Abstract
Oxidation of membrane phospholipids is associated with inflammation, neurodegenerative disease, and cancer. Oxyradical damage to phospholipids results in the production of reactive aldehydes that adduct proteins and modulate their function. 4-Hydroxynonenal (HNE), a common product of oxidative damage to lipids, adducts proteins at exposed Cys, His, or Lys residues. Here, we demonstrate that peptidyl-prolyl cis/trans-isomerase A1 (Pin1), an enzyme that catalyzes the conversion of the peptide bond of pSer/pThr-Pro moieties in signaling proteins from cis to trans, is highly susceptible to HNE modification. Incubation of purified Pin1 with HNE followed by MALDI-TOF/TOF mass spectrometry resulted in detection of Michael adducts at the active site residues His-157 and Cys-113. Time and concentration dependencies indicate that Cys-113 is the primary site of HNE modification. Pin1 was adducted in MDA-MB-231 breast cancer cells treated with 8-alkynyl-HNE as judged by click chemistry conjugation with biotin followed by streptavidin-based pulldown and Western blotting with anti-Pin1 antibody. Furthermore, orbitrap MS data support the adduction of Cys-113 in the Pin1 active site upon HNE treatment of MDA-MB-231 cells. siRNA knockdown of Pin1 in MDA-MB-231 cells partially protected the cells from HNE-induced toxicity. Recent studies indicate that Pin1 is an important molecular target for the chemopreventive effects of green tea polyphenols. The present study establishes that it is also a target for electrophilic modification by products of lipid peroxidation.
Introduction
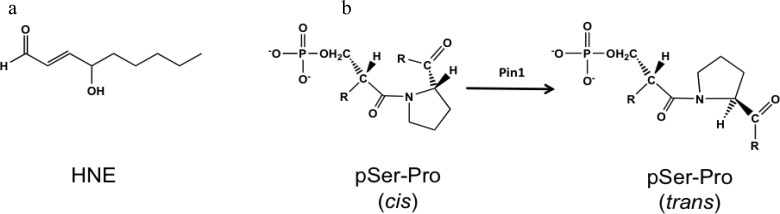
Oxidation of lipids is widely regarded as a contributing factor to neurodegeneration, inflammation, and cancer.1−3 The polyunsaturated fatty acid residues of membrane phospholipids are prime targets for oxidative attack, as these molecules possess multiple bis-allylic hydrogens, which are sensitive to abstraction by oxidizing agents. Abstraction of a labile hydrogen in the presence of O2 leads to a radical chain oxidation mediated by lipid peroxyl radicals, which consumes multiple fatty acid moieties and generates a panoply of products. Among these are reactive aldehydes [e.g., 4-hydroxynonenal (HNE), Figure 1a], which are capable of protein, DNA, and RNA modification. Lipid electrophile modification of nucleic acids can induce base-pair substitutions, frameshift mutations, and strand breaks, whereas modification of proteins distorts tertiary structure and alters protein function. Furthermore, free radical species can recycle to oxidize neighboring phospholipids in a continuum until the radical is intercepted.4
Figure 1.
(a) Structure of HNE. (b) Reaction catalyzed by Pin1. Pin1 binds the cis-phosphorylated serine-proline moiety via a WW binding domain. The PPIase domain of Pin1 catalyzes the conversion of the prolyl bond from cis to trans.
Much attention has been focused on the cellular consequences of HNE production. HNE is generated in vivo at low micromolar levels endogenously and is elevated in tissues experiencing an oxidative insult, such as the brains of Alzheimer’s disease (AD) patients and the lungs of patients with chronic obstructive pulmonary disease.5−7 In these and other conditions, HNE modification of cellular constituents is associated with disease pathogenesis. However, the contribution of HNE to cellular fate is complex and incompletely understood. To gain an understanding of the spectrum of biochemical consequences of HNE production, our laboratory used a multipronged approach to compile an inventory of protein signaling networks activated by HNE.8,9 Using click chemistry and mass spectrometry, an inventory of protein targets of HNE was generated;10 separately, microarray analysis of HNE-treated cells revealed up- or down-regulated genes in response to treatment.11 Using bioinformatic analysis, protein targets of HNE were linked to differentially regulated genes via specific transcription factors.8 This approach resulted in multiple protein targets and altered signaling networks to investigate.
Analysis of the protein adduction inventory revealed that peptidyl-prolyl cis/trans-isomerases (PPIases) are a class of proteins modified by HNE. PPIases catalyze proline-directed isomerizations in proteins. Peptidylprolyl cis/trans-isomerase A1 (Pin1) is a unique PPIase that catalyzes only phosphoserine- and phosphothreonine-proline conversions from cis to trans (Figure 1b).12 This enzyme is particularly important in phosphorylation-dependent signaling pathways, as proline-directed phosphatases are trans-specific.13 Molecules that covalently modify Pin1’s catalytic or binding site residues have been previously demonstrated to induce apoptosis and inhibit cell proliferation, possibly due to inhibition of Pin1’s actions on the cell cycle. Epigallocatechin gallate (EGCG), an anticancer polyphenol in green tea, inhibits cancer cell growth by interacting with Arg-17 of Pin1 in the WW domain, preventing Pin1–substrate interactions. EGCG also interacts with the PPIase catalytic domain, inhibiting enzyme action.14 Additionally, PiB, a derivative of juglone (1,4-naphthoquinone), has been reported to induce cell death in a Pin1-dependent fashion.15
The chemistry and biology of Pin1 modification by lipid electrophiles have not been investigated. Pin1 contains multiple exposed Cys, His, and Lys residues capable of reacting with HNE. Using MALDI-TOF mass spectrometry, we mapped the HNE adduction sites on Pin1. Data from MALDI-TOF and MALDI-TOF/TOF analysis with purified Pin1 treated with HNE support the adduction of critical active site residues, with Cys-113 being the most reactive. Treatment of MDA-MB-231 cells with HNE, followed by immunoprecipitation and high-resolution MS/MS analysis, also revealed the presence of a Cys-113–HNE Michael adduct within the active site of the protein. siRNA-mediated knockdown of Pin1 afforded protection from HNE-induced cell death. These studies demonstrate that Pin1 is an important target for modification by lipid electrophiles, and this leads to alteration in cell signaling and viability.
