubMed] [Google Scholar]
- Huang W, Chung UI, Kronenberg HM, de Crombrugghe B 2001. The chondrogenic transcription factor Sox9 is a target of signaling by the parathyroid hormone-related peptide in the growth plate of endochondral bones. Proc Natl Acad Sci 98: 160–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV 2006. Signaling from Smo to Ci/Gli: Conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development 133: 3–14 [DOI] [PubMed] [Google Scholar]
- Hung IH, Yu K, Lavine KJ, Ornitz DM 2007. FGF9 regulates early hypertrophic chondrocyte differentiation and skeletal vascularization in the developing stylopod. Dev Biol 307: 300–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada M, Yasui T, Nomura S, Miyake S, Deguchi K, Himeno M, Sato M, Yamagiwa H, Kimura T, Yasui N, et al. 1999. Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Dev Dyn 214: 279–290 [DOI] [PubMed] [Google Scholar]
- Inada M, Wang Y, Byrne MH, Rahman MU, Miyaura C, Lopez-Otin C, Krane SM 2004. Critical roles for collagenase-3 (Mmp13) in development of growth plate cartilage and in endochondral ossification. Proc Natl Acad Sci 101: 17192–17197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP 2001. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev 15: 3059–3087 [DOI] [PubMed] [Google Scholar]
- Ionescu A, Kozhemyakina E, Nicolae C, Kaestner KH, Olsen BR, Lassar AB 2012. FoxA family members are crucial regulators of the hypertrophic chondrocyte differentiation program. Dev Cell 22: 927–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isidor B, Lindenbaum P, Pichon O, Bezieau S, Dina C, Jacquemont S, Martin-Coignard D, Thauvin-Robinet C, Le Merrer M, Mandel JL, et al. 2011. Truncating mutations in the last exon of NOTCH2 cause a rare skeletal disorder with osteoporosis. Nat Genet 43: 306–308 [DOI] [PubMed] [Google Scholar]
- Itoh N, Ornitz DM 2008. Functional evolutionary history of the mouse Fgf gene family. Dev Dyn 237: 18–27 [DOI] [PubMed] [Google Scholar]
- Iwata T, Chen L, Li C, Ovchinnikov DA, Behringer RR, Francomano CA, Deng CX 2000. A neonatal lethal mutation in FGFR3 uncouples proliferation and differentiation of growth plate chondrocytes in embryos. Hum Mol Genet 9: 1603–1613 [DOI] [PubMed] [Google Scholar]
- Iwata T, Li CL, Deng CX, Francomano CA 2001. Highly activated Fgfr3 with the K644M mutation causes prolonged survival in severe dwarf mice. Hum Mol Genet 10: 1255–1264 [DOI] [PubMed] [Google Scholar]
- Jacob AL, Smith C, Partanen J, Ornitz DM 2006. Fibroblast growth factor receptor 1 signaling in the osteo-chondrogenic cell lineage regulates sequential steps of osteoblast maturation. Dev Biol 296: 315–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobert AS, Zhang P, Couvineau A, Bonaventure J, Roume J, Le Merrer M, Silve C 1998. Absence of functional receptors for parathyroid hormone and parathyroid hormone-related peptide in Blomstrand chondrodysplasia. J Clin Invest 102: 34–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochum W, David JP, Elliott C, Wutz A, Plenk H Jr, Matsuo K, Wagner EF 2000. Increased bone formation and osteosclerosis in mice overexpressing the transcription factor Fra-1. Nat Med 6: 980–984 [DOI] [PubMed] [Google Scholar]
- Joeng KS, Long F 2009. The Gli2 transcriptional activator is a crucial effector for Ihh signaling in osteoblast development and cartilage vascularization. Development 136: 4177–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joeng KS, Schumacher CA, Zylstra-Diegel CR, Long F, Williams BO 2011. Lrp5 and Lrp6 redundantly control skeletal development in the mouse embryo. Dev Biol 359: 222–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DC, Wein MN, Oukka M, Hofstaetter JG, Glimcher MJ, Glimcher LH 2006. Regulation of adult bone mass by the zinc finger adapter protein Schnurri-3. Science 312: 1223–1227 [DOI] [PubMed] [Google Scholar]
