Abstract
Viruses are obligate intracellular parasites, and their replication requires host cell functions. Although the size, composition, complexity, and functions encoded by their genomes are remarkably diverse, all viruses rely absolutely on the protein synthesis machinery of their host cells. Lacking their own translational apparatus, they must recruit cellular ribosomes in order to translate viral mRNAs and produce the protein products required for their replication. In addition, there are other constraints on viral protein production. Crucially, host innate defenses and stress responses capable of inactivating the translation machinery must be effectively neutralized. Furthermore, the limited coding capacity of the viral genome needs to be used optimally. These demands have resulted in complex interactions between virus and host that exploit ostensibly virus-specific mechanisms and, at the same time, illuminate the functioning of the cellular protein synthesis apparatus.
Viruses commandeer and control translation in the host cell. Mechanisms range from manipulating key translation factors to using cis-acting elements that recruit ribosomes or modify genome-coding capacity.
The dependence of viruses on the host translation system imposes constraints that are central to virus biology and have led to specialized mechanisms and intricate regulatory interactions. Failure to translate viral mRNAs and to modulate host mRNA translation would have catastrophic effects on virus replication, spread, and evolution. Accordingly, a wide assortment of virus-encoded functions is dedicated to commandeering and controlling the cellular translation apparatus. Viral strategies to dominate the host translation machinery target the initiation, elongation, and termination steps and include mechanisms ranging from the manipulation of key eukaryotic translation factors to the evolution of specialized cis-acting elements that recruit ribosomes or modify genome-coding capacity. Because many of these strategies have likely been pirated from their hosts and because virus genetic systems can be manipulated with relative ease, the study of viruses has been a preeminent source of information on the mechanism and regulation of the protein synthesis machinery. In this article, we focus on select viruses that infect mammalian or plant cells and review the mechanisms they use to exploit and control the cellular protein synthesis machinery.
VIRAL REPLICATION AND TRANSLATION STRATEGIES
The prodigious diversity of viruses, their unparalleled rate of evolution, and the wide repertoire of their interactions with their hosts, all contribute to their utility as biological tools to study the translation system. Thus, viral genomes are composed of DNA or RNA, either of which may be single- or double-stranded; replication may take place in the nucleus or cytoplasm of the cell; infection may give rise to acute or persistent infection; viruses may be agents of disease or innocuous; and the consequences of infection may vary in different organisms or cell types. For a discussion of the underlying biological principles, you are referred to virology reviews (e.g., Pe’ery and Mathews 2007). Here, we cannot do more than briefly introduce the relationship between virus genome structure and mRNA biogenesis.
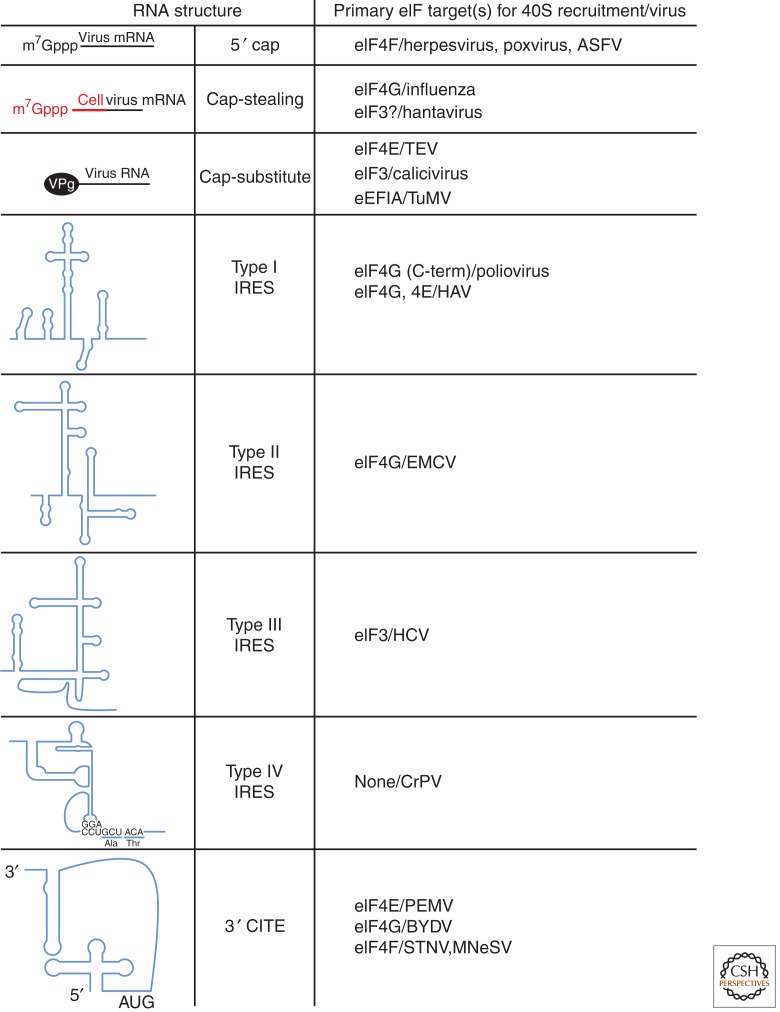
Viruses are classified into families according to their genome structure, which dictates the mode of synthesis of their mRNA, its structure, and often its translation. In some viral families, the viral RNA is single-stranded and of the same sense as the mRNA, termed (+)-stranded, and thus it can be directly translated. Viruses with RNA genomes that are of the opposite polarity, that is, (−)-stranded or double-stranded (ds), require virus-encoded RNA-dependent RNA polymerases to generate their mRNA. Exceptionally, retroviruses use viral reverse transcriptase to convert their (+)-stranded RNA genomes into a dsDNA form, which then serves as template for mRNA synthesis via cellular pathways. Most viruses with DNA genomes take advantage of the cellular machinery for mRNA production, but others (such as the poxviruses) use their own enzyme systems. As a consequence, virus families are characterized by mRNAs that have a range of structures at their 5′ ends (capped, not capped, cap-substituted) and 3′ ends (polyadenylated or not), and carry various cis-acting sequences and elements (internal ribosome entry sites [IRES], protein binding sites, etc.)—features that determine many aspects of viral mRNA translation (Fig. 1).
Figure 1.
Recruitment of 40S ribosome subunits to viral mRNAs: structural features and initiation factor targets. RNA structural elements involved in recruiting cellular 40S ribosome subunits to the 5′ end of viral mRNAs are shown on the left, with their corresponding name immediately to the right. The primary eukaryotic translation initiation factor (eIF) targets involved in recognizing each structural element and recruiting additional factors or mediating 40S subunit binding are indicated, with representative viruses that use them. (?) indicates that the precise factor targeted by hantavirus N protein remains unknown. Abbreviations: MNeSV, Maize necrotic streak virus; BYDV, Barley yellow dwarf virus; PEMV, Pea enation mosaic virus.
IMPAIRING HOST TRANSLATION
The vast majority of eukaryotic mRNAs are translated in a cap-dependent manner that involves regulated recruitment of a 40S ribosome-containing pre-initiation complex (43S complex) by eukaryotic translation initiation factors (eIFs) (Hershey et al. 2012; Hinnebusch and Lorsch 2012). By impairing cap-dependent ribosome recruitment to host mRNAs, many viruses globally interfere with host mRNA translation—a phenomenon termed “host shut-off”—thereby crippling host antiviral responses and favoring viral protein synthesis. Viral mRNA translation in these instances proceeds via an alternative initiation strategy that relies on cis-acting RNA elements. Other viruses target host mRNA metabolism, impairing host mRNA processing, stability, and/or export to the cytoplasm.
Direct Effects on Cellular Translation Factors
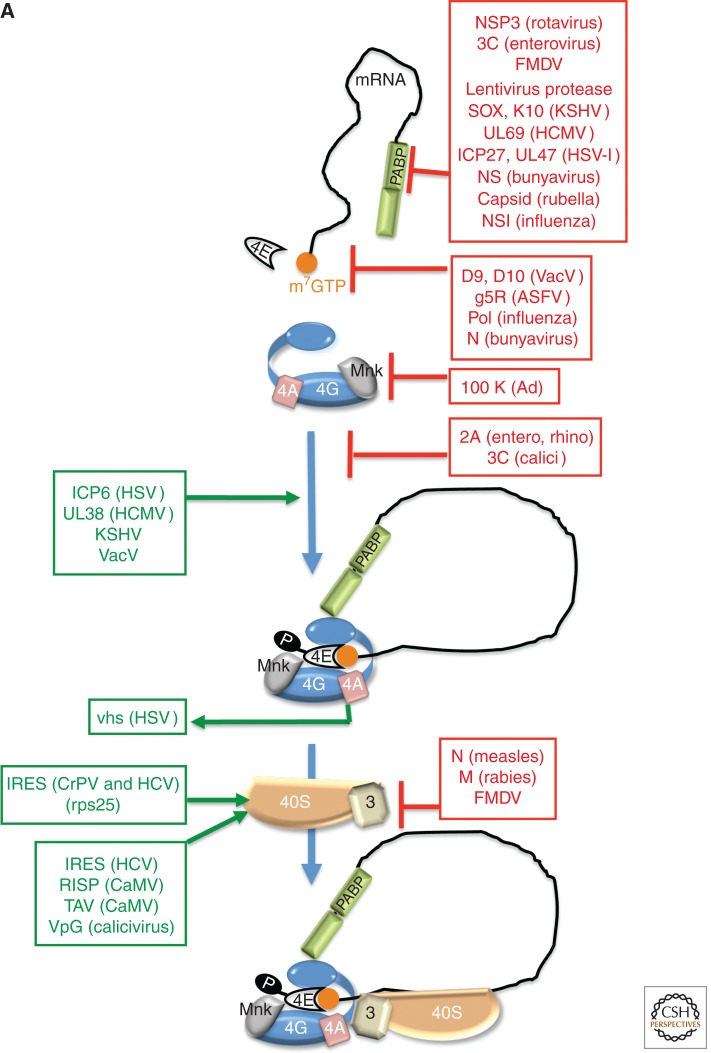
Some viruses directly target cellular translation factors to prevent ribosome recruitment by host mRNAs (Fig. 2A). Poliovirus (an enterovirus), feline calicivirus, and retroviruses encode proteases that cleave eIF4G, separating its (amino-terminal) eIF4E-interacting domain from its eIF4A- and eIF3-binding segment (Etchison et al. 1982; Ventoso et al. 2001; Alvarez et al. 2003; Willcocks et al. 2004). In poliovirus-infected cells, both eIF4GI and eIF4GII are cleaved, and host shut-off correlates more closely with the protracted cleavage of eIF4GII (Gradi et al. 1998). In addition to eIF4G proteolysis, enterovirus 71 infection induces host micro-RNA (miRNA) miR-141 expression, which reduces eIF4E abundance and suppresses host protein synthesis (Ho et al. 2010). Vesicular stomatitis virus (VSV), influenza virus, and adenovirus (Ad) decrease eIF4E phosphorylation (Huang and Schneider 1991; Feigenblum and Schneider 1993; Connor and Lyles 2002). By binding to eIF4G, the Ad 100K protein displaces the kinase Mnk1 (see below), resulting in the accumulation of unphosphorylated eIF4E late in infection (Cuesta et al. 2000b, 2004). This stimulates selective late viral mRNA translation (discussed below) and may contribute to host shut-off (Huang and Schneider 1991). During infection with some RNA viruses, however, accumulation of unphosphorylated eIF4E represents a host antiviral response, which induces interferon (IFN)-β production through translational activation of NF-κB (Herdy et al. 2012).
Figure 2.
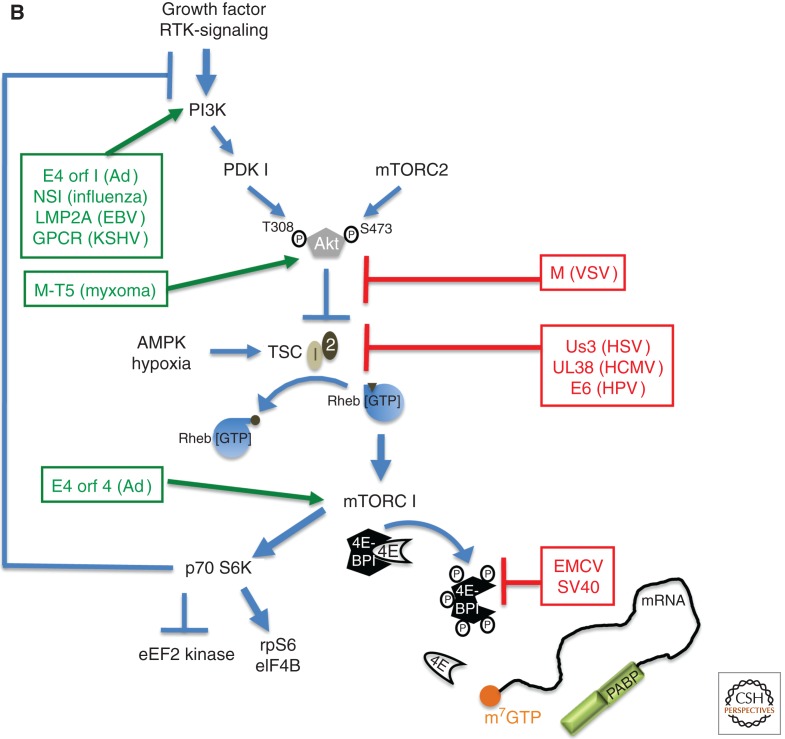
Control of translation by virus-encoded functions that regulate assembly of eIF4F. (A) eIF4E (4E) is able to interact with eIF4G and assemble the multi-subunit eIF4F complex (4E, 4A, 4G) on the m7GTP-capped (orange ball) mRNA 5′ end. eIF4F assembly typically results in eIF4E phosphorylation by the eIF4G-associated kinase Mnk and recruits eIF3 bound to the 40S ribosome subunit together with associated factors in the 43S complex. PABP is depicted bound to the 3′-poly(A) tail and associates with eIF4G to stimulate translation. Virus-encoded factors that activate (green, on the left) and repress (red, to the right) cellular functions are shown. (B) Cell signaling pathways that control the activity of the translational repressor 4E-BP1 and regulate eIF4F assembly allow for rapid changes in gene expression programs in response to a variety of physiological cues, including viral infection. In response to growth factor–stimulated receptor tyrosine kinases (RTK), PI-3-kinase (PI3K) and mTORC2 activate Akt by phosphorylation on T308 and S473, which, in turn, represses the tuberous sclerosis complex (TSC) by phosphorylating the TSC2 subunit. TSC is a GTPase-activating protein (GAP) that limits the activity of Rheb by promoting Rheb·GDP accumulation, which represses mTORC1. Whereas AMPK and hypoxia stimulate TSC GAP activity to inhibit mTORC1, Receptor Tyrosine Kinase (RTK)–mediated Akt activation inhibits TSC, resulting in mTORC1 activation. Inhibiting TSC allows rheb·GTP accumulation and mTORC1 activation, and results in p70S6K and 4E-BP1 phosphorylation. By stimulating ribosomal protein S6 phosphorylation (rpS6), p70 S6K activation by mTORC1 stimulates the eIF4A-accessory factor eIF4B and inhibits eukaryotic elongation factor 2 (eEF2) kinase, stimulating elongation. Normally, p70 S6K activation represses PI3-kinase (PI3K) to feedback and limit mTORC1 activation. 4E-BP1 hyperphosphorylation (depicted as circled P) relieves translational repression and releases eIF4E.
Enteroviral, apthoviral, caliciviral, and retroviral proteases also cleave the poly(A)-binding protein (PABP) (Kuyumcu-Martinez et al. 2004a,b; Alvarez et al. 2006; Rodriguez Pulido et al. 2007; Zhang et al. 2007; Bonderoff et al. 2008). PABP cleavage in enterovirus-infected cells does not always correlate with host shut-off, however (discussed below). Rather than cleaving PABP, the rotavirus NSP3 protein disrupts the PABP–eIF4G association (Piron et al. 1998; Groft and Burley 2002), allowing viral mRNA translation to proceed while host mRNA translation, which requires PABP, is suppressed (Montero et al. 2006). The large DNA-containing poxviruses produce small non-coding polyadenylated RNAs (POLADs) that have been reported to impair host PABP-dependent translation, but their precise function in infected cell biology remains unknown (Cacoullos and Bablanian 1991; Lu and Bablanian 1996).
Some RNA viruses also induce eIF2α phosphorylation to impair host responses (see below), or—in the case of rabies virus (M protein) (Komarova et al. 2007), measles virus (N protein) (Sato et al. 2007), and coronavirus (spike protein) (Xiao et al. 2008)—impair cap-dependent translation via virus-encoded eIF3-interacting proteins. How viral mRNAs are translated under the latter conditions remains unknown, as are the contributions of the eIF3-binding proteins to viral biology.
Indirect Effects on Cellular Translation Factors
Other viruses impact initiation factors indirectly (Fig. 2B). Many RNA viruses, including encephalomyocarditis virus (EMCV), poliovirus, cricket paralysis virus (CrPV), VSV, Sindbis virus (SINV), Dengue virus (DENV), and reovirus (Gingras et al. 1996; Connor and Lyles 2002; Villas-Boas et al. 2009; Garrey et al. 2010; Mohankumar et al. 2011), as well as small DNA viruses such as SV40 (Yu et al. 2005), induce the accumulation of hypophosphorylated 4E-BP1, which sequesters the cap-binding subunit eIF4E and prevents eIF4F assembly (Hinnebusch and Lorsch 2012; Roux and Topisirovic 2012). In the case of VSV, this requires the viral M protein, which suppresses Akt-signaling to prevent mTORC1-mediated inactivation of 4E-BP1 (Dunn and Connor 2011). SV40 small T-antigen, however, uses a protein phosphatase 2A (PP2A)–dependent mechanism to dephosphorylate 4E-BP1 (Yu et al. 2005). Inhibition of eIF4F via 4E-BP1 can also suppress host antiviral responses. Mouse embryo fibroblasts (MEFs) lacking 4E-BP1 and 4E-BP2 produce high levels of type I IFN because of increased IRF7 mRNA translation, and 4E-BP1/2-deficient knockout mice are more resistant to EMCV, VSV, influenza, or SINV infection (Colina et al. 2008).
Changing the subcellular distribution of translation factors in virus-infected cells represents another way in which host cap-dependent mRNA translation can be altered by viruses. SINV redistributes eIF3 and eEF2 to cytoplasmic replication compartments while excluding eIF4G (Sanz et al. 2009). Because SINV mRNAs contain an IRES, this selective redistribution likely suppresses host translation and fosters selective viral protein synthesis. eIF4E is redistributed to the nucleus by poliovirus (Sukarieh et al. 2010). Although the 2A protein encoded by EMCV, another picornavirus, has a nuclear localization signal and binds to eIF4E (Groppo et al. 2010), whether eIF4E accumulates within the nuclei of infected cells remains unknown. PABP distribution can be similarly altered upon virus infection. Bunyavirus NSS (Blakqori et al. 2009) and rotavirus NSP3 proteins (Harb et al. 2008) cause nuclear PABP accumulation. Herpes simplex virus 1 (HSV-1) and Kaposi’s sarcoma–associated herpesvirus (KSHV) do not stimulate PABP recruitment into eIF4F complexes but redistribute it to the nucleus, which may contribute to host shut-off by some herpesviruses. PABP redistribution in HSV-1-infected cells involves the viral proteins ICP27 and UL47 (Dobrikova et al. 2010; Salaun et al. 2010), and SOX and/or K8.1 in KSHV-infected cells (Arias et al. 2009; Covarrubias et al. 2009; Lee and Glaunsinger 2009; Kumar and Glaunsinger 2010; Kumar et al. 2011). Notably, both viruses induce host shut-off and, in the case of KSHV, SOX mutants that fail to redistribute PABP to the nucleus do not impair host translation (Covarrubias et al. 2009). In contrast, human cytomegalovirus (HCMV), a β-herpesvirus that does not impair host translation, does not redistribute PABP to the nucleus but does recruit it into eIF4F complexes (Walsh et al. 2005; Perez et al. 2011). Redistribution of host factors also occurs in cells infected with vaccinia virus (VacV), a poxvirus, or African swine fever virus (ASFV), an asfarvirus (Katsafanas and Moss 2007; Walsh et al. 2008; Castelló et al. 2009). Both of these large DNA viruses replicate in the cytoplasm within specialized compartments termed viral factories. Redistribution of a number of eIFs to these sites may contribute to the suppression of host protein synthesis while at the same time favoring viral mRNA translation.
Controlling Translation by Manipulating RNA
Beyond targeting translation factors, mRNA structures (Fig. 1), metabolism, and trafficking have all been targeted by viruses to interfere with cellular protein production. Capped mRNAs produced by influenza virus, hantavirus, and the yeast L-A virus contain m7GTP caps derived from host mRNAs (Plotch et al. 1981; Mir et al. 2008; Reguera et al. 2010; Fujimura and Esteban 2011). Although L-A virus transfers only the host cap to the viral mRNA 5′ end, influenza virus and hantavirus use a viral endonuclease to cleave host mRNAs 10–18 nucleotides downstream from the m7GTP cap (Plotch et al. 1981; Shih and Krug 1996; Guilligay et al. 2008; Mir et al. 2008; Dias et al. 2009). The resulting capped oligonucleotides prime viral RNA synthesis (Plotch et al. 1981; Garcin et al. 1995; Reguera et al. 2010). At the same time, this process destabilizes host mRNAs and inhibits cellular mRNA translation. Notably, the severe acute respiratory virus (SARS) coronavirus protein Nsp1 associates with 40S ribosomes and selectively cleaves host mRNAs to induce host shut-off (Kamitani et al. 2006, 2009; Huang et al. 2011). An alternate strategy used by VSV, which replicates in the cytoplasm, suppresses nuclear export of cellular mRNAs to preclude the synthesis of host defense-related proteins (Faria et al. 2005).
Among DNA viruses that produce capped, polyadenylated mRNAs, poxvirus and asfarvirus decapping enzymes destabilize mRNAs (Parrish and Moss 2007; Parrish et al. 2007). In Ad-infected cells, cellular mRNA nuclear export is inhibited, and the E1B 55K and E4 ORF6 proteins selectively export viral mRNAs from the nucleus via an NXF1/TAP-dependent mechanism (Kratzer et al. 2000; Yatherajam et al. 2011). The endoribonuclease encoded by the HSV-1 virion host shut-off (vhs) gene associates with eIF4F by interacting with eIF4A and eIF4B/eIF4H to increase mRNA turnover of host and viral mRNAs (Feng et al. 2005). Although stimulating mRNA decay impairs translation of host mRNAs, including those encoding host defense functions, accelerating viral mRNA turnover helps demarcate different kinetic populations of virus-encoded mRNAs expressed at immediate-early, early, or late times post-infection. The KSHV SOX protein also cleaves mRNA but recruits XRN1 to mediate target mRNA degradation (Covarrubias et al. 2011; Richner et al. 2011). The HSV-1-encoded protein ICP27 inhibits host mRNA splicing and transport, while promoting intronless viral mRNA export (Sandri-Goldin 2011). ICP27 also reportedly interacts with PABP to mediate selective translation of a small subset of viral mRNAs (Ellison et al. 2005; Larralde et al. 2006; Fontaine-Rodriguez and Knipe 2008), but the precise mechanism is unclear. A related protein encoded by KSHV ORF57 binds a host factor, PYM, to load ribosomes onto viral mRNAs, potentially coupling viral mRNA export and translation (Boyne et al. 2010; Jackson et al. 2011). Herpesviruses also encode miRNAs that selectively suppress translation of cellular and viral target mRNAs (Pfeffer et al. 2004; Cai et al. 2005; Murphy et al. 2008; Tang et al. 2008; Umbach et al. 2008; Wang et al. 2008; Bellare and Ganem 2009; Nachmani et al. 2009; Santhakumar et al. 2010). For HSV-1, these miRNAs may prevent lytic transcript accumulation in latently infected cells.
A key component of the cellular antiviral response relies on RNase L, which inactivates both rRNA and mRNA. In response to dsRNA, a molecular signature associated with virus infection, oligoadenylate (OA) polynucleotide chains with a unique 2′–5′ linkage, are generated by the IFN-inducible OA synthetase (OAS). 2′–5′ OA is a potent RNase L activator. To preserve the integrity of viral mRNAs and cellular rRNAs, viruses have evolved an array of countermeasures discussed below (Banerjee et al. 2000; Sanchez and Mohr 2007; Silverman 2007; Chakrabarti et al. 2011). Cytoplasmic structures associated with mRNA degradation may also play roles in infection, although they remain poorly understood. Poliovirus and CrPV suppress processing (P)–body formation (Dougherty et al. 2011; Khong and Jan 2011), which could represent another strategy to antagonize host responses because P-bodies restrict human immunodeficiency virus (HIV) infection (Nathans et al. 2009). It should be noted, however, that P-bodies facilitate the replication of brome mosaic virus (Beckham et al. 2007).
CAP-INDEPENDENT TRANSLATION
For viruses that interfere with eIF4F to suppress host protein synthesis and antiviral responses, an alternative, non-canonical mode of translation initiation is required, releasing viral mRNAs from regulatory constraints that normally control cellular mRNA translation.
Protein-Linked 5′ Ends
Instead of a m7GTP cap, some viral mRNAs contain a small virus-encoded protein, VPg (viral protein genome-linked), covalently attached to the 5′ end of their (+)-strand RNA genomes (Fig. 1). Calicivirus VPg interacts with eIF4E or eIF3 to mediate ribosome recruitment to viral mRNAs (Daughenbaugh et al. 2003, 2006; Goodfellow et al. 2005; Chaudhry et al. 2006). Interestingly, distinct plant virus-encoded VPg proteins selectively associate with different translation factors (Khan et al. 2006, 2008; Beauchemin et al. 2007). Tobacco mosaic virus (TMV) VPg binds eEF1A and concentrates it in membrane-associated viral replication sites (Thivierge et al. 2008), which may promote viral protein synthesis and/or impair host translation, whereas potyvirus potato virus A VPg promotes both viral mRNA stability and translation, effectively functioning as a proteinaceous cap (Eskelin et al. 2011). Free VPg can also suppress reporter mRNA translation, suggesting that it may sequester host factors to dampen host translation (Khan et al. 2008; Eskelin et al. 2011).
IRES-Dependent Mechanisms in Picornaviruses
Several RNA viruses contain an IRES within their 5′-untranslated regions (UTRs) that directs initiation through interactions with eIFs and/or ribosomal proteins (Fig. 1) (Jackson 2012). First discovered in poliovirus (Pelletier and Sonenberg 1988; Trono et al. 1988) and subsequently identified in other viruses including related picornaviruses (rhinovirus, hepatitis A [HAV], EMCV) (Jang et al. 1988) and hepatitis C virus (HCV), IRESs are divided into four structurally distinct classes, although a fifth related to class I and II IRESs has recently been proposed (Sweeney et al. 2011). Both class I and class II IRESs interact with the carboxy-terminal half of eIF4G (Kolupaeva et al. 1998; Bordeleau et al. 2006; de Breyne et al. 2009), which recruits the 40S pre-initiation complex through its binding to eIF3. Class I and II IRESs also use specific host factors, termed IRES trans-activating factors (ITAFs), which alter IRES conformation and enhance eIF4G binding to facilitate ribosome recruitment (Kolupaeva et al. 1996; Kafasla et al. 2010; Yu et al. 2011a). During infection with poliovirus, the viral 2A protease cleaves eIF4G, inhibiting host cap-dependent translation, whereas its class I IRES uses the large eIF4G fragment that contains eIF4A and eIF3 binding sites to promote selective viral mRNA translation (Krausslich et al. 1987; Kempf and Barton 2008; Willcocks et al. 2011). The related picornaviruses EMCV and HAV contain a class II IRES whose function does not involve eIF4G cleavage. Instead, hypophosphorylated 4E-BP1 accumulates in EMCV-infected cells, which, in turn, sequester the cap-binding protein eIF4E and suppresses cap-dependent translation (Gingras et al. 1996). The HAV IRES is unusual because it uses intact eIF4F. However, the eIF4E cap-binding slot must be unoccupied, rendering HAV IRES initiation cap-independent and suggesting that it interacts with eIF4E or that eIF4E induces conformational changes in eIF4G that facilitate interaction with the IRES (Ali et al. 2001; Borman et al. 2001). The Aichivirus IRES is unusual in its absolute requirement for the helicase DHX59 to expose the start codon buried in a stable hairpin (Yu et al. 2011b).
Translation Mediated by Class III and IV IRES
The class III IRES in HCV and pestivirus RNAs uses a prokaryote-like mode of translation initiation by positioning ribosomes on the mRNA through interactions with both eIF3 and the ribosome itself, thereby circumventing any need for eIF4F (Pestova et al. 1998; Sizova et al. 1998; Ji et al. 2004; Otto and Puglisi 2004; Pisarev et al. 2004; Fraser and Doudna 2007; Locker et al. 2007; Babaylova et al. 2009; Berry et al. 2010). Translation of HCV RNA is also enhanced by the binding of host miR-122 to two target sites in the 5′ UTR, thereby promoting ribosome recruitment (Jopling et al. 2005, 2008; Henke et al. 2008; Jangra et al. 2010). In HIV-1, an IRES within the Gag open reading frame (ORF) also interacts with eIF3 and the 40S ribosome (Locker et al. 2010). The simian picornavirus (SPV9) IRES binds eIF3 and the 40S ribosome, yet its activity is enhanced by eIF4F (de Breyne et al. 2008b). In a striking departure, the class IV IRESs of the Dicistroviridae, including CrPV, recruit 40S subunits, assemble 80S ribosomes, and direct initiation in the absence of any eIFs and even of the initiator Met-tRNAi, requiring only eEFs for polypeptide chain formation (Wilson et al. 2000; Jan and Sarnow 2002; Spahn et al. 2004; Cevallos and Sarnow 2005; Kamoshita et al. 2009). By positioning a CCU codon that is not decoded into the P-site, the CrPV IRES is able to initiate protein synthesis from an alanine codon positioned in the A-site after initial pseudotranslocation of the ribosome. The small ribosomal subunit protein RPS25 is essential for initiation from the CrPV and HCV IRES (Landry et al. 2009). RPS25 deletion in yeast or depletion in mammalian cells has minimal effects on cellular protein synthesis, implying that a ribosomal protein can be selectively required for IRES-mediated translation. Moreover, deficiencies in rRNA pseudouridylation reduce CrPV IRES binding to 40S ribosome subunits and inhibit IRES-dependent translation (Jack et al. 2011).
IRES-Like Mechanisms in Other Viruses
The 5′ UTRs of some plant potyviruses contain IRES-like structures that bind initiation factors (Fig. 1). For example, tobacco etch virus (TEV) interacts with eIF4G (Gallie 2001). Plant viruses also contain cis-acting cap-independent translational elements (CITEs) in their 3′ UTRs that interact with the 5′ UTR, bind translation factors, and place them proximal to the 5′ initiation site (Miller et al. 2007; Treder et al. 2008). The 3′ CITE in RNA2 of pea enation mosaic virus contains a pseudoknot that occupies the cap-binding slot in eIF4E, showing another mode by which the cap-binding protein can recognize mRNA (Wang et al. 2011). The turnip crinkle virus (TCV) CITE contains a 102-bp tRNA-like structural element that recruits ribosomes (Zuo et al. 2010). A related strategy for recruiting factors to the 3′ UTR is used by DENV, where non-polyadenylated 3′-UTR sequences bind PABP to promote viral mRNA translation (Polacek et al. 2009).
Some RNA viruses that produce mRNAs with m7GTP-capped 5′ ends—either by encoding methyltransferases and other capping enzymes or by acquiring caps from host mRNAs— can still use cap-independent modes of translation initiation. SINV provides an example of this latter strategy. In contrast to many RNA viruses, all known DNA viruses produce capped, polyadenylated, predominantly monocistronic mRNAs and generally use canonical cap-dependent modes of translation initiation. Rare instances of cap-independent strategies occur, however, typically involving polycistronic viral mRNAs. Thus, initiation on the SV40 late 19S mRNA is mediated by an IRES (Yu and Alwine 2006), and IRES-mediated translation of several herpesvirus mRNAs has also been reported (Bieleski and Talbot 2001; Grundhoff and Ganem 2001; Low et al. 2001; Griffiths and Coen 2005; Grainger et al. 2010).
MODULATION OF CAP-DEPENDENT INITIATION
As noted above, most dsDNA virus mRNAs are translated by conventional mechanisms used by the majority of host mRNAs, and many, but not all, of these viruses suppress host mRNA translation by altering mRNA metabolism (Clyde and Glaunsinger 2010). Unlike many RNA viruses, numerous DNA viruses stimulate factors, such as eIF4F to foster viral protein synthesis and replication irrespective of whether they inhibit host protein synthesis (Kudchodkar et al. 2004; Walsh and Mohr 2004; Walsh et al. 2005, 2008; Arias et al. 2009; Castelló et al. 2009). The mechanisms used in several viral families are discussed here.
Polyoma-, Papilloma-, and Adenoviruses
To activate mTORC1 and stimulate 4E-BP1 hyperphosphorylation, human papilloma virus (HPV) E6 inhibits the tuberous sclerosis complex (TSC) (Lu et al. 2004; Zheng et al. 2008; Spangle and Munger 2010). Merkel cell polyomavirus small T antigen also activates mTORC1, and its activity is required for transformation (Shuda et al. 2011). Ad E4-ORF1 stimulates phosphatidylinositol 3-kinase (PI3K) (Feigenblum and Schneider 1996; Gingras and Sonenberg 1997; O’Shea et al. 2005). In addition, Ad E4-ORF4 stimulates mTORC1 via PP2A independently of TSC (Fig. 2B) (O’Shea et al. 2005).
Late Ad mRNAs contain a common 200-nucleotide sequence at their 5′ ends. Termed the tripartite leader (TPL), it contains sequences complementary to 18S rRNA that are critical for ribosome shunting (Yueh and Schneider 1996, 2000). After loading onto the capped mRNA, the 40S ribosome is able to bypass large segments of the 5′ UTR via a nonlinear translocation mechanism before recognizing the AUG. Shunting is stimulated by the Ad 100 K protein, which binds TPL sequences and eIF4G (Xi et al. 2004, 2005). A shunting mechanism also controls production of the HPV E1 replication protein from a polycistronic mRNA (Remm et al. 1999).
Herpesviruses
Among the largest of the DNA viruses, herpesviruses replicate in the nucleus and exist in two discrete developmental states. Infection results in lifelong latency characterized by a restricted gene expression program, punctuated by episodic reactivation events in which productive, lytic viral replication ensues. Different herpesvirus subfamily members colonize specialized differentiated cell types. For example, α-herpesviruses such as HSV-1 are neurotrophic; HCMV, a β-herpesvirus, colonizes myeloid progenitors; and KHSV and Epstein-Barr virus (EBV), both γ-herpesviruses, colonize lymphoid cells and are associated with malignancies. During their productive or lytic replication cycle, these viruses induce 4E-BP1 hyperphosphorylation, eIF4F assembly, and eIF4E phosphorylation to promote viral protein production and virus replication (Kudchodkar et al. 2004; Walsh and Mohr 2004; Walsh et al. 2005; Arias et al. 2009). Remarkably, different mechanistic strategies are enlisted to achieve this goal (Fig. 2A,B). For example, both HSV-1 and HCMV stimulate mTORC1 by targeting TSC. The HSV-1 Us3 ser/thr kinase directly phosphorylates substrates on sites targeted by Akt, including TSC2, functioning as an Akt mimic (Chuluunbaatar et al. 2010). The HCMV UL38 protein, however, physically associates with TSC2 and inactivates it via an unknown mechanism that does not involve direct phosphorylation (Moorman et al. 2008). HCMV also alters mTOR substrate specificity and subcellular localization (Kudchodkar et al. 2006; Clippinger et al. 2011). Two γ-herpesvirus proteins, KSHV vGPCR (Sodhi et al. 2006; Nichols et al. 2011) and EBV LMP2A (Moody et al. 2005), also activate signaling pathways upstream of mTORC1. mTOR active-site inhibitors PP242 and Torin1, which target both mTORC1 and 2 (Feldman et al. 2009; Thoreen et al. 2009), impair viral protein synthesis and replication (Moorman and Shenk 2010; McMahon et al. 2011), and their effect requires the translation repressor 4E-BP1 (Chuluunbaatar et al. 2010; Moorman and Shenk 2010; Perez et al. 2011). Besides stimulating cap-dependent initiation, mTORC1 activation during infection likely coordinately increases elongation rates by inhibiting eEF2 kinase (Browne and Proud 2002, 2004; Browne et al. 2004). Viruses can also directly modify elongation factors. A kinase conserved among different herpesviruses stimulates eEF1Bα phosphorylation (Kawaguchi et al. 1999); however, the significance to host or viral protein synthesis remains unknown.
In addition to its phosphorylation, 4E-BP1 is degraded by the proteasome in cells infected with HSV-1 or VacV (Walsh and Mohr 2004; Walsh et al. 2008). Ubiquitination and proteasomal degradation of 4E-BP1 are also observed in uninfected cells (Elia et al. 2008; Braunstein et al. 2009). Inactivation of 4E-BP1 is not sufficient to promote eIF4F assembly in HSV-1-infected cells, however. During virus-induced stress, binding of eIF4E to eIF4G requires the virus-encoded ICP6 protein (Walsh and Mohr 2006), which shares a small domain with homology to hsp27, a cellular chaperone that regulates eIF4F formation during stress and recovery (Carper et al. 1997; Cuesta et al. 2000a). Thus ICP6 may represent a strategy to foster eIF4F formation under the stress of infection.
HCMV uses different strategies to control eIF4F activity. Although unchanged in HSV1-infected cells, the steady-state levels of eIF4E, eIF4G, eIF4A, and PABP are increased by HCMV (Walsh et al. 2005). Increased eIF4E, eIF4G, and eIF4A mRNA levels accompany the rise in abundance of the corresponding proteins, whereas PABP abundance, along with other TOP mRNA-encoded host proteins, increases through an mTORC1-dependent translational control mechanism that requires UL38 (Perez et al. 2011; McKinney et al. 2012) and promotes viral protein accumulation, eIF4F assembly, and viral replication. This represents the first example of a virus-directed increase in translation initiation factor concentration that contributes to viral replication, and it contrasts with poliovirus biology, where full-length translation initiation factor abundance is selectively reduced by virus-encoded functions. Additionally, an HCMV RNA-binding protein (UL69) important for viral mRNA export from the nucleus (Lischka et al. 2006) reportedly interacts with eIF4A and PABP and stimulates eIF4E release from 4E-BP1 (Aoyagi et al. 2010), although the underlying mechanism remains unknown.
Poxviruses and Asfarviruses
Unlike herpesviruses, poxviruses and asfarviruses (ASFV) cause acute infections and replicate in the cytoplasm. Both viruses encode their own mRNA capping enzymes (Ensinger et al. 1975; Dixon et al. 1994). Notably, VacV and ASFV redistribute eIF4E and eIF4G to cytoplasmic viral factories, where replication occurs (Katsafanas and Moss 2007; Walsh et al. 2008; Castelló et al. 2009). As neither virus increases eIF4F subunit abundance (unlike HCMV), redistribution raises the effective local concentration of these factors to promote eIF4F assembly, perhaps by altering the eIF4E:eIF4G interaction equilibrium. Consistent with this, inhibiting viral factory formation and eIF4G redistribution suppressed eIF4F assembly in poxvirus-infected cells (Zaborowska et al. 2012). In addition, 4EGi-1, an inhibitor of eIF4F and ribosome binding, has potent antiviral activity against VacV and HSV-1 (McMahon et al. 2011), and siRNA-mediated eIF4G depletion suppresses VacV infection (Welnowska et al. 2009). Concentrating eIF4G within replication factories may involve the VacV ssDNA-binding protein I3, which binds eIF4G and accumulates within replication factories (Zaborowska et al. 2012). VacV and ASFV infection also promotes 4E-BP1 hyperphosphorylation by mTORC1 (Walsh et al. 2008; Castelló et al. 2009). Although the mechanism by which VacV and ASFV stimulate mTORC1 signaling remains unknown, the rabbit poxvirus myxoma M-T5 protein directly activates Akt signaling (Werden et al. 2007), yet its potential role in translational control has not been explored.
Mimi- and Megaviruses
The recently discovered giant viruses that infect Acanthamoeba (mimivirus, megavirus) and zooplankton (Cafeteria roenbergensis virus) encode orthologs of translation factors, including eIF4E (Raoult et al. 2004; Saini and Fischer 2007; Fischer et al. 2010; Arslan et al. 2011). Whether these viral orthologs function to control translation in infected cells is unknown.
eIF4E Phosphorylation Promotes DNA Virus Replication
By stimulating eIF4F assembly, the eIF4G-associated kinase Mnk1 is positioned near its substrate eIF4E in poxvirus-infected (Walsh et al. 2008; Zaborowska and Walsh 2009), asfarvirus-infected (Castelló et al. 2009), and herpesvirus-infected (Walsh and Mohr 2004, 2006; Walsh et al. 2005; Arias et al. 2009) cells, resulting in eIF4E phosphorylation. Mnk1 activation and subsequent eIF4E phosphorylation is dependent on ERK and/or p38 MAPK signaling. The small molecule Mnk1 inhibitor, CGP57380, impairs HSV1 (Walsh and Mohr 2004), HCMV (Walsh et al. 2005), ASFV (Castelló et al. 2009), and VacV (Walsh et al. 2008) protein synthesis, and VacV replication is reduced in Mnk1-deficient MEFs (Walsh et al. 2008), indicating that this kinase plays an important role in controlling viral protein synthesis and replication. HSV-1 and VacV replication is also reduced in MEFs containing an eIF4E-substituted allele in which eIF4E cannot be phosphorylated, directly establishing that eIF4E phosphorylation regulates virus replication (Herdy et al. 2012). For KSHV, Mnk1 activity is important in reactivation from latency because CGP57380 inhibits the transition from latency to lytic replication, suggesting a role for eIF4E phosphorylation in this developmental switch (Arias et al. 2009). Because Mnk1 is not essential for translation initiation but likely plays a regulatory role, it represents a potential target for therapeutic intervention against a wide range of large DNA viruses.
Cap-Dependent Translation in RNA Viruses
Some RNA viruses, including coronaviruses (SARS), orthomyxoviruses (influenza), rhabdoviruses (VSV, rabies virus), reoviruses, hantavirus, and alphaviruses, also produce capped mRNAs. Coronavirus mRNAs, which share a common capped 5′-leader and are polyadenylated, rely on eIF4F, because chemical inhibitors of the eIF4E·eIF4G interaction impair coronavirus replication in cultured cells (Cencic et al. 2011). Furthermore, coronavirus infection stimulates eIF4E phosphorylation (Banerjee et al. 2002; Mizutani et al. 2004). In influenza virus-infected cells, the viral cap-binding polymerase subunit proteins PB2 and NS1 reportedly interact with eIF4G and PABP (Aragon et al. 2000; Burgui et al. 2003, 2007; Yángüez et al. 2012), recruiting them to viral mRNAs. NS1 is not absolutely required for viral protein synthesis, however (Salvatore et al. 2002). Remarkably, the hantavirus N protein has been reported to have cap-binding, RNA-binding, RNA helicase, and ribosome-recruiting activities that functionally substitute for the entire eIF4F complex (Mir and Panganiban 2008, 2010). Finally, the dsRNA reoviruses and rotaviruses encode capping enzymes, and reovirus mRNAs are translated by a cap-dependent mechanism in vitro (Muthukrishnan et al. 1976; Sonenberg et al. 1979). Recently, modest reductions in eIF4E, eIF4G, eIF4B, and 4E-BP1 phosphorylation, along with enhanced eEF2 phosphorylation, have been reported in avian reovirus-infected cells or in cells expressing the viral p17 protein (Ji et al. 2009; Chulu et al. 2010). These modifications, which suppress translation, have been proposed to selectively impair translation of host mRNAs, although further investigation is required to test this hypothesis.
eIF2: A CENTRAL PLAYER IN THE INNATE IMMUNE RESPONSE TO INFECTION
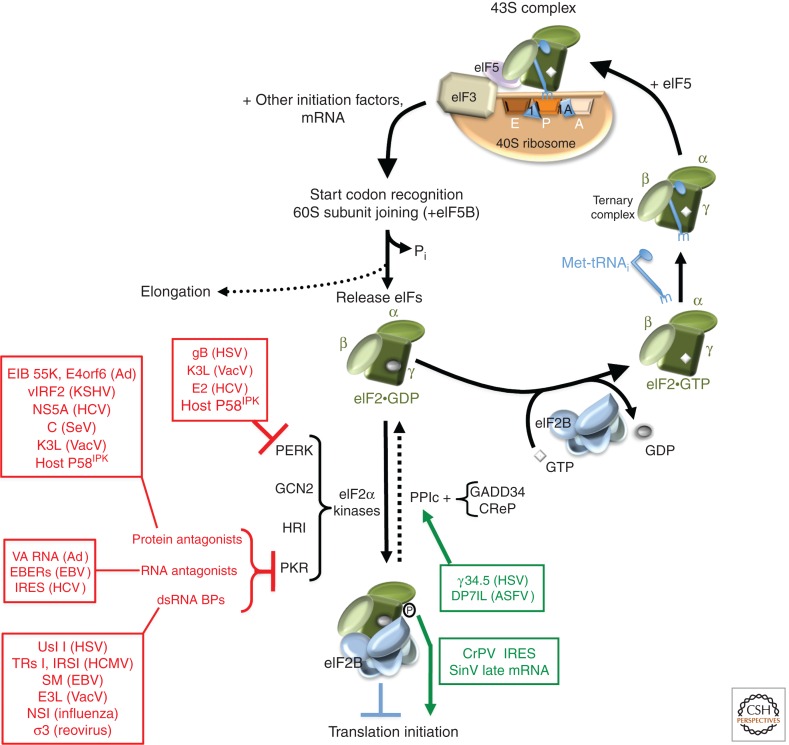
For most host and viral mRNAs, the methionine-charged initiator tRNA is loaded into the ribosome by a ternary complex (TC) composed of eIF2, GTP, and an initiator tRNA (Met-tRNAi) (Fig. 3). In addition to its critical role in initiation and AUG selection (Hinnebusch and Lorsch 2012), TC formation is targeted by innate host antiviral immune defenses. Like OAS (discussed above), the IFN-induced eIF2α kinase PKR acts as a pattern recognition sentinel that detects dsRNA or viral replication intermediates. Once activated, phosphorylated eIF2 accumulates and binds with high affinity to the eIF2B guanine nucleotide exchange factor (GEF), preventing exchange of GDP for GTP and inhibiting translation initiation. Small changes in eIF2α phosphorylation can result in a large suppression of ongoing protein synthesis and even incapacitate the cellular translation machinery because eIF2B is present in limiting quantities.
Figure 3.
Antagonism of host antiviral defenses that inactivate eIF2 by viral functions. eIF2·GTP forms a ternary complex (TC) with Met-tRNAi and is loaded into the P-site in the 40S ribosome to form a 43S pre-initiation complex (top right). Upon engaging mRNA via other initiation factors (e.g., eIF4F) or via a specialized cis-acting RNA element (e.g., an IRES), the AUG start codon is identified (typically via scanning), the GTPase-activating protein eIF5 stimulates GTP hydrolysis, and 60S subunit joining triggers eIF2·GDP + phosphate (Pi) release. Translation elongation is executed by the resulting 80S ribosome. Inactive eIF2 (α, β, γ subunits depicted in center) bound to GDP (eIF2·GDP) is recycled to the active GTP-bound form (eIF2·GTP) by the five-subunit guanine nucleotide exchange factor eIF2B. Site-specific eIF2 phosphorylation on its α-subunit (S51) by either of four different cellular eIF2α-kinases (see text), each of which is activated by discrete stress, prevents eIF2 recycling. Phosphorylated eIF2 (bottom) binds tightly to and inhibits eIF2B, blocking translation initiation. The host protein phosphatase 1 catalytic (PP1c) subunit can dephosphorylate eIF2 when partnered with either an inducible (GADD34) or constitutively active (CReP) regulatory component. Virus-encoded factors that antagonize (red, to the left) specific host eIF2α kinases are indicated to the left. Viral PKR antagonists have been subdivided according to their mechanisms of action (see text; [dsRNA BPs] double-stranded RNA binding proteins). Virus-encoded factors that activate cellular phosphatases to dephosphorylate eIF2α, or allow translation to proceed without eIF2 or in the presence of phosphorylated eIF2α, are shown (green, to the right).
Eukaryotes contain three additional eIF2α kinases (Fig. 3), each of which responds to a discrete set of stresses. Both GCN2, which responds to UV irradiation and amino acid deprivation, and PERK, which can be triggered by ER overload in infected cells (Pavitt and Ron 2012), can have antiviral effects (Berlanga et al. 2006; Won et al. 2011). eIF2α kinases have also been implicated in autophagy induction (Talloczy et al. 2002). This powerful host response has forced viruses to evolve countermeasures such that they can sustain protein synthesis and complete their growth cycles in infected cells. Although many of these functions target PKR, others have broader capabilities and can act directly on eIF2 or other eIF2α kinases. Viral functions that hinder eIF2α phosphorylation are key components of viral pathogenesis, and mutations that compromise these functions often result in attenuated variants. Adaptive evolutionary changes in PKR, under pressure to avoid viral substrate mimicry while maintaining natural substrate (eIF2α) recognition, emphasizes the ongoing nature of this struggle between host defenses and viral countermeasures for control over eIF2 (Elde et al. 2009; Rothenburg et al. 2009). eIF2 itself is regulated by IFN-stimulated genes (ISGs) 54 and 56, which antagonize ribosome binding to eIF4F and TC loading by binding to eIF3e and eIF3c (Guo et al. 2000; Hui et al. 2003, 2005; Terenzi et al. 2006). Viral ISG54 and 56 antagonists, however, have not been reported.
Viral Inhibitors of eIF2α Phosphorylation
Perhaps the most common strategy for preventing PKR activation and safeguarding eIF2 in virus-infected cells involves virus-encoded dsRNA-binding proteins that sequester dsRNA, preventing PKR from recognizing its activating ligand. In addition to dsRNA binding, many of these proteins also associate with and inhibit PKR, and prevent OAS activation (for review, see Walsh and Mohr 2011). A variation on this theme uses a structured, non-coding RNA lure to inhibit PKR activation, such as Ad VA RNAI, HIV-1 TAR, and EBV EBER. VacV also encodes a pseudosubstrate, K3L, that effectively deflects PKR and PERK away from its substrate, eIF2α. The host molecular chaperone p58IPK, a PKR/PERK inhibitor, is recruited to prevent eIF2α phosphorylation in cells infected with influenza virus, TMV, or TEV (Yan et al. 2002; Bilgin et al. 2003; Goodman et al. 2007, 2009). Finally, eIF2α itself can be targeted for dephosphorylation by viral functions. The ASFV DP17L-, HPV E6-, and HSV1 γ34.5-encoded phosphatase regulatory subunits target cellular catalytic PP1 subunits to phosphorylated eIF2α (He et al. 1997; Kazemi et al. 2004; Zhang et al. 2010; Li et al. 2011). Recruiting a phosphatase to dephosphorylate eIF2α is advantageous, conferring resistance to any cellular eIF2α kinase, not just PKR.
Combinatorial Strategies to Prevent eIF2α Phosphorylation
Some viruses use multiple, independent strategies to prevent eIF2α phosphorylation. VacV, for example, combines the dsRNA-binding protein/PKR inhibitor E3L with the PERK and PKR pseudosubstrate K3L (Chang et al. 1992; Davies et al. 1992, 1993; Kawagishi-Kobayashi et al. 1997; Sood et al. 2000; Ramelot et al. 2002; Pavio et al. 2003; Seo et al. 2008; Rothenburg et al. 2011). HSV-1, on the other hand, relies on the Us11 dsRNA-binding/PKR antagonist (Mulvey et al. 1999), the γ34.5 eIF2α phosphatase subunit, and a viral glycoprotein B (gB) PERK antagonist that prevents ER stress-induced eIF2α phosphorylation (Mulvey et al. 2003, 2007). Genomic elements can also play a role in inhibiting PKR. Although HCV E2 is a PKR and PERK pseudosubstrate (Taylor et al. 1999, 2001; Pavio et al. 2003), both the HCV NS5A protein (Gale et al. 1998; Gale and Foy 2005) and the HCV IRES itself (Vyas et al. 2003) directly inhibit PKR. Additionally, HIV-1 Tat and the cellular TAR RNA-binding protein (TRBP) bind the viral TAR element, and TRBP inhibits PKR (Gunnery et al. 1992).
Exploiting eIF2α Phosphorylation and Bypassing eIF2
Instead of preventing eIF2α phosphorylation, some RNA viruses induce eIF2α phosphorylation (O’Malley et al. 1989; Jordan et al. 2002; Gorchakov et al. 2004; McInerney et al. 2005; Smith et al. 2006; Montero et al. 2008; Garrey et al. 2010; White et al. 2010). Although stimulating the phosphorylation of eIF2α facilitates the host antiviral response by globally inhibiting protein synthesis in infected cells, some viruses use an alternative mode of translation initiation that does not absolutely require eIF2. These viruses derive benefit from eIF2α phosphorylation, which can effectively cause host shut-off and prevent IFN synthesis. Classical swine fever virus (CSFV) uses both eIF2-dependent and -independent modes of translation initiation (Pestova et al. 2008). In SINV-infected cells, a stem–loop structure in the late 26S mRNA stalls the ribosome at the initiation site and requires eIF2A to initiate translation in the presence of phosphorylated eIF2α (Ventoso et al. 2006). As noted above, the CrPV IRES loads ribosomes and initiates translation without eIF2, other eIFs, or initiator tRNA. With high magnesium concentrations in vitro, the HCV IRES also directs eIF2-independent initiation (Lancaster et al. 2006), whereas, it can use eIF2A-mediated Met-tRNAi delivery under stress conditions (Kim et al. 2011). However, although eIF2α phosphorylation is induced during infection (Garaigorta and Chisari 2009; Kang et al. 2009; Arnaud et al. 2010), the HCV IRES has been suggested to compete efficiently for eIF2 and Met-tRNAi under limiting conditions (Robert et al. 2006). Furthermore, as discussed above, HCV encodes three distinct functions to inhibit PKR and eIF2α phosphorylation. As with Ad (O’Malley et al. 1989), one possible explanation posits that HCV prevents eIF2 phosphorylation specifically at intracellular viral replication sites while allowing eIF2α phosphorylation to occur in the cytoplasm to impair host protein synthesis.
eIF2α is also phosphorylated as poliovirus infection progresses, and this correlates with declining viral protein synthesis (O’Neill and Racaniello 1989). Although poliovirus mRNA translation is sensitive to eIF2α phosphorylation in vitro, viral mRNA translation late in infection is partially resistant to stress-induced eIF2α phosphorylation (Redondo et al. 2011; Welnowska et al. 2011; White et al. 2011). The underlying mechanism is unknown but may involve cleavage of eIF5B (de Breyne et al. 2008a; White et al. 2011) and local control of active eIF2 pools or cellular factors. Indeed, host factors such as ligatin (eIF2D) and MCT1-DENR can recruit Met-tRNAi to ribosomes and function when the AUG start codon is directly positioned in the P-site (Dmitriev et al. 2010; Skabkin et al. 2010), potentially explaining eIF2-independent initiation mechanisms used by HCV and SINV.
eIF2α phosphorylation is also implicated in the formation of stress granules (SGs), stress-induced cytoplasmic sites of translationally inactive mRNA accumulation. Many viruses disrupt SG formation to evade translational inhibition (Emara and Brinton 2007; White et al. 2007; Montero et al. 2008; Khong and Jan 2011; Simpson-Holley et al. 2011), whereas others induce their formation to impair host translation or even facilitate virus assembly (McInerney et al. 2005; Raaben et al. 2007; Qin et al. 2009; Lindquist et al. 2010; Piotrowska et al. 2010). Notably, mammalian orthoreovirus escapes translational suppression by disrupting SGs, but does so independently of the status of eIF2α phosphorylation or PKR (Qin et al. 2011).
REGULATED TERMINATION AND REINITIATION: TRANSLATIONAL STRATEGIES FOR MAXIMIZING GENOME CODING CAPACITY
The small genome size of many RNA viruses constrains their coding capacity. Consequently, many viral mRNAs contain overlapping ORFs regulated by frameshifting or multiple ORFs regulated by termination and reinitiation events. Thus, translational control of ORF decoding plays an indispensable role in regulating viral protein production from polycistronic mRNAs. Influenza and feline calicivirus (FCV) use 45- to 87-nucleotide stretches termed TURBS (termination upstream ribosome binding site) that base-pair with terminating ribosomes to promote reinitiation at nearby downstream ORFs encoding viral BM2 and VP2 proteins, respectively (Horvath et al. 1990; Luttermann and Meyers 2009; Powell et al. 2011), as well as binding the ribosome and eIF3 for FCV VP1 reinitiation (Poyry et al. 2007). Murine norovirus VP2 is also translated by a coupled termination and reinitiation process (Napthine et al. 2009).
Viral proteins also regulate reinitiation; the retroviral Gag protein of Moloney murine leukemia virus (M-MuLV) binds eRF1 to enhance reinitiation or translational readthrough to synthesize the Gag-Pol precursor protein (Orlova et al. 2003). Reinitiation frequency is fine-tuned by pH-dependent conformational changes in the M-MuLV recoding signal sequence to ensure the balance of gag versus gag-pol fusion protein synthesis (Houck-Loomis et al. 2011). Reinitiation also protects HIV-1 retrovirus mRNA from nonsense-mediated decay (Hogg and Goff 2010). In an intricate regulatory mechanism, Borna virus X and P proteins are encoded by an mRNA that contains an additional uORF. X translation is regulated by a coupled termination and reinitiation event at the uORF, and the efficiency of reinitiation on the X ORF is enhanced by P through interactions with a host helicase, DDX21 (Watanabe et al. 2009).
Reinitiation on cauliflower mosaic virus (CaMV) polycistronic mRNA involves viral and host factors that manipulate eIF3. The viral transactivator viroplasmin (TAV) associates with the plant protein RISP, which supports reinitiation, and subsequently binds to eIF3g (Thiebeauld et al. 2009). Interestingly, eIF4B and TAV-bound RISP bind to the same eIF3g site. RISP associates with eIF3a, eIF3c, and ribosomal protein L24 to link TAV with 60S and eIF3-bound 40S subunits. TAV also recruits the plant cell TOR, which phosphorylates RISP, promoting reinitiation and viral replication (Schepetilnikov et al. 2011).
Most mRNAs encoded by large DNA viruses are not polycistronic, limiting the need for termination/reinitiation strategies. Nevertheless, termination within a small HCMV uORF plays a regulatory role restricting reinitiation at downstream cistrons. Sequence-dependent ribosomal stalling induced by the uORF2 peptide prevents scanning ribosomes from reaching the downstream UL4 ORF (Janzen et al. 2002). By binding eRF1, the uORF2 peptide inhibits translation at its own stop codon, and the stalled ribosome disengages the mRNA. Cryo-EM reconstruction showed uORF2 stabilized at the ribosomal exit tunnel constriction, where it directly interacts with ribosomal proteins L4 and L17. Peptide-induced conformational changes in the peptidyl transfer center were also observed, potentially contributing to translational stalling (Bhushan et al. 2010).
BALANCING TRANSLATION, GENOME REPLICATION, AND ENCAPSIDATION
Although (+)-strand RNA virus genomes such as those of poliovirus and HCV serve immediately as mRNAs to produce proteins required for viral replication, they are also genome replication templates. Production of (−)-strand RNA replication intermediates from (+)-strand templates is incompatible with ongoing (+)-strand translation (Andino et al. 1999). By cleaving PABP, which stimulates some IRESs (Michel et al. 2001; Bradrick et al. 2007), enteroviruses suppress mRNA translation to stimulate RNA replication. Rubella virus capsid protein similarly binds PABP to inhibit viral translation (Ilkow et al. 2008). Alternatively, the potyvirus 3′CITE can also suppress translation when genome replication begins (Miller et al. 2007).
Packaging translation-competent (+)-strand RNA genomes creates a different set of obstacles. Following reverse transcription and integration into the host cell genome, retroviruses such as HIV use the host RNA polymerase II transcription machinery to produce viral mRNAs. Reactivation and replication of HIV-1 are regulated by cyclin T1, which itself is translationally controlled by nuclear factor 90 (NF90) (Hoque et al. 2011). Although some of these genome-length viral mRNAs are translated, others are encapsidated into infectious particles. Importantly, binding of the HIV-1 Gag protein to eEF1A impairs viral mRNA translation to promote RNA packaging (Cimarelli and Luban 1999).
CONCLUDING REMARKS
Because of their reliance on the host for protein synthesis, the intimate relationship between viral genomes and their mRNAs, and the exigencies imposed by host defense mechanisms, viruses have evolved a vast array of strategies for exploiting and dominating the cellular translation machinery. Studies of translational regulation during virus infection have illuminated many features of the cellular translation system. They have also led to the discovery of translation mechanisms that figure prominently in virus-infected cells, and some that may even be virus specific. Notwithstanding the wealth of detail already amassed, there are many open questions, some of them noted above, and there is every reason to believe that we have only scratched the surface.
One prediction is that new principles will continue to emerge as the exploration of virus–host cell interactions expands in breadth and depth. For example, the control of translation in virus-infected cells shares notable similarities with stress-induced translational control mechanisms operative in uninfected cells. To prevent the accumulation of phosphorylated eIF2α, HSV1 encodes a phosphatase regulatory subunit similar to the ER stress-induced GADD34 (He et al. 1997). Cap-dependent translation is impaired in heat-shocked cells, necessitating a mechanism for heat shock protein synthesis much in the same way that viral translation proceeds in infected cells (Yueh and Schneider 1996, 2000). Indeed, both Ad late RNA and HSP70 mRNA contain a ribosome shunt to enable translation when eIF4F is limiting. Many of the key integrators controlling translation in response to nutrient availability, growth factors, and energy supplies are hijacked in infected cells to ensure high-level synthesis of viral proteins.
Another strong prediction is that the intensive analysis of virus–host interactions will have practical value, leading to therapeutics. Idiosyncratic mechanisms evolved by viruses, as well as the interplay between the virus and its host’s translation system, represent potentially exploitable weaknesses. Remarkably, most, if not all, recessive virus resistance alleles in plants map to eIF4E or eIF4G, highlighting the importance of translation factors in the biology of plant RNA viruses (Nieto et al. 2007; Yeam et al. 2007; Truniger and Aranda 2009). As a potential host defense mechanism, pokeweed antiviral protein associates with eIF4G to access and depurinate viral RNA (Wang and Hudak 2006). Virus-specific mechanisms such as IRESs and frameshifting present direct targets for small-molecule drugs (Novac et al. 2004; Parsons et al. 2009; Gasparian et al. 2010; Paulsen et al. 2010). Cellular molecules such as ITAFs (Fontanes et al. 2009) and signaling pathways are also susceptible. Inhibitors of mTOR and eIF4F effectively reduce infection by many DNA as well as some RNA viruses, and may also serve to treat the tumors associated with some infections (Cen and Longnecker 2011), whereas chemical inducers of eIF2α phosphorylation reduce HSV-1 infection in vitro and in vivo (Boyce et al. 2005). Looking somewhat further afield, translational control is also being exploited in the development of new viral oncolytic therapies (Taneja et al. 2001; Mohr 2005; Stanford et al. 2007; Oliere et al. 2008; Thomas et al. 2009; Alain et al. 2010; Goetz et al. 2010).
Footnotes
Editors: John W.B. Hershey, Nahum Sonenberg, and Michael B. Mathews
Additional Perspectives on Protein Synthesis and Translational Control available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Alain T, Lun X, Martineau Y, Sean P, Pulendran B, Petroulakis E, Zemp FJ, Lemay CG, Roy D, Bell JC, et al. 2010. Vesicular stomatitis virus oncolysis is potentiated by impairing mTORC1-dependent type I IFN production. Proc Natl Acad Sci 107: 1576–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali IK, McKendrick L, Morley SJ, Jackson RJ 2001. Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J Virol 75: 7854–7863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez E, Menendez-Arias L, Carrasco L 2003. The eukaryotic translation initiation factor 4GI is cleaved by different retroviral proteases. J Virol 77: 12392–12400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez E, Castello A, Menendez-Arias L, Carrasco L 2006. HIV protease cleaves poly(A)-binding protein. Biochem J 396: 219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andino R, Boddeker N, Silvera D, Gamarnik AV 1999. Intracellular determinants of picornavirus replication. Trends Microbiol 7: 76–82 [DOI] [PubMed] [Google Scholar]
- Aoyagi M, Gaspar M, Shenk TE 2010. Human cytomegalovirus UL69 protein facilitates translation by associating with the mRNA cap-binding complex and excluding 4EBP1. Proc Natl Acad Sci 107: 2640–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon T, de la Luna S, Novoa I, Carrasco L, Ortin J, Nieto A 2000. Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol Cell Biol 20: 6259–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias C, Walsh D, Harbell J, Wilson AC, Mohr I 2009. Activation of host translational control pathways by a viral developmental switch. PLoS Pathog 5: e1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud N, Dabo S, Maillard P, Budkowska A, Kalliampakou KI, Mavromara P, Garcin D, Hugon J, Gatignol A, Akazawa D, et al. 2010. Hepatitis C virus controls interferon production through PKR activation. PLoS ONE 5: e10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arslan D, Legendre M, Seltzer V, Abergel C, Claverie JM 2011. Distant mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc Natl Acad Sci 108: 17486–17491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babaylova E, Graifer D, Malygin A, Stahl J, Shatsky I, Karpova G 2009. Positioning of subdomain IIId and apical loop of domain II of the hepatitis C IRES on the human 40S ribosome. Nucleic Acids Res 37: 1141–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, An S, Zhou A, Silverman RH, Makino S 2000. RNase L-independent specific 28S rRNA cleavage in murine coronavirus-infected cells. J Virol 74: 8793–8802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Narayanan K, Mizutani T, Makino S 2002. Murine coronavirus replication-induced p38 mitogen-activated protein kinase activation promotes interleukin-6 production and virus replication in cultured cells. J Virol 76: 5937–5948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchemin C, Boutet N, Laliberte JF 2007. Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in Planta. J Virol 81: 775–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham CJ, Light HR, Nissan TA, Ahlquist P, Parker R, Noueiry A 2007. Interactions between brome mosaic virus RNAs and cytoplasmic processing bodies. J Virol 81: 9759–9768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellare P, Ganem D 2009. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: An evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe 6: 570–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlanga JJ, Ventoso I, Harding HP, Deng J, Ron D, Sonenberg N, Carrasco L, de Haro C 2006. Antiviral effect of the mammalian translation initiation factor 2α kinase GCN2 against RNA viruses. EMBO J 25: 1730–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry KE, Waghray S, Doudna JA 2010. The HCV IRES pseudoknot positions the initiation codon on the 40S ribosomal subunit. RNA 16: 1559–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan S, Meyer H, Starosta AL, Becker T, Mielke T, Berninghausen O, Sattler M, Wilson DN, Beckmann R 2010. Structural basis for translational stalling by human cytomegalovirus and fungal arginine attenuator peptide. Mol Cell 40: 138–146 [DOI] [PubMed] [Google Scholar]
- Bieleski L, Talbot SJ 2001. Kaposi’s sarcoma-associated herpesvirus vCyclin open reading frame contains an internal ribosome entry site. J Virol 75: 1864–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgin DD, Liu Y, Schiff M, Dinesh-Kumar SP 2003. P58(IPK), a plant ortholog of double-stranded RNA-dependent protein kinase PKR inhibitor, functions in viral pathogenesis. Dev Cell 4: 651–661 [DOI] [PubMed] [Google Scholar]
- Blakqori G, van Knippenberg I, Elliott RM 2009. Bunyamwera orthobunyavirus S-segment untranslated regions mediate poly(A) tail-independent translation. J Virol 83: 3637–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonderoff JM, Larey JL, Lloyd RE 2008. Cleavage of poly(A)-binding protein by poliovirus 3C proteinase inhibits viral internal ribosome entry site-mediated translation. J Virol 82: 9389–9399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, Pelletier J 2006. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol 2: 213–220 [DOI] [PubMed] [Google Scholar]
- Borman AM, Michel YM, Kean KM 2001. Detailed analysis of the requirements of hepatitis A virus internal ribosome entry segment for the eukaryotic initiation factor complex eIF4F. J Virol 75: 7864–7871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D, et al. 2005. A selective inhibitor of eIF2α dephosphorylation protects cells from ER stress. Science 307: 935–939 [DOI] [PubMed] [Google Scholar]
- Boyne JR, Jackson BR, Taylor A, Macnab SA, Whitehouse A 2010. Kaposi’s sarcoma-associated herpesvirus ORF57 protein interacts with PYM to enhance translation of viral intronless mRNAs. EMBO J 29: 1851–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradrick SS, Dobrikova EY, Kaiser C, Shveygert M, Gromeier M 2007. Poly(A)-binding protein is differentially required for translation mediated by viral internal ribosome entry sites. RNA 13: 1582–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein S, Badura ML, Xi Q, Formenti SC, Schneider RJ 2009. Regulation of protein synthesis by ionizing radiation. Mol Cell Biol 29: 5645–5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne GJ, Proud CG 2002. Regulation of peptide-chain elongation in mammalian cells. Eur J Biochem 269: 5360–5368 [DOI] [PubMed] [Google Scholar]
- Browne GJ, Proud CG 2004. A novel mTOR-regulated phosphorylation site in elongation factor 2 kinase modulates the activity of the kinase and its binding to calmodulin. Mol Cell Biol 24: 2986–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne GJ, Finn SG, Proud CG 2004. Stimulation of the AMP-activated protein kinase leads to activation of eukaryotic elongation factor 2 kinase and to its phosphorylation at a novel site, serine 398. J Biol Chem 279: 12220–12231 [DOI] [PubMed] [Google Scholar]
- Burgui I, Aragon T, Ortin J, Nieto A 2003. PABP1 and eIF4GI associate with influenza virus NS1 protein in viral mRNA translation initiation complexes. J Gen Virol 84: 3263–3274 [DOI] [PubMed] [Google Scholar]
- Burgui I, Yanguez E, Sonenberg N, Nieto A 2007. Influenza virus mRNA translation revisited: Is the eIF4E cap-binding factor required for viral mRNA translation? J Virol 81: 12427–12438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacoullos N, Bablanian R 1991. Polyadenylated RNA sequences produced in vaccinia virus-infected cells under aberrant conditions inhibit protein synthesis in vitro. Virology 184: 747–751 [DOI] [PubMed] [Google Scholar]
- Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR 2005. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci 102: 5570–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carper SW, Rocheleau TA, Cimino D, Storm FK 1997. Heat shock protein 27 stimulates recovery of RNA and protein synthesis following a heat shock. J Cell Biochem 66: 153–164 [PubMed] [Google Scholar]
- Castelló A, Quintas A, Sánchez EG, Sabina P, Nogal M, Carrasco L, Revilla Y 2009. Regulation of host translational machinery by African Swine Fever Virus. PLoS Pathog 5: e1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen O, Longnecker R 2011. Rapamycin reverses splenomegaly and inhibits tumor development in a transgenic model of Epstein-Barr virus-related Burkitt’s lymphoma. Mol Cancer Ther 10: 679–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R, Desforges M, Hall DR, Kozakov D, Du Y, Min J, Dingledine R, Fu H, Vajda S, Talbot PJ, et al. 2011. Blocking eIF4E–eIF4G interaction as a strategy to impair coronavirus replication. J Virol 85: 6381–6389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevallos RC, Sarnow P 2005. Factor-independent assembly of elongation-competent ribosomes by an internal ribosome entry site located in an RNA virus that infects penaeid shrimp. J Virol 79: 677–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti A, Jha BK, Silverman RH 2011. New insights into the role of RNase L in innate immunity. J Interferon Cytokine Res 31: 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HW, Watson JC, Jacobs BL 1992. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc Natl Acad Sci 89: 4825–4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry Y, Nayak A, Bordeleau ME, Tanaka J, Pelletier J, Belsham GJ, Roberts LO, Goodfellow IG 2006. Caliciviruses differ in their functional requirements for eIF4F components. J Biol Chem 281: 25315–25325 [DOI] [PubMed] [Google Scholar]
- Chulu JL, Huang WR, Wang L, Shih WL, Liu HJ 2010. Avian reovirus nonstructural protein p17–induced G2/M cell cycle arrest and host cellular protein translation shutoff involve activation of p53-dependent pathways. J Virol 84: 7683–7694 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chuluunbaatar U, Roller R, Feldman ME, Brown S, Shokat KM, Mohr I 2010. Constitutive mTORC1 activation by a herpesvirus Akt surrogate stimulates mRNA translation and viral replication. Genes Dev 24: 2627–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimarelli A, Luban J 1999. Translation elongation factor 1-α interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J Virol 73: 5388–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clippinger AJ, Maguire TG, Alwine JC 2011. The changing role of mTOR kinase in the maintenance of protein synthesis during human cytomegalovirus infection. J Virol 85: 3930–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyde K, Glaunsinger BA 2010. Getting the message direct manipulation of host mRNA accumulation during gammaherpesvirus lytic infection. Adv Virus Res 78: 1–42 [DOI] [PubMed] [Google Scholar]
- Colina R, Costa-Mattioli M, Dowling RJ, Jaramillo M, Tai LH, Breitbach CJ, Martineau Y, Larsson O, Rong L, Svitkin YV, et al. 2008. Translational control of the innate immune response through IRF-7. Nature 452: 323–328 [DOI] [PubMed] [Google Scholar]
- Connor JH, Lyles DS 2002. Vesicular stomatitis virus infection alters the eIF4F translation initiation complex and causes dephosphorylation of the eIF4E binding protein 4E-BP1. J Virol 76: 10177–10187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias S, Richner JM, Clyde K, Lee YJ, Glaunsinger BA 2009. Host shutoff is a conserved phenotype of gammaherpesvirus infection and is orchestrated exclusively from the cytoplasm. J Virol 83: 9554–9566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias S, Gaglia MM, Kumar GR, Wong W, Jackson AO, Glaunsinger BA 2011. Coordinated destruction of cellular messages in translation complexes by the gammaherpesvirus host shutoff factor and the mammalian exonuclease Xrn1. PLoS Pathog 7: e1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta R, Laroia G, Schneider RJ 2000a. Chaperone hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes Dev 14: 1460–1470 [PMC free article] [PubMed] [Google Scholar]
- Cuesta R, Xi Q, Schneider RJ 2000b. Adenovirus-specific translation by displacement of kinase Mnk1 from cap-initiation complex eIF4F. EMBO J 19: 3465–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta R, Xi Q, Schneider RJ 2004. Structural basis for competitive inhibition of eIF4G–Mnk1 interaction by the adenovirus 100-kilodalton protein. J Virol 78: 7707–7716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughenbaugh KF, Fraser CS, Hershey JW, Hardy ME 2003. The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment. EMBO J 22: 2852–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughenbaugh KF, Wobus CE, Hardy ME 2006. VPg of murine norovirus binds translation initiation factors in infected cells. Virol J 3: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MV, Elroy-Stein O, Jagus R, Moss B, Kaufman RJ 1992. The vaccinia virus K3L gene product potentiates translation by inhibiting double-stranded-RNA-activated protein kinase and phosphorylation of the alpha subunit of eukaryotic initiation factor 2. J Virol 66: 1943–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MV, Chang HW, Jacobs BL, Kaufman RJ 1993. The E3L and K3L vaccinia virus gene products stimulate translation through inhibition of the double-stranded RNA-dependent protein kinase by different mechanisms. J Virol 67: 1688–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Breyne S, Bonderoff JM, Chumakov KM, Lloyd RE, Hellen CU 2008a. Cleavage of eukaryotic initiation factor eIF5B by enterovirus 3C proteases. Virology 378: 118–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Breyne S, Yu Y, Pestova TV, Hellen CU 2008b. Factor requirements for translation initiation on the Simian picornavirus internal ribosomal entry site. RNA 14: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CU 2009. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc Natl Acad Sci 106: 9197–9202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias A, Bouvier D, Crepin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RW 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458: 914–918 [DOI] [PubMed] [Google Scholar]
- Dixon LK, Twigg SR, Baylis SA, Vydelingum S, Bristow C, Hammond JM, Smith GL 1994. Nucleotide sequence of a 55 kbp region from the right end of the genome of a pathogenic African Swine Fever Virus isolate (Malawi LIL20/1). J Gen Virol 75: 1655–1684 [DOI] [PubMed] [Google Scholar]
- Dmitriev SE, Terenin IM, Andreev DE, Ivanov PA, Dunaevsky JE, Merrick WC, Shatsky IN 2010. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol Chem 285: 26779–26787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrikova E, Shveygert M, Walters R, Gromeier M 2010. Herpes simplex virus proteins ICP27 and UL47 associate with polyadenylate-binding protein and control its subcellular distribution. J Virol 84: 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty JD, White JP, Lloyd RE 2011. Poliovirus-mediated disruption of cytoplasmic processing bodies. J Virol 85: 64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn EF, Connor JH 2011. Dominant inhibition of Akt/protein kinase B signaling by the matrix protein of a negative-strand RNA virus. J Virol 85: 422–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elde NC, Child SJ, Geballe AP, Malik HS 2009. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature 457: 485–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia A, Constantinou C, Clemens MJ 2008. Effects of protein phosphorylation on ubiquitination and stability of the translational inhibitor protein 4E-BP1. Oncogene 27: 811–822 [DOI] [PubMed] [Google Scholar]
- Ellison KS, Maranchuk RA, Mottet KL, Smiley JR 2005. Control of VP16 translation by the herpes simplex virus type 1 immediate-early protein ICP27. J Virol 79: 4120–4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara MM, Brinton MA 2007. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc Natl Acad Sci 104: 9041–9046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensinger MJ, Martin SA, Paoletti E, Moss B 1975. Modification of the 5′-terminus of mRNA by soluble guanylyl and methyl transferases from vaccinia virus. Proc Natl Acad Sci 72: 2525–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelin K, Hafren A, Rantalainen KI, Makinen K 2011. Potyviral VPg enhances viral RNA translation and inhibits reporter mRNA translation in planta. J Virol 85: 9210–9221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchison D, Milburn SC, Edery I, Sonenberg N, Hershey JW 1982. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem 257: 14806–14810 [PubMed] [Google Scholar]
- Faria PA, Chakraborty P, Levay A, Barber GN, Ezelle HJ, Enninga J, Arana C, van Deursen J, Fontoura BM 2005. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol Cell 17: 93–102 [DOI] [PubMed] [Google Scholar]
- Feigenblum D, Schneider RJ 1993. Modification of eukaryotic initiation factor 4F during infection by influenza virus. J Virol 67: 3027–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigenblum D, Schneider RJ 1996. Cap-binding protein (eukaryotic initiation factor 4E) and 4E-inactivating protein BP-1 independently regulate cap-dependent translation. Mol Cell Biol 16: 5450–5457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM 2009. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol 7: e1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng P, Everly DN Jr, Read GS 2005. mRNA decay during herpes simplex virus (HSV) infections: Protein–protein interactions involving the HSV virion host shutoff protein and translation factors eIF4H and eIF4A. J Virol 79: 9651–9664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer MG, Allen MJ, Wilson WH, Suttle CA 2010. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc Natl Acad Sci 107: 19508–19513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine-Rodriguez EC, Knipe DM 2008. Herpes simplex virus ICP27 increases translation of a subset of viral late mRNAs. J Virol 82: 3538–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanes V, Raychaudhuri S, Dasgupta A 2009. A cell-permeable peptide inhibits hepatitis C virus replication by sequestering IRES transacting factors. Virology 394: 82–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CS, Doudna JA 2007. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat Rev 5: 29–38 [DOI] [PubMed] [Google Scholar]
- Fujimura T, Esteban R 2011. Cap-snatching mechanism in yeast L-A double-stranded RNA virus. Proc Natl Acad Sci 108: 17667–17671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale M Jr, Foy EM 2005. Evasion of intracellular host defence by hepatitis C virus. Nature 436: 939–945 [DOI] [PubMed] [Google Scholar]
- Gale M Jr, Blakely CM, Kwieciszewski B, Tan SL, Dossett M, Tang NM, Korth MJ, Polyak SJ, Gretch DR, Katze MG 1998. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: Molecular mechanisms of kinase regulation. Mol Cell Biol 18: 5208–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR 2001. Cap-independent translation conferred by the 5′ leader of tobacco etch virus is eukaryotic initiation factor 4G dependent. J Virol 75: 12141–12152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaigorta U, Chisari FV 2009. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe 6: 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcin D, Lezzi M, Dobbs M, Elliott RM, Schmaljohn C, Kang CY, Kolakofsky D 1995. The 5′ ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J Virol 69: 5754–5762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrey JL, Lee YY, Au HH, Bushell M, Jan E 2010. Host and viral translational mechanisms during cricket paralysis virus infection. J Virol 84: 1124–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparian AV, Neznanov N, Jha S, Galkin O, Moran JJ, Gudkov AV, Gurova KV, Komar AA 2010. Inhibition of encephalomyocarditis virus and poliovirus replication by quinacrine: Implications for the design and discovery of novel antiviral drugs. J Virol 84: 9390–9397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Sonenberg N 1997. Adenovirus infection inactivates the translational inhibitors 4E-BP1 and 4E-BP2. Virology 237: 182–186 [DOI] [PubMed] [Google Scholar]
- Gingras AC, Svitkin Y, Belsham GJ, Pause A, Sonenberg N 1996. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc Natl Acad Sci 93: 5578–5583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz C, Everson RG, Zhang LC, Gromeier M 2010. MAPK signal-integrating kinase controls cap-independent translation and cell type-specific cytotoxicity of an oncolytic poliovirus. Mol Ther 18: 1937–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I, Chaudhry Y, Gioldasi I, Gerondopoulos A, Natoni A, Labrie L, Laliberte JF, Roberts L 2005. Calicivirus translation initiation requires an interaction between VPg and eIF 4 E. EMBO Rep 6: 968–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AG, Smith JA, Balachandran S, Perwitasari O, Proll SC, Thomas MJ, Korth MJ, Barber GN, Schiff LA, Katze MG 2007. The cellular protein P58IPK regulates influenza virus mRNA translation and replication through a PKR-mediated mechanism. J Virol 81: 2221–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AG, Fornek JL, Medigeshi GR, Perrone LA, Peng X, Dyer MD, Proll SC, Knoblaugh SE, Carter VS, Korth MJ, et al. 2009. P58IPK: A novel “CIHD” member of the host innate defense response against pathogenic virus infection. PLoS Pathog 5: e1000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R, Frolova E, Williams BR, Rice CM, Frolov I 2004. PKR-dependent and -independent mechanisms are involved in translational shutoff during Sindbis virus infection. J Virol 78: 8455–8467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradi A, Svitkin YV, Imataka H, Sonenberg N 1998. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci 95: 11089–11094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger L, Cicchini L, Rak M, Petrucelli A, Fitzgerald KD, Semler BL, Goodrum F 2010. Stress-inducible alternative translation initiation of human cytomegalovirus latency protein pUL138. J Virol 84: 9472–9486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths A, Coen DM 2005. An unusual internal ribosome entry site in the herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci 102: 9667–9672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groft CM, Burley SK 2002. Recognition of eIF4G by rotavirus NSP3 reveals a basis for mRNA circularization. Mol Cell 9: 1273–1283 [DOI] [PubMed] [Google Scholar]
- Groppo R, Brown BA, Palmenberg AC 2010. Mutational analysis of the EMCV 2A protein identifies a nuclear localization signal and an eIF4E binding site. Virology 410: 257–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundhoff A, Ganem D 2001. Mechanisms governing expression of the v-FLIP gene of Kaposi’s sarcoma-associated herpesvirus. J Virol 75: 1857–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilligay D, Tarendeau F, Resa-Infante P, Coloma R, Crepin T, Sehr P, Lewis J, Ruigrok RW, Ortin J, Hart DJ, et al. 2008. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat Struct Mol Biol 15: 500–506 [DOI] [PubMed] [Google Scholar]
- Gunnery S, Green SR, Mathews MB 1992. Tat-responsive region RNA of human immunodeficiency virus type 1 stimulates protein synthesis in vivo and in vitro: Relationship between structure and function. Proc Natl Acad Sci 89: 11557–11561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Hui DJ, Merrick WC, Sen GC 2000. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J 19: 6891–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harb M, Becker MM, Vitour D, Baron CH, Vende P, Brown SC, Bolte S, Arold ST, Poncet D 2008. Nuclear localization of cytoplasmic poly(A)-binding protein upon rotavirus infection involves the interaction of NSP3 with eIF4G and RoXaN. J Virol 82: 11283–11293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Gross M, Roizman B 1997. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the α subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci 94: 843–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M 2008. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J 27: 3300–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdy B, Jaramillo M, Svitkin YV, Rosenfeld AB, Kobayashi M, Walsh D, Alain T, Sean P, Robichaud N, Topisirovic I, et al. 2012. Translational control of the activation of transcription factor NF-κB and production of type I Interferon by phosphorylation of the translation factor eIF4E. Nat Immunol 10.1038/ni.2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hershey WB, Sonenberg N, Mathews MB. 2012. Principles of translational control: An overview. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a011528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hinnebusch AG, Lorsch JR 2012. The mechanism of eukaryotic translation initiation: New insights and challenges. Cold Spring Harb Perspect Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho BC, Yu SL, Chen JJ, Chang SY, Yan BS, Hong QS, Singh S, Kao CL, Chen HY, Su KY, et al. 2010. Enterovirus-induced miR-141 contributes to shutoff of host protein translation by targeting the translation initiation factor eIF4E. Cell Host Microbe 9: 58–69 [DOI] [PubMed] [Google Scholar]
- Hogg JR, Goff SP 2010. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143: 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M, Shamanna RA, Guan D, Pe’ery T, Mathews MB 2011. HIV-1 replication and latency are regulated by translational control of cyclin T1. J Mol Biol 410: 917–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath CM, Williams MA, Lamb RA 1990. Eukaryotic coupled translation of tandem cistrons: Identification of the influenza B virus BM2 polypeptide. EMBO J 9: 2639–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houck-Loomis B, Durney MA, Salguero C, Shankar N, Nagle JM, Goff SP, D’Souza VM 2011. An equilibrium-dependent retroviral mRNA switch regulates translational recoding. Nature 480: 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JT, Schneider RJ 1991. Adenovirus inhibition of cellular protein synthesis involves inactivation of cap-binding protein. Cell 65: 271–280 [DOI] [PubMed] [Google Scholar]
- Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL, Makino S 2011. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: Viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog 7: e1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui DJ, Bhasker CR, Merrick WC, Sen GC 2003. Viral stress-inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2·GTP·Met-tRNAi. J Biol Chem 278: 39477–39482 [DOI] [PubMed] [Google Scholar]
- Hui DJ, Terenzi F, Merrick WC, Sen GC 2005. Mouse p56 blocks a distinct function of eukaryotic initiation factor 3 in translation initiation. J Biol Chem 280: 3433–3440 [DOI] [PubMed] [Google Scholar]
- Ilkow CS, Mancinelli V, Beatch MD, Hobman TC 2008. Rubella virus capsid protein interacts with poly(A)-binding protein and inhibits translation. J Virol 82: 4284–4294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack K, Bellodi C, Landry DM, Niederer RO, Meskauskas A, Musalgaonkar S, Kopmar N, Krasnykh O, Dean AM, Thompson SR, et al. 2011. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol Cell 44: 660–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Jackson RJ 2012. The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb Perspect Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson BR, Boyne JR, Noerenberg M, Taylor A, Hautbergue GM, Walsh MJ, Wheat R, Blackbourn DJ, Wilson SA, Whitehouse A 2011. An interaction between KSHV ORF57 and UIF provides mRNA-adaptor redundancy in herpesvirus intronless mRNA export. PLoS Pathog 7: e1002138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E, Sarnow P 2002. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol 324: 889–902 [DOI] [PubMed] [Google Scholar]
- Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol 62: 2636–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra RK, Yi M, Lemon SM 2010. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J Virol 84: 6615–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen DM, Frolova L, Geballe AP 2002. Inhibition of translation termination mediated by an interaction of eukaryotic release factor 1 with a nascent peptidyl-tRNA. Mol Cell Biol 22: 8562–8570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Fraser CS, Yu Y, Leary J, Doudna JA 2004. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci 101: 16990–16995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji WT, Wang L, Lin RC, Huang WR, Liu HJ 2009. Avian reovirus influences phosphorylation of several factors involved in host protein translation including eukaryotic translation elongation factor 2 (eEF2) in Vero cells. Biochem Biophys Res Commun 384: 301–305 [DOI] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309: 1577–1581 [DOI] [PubMed] [Google Scholar]
- Jopling CL, Schutz S, Sarnow P 2008. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe 4: 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan R, Wang L, Graczyk TM, Block TM, Romano PR 2002. Replication of a cytopathic strain of bovine viral diarrhea virus activates PERK and induces endoplasmic reticulum stress-mediated apoptosis of MDBK cells. J Virol 76: 9588–9599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafasla P, Morgner N, Robinson CV, Jackson RJ 2010. Polypyrimidine tract-binding protein stimulates the poliovirus IRES by modulating eIF4G binding. EMBO J 29: 3710–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W, Narayanan K, Huang C, Lokugamage K, Ikegami T, Ito N, Kubo H, Makino S 2006. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc Natl Acad Sci 103: 12885–12890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W, Huang C, Narayanan K, Lokugamage KG, Makino S 2009. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat Struct Mol Biol 16: 1134–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoshita N, Nomoto A, RajBhandary UL 2009. Translation initiation from the ribosomal A site or the P site, dependent on the conformation of RNA pseudoknot I in dicistrovirus RNAs. Mol Cell 35: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JI, Kwon SN, Park SH, Kim YK, Choi SY, Kim JP, Ahn BY 2009. PKR protein kinase is activated by hepatitis C virus and inhibits viral replication through translational control. Virus Res 142: 51–56 [DOI] [PubMed] [Google Scholar]
- Katsafanas GC, Moss B 2007. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe 2: 221–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagishi-Kobayashi M, Silverman JB, Ung TL, Dever TE 1997. Regulation of the protein kinase PKR by the vaccinia virus pseudosubstrate inhibitor K3L is dependent on residues conserved between the K3L protein and the PKR substrate eIF2α. Mol Cell Biol 17: 4146–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Matsumura T, Roizman B, Hirai K 1999. Cellular elongation factor 1δ is modified in cells infected with representative alpha-, beta-, or gammaherpesviruses. J Virol 73: 4456–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazemi S, Papadopoulou S, Li S, Su Q, Wang S, Yoshimura A, Matlashewski G, Dever TE, Koromilas AE 2004. Control of α subunit of eukaryotic translation initiation factor 2 (eIF2α) phosphorylation by the human papillomavirus type 18 E6 oncoprotein: Implications for eIF2 α-dependent gene expression and cell death. Mol Cell Biol 24: 3415–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf BJ, Barton DJ 2008. Poliovirus 2A(Pro) increases viral mRNA and polysome stability coordinately in time with cleavage of eIF4G. J Virol 82: 5847–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Miyoshi H, Ray S, Natsuaki T, Suehiro N, Goss DJ 2006. Interaction of genome-linked protein (VPg) of turnip mosaic virus with wheat germ translation initiation factors eIFiso4E and eIFiso4F. J Biol Chem 281: 28002–28010 [DOI] [PubMed] [Google Scholar]
- Khan MA, Miyoshi H, Gallie DR, Goss DJ 2008. Potyvirus genome-linked protein, VPg, directly affects wheat germ in vitro translation: Interactions with translation initiation factors eIF4F and eIFiso4F. J Biol Chem 283: 1340–1349 [DOI] [PubMed] [Google Scholar]
- Khong A, Jan E 2011. Modulation of stress granules and P bodies during dicistrovirus infection. J Virol 85: 1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Park SM, Park JH, Keum SJ, Jang SK 2011. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J 30: 2454–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Hellen CU, Shatsky IN 1996. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA 2: 1199–1212 [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Pestova TV, Hellen CU, Shatsky IN 1998. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J Biol Chem 273: 18599–18604 [DOI] [PubMed] [Google Scholar]
- Komarova AV, Real E, Borman AM, Brocard M, England P, Tordo N, Hershey JW, Kean KM, Jacob Y 2007. Rabies virus matrix protein interplay with eIF3, new insights into rabies virus pathogenesis. Nucleic Acids Res 35: 1522–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratzer F, Rosorius O, Heger P, Hirschmann N, Dobner T, Hauber J, Stauber RH 2000. The adenovirus type 5 E1B-55K oncoprotein is a highly active shuttle protein and shuttling is independent of E4orf6, p53 and Mdm2. Oncogene 19: 850–857 [DOI] [PubMed] [Google Scholar]
- Krausslich HG, Nicklin MJ, Toyoda H, Etchison D, Wimmer E 1987. Poliovirus proteinase 2A induces cleavage of eucaryotic initiation factor 4F polypeptide 220 J Virol 61: 2711–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudchodkar SB, Yu Y, Maguire TG, Alwine JC 2004. Human cytomegalovirus infection induces rapamycin-insensitive phosphorylation of downstream effectors of mTOR kinase. J Virol 78: 11030–11039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudchodkar SB, Yu Y, Maguire TG, Alwine JC 2006. Human cytomegalovirus infection alters the substrate specificities and rapamycin sensitivities of raptor- and rictor-containing complexes. Proc Natl Acad Sci 103: 14182–14187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar GR, Glaunsinger BA 2010. Nuclear import of cytoplasmic poly(A) binding protein restricts gene expression via hyperadenylation and nuclear retention of mRNA. Mol Cell Biol 30: 4996–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar GR, Shum L, Glaunsinger BA 2011. Importin α-mediated nuclear import of cytoplasmic poly(A) binding protein occurs as a direct consequence of cytoplasmic mRNA depletion. Mol Cell Biol 31: 3113–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyumcu-Martinez M, Belliot G, Sosnovtsev SV, Chang KO, Green KY, Lloyd RE 2004a. Calicivirus 3C-like proteinase inhibits cellular translation by cleavage of poly(A)-binding protein. J Virol 78: 8172–8182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Van Eden ME, Younan P, Lloyd RE 2004b. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: A novel mechanism for host translation shutoff. Mol Cell Biol 24: 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster AM, Jan E, Sarnow P 2006. Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA 12: 894–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry DM, Hertz MI, Thompson SR 2009. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev 23: 2753–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larralde O, Smith RW, Wilkie GS, Malik P, Gray NK, Clements JB 2006. Direct stimulation of translation by the multifunctional herpesvirus ICP27 protein. J Virol 80: 1588–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Glaunsinger BA 2009. Aberrant herpesvirus-induced polyadenylation correlates with cellular messenger RNA destruction. PloS Biol 7: e1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang C, Chen X, Yu J, Wang Y, Yang Y, Du M, Jin H, Ma Y, He B, et al. 2011. ICP34.5 protein of herpes simplex virus facilitates the initiation of protein translation by bridging eukaryotic initiation factor 2α (eIF2α) and protein phosphatase 1. J Biol Chem 286: 24785–24792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist ME, Lifland AW, Utley TJ, Santangelo PJ, Crowe JE Jr 2010. Respiratory syncytial virus induces host RNA stress granules to facilitate viral replication. J Virol 84: 12274–12284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lischka P, Toth Z, Thomas M, Mueller R, Stamminger T 2006. The UL69 transactivator protein of human cytomegalovirus interacts with DEXD/H-Box RNA helicase UAP56 to promote cytoplasmic accumulation of unspliced RNA. Mol Cell Biol 26: 1631–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker N, Easton LE, Lukavsky PJ 2007. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J 26: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker N, Chamond N, Sargueil B 2010. A conserved structure within the HIV gag open reading frame that controls translation initiation directly recruits the 40S subunit and eIF3. Nucleic Acids Res 39: 2367–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low W, Harries M, Ye H, Du MQ, Boshoff C, Collins M 2001. Internal ribosome entry site regulates translation of Kaposi’s sarcoma-associated herpesvirus FLICE inhibitory protein. J Virol 75: 2938–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Bablanian R 1996. Characterization of small nontranslated polyadenylylated RNAs in vaccinia virus-infected cells. Proc Natl Acad Sci 93: 2037–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Hu X, Li Y, Zheng L, Zhou Y, Jiang H, Ning T, Basang Z, Zhang C, Ke Y 2004. Human papillomavirus 16 E6 oncoprotein interferences with insulin signaling pathway by binding to tuberin. J Biol Chem 279: 35664–35670 [DOI] [PubMed] [Google Scholar]
- Luttermann C, Meyers G 2009. The importance of inter- and intramolecular base pairing for translation reinitiation on a eukaryotic bicistronic mRNA. Genes Dev 23: 331–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney GM, Kedersha NL, Kaufman RJ, Anderson P, Liljestrom P 2005. Importance of eIF2α phosphorylation and stress granule assembly in alphavirus translation regulation. Mol Biol Cell 16: 3753–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney C, Perez C, Mohr I 2012. Poly(A) binding protein abundance regulates eukaryotic translation initiation factor 4F assembly in human cytomegalovirus-infected cells. Proc Natl Acad Sci 109: 5627–5632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon R, Zaborowska I, Walsh D 2011. Noncytotoxic inhibition of viral infection through eIF4F-independent suppression of translation by 4EGi-1. J Virol 85: 853–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel YM, Borman AM, Paulous S, Kean KM 2001. Eukaryotic initiation factor 4G-poly(A) binding protein interaction is required for poly(A) tail-mediated stimulation of picornavirus internal ribosome entry segment-driven translation but not for X-mediated stimulation of hepatitis C virus translation. Mol Cell Biol 21: 4097–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WA, Wang Z, Treder K 2007. The amazing diversity of cap-independent translation elements in the 3′-untranslated regions of plant viral RNAs. Biochem Soc Trans 35: 1629–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir MA, Panganiban AT 2008. A protein that replaces the entire cellular eIF4F complex. EMBO J 27: 3129–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir MA, Panganiban AT 2010. The triplet repeats of the Sin Nombre hantavirus 5′ untranslated region are sufficient in cis for nucleocapsid-mediated translation initiation. J Virol 84: 8937–8944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir MA, Duran WA, Hjelle BL, Ye C, Panganiban AT 2008. Storage of cellular 5′ mRNA caps in P bodies for viral cap-snatching. Proc Natl Acad Sci 105: 19294–19299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani T, Fukushi S, Saijo M, Kurane I, Morikawa S 2004. Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus-infected cells. Biochem Biophys Res Commun 319: 1228–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohankumar V, Dhanushkodi NR, Raju R 2011. Sindbis virus replication, is insensitive to rapamycin and torin1, and suppresses Akt/mTOR pathway late during infection in HEK cells. Biochem Biophys Res Commun 406: 262–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr I 2005. To replicate or not to replicate: Achieving selective oncolytic virus replication in cancer cells through translational control. Oncogene 24: 7697–7709 [DOI] [PubMed] [Google Scholar]
- Montero H, Arias CF, Lopez S 2006. Rotavirus nonstructural protein NSP3 is not required for viral protein synthesis. J Virol 80: 9031–9038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero H, Rojas M, Arias CF, Lopez S 2008. Rotavirus infection induces the phosphorylation of eIF2α but prevents the formation of stress granules. J Virol 82: 1496–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody CA, Scott RS, Amirghahari N, Nathan CA, Young LS, Dawson CW, Sixbey JW 2005. Modulation of the cell growth regulator mTOR by Epstein-Barr virus-encoded LMP2A. J Virol 79: 5499–5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman NJ, Shenk T 2010. Rapamycin-resistant mTORC1 activity is required for herpesvirus replication. J Virol 84: 5260–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman NJ, Cristea IM, Terhune SS, Rout MP, Chait BT, Shenk T 2008. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 3: 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M, Poppers J, Ladd A, Mohr I 1999. A herpesvirus ribosome-associated, RNA-binding protein confers a growth advantage upon mutants deficient in a GADD34-related function. J Virol 73: 3375–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M, Poppers J, Sternberg D, Mohr I 2003. Regulation of eIF2α phosphorylation by different functions that act during discrete phases in the herpes simplex virus type 1 life cycle. J Virol 77: 10917–10928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M, Arias C, Mohr I 2007. Maintenance of endoplasmic reticulum (ER) homeostasis in herpes simplex virus type 1-infected cells through the association of a viral glycoprotein with PERK, a cellular ER stress sensor. J Virol 81: 3377–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, Vanicek J, Robins H, Shenk T, Levine AJ 2008. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: Implications for latency. Proc Natl Acad Sci 105: 5453–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthukrishnan S, Morgan M, Banerjee AK, Shatkin AJ 1976. Influence of 5′-terminal m7G and 2′-O-methylated residues on messenger ribonucleic acid binding to ribosomes. Biochemistry 15: 5761–5768 [DOI] [PubMed] [Google Scholar]
- Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O 2009. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 5: 376–385 [DOI] [PubMed] [Google Scholar]
- Napthine S, Lever RA, Powell ML, Jackson RJ, Brown TD, Brierley I 2009. Expression of the VP2 protein of murine norovirus by a translation termination-reinitiation strategy. PLoS ONE 4: e8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM 2009. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell 34: 696–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols LA, Adang LA, Kedes DH 2011. Rapamycin blocks production of KSHV/HHV8: Insights into the anti-tumor activity of an immunosuppressant drug. PLoS ONE 6: e14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto C, Piron F, Dalmais M, Marco CF, Moriones E, Gomez-Guillamon ML, Truniger V, Gomez P, Garcia-Mas J, Aranda MA, et al. 2007. EcoTILLING for the identification of allelic variants of melon eIF4E, a factor that controls virus susceptibility. BMC Plant Biol 7: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novac O, Guenier AS, Pelletier J 2004. Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res 32: 902–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliere S, Arguello M, Mesplede T, Tumilasci V, Nakhaei P, Stojdl D, Sonenberg N, Bell J, Hiscott J 2008. Vesicular stomatitis virus oncolysis of T lymphocytes requires cell cycle entry and translation initiation. J Virol 82: 5735–5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley RP, Duncan RF, Hershey JW, Mathews MB 1989. Modification of protein synthesis initiation factors and the shut-off of host protein synthesis in adenovirus-infected cells. Virology 168: 112–118 [DOI] [PubMed] [Google Scholar]
- O’Neill RE, Racaniello VR 1989. Inhibition of translation in cells infected with a poliovirus 2Apro mutant correlates with phosphorylation of the α subunit of eucaryotic initiation factor 2. J Virol 63: 5069–5075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova M, Yueh A, Leung J, Goff SP 2003. Reverse transcriptase of Moloney murine leukemia virus binds to eukaryotic release factor 1 to modulate suppression of translational termination. Cell 115: 319–331 [DOI] [PubMed] [Google Scholar]
- O’Shea C, Klupsch K, Choi S, Bagus B, Soria C, Shen J, McCormick F, Stokoe D 2005. Adenoviral proteins mimic nutrient/growth signals to activate the mTOR pathway for viral replication. EMBO J 24: 1211–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto GA, Puglisi JD 2004. The pathway of HCV IRES-mediated translation initiation. Cell 119: 369–380 [DOI] [PubMed] [Google Scholar]
- Parrish S, Moss B 2007. Characterization of a second vaccinia virus mRNA-decapping enzyme conserved in poxviruses. J Virol 81: 12973–12978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish S, Resch W, Moss B 2007. Vaccinia virus D10 protein has mRNA decapping activity, providing a mechanism for control of host and viral gene expression. Proc Natl Acad Sci 104: 2139–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons J, Castaldi MP, Dutta S, Dibrov SM, Wyles DL, Hermann T 2009. Conformational inhibition of the hepatitis C virus internal ribosome entry site RNA. Nat Chem Biol 5: 823–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen RB, Seth PP, Swayze EE, Griffey RH, Skalicky JJ, Cheatham TE III, Davis DR 2010. Inhibitor-induced structural change in the HCV IRES domain IIa RNA. Proc Natl Acad Sci 107: 7263–7268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavio N, Romano PR, Graczyk TM, Feinstone SM, Taylor DR 2003. Protein synthesis and endoplasmic reticulum stress can be modulated by the hepatitis C virus envelope protein E2 through the eukaryotic initiation factor 2α kinase PERK. J Virol 77: 3578–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Pavitt GD, Ron D 2012. New insights into translational regulation in the endoplasmic reticulum unfolded protein response. Cold Spring Harb Perspect Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pe’ery T, Mathews MB 2007. Viral conquest of the host cell. In Fields virology (ed. Knipe DM, PM Howley), pp. 168–208 Lippincott Williams and Wilkins, Philadelphia [Google Scholar]
- Pelletier J, Sonenberg N 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334: 320–325 [DOI] [PubMed] [Google Scholar]
- Perez C, McKinney C, Chulunbaatar U, Mohr I 2011. Translational control of the abundance of cytoplasmic poly(A) binding protein in human cytomegalovirus-infected cells. J Virol 85: 156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev 12: 67–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CU 2008. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: A common role of domain II. EMBO J 27: 1060–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, et al. 2004. Identification of virus-encoded microRNAs. Science 304: 734–736 [DOI] [PubMed] [Google Scholar]
- Piotrowska J, Hansen SJ, Park N, Jamka K, Sarnow P, Gustin KE 2010. Stable formation of compositionally unique stress granules in virus-infected cells. J Virol 84: 3654–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piron M, Vende P, Cohen J, Poncet D 1998. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J 17: 5811–5821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Chard LS, Kaku Y, Johns HL, Shatsky IN, Belsham GJ 2004. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J Virol 78: 4487–4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotch SJ, Bouloy M, Ulmanen I, Krug RM 1981. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23: 847–858 [DOI] [PubMed] [Google Scholar]
- Polacek C, Friebe P, Harris E 2009. Poly(A)-binding protein binds to the non-polyadenylated 3′ untranslated region of dengue virus and modulates translation efficiency. J Gen Virol 90: 687–692 [DOI] [PubMed] [Google Scholar]
- Powell ML, Leigh KE, Poyry TA, Jackson RJ, Brown TD, Brierley I 2011. Further characterisation of the translational termination-reinitiation signal of the influenza B virus segment 7 RNA. PLoS ONE 6: e16822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyry TA, Kaminski A, Connell EJ, Fraser CS, Jackson RJ 2007. The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation. Genes Dev 21: 3149–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q, Hastings C, Miller CL 2009. Mammalian orthoreovirus particles induce and are recruited into stress granules at early times postinfection. J Virol 83: 11090–11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q, Carroll K, Hastings C, Miller CL 2011. Mammalian orthoreovirus escape from host translational shutoff correlates with stress granule disruption and is independent of eIF2α phosphorylation and PKR. J Virol 85: 8798–8810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaben M, Groot Koerkamp MJ, Rottier PJ, de Haan CA 2007. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell Microbiol 9: 2218–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramelot TA, Cort JR, Yee AA, Liu F, Goshe MB, Edwards AM, Smith RD, Arrowsmith CH, Dever TE, Kennedy MA 2002. Myxoma virus immunomodulatory protein M156R is a structural mimic of eukaryotic translation initiation factor eIF2α. J Mol Biol 322: 943–954 [DOI] [PubMed] [Google Scholar]
- Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM 2004. The 1.2-megabase genome sequence of Mimivirus. Science 306: 1344–1350 [DOI] [PubMed] [Google Scholar]
- Redondo N, Sanz MA, Welnowska E, Carrasco L 2011. Translation without eIF2 promoted by poliovirus 2A protease. PLoS ONE 6: e25699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera J, Weber F, Cusack S 2010. Bunyaviridae RNA polymerases (L-protein) have an N-terminal, influenza-like endonuclease domain, essential for viral cap-dependent transcription. PLoS Pathog 6: e1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remm M, Remm A, Ustav M 1999. Human papillomavirus type 18 E1 protein is translated from polycistronic mRNA by a discontinuous scanning mechanism. J Virol 73: 3062–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richner JM, Clyde K, Pezda AC, Cheng BY, Wang T, Kumar GR, Covarrubias S, Coscoy L, Glaunsinger B 2011. Global mRNA degradation during lytic gammaherpesvirus infection contributes to establishment of viral latency. PLoS Pathog 7: e1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Kapp LD, Khan SN, Acker MG, Kolitz S, Kazemi S, Kaufman RJ, Merrick WC, Koromilas AE, Lorsch JR, et al. 2006. Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2·GTP·Met-tRNAiMet ternary complex availability. Mol Biol Cell 17: 4632–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez Pulido M, Serrano P, Saiz M, Martinez-Salas E 2007. Foot-and-mouth disease virus infection induces proteolytic cleavage of PTB, eIF3a,b, and PABP RNA-binding proteins. Virology 364: 466–474 [DOI] [PubMed] [Google Scholar]
- Rothenburg S, Seo EJ, Gibbs JS, Dever TE, Dittmar K 2009. Rapid evolution of protein kinase PKR alters sensitivity to viral inhibitors. Nat Struct Mol Biol 16: 63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenburg S, Chinchar VG, Dever TE 2011. Characterization of a ranavirus inhibitor of the antiviral protein kinase PKR. BMC Microbiol 11: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Roux PP, Topisirovic I 2012. Regulation of mRNA translation by signaling pathways. Cold Spring Harb Perspect Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini HK, Fischer D 2007. Structural and functional insights into Mimivirus ORFans. BMC Genomics 8: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salaun C, Macdonald AI, Larralde O, Howard L, Lochtie K, Burgess HM, Brook M, Malik P, Gray NK, Graham SV 2010. Poly(A)-binding protein 1 (PABP1) partially relocalises to the nucleus during HSV-1 infection in an ICP27-independent manner and does not inhibit virus replication. J Virol 84: 8539–8548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatore M, Basler CF, Parisien JP, Horvath CM, Bourmakina S, Zheng H, Muster T, Palese P, Garcia-Sastre A 2002. Effects of influenza A virus NS1 protein on protein expression: The NS1 protein enhances translation and is not required for shutoff of host protein synthesis. J Virol 76: 1206–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Mohr I 2007. Inhibition of cellular 2′–5′ oligoadenylate synthetase by the herpes simplex virus type 1 Us11 protein. J Virol 81: 3455–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri-Goldin RM 2011. The function and activities of HSV-1 ICP27, a multifunctional regulator of gene expression. In Alphaherpesviruses: Molecular virology (ed. Weller S), pp. 39–50 Caister Academic Press, Norfolk, UK [Google Scholar]
- Santhakumar D, Forster T, Laqtom NN, Fragkoudis R, Dickinson P, Abreu-Goodger C, Manakov SA, Choudhury NR, Griffiths SJ, Vermeulen A, et al. 2010. Combined agonist–antagonist genome-wide functional screening identifies broadly active antiviral microRNAs. Proc Natl Acad Sci 107: 13830–13835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz MA, Castello A, Ventoso I, Berlanga JJ, Carrasco L 2009. Dual mechanism for the translation of subgenomic mRNA from Sindbis virus in infected and uninfected cells. PLoS ONE 4: e4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Masuda M, Kanai M, Tsukiyama-Kohara K, Yoneda M, Kai C 2007. Measles virus N protein inhibits host translation by binding to eIF3-p40. J Virol 81: 11569–11576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Kobayashi K, Geldreich A, Caranta C, Robaglia C, Keller M, Ryabova LA 2011. Viral factor TAV recruits TOR/S6K1 signalling to activate reinitiation after long ORF translation. EMBO J 30: 1343–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo EJ, Liu F, Kawagishi-Kobayashi M, Ung TL, Cao C, Dar AC, Sicheri F, Dever TE 2008. Protein kinase PKR mutants resistant to the poxvirus pseudosubstrate K3L protein. Proc Natl Acad Sci 105: 16894–16899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih SR, Krug RM 1996. Surprising function of the three influenza viral polymerase proteins: Selective protection of viral mRNAs against the cap-snatching reaction catalyzed by the same polymerase proteins. Virology 226: 430–435 [DOI] [PubMed] [Google Scholar]
- Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS 2011. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J Clin Invest 121: 3623–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman RH 2007. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol 81: 12720–12729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson-Holley M, Kedersha N, Dower K, Rubins KH, Anderson P, Hensley LE, Connor JH 2011. Formation of antiviral cytoplasmic granules during orthopoxvirus infection. J Virol 85: 1581–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU 1998. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol 72: 4775–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV 2010. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev 24: 1787–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, Schmechel SC, Raghavan A, Abelson M, Reilly C, Katze MG, Kaufman RJ, Bohjanen PR, Schiff LA 2006. Reovirus induces and benefits from an integrated cellular stress response. J Virol 80: 2019–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi A, Chaisuparat R, Hu J, Ramsdell AK, Manning BD, Sausville EA, Sawai ET, Molinolo A, Gutkind JS, Montaner S 2006. The TSC2/mTOR pathway drives endothelial cell transformation induced by the Kaposi’s sarcoma-associated herpesvirus G protein-coupled receptor. Cancer Cell 10: 133–143 [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Rupprecht KM, Hecht SM, Shatkin AJ 1979. Eukaryotic mRNA cap binding protein: Purification by affinity chromatography on Sepharose-coupled m7GDP. Proc Natl Acad Sci 76: 4345–4349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood R, Porter AC, Ma K, Quilliam LA, Wek RC 2000. Pancreatic eukaryotic initiation factor-2α kinase (PEK) homologues in humans, Drosophila melanogaster and Caenorhabditis elegans that mediate translational control in response to endoplasmic reticulum stress. Biochem J 346: 281–293 [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J 2004. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: The IRES functions as an RNA-based translation factor. Cell 118: 465–475 [DOI] [PubMed] [Google Scholar]
- Spangle JM, Munger K 2010. The human papillomavirus type 16 E6 oncoprotein activates mTORC1 signaling and increases protein synthesis. J Virol 84: 9398–9407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford MM, Barrett JW, Nazarian SH, Werden S, McFadden G 2007. Oncolytic virotherapy synergism with signaling inhibitors: Rapamycin increases myxoma virus tropism for human tumor cells. J Virol 81: 1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukarieh R, Sonenberg N, Pelletier J 2010. Nuclear assortment of eIF4E coincides with shut-off of host protein synthesis upon poliovirus infection. J Gen Virol 91: 1224–1228 [DOI] [PubMed] [Google Scholar]
- Sweeney TR, Dhote V, Yu Y, Hellen CU 2011. A distinct class of internal ribosomal entry site (IRES) in members of the Kobuvirus and proposed Salivirusand Paraturdivirus genera of Picornaviridae. J Virol 10.1128/JVI.05862-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talloczy Z, Jiang W, Virgin HWt, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B 2002. Regulation of starvation- and virus-induced autophagy by the eIF2α kinase signaling pathway. Proc Natl Acad Sci 99: 190–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja S, MacGregor J, Markus S, Ha S, Mohr I 2001. Enhanced antitumor efficacy of a herpes simplex virus mutant isolated by genetic selection in cancer cells. Proc Natl Acad Sci 98: 8804–8808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Bertke AS, Patel A, Wang K, Cohen JI, Krause PR 2008. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc Natl Acad Sci 105: 10931–10936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DR, Shi ST, Romano PR, Barber GN, Lai MM 1999. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285: 107–110 [DOI] [PubMed] [Google Scholar]
- Taylor DR, Tian B, Romano PR, Hinnebusch AG, Lai MM, Mathews MB 2001. Hepatitis C virus envelope protein E2 does not inhibit PKR by simple competition with autophosphorylation sites in the RNA-binding domain. J Virol 75: 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenzi F, Hui DJ, Merrick WC, Sen GC 2006. Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56. J Biol Chem 281: 34064–34071 [DOI] [PubMed] [Google Scholar]
- Thiebeauld O, Schepetilnikov M, Park HS, Geldreich A, Kobayashi K, Keller M, Hohn T, Ryabova LA 2009. A new plant protein interacts with eIF3 and 60S to enhance virus-activated translation re-initiation. EMBO J 28: 3171–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thivierge K, Cotton S, Dufresne PJ, Mathieu I, Beauchemin C, Ide C, Fortin MG, Laliberte JF 2008. Eukaryotic elongation factor 1A interacts with Turnip mosaic virus RNA-dependent RNA polymerase and VPg-Pro in virus-induced vesicles. Virology 377: 216–225 [DOI] [PubMed] [Google Scholar]
- Thomas MA, Broughton RS, Goodrum FD, Ornelles DA 2009. E4orf1 limits the oncolytic potential of the E1B-55K deletion mutant adenovirus. J Virol 83: 2406–2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 284: 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treder K, Kneller EL, Allen EM, Wang Z, Browning KS, Miller WA 2008. The 3′ cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA 14: 134–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trono D, Pelletier J, Sonenberg N, Baltimore D 1988. Translation in mammalian cells of a gene linked to the poliovirus 5′ noncoding region. Science 241: 445–448 [DOI] [PubMed] [Google Scholar]
- Truniger V, Aranda MA 2009. Recessive resistance to plant viruses. Adv Virus Res 75: 119–159 [DOI] [PubMed] [Google Scholar]
- Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454: 780–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventoso I, Blanco R, Perales C, Carrasco L 2001. HIV-1 protease cleaves eukaryotic initiation factor 4G and inhibits cap-dependent translation. Proc Natl Acad Sci 98: 12966–12971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventoso I, Sanz MA, Molina S, Berlanga JJ, Carrasco L, Esteban M 2006. Translational resistance of late alphavirus mRNA to eIF2α phosphorylation: A strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev 20: 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villas-Boas CS, Conceicao TM, Ramirez J, Santoro AB, Da Poian AT, Montero-Lomeli M 2009. Dengue virus-induced regulation of the host cell translational machinery. Braz J Med Biol Res 42: 1020–1026 [DOI] [PubMed] [Google Scholar]
- Vyas J, Elia A, Clemens MJ 2003. Inhibition of the protein kinase PKR by the internal ribosome entry site of hepatitis C virus genomic RNA. RNA 9: 858–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Mohr I 2004. Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev 18: 660–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Mohr I 2006. Assembly of an active translation initiation factor complex by a viral protein. Genes Dev 20: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Mohr I 2011. Viral subversion of the host protein synthesis machinery. Nat Rev Microbiol 9: 860–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Perez C, Notary J, Mohr I 2005. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J Virol 79: 8057–8064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Arias C, Perez C, Halladin D, Escandon M, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Mohr I 2008. Eukaryotic translation initiation factor 4F architectural alterations accompany translation initiation factor redistribution in poxvirus-infected cells. Mol Cell Biol 28: 2648–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Hudak KA 2006. A novel interaction of pokeweed antiviral protein with translation initiation factors 4G and iso4G: A potential indirect mechanism to access viral RNAs. Nucleic Acids Res 34: 1174–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FZ, Weber F, Croce C, Liu CG, Liao X, Pellett PE 2008. Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J Virol 82: 9065–9074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Parisien M, Scheets K, Miller WA 2011. The cap-binding translation initiation factor, eIF4E, binds a pseudoknot in a viral cap-independent translation element. Structure 19: 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Ohtaki N, Hayashi Y, Ikuta K, Tomonaga K 2009. Autogenous translational regulation of the Borna disease virus negative control factor X from polycistronic mRNA using host RNA helicases. PLoS Pathog 5: e1000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welnowska E, Castelló A, Moral P, Carrasco L 2009. Translation of mRNAs from vesicular stomatitis virus and vaccinia virus is differentially blocked in cells with depletion of eIF4GI and/or eIF4GII. J Mol Biol 394: 506–521 [DOI] [PubMed] [Google Scholar]
- Welnowska E, Sanz MA, Redondo N, Carrasco L 2011. Translation of viral mRNA without active eIF2: The case of picornaviruses. PLoS ONE 6: e22230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werden SJ, Barrett JW, Wang G, Stanford MM, McFadden G 2007. M-T5, the ankyrin repeat, host range protein of myxoma virus, activates Akt and can be functionally replaced by cellular PIKE-A. J Virol 81: 2340–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JP, Cardenas AM, Marissen WE, Lloyd RE 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2: 295–305 [DOI] [PubMed] [Google Scholar]
- White LK, Sali T, Alvarado D, Gatti E, Pierre P, Streblow D, Defilippis VR 2010. Chikungunya virus induces IPS-1-dependent innate immune activation and protein kinase R-independent translational shutoff. J Virol 85: 606–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JP, Reineke LC, Lloyd RE 2011. Poliovirus switches to an eIF2-independent mode of translation during infection. J Virol 85: 8884–8893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcocks MM, Carter MJ, Roberts LO 2004. Cleavage of eukaryotic initiation factor eIF4G and inhibition of host-cell protein synthesis during feline calicivirus infection. J Gen Virol 85: 1125–1130 [DOI] [PubMed] [Google Scholar]
- Willcocks MM, Locker N, Gomwalk Z, Royall E, Bakhshesh M, Belsham GJ, Idamakanti N, Burroughs KD, Reddy PS, Hallenbeck PL, et al. 2011. Structural features of the Seneca Valley virus internal ribosome entry site element; a picornavirus with a pestivirus-like IRES. J Virol 85: 4452–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Pestova TV, Hellen CU, Sarnow P 2000. Initiation of protein synthesis from the A site of the ribosome. Cell 102: 511–520 [DOI] [PubMed] [Google Scholar]
- Won S, Eidenschenk C, Arnold CN, Siggs OM, Sun L, Brandl K, Mullen TM, Nemerow GR, Moresco EM, Beutler B 2011. Increased susceptibility to DNA virus infection in mice with a GCN2 mutation. J Virol 86: 1802–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Q, Cuesta R, Schneider RJ 2004. Tethering of eIF4G to adenoviral mRNAs by viral 100k protein drives ribosome shunting. Genes Dev 18: 1997–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Q, Cuesta R, Schneider RJ 2005. Regulation of translation by ribosome shunting through phosphotyrosine-dependent coupling of adenovirus protein 100k to viral mRNAs. J Virol 79: 5676–5683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Xu LH, Yamada Y, Liu DX 2008. Coronavirus spike protein inhibits host cell translation by interaction with eIF3f. PLoS ONE 3: e1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Frank CL, Korth MJ, Sopher BL, Novoa I, Ron D, Katze MG 2002. Control of PERK eIF2α kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc Natl Acad Sci 99: 15920–15925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yángüez E, Rodriguez P, Goodfellow I, Nieto A 2012. Influenza virus polymerase confers independence of the cellular cap-binding factor eIF4E for viral mRNA translation. Virology 422: 297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatherajam G, Huang W, Flint SJ 2011. Export of adenoviral late mRNA from the nucleus requires the Nxf1/Tap export receptor. J Virol 85: 1429–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeam I, Cavatorta JR, Ripoll DR, Kang BC, Jahn MM 2007. Functional dissection of naturally occurring amino acid substitutions in eIF4E that confers recessive potyvirus resistance in plants. Plant Cell 19: 2913–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Alwine JC 2006. 19S late mRNAs of simian virus 40 have an internal ribosome entry site upstream of the virion structural protein 3 coding sequence. J Virol 80: 6553–6558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Kudchodkar SB, Alwine JC 2005. Effects of simian virus 40 large and small tumor antigens on mammalian target of rapamycin signaling: Small tumor antigen mediates hypophosphorylation of eIF4E-binding protein 1 late in infection. J Virol 79: 6882–6889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Abaeva IS, Marintchev A, Pestova TV, Hellen CU 2011a. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res 39: 4851–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Sweeney TR, Kafasla P, Jackson RJ, Pestova TV, Hellen CU 2011b. The mechanism of translation initiation on Aichivirus RNA mediated by a novel type of picornavirus IRES. EMBO J 30: 4423–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yueh A, Schneider RJ 1996. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev 10: 1557–1567 [DOI] [PubMed] [Google Scholar]
- Yueh A, Schneider RJ 2000. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev 14: 414–421 [PMC free article] [PubMed] [Google Scholar]
- Zaborowska I, Walsh D 2009. PI3K signaling regulates rapamycin-insensitive translation initiation complex formation in vaccinia virus-infected cells. J Virol 83: 3988–3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborowska I, Kellner K, Henry M, Meleady P, Walsh D 2012. Recruitment of host translation initiation factor eIF4G by the vaccinia virus ssDNA binding protein I3. Virology 425: 11–22 [DOI] [PubMed] [Google Scholar]
- Zhang B, Morace G, Gauss-Muller V, Kusov Y 2007. Poly(A) binding protein, C-terminally truncated by the hepatitis A virus proteinase 3C, inhibits viral translation. Nucleic Acids Res 35: 5975–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Moon A, Childs K, Goodbourn S, Dixon LK 2010. The African Swine Fever Virus DP71L protein recruits the protein phosphatase 1 catalytic subunit to dephosphorylate eIF2α and inhibits CHOP induction but is dispensable for these activities during virus infection. J Virol 84: 10681–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Ding H, Lu Z, Li Y, Pan Y, Ning T, Ke Y 2008. E3 ubiquitin ligase E6AP-mediated TSC2 turnover in the presence and absence of HPV16 E6. Genes Cells 13: 285–294 [DOI] [PubMed] [Google Scholar]
- Zuo X, Wang J, Yu P, Eyler D, Xu H, Starich MR, Tiede DM, Simon AE, Kasprzak W, Schwieters CD, et al. 2010. Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3′ UTR of turnip crinkle virus. Proc Natl Acad Sci 107: 1385–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]