Abstract
Background and Aims
Although the large variation in genome size among different species is widely acknowledged, the occurrence and extent of variation below the species level are still controversial and have not yet been satisfactorily analysed. The aim of this study was to assess genome size variation in six ploidy levels (2n = 3x–8x) of the polyploid Allium oleraceum over a large geographical gradient and to search for potential interpretations of the size variation.
Methods
The genome sizes of 407 individuals of A. oleraceum collected from 114 populations across Europe were determined by flow cytometry using propidium iodide staining. The genome size variation was correlated with spatial, climatic and habitat variables.
Key Results
The mean holoploid genome size (2C DNA) was 42·49, 52·14, 63·34, 71·94, 85·51 and 92·12 pg at the tri-, tetra-, penta-, hexa-, hepta- and octoploid levels, respectively. Genome size varied from a minimum of 2·3 % in the octoploids to a maximum of 18·3 % in the tetraploids. Spatial structuring of genome size was observed within the tetra- and pentaploids, where 2C DNA significantly increased with both latitude and longitude, and correlated with several climatic variables, suggesting a gradient of continentality. Genome size in hexaploids showed low variation, weak correlation with climatic variables and no spatial structuring. Downsizing in monoploid genome size was observed between all cytotypes except for heptaploids. Splitting populations into western and eastern European groups resulted in strong differences in monoploid genome size between groups in tetra- and pentaploids but not in hexaploids. The monoploid genome sizes of the cytotypes were similar in the western group but diverged in the eastern group.
Conclusions
Complex patterns of holoploid and monoploid genome size variation found both within and between A. oleraceum cytotypes are most likely the result of several interacting factors, including different evolutionary origins of cytotypes via hybridization of parental combinations with different genome sizes in the south-western and south-eastern part of Europe, introgression between cytotypes, and antropic dispersal. The role of broad-scale and fine-scale environmental variables in shaping genome size is probably of minor importance in A. oleraceum.
Keywords: Allium oleraceum, Amaryllidaceae, climate, Europe, flow cytometry, genome size variation, geophytes, polyploidy, spatial distribution
INTRODUCTION
The nuclear DNA content (2C-value; holoploid genome size) in angiosperms ranges more than 2300-fold from only 0·130 pg for Genlisea margaretae Hutch. (Greilhuber et al., 2006) to 304·5 pg for Paris japonica (Franch. & Sav.) Franch. (Pellicer et al., 2010). This range in C-value is not entirely related to the complexity of the organism or the number of genes contained in its genome (Gregory, 2005). This variation in nuclear DNA content and its lack of correlation with gene number/organism complexity was deemed wholly counterintuitive before the discovery of non-coding DNA and became known as the ‘C-value paradox’ (Gregory, 2001).
The amount of nuclear DNA and genome size are important adaptively relevant biodiversity parameters with fundamental biological importance (Bennett and Leitch, 2011). Diverse comparative studies based on large data samples at the species level showed that genome size correlates with a range of features at many hierarchical levels, ranging from the nucleus and cell to the organism (i.e. ‘nucleotype theory’; Bennett, 1972), including cell size (Cavalier-Smith, 2005), cell division rate (Francis et al., 2008), seed size and mass (Beaulieu et al., 2007b), photosynthetic rate (Beaulieu et al., 2007a), leaf cell size and stomatal density (Beaulieu et al., 2008; Hodgson et al., 2010), life forms and geographical distribution (Ohri, 2005; Veselý et al., 2012), and variation in plant phenology (Labani and Elkington, 1987; Veselý et al., 2012). Based on an extensive data analysis across environmental gradients, Knight et al. (2005) postulated that genome size does not generate a consistent causative effect across the whole genomic size range and that only species with large genomes are under-represented in extreme environments. Knight et al. (2005) also encouraged further examination of the genome size distribution across abiotic gradients, perhaps even including analyses across gradients of elevation and latitude over a range of habitat types. Using the same concept, small genomes have been suggested as a prerequisite for plant invasiveness because species with low nuclear DNA content usually produce many light seeds and are often quickly established in disturbed habitats (Bennett, 1987; Rejmánek, 1996).
The occurrence and extent of the genome size variation below the species level are still controversial and have not yet been satisfactorily analysed (Ceccarelli et al., 2011). Initially, a high degree of genome constancy was expected within species (Swift, 1950). The variation in genome size within species was attributed to chromosomal variations and rearrangements (e.g. polyploidy, aneuploidy, B-chromosomes, sex chromosomes; Greilhuber, 1998). However, more than 50 early studies, often based on densitometry or cytofluorometry techniques, observed intraspecific variation in genome size other than that due to chromosome polymorphisms. The majority of these reports were not confirmed by subsequent investigations using best-practice methodology (reviewed by Greilhuber, 1998, 2005). Therefore, despite the dynamic nature of the evolution of genome size (Lysák et al., 2009), its size within the same evolutionary unit seems to be stabilized by internal mechanisms, and as a consequence, intraspecific variation in genome size is quite rare (Loureiro et al., 2010). However, using improved methodology, the existence of intraspecific or even intrapopulation variation and differences in genome size has been clearly documented for several species during the last decade (Šmarda and Bureš, 2010, and references therein). Due to the nucleotypic effect, a non-random distribution of genome sizes may also exist within species (Knight et al., 2005), disadvantaging larger genomes in environments limited by the duration of the period with favourable temperatures, e.g. at higher latitudes and altitudes. Šmarda and Bureš (2010), however, argued that the small differences in genome size within species are most likely of minor importance compared with other components of plant fitness.
Polyploidy, a widespread phenomenon among flowering plants (Wendel, 2000), has been viewed as a major source of increasing genome size. Several studies have shown that although the total DNA content increases on average with an increase in ploidy level, the DNA content of the non-replicated monoploid genome (i.e. the 1Cx value sensu Greilhuber et al., 2005) can respond to polyploidization in different ways, including frequent decreases and increases or constancy within a polyploid series (Leitch and Bennett, 2004). Therefore, detecting the patterns of change in the 1Cx values within a given taxonomic group may be useful in comparisons across polyploid complexes (Šmarda et al., 2008a; Balao et al., 2009) and may allow for inferences about phylogenetic history. The holoploid and monoploid genome sizes also have potential as taxonomically significant traits at the lower taxonomic levels (Murray, 2005; Šmarda et al., 2008b) or for the identification of both heteroploid and homoploid hybrids (e.g. Mahelka et al., 2005; Ekrt et al., 2010; Loureiro et al., 2010).
In the present study, an analysis of the genome size variation (both 2C and 1Cx) of cytotypes of Allium oleraceum is performed using flow cytometry. Allium oleraceum belongs to section Codonoprasum Reichenb., which represents an evolutionarily young group of bulbous geophytes of the genus Allium (Friesen et al., 2006) consisting of diploid and polyploid species. Based on morphological similarity, A. oleraceum is considered to be a member of an informal complex of A. paniculatum L. Species of this complex occur from the westernmost part of Macaronesia, northern Africa and the Iberian Peninsula through the whole Mediterranean area and Europe to Iran and south-western Siberia (Stearn, 1980; Friesen, 1987). Most of the species considered to be members of this complex consist of either diploid or diploid and polyploid cytotypes and occur in the southern latitudes. The only exclusively polyploid species with an extreme diversity of cytotypes of this complex is A. oleraceum. This species is represented by six ploidy levels ranging from tri- to octoploids (2n = 24, 32, 40, 48, 56, 64; Duchoslav et al., 2010; M. Duchoslav et al., unpubl. res.). It is common in the northern and central parts of Europe and extends southwards to the northern part of the Mediterranean (Meusel et al., 1965). It is the only species covering the whole northern part of the distribution area of this complex. Levan (1937) considered A. oleraceum to be an autopolyploid form of the diploid Allium paniculatum. However, reinspection of the crossing experiments in Levan's paper showed that the ‘synthetic’ tetraploid A. oleraceum obtained by Levan may be a result of interspecific hybridization through unreduced gametes, with several existing diploid candidates (see Duchoslav et al., 2010). A detailed screen of the distribution and ecology of the cytotypes of A. oleraceum in Central Europe (Duchoslav et al., 2010; Šafářová et al., 2011) showed a complex spatial pattern of cytotypes with a high representation of mixed populations. The results also provided evidence for ecological differentiation among cytotypes that together with the prevalence of asexual reproduction and localized dispersal contributed to the observed spatial patterns (Šafářová and Duchoslav, 2010). Hence, knowledge of the holoploid and monoploid genome size variation over a large geographical area, especially over contact zones between A. oleraceum and its presupposed progenitors (i.e. other members of the A. paniculatum group), may allow for inferences about the ecogeographical relationships and evolutionary history of polyploidy in A. oleraceum.
The main goals of this study were: (1) to estimate the genome size of the cytotypes and evaluate the existence of intra- and interpopulation variations in genome size, (2) to investigate relationships with geographical and environmental parameters, and (3) to make reasonable inferences about the ecological or evolutionary interpretation of genome size variation in this complex. This study is exceptional because it simultaneously considers many cytotypes over a large geographical area.
MATERIALS AND METHODS
Plant sampling and environmental data
To estimate genome size variation, 114 populations and 407 individuals of tri-, tetra-, penta-, hexa-, hepta- and octoploid cytotypes were selected using a stratified random procedure from our collection of Allium oleraceum L. populations obtained during 2001–2009 throughout Europe and planted in the garden of the Palacký University in Olomouc, Czech Republic (Supplementary Data Table S1). We attempted to cover most of the distribution areas, elevations, and climate and habitat types in which each cytotype grows. At least three (in some cases only two) randomly selected plants per population were analysed (mean ± s.d. 3·6 ± 1·9; range 2–10 plants per population). Before the measurements, all samples were cultivated under the standard homogeneous conditions of the experimental garden for two or more years, followed by 3 months in a cold glasshouse.
At each sampled site, the following environmental variables that were identical to those used in previous study (Duchoslav et al., 2010) were recorded. (1) Habitat naturalness, i.e. vegetation at the site, was classified according to the degree of anthropic impact as either ‘human-impacted’ (vegetation strongly influenced or created by humans, typically with higher proportions of ruderal or alien species; coded as 0) or ‘natural’ (natural and seminatural vegetation without a strong anthropic influence; coded as 1). (2) The altitude was estimated using a GPS instrument on-site or later according to the coordinates based on the Google Earth application (Google Inc.).
To characterize each population climatically, a set of climate layers – such as annual mean temperature, maximal temperature of warmest month, minimal temperature of coldest month, annual precipitation, precipitation of wettest month and precipitation of driest month – with a spatial resolution of 1 km2 was obtained from the WORLDCLIM database (Hijmans et al., 2005). For each population, the climatic data were derived from this database using the ArcView GIS 3.1 software (Environmental Systems Research Institute, Inc., Redlands, CA, USA).
Flow cytometry and chromosome counts
The absolute and relative (DNA ploidy level) nuclear DNA contents were determined by flow cytometry (FCM) using the method of internal standardization. The nuclei of the standard and sample were isolated, stained and analysed together. Triticum aestivum ‘Saxana’ was used as an internal standard, and it was calibrated against the plant reference standard Secale cereale ‘Daňkovské’ with 2C DNA 16·19 pg (Doležel et al., 1998). The relative fluorescence intensity of the stained nuclei was analysed using a Partec PAS instrument (Partec GmbH, Münster, Germany) equipped with an argon ion laser (488 nm). Young fresh leaves of the internal standard and the Allium sample were chopped with a new razor blade in a Petri dish containing 1 mL of ice cold LB01 buffer (Doležel et al., 2007). The suspension was filtered through a 42-μm nylon mesh. The nuclei suspension was supplemented with RNase and stained with propidium iodide (PI) (both 50 µg ml−1). Histograms of the fluorescence intensity were registered over 512 channels. The plants with counted chromosomes were used for specification of the internal standard-sample position. The fluorescence ratios between the positions of the sample and internal reference standard peaks were 1·18–1·29, 1·40–1·69, 1·74–1·98, 2·02–2·23, 2·37–2·59 and 2·64–2·77 for the 3x–8x cytotypes, respectively.
The following measurement strategy was chosen to ensure the maximum validity of the absolute nuclear DNA content estimation: (1) each sample was measured by the same operator at least three different times on different days to minimize potential instrumental drift; (2) all measurements were made over 1 month when the plants were in an identical phenological phase of development (April–May 2007–2011); (3) at least 5000 nuclei per sample were recorded; (4) if an outlier was detected, the most remote value was discarded, and the sample was re-analysed. Because of the huge content of the cytosolic compounds, CV for the G0/G1 peaks of the standard and Allium samples varied between 3 and 5 % (mean CV ± SE of the standard was 4·11 ± 0·02 %, mean CV ± SE of the samples were 4·03 ± 0·06, 4·10 ± 0·04, 4·09 ± 0·02, 4·31 ± 0·03, 4·09 ± 0·31 and 6·02 ± 0·93 % for 3x–8x, respectively). The analysis of variation of repeated measurements showed that the approximate measurement inaccuracy did not exceed ±2·5 % in 95 % of the accessions for all the ploidy levels studied. Therefore, any variation beyond the arbitrary fluctuation (5·0 %) should be considered as truly existing. The absolute 2C DNA content (= holoploid genome size) of a sample was calculated based on the values of the G0/G1 peak means {[(sample G0/G1 peak mean)/(standard G0/G1 peak mean)] × standard 2C DNA content; pg DNA} (Doležel and Bartoš, 2005). The DNA content of the non-replicated monoploid genome (i.e. the 1Cx value = monoploid genome size, sensu Greilhuber et al., 2005) was estimated as the 2C DNA content divided by the ploidy level. To verify the intraspecific DNA variation and avoid artefacts in the measurements, we simultaneously analysed two samples with markedly different DNA contents within the same ploidy level. The appearance of separate peaks was considered to be proof of true differences in the amount of nuclear DNA (Greilhuber et al., 2007). Chromosome number determination followed the same procedure as described by Duchoslav et al. (2010). Chromosomes were counted in 25 plants covering the genome size range of each cytotype (cytotype/number of counted plants: 3x/4; 4x/4; 5x/9; 6x/4, 7x/3; 8/1).
Statistical analyses
The cytotypes (except for octoploids) were tested for intra- and interpopulation variations in DNA content employing mixed model ANOVA. An overall test on the mean of repeated measurements of the genome size of individuals was performed with cytotype as a fixed factor and population as a random factor nested within cytotype. The proportion of total variance in genome size attributable to interpopulation variation was calculated for each cytotype separately in a mixed model ANOVA as estimated variance components in terms of percentages of total variance. The Tukey HSD multiple comparison test was used for comparison of the mean genome sizes of the cytotypes. The range of the DNA content (‘intra-ploidy variation’) was calculated as the highest/lowest DNA content ratio found in individuals of the respective cytotype. ANOVAs were performed using Statistica 10 (StatSoft Inc., Tulsa, OK, USA).
Independence of geographical distance and genome size differences between the populations was tested using the Mantel test (Legendre and Legendre, 1998) for each cytotype (3x–7x) separately. The spatial relationship of the monoploid genome size (1Cx) of the cytotypes with large ni (4x–6x) was tested with the Procrustes analysis using the PROTEST software (Peres-Neto and Jackson, 2000) with the significance test by Jackson (1995) based on 9999 permutations. The data settings followed the approach used by Šmarda and Bureš (2006). Significant results of this test indicate that there is a geographical relationship between the genome sizes of the cytotypes under comparison.
The relationship between genome size and each geographical and environmental variable was first tested using Spearman's correlation coefficient and the Kruskal–Wallis test. Due to the frequent interrelationships between geographical and climatic variables and possible existence of spatial autocorrelation typical of ecological data (Legendre and Fortin, 1989), we evaluated how the importance of different environmental variables changed depending on scales of spatial dependence accounted for by the regression models using procedures explained in detail by Lichstein et al. (2002). In all regression models mentioned below, the response variable was holoploid genome size (2C) for each cytotype with large ni (i.e. 4x, 5x and 6x) to fulfil the conditions of the statistical tests. Preliminary tests showed strong collinearity between temperature variables and between precipitation variables (correlation coefficients were >0·80 in all cases) that confounded the analyses. Therefore, only three climatic and habitat variables were selected and used as explanatory variables in all regression models: mean annual temperature, annual precipitation and habitat naturalness. First, ordinary least squares multiple regression (OLS) was fitted to environmental variables (hereafter, ‘OLS environment’) ignoring both broadscale spatial trend and fine-scale autocorrelation. Secondly, we used OLS including both environmental variables and trend variables (latitude, longitude) to analyse how OLS environment models changed after accounting for broadscale trend (‘OLS trend/environment’). Just first-degree polynomial terms of the spatial coordinates of the sites were used in the analyses. Finally, we examined how OLS trend/environment models changed when we accounted for fine-scale autocorrelation (‘CAR trend/environment’) using the conditional autoregressive (CAR) model (Cressie, 1993), which allows us to analyse fine-scale autocorrelation, i.e. phenomena occurring in a geographical area immediately surrounding the site analysed. An appropriate weight function was selected based on model fit and how well the model accounted for autocorrelation in the residuals. Annual precipitation was square-root transformed before analyses. One tetraploid population (No. 31) was excluded from analyses due to extreme values of climatic variables that distorted analyses.
Non-hierarchical K-mean clustering (Legendre and Legendre, 1998) was performed to obtain insights into the relationships between populations characterized by average monoploid genome sizes. Regression and clustering analyses were performed using SAM 4·0 (Rangel et al., 2010).
RESULTS
Genome size variation
Significant effects of cytotype (F = 586·2, P < 0·001) and population (F = 19·9, P < 0·001) on the holoploid genome size were found by mixed model ANOVA. As expected, the mean 2C DNA content significantly differed between any two cytotypes (Table 1, Fig. 1A). The monoploid genome size was significantly dependent on the ploidy level (F = 130·4, P < 0·001), and the genome downsizing between two successive cytotypes was 8·0, 2·8, 5·4 and 5·9 % in the 3x/4x, 4x/5x, 5x/6x and 7x/8x cytotypes, respectively. The only exception was the heptaploids, which did not differ in 1Cx from hexaploids (Fig. 1B).
Table 1.
Variation in the holoploid (2C DNA) and monoploid genome sizes (1Cx DNA) for the six ploidy levels of Allium oleraceum using standard Triticum aestivum ‘Saxana’ (2C = 34·2388 pg)
| Ploidy level (2n) | No. of populations/no. of individuals analysed | 2C DNA (pg; mean ± s.d.) | 2C DNA range (pg; min/max) | 1Cx DNA (pg; mean) | Intraploidy variation (%) | Proportion of total variance in genome size attributable to interpopulation variation (%) |
|---|---|---|---|---|---|---|
| 3x | 8/23 | 42·49 ± 0·92 | 40·52/43·55 | 14·16 | 7·5 | 74·9* |
| 4x | 26/77 | 52·14 ± 2·65 | 47·96/56·74 | 13·03 | 18·3 | 92·5*** |
| 5x | 50/195 | 63·34 ± 1·65 | 59·59/66·15 | 12·67 | 11·0 | 74·8*** |
| 6x | 24/96 | 71·94 ± 0·99 | 69·10/73·66 | 11·99 | 6·6 | 45·8* |
| 7x | 5/12 | 85·51 ± 1·96 | 82·98/88·66 | 12·22 | 6·8 | 52·4* |
| 8x | 1/4 | 92·12 ± 0·99 | 91·12/93·25 | 11·50 | 2·3 | – |
Significance of the interpopulation variation in 2C DNA: *P ≤ 0·05, **P ≤ 0·01, ***P ≤ 0·001.
Fig. 1.
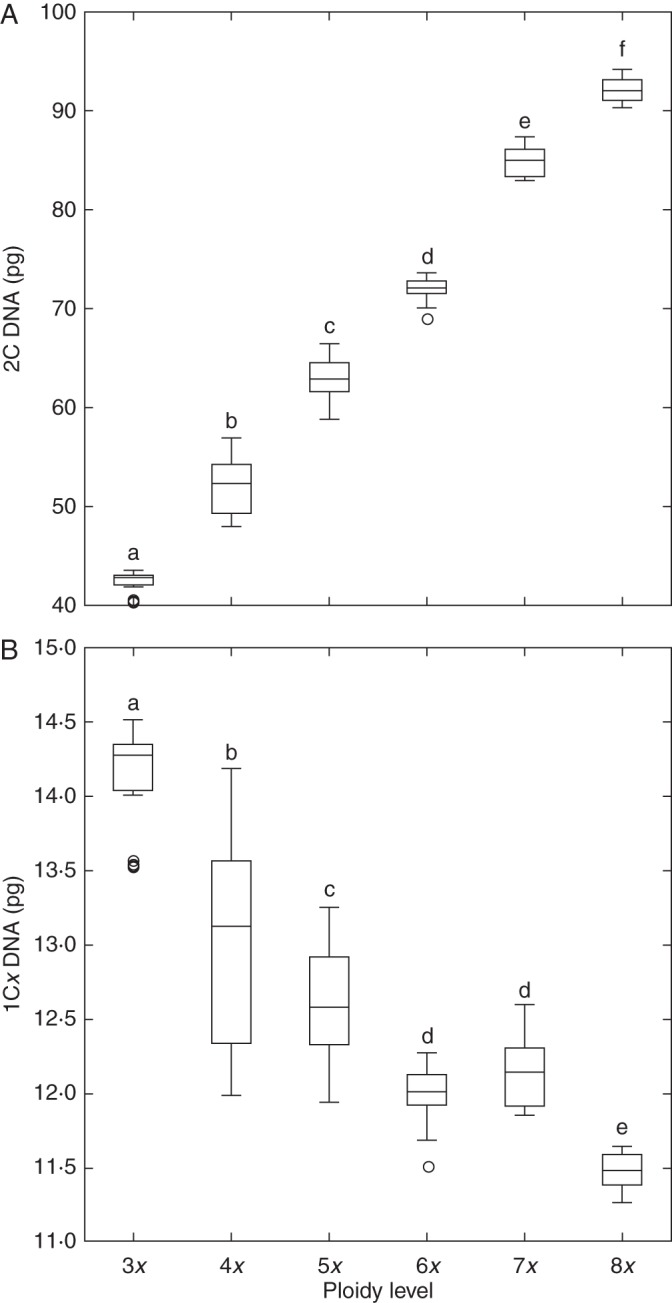
Box plot of (A) the holoploid (2C DNA) and (B) monoploid (1Cx DNA) genome sizes in different cytotypes of Allium oleraceum. Those means not significantly different at P ≤ 0·05 are indicated by the same letter (Tukey's HSD test).
The intra-ploidy variation in genome size was relatively large in the tetraploids (18·3 %) and pentaploids (11·0 %), and no outliers were observed. The divergence in the estimated genome size between the populations was confirmed by simultaneous FCM analyses (Supplementary Data Fig. S1). A smaller intra-ploidy variation in genome size was discovered in the other cytotypes (≤7·5 %), which decreased further after the elimination of populations with extreme DNA content, e.g. in the hexaploids and triploids. Within the tri-, tetra- and pentaploid cytotypes, the majority (≥74 %) of the total variation in genome size was attributable to interpopulation variation. However, the interpopulation variation was apparently less pronounced in the hexa- (45·8 %) and heptaploids (52·4 %) than in the other cytotypes (Table 1). Intrapopulation variation in 2C DNA content was negligible.
Chromosome counts of 25 plants agreed with DNA-ploidy levels inferred from holoploid genome size measurements, even in plants with extreme values of genome size. No aneuploids were observed (Supplementary Data Table S1).
Ecogeographical pattern of genome size variation
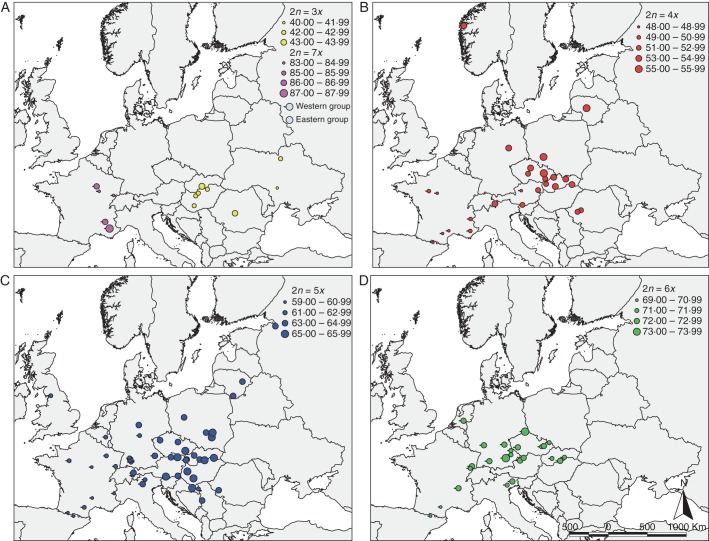
The dissimilarities in genome size were significantly related to geographical distance between the populations in the tetra- (Mantel test statistics, rM = 0·197, P = 0·031) and pentaploids (rM = 0·234, P = 0·013), which indicates that populations situated at short distances from each other have similar genome sizes. However, no spatial structuring of the genome size was found in the triploids (rM = 0·058, P = 0·277) and hexaploids (rM = 0·140, P = 0·116). The negative correlation between dissimilarities in genome size and geographical distance in the heptaploids (rM = –0·397, P = 0·043) is probably a sampling artefact due to the low number of populations analysed. A strong spatial coincidence of the genome size of the tetra- and pentaploids and the penta- and hexaploids was revealed with the Procrustean analysis (both P < 0·001). However, no spatial coincidence of the genome size was found between the tetra- and hexaploids (P = 0·242).
The relationships between the geographical variables, ecological variables, climatic variables and genome size of the populations were analysed (Table 2, Figs 2 and 3). In the triploids, the positive correlations between genome size and precipitation and altitude are most likely the result of confounding altitude and geography. Significant correlations between temperature and genome size in the heptaploids are most likely the result of a low sample size. In the tetra- and pentaploids, genome size was significantly correlated with several climatic variables, with the tendency of plants with larger genomes to occur in drier conditions with lower minimal winter temperatures at lower altitudes. In contrast, genome size in the hexaploids was not correlated with any variable. With regard to local-scale environmental variables, only pentaploids with a smaller genome size occurred in natural habitats, whereas those with a larger genome size occurred in human-impacted habitats (Table 2).
Table 2.
Relationship between the holoploid genome size (2C DNA) in the tri-, tetra-, penta-, hexa- and heptaploid cytotypes of Allium oleraceum and geographical, climatic and ecological variables. Values are for Spearman correlation coefficient (rS) except habitat naturalness, which give results of Kruskal–Wallis test
| Latitude (°N) | Longitude (°E) | Altitude (m) | Annual mean temperature (°C) | Max. temp. of warmest month (°C) | Min. temp. of coldest month (°C) | Annual precipitation (mm) | Precip. of wettest month (mm) | Precip. of driest month (mm) | Habitat naturalness | |
|---|---|---|---|---|---|---|---|---|---|---|
| 3x; n = 8 | –0·48 | –0·33 | 0·95 | 0·12 | –0·38 | –0·05 | 0·79 | 0·68 | 0·17 | 0·44 |
| P | 0·233 | 0·420 | < 0·001 | 0·779 | 0·349 | 0·911 | 0·021 | 0·062 | 0·690 | 0·505 |
| 4x; n = 26 | 0·75 | 0·64 | –0·24 | –0·27 | –0·16 | –0·37 | –0·36 | –0·10 | –0·47 | 2·22 |
| P | < 0·001 | < 0·001 | 0·234 | 0·183 | 0·430 | 0·059 | 0·094 | 0·632 | 0·015 | 0·136 |
| 5x; n = 50 | 0·37 | 0·74 | –0·35 | –0·04 | 0·35 | –0·62 | –0·60 | –0·19 | –0·67 | 7·12 |
| P | 0·009 | < 0·001 | 0·013 | 0·804 | 0·014 | < 0·001 | < 0·001 | 0·189 | < 0·001 | 0·010 |
| 6x; n = 24 | 0·34 | –0·11 | 0·167 | –0·10 | –0·31 | 0·20 | 0·12 | –0·06 | 0·22 | 1·52 |
| P | 0·103 | 0·616 | 0·437 | 0·639 | 0·140 | 0·354 | 0·582 | 0·768 | 0·296 | 0·230 |
| 7x; n = 5 | –0·70 | 0·20 | 0·70 | 0·90 | 1·00 | 0·41 | –0·30 | 0·10 | –0·80 | 0·23 |
| P | 0·188 | 0·747 | 0·188 | 0·037 | < 0·001 | 0·493 | 0·624 | 0·873 | 0·104 | 0·667 |
Significant results (P ≤ 0·05) are in bold.
Fig. 2.
Observed landscape pattern of the holoploid genome size (2C DNA) of the Allium oleraceum populations studied: (A) triploids and heptaploids, (B) tetraploids, (C), pentaploids and (D) hexaploids. The size of the symbol reflects mean genome size of population samples. Note the non-equidistant intervals of 2C DNA in some cases due to missing values. Two groups of populations (western and eastern) based on results of non-hierarchical K-mean clustering of monoploid genome size are depicted by different symbols, as indicated in (A).
Fig. 3.
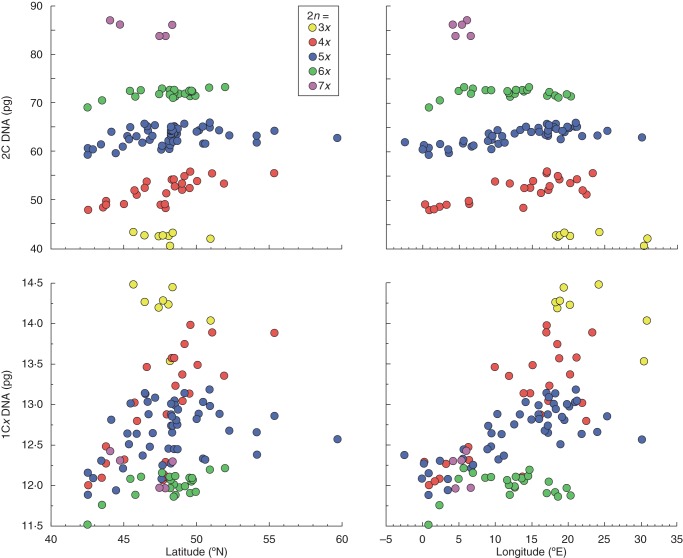
Relationships between geographical variables (latitude, longitude) and genome size (2C DNA, 1Cx DNA) in different cytotypes of Allium oleraceum.
OLS environmental models were significant in tetra-, penta- and hexaploids (Table 3). In tetra- and pentaploids, genome size increased with decreasing annual precipitation and annual temperature (only in tetraploids) and was lower in natural than in anthropogenic habitats. On the other hand, genome size in hexaploids increased with increasing precipitation. After incorporating spatial trend variables (latitude, longitude; OLS trend/environment model), east–west and south–north trends in genome size were significant in tetraploids, but only an east–west trend was significant in pentaploids. However, the effect of climate mostly disappears, suggesting that climatic variables were correlated with the spatial trend, i.e. represent a gradient of continentality. Trend variables also explain sufficient additional variation uncorrelated with environmental variables. A significant influence of habitat naturalness suggests a non-spatially structured habitat effect. Despite the weakly significant overall test (P = 0·043), no variable had a significant partial effect on genome size in the OLS trend/environment model in hexaploids. Incorporating trend resulted in disappearance of the significance of annual precipitation, suggesting its correlation with spatial trend (Table 3). CAR trend/environment models were significant only in tetra- and pentaploids with just trend variables (tetraploids: latitude, longitude; pentaploids: longitude) remaining significant. The influence of habitat naturalness on genome size appeared to be non-significant due to autocorrelation of genome size between neighbouring populations (Table 3).
Table 3.
Parameter estimates (standardized regression coefficients) for climatic, habitat and geographic variables in regression models of the variation in the holoploid genome size (2C DNA) in the tetra-, penta- and hexaploid Allium oleraceum populations ignoring both broad-scale spatial trend (latitude, longitude) and fine-scale autocorrelation (OLS environment model), accounting for broad-scale trend (OLS trend/environment model) and for both broad-scale trend and fine-scale autocorrelation (CAR trend/environment model)
| Variable | OLS environment model | OLS trend/environment model | CAR trend/environment model |
|---|---|---|---|
| 2n = 4x | |||
| Habitat naturalness | –0·335* | –0·277* | –0·204 |
| Annual precipitation | –0·765*** | –0·228 | –0·083 |
| Annual mean temperature | –0·579** | –0·245 | –0·099 |
| Latitude | – | 0·354* | 0·471* |
| Longitude | – | 0·430** | 0·453* |
| R2 | 0·526 | 0·810 | 0·722 |
| 2n = 5x | |||
| Habitat naturalness | –0·273* | –0·254* | –0·179 |
| Annual precipitation | –0·498*** | 0·010 | 0·001 |
| Annual mean temperature | –0·055 | 0·163 | 0·011 |
| Latitude | – | 0·115 | 0·081 |
| Longitude | – | 0·692*** | 0·758*** |
| R2 | 0·366 | 0·650 | 0·922 |
| 2n = 6x | |||
| Habitat naturalness | 0·300 | 0·182 | 0·038 |
| Annual precipitation | 0·515* | 0·390 | 0·467 |
| Annual mean temperature | –0·423 | –0·336 | –0·445 |
| Latitude | – | 0·254 | 0·315 |
| Longitude | – | –0·430 | –0·296 |
| R2 | 0·344 | 0·524 | 0·677 |
Significant regression coefficients (P ≤ 0·05) are in bold. *P ≤ 0·05, **P ≤ 0·01, ***P ≤ 0·001; R2 = adjusted coefficient of determination.
The monoploid genome size of tetra-, penta- and hexaploid cytotypes was similar in the western part of Europe but diverged towards the eastern part of Europe (Fig. 3). Combining the cytotypes and geographical structuring along the main spatial trend (longitude) resulted in eight groups (Fig. 4) differing in monoploid genome size (ANOVA, F = 540·9, P < 0·001). The monoploid genome size of the tetra- and pentaploid cytotypes was similar in the western part of Europe but differed in the eastern part of Europe. In contrast, the western and eastern hexaploids did not differ in their 1Cx values (Figs 3 and 4). Non-hierarchical K-mean clustering of monoploid genome size revealed two groups of populations differing clearly in east–west geographical direction with the contact zone running through Central Europe (Fig. 2).
Fig. 4.
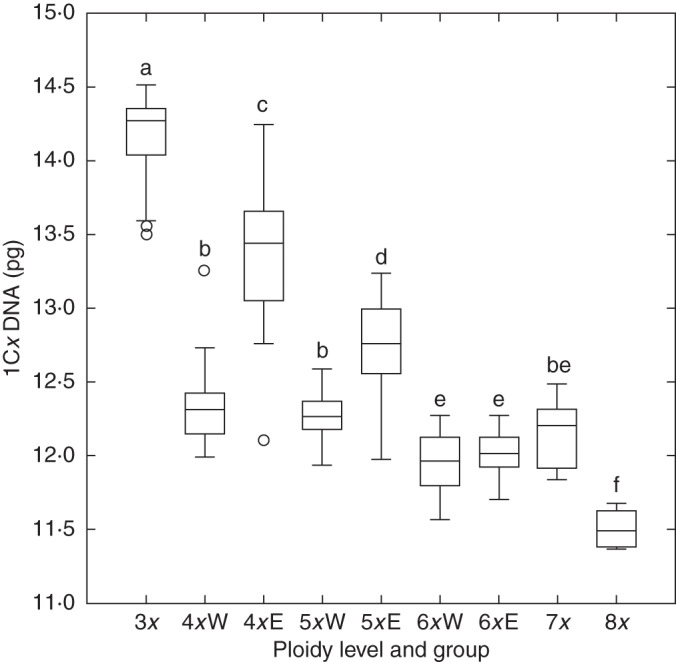
Box plot of the monoploid genome size (1Cx DNA) in Allium oleraceum cytotypes from the western (‘W’) and eastern (‘E’) part of the European range. The border between the regions was set arbitrarily at 8°E. Note that triploids occur only in eastern Europe and the hepta- and octoploids only in western Europe. Those means not significantly different at P ≤ 0·05 are indicated by the same letter (Tukey's HSD test).
DISCUSSION
Our nuclear DNA values of five out of six cytotypes represent primary estimates. However, the estimation of genome size in octoploids must be considered to be preliminary because of the high coefficient of variation obtained. Previously, only three nuclear DNA values for A. oleraceum had been gathered, with two determined for pentaploids using Feulgen densitometry. Although the estimate of Baranyi and Greilhuber (1999; 2C = 60·37 pg) was in good agreement with our estimates, the primary estimate 2C = 52·78 pg for pentaploids by Labani and Elkington (1987) is suspicious and should be discarded. The recent estimate of genome size by FCM in plants of unknown ploidy level sampled at one locality in the Czech Republic (Veselý et al., 2012) resulted in 2C = 57·88 pg, which is just between our 2C ranges for tetra- and pentaploids.
Our study revealed considerable intraspecific variation in genome size. In addition to confirmation of the expected intercytotype differences, the major part of the variation in genome size within each of the 3x–7x cytotypes was attributable to interpopulation variation, although this variation was apparently lower in the tri-, hexa- and heptaploids (approx. 6–8 %) than in the tetra- and pentaploid cytotypes (approx. 11–18 %). However, the within-population variation was not proven to be significant in any of the cytotypes studied. The simple explanation of the intracytotype genome size variation is based on methodological artefacts, usually associated with instrumental drift and/or dissimilar levels of secondary metabolites that may interfere with DNA fluorochromes (Greilhuber, 2005; Loureiro et al., 2006). Allium oleraceum is a difficult-to-measure plant with a huge mucilage content that is mirrored in the coefficients of variance for the histograms of fluorescence intensity, which were slightly higher than those proposed by Doležel et al. (2007) for the genome size estimation. Consequently, the higher coefficient of variance causes insufficient sensitivity for this method to distinguish two individual peaks when individuals from the same population are measured simultaneously (Benson and Braylan, 1994; Balao et al., 2009). Another likely reason for the lack of within-population variation is the predominant asexual reproduction being most pronounced in the tri- and hexaploids (Ohryzek, 2007; Karpavičienė, 2012).
However, we are convinced that the intracytotype variation reported in the present study should be considered genuine as simultaneous analyses of samples from the same cytotype but with different 2C DNA always yielded histograms with a single bifurcated or two separate peaks, which is considered the most convincing evidence for real differences in the nuclear DNA content (Greilhuber, 2005). Intraspecific or even intrapopulation genome size variation has been recently documented in several plant species using appropriate methodology (e.g. Šmarda and Bureš, 2006, 2010; Šmarda et al., 2008a; Balao et al., 2009; Slovák et al., 2009; Cires et al., 2010; Benor et al., 2011; Kim et al., 2012; Olšavská et al., 2012).
Possible sources of inter- and intracytotype variations in genome size and correlates of genome size with environment and space on a large spatial scale: role of adaptive and non-adaptive explanations
Apart from polyploidy, variation in intraspecific genome size could be attributed to several sources specific to respective species but may also mutually interact. Regarding A. oleraceum, part of the intracytotype variation in the 2C DNA can be explained by aneuploidy (Greilhuber, 1998), which has been rarely found in chromosome counting in seedlings from pentaploids by Fialová (1996) and Jandová (2010). However, in adult A. oleraceum plants, only euploid chromosome numbers are reported in the literature (Fialová, 1996; Karpavičienė, 2007; Duchoslav et al., 2010), and these numbers have also been found in all the individuals analysed here, even in the simultaneously measured plants with the greatest differences in genome size. Moreover, the geographical component of genome size variation seems to contradict this scenario, at least on a continental scale.
Alternatively, genome size variation may be related to the amount of repetitive and non-coding DNA, which is considered the major mechanism responsible for changes of genome size (Leitch and Bennett, 2004; Bennetzen et al., 2005). Given previous evidence for chromosome structural variation (i.e. length of chromosomes) in the related species of section Codonoprasum, which was related to varying amount of heterochromatin (Loidl, 1983), we suggest that variation in heterochromatin composition (Meagher and Vassiliadis, 2005; Biémont, 2008; Šmarda and Bureš, 2010) is a probable candidate to explain the variation in genome size in A. oleraceum (see also Ohri et al., 1998). While genome size variation could be generated by lineage-specific differences in the molecular mechanisms of DNA amplification and removal, variation in nuclear DNA content can then serve as the substrate for fitness-based selection (Bennetzen et al., 2005). Hence, the variation in inter- and intraspecific genome size could be a product of local adaptation along ecogeographical gradients (Knight et al., 2005, and references therein). This is a consequence of the fact that amplification of large genomes requires more energy and longer periods of time (Bennett, 1987), which could be disadvantageous in time-limited habitats with shorter vegetation periods, e.g. at higher latitudes or altitudes, or in dry environments (Šmarda and Bureš, 2010). With regard to the spatial and ecological correlates, the correlation of geographical location and climatic variables with genome size was strong in the tetra- and pentaploids but small in the other cytotypes of A. oleraceum. Such differences in interpopulation genome size variation between cytotypes may be partly influenced by the different sizes of the studied geographical ranges, with the tri- and heptaploids seeming to have smaller geographical ranges and occurring only in either eastern or western Europe, respectively (Fig. 2; see also Duchoslav et al., 2010, M. Duchoslav et al., pers. observ.). Hence, the observed correlations may have limited value in the triploids and heptaploids. In the tetra- and pentaploids, genome size increased with decreasing precipitation and temperature, which were spatially structured along a SW–NE geographical gradient. Potentially, genome size mirrors complex gradients in the climatic conditions from oceanic to more continental zones. Moreover, the variation seems more or less continuous along these gradients at first glance. This observation seems to fit well with the results of a study by MacGillivray and Grime (1995), who found a positive relationship between genome size and frost resistance in British herbaceous plants This correlation is related to the advantage of growth by preformation of cells and whole organs by cell division conducted in the rest period of the preceding season (i.e. summer, autumn) and cell expansion early in the spring at low temperatures, that is typical of many geophytes (Veselý et al., 2012).
Most surprising is the relatively low variation in the genome size of hexaploids. Low intraploidy variation has also been observed in other plants studied in detail (e.g. Lysák et al., 2000), but despite the large variation found in the tetra- and pentaploids, the genome size stability in the hexaploids is striking. Thus, we expected and also corroborated almost no relationships between genome size and ecogeographical variables. However, in the regression analysis, weak but significant reverse correlations of the genome size of the hexaploids were found with annual precipitation, which was in contrast to that observed in the tetra- and pentaploids. However, this trend was weak and cannot have any biological meaning. Alternatively, under similar ecoclimatic conditions, selection may operate on hexaploids in a different way than on the tetra- and pentaploids.
Correlations between genome size and ecogeographical variables have also been reported for several plant species from Europe, but no general relationship has emerged. Similar to our results, Pečinka et al. (2006) and Šmarda and Bureš (2006) found an increase in DNA content towards the eastern and south-eastern part of Central Europe in Koeleria macrantha var. majoriflora and Festuca pallens, respectively. However, the authors explained the observed trends either by the different evolutionary history of eastern and western populations or by the tendency of plants with larger genomes to persist in the south-eastern areas where periglacial steppes occurred during harsh conditions of the Glacial 20 000 years ago, respectively. Similarly, two recent studies also found correlations between genome size and longitude in the European context at the intrageneric (Chrtek et al., 2009) and intraspecific (Slovák et al., 2009) levels but explained them by the basal phylogenetic divergence into species of eastern and western European origins and the existence of several independent lineages with unique evolutionary histories, respectively. In contrast, the fluorescence intensity of the tetraploid Lythrum salicaria was negatively correlated with population location along an west–east gradient in Europe (Kubátová et al., 2008) and plants of the Centaurea triumfetti and C. montana groups with larger genomes appeared in the southern parts of Central Europe at higher elevations (Olšavská et al., 2012). But the authors of these papers did not draw decisive conclusions on the cause of these spatial variations. Such contradictory results may potentially arise because most studies do not consider the full geographical and/or ecological ranges of the studied plants, which may show a unimodal trend of genome size over entire environmental gradients (Knight et al., 2005). Moreover, our analysis did not account for the full geographical ranges of the cytotypes because of the limited availability of samples from eastern Europe and because the weak lack of fit for the genome size of pentaploids (and partly also of tetraploids), which is visible at higher latitudes/longitudes, suggests a partly divergent trend from the one fitted by linear regression (Fig. 3).
There is, however, an alternative explanation for the genome size variations observed, supposing a complex evolutionary history of the studied species (Loureiro et al., 2010; Šmarda and Bureš, 2010) due to non-recognized phylogenetic components (i.e. the so-called ‘orthodox’ variation; Greilhuber, 1998, 2005). We think that the complex pattern of genome size variation in the cytotypes of A. oleraceum could be the result of multiple origins via independent crosses between different members of the A. paniculatum complex with at least two independent lineages – eastern and western – and various levels of gene flow within and between the cytotypes, as suggested by Duchoslav et al. (2010). At present, this hypothesis is supported by (1) the contrasting ecogeographical patterns and amount of genome size variation in tetra- and pentaploids vs. hexaploids; (2) the lower variation of genome size in the tetra- and pentaploids in western Europe, which contrasts with the apparent variation in their genome size in Central Europe; and (3) divergence in monoploid genome size along a west–east longitudinal gradient in common cytotypes. We discuss several important points here in detail.
Longitude appears to be an important factor affecting genome size in tetra- and pentaploids. Consequently, the dominant east–west gradient in genome size observed by us in tetraploid and pentaploid A. oleraceum may potentially fit with the hypothesis of Bogdanovič et al. (2009), who suggest that there are two speciation centres in Europe in the autumn-flowering species of Allium sect. Codonoprasum: one in the western Mediterranean and the other in the eastern Mediterranean area. Adopting this concept for summer-flowering species of Allium sect. Codonoprasum, tetraploids (and perhaps also pentaploids) may thus have originated independently in the western and eastern Mediterranean from either the same or different progenitors differing also in genome size. The high genome size variation found by us in the tetraploids and partly also in the pentaploids in Central Europe suggests that those cytotypes are composed of fairly heterogeneous units potentially originating via various mechanisms, including: hybridization of different diploid progenitors, as experimentally documented by Levan (1937), and diversity of which is greater in eastern than in the western Europe, although their distribution is generally quite localized (Stearn, 1980; Brullo et al., 1991); secondary contacts between partially genetically different tetraploids as mentioned by Šafářová et al. (2011); and gene flow between cytotypes via the fusion of various forms of gametes (unreduced, reduced and partially reduced), which was experimentaly confirmed by Jírová (2007). Observed spatial relationships of genome size in the tetra- and pentaploids and the penta- and hexaploids may thus indicate an evolutionary link between ploidy levels, i.e. the possibility of repeated (recent) autopolyploidization events or gene flow between respective pairs of cytotypes. Yet, we cannot exclude the possibility of the selective pressure of climatic variables on genome size, and so the pattern observed by us at the continental scale may represent the result of interactions between non-adaptive (different evolutionary origins and spatial dynamics) and adaptive forces.
Concerning hexaploids, Duchoslav et al. (2010) suggest that their common occurrence and narrow ecological niche in the Czech Republic represent evidence of a recent range expansion of a newly established type. Except for two exceptionally low 2C values measured in plants from hexaploid populations in Spain and France, which may suggest a different polyploidization event, our results support this hypothesis. The low genome size variation in the hexaploids is most likely maintained almost exclusively by asexual reproduction (Ohryzek, 2007) and coincides with their low genetic (Staňková, 2005) and morphological variation (Burešová, 2012). A similar relationship between genome size variation and the amount of genetic variation was observed in, for example, apomictic triploids of Hieracium alpinum (Mráz et al., 2009).
When examining the variation in the monoploid genome size, we found that, with one exception, the ploidy levels of A. oleraceum differ on average from each other in their monoploid genome size, with 1Cx gradually decreasing (except for 7x) from the tri- to octoploid cytotypes. This finding might indicate the downsizing of genomes after polyploidization, a general trend observed in angiosperms, including various representatives of the genus Allium (Leitch and Bennett, 2004) due to several mechanisms, of which the deletion of repeated DNA sequences and the dynamics of transposable elements are considered to be the most important (Bennetzen et al., 2005). However, there is no general trend of either downsizing or upsizing within polyploid species, and the absence of downsizing in polyploids usually suggests (very) young neopolyploids (Bancheva and Greilhuber, 2006; Weiss-Schneeweiss et al., 2006; Cosendai et al., 2011; but see Suda et al., 2007), which are most likely of autopolyploid origin (Balao et al., 2009). Therefore, the differences in the monoploid genome size between cytotypes in A. oleraceum probably indicate some genomic differentiation and an ancestral origin rather than recent autopolyploidy. However, when the geographical scale of the variation of the monoploid genome size was taken into account, a more complicated pattern emerged. In short, cytotypes have similarly sized monoploid genome sizes in western but not in eastern Europe (see Figs 3 and 4). The most parsimonious explanation for the similarly sized 1Cx values of the 4x–7x cytotypes from (south-)western Europe is their (recent) autopolyploid origin, whereas the differently sized genomes of the eastern 3x–6x cytotypes could be the result of independent polyploidization events, including introgression with other taxa of the A. paniculatum group, and/or ancient autopolyploidization. Unfortunately, rigorously testing this hypothesis is difficult because no reliable data concerning the genome size of the potential diploid (or lower polyploid) progenitors of the A. paniculatum group are presently available from the literature (Bennett and Leitch, 2011).
The pattern of genome size in cytotypes and the results of clustering also provide a preliminary idea of Central Europe as a ‘hot zone’, i.e. a zone where different types have come into secondary contact. Such an important biogeographical contact zone may have occurred as a result of postglacial range re-colonization from different refugia (Hewitt, 1999, 2004; Schmitt, 2007). However, we are aware of the possibility that the wide expansion of cytotypes can also be connected to the more recent creation of secondary, human-made habitats, including deforestation, introduction of grazing and cultivation of crop-plants.
Ploidy level, genome size and invasion potential
There is recent evidence suggesting that species or genotypes with smaller monoploid genome sizes are more invasive due to the effects of genome size on key life-history traits that enhance colonization potential, e.g. rapid plant development and generation time and the production of propagules with greater dispersal potential (Chen, 2010; Kubešová et al., 2010; Lavergne et al., 2010; te Beest et al., 2012). Contrary to expectations, our data concerning the intracytotype variation suggest that the populations of tetra- and pentaploids occurring in anthropogenic habitats have a larger genome size than populations inhabiting natural habitats, whereas no relationships between genome size and habitat type were found in the other cytotypes. Within all ploidy levels studied, we therefore did not corroborate the hypothesis of genome size reduction enhancing invasive ability. Similar results were obtained by Kubátová et al. (2008) for native and invasive populations of Lythrum salicaria. At the interploidy level, the high or low colonization potential of cytotypes with either moderate (i.e. tetra- and pentaploids) or high/low (i.e. tri-/hexaploids) monoploid genome sizes, based on their present ranges, is also not consistent with this hypothesis. Therefore, the evolutionary history of the respective cytotypes is more probably responsible for their present pattern of distribution.
Conclusions
The large-scale geographical pattern of genome size variation in A. oleraceum cytotypes was found to be remarkably complex. This finding suggests that none of the suggested explanations frequently reported in the recent literature alone is applicable to A. oleraceum. The present data show that the observed genome size variations are most likely the result of several interacting factors. The high genome size variation found in the tetraploids suggests that this cytotype is composed of fairly heterogeneous units which have potentially originated via various mechanisms, including the hybridization of different diploid progenitors, secondary contacts between partially genetically different tetraploids, and selective pressure of broad- and fine-scale environmental variables. The spatial relationships of genome size in the tetra- and pentaploids and the penta- and hexaploids most likely indicate an evolutionary link between the respective ploidy levels. The different pattern of genome size variation observed in the hexaploids suggests that the role of adaptation in shaping the pattern of genome size in these cytotypes may be more relaxed in A. oleraceum. Four important confounding factors which blur the spatial patterns in genome size variation are: (1) probably different evolutionary ages of cytotypes, (2) various levels of gene flow within and between cytotypes, (3) complex interactions between spatial and environmental variables and (4) anthropogenic impact on the establishment and spread of the cytotypes. The present study clearly shows the urgent need to incorporate extensive sampling throughout the species range to gain reliable insights into variation in genome size at the intraspecific level but also the necessity of a multi-method approach, with an emphasis on molecular analyses, to gain deeper insights into the evolutionary history of heteroploid species.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank J. Ohryzek, M. Fialová, A. Jírová and J. Duchoslavová for help with fieldwork and maintenance of the samples in the experimental garden, and F. Krahulec, D. Oreshkin, O. Rauch, Z. Skála, J. Ohryzek, M. Fialová, B. Karpavičienė, L. Medina and A. Jírová for sampling some of the populations. Our thanks also go to F. Krahulec and two anonymous reviewers for their comments and advice on previous versions of the manuscript. The English text was corrected by American Journal Experts and J. W. Jongepier. This work was supported by the Grant Agency of the Czech Republic (grant numbers 206/04/P115 and 206/09/1126) and final completion by an internal grant of Palacký University (PrF 2012/001).
LITERATURE CITED
- Balao F, Casimiro-Soriguer R, Talavera M, Herrera J, Talavera S. Distribution and diversity of cytotypes in Dianthus broteri as evidenced by genome size variations. Annals of Botany. 2009;104:965–973. doi: 10.1093/aob/mcp182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancheva S, Greilhuber J. Genome size in Bulgarian Centaurea s.l. (Asteraceae) Plant Systematics and Evolution. 2006;257:95–117. [Google Scholar]
- Baranyi M, Greilhuber J. Genome size in Allium: in quest of reproducible data. Annals of Botany. 1999;83:687–695. [Google Scholar]
- Beaulieu JM, Leitch IJ, Knight CA. Genome size evolution in relation to leaf strategy and metabolic rates revisited. Annals of Botany. 2007a;99:495–505. doi: 10.1093/aob/mcl271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JM, Moles AT, Leitch IJ, Bennett MD, Dickie JB, Knight CA. Correlated evolution of genome size and seed mass. New Phytologist. 2007b;173:422–437. doi: 10.1111/j.1469-8137.2006.01919.x. [DOI] [PubMed] [Google Scholar]
- Beaulieu JM, Leitch IJ, Patel S, Pendharkar A, Knight CA. Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytologist. 2008;179:975–986. doi: 10.1111/j.1469-8137.2008.02528.x. [DOI] [PubMed] [Google Scholar]
- te Beest M, Le Roux JJ, Richardson DM, et al. The more the better? The role of polyploidy in facilitating plant invasions. Annals of Botany. 2012;109:19–45. doi: 10.1093/aob/mcr277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD. Nuclear DNA content and minimum generation time in herbaceous plants. Proceedings of the Royal Society of London Series B. 1972;181:109–135. doi: 10.1098/rspb.1972.0042. [DOI] [PubMed] [Google Scholar]
- Bennett MD. Variation in genomic form in plants and its ecological implications. New Phytologist. 1987;106:177–200. [Google Scholar]
- Bennett MD, Leitch IJ. Nuclear DNA amounts in angiosperms: targets, trends and tomorrow. Annals of Botany. 2011;107:467–590. doi: 10.1093/aob/mcq258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Ma J, Devos KM. Mechanisms of recent genome size variation in flowering plants. Annals of Botany. 2005;95:127–132. doi: 10.1093/aob/mci008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benor S, Fuchs J, Blattner FR. Genome size variation in Corchorus olitorius (Malvaceae s.l.) and its correlation with elevation and phenotypic traits. Genome. 2011;54:575–585. doi: 10.1139/g11-021. [DOI] [PubMed] [Google Scholar]
- Benson NA, Braylan RC. Evaluation of sensitivity in DNA aneuploidy detection using a mathematical model. Cytometry. 1994;15:53–58. doi: 10.1002/cyto.990150109. [DOI] [PubMed] [Google Scholar]
- Biémont C. Within-species variation in genome size. Heredity. 2008;101:297–298. doi: 10.1038/hdy.2008.80. [DOI] [PubMed] [Google Scholar]
- Bogdanovič S, Brullo S, del Galdo GG, Salmeri C. A new autumn-flowering species of Allium (Alliaceae) from Croatia. Folia Geobotanica. 2009;44:83–93. [Google Scholar]
- Brullo S, Pavone P, Salmeri C. Cytotaxonomical notes on Allium dentiferum Webb & Bethelot, an unknown species of the Mediterranean flora. Botanica Chronika. 1991;10:785–796. [Google Scholar]
- Burešová V. Comparative morphology of cytotypes of Allium oleraceum. Olomouc, Czech Republic: Department of Botany, Palacký University; 2012. MSc thesis. [Google Scholar]
- Cavalier-Smith T. Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Annals of Botany. 2005;95:147–175. doi: 10.1093/aob/mci010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli M, Sarri V, Caceres ME, Cionini PG. Intraspecific genotypic diversity in plants. Genome. 2011;54:701–709. doi: 10.1139/g11-039. [DOI] [PubMed] [Google Scholar]
- Chen ZJ. Molecular mechanisms of polyploidy and hybrid vigor. Trends in Plant Science. 2010;15:57–71. doi: 10.1016/j.tplants.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrtek J, Zahradníček J, Krak K, Fehrer J. Genome size in Hieracium subgenus Hieracium (Asteraceae) is strongly correlated with major phylogenetic groups. Annals of Botany. 2009;104:161–178. doi: 10.1093/aob/mcp107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cires E, Cuesta C, Revilla MA, Prieto JAD. Intraspecific genome size variation and morphological differentiation of Ranunculus parnassifolius (Ranunculaceae), an Alpine–Pyrenean–Cantabrian polyploid group. Biological Journal of the Linnean Society. 2010;101:251–271. [Google Scholar]
- Cosendai AC, Rodewald J, Hörandl E. Origin and evolution of apomixis via autopolyploidy in the alpine plant species Ranunculus kuepferi. Taxon. 2011;60:355–364. [Google Scholar]
- Cressie NA. Statistics for spatial data. Revised edition. New York: Wiley; 1993. [Google Scholar]
- Doležel J, Bartoš J. Plant DNA flow cytometry and estimation of nuclear genome size. Annals of Botany. 2005;95:99–110. doi: 10.1093/aob/mci005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J, Greilhuber J, Lucretti S, et al. Plant genome size estimation by flow cytometry: inter-laboratory comparison. Annals of Botany. 1998;82(Suppl. A):17–26. [Google Scholar]
- Doležel J, Greilhuber J, Suda J. Estimation of nuclear DNA content in plants using flow cytometry. Nature Protocols. 2007;2:2233–2244. doi: 10.1038/nprot.2007.310. [DOI] [PubMed] [Google Scholar]
- Duchoslav M, Šafářová L, Krahulec F. Complex distribution patterns, ecology and coexistence of ploidy levels of Allium oleraceum (Alliaceae) in the Czech Republic. Annals of Botany. 2010;105:719–735. doi: 10.1093/aob/mcq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekrt L, Holubová R, Trávníček P, Suda J. Species boundaries and frequency of hybridization in the Dryopteris carthusiana (Dryopteridaceae) complex: a taxonomic puzzle resolved using genome size data. American Journal of Botany. 2010;97:1208–1219. doi: 10.3732/ajb.0900206. [DOI] [PubMed] [Google Scholar]
- Fialová R. Polyploid complexes in the genus Allium. Olomouc, Czech Republic: Department of Botany, Faculty of Science, Palacký University; 1996. PhD thesis. [Google Scholar]
- Francis D, Stuart-Davies M, Barlow PW. A strong nucleotypic effect on the cell cycle regardless of ploidy level. Annals of Botany. 2008;101:747–757. doi: 10.1093/aob/mcn038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen N. The genera Allium L. and Caloscordum Herbert. 1987. In: Malyshev L, Peshkova G, editors. Flora of Siberia: Araceae-Orchidaceae. Novosibirsk: Nauka, 177–195; 1987. (in Russian) [Google Scholar]
- Friesen N, Fritsch R, Blattner F. Phylogeny and new intragenetic classification of Allium (Alliaceae) based on nuclear ribosomal DNA ITS sequences. Aliso. 2006;22:372–395. [Google Scholar]
- Gregory TR. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biological Reviews. 2001;76:65–101. doi: 10.1017/s1464793100005595. [DOI] [PubMed] [Google Scholar]
- Gregory TR. The C-value enigma in plants and animals: a review of parallels and an appeal for partnership. Annals of Botany. 2005;95:133–146. doi: 10.1093/aob/mci009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greilhuber J. Intraspecific variation in genome size: a critical reassessment. Annals of Botany. 1998;82(Suppl. A):27–35. [Google Scholar]
- Greilhuber J. Intraspecific variation in genome size in angiosperms: identifying its existence. Annals of Botany. 2005;95:91–98. doi: 10.1093/aob/mci004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greilhuber J, Doležel J, Lysák MA, Bennett MD. The origin, evolution and proposed stabilization of the terms ‘Genome Size’ and ‘C-Value’ to describe nuclear DNA contents. Annals of Botany. 2005;95:255–260. doi: 10.1093/aob/mci019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greilhuber J, Borsch T, Muller K, Worberg A, Porembski S, Barthlott W. Smallest angiosperm genomes found in Lentibulariaceae, with chromosomes of bacterial size. Plant Biology. 2006;8:770–777. doi: 10.1055/s-2006-924101. [DOI] [PubMed] [Google Scholar]
- Greilhuber J, Temsch EM, Loureiro JCM. Nuclear DNA content measurement. In: Doležel J, Greilhuber J, Suda J, editors. Flow cytometry with plant cells: analysis of genes, chromosomes and genomes. Weinheim: Wiley; 2007. pp. 67–101. [Google Scholar]
- Hewitt GM. Postglacial recolonization of European Biota. Biological Journal of the Linnean Society. 1999;68:87–112. [Google Scholar]
- Hewitt GM. Genetic consequences of climatic oscillations in the quaternary. Philosophical Transactions of the Royal Society B: Biological Sciences. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology. 2005;25:1965–1978. [Google Scholar]
- Hodgson JG, Sharafi M, Jalili A, et al. Stomatal vs. genome size in angiosperms: the somatic tail wagging the genomic dog? Annals of Botany. 2010;105:573–584. doi: 10.1093/aob/mcq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA. Protest: a procrustean randomization test of community environment concordance. Ecoscience. 1995;2:297–303. [Google Scholar]
- Jandová M. Cytogenetic study of generative offsprings in polyploid Allium oleraceum. Olomouc, Czech Republic: Department of Botany, Faculty of Science, Palacký University; 2010. MSc thesis. [Google Scholar]
- Jírová A. Reproductive biology and phenology in polyploid Allium oleraceum. Olomouc, Czech Republic: Department of Botany, Faculty of Science, Palacký University; 2007. MSc thesis. [Google Scholar]
- Karpavičienė B. Chromosome numbers of Allium from Lithuania. Annales Botanici Fennici. 2007;44:345–352. [Google Scholar]
- Karpavičienė B. Morphological, reproductive and karyological variability in Allium oleraceum in Lithuania. Biologia. 2012;67:278–283. [Google Scholar]
- Kim S, Rayburn AL, Parrish A, Lee DK. Cytogeographic distribution and genome size variation in prairie cordgrass (Spartina pectinata Bosc ex Link) Plant Molecular Biology Reporter. 2012;30:1073–1079. [Google Scholar]
- Knight CA, Molinari NA, Petrov DA. The large genome constraint hypothesis: evolution, ecology and phenotype. Annals of Botany. 2005;95:177–190. doi: 10.1093/aob/mci011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubátová B, Trávníček P, Bastlová D, Čurn V, Jarolímová V, Suda J. DNA ploidy-level variation in native and invasive populations of Lythurum salicaria at a large geographical scale. Journal of Biogeography. 2008;35:167–176. [Google Scholar]
- Kubešová M, Moravcová L, Suda J, Jarošík V, Pyšek P. Naturalized plants have smaller genomes than their non-invading relatives: a flow cytometric analysis of the Czech alien flora. Preslia. 2010;82:81–96. [Google Scholar]
- Labani RM, Elkington TT. Nuclear DNA variation in the genus Allium L. (Liliaceae) Heredity. 1987;59:119–128. [Google Scholar]
- Lavergne S, Muenke NJ, Molofsky J. Genome size reduction can trigger rapid phenotypic evolution in invasive plants. Annals of Botany. 2010;105:109–116. doi: 10.1093/aob/mcp271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P, Fortin MJ. Spatial patterns and ecological analysis. Vegetatio. 1989;80:107–138. [Google Scholar]
- Legendre P, Legendre L. Numerical ecology. 2nd English edition. Amsterdam: Elsevier Science BV; 1998. [Google Scholar]
- Leitch IJ, Bennett MD. Genome downsizing in polyploid plants. Biological Journal of the Linnean Society. 2004;82:651–663. [Google Scholar]
- Levan A. Cytological studies in Allium paniculatum group. Hereditas. 1937;23:317–370. [Google Scholar]
- Lichstein JW, Simons TR, Shriner SA, Franzreb KE. Spatial autocorrelation and autoregressive models in ecology. Ecological Monographs. 2002;72:445–463. [Google Scholar]
- Loidl J. Some features of heterochromatin in wild Allium species. Plant Systematics and Evolution. 1983;143:117–131. [Google Scholar]
- Loureiro J, Rodriguez E, Doležel J, Santos C. Flowcytometric and microscopic analysis of the effect of tannic acid on plant nuclei and estimation of DNA content. Annals of Botany. 2006;98:515–527. doi: 10.1093/aob/mcl140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro J, Trávníček P, Rauchová J, et al. The use of flow cytometry in the biosystematics, ecology and population biology of homoploid plants. Preslia. 2010;82:3–21. [Google Scholar]
- Lysák MA, Rostková A, Dixon JM, Rossi G, Doležel J. Limited genome size variation in Sesleria albicans. Annals of Botany. 2000;86:399–403. [Google Scholar]
- Lysák MA, Koch MA, Beaulieu JM, Meister A, Leitch IJ. The dynamic ups and downs of genome size evolution in Brassicaceae. Molecular Biology and Evolution. 2009;26(85–98) doi: 10.1093/molbev/msn223. [DOI] [PubMed] [Google Scholar]
- MacGillivray CW, Grime JP. Genome size predicts frost resistance in British herbaceous plants: implications for rates of vegetation response to global warming. Functional Ecology. 1995;9:320–325. [Google Scholar]
- Mahelka V, Suda J, Jarolímová V, Trávníček P, Krahulec F. Genome size discriminates between closely related taxa Elytrigia repens and E. intermedia (Poaceae: Triticeae) and their hybrid. Folia Geobotanica. 2005;40:367–384. [Google Scholar]
- Meagher TR, Vassiliadis C. Phenotypic impacts of repetitive DNA in flowering plants. New Phytologist. 2005;168:71–80. doi: 10.1111/j.1469-8137.2005.01527.x. [DOI] [PubMed] [Google Scholar]
- Meusel H, Jäger E, Weinert E. Vergleichende Chorologie der zentraleuropäischen Flora. Jena: Gustav Fischer Verlag; 1965. [Google Scholar]
- Mráz P, Chrtek J, Šingliarová B. Geographical parthenogenesis, genome size variation and pollen production in the arctic–alpine species Hieracium alpinum. Botanica Helvetica. 2009;119:41–51. [Google Scholar]
- Murray BG. When does intraspecific C-value variation become taxonomically significant? Annals of Botany. 2005;95:119–125. doi: 10.1093/aob/mci007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohri D. Climate and growth form: the consequences for genome size in plants. Plant Biology. 2005;7:449–458. doi: 10.1055/s-2005-865878. [DOI] [PubMed] [Google Scholar]
- Ohri D, Fritsch RM, Hanelt P. Evolution of genome size in Allium (Alliaceae) Plant Systematics and Evolution. 1998;210:57–86. [Google Scholar]
- Ohryzek J. Comparative biology of cytotypes of Allium oleraceum. Olomouc, Czech Republic: Department of Botany, Faculty of Science, Palacký University; 2007. MSc thesis. [Google Scholar]
- Olšavská K, Perný M, Španiel S, Šingliarová B. Nuclear DNA content variation among perennial taxa of the genus Cyanus (Asteraceae) in Central Europe and adjacent areas. Plant Systematics and Evolution. 2012;298:1463–1482. [Google Scholar]
- Pečinka A, Suchánková P, Lysák MA, Trávníček B, Doležel J. Nuclear DNA content variation among Central European Koeleria taxa. Annals of Botany. 2006;98:117–122. doi: 10.1093/aob/mcl077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicer J, Fay MF, Leitch IJ. The largest eukaryotic genome of them all? Botanical Journal of the Linnean Society. 2010;164:10–15. [Google Scholar]
- Peres-Neto PR, Jackson DA. How well do multivariate data sets match? The advantages of a Procrustean superimposition approach over the Mantel test. Oecologia. 2000;129:169–178. doi: 10.1007/s004420100720. [DOI] [PubMed] [Google Scholar]
- Rangel TF, Diniz-Filho JAF, Bini LM. SAM: a comprehensive application for spatial analysis in macroecology. Ecography. 2010;33:1–5. [Google Scholar]
- Rejmánek M. A theory of seed plant invasiveness: the first sketch. Biological Conservation. 1996;78:171–181. [Google Scholar]
- Šafářová L, Duchoslav M. Cytotype distribution in mixed populations of polyploid Allium oleraceum measured at a microgeographic scale. Preslia. 2010;82:107–126. [Google Scholar]
- Šafářová L, Duchoslav M, Jandová M, Krahulec F. Allium oleraceum in Slovakia: cytotype distribution and ecology. Preslia. 2011;83:513–527. [Google Scholar]
- Schmitt T. Molecular biogeography of Europe: Pleistocene cycles and postglacial trends. Frontiers in Zoology. 2007;4(11) doi: 10.1186/1742-9994-4-11. http://dx.doi.org/10.1186/1742-9994-4-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slovák M, Vít P, Urfus T, Suda J. Complex pattern of genome size variation in a polymorphic member of the Asteraceae. Journal of Biogeography. 2009;36:372–384. [Google Scholar]
- Šmarda P, Bureš P. Intraspecific DNA content variability in Festuca pallens on different geographical scales and ploidy levels. Annals of Botany. 2006;98:665–678. doi: 10.1093/aob/mcl150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmarda P, Bureš P. Understanding intraspecific variation in genome size in plants. Preslia. 2010;82:41–61. [Google Scholar]
- Šmarda P, Bureš P, Horová L, Rotreklová O. Intrapopulation genome size dynamics in Festuca pallens. Annals of Botany. 2008a;102:599–607. doi: 10.1093/aob/mcn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmarda P, Bureš P, Horová L. Genome size and GC content evolution of Festuca: ancestral expansion and subsequent reduction. Annals of Botany. 2008b;101:421–433. doi: 10.1093/aob/mcm307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staňková H. Population genetics of the polyploid complex Allium oleraceum in the Czech Republic. Olomouc, Czech Republic: Department of Botany, Faculty of Science, Palacký University; 2005. MSc thesis. [Google Scholar]
- Stearn WT. In: Allium L. In. Tutin TG, Heywood VH, Burges NA, et al., editors. Cambridge: Cambridge University Press; 1980. pp. 49–69. Flora Europaea 5. [Google Scholar]
- Suda J, Krahulcová A, Trávníček P, Rosenbaumová R, Peckert T, Krahulec F. Genome size variation and species relationships in Hieracium sub-genus Pilosella (Asteraceae) as inferred by flow cytometry. Annals of Botany. 2007;100:1323–1335. doi: 10.1093/aob/mcm218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift HH. The constancy of desoxyribose nucleic acid in plant nuclei. Proceedings of the National Academy of Sciences. 1950;36:643–654. doi: 10.1073/pnas.36.11.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veselý P, Bureš P, Šmarda P, Pavlíček T. Genome size and DNA base composition of geophytes: the mirror of phenology and ecology? Annals of Botany. 2012;109:65–75. doi: 10.1093/aob/mcr267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss-Schneeweiss H, Greilhuber J, Schneeweiss GM. Genome size evolution in holoparasitic Orobanche (Orobanchaceae) and related genera. American Journal of Botany. 2006;93:148–156. doi: 10.3732/ajb.91.3.439. [DOI] [PubMed] [Google Scholar]
- Wendel J. Genome evolution in polyploids. Plant Molecular Biology. 2000;42:225–249. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.