- Kamiya N, Ye L, Kobayashi T, Lucas DJ, Mochida Y, Yamauchi M, Kronenberg HM, Feng JQ, Mishina Y 2008a. Disruption of BMP signaling in osteoblasts through type IA receptor (BMPRIA) increases bone mass. J Bone Miner Res 23: 2007–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya N, Ye L, Kobayashi T, Mochida Y, Yamauchi M, Kronenberg HM, Feng JQ, Mishina Y 2008b. BMP signaling negatively regulates bone mass through sclerostin by inhibiting the canonical Wnt pathway. Development 135: 3801–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya N, Kobayashi T, Mochida Y, Yu PB, Yamauchi M, Kronenberg HM, Mishina Y 2010. Wnt inhibitors Dkk1 and Sost are downstream targets of BMP signaling through the type IA receptor (BMPRIA) in osteoblasts. J Bone Miner Res 25: 200–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzler B, Kuschert SJ, Liu YH, Mallo M 1998. Hoxa-2 restricts the chondrogenic domain and inhibits bone formation during development of the branchial area. Development 125: 2587–2897 [DOI] [PubMed] [Google Scholar]
- Karaplis AC, Luz A, Glowacki J, Bronson RT, Tybulewicz VL, Kronenberg HM, Mulligan RC 1994. Lethal skeletal dysplasia from targeted disruption of the parathyroid hormone-related peptide gene. Genes Dev 8: 277–289 [DOI] [PubMed] [Google Scholar]
- Karaplis AC, He B, Nguyen MT, Young ID, Semeraro D, Ozawa H, Amizuka N 1998. Inactivating mutation in the human parathyroid hormone receptor type 1 gene in Blomstrand chondrodysplasia. Endocrinology 139: 5255–5258 [DOI] [PubMed] [Google Scholar]
- Karp SJ, Schipani E, St-Jacques B, Hunzelman J, Kronenberg H, McMahon AP 2000. Indian Hedgehog coordinates endochondral bone growth and morphogenesis via parathyroid hormone related-protein-dependent and -independent pathways. Development 127: 543–548 [DOI] [PubMed] [Google Scholar]
- Karperien M, van der Harten HJ, van Schooten R, Farih-Sips H, den Hollander NS, Kneppers SL, Nijweide P, Papapoulos SE, Lowik CW 1999. A frame-shift mutation in the type I parathyroid hormone (PTH)/PTH-related peptide receptor causing Blomstrand lethal osteochondrodysplasia. J Clin Endocrinol Metab 84: 3713–3720 [DOI] [PubMed] [Google Scholar]
- Karsenty G 2009. Transcriptional control of osteoblast differentiation. In The skeletal system (ed. Pourquie O), pp. 205–218 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Kato M, Patel MS, Levasseur R, Lobov I, Chang BH, Glass DA II, Hartmann C, Li L, Hwang TH, Brayton CF, et al. 2002. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol 157: 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenner L, Hoebertz A, Beil T, Keon N, Karreth F, Eferl R, Scheuch H, Szremska A, Amling M, Schorpp-Kistner M, et al. 2004. Mice lacking JunB are osteopenic due to cell-autonomous osteoblast and osteoclast defects. J Cell Biol 164: 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IS, Otto F, Zabel B, Mundlos S 1999. Regulation of chondrocyte differentiation by Cbfa1. Mech Dev 80: 159–170 [DOI] [PubMed] [Google Scholar]
- Kim S, Koga T, Isobe M, Kern BE, Yokochi T, Chin YE, Karsenty G, Taniguchi T, Takayanagi H 2003. Stat1 functions as a cytoplasmic attenuator of Runx2 in the transcriptional program of osteoblast differentiation. Genes Dev 17: 1979–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kist R, Schrewe H, Balling R, Scherer G 2002. Conditional inactivation of Sox9: A mouse model for campomelic dysplasia. Genesis 32: 121–123 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Soegiarto DW, Yang Y, Lanske B, Schipani E, McMahon AP, Kronenberg HM 2005. Indian hedgehog stimulates periarticular chondrocyte differentiation to regulate growth plate length independently of PTHrP. J Clin Invest 115: 1734–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga T, Matsui Y, Asagiri M, Kodama T, de Crombrugghe B, Nakashima K, Takayanagi H 2005. NFAT and Osterix cooperatively regulate bone formation. Nat Med 11: 880–885 [DOI] [PubMed] [Google Scholar]
- Kohn A, Dong Y, Mirando AJ, Jesse AM, Honjo T, Zuscik MJ, O’Keefe RJ, Hilton MJ 2012. Cartilage-specific RBPjκ-dependent and -independent Notch signals regulate cartilage and bone development. Development 139: 1198–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokabu S, Gamer L, Cox K, Lowery J, Tsuji K, Raz R, Economides A, Katagiri T, Rosen V 2012. BMP3 suppresses osteoblast differentiation of bone marrow stromal cells via interaction with Acvr2b. Mol Endocrinol 26: 87–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, et al. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89: 755–764 [DOI] [PubMed] [Google Scholar]
- Kopan R, Goate A 2000. A common enzyme connects Notch signaling and Alzheimer’s disease. Genes Dev 14: 2799–2806 [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX 2009. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 137: 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozhemyakina E, Cohen T, Yao TP, Lassar AB 2009. Parathyroid hormone-related peptide represses chondrocyte hypertrophy through a protein phosphatase 2A/histone deacetylase 4/MEF2 pathway. Mol Cell Biol 29: 5751–5762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziel L, Wuelling M, Schneider S, Vortkamp A 2005. Gli3 acts as a repressor downstream of Ihh in regulating two distinct steps of chondrocyte differentiation. Development 132: 5249–5260 [DOI] [PubMed] [Google Scholar]
- Kramer I, Halleux C, Keller H, Pegurri M, Gooi JH, Weber PB, Feng JQ, Bonewald LF, Kneissel M 2010. Osteocyte Wnt/β-catenin signaling is required for normal bone homeostasis. Mol Cell Biol 30: 3071–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg HM 2003. Developmental regulation of the growth plate. Nature 423: 332–336 [DOI] [PubMed] [Google Scholar]
- Kveiborg M, Sabatakos G, Chiusaroli R, Wu M, Philbrick WM, Horne WC, Baron R 2004. ΔFosB induces osteosclerosis and decreases adipogenesis by two independent cell-autonomous mechanisms. Mol Cell Biol 24: 2820–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanske B, Karaplis AC, Lee K, Luz A, Vortkamp A, Pirro A, Karperien M, Defize LH, Ho C, Mulligan RC, et al. 1996. PTH/PTHrP receptor in early development and Indian hedgehog-regulated bone growth. Science 273: 663–666 [DOI] [PubMed] [Google Scholar]
- Lazarus JE, Hegde A, Andrade AC, Nilsson O, Baron J 2007. Fibroblast growth factor expression in the postnatal growth plate. Bone 40: 577–586 [DOI] [PubMed] [Google Scholar]
- Lee K, Deeds JD, Segre GV 1995. Expression of parathyroid hormone-related peptide and its receptor messenger ribonucleic acids during fetal development of rats. Endocrinology 136: 453–463 [DOI] [PubMed] [Google Scholar]
- Lee B, Thirunavukkarasu K, Zhou L, Pastore L, Baldini A, Hecht J, Geoffroy V, Ducy P, Karsenty G 1997. Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nat Genet 16: 307–310 [DOI] [PubMed] [Google Scholar]
- Lee MH, Kim YJ, Yoon WJ, Kim JI, Kim BG, Hwang YS, Wozney JM, Chi XZ, Bae SC, Choi KY, et al. 2005. Dlx5 specifically regulates Runx2 type II expression by binding to homeodomain-response elements in the Runx2 distal promoter. J Biol Chem 280: 35579–35587 [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Li P, de Crombrugghe B 1998. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J 17: 5718–5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Chen L, Iwata T, Kitagawa M, Fu XY, Deng CX 1999. A Lys644Glu substitution in fibroblast growth factor receptor 3 (FGFR3) causes dwarfism in mice by activation of STATs and ink4 cell cycle inhibitors. Hum Mol Genet 8: 35–44 [DOI] [PubMed] [Google Scholar]
- Little RD, Carulli JP, Del Mastro RG, Dupuis J, Osborne M, Folz C, Manning SP, Swain PM, Zhao SC, Eustace B, et al. 2002. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am J Hum Genet 70: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Pouponnot C, Massague J 1997. Dual role of the Smad4/DPC4 tumor suppressor in TGFβ-inducible transcriptional complexes. Genes Dev 11: 3157–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Xu J, Colvin JS, Ornitz DM 2002. Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes Dev 16: 859–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Lavine KJ, Hung IH, Ornitz DM 2007. FGF18 is required for early chondrocyte proliferation, hypertrophy and vascular invasion of the growth plate. Dev Biol 302: 80–91 [DOI] [PubMed] [Google Scholar]
- Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ 2002. Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 33: 77–80 [DOI] [PubMed] [Google Scholar]
- Long F 2012a. Building strong bones: Molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol 13: 27–38 [DOI] [PubMed] [Google Scholar]
- Long F 2012b. Prenatal bone development. In Pediatric bone, 2nd ed (ed. Glorieux FH, Pettifor JM, Juppner H), pp. 39–54 Academic, San Diego [Google Scholar]
- Long F, Zhang XM, Karp S, Yang Y, McMahon AP 2001. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development 128: 5099–5108 [DOI] [PubMed] [Google Scholar]
- Long F, Chung UI, Ohba S, McMahon J, Kronenberg HM, McMahon AP 2004. Ihh signaling is directly required for the osteoblast lineage in the endochondral skeleton. Development 131: 1309–1318 [DOI] [PubMed] [Google Scholar]
- Lu Y, Xie Y, Zhang S, Dusevich V, Bonewald LF, Feng JQ 2007. DMP1-targeted Cre expression in odontoblasts and osteocytes. J Dent Res 86: 320–325 [DOI] [PubMed] [Google Scholar]
- Lu P, Yu Y, Perdue Y, Werb Z 2008. The apical ectodermal ridge is a timer for generating distal limb progenitors. Development 135: 1395–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes C, Kobayashi T, Selig MK, Torrekens S, Roth SI, Mackem S, Carmeliet G, Kronenberg HM 2010. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev Cell 19: 329–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak KK, Chen MH, Day TF, Chuang PT, Yang Y 2006. Wnt/β-catenin signaling interacts differentially with Ihh signaling in controlling endochondral bone and synovial joint formation. Development 133: 3695–3707 [DOI] [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D 2005. Smad transcription factors. Genes Dev 19: 2783–2810 [DOI] [PubMed] [Google Scholar]
- McMahon AP, Ingham PW, Tabin CJ 2003. Developmental roles and clinical significance of Hedgehog signaling. Curr Top Dev Biol 53: 1–114 [DOI] [PubMed] [Google Scholar]
- Mead TJ, Yutzey KE 2009. Notch pathway regulation of chondrocyte differentiation and proliferation during appendicular and axial skeleton development. Proc Natl Acad Sci 106: 14420–14425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina Y, Starbuck MW, Gentile MA, Fukuda T, Kasparcova V, Seedor JG, Hanks MC, Amling M, Pinero GJ, Harada S, et al. 2004. Bone morphogenetic protein type IA receptor signaling regulates postnatal osteoblast function and bone remodeling. J Biol Chem 279: 27560–27566 [DOI] [PubMed] [Google Scholar]
- Mohamed SA, Aherrahrou Z, Liptau H, Erasmi AW, Hagemann C, Wrobel S, Borzym K, Schunkert H, Sievers HH, Erdmann J 2006. Novel missense mutations (p.T596M and p.P1797H) in NOTCH1 in patients with bicuspid aortic valve. Biochem Biophys Res Commun 345: 1460–1465 [DOI] [PubMed] [Google Scholar]
- Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JH, et al. 1997. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 89: 773–779 [DOI] [PubMed] [Google Scholar]
- Murakami S, Balmes G, McKinney S, Zhang Z, Givol D, De Crombrugghe B 2004. Constitutive activation of MEK1 in chondrocytes causes Stat1-independent achondroplasia-like dwarfism and rescues the Fgfr3-deficient mouse phenotype. Genes Dev 18: 290–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B 2002. The novel zinc finger-containing transcription factor Osterix is required for osteoblast differentiation and bone formation. Cell 108: 17–29 [DOI] [PubMed] [Google Scholar]
- Naski MC, Colvin JS, Coffin JD, Ornitz DM 1998. Repression of hedgehog signaling and BMP4 expression in growth plate cartilage by fibroblast growth factor receptor 3. Development 125: 4977–4988 [DOI] [PubMed] [Google Scholar]
- Nishikawa K, Nakashima T, Takeda S, Isogai M, Hamada M, Kimura A, Kodama T, Yamaguchi A, Owen MJ, Takahashi S, et al. 2010. Maf promotes osteoblast differentiation in mice by mediating the age-related switch in mesenchymal cell differentiation. J Clin Invest 120: 3455–3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba S, Kawaguchi H, Kugimiya F, Ogasawara T, Kawamura N, Saito T, Ikeda T, Fujii K, Miyajima T, Kuramochi A, et al. 2008. Patched1 haploinsufficiency increases adult bone mass and modulates Gli3 repressor activity. Dev Cell 14: 689–699 [DOI] [PubMed] [Google Scholar]
- Ohbayashi N, Shibayama M, Kurotaki Y, Imanishi M, Fujimori T, Itoh N, Takada S 2002. FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev 16: 870–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Marie PJ 2002. FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev 16: 1446–1465 [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, et al. 1997. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89: 765–771 [DOI] [PubMed] [Google Scholar]
- Ovchinnikov DA, Deng JM, Ogunrinu G, Behringer RR 2000. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis 26: 145–146 [PubMed] [Google Scholar]
- Pizette S, Niswander L 2000. BMPs are required at two steps of limb chondrogenesis: Formation of prechondrogenic condensations and their differentiation into chondrocytes. Dev Biol 219: 237–249 [DOI] [PubMed] [Google Scholar]
- Pogue R, Lyons K 2006. BMP signaling in the cartilage growth plate. Curr Top Dev Biol 76: 1–48 [DOI] [PubMed] [Google Scholar]
- Poole AR 1991. The growth plate: Cellular physiology, cartilage assembly and mineralization. In Cartilage: Molecular aspects (ed. Hall BK, Newman SA), pp. 179–212 CRC Press, Boca Raton, FL [Google Scholar]
- Priore R, Dailey L, Basilico C 2006. Downregulation of Akt activity contributes to the growth arrest induced by FGF in chondrocytes. J Cell Physiol 207: 800–808 [DOI] [PubMed] [Google Scholar]
- Rached MT, Kode A, Xu L, Yoshikawa Y, Paik JH, Depinho RA, Kousteni S 2010. FoxO1 is a positive regulator of bone formation by favoring protein synthesis and resistance to oxidative stress in osteoblasts. Cell Metab 11: 147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radice GL, Rayburn H, Matsunami H, Knudsen KA, Takeichi M, Hynes RO 1997. Developmental defects in mouse embryos lacking N-cadherin. Dev Biol 181: 64–78 [DOI] [PubMed] [Google Scholar]
- Retting KN, Song B, Yoon BS, Lyons KM 2009. BMP canonical Smad signaling through Smad1 and Smad5 is required for endochondral bone formation. Development 136: 1093–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo RF, Rajan L, Li X, Lufkin T 2002. The Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial, and appendicular skeletal development. Genes Dev 16: 1089–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda SJ, McMahon AP 2006. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 133: 3231–3244 [DOI] [PubMed] [Google Scholar]
- Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, MacDougald OA 2000. Inhibition of adipogenesis by Wnt signaling. Science 289: 950–953 [DOI] [PubMed] [Google Scholar]
- Rudnicki JA, Brown AM 1997. Inhibition of chondrogenesis by Wnt gene expression in vivo and in vitro. Dev Biol 185: 104–118 [DOI] [PubMed] [Google Scholar]
- Sabatakos G, Sims NA, Chen J, Aoki K, Kelz MB, Amling M, Bouali Y, Mukhopadhyay K, Ford K, Nestler EJ, et al. 2000. Overexpression of ΔFosB transcription factor(s) increases bone formation and inhibits adipogenesis. Nat Med 6: 985–990 [DOI] [PubMed] [Google Scholar]
- Salie R, Kneissel M, Vukevic M, Zamurovic N, Kramer I, Evans G, Gerwin N, Mueller M, Kinzel B, Susa M 2010. Ubiquitous overexpression of Hey1 transcription factor leads to osteopenia and chondrocyte hypertrophy in bone. Bone 46: 680–694 [DOI] [PubMed] [Google Scholar]
- Sasagawa S, Takemori H, Uebi T, Ikegami D, Hiramatsu K, Ikegawa S, Yoshikawa H, Tsumaki N 2012. SIK3 is essential for chondrocyte hypertrophy during skeletal development in mice. Development 139: 1153–1163 [DOI] [PubMed] [Google Scholar]
- Satokata I, Ma L, Ohshima H, Bei M, Woo I, Nishizawa K, Maeda T, Takano Y, Uchiyama M, Heaney S, et al. 2000. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat Genet 24: 391–395 [DOI] [PubMed] [Google Scholar]
- Schipani E, Kruse K, Juppner H 1995. A constitutively active mutant PTH-PTHrP receptor in Jansen-type metaphyseal chondrodysplasia. Science 268: 98–100 [DOI] [PubMed] [Google Scholar]
- Schipani E, Langman CB, Parfitt AM, Jensen GS, Kikuchi S, Kooh SW, Cole WG, Juppner H 1996. Constitutively activated receptors for parathyroid hormone and parathyroid hormone-related peptide in Jansen’s metaphyseal chondrodysplasia. N Engl J Med 335: 708–714 [DOI] [PubMed] [Google Scholar]
- Schipani E, Lanske B, Hunzelman J, Luz A, Kovacs CS, Lee K, Pirro A, Kronenberg HM, Juppner H 1997. Targeted expression of constitutively active receptors for parathyroid hormone and parathyroid hormone-related peptide delays endochondral bone formation and rescues mice that lack parathyroid hormone-related peptide. Proc Natl Acad Sci 94: 13689–13694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeter EH, Kisslinger JA, Kopan R 1998. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393: 382–386 [DOI] [PubMed] [Google Scholar]
- Semenov MV, He X 2006. LRP5 mutations linked to high bone mass diseases cause reduced LRP5 binding and inhibition by SOST. J Biol Chem 281: 38276–38284 [DOI] [PubMed] [Google Scholar]
- Seriwatanachai D, Densmore MJ, Sato T, Correa D, Neff L, Baron R, Lanske B 2011. Deletion of Zfp521 rescues the growth plate phenotype in a mouse model of Jansen metaphyseal chondrodysplasia. FASEB J 25: 3057–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono K, Tung WE, Macolino C, Chi AH, Didizian JH, Mundy C, Chandraratna RA, Mishina Y, Enomoto-Iwamoto M, Pacifici M, et al. 2011. Potent inhibition of heterotopic ossification by nuclear retinoic acid receptor-γ agonists. Nat Med 17: 454–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoyama A, Wada M, Ikeda F, Hata K, Matsubara T, Nifuji A, Noda M, Amano K, Yamaguchi A, Nishimura R, et al. 2007. Ihh/Gli2 signaling promotes osteoblast differentiation by regulating Runx2 expression and function. Mol Biol Cell 18: 2411–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson MA, Irving MD, Asilmaz E, Gray MJ, Dafou D, Elmslie FV, Mansour S, Holder SE, Brain CE, Burton BK, et al. 2011. Mutations in NOTCH2 cause Hajdu-Cheney syndrome, a disorder of severe and progressive bone loss. Nat Genet 43: 303–305 [DOI] [PubMed] [Google Scholar]
- Smits P, Li P, Mandel J, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B, Lefebvre V 2001. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev Cell 1: 277–290 [DOI] [PubMed] [Google Scholar]
- Stadler HS, Higgins KM, Capecchi MR 2001. Loss of Eph-receptor expression correlates with loss of cell adhesion and chondrogenic capacity in Hoxa13 mutant limbs. Development 128: 4177–4188 [DOI] [PubMed] [Google Scholar]
- Staehling-Hampton K, Proll S, Paeper BW, Zhao L, Charmley P, Brown A, Gardner JC, Galas D, Schatzman RC, Beighton P, et al. 2002. A 52-kb deletion in the SOST-MEOX1 intergenic region on 17q12-q21 is associated with van Buchem disease in the Dutch population. Am J Med Genet 110: 144–152 [DOI] [PubMed] [Google Scholar]
- Stickens D, Behonick DJ, Ortega N, Heyer B, Hartenstein B, Yu Y, Fosang AJ, Schorpp-Kistner M, Angel P, Werb Z 2004. Altered endochondral bone development in matrix metalloproteinase 13-deficient mice. Development 131: 5883–5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Jacques B, Hammerschmidt M, McMahon AP 1999. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev 13: 2072–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott NS, Jiang TX, Chuong CM 1999. Successive formative stages of precartilaginous mesenchymal condensations in vitro: Modulation of cell adhesion by Wnt-7A and BMP-2. J Cell Physiol 180: 314–324 [DOI] [PubMed] [Google Scholar]
- Su N, Du X, Chen L 2008. FGF signaling: Its role in bone development and human skeleton diseases. Front Biosci 13: 2842–2865 [DOI] [PubMed] [Google Scholar]
- Takada I, Mihara M, Suzawa M, Ohtake F, Kobayashi S, Igarashi M, Youn MY, Takeyama K, Nakamura T, Mezaki Y, et al. 2007. A histone lysine methyltransferase activated by non-canonical Wnt signalling suppresses PPAR-γ transactivation. Nat Cell Biol 9: 1273–1285 [DOI] [PubMed] [Google Scholar]
- Takeda S, Bonnamy JP, Owen MJ, Ducy P, Karsenty G 2001. Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Genes Dev 15: 467–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Weng T, Zhang J, Wang J, Li W, Wan H, Lan Y, Cheng X, Hou N, Liu H, et al. 2007. Smad4 is required for maintaining normal murine postnatal bone homeostasis. J Cell Sci 120: 2162–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Chen S, Yang T, Dawson B, Munivez E, Bertin T, Lee B 2010. Osteosclerosis owing to Notch gain of function is solely Rbpj-dependent. J Bone Miner Res 25: 2175–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge D, Brugmann SA, Helms JA, Nusse R 2008. Wnt and FGF signals interact to coordinate growth with cell fate specification during limb development. Development 135: 3247–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DM, Carty SA, Piscopo DM, Lee JS, Wang WF, Forrester WC, Hinds PW 2001. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell 8: 303–316 [DOI] [PubMed] [Google Scholar]
- Thorogood PV, Hinchliffe JR 1975. An analysis of the condensation process during chondrogenesis in the embryonic chick hind limb. J Embryol Exp Morphol 33: 581–606 [PubMed] [Google Scholar]
- Tribioli C, Lufkin T 1999. The murine Bapx1 homeobox gene plays a critical role in embryonic development of the axial skeleton and spleen. Development 126: 5699–5711 [DOI] [PubMed] [Google Scholar]
- Tsuji K, Bandyopadhyay A, Harfe BD, Cox K, Kakar S, Gerstenfeld L, Einhorn T, Tabin CJ, Rosen V 2006. BMP2 activity, although dispensable for bone formation, is required for the initiation of fracture healing. Nat Genet 38: 1424–1429 [DOI] [PubMed] [Google Scholar]
- Tu X, Joeng KS, Nakayama KI, Nakayama K, Rajagopal J, Carroll TJ, McMahon AP, Long F 2007. Noncanonical Wnt signaling through G protein-linked PKCδ activation promotes bone formation. Dev Cell 12: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu X, Chen J, Lim J, Karner CM, Lee S-Y, Heisig J, Wiese C, Surendran K, Kopan R, Gessler M, et al. 2012a. Physiological Notch signaling maintains bone homeostasis via RBPjk and Hey upstream of NFATc1. PLoS Genet 8: e1002577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu X, Joeng KS, Long F 2012b. Indian hedgehog requires additional effectors besides Runx2 to induce osteoblast differentiation. Dev Biol 362: 76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N, Grose R 2010. Fibroblast growth factor signalling: From development to cancer. Nat Rev Cancer 10: 116–129 [DOI] [PubMed] [Google Scholar]
- Ueta C, Iwamoto M, Kanatani N, Yoshida C, Liu Y, Enomoto-Iwamoto M, Ohmori T, Enomoto H, Nakata K, Takada K, et al. 2001. Skeletal malformations caused by overexpression of Cbfa1 or its dominant negative form in chondrocytes. J Cell Biol 153: 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde-Franco G, Liu H, Davidson D, Chai S, Valderrama-Carvajal H, Goltzman D, Ornitz DM, Henderson JE 2004. Defective bone mineralization and osteopenia in young adult FGFR3–/– mice. Hum Mol Genet 13: 271–284 [DOI] [PubMed] [Google Scholar]
- Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, McAnally J, Pomajzl C, Shelton JM, Richardson JA, et al. 2004. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 119: 555–566 [DOI] [PubMed] [Google Scholar]
- Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ 1996. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science 273: 613–622 [DOI] [PubMed] [Google Scholar]
- Vu TH, Shipley JM, Bergers G, Berger JE, Helms JA, Hanahan D, Shapiro SD, Senior RM, Werb Z 1998. MMP-9/gelatinase B is a key regulator of growth plate angiogenesis and apoptosis of hypertrophic chondrocytes. Cell 93: 411–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada N, Kimura I, Tanaka H, Ide H, Nohno T 1998. Glycosylphosphatidylinositol-anchored cell surface proteins regulate position-specific cell affinity in the limb bud. Dev Biol 202: 244–252 [DOI] [PubMed] [Google Scholar]
- Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli FD, Keutel J, Hustert E, et al. 1994. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 79: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Wang X, Kua HY, Hu Y, Guo K, Zeng Q, Wu Q, Ng HH, Karsenty G, de Crombrugghe B, Yeh J, et al. 2006. p53 functions as a negative regulator of osteoblastogenesis, osteoblast-dependent osteoclastogenesis, and bone remodeling. J Cell Biol 172: 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lian N, Ma Y, Li L, Gallant RC, Elefteriou F, Yang X 2012. Chondrocytic Atf4 regulates osteoblast differentiation and function via Ihh. Development 139: 601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir EC, Philbrick WM, Amling M, Neff LA, Baron R, Broadus AE 1996. Targeted overexpression of parathyroid hormone-related peptide in chondrocytes causes chondrodysplasia and delayed endochondral bone formation. Proc Natl Acad Sci 93: 10240–10245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston AD, Rosen V, Chandraratna RA, Underhill TM 2000. Regulation of skeletal progenitor differentiation by the BMP and retinoid signaling pathways. J Cell Biol 148: 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winslow MM, Pan M, Starbuck M, Gallo EM, Deng L, Karsenty G, Crabtree GR 2006. Calcineurin/NFAT signaling in osteoblasts regulates bone mass. Dev Cell 10: 771–782 [DOI] [PubMed] [Google Scholar]
- Wodarz A, Nusse R 1998. Mechanisms of Wnt signaling in development. Annu Rev Cell Dev Biol 14: 59–88 [DOI] [PubMed] [Google Scholar]
- Wu X, Tu X, Joeng KS, Hilton MJ, Williams DA, Long F 2008. Rac1 activation controls nuclear localization of β-catenin during canonical Wnt signaling. Cell 133: 340–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Onal M, Jilka RL, Weinstein RS, Manolagas SC, O’Brien CA 2011. Matrix-embedded cells control osteoclast formation. Nat Med 17: 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Lawshe A, MacArthur CA, Ornitz DM 1999. Genomic structure, mapping, activity and expression of fibroblast growth factor 17. Mech Dev 83: 165–178 [DOI] [PubMed] [Google Scholar]
- Yadav VK, Ryu JH, Suda N, Tanaka KF, Gingrich JA, Schutz G, Glorieux FH, Chiang CY, Zajac JD, Insogna KL, et al. 2008. Lrp5 controls bone formation by inhibiting serotonin synthesis in the duodenum. Cell 135: 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav VK, Arantes HP, Barros ER, Lazaretti-Castro M, Ducy P 2010. Genetic analysis of Lrp5 function in osteoblast progenitors. Calcif Tissue Int 86: 382–388 [DOI] [PubMed] [Google Scholar]
- Yang Y, Topol L, Lee H, Wu J 2003. Wnt5a and Wnt5b exhibit distinct activities in coordinating chondrocyte proliferation and differentiation. Development 130: 1003–1015 [DOI] [PubMed] [Google Scholar]
- Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, et al. 2004. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology: Implication for Coffin-Lowry syndrome. Cell 117: 387–398 [DOI] [PubMed] [Google Scholar]
- Yoon BS, Ovchinnikov DA, Yoshii I, Mishina Y, Behringer RR, Lyons KM 2005. Bmpr1a and Bmpr1b have overlapping functions and are essential for chondrogenesis in vivo. Proc Natl Acad Sci 102: 5062–5067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon BS, Pogue R, Ovchinnikov DA, Yoshii I, Mishina Y, Behringer RR, Lyons KM 2006. BMPs regulate multiple aspects of growth-plate chondrogenesis through opposing actions on FGF pathways. Development 133: 4667–4678 [DOI] [PubMed] [Google Scholar]
- Yoshida CA, Yamamoto H, Fujita T, Furuichi T, Ito K, Inoue K, Yamana K, Zanma A, Takada K, Ito Y, et al. 2004. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev 18: 952–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Ornitz DM 2008. FGF signaling regulates mesenchymal differentiation and skeletal patterning along the limb bud proximodistal axis. Development 135: 483–491 [DOI] [PubMed] [Google Scholar]
- Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, Towler DA, Ornitz DM 2003. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development 130: 3063–3074 [DOI] [PubMed] [Google Scholar]
- Yu VW, Ambartsoumian G, Verlinden L, Moir JM, Prud’homme J, Gauthier C, Roughley PJ, St-Arnaud R 2005. FIAT represses ATF4-mediated transcription to regulate bone mass in transgenic mice. J Cell Biol 169: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Liu H, Yan M, Yang J, Long F, Muneoka K, Chen Y 2007. Shox2 is required for chondrocyte proliferation and maturation in proximal limb skeleton. Dev Biol 306: 549–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti S, Smerdel-Ramoya A, Stadmeyer L, Durant D, Radtke F, Canalis E 2008. Notch inhibits osteoblast differentiation and causes osteopenia. Endocrinology 149: 3890–3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelzer E, McLean W, Ng YS, Fukai N, Reginato AM, Lovejoy S, D’Amore PA, Olsen BR 2002. Skeletal defects in VEGF120/120mice reveal multiple roles for VEGF in skeletogenesis. Development 129: 1893–1904 [DOI] [PubMed] [Google Scholar]
- Zhang J, Tan X, Li W, Wang Y, Wang J, Cheng X, Yang X 2005. Smad4 is required for the normal organization of the cartilage growth plate. Dev Biol 284: 311–322 [DOI] [PubMed] [Google Scholar]
- Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM 2006. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem 281: 15694–15700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Zhang Z, Feng JQ, Dusevich VM, Sinha K, Zhang H, Darnay BG, de Crombrugghe B 2010. Multiple functions of Osterix are required for bone growth and homeostasis in postnatal mice. Proc Natl Acad Sci 107: 12919–12924 [DOI] [PMC free article] [PubMed] [Google Scholar]