Abstract
Epstein–Barr virus (EBV) is a paradigm for human tumor viruses: it is the first virus recognized to cause cancer in people; it causes both lymphomas and carcinomas; yet these tumors arise infrequently given that most people in the world are infected with the virus. EBV is maintained extrachromosomally in infected normal and tumor cells. Eighty-four percent of these viral plasmids replicate each S phase, are licensed, require a single viral protein for their synthesis, and can use two functionally distinct origins of DNA replication, oriP, and Raji ori. Eighty-eight percent of newly synthesized plasmids are segregated faithfully to the daughter cells. Infectious viral particles are not synthesized under these conditions of latent infection. This plasmid replication is consistent with survival of EBV’s host cells. Rare cells in an infected population either spontaneously or following exogenous induction support EBV’s lytic cycle, which is lethal for the cell. In this case, the viral DNA replicates 100-fold or more, uses a third kind of viral origin of DNA replication, oriLyt, and many viral proteins. Here we shall describe the three modes of EBV’s replication as a function of the viral origins used and the viral and cellular proteins that mediate the DNA synthesis from these origins focusing, where practical, on recent advances in our understanding.
Epstein–Barr virus has infected more than 6.5 billion people in the world. It has distinct modes of replication.
oriP
A single contiguous fragment of EBV DNA with a length of 1.7 kbps supports autonomous extrachromosomal replication and maintenance of recombinant plasmids in human cells (Yates 1996) when the Epstein–Barr nuclear protein 1 (EBNA1) is provided in trans (Yates 1996) (Fig. 1). This subgenomic EBV DNA is termed oriP for “origin of plasmid replication.” Similar to the parental EBV genome (Yates 1996), initiation of DNA replication within oriP is synchronized with the timed and strictly regulated replication of chromosomal DNA (Yates 1996; Shirakata et al. 1999), and mediated by the cellular replication machinery acting at oriP. The viral protein EBNA1 binds to oriP site-specifically and recruits the cellular DNA replication machinery (Chaudhuri et al. 2001; Dhar et al. 2001; Schepers et al. 2001; Ritzi et al. 2003). Thus, EBNA1 is essential but not sufficient for oriP’s function. In resting and proliferating cells, oriP confers stable extrachromosomal maintenance of recombinant plasmids, which also relies on EBNA1 in trans independent of its role in DNA replication. An EBV genome with a deletion in EBNA1 has been found only to be integrated into the cellular chromosome in virus-infected B cells, consistent with its being unable to be maintained extrachromosomally (Humme et al. 2003).
Figure 1.
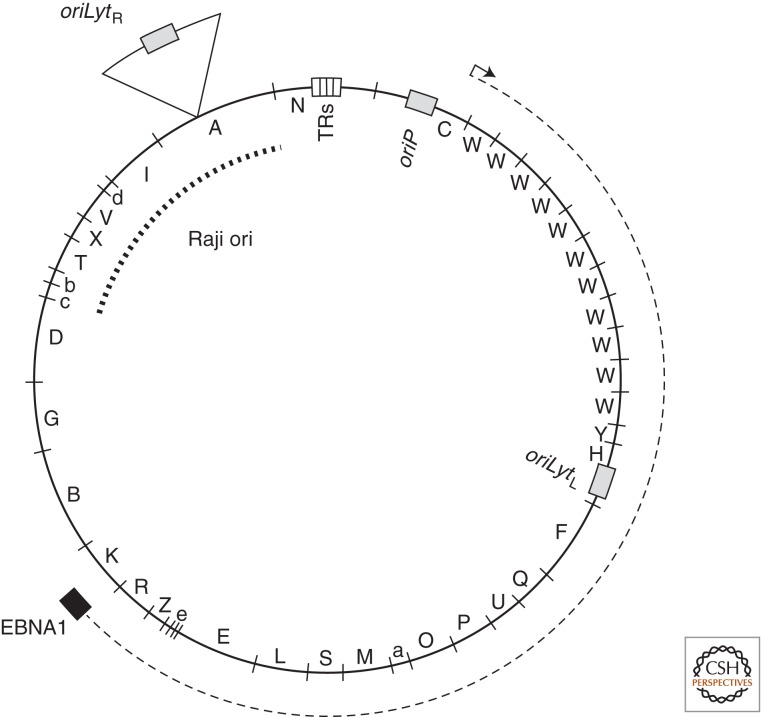
A physical map of the B95-8 laboratory strain of EBV DNA of 165 kbp is shown. The letters on the inner surface of the circle denote the fragments generated by digestion of this DNA with the BamH1 endonuclease and used to mark its transcripts (Baer et al. 1984). The primary transcript for EBNA1 is denoted by the dashed line. The white box marked TRs indicates the terminal repeats of linear virion DNA that mediate circularization of the viral DNA on infection of cells. The sites for oriP and oriLyt (L, left) are shown with the approximate location of DNA deleted from the B95-8 strain, present in all other analyzed strains, that contains a second copy of oriLyt (R, right). Raji ori, a region that is defined only approximately maps to sequences spanning the deletion found in the B95-8 strain.
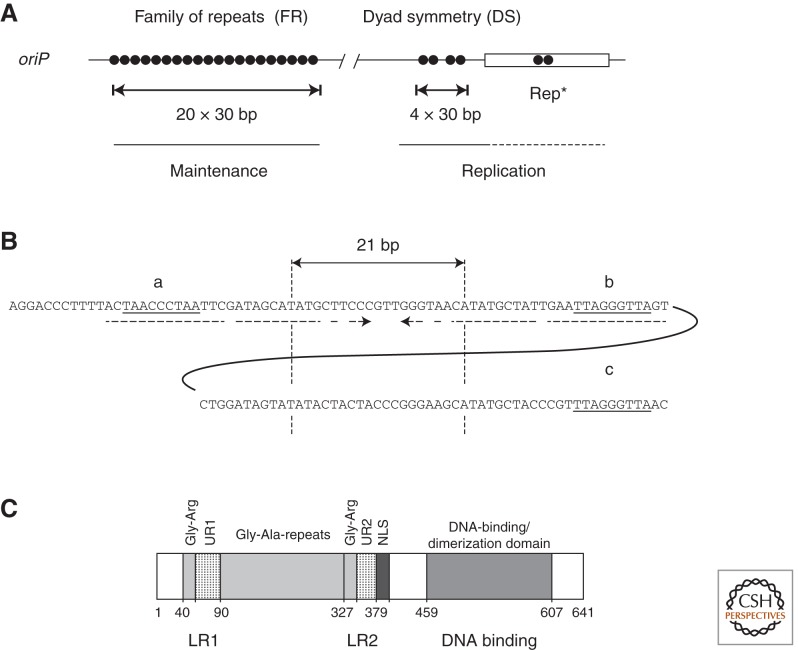
oriP’s replicator is coincident with or located near the origin of DNA replication (Yates 1996) and is operationally termed the “dyad symmetry” (DS) element (Fig. 2A). About 1 kbp upstream of DS, an array of tandem repeats 650 bps in total size, termed the “family of repeats” (FR), provides 20 sequence motifs to which EBNA1 molecules bind with high affinity. The FR element does not contribute to DNA synthesis but is mandatory for the stable maintenance and nuclear retention of oriP plasmids and genomic EBV DNA.