Materials and Methods
Cell Culture and Treatment
MDA-MB-231 cells, a triple-negative human breast carcinoma cell line, were obtained from the American Type Culture Collection (ATCC) and were cultured in RPMI1640 medium (Gibco) supplemented with 10% fetal bovine serum (FBS). MDA-MB-231 cells were maintained at 37 °C in a humidified cell culture incubator under 5% CO2 and 95% air. HNE and alkynyl-4-hydroxynonenal (aHNE) were synthesized as previously described.10,16 aHNE, HNE, or DMSO (vehicle control) was added to cell culture medium to achieve a final DMSO concentration of less than 1%. Electrophile concentrations and times of incubation are indicated in the figure legends.
In-Solution Modification of Purified Pin1
Purified Pin1 was obtained from Genway Biosciences (GWB-523EFE) and was buffer-exchanged once with Dulbecco’s-modified PBS (Gibco) before use. Protein (2.5 μg) was diluted to 25 μL with PBS and incubated for the indicated times with HNE at 37 °C with agitation. Reactions were terminated with NaBH4 and rotated end-over-end for 30 min at room temperature (RT). Samples were dried with a SpeedVac and reconstituted in 10 μL of 6 M guanidine hydrochloride for 45 min at RT. Each sample was incubated with DTT (150 μM) for 30 min at 37 °C, followed by 750 μM iodoacetamide for 15 min at RT in the dark. Samples were diluted 20-fold with 20 mM NH4HCO3 and digested with 500 ng of chymotrypsin for 24 h at 37 °C. The following day, chymotryptic digests were desalted using C18 ZipTips, followed by elution with 60% acetonitrile/0.1% trifluoroacetic acid. Samples were mixed 1:1 by volume with matrix [20 mg/mL α-cyano-4-hydroxycinnamic acid (CHCA) in 60% acetonitrile] and subjected to MALDI mass spectrometry.
Click Chemistry
MDA-MB-231 cells were treated with aHNE for 1 h in serum-free medium. Following treatment, cells were washed with PBS and collected by scraping and centrifugation for 5 min at 1000g. Cell pellets were lysed in buffer containing 50 mM HEPES, pH 7.5, 150 mM NaCl, 0.5% Igepal, and mammalian protease inhibitor cocktail (Sigma-Aldrich). Suspended cell pellets were sonicated by 10 1 s pulses with a Virsonic Cell Disruptor and then cleared by centrifugation at 16000g for 10 min. Bicinchoninic acid (BCA) assay was used to determine the protein concentration and was performed according to the manufacturer’s instructions (Thermo). Each sample was diluted to 2 mg/mL in 3 mL of lysis buffer and reduced with 2 mM NaBH4 for 1 h at RT with agitation to stabilize adducts. Unreacted NaBH4 was quenched with 3 μL of acetone, and reduced lysates were subjected to Huisgen 1,3-cycloaddition chemistry by the addition of 0.2 mM biotin benzoin (N3-biotin linker, Porter laboratory), 1 mM tris(2-carboxyethyl)phosphine (TCEP), 0.1 mM ligand tris((1-benzyl-1H-1,2,3-triazol-4-yl)methyl)amine (triazole ligand, Porter laboratory), and 1 mM CuSO4. Tubes containing samples were covered with foil and rotated end-over-end for 2 h at RT. Proteins were precipitated by the addition of 6 mL (twice the volume of sample) of ice-cold methanol for 30 min on ice and pelleted by centrifugation. Pellets were washed twice with cold methanol, resuspended in 1 mL of 0.5% SDS, and boiled for 5 min. After they were boiled, 50 μL of sample was aliquotted to serve as the input fraction to demonstrate equal protein loading onto streptavidin beads. The remaining 950 μL was diluted to 10 mL with PBS containing 1 mL of streptavidin-agarose beads and 0.1% SDS. Samples were incubated overnight at 4 °C with rotation. The following day, beads were washed with 1% SDS, 4 M urea, PBS containing 1 M NaCl, and PBS each. To elute aHNE-modified proteins from the streptavidin beads, samples were incubated under 365 nm UV light with rotation for 90 min at RT. During this step, samples were placed approximately 1 in. from a Spectroline ENF-280c UV lamp containing one 8 W long wave tube. Eluted proteins were separated from beads by centrifugation, concentrated, and subjected to SDS-PAGE and Western blot.
SDS-PAGE and Western Blotting
Samples for SDS-PAGE were mixed 1:1 by volume with Laemmli buffer containing 5% (w/v) β-mercaptoethanol and boiled for 5 min. A 4–20% gradient Tris-HCl gel was used to separate proteins. Proteins in the gel were transferred onto a 0.45 μm nitrocellulose membrane and blocked with 5% nonfat dry milk for 1 h. Primary antibodies were incubated (1:1000 for Pin1, FLAG, and GAPDH) with membranes overnight at 4 °C. The following day, blots were washed with Tris-buffered saline containing 0.1% Tween-20 (TBST) three times and then incubated with secondary antibody for 1 h at RT with shaking. Blots were washed three times with TBST and developed using luminol-based detection. FLAG and Pin1 antibodies were obtained from Cell Signaling, while GAPDH antibody was obtained from Santa Cruz Biotechnology.
MALDI-TOF and MALDI-TOF/TOF
All spectra were acquired in positive ion mode on either an Autoflex Speed TOF MS or an Ultraflextreme TOF/TOF MS (Bruker Daltonics), both equipped with a Nd:YAG (solid state) laser operating at 355 nm. Peptide-CHCA solutions (1 μL) were deposited on MALDI target plates and air-dried prior to analysis. Full mass spectra of digested peptides were obtained in reflectron mode on the Ultraflextreme, using a 500–4000 mass range. Peptide masses and sequence ions were manually examined for mass shifts of +158 m/z or +141 m/z, corresponding to reduced HNE–Michael adducts or reduced Schiff base adducts, respectively. Selected peptide ions were dissociated using LIFT on the TOF/TOF. TOF/TOF fragmentation data were interrogated using FlexAnalysis software and analyzed against a theoretical peptide digest using Protein Prospector.
Plasmid Construct and Transfection
A cDNA clone of Pin1 (Mammalian Gene Collection accession number BC002899) was obtained from the Vanderbilt Microarray Shared Resource cDNA clone collection. Pin1 was PCR-amplified from pOTB7 using the primers 5′-CCTGCTAGCTCCACCATGGATTACAAGGATGACGACGATAAGGCGGACGAGGAGAAGCTGCCGC-3′ (includes an N-terminal FLAG tag) and 5′-CCTGGTACCTCACTCAGTGCGGAGGATGA-3′. The PCR product was digested with NheI and KpnI and ligated into pcDNA3.1 Hygro(+) (Invitrogen). MDA-MB-231 cells at 80% confluency were transfected using 10 μg of expression construct and 20 μL of lipofectamine (Thermo-Scientific, Lafayette, CO) for 24 h. After 24 h, cells were incubated with fresh medium for an additional 48 h at 37 °C before treatment and collection.
Immunoprecipitation
The Direct Immunoprecipitation Kit (Pierce) was used to immunoprecipitate FLAG-Pin1 from MDA-MB-231 cells. Eighty microliters of anti-FLAG resin (Sigma-Aldrich) was loaded onto columns and washed once with PBS. One milligram of protein lysate was added per column in 500 μL of total volume. Columns were plugged and rotated end-over-end overnight at 4 °C. The following day, columns were unplugged and centrifuged at 1000 rpm to elute nonspecific proteins. Beads containing FLAG-Pin1 were washed three times with IP/Lysis wash buffer and once with conditioning buffer containing 0.5% protease inhibitors (Pierce). Bound proteins were eluted from the beads using elution buffer. Eluents were neutralized with the addition of 1 M NaOH and treated with NaBH4 to stabilize HNE-adducted proteins. Because multiple columns were necessary to achieve sufficient protein quantities for proteomics, samples were pooled and concentrated to 50 μL using 3 kDa molecular weight cutoff filters immediately prior to SDS-PAGE.
LC-MS/MS Analysis of Pin1
Following SDS-PAGE of immunoprecipitated FLAG-Pin1, the gel was stained with colloidal Coomassie Blue, and the Pin1 band was excised from the gel and cut into 1 mm3 pieces. The gel pieces were treated with 45 mM DTT for 45 min, and available Cys residues were carbamidomethylated with 100 mM iodoacetamide for 45 min. After the gel pieces were destained with 50% acetonitrile in 25 mM ammonium bicarbonate, Pin1 was digested with endoproteinase AspN (10 ng/μL) in 25 mM NH4HCO3 overnight at 37 °C. Peptides were extracted by gel dehydration (60% acetonitrile, 0.1% TFA), the extract was dried by vacuum centrifugation, and the peptides were reconstituted in 0.1% formic acid. The peptide solution was loaded onto a capillary reversed-phase analytical column (360 μm o.d. × 100 μm i.d.) using an Eksigent NanoLC Ultra HPLC and autosampler. The analytical column was packed with 20 cm of C18 reversed-phase material (Jupiter, 3 μm beads, 300 Å, Phenomenex), directly into a laser-pulled emitter tip. Peptides were gradient-eluted at a flow rate of 500 nL/min, and the mobile phase solvents consisted of water containing 0.1% formic acid (solvent A) and acetonitrile containing 0.1% formic acid (solvent B). A 90 min gradient was performed, consisting of the following: 0–10 min, 2% B; 10–50 min, 2–45% B; 50–60 min, 45–90% B; 60–65 min, 95% B; 65–70 min 95–2% B; and 70–90 min, 2% B. Eluting peptides were mass analyzed on an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific), equipped with a nanoelectrospray ionization source. The instrument was operated using a data-dependent method with dynamic exclusion enabled. Full-scan (m/z 300–2000) spectra were acquired with the Orbitrap (resolution 60,000), and the top 16 most abundant ions in each MS scan were selected for fragmentation in the LTQ. An isolation width of 2 m/z, activation time of 10 ms, and 35% normalized collision energy were used to generate MS2 spectra. Dynamic exclusion settings allowed for a repeat count of 2 within a repeat duration of 10 s, and the exclusion duration time was set to 15 s. For identification of Pin1 peptides, tandem mass spectra were searched with Sequest (Thermo Fisher Scientific) against a human subset database created from the UniprotKB protein database (www.uniprot.org). Variable modifications of +57.0214 on Cys (carbamidomethylation), +15.9949 on Met (oxidation), +141.1279 on Lys and Arg (corresponding to reduced Schiff base), and +158.1306 on Cys, Lys, and His residues (corresponding to reduced HNE modification) were included for database searching. Search results were assembled using Scaffold 3.0 (Proteome Software). Spectra acquired of Pin1 peptides of interest were then inspected using Xcalibur 2.1 Qual Browser software (Thermo Scientific). Tandem mass spectra of HNE-modified peptide precursors and the spectra acquired of the corresponding unmodified peptide forms were examined by manual interrogation.
Pin1 siRNA Knockdown
siRNA for Pin1 (CCGUGUUCACGGAUUCCGCAUCCA) and scrambled control were obtained from Invitrogen. siRNA was diluted 1:200 in Optimem (Gibco) for 5 min, and separately, Dharmafect was diluted 1:40 in Optimem. After 5 min, solutions were combined and incubated for 20 min at RT. The solution containing Optimem, Dharmafect, and siRNA was added to 4 mL of medium containing 2 × 106 MDA-MB-231 cells and incubated overnight at 37 °C. The medium was changed the following day, and cells were incubated for an additional 48 h at 37 °C. Cells were lysed and processed for Western blot. For viability assays, 7500 cells/well were seeded in 96-well plates, and wells were incubated with 3.75 μL of siRNA/Dharmafect/Optimem solution for 24 h before the medium was changed. After an additional 48 h, cells were treated with HNE for 48 h for viability assay.
Viability Assay
The cell viability was assessed using the Calcein AM assay, whereby a cell-permeable substrate (calcein AM) penetrates live cells and is converted to fluorescent calcein by intracellular esterases. Just prior to the assay, MDA-MB-231 medium was removed, and cells were washed twice with PBS. Calcein AM (Invitrogen) was dissolved in DMSO for a stock concentration of 2 mM. Calcein AM stock was diluted to 2 μM in PBS and added to cells for 30 min at RT. After incubation, the solution was removed from the wells, and fluorescence was quantified using a Spectramax plate reader (excitation = 494 nm, and emission = 517 nm).
Statistical Analysis
Viability data were analyzed using GraphPad Prism Statistical Software. A one-way ANOVA for treatment followed by Tukey posthoc test was used to assess statistical significance. p < 0.05 was considered significant.