Figure 2.
Shown are depictions of oriP and EBNA1. (A) oriP consists of multiple sets of binding sites for EBNA1. The family of repeats in the B95-8 strain of EBV has 20 binding sites for EBNA1, which together mediate maintenance of oriP plasmids. DS has two pairs of binding sites and is the site or close to the site at which DNA synthesis initiates within oriP. Rep* can substitute for DS functionally but inefficiently. It too contains a pair of EBNA1-binding sites with the same spacing as each pair in DS. When multimerized eightfold, Rep* functions as efficiently as does DS to support initiation of DNA synthesis. (B) The sequence and spacing of EBNA1-binding sites in DS of oriP are shown. The two sites of each pair are separated by 21 bps allowing similar contacts on one face of helical DNA. The nonomer repeats that can bind telomere-associated proteins are underlined and denoted a, b, and c. (C) The domains of EBNA1 from the B95-8 strain of EBV are represented and are composed of 641 residues. LR1 and LR2 are linking regions rich in Gly-, Arg residues, which when fused to protein DNA-binding domains, can link the DNAs presumably by the AT-hook activities intrinsic to LR1 and LR2. LR1 and LR2 each contain unique sequences, UR1 and UR2. UR1 is involved in transcription regulated by EBNA1; UR2 does not have a known function. The Gly, Ala repeats have various activities ascribed to them but little or no effect on EBNA1’s functions in cell culture. EBNA1 has one identified nuclear localization sequence (NLS) (Yates 1996). EBNA1’s carboxy-terminal one-third comprises a dimerization and DNA-binding domain.
FR, the Plasmid Maintenance Element
The family of repeats, termed FR, is an array of 21 imperfectly conserved, 30 bp, direct repeats (Fig. 2A). EBNA1 binds as homotypic dimers to 20 sequence motifs within FR although only seven EBNA1-binding sites are minimally needed to function efficiently (Yates 1996). One essential role of FR is to prevent plasmid loss from proliferating cells. This plasmid maintenance/nuclear retention function appears to act by tethering the FR element via EBNA1 to condensed mitotic chromosomes as they segregate during mitosis leading to comigration of FR-carrying DNA molecules (Yates 1996; Marechal et al. 1999; Hung et al. 2001). This tethering by EBNA1 is accomplished through its carboxyl terminus binding site, specifically to FR and its linking regions in its amino terminus binding AT-rich sequences in host chromosomes (see below). Live-cell imaging has shown that newly synthesized daughter molecules remain colocalized as pairs until anaphase when 88% segregate one-to-one to daughter cells and the remaining 12% segregate randomly (Nanbo et al. 2007). The mechanism of this nonrandom segregation, which must be established in S phase, is not understood. FR in conjunction with EBNA1 also can act as an EBNA1-dependent, transcriptional enhancer (Kennedy and Sugden 2003).
DS, the Origin of DNA Replication
The DS element is an EBNA1-dependent replicator for the EBV genome. An approximately 120 bp region of oriP containing the minimal replicator has been termed DS because it contains an element of dyad symmetry (DS) 65 bp in length (Fig. 2B). Bidirectional DNA synthesis initiates at or close to DS (Yates 1996) and DS does support EBNA1-dependent DNA replication in the absence of the FR element (Yates et al. 2000). The DS element consists of two pairs of EBNA1-binding sites (Fig. 2B). The EBNA1-binding sites within one pair are separated by 21 bp such that they are in the same helical phase, which is essential for their acting as a replicator (Bashaw and Yates 2001). The binding sites within FR for EBNA1 do not require the same spatially strict arrangement for FR’s functions.
It has been shown that one pair of correctly spaced EBNA1-binding sites is sufficient to recruit the origin recognition complex (see below) to the replicator (Julien et al. 2004) indicating that the correct binding of EBNA1 is a critical parameter for origin activation. This minimal replicator shows only a fraction of the activity of the intact DS element. Auxiliary elements are likely to contribute to the efficiency of initiation of DNA synthesis, but their genetic dissection is still incomplete. These auxiliary elements include 14 bp repeats in which are embedded nonamer motifs (Fig. 2B) that bind some telomere-associated proteins (see below). The binding of these proteins increases the efficiency of DS as a replicator (Deng et al. 2002, 2003; Lindner et al. 2008).
An element termed Rep* of about 300 bps, which is located downstream from DS was identified to be an alternative EBNA1-dependent replicator with reduced activity (Fig. 2A) (Kirchmaier and Sugden 1998). Although Rep* supports only short-term plasmid replication, it may contribute to the overall replication efficiency of oriP because multimers of Rep* support long-term plasmid replication as efficiently as does oriP (Wang et al. 2006). Initiation of DNA replication within an oriP replicon is likely to be influenced by its chromatin structure. Nucleosomes surrounding DS appear to be phased and modified in a cell-cycle-dependent manner (Wensing et al. 2001; Avolio-Hunter and Frappier 2003; Zhou et al. 2005; Tempera and Lieberman 2010). The extent to which the chromatin structure controls the replicator or is controlled by the machinery at the replicator is not yet clear.
Proteins that Support the Functions of oriP
oriP consists largely of two stretches of binding sites for EBV’s EBNA1 protein that differ in their numbers, spacing, and affinities. EBNA1 is an essential participant in oriP’s functions. EBNA1 can now be viewed as an assembly of at least four functional elements that support EBV as an extrachromosomal replicon (Fig. 2C).
The carboxy-terminal quarter of EBNA1 encodes two functions essential for all trans-acting phenotypes ascribed to EBNA1. This region provides a structure that forms a dimer that binds DNA site-specifically. This structure closely resembles that of the dimerization and DNA-binding domain of the E2 protein encoded by papillomaviruses (Hegde et al. 1992; Bochkarev et al. 1996). The dimerization and DNA-binding domain of EBNA1 alone bends DNAs it binds site-specifically as does that of the E2 proteins (Hegde et al. 1992; Kim et al. 2000; Bashaw and Yates 2001; Wang et al. 2006). It also acts as a dominant negative derivative for EBNA1’s known functions such that its expression in EBV-positive tumors forces the loss of the virus from cells to reveal the phenotypes provided them by EBV (Vereide and Sugden 2011).
A stretch of glycine, alanine repeats that spans more than 200 residues in EBNA1 of the B95-8 laboratory strain of EBV has been found to inhibit antigen processing (Levitskaya et al. 1997). This same repeated element has been found to limit translation in cis, which might also underlie EBNA1’s being poorly recognized by cytotoxic T-cells (Yin et al. 2003). These repeats may contribute to EBNA1’s being recognized by CD4+ T cells following its autophagic degradation and the display of its resulting peptides on MHC class II molecules (Paludan et al. 2005). EBNA-1 is the only viral protein consistently expressed in proliferating, infected cells. Its atypical immune recognition may be essential for EBV’s long-term success as a human parasite.
A third functional element of EBNA1 consists of a stretch of 25 amino acids termed UR1 (unique region 1) positioned between a stretch of glycine, arginine and the glycine, alanine repeats. It is essential for EBV’s transformation of human B cells (Altmann et al. 2006). UR1 supports EBNA1’s activation of transcription but not replication (Wu et al. 2002; Kennedy and Sugden 2003). This element contains two cysteines, which allow two monomers of EBNA1 to bind Zinc coordinately. This zinc coordination is necessary for EBNA1’s transcriptional transactivation (Aras et al. 2009) and is reminiscent of the amino terminus of the E2 protein of papillomaviruses, which can form intradimers within one pair of monomers or interdimers between two pairs of monomers (Antson et al. 2000).
Another functional moiety of EBNA1 consists of two redundant elements termed LR1 and LR2 (linking region 1 and 2). LR1 and LR2 are rich in arginine and glycine residues and were identified by their ability to loop regions of DNA together to which they were bound in cis (Yates 1996) or to link such elements in trans (Yates 1996). Genetic analyses of LR1 and LR2 indicate that both contribute to EBNA1’s support of transcription and of replication (Mackey and Sugden 1999). One mechanism by which these linking regions contribute to EBNA1’s support of replication of oriP has emerged. Both LR1 and LR2 have AT-hook activities derived from their arginine, glycine repeats, which promote the association of EBNA1 to AT-rich stretches of DNA (Sears et al. 2004). Importantly, fusions of the cellular protein HMGA1a, which has three AT-hooks to the dimerization and DNA-binding domain of EBNA1 support the replication of oriP plasmids in human cells stably (Hung et al. 2001; Sears et al. 2003). Fusions of EBNA1’s dimerization and DNA-binding domain to HMG1 or EBP2, both of which appear to lack AT-hooks, fail to support the replication of oriP stably in human cells (Sears et al. 2003, 2004). The AT-hooks of EBNA1’s LR1 and LR2 thus are important for its support of the maintenance of oriP in proliferating cells. They likely provide EBV another, associated function. The tethering of EBV plasmid DNAs to AT-rich chromosomal sites may insure that the viral replicators home to special sites in the nucleus where they can function when their attached chromosomal replicators initiate synthesis. A third proposed contribution of the linking regions to replication is their binding G-rich RNA and through this binding apparently recruiting ORC to DS bound by EBNA1 (Norseen et al. 2008). This role is supported by GST-fusions to LR1 and LR2 binding ORC from cell extracts in an RNAse-susceptible manner (Norseen et al. 2008). This described role is perplexing though, because it is not easily reconciled with the requirement for the specific spacing of EBNA1’s DNA-binding sites needed to recruit ORC (Bashaw and Yates 2001; Wang et al. 2006). In addition, Cdc6 binds EBNA1 directly in vitro in an RNAse-resistant manner and contributes to EBNA1’s recruitment of ORC to DS (Moriyama et al. 2012).
Proteins other than EBNA1 and ORC can bind to or associate with oriP to facilitate its functions. The MCM complex associates with DS in a cell cycle-dependent manner; MCM2, 3, and 7 have been detected at DS by chromatin immunoprecipitation and to be enriched there during G1 and early S phases (Chaudhuri et al. 2001; Ritzi et al. 2003). Additional cellular proteins have been found to associate with DS dependent on EBNA1 in cell extracts and in vivo. The three nonamer repeats (TTAGGGTTA) abutting EBNA1-binding sites in DS are similar to telomeric repeats (TTAGGG) and bind proteins in vivo as measured by genomic footprinting (Fig. 2B) (Niller et al. 1995). DNA-affinity chromatography with DS coupled to a matrix enriches for several telomere-associated proteins only from EBNA1-positive cells (Deng et al. 2002, 2003). These enriched proteins include TRF1 and TRF2 (telomeric repeat binding factors 1 and 2) and hRap1 (homolog of repressor activator protein 1), which bind the nonamers directly (TRF1 and TRF2) or indirectly by binding TRF2 (Deng et al. 2002, 2003). The roles these proteins have in oriP-mediated replication are not clear, though. Derivatives of oriP with mutated nonamers that fail to bind these telomere-associated proteins are maintained stably in cells at copy numbers one-half of that of wild-type oriP (Deng et al. 2003). One simple model for TRF2, is based on its binding ORC1 directly (Atanasiu et al. 2006). Binding of TRF2 to DS should aid in recruiting ORC to DS. This simple model is consistent with analyses of Rep*. The two binding sites for EBNA1 in Rep* do not have neighboring nonamer sequences and support DNA replication poorly (Wang et al. 2006). When these EBNA1-binding sites are embedded in neutral lambda DNA and octamerized, they support DNA synthesis as efficiently as does wild-type DS (Wang et al. 2006). This result is consistent with the finding that the overall avidity of EBNA1 for derivatives of DS correlates with the efficiency of replication of those derivatives (Lindner et al. 2008).
Chromatin immunoprecipitation experiments have identified additional proteins present at oriP in cells that likely affect its replication. Two proteins implicated in yeast in stabilizing replication forks, Timeless (Tim) and Timeless interacting protein, have been detected there (Dheekollu and Lieberman 2011). Inhibiting Tim leads to the accumulation of linear EBV DNA in cells, consistent with these proteins stabilizing replication forks at oriP (Dheekollu and Lieberman 2011). The deubiquitylating enzyme, USP7, binds EBNA1 in vitro and localizes to oriP in vivo (Sarkari et al. 2009). This association affects the levels of monoubiquitylated histone H2B in the vicinity of oriP and is thought to affect transactivation of transcription by EBNA1 bound to FR (Sarkari et al. 2009).
Raji ori, AN ALTERNATE LICENSED ORIGIN OF EBV
Although DS of oriP was the only origin of plasmid DNA synthesis identified by screening cloned fragments of EBV DNA for “ARS” activity in EBV-positive cells (Yates 1996), work of Carl Schildkraut and his colleagues has identified a new class of origins that function in extrachromosomal EBV (Yates 1996; Norio and Schildkraut 2004). They studied replicative intermediates of EBV DNA in Raji cells with two dimensional gel analyses and showed that DNA synthesis in Raji EBV often originated 25 kbp away from its oriP (Fig. 1) (Yates 1996). Multiple observations indicate that this origin, which we shall refer to as “Raji ori,” is akin to chromosomal origins consisting of “zones of replication” such as that in the DHFR locus, but Raji ori functions in an extrachromosomal replicon.
Raji ori can support DNA synthesis efficiently and is not unique to the EBV strain in Raji cells. EBV DNA synthesis has been characterized in Raji cells and in the Mutu1 cell line using “single molecule analysis of replicated DNA” (Norio and Schildkraut 2004). This approach has revealed that DNA synthesis can originate throughout much or all of EBV DNA in both Raji cells and Mutu1 cells, that initiation is most common at Raji ori in Raji cells, and is less frequent but does occur in the Raji ori region of the EBV DNA in Mutu1 cells. The frequency of initiation within Raji ori in Raji cells does not reflect a genetic defect in the DS element of its oriP. This DS has been isolated, sequenced, and found not to have significant differences from the wild type sequence (Koons et al. 2001). DS has been deleted from a third strain of EBV, the recombinant virus introduced into an EBV-negative, B-cell line, and replicative intermediates analyzed by two dimensional gel analysis. The Raji ori of this recombinant strain was found to support the initiation of DNA synthesis efficiently, although other regions showed the presence of bubble arcs too (Norio et al. 2000).
Raji ori shares at least two properties with some cellular origins. First, it is licensed. The EBV DNA in Raji cells was studied in density shift experiments and shown to incorporate BrdU at the same rate as did host chromosomal DNA showing that EBV DNA replicates semi-conservatively, once per cell cycle in these cells (Yates 1996), consistent with its being licensed. Second, Raji ori also appears to have multiple sites at which DNA synthesis can initiate (Yates 1996) as has been found, for example, with the cellular DHFR locus (Kobayashi et al. 1998).
Both EBV and the host cell contribute trans-acting factors necessary for the function of Raji ori. Multiple observations indicate that EBNA1 in trans in conjunction with FR in cis is essential for the maintenance of replicons using Raji ori in proliferating cells (Yates 1996). EBNA1 does not contribute directly to origin function of Raji ori, though. EBNA1 does not bind detectably to Raji ori as measured by gel shift assays using 40 overlapping fragments of 600 bp that span this zone of initiation (Wang and Sugden 2008). Given that Raji ori is a zone with multiple sites for the initiation of licensed DNA replication, ORC likely binds those sites but the means by which it does so is unknown.
One question, “Why does EBV have oriP given it has Raji ori?” has been answered. The DNA replication mediated by DS at oriP is more efficient than that of Raji ori with FR in cis and EBNA1 in trans when initially introduced into Raji cells. This more efficient replication allows plasmids with DS plus FR to become “established” (Wang and Sugden 2008). Only 1%–10% of EBV’s plasmid replicons that are introduced into cells and initially replicate progress to be maintained stably after 15 generations or so, a process termed “establishment” (Leight and Sugden 2001). EBV plasmids are present in all examined clones of cells in a wide distribution of numbers of plasmids per cell, an equilibrium that reflects both defects in their synthesis and partitioning (Nanbo et al. 2007). The efficient initial replication of replicons with DS plus FR allows them to achieve this distribution needed to be established. Replicons with Raji ori and FR fail to be established but can be maintained in Raji cells once established by virtue of having had DS in cis (Wang and Sugden 2008). Thus, the DS replicator is peculiarly efficient on being introduced into cells, a function required during virus infection to allow EBV to be established. Once established chromosomal-like replicators can take over and sustain replication of this viral extrachromosomal replicon.
oriLyt
DNA replication during the lytic phase of EBV’s life cycle is uncoupled from and independent of that during its latent phase. Both the cis-acting elements and trans-acting factors involved in latent and lytic viral DNA replication differ, indicative of their different mechanisms. Although DNA replication during the latent phase ensures the faithful duplication of 84% of the viral genomes in each cell cycle (Nanbo et al. 2007), viral genomes replicate independently of such constraints during EBV’s lytic cycle and are amplified several hundred-fold within one to two days (Hammerschmidt and Sugden 1988). The products of lytic DNA replication are long concatemers (Bloss and Sugden 1994). They become the substrate for further processing eventually yielding cleaved, packaged, linear genome units bound in their preformed capsids with polyamines (Gibson and Roizman 1971). These “naked” viral DNAs are unmethylated, circularize in the recipient cell following infection, and eventually reside in the nucleus in which they are organized into chromatin and become methylated at cytosines in CpG dinucleotides (Shaw et al. 1979; Fernandez et al. 2009; Kalla et al. 2010).
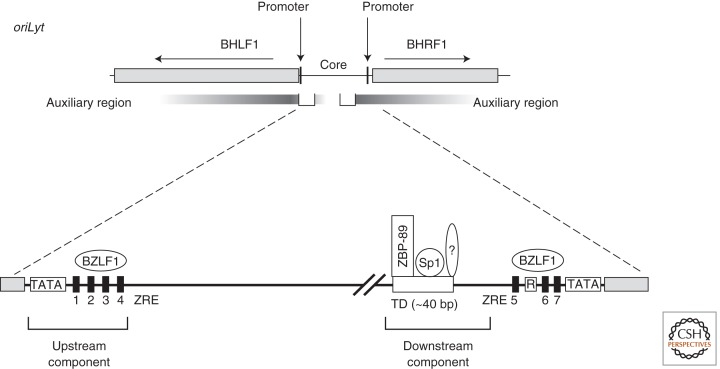
The cis-acting element, which acts as the lytic origin of replication of EBV, is termed oriLyt (Yates 1996). Two copies, which are about 100 kbps apart and therefore located opposite to each other in the circular genome, are present in all EBV strains examined with the exception of the B95-8 laboratory strain (Fig. 1). EBV’s oriLyt shares limited sequence homology with lytic origins of members of the β- and γ-herpesvirus family but not with those of α-herpesviruses such as herpes simplex virus type 1 (HSV-1). oriLyt is characterized by a duplicated 1055-bp long core element, which is virtually identical in both copies of oriLyt (Fig. 3). Within this core element there are two essential components separated by about 530 bps. Loosely defined auxiliary components that flank the two essential upstream and downstream components also contribute to oriLyt’s activity (Hammerschmidt and Sugden 1988; Schepers et al. 1993b). These nonessential but auxiliary components greatly enhance the activity of the core components (Hammerschmidt and Sugden 1988; Schepers et al. 1993b).
Figure 3.
oriLyt and its expanded core domain are shown. oriLyt spans about 7700 bp of the B95-8 strain of EBV including two genes BHLF1 and BHRF1 and their promoters. Sequences in these genes contribute to but are not essential for the function of oriLyt. The essential or core domain of oriLyt includes the promoters for and intergenic region of these genes. Two essential components (upstream and downstream) of oriLyt bind viral (BZLF1) at BZLF1-responsive elements (ZRE) and cellular factors (ZBP-89, Sp1, and likely additional unknown proteins) are required for oriLyt’s function. The BZLF1 protein also binds to promoter elements of the BHRF1 gene to enhance oriLyt’s activity. This domain (ZRE sites 5, 6, and 7) can be replaced by a heterologous enhancer to support oriLyt’s function. The boxed element marked R binds the viral BRLF1 transactivator, which does not contribute directly to EBV’s lytic DNA replication.
The products of lytic DNA replication of all herpesviruses are DNA concatemers in which the single genome units are arranged head to tail. This arrangement has also been shown for EBV (Hammerschmidt and Sugden 1988; Schepers et al. 1993b) and is consistent with circularized monomeric DNA molecules serving as templates for DNA replication via a rolling-circle mechanism. However, a rolling-circle mode alone poorly accounts for the rapid accumulation of progeny DNA. For example, in HSV-1-infected cells, about 30 min is needed to complete the replication of one viral molecule, but this herpesviral DNA is replicated several hundredfold during a short period of time to yield up to 1000 genomic copies per cell (Jacob and Roizman 1977). A pure rolling-circle mechanism synthesizes progeny DNA only linearly, not exponentially with time, and appears inadequate to explain the rapid amplification of herpesviral DNA. In fact, early during EBV’s lytic cycle viral replicative DNA intermediates were found to replicate semi-conservatively, to be amplified exponentially, and to be covalently closed, circular DNAs of parental length. These products of the first phase of EBV’s lytic DNA synthesis likely provide the many circular templates for DNA synthesis via a rolling-circle mechanism needed to yield efficient DNA amplification during the second phase (Pfuller and Hammerschmidt 1996). This biphasic mode of herpesviral DNA replication is consistent with the notion that the origin-binding proteins of herpesviruses are needed only to initiate but not to support continued lytic DNA synthesis (Schildgen et al. 2005).
Proteins that Support the Functions of oriLyt
All herpesviruses encode replication proteins that specifically interact with their origins of lytic DNA replication to provide key functions, including polymerases, helicases, primases, DNA-binding proteins, and associated factors, as well as enzymatic activities involved in nucleotide synthesis and phosphorylation (Challberg 1986; Challberg and Kelly 1989). Six EBV genes, which encode essential lytic functions, have been identified (Challberg 1986; Tsurumi 2001). With the exception of the virus-specific DNA-binding proteins essential for the activation of lytic origins of herpesviral DNA replication, these lytic gene products share considerable primary amino acid sequence and function among herpesviruses (Table 1) (Tsurumi 2001). These genes of EBV are under the control of two viral transactivators, BZLF1 and BRLF1, which thus orchestrate transcriptionally the expression of EBV’s replication machinery (Feederle et al. 2000).
Table 1.
Viral proteins acting at EBV’s characterized origins and their functions
| Origin | Protein | Function |
|---|---|---|
| oriP | EBNA1 | 1. Origin-binding protein at the DS element; recruits ORC for licensed DNA replication 2. Binds FR element to mediate maintenance of the plasmid replicon |
| oriLyt | BZLF1 | 1. Origin-binding protein of oriLyt that can bind multiple viral replication proteins 2. Viral transcription factor required to initiate the lytic phase of EBV and support expression of viral factors of lytic DNA amplification |
| BALF5 | DNA polymerase catalytic subunit; interacts with the helicase–primase complex (Fujii et al. 2000) | |
| BALF2 | Single-stranded DNA-binding protein | |
| BMRF1 | DNA polymerase accessory subunit with a PCNA-like structure (Murayama et al. 2009; Nakayama et al. 2010); interacts with BALF5 (Tsurumi et al. 1993); can act as a transcriptional coactivator of BZLF1 (Nakayama et al. 2009) | |
| BBLF4 | DNA helicase | |
| BSLF1 | Primase | |
| BBLF2/3 | Primase-associated factor. BBLF4, BSLF1, and BBLF2/3 form the helicase–primase complex |
BZLF1, a key transcriptional regulator, is EBV’s lytic origin-binding protein (Schepers et al. 1993a, 1996). BZLF1 has been found to be required for wild-type levels of lytic DNA replication when it is bound to four sites within the essential upstream component of oriLyt (Schepers et al. 1993a, 1996), although it binds additional sites within the oriLyt sequence (Fig. 3). These four sites are an intrinsic part of the BHLF1 promoter; deletion experiments indicated that promoter elements in addition to the BZFL1-binding motifs contribute to lytic DNA replication (Schepers et al. 1993b; Rennekamp and Lieberman 2011). One of the putative factors could be the BHLF1 transcript itself that is expressed from this promoter (Fig. 3) (Rennekamp et al. 2010; Rennekamp and Lieberman 2011), but efficient transcription from this promoter is not sufficient to support replication (Schepers et al. 1993a, 1996) because a viral mutant lacking all BZLF1-binding-sites within oriLyt supported a low level of lytic replication (Feederle and Delecluse 2004).
BZLF1 may act independently of EBV’s six lytic gene products to support the initiation of DNA replication at oriLyt (Yates 1996). This hypothesis suggests that cellular proteins contribute to the formation of early replicative DNA intermediates, which subsequently become the substrate for lytic DNA amplification. In striking contrast to the origin-binding protein of HSV-1, UL9, which is a sequence specific helicase, BZLF1 does not contribute an intrinsic enzymatic function to DNA synthesis.
BZLF1 contributes to the replication complex at oriLyt presumably in part by associating with some members of the complex. BZLF1 has been reported to interact with the viral helicase–primase complex (Table 1) (Liao et al. 2001, 2005; El-Guindy et al. 2010) and the viral polymerase accessory factor BMRF1 (Takagi et al. 1991; Daikoku et al. 2005; Nakayama et al. 2009). BMRF1 bears structural similarities with cellular PCNA (Murayama et al. 2009; Nakayama et al. 2010) and could potentially provide an additional tethering function for the replication complex (Zhang et al. 1997; Baumann et al. 1999). In addition, the primase-associated factor might serve a similar tethering function in conjunction with cellular, oriLyt-binding proteins (see below and Liao et al. 2005).
The essential downstream component of oriLyt is highly sensitive to sequence alterations within a stretch of about 40 bps, termed the “TD” element (Gruffat et al. 1995). TD was found to be the binding site for several cellular proteins (Gruffat et al. 1995). The transcription factors Sp1, Sp3, and ZBP-89, have been identified and shown to make essential and direct contributions to oriLyt’s function(s) (Gruffat et al. 1995; Baumann et al. 1999). They interact with EBV’s DNA polymerase and its processivity factor and likely tether viral replication proteins to oriLyt via direct protein–protein interactions at TD. These findings have a parallel in ZBRK1 and KAP-1, a zinc-finger DNA binding protein and its corepressor, which bind to a site located about 200 bps downstream of TD (Liao et al. 2005) within the previously identified oriLyt enhancer region (Yates 1996) and colocalize EBV’s helicase–primase complex to operationally defined replication compartments in lytically induced cells. Replication compartments are sites to which components of the cellular homologous recombination pathway are recruited (Wilkinson and Weller 2004, and references therein) together with various cis-acting elements of lytic replicons of herpesviruses. Lytic DNA replication involves both replication and homologous recombination of DNA, which are two interdependent processes active during the lytic phase of EBV’s life cycle (Pfuller and Hammerschmidt 1996). Several cellular recombination and DNA repair factors have been recently found recruited to EBV’s replication compartments (Daikoku et al. 2006; Kudoh et al. 2009; Sugimoto et al. 2011) and induction of EBV’s lytic phase induces a genuine DNA-damage response signal (Kudoh et al. 2005; Sato et al. 2010). It now seems that oriLyt is complex not only in mediating mechanistically distinct biphasic DNA replication but also in using a complex repertoire of viral and cellular proteins to carry out these two modes of replication.
IN SUMMARY
EBV is an enormously successful human parasite, having infected more than 6.5 billion people in the world today. Its success is intimately tied to its having evolved as an extrachromosomal replicon, inducing the infected cell to cycle, and successfully usurping the cell’s machinery to carry out its licensed DNA synthesis. It has built on a common herpesviral property of amplifying viral DNA during productive infections through dedicated viral origins of DNA synthesis and an array of viral proteins that mediate this unlicensed synthesis. It carries out these different modes of DNA replication in human B cells by affecting the differentiated state of it B-cell host.
Footnotes
Editors: Stephen D. Bell, Marcel Méchali, and Melvin L. DePamphilis
Additional Perspectives on DNA Replication available at www.cshperspectives.org
REFERENCES
- Altmann M, Pich D, Ruiss R, Wang J, Sugden B, Hammerschmidt W 2006. Transcriptional activation by EBV nuclear antigen 1 is essential for the expression of EBV’s transforming genes. Proc Natl Acad Sci 103: 14188–14193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antson AA, Burns JE, Moroz OV, Scott DJ, Sanders CM, Bronstein IB, Dodson GG, Wilson KS, Maitland NJ 2000. Structure of the intact transactivation domain of the human papillomavirus E2 protein. Nature 403: 805–809 [DOI] [PubMed] [Google Scholar]
- Aras S, Singh G, Johnston K, Foster T, Aiyar A 2009. Zinc coordination is required for and regulates transcription activation by Epstein–Barr nuclear antigen 1. PLoS Pathog 5: e1000469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasiu C, Deng Z, Wiedmer A, Norseen J, Lieberman PM 2006. ORC binding to TRF2 stimulates OriP replication. EMBO Rep 7: 716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avolio-Hunter TM, Frappier L 2003. EBNA1 efficiently assembles on chromatin containing the Epstein–Barr virus latent origin of replication. Virology 315: 398–408 [DOI] [PubMed] [Google Scholar]
- Baer R, Bankier AT, Biggin MD, Deininger PL, Farrell PJ, Gibson TJ, Hatfull G, Hudson GS, Satchwell SC, Seguin C, et al. 1984. DNA sequence and expression of the B95-8 Epstein–Barr virus genome. Nature 310: 207–211 [DOI] [PubMed] [Google Scholar]
- Bashaw JM, Yates JL 2001. Replication from oriP of Epstein–Barr virus requires exact spacing of two bound dimers of EBNA1 which bend DNA. J Virol 75: 10603–10611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann M, Feederle R, Kremmer E, Hammerschmidt W 1999. Cellular transcription factors recruit viral replication proteins to activate the Epstein–Barr virus origin of lytic DNA replication, oriLyt. EMBO J 18: 6095–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloss TA, Sugden B 1994. Optimal lengths for DNAs encapsidated by Epstein–Barr virus. J Virol 68: 8217–8222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkarev A, Barwell JA, Pfuetzner RA, Bochkareva E, Frappier L, Edwards AM 1996. Crystal structure of the DNA-binding domain of the Epstein–Barr virus origin-binding protein, EBNA1, bound to DNA. Cell 84: 791–800 [DOI] [PubMed] [Google Scholar]
- Challberg MD 1986. A method for identifying the viral genes required for herpesvirus DNA replication. Proc Natl Acad Sci 83: 9094–9098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challberg MD, Kelly TJ 1989. Animal virus DNA replication. Annu Rev Biochem 58: 671–717 [DOI] [PubMed] [Google Scholar]
- Chaudhuri B, Xu H, Todorov I, Dutta A, Yates JL 2001. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein–Barr virus. Proc Natl Acad Sci 98: 10085–10089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daikoku T, Kudoh A, Fujita M, Sugaya Y, Isomura H, Shirata N, Tsurumi T 2005. Architecture of replication compartments formed during Epstein–Barr virus lytic replication. J Virol 79: 3409–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daikoku T, Kudoh A, Sugaya Y, Iwahori S, Shirata N, Isomura H, Tsurumi T 2006. Postreplicative mismatch repair factors are recruited to Epstein–Barr virus replication compartments. J Biol Chem 281: 11422–11430 [DOI] [PubMed] [Google Scholar]
- Deng Z, Lezina L, Chen CJ, Shtivelband S, So W, Lieberman PM 2002. Telomeric proteins regulate episomal maintenance of Epstein–Barr virus origin of plasmid replication. Mol Cell 9: 493–503 [DOI] [PubMed] [Google Scholar]
- Deng Z, Atanasiu C, Burg JS, Broccoli D, Lieberman PM 2003. Telomere repeat binding factors TRF1, TRF2, and hRAP1 modulate replication of Epstein–Barr virus OriP. J Virol 77: 11992–12001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SK, Yoshida K, Machida Y, Khaira P, Chaudhuri B, Wohlschlegel JA, Leffak M, Yates J, Dutta A 2001. Replication from oriP of Epstein–Barr virus requires human ORC and is inhibited by geminin. Cell 106: 287–296 [DOI] [PubMed] [Google Scholar]
- Dheekollu J, Lieberman PM 2011. The replisome pausing factor Timeless is required for episomal maintenance of latent Epstein–Barr virus. J Virol 85: 5853–5863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Guindy A, Heston L, Miller G 2010. A subset of replication proteins enhances origin recognition and lytic replication by the Epstein–Barr virus ZEBRA protein. PLoS Pathog 6: e1001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feederle R, Delecluse HJ 2004. Low level of lytic replication in a recombinant Epstein–Barr virus carrying an origin of replication devoid of BZLF1-binding sites. J Virol 78: 12082–12084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feederle R, Kost M, Baumann M, Janz A, Drouet E, Hammerschmidt W, Delecluse HJ 2000. The Epstein–Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J 19: 3080–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AF, Rosales C, Lopez-Nieva P, Grana O, Ballestar E, Ropero S, Espada J, Melo SA, Lujambio A, Fraga MF, et al. 2009. The dynamic DNA methylomes of double-stranded DNA viruses associated with human cancer. Genome Res 19: 438–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii K, Yokoyama N, Kiyono T, Kuzushima K, Homma M, Nishiyama Y, Fujita M, Tsurumi T 2000. The Epstein–Barr virus pol catalytic subunit physically interacts with the BBLF4-BSLF1-BBLF2/3 complex. J Virol 74: 2550–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W, Roizman B 1971. Compartmentalization of spermine and spermidine in the herpes simplex virion. Proc Natl Acad Sci 68: 2818–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruffat H, Renner O, Pich D, Hammerschmidt W 1995. Cellular proteins bind to the downstream component of the lytic origin of DNA replication of Epstein–Barr virus. J Virol 69: 1878–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt W, Sugden B 1988. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein–Barr virus. Cell 55: 427–433 [DOI] [PubMed] [Google Scholar]
- Hegde RS, Grossman SR, Laimins LA, Sigler PB 1992. Crystal structure at 1.7 A of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature 359: 505–512 [DOI] [PubMed] [Google Scholar]
- Humme S, Reisbach G, Feederle R, Delecluse HJ, Bousset K, Hammerschmidt W, Schepers A 2003. The EBV nuclear antigen 1 (EBNA1) enhances B cell immortalization several thousandfold. Proc Natl Acad Sci 100: 10989–10994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung SC, Kang MS, Kieff E 2001. Maintenance of Epstein–Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc Natl Acad Sci 98: 1865–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob RJ, Roizman B 1977. Anatomy of herpes simplex virus DNA VIII. Properties of the replicating DNA. J Virol 23: 394–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien MD, Polonskaya Z, Hearing J 2004. Protein and sequence requirements for the recruitment of the human origin recognition complex to the latent cycle origin of DNA replication of Epstein–Barr virus oriP. Virology 326: 317–328 [DOI] [PubMed] [Google Scholar]
- Kalla M, Schmeinck A, Bergbauer M, Pich D, Hammerschmidt W 2010. AP-1 homolog BZLF1 of Epstein–Barr virus has two essential functions dependent on the epigenetic state of the viral genome. Proc Natl Acad Sci 107: 850–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy G, Sugden B 2003. EBNA-1, a bifunctional transcriptional activator. Mol Cell Biol 23: 6901–6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SS, Tam JK, Wang AF, Hegde RS 2000. The structural basis of DNA target discrimination by papillomavirus E2 proteins. J Biol Chem 275: 31245–31254 [DOI] [PubMed] [Google Scholar]
- Kirchmaier AL, Sugden B 1998. Rep*: A viral element that can partially replace the origin of plasmid DNA synthesis of Epstein–Barr virus. J Virol 72: 4657–4666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Rein T, DePamphilis ML 1998. Identification of primary initiation sites for DNA replication in the hamster dihydrofolate reductase gene initiation zone. Mol Cell Biol 18: 3266–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koons MD, Van Scoy S, Hearing J 2001. The replicator of the Epstein–Barr virus latent cycle origin of DNA replication, oriP, is composed of multiple functional elements. J Virol 75: 10582–10592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudoh A, Fujita M, Zhang L, Shirata N, Daikoku T, Sugaya Y, Isomura H, Nishiyama Y, Tsurumi T 2005. Epstein–Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J Biol Chem 280: 8156–8163 [DOI] [PubMed] [Google Scholar]
- Kudoh A, Iwahori S, Sato Y, Nakayama S, Isomura H, Murata T, Tsurumi T 2009. Homologous recombinational repair factors are recruited and loaded onto the viral DNA genome in Epstein–Barr virus replication compartments. J Virol 83: 6641–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leight ER, Sugden B 2001. Establishment of an oriP replicon is dependent upon an infrequent, epigenetic event. Mol Cell Biol 21: 4149–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitskaya J, Sharipo A, Leonchiks A, Ciechanover A, Masucci MG 1997. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein–Barr virus nuclear antigen 1. Proc Natl Acad Sci 94: 12616–12621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G, Wu FY, Hayward SD 2001. Interaction with the Epstein–Barr virus helicase targets Zta to DNA replication compartments. J Virol 75: 8792–8802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G, Huang J, Fixman ED, Hayward SD 2005. The Epstein–Barr virus replication protein BBLF2/3 provides an origin-tethering function through interaction with the zinc finger DNA binding protein ZBRK1 and the KAP-1 corepressor. J Virol 79: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner SE, Zeller K, Schepers A, Sugden B 2008. The affinity of EBNA1 for its origin of DNA synthesis is a determinant of the origin’s replicative efficiency. J Virol 82: 5693–6702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Sugden B 1999. The linking regions of EBNA1 are essential for its support of replication and transcription. Mol Cell Biol 19: 3349–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal V, Dehee A, Chikhi-Brachet R, Piolot T, Coppey-Moisan M, Nicolas JC 1999. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J Virol 73: 4385–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama K, Yoshizawa-Sugata N, Obuse C, Tsurimoto T, Masai H 2012. Epstein–Barr nuclear antigen 1 (EBNA-1)- dependent recruitment of origin recognition complex (Orc) on oriP of Epstein–Barr virus with purified proteins: Stimulation by Cdc6 through its direct interaction with EBNA1. J Biol Chem 287: 23977–23994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama K, Nakayama S, Kato-Murayama M, Akasaka R, Ohbayashi N, Kamewari-Hayami Y, Terada T, Shirouzu M, Tsurumi T, Yokoyama S 2009. Crystal structure of Epstein–Barr virus DNA polymerase processivity factor BMRF1. J Biol Chem 284: 35896–35905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama S, Murata T, Murayama K, Yasui Y, Sato Y, Kudoh A, Iwahori S, Isomura H, Kanda T, Tsurumi T 2009. Epstein–Barr virus polymerase processivity factor enhances BALF2 promoter transcription as a coactivator for the BZLF1 immediate-early protein. J Biol Chem 284: 21557–21568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama S, Murata T, Yasui Y, Murayama K, Isomura H, Kanda T, Tsurumi T 2010. Tetrameric ring formation of Epstein–Barr virus polymerase processivity factor is crucial for viral replication. J Virol 84: 12589–12598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbo A, Sugden A, Sugden B 2007. The coupling of synthesis and partitioning of EBV’s plasmid replicon is revealed in live cells. EMBO J 26: 4252–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niller HH, Glaser G, Knuchel R, Wolf H 1995. Nucleoprotein complexes and DNA 5′-ends at oriP of Epstein–Barr virus. J Biol Chem 270: 12864–12868 [DOI] [PubMed] [Google Scholar]
- Norio P, Schildkraut CL 2004. Plasticity of DNA replication initiation in Epstein–Barr virus episomes. PLoS Biol 2: e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio P, Schildkraut CL, Yates JL 2000. Initiation of DNA replication within oriP is dispensable for stable replication of the latent Epstein–Barr virus chromosome after infection of established cell lines. J Virol 74: 8563–8574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norseen J, Thomae A, Sridharan V, Aiyar A, Schepers A, Lieberman PM 2008. RNA-dependent recruitment of the origin recognition complex. EMBO J 27: 3024–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Münz C 2005. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 307: 593–596 [DOI] [PubMed] [Google Scholar]
- Pfuller R, Hammerschmidt W 1996. Plasmid-like replicative intermediates of the Epstein–Barr virus lytic origin of DNA replication. J Virol 70: 3423–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennekamp AJ, Lieberman PM 2011. Initiation of Epstein–Barr virus lytic replication requires transcription and the formation of a stable RNA–DNA hybrid molecule at OriLyt. J Virol 85: 2837–2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennekamp AJ, Wang P, Lieberman PM 2010. Evidence for DNA hairpin recognition by Zta at the Epstein–Barr virus origin of lytic replication. J Virol 84: 7073–7082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzi M, Tillack K, Gerhardt J, Ott E, Humme S, Kremmer E, Hammerschmidt W, Schepers A 2003. Complex protein-DNA dynamics at the latent origin of DNA replication of Epstein–Barr virus. J Cell Sci 116: 3971–3984 [DOI] [PubMed] [Google Scholar]
- Sarkari F, Sanchez-Alcaraz T, Wang S, Holowaty MN, Sheng Y, Frappier L 2009. EBNA1-mediated recruitment of a histone H2B deubiquitylating complex to the Epstein–Barr virus latent origin of DNA replication. PLoS Pathog 5: e1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Shirata N, Murata T, Nakasu S, Kudoh A, Iwahori S, Nakayama S, Chiba S, Isomura H, Kanda T, et al. 2010. Transient increases in p53-responsible gene expression at early stages of Epstein–Barr virus productive replication. Cell Cycle 9: 807–814 [DOI] [PubMed] [Google Scholar]
- Schepers A, Pich D, Hammerschmidt W 1993a. A transcription factor with homology to the AP-1 family links RNA transcription and DNA replication in the lytic cycle of Epstein–Barr virus. EMBO J 12: 3921–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers A, Pich D, Mankertz J, Hammerschmidt W 1993b. cis-acting elements in the lytic origin of DNA replication of Epstein–Barr virus. J Virol 67: 4237–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers A, Pich D, Hammerschmidt W 1996. Activation of oriLyt, the lytic origin of DNA replication of Epstein–Barr virus, by BZLF1. Virology 220: 367–376 [DOI] [PubMed] [Google Scholar]
- Schepers A, Ritzi M, Bousset K, Kremmer E, Yates JL, Harwood J, Diffley JF, Hammerschmidt W 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein–Barr virus. EMBO J 20: 4588–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildgen O, Graper S, Blumel J, Matz B 2005. Genome replication and progeny virion production of herpes simplex virus type 1 mutants with temperature-sensitive lesions in the origin-binding protein. J Virol 79: 7273–7278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears J, Kolman J, Wahl GM, Aiyar A 2003. Metaphase chromosome tethering is necessary for the DNA synthesis and maintenance of oriP plasmids but is insufficient for transcription activation by Epstein–Barr nuclear antigen 1. J Virol 77: 11767–11780 Erratum in J Virol78: 5531, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears J, Ujihara M, Wong S, Ott C, Middeldorp J, Aiyar A 2004. The amino terminus of Epstein–Barr virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J Virol 78: 11487–11505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JE, Levinger LF, Carter CW Jr 1979. Nucleosomal structure of Epstein–Barr virus DNA in transformed cell lines. J Virol 29: 657–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakata M, Imadome KI, Hirai K 1999. Requirement of replication licensing for the dyad symmetry element-dependent replication of the Epstein–Barr virus oriP minichromosome. Virology 263: 42–54 [DOI] [PubMed] [Google Scholar]
- Sugimoto A, Kanda T, Yamashita Y, Murata T, Saito S, Kawashima D, Isomura H, Nishiyama Y, Tsurumi T 2011. Spatiotemporally different DNA repair systems participate in Epstein–Barr virus genome maturation. J Virol 85: 6127–6135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi S, Takada K, Sairenji T 1991. Formation of intranuclear replication compartments of Epstein–Barr virus with redistribution of BZLF1 and BMRF1 gene products. Virology 185: 309–315 [DOI] [PubMed] [Google Scholar]
- Tempera I, Lieberman PM 2010. Chromatin organization of gammaherpesvirus latent genomes. Biochim Biophys Acta 1799: 236–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumi T 2001. EBV replication enzymes. Curr Top Microbiol Immunol 258: 65–87 [DOI] [PubMed] [Google Scholar]
- Tsurumi T, Kobayashi A, Tamai K, Daikoku T, Kurachi R, Nishiyama Y 1993. Functional expression and characterization of the Epstein–Barr virus DNA polymerase catalytic subunit. J Virol 67: 4651–4658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vereide DT, Sugden B 2011. Lymphomas differ in their dependence on Epstein–Barr virus. Blood 117: 1977–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Sugden B 2008. Identifying a property of origins of DNA synthesis required to support plasmids stably in human cells. Proc Natl Acad Sci 105: 9639–9644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lindner SE, Leight ER, Sugden B 2006. Essential elements of a licensed, mammalian plasmid origin of DNA synthesis. Mol Cell Biol 26: 1124–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensing B, Stuhler A, Jenkins P, Hollyoake M, Karstegl CE, Farrell PJ 2001. Variant chromatin structure of the oriP region of Epstein–Barr virus and regulation of EBER1 expression by upstream sequences and oriP. J Virol 75: 6235–6241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DE, Weller SK 2004. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J Virol 78: 4783–4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Kapoor P, Frappier L 2002. Separation of the DNA replication, segregation, and transcriptional activation functions of Epstein–Barr nuclear antigen 1. J Virol 76: 2480–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JL 1996. Epstein–Barr virus DNA replication. In DNA replication in eukaryotic cells (ed. ML DePamphilis), pp. 751–774 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Yates JL, Camiolo SM, Bashaw JM 2000. The minimal replicator of Epstein–Barr virus oriP. J Virol 74: 4512–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Manoury B, Fahraeus R 2003. Self-inhibition of synthesis and antigen presentation by Epstein–Barr virus-encoded EBNA1. Science 301: 1371–1374 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Holley-Guthrie E, Ge JQ, Dorsky D, Kenney S 1997. The Epstein–Barr virus (EBV) DNA polymerase accessory protein, BMRF1, activates the essential downstream component of the EBV oriLyt. Virology 230: 22–34 [DOI] [PubMed] [Google Scholar]
- Zhou J, Chau CM, Deng Z, Shiekhattar R, Spindler MP, Schepers A, Lieberman PM 2005. Cell cycle regulation of chromatin at an origin of DNA replication. EMBO J 24: 1406–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]