Results
Incubation of Purified Pin1 with HNE Results in Covalent Adduction at Critical Active Site Residues
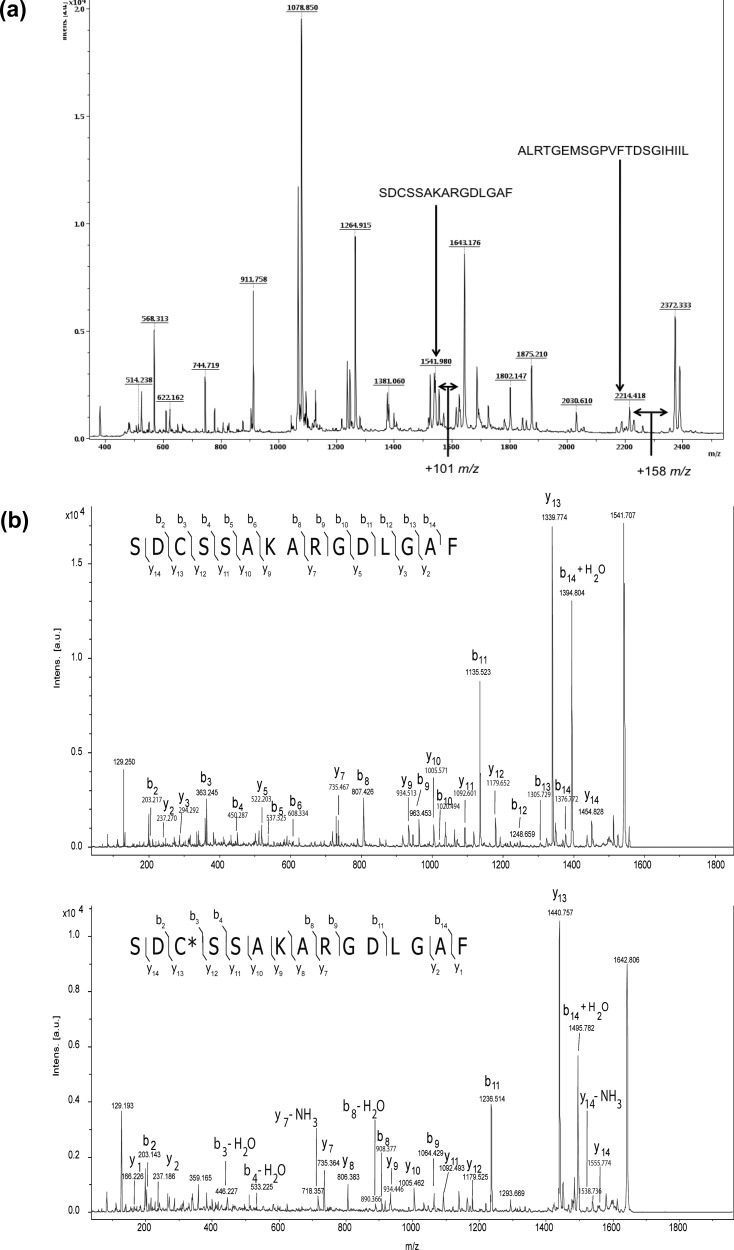
To map the adduction sites of Pin1 by HNE, we adapted a previously reported protocol for determining the adduction sites of cytochrome c.17 Purified Pin1 was treated with a high concentration of HNE (2 mM) for 3 h at 37 °C, followed by NaBH4-mediated aldehyde reduction. Although 2 mM greatly exceeds the pathophysiological range of HNE concentrations reported in cells, the aim of this experiment was to compile all of the potential Pin1 modification sites. Samples were carboxamidomethylated by treatment with iodoacetamide, then digested with chymotrypsin, and analyzed by MALDI-TOF MS. Several peaks with mass shifts of +158 m/z relative to theoretical Pin1 chymotryptic peptides were present in the digest, corresponding to reduced Michael adducts of HNE (Figure 2a). In the case of HNE modification at a Cys residue, the peak shift occurs +101 m/z relative to the alkylated peptide because of the replacement of the carboxamidomethyl group (Figure 2a). No mass shifts corresponding to reduced or unreduced Schiff bases were observed. Peaks corresponding to reduced HNE–Michael adducts, as well as unmodified peptides, were fragmented by MALDI-TOF/TOF to confirm peptide identification and identify the site of HNE modification. TOF/TOF analysis of peak 1643 m/z revealed the presence of an HNE–Cys reduced Michael adduct occurring at Cys-113 (Figure 2b, bottom panel), while His-157 adduction was observed upon fragmentation of peak 2372 m/z (Figure S1, bottom panel, in the Supporting Information). Interestingly, both Cys-113 and His-157 are located in the Pin1 active site and are involved in its catalytic function. In addition, an HNE–Lys-132 adduct was also detected (Table 1 and Figure S2 in the Supporting Information). Because peaks of modified and corresponding unmodified Pin1 peptides are detectable simultaneously in the MALDI-TOF spectra, the relative reactivity of recovered peptides can be ascertained.18As shown in Figure 2a and Table 1, the intensities of peaks corresponding to Cys-113 and His-157 are increased approximately 3-fold relative to their respective unmodified peaks. By contrast, the adducted to unadducted peak ratio corresponding to Lys-132 adduction was 0.23, indicating a relatively lower sensitivity to HNE modification. These data support the hypothesis that active site residues in Pin1 are particularly susceptible to modification by HNE.
Figure 2.
Pin1 is modified by HNE at active site residues in vitro. (a) MALDI-TOF spectrum of chymotryptic digest from HNE-treated Pin1. Two peptides are indicated, and arrows represent peaks with shifts of +158 m/z from native Pin1 chymotryptic digests. Peak 1643 is shifted 101 m/z from the cysteine-containing alkylated peptide indicated. (b) MALDI-TOF/TOF spectrum of peaks 1542 (top) and 1643 m/z (bottom) from HNE-treated Pin1 chymotryptic digest. Spectral interrogation reveals the presence of a mass shift of +158 m/z between y12 and y13 (or +101 m/z as compared to top spectrum containing alkylated Cys-113) in the bottom spectrum corresponding to a reduced Michael adduct occurring at Cys-113. Matched masses of b and y ions in the spectrum to corresponding theoretical masses of Pin1 peptide SDCSSAKARGDLGAF and SDC(+158 m/z)SSAKARGDLGAF are indicated by arrows. The asterisk within the peptide sequence indicates the site of modification.
Table 1. Identification of Adducted Pin1 Amino Acids by HNE as Detected by MALDI-TOF/TOF.
| sequence | modified residue | type of modification | ratio (intensity of adducted peak/intensity of unadducted peak) |
|---|---|---|---|
| SRGQMQKPF | K132 | Michael adduct | 0.23 |
| SDCSSAKARGDLGAF | C113 | Michael adduct | 3.56 |
| ALRTGEMSGPVFTDSGIHIIL | H157 | Michael adduct | 3.03 |
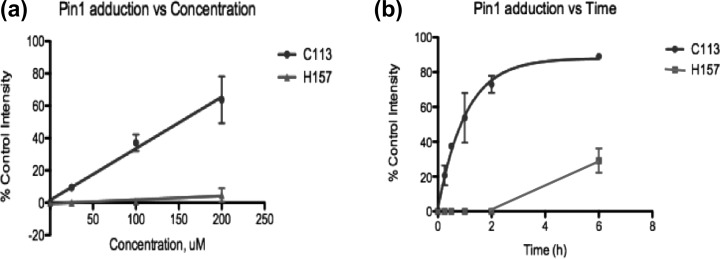
We investigated the relative rate and extent of adduction of the two active site residues. Purified Pin1 was incubated with 25, 100, or 200 μM HNE for 3 h, followed by NaBH4 reduction, digestion, and analysis by MALDI-TOF MS. Spectral investigation revealed that the formation of the Cys-113 adduct increased with HNE concentration, whereas His-157 adduct formation was minimal (Figure 3a). Incubation of Pin1 with HNE for increasing times revealed complete modification of Cys-113 by HNE before any His-157 modification was observed (Figure 3b). This indicates that the catalytic cysteine of Pin1 is the primary target for HNE modification.
Figure 3.
Relative rate of formation of Michael adducts with C113 vs H157 (a) as a function of HNE concentration and (b) as a function of time of HNE incubation. For the graph in panel a, various concentrations were incubated for 3 h before termination of the reaction with NaBH4. For function of time experiments, 200 μM HNE was incubated with Pin1 for indicated times. Cys-113 becomes saturated with HNE before the detection of the His-157 Michael adduct. ± standard deviation, n = 3/time point or concentration.
Treatment of MDA-MB-231 Cells with aHNE Leads to Covalent Pin1 Adduction as Judged by Click Chemistry
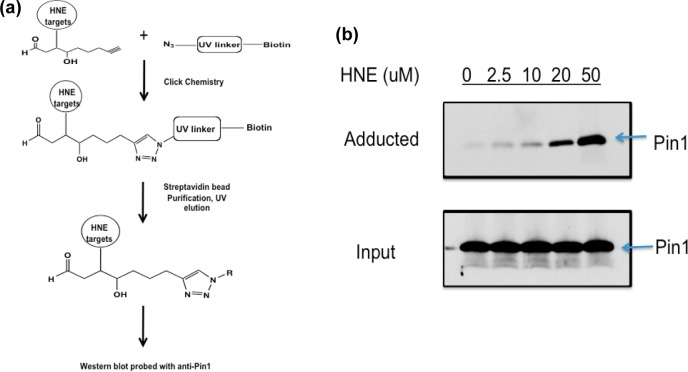
Previous studies have demonstrated that administration of HNE to cells results in extensive protein adduction.10,19 The use of alkynylated analogues as taggable surrogates for electrophiles allows for the separation of adducted proteins from unadducted proteins. A schematic of our method is shown in Figure 4a. Cells treated with aHNE are lysed and exposed to click chemistry, in which the alkyne at positions 8–9 of aHNE reacts with an azido-biotin tag bearing a UV-cleavable linker. Only HNE-adducted proteins are biotinylated, so these targets can be purified by binding to streptavidin-coated beads, followed by elution from the beads with UV light.20 Western blotting of eluted fractions with antibodies allows for detection of specific proteins that are adducted by aHNE. MDA-MB-231 cells were chosen as the model cell line for this study because of a previously reported dependence on Pin1 for cell proliferation.21 Western blots of lysates from MDA-MB-231 cells dosed with 2.5–50 μM aHNE for 1 h revealed a dose-dependent increase in Pin1 adduction as a function of concentration (Figure 4b). A blot of the input fraction indicated equal loading of cell lysate onto streptavidin beads, as well as an absence of changes in Pin1 protein levels with aHNE treatment (Figure 4b).
Figure 4.
Pin1 is dose dependently modified by aHNE in MDA-MB-231 cells. (a) Schematic of detection of HNE-adducted proteins in vitro, whereby targets of aHNE are separated from nontargets via click chemistry, followed by streptavidin purification, UV light-induced elution, subject to Western blot, and probed with anti-Pin1 antibody. (b) Western blot of adducted Pin1. Treatment of MDA-MB-231 cells with aHNE and subjection to click-mediated separation of electrophile-modified proteins followed by Western blot with anti-Pin1 antibody reveal a concentration-dependent adduction of Pin1 in this cell line (top blot); the lower blot is shown to demonstrate equal protein loading onto Streptavidin beads.
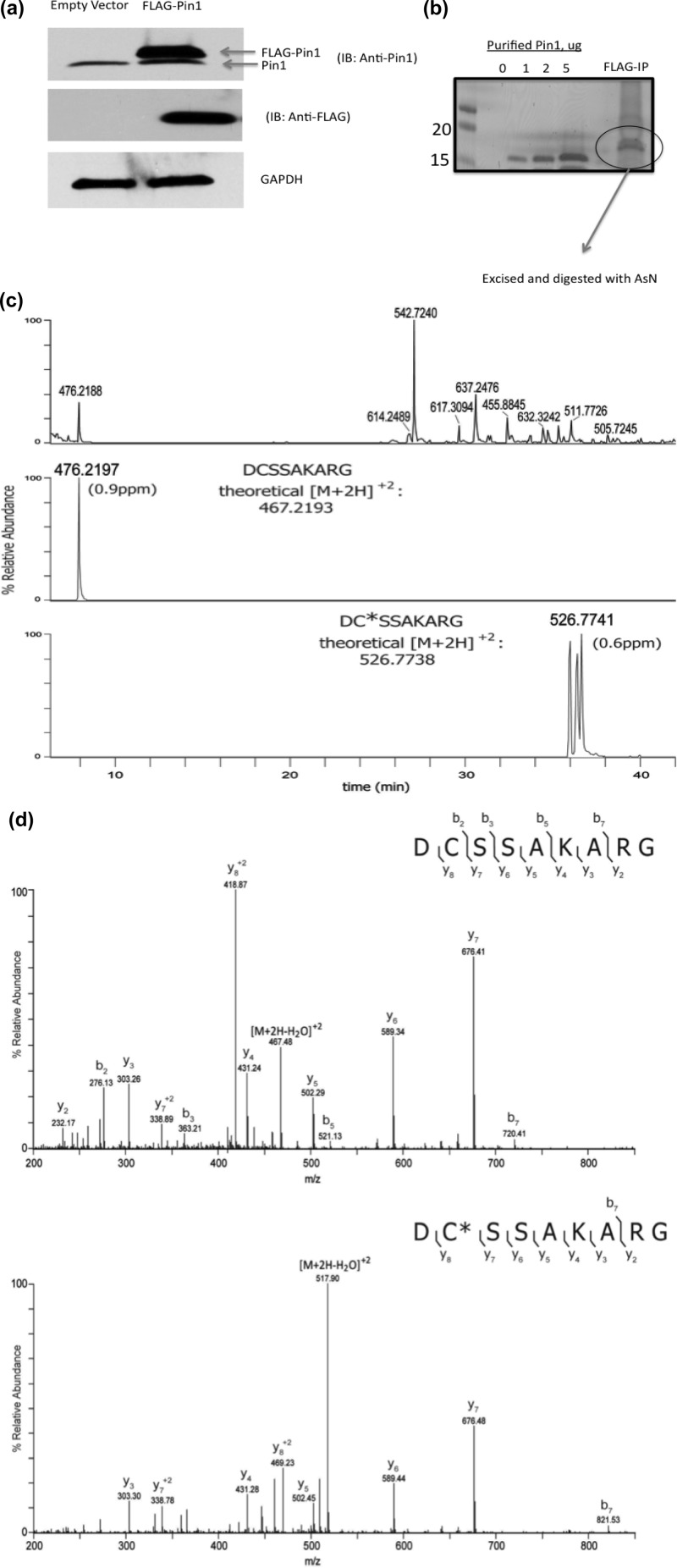
Cys-113 of Pin1 Is Adducted in MDA-MB-231 Cells Treated with HNE
Identification of electrophile-modified residues of Pin1 in cells treated with HNE would provide critical information into the role the adduct plays in altering Pin1 biochemical functions. Our laboratory previously elucidated the HNE adduction sites of HSP90 via geldanamycin-biotin affinity capture and LC-coupled tandem mass spectrometry (LC-MS/MS).22 In the current study, we employed a similar strategy of Pin1 affinity purification and LC-MS/MS analysis to examine HNE adduction of Pin1. To facilitate isolation of Pin1 from cells treated with HNE, a FLAG-Pin1 plasmid was generated and transfected into MDA-MB-231 cells (Figure 5a). Immunoprecipitation using anti-FLAG resin enabled purification of sufficient protein to observe with SDS-PAGE analysis and subsequent colloidal Coomassie staining (Figure 5b). The resolved Pin1 band was excised and in-gel digested with endoproteinase AspN, and then, peptides were extracted and analyzed on an LTQ-Orbitrap Velos mass spectrometer. The LC-MS base peak chromatogram demonstrates thorough proteolytic digestion of the excised Pin1 gel band (Figure 5c, top panel). Following LC-MS/MS analysis, the resulting tandem mass spectra were searched via Sequest against a human database, and assigned spectra corresponding to AspN-derived Pin1 peptides of interest were validated via manual examination. Among those peptides detected, Pin1 peptides containing Cys-113 (DCSSAKARG, residues 112–120) and His-157 (DSGIHIILRTE-, residues 153–163) were identified (Figure 5c and Figure S3 in the Supporting Information). Upon mass spectral interrogation, no Michael adducts of the peptide containing His-157 were detected (Figure S3 in the Supporting Information). However, in addition to detecting the unmodified peptide containing Cys-113, an HNE-adducted species of this peptide was also observed (Figure 5c). Relative to the unmodified (and reduced) theoretical mass of the peptide, the adducted peptide contained a mass shift of 158.13 Da, corresponding to reduced HNE adduction having an elemental composition C9H18O2. The theoretical m/z values of the observed peptide precursors were used to generate extracted ion chromatograms (XICs Figure 5c) for each peptide form. The calculated mass errors of the unmodified and HNE-adducted peptide were each within 1 part per million (ppm) of theoretical values, adding high confidence in the identification of the HNE-adducted peptide. In addition, the HNE-adducted peptide displayed the expected shift in retention time due to increased hydrophobicity, relative to the unmodified peptide (Figure 5c). Tandem mass spectra acquired from the modified peptides were manually interrogated to determine the site of adduction, and for comparison, the acquired MS/MS spectrum of the unmodified peptide was also examined. Tandem MS analysis revealed the presence of an HNE–Michael adduct localized to Cys-113 (Figure 5d). Further interrogation of other recovered Pin1 peptides failed to reveal any other HNE–Michael adducts or Schiff bases, further demonstrating the sensitivity of the catalytic cysteine of Pin1 to electrophile adduction.
Figure 5.
Pin1 is covalently modified at Cys-113 in MDA-MB-231 cells treated with HNE. (a) Western blot of Pin1 (top) and FLAG from MDA-MB-231 cells transfected with FLAG-Pin1. (b) 1D gel of FLAG-Pin1 immunoprecipitation from MDA-MB-231 cells treated with HNE. (c) Base peak chromatogram of peptides derived from immunoprecipitated and in-gel-digested flag-Pin1 and extracted ion chromatograms of observed forms of Pin1 peptide, DCSSAKARG. The base peak chromatogram (top) shows the LC-MS elution profile of peptides generated with endoproteinase AspN digestion of the excised Pin1 band. The observed m/z values of a selection of peptides are provided above their corresponding chromatographic peaks. Extracted ion chromatograms (XICs) of the in vivo unmodified and HNE-modified peptide DCSSAKARG, residues 112–120, are shown in middle and lower panels, respectively. A tolerance of ±10 ppm around the theoretical m/z values of the precursor ions observed for peptide forms of DCSSAKARG was used to generate the XICs. The observed m/z values are provided above the XIC peaks. The theoretical values of these precursor ions are adjacent to the observed peaks and were used to calculate ppm mass errors (shown in parentheses) for the monoisotopic ions of the identified peptide forms. Note that the Pin1 gel band was treated with DTT and iodoacetamide, resulting in carbamidomethylation of available Cys residues. Thus, the in vivo unmodified form of the peptide DCSSAKARG contains a carbamidomethyl group on Cys-113. (d) MS/MS spectra of in vivo unmodified (top) and HNE-modified peptide (bottom), DCSSAKARG. The [M + 2H]+2 precursor ions with m/z values 476.21 and 526.77, respectively, were selected for fragmentation. The observed singly and doubly protonated b- and y-type product ions are assigned to their corresponding m/z peaks in the tandem mass spectra. The amino acid sequence is provided above the annotated spectra, and the interresidue-placed brackets denote sites of fragmentation that occurred with collision-induced dissociation (CID) to produce the observed product ions. An asterisk is used to indicate the localization of the cysteine residue (Cys-113) modified by HNE.
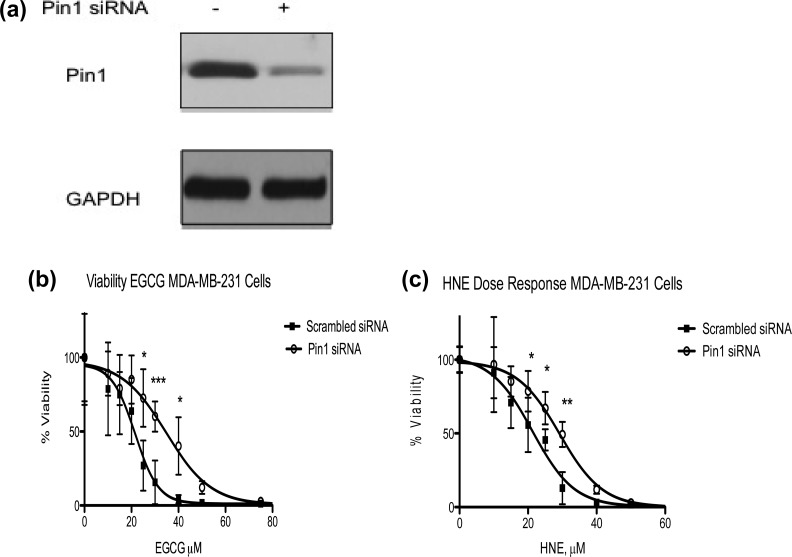
Knockdown of Pin1 Desensitizes MDA-MB-231 Cells to HNE-Induced Growth Inhibition
EGCG and PiB are pharmacological agents capable of suppressing proliferation of some cell types in a Pin1-dependent manner.14,15 HNE, which is produced endogenously and appears to modify the catalytic cysteine of Pin1, is also cytotoxic. Although HNE has many targets besides Pin1, we reasoned that the toxicity of HNE in MDA-MB-231 cells may be partly due to modification of this protein. We used siRNA to knock down Pin1 in cells and verified the knockdown by Western blot (Figure 6a). Cells transfected with scrambled siRNA and Pin1 siRNA were treated with increasing doses of HNE or the Pin1 inhibitor EGCG for 48 h, at which point cell viability was measured. Unsurprisingly, Pin1 siRNA-transfected MDA-MB-231 cells were less sensitive to EGCG-induced growth inhibition as compared to scrambled siRNA-transfected cells (IC50 values for scrambled and Pin1 siRNA-tranfected cells were 21.53 and 34.80, respectively) (Figure 6b). Cell viability was statistically higher in EGCG-treated Pin1 siRNA cells as compared to scrambled control cells at 25, 30, and 40 μM (Figure 6b). Interestingly, Pin1 knockdown also afforded protection from HNE toxicity relative to scrambled siRNA-transfected (control) cells. The percent viability of Pin1 siRNA cells treated with HNE was statistically increased at 20, 25, and 30 μM as compared to scrambled control siRNA cells, resulting in an increase in the IC50 for HNE (21.29 μM for scrambled vs 29.24 μM HNE for Pin1 siRNA cells, respectively) (Figure 6c). These data support the hypothesis that HNE modification of Pin1 at Cys-113 plays a role in the cellular response to lipid electrophile production during conditions of oxidative stress.
Figure 6.
Knockdown of Pin1 partially protects MDA-MB-231 cells against HNE-induced growth inhibition. (a) Western blot of Pin1 siRNA and scrambled control (GAPDH loading control). (b) Viability assay of scrambled and Pin1 siRNA-transfected MDA-MB-231 cells treated with the Pin1 inhibitor EGCG. Knockdown of Pin1 in MDA-MB-231 cells results in protection from EGCG-induced growth inhibition (21.53 μM for scrambled vs 34.80 μM for Pin1-siRNA transfected cells). (c) Viability assay of scrambled and Pin1 siRNA-transfected MDA-MB-231 cells treated with HNE. Knockdown of Pin1 in MDA-MB-231 cells results in a shift in the IC50 of HNE (21.29 μM for scrambled vs 29.24 μM for Pin1-siRNA transfected cells). n = 8/group; error bars represent standard deviation; *p < 0.05, **p < 0.01, and ***p < 0.001.
Discussion
The cis isomer of peptidyl-prolyl motifs in protein sequences occurs with a frequency of approximately 5–6%,23,24 a large majority of which appear at surface-exposed bend, coil, or turn conformations.25−27 Phosphorylation of serine or threonine preceding proline renders the peptide bond resistant to conventional PPIases, such as cyclophilin (Cyp18) and FKBP (FKBP12) enzymes, while simultaneously generating a Pin1-bindable motif.12 The tertiary structure and activity of proteins containing multiple pS-P or pT-P motifs can be largely dictated by whether these bonds are present in cis or trans. Therefore, Pin1 maintenance in cells is of great importance, as protein substrate activity and/or stability can be directly dependent on Pin1 catalysis of pSer-Pro and pThr-Pro bonds.
Here, we investigated the susceptibility of Pin1 to modification by HNE. HNE is a reactive aldehyde generated from the nonenzymatic oxidation of arachidonic acid or linoleic acid. Although HNE is produced endogenously at low micromolar concentrations, elevated levels of HNE are associated with a variety of diseases such as AD,28 carcinogenesis,29 and diabetes,30,31 among others. Previous work from our laboratory has shown that HNE administration to cells activates multiple signaling networks, such as the DNA damage, antioxidant, and heat shock response pathways.11 Additionally, HNE modifies proteins at exposed nucleophilic sites, altering tertiary structure and ultimately modifying activity. Because of the importance of Pin1 to the stability of transcription factors (e.g., p53, β-catenin, c-myc) as well as its effects on cell cycle checkpoint kinetics, the susceptibility of Pin1 to modification during conditions of oxidative stress warrants investigation.
Results of our experiments using purified protein incubated with HNE, as well as from cells treated with HNE, support that Pin1 is sensitive to electrophile adduction. We chose MDA-MB-231 breast cancer cells as our model to study the susceptibility of Pin1 to HNE modification, as previous studies have reported suppressed growth rates of small interfering RNA (siRNA)-silenced Pin1 MDA-MB-231 cells in vitro and decreased volume of tumors following orthotopic injection of short hairpin RNA (shRNA)-silenced Pin1 MDA-MB-231 cells into the mammary fat pad of immunodeficient mice.21 MDA-MB-231 cells incubated with aHNE and subject to click chemistry revealed a concentration-dependent increase in aHNE adduction of Pin1, with increasing modification observed as low as 2.5 μM. In resting cells, concentrations of HNE are reported to range from 0.5 to 3 μM;32 however, in tissues and fluids experiencing oxidative stress, levels of HNE are elevated as much as 10-fold.33 Levels of HNE in AD ventricular fluid have been reported at 15.2 μM,34 and coincidentally, Pin1 has been identified as an excessively carbonylated (2,4-dinitrophyenylhydrazine-reactive, DNPH-reactive) protein in brains of patients with AD and mild cognitive impairment (MCI).35,36 Although the site of Pin1 modification and adducting carbonyl-containing species was not determined in these studies, HNE adduction of Pin1-active site residues is a possible scenario. Nevertheless, our data support that Pin1 is covalently modified by lipid electrophiles under conditions that mimic a pathogenic level of oxidative stress.
Elucidation of amino acid sites of modification by HNE is critical to determining the effect the modification has on cellular signaling pathways. Experiments incubating purified protein with electrophiles have been valuable in categorizing proteins with HNE-sensitive active sites (e.g., thioredoxin and thioredoxin reductase)37 and those whose modification occurs at residues outside of the active site (e.g., HSP90).22 Active site modifications of proteins by HNE would theoretically have a greater impact on protein function relative to modifications that occur outside of the active site. Our data from experiments incubating purified Pin1 with 2 mM HNE revealed the presence of three Michael adducts, with two (Cys-113 and His-157) occurring at surface-exposed active site residues. No Schiff base modifications were observed, and although a third Michael adduct was detected (Lys-132), the relative extent of modification was significantly lower as compared to Cys-113 and His-157 adduction. Data from time-dependent experiments support the saturation of Cys-113 by HNE before quantifyable levels of His-157 adduction were observed. Furthermore, Cys-113 adduction was detectable at lower concentrations of HNE incubated with Pin1. We suspect that the equivalent relative reactivity of Cys-113 and His-157 observed in mapping the amino acid modification sites (Figure 2a and Table 1) is due to a stepwise saturation of Cys-113, followed by modification of His-157, resulting from the large amount of HNE used (2 mM) in this experiment. This suggestion is supported by results of the concentration-dependent HNE incubation experiments, in which the maximal concentration was 10-fold lower (200 μM) and revealed minimal His-157 adduction.
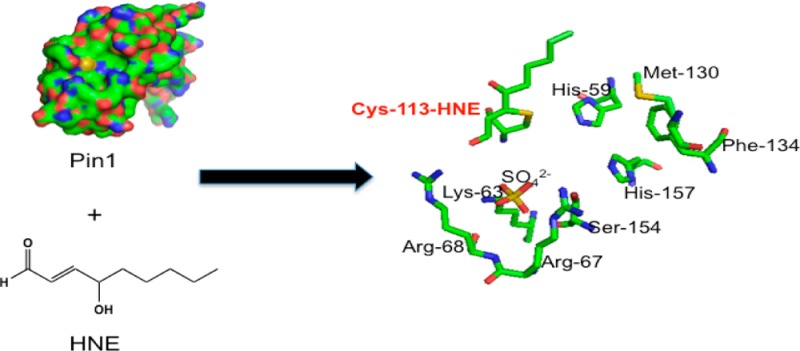
Cys-113 is the most important amino acid to Pin1 function, and cysteine residues are the most reactive amino acids with HNE. Cys-113 of Pin1 is surface exposed, thereby accessible to electrophiles such as HNE (Figure 7a). A proposed mechanism of isomerase action has suggested that His-59 abstracts a proton from Cys-113, and the resulting thiolate interacts with the carbonyl carbon of proline from the substrate,38 although this mechanism has been challenged.39 Mutagenesis experiments have revealed that mutation of Cys-113 to Ala results in 123-fold loss of protein activity.12 Furthermore, this conserved cysteine is essential for inhibition of parvulin PPIases by juglone.40 Upon observing that Cys-113 is the primary site of adduction of purified Pin1 incubated with HNE, we examined whether similar modifications to Pin1 would be observed in cells treated with HNE. Our proteomics data of Pin1 immunoprecipitated from FLAG-Pin1 transfected MDA-MB-231 cells treated with HNE support that Cys-113 is the primary site of modification of Pin1 upon cellular generation of HNE. Evidence of adduction of this peptide is supported by low ppm (1.1) mass error as well as a large shift in retention time of peptide elution (resulting from increased hydrophobicity of the reduced HNE-bound peptide relative to the unadducted peptide). Furthermore, multiple chromatographic peaks each having equivalent mass (Figure 5c) were observed. It is likely that this is due to chiralty conferred on the modified peptide by the reduced HNE–Michael adduct, causing slight but noticeable differences in retention time, as previously reported.41,42 No other modified Pin1 peptides were observed, including His-157, although the unmodified species of this peptide was recovered (Figure S3 in the Supporting Information). Our data support that Pin1 is covalently modified in cells primarily at the catalytic cysteine upon generation of HNE.
Figure 7.
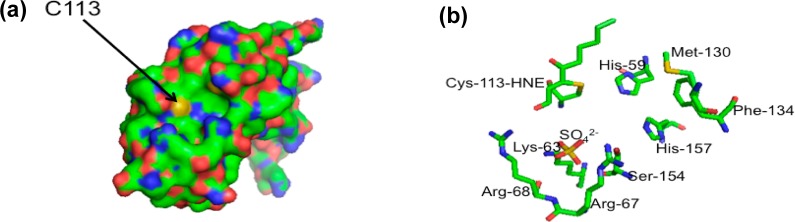
PyMol image of Pin1 surface (left) and active site (right) including HNE adducted to Cys-113. (a) Cys-113, a surface exposed residue, is indicated by the arrow and labeled in orange. (b) Pymol image of the Pin1 active site, containing HNE–Michael adduct at Cys-113.
Pin1 inhibition leads to neurofibrillary tangle formation and neuronal death in AD. Also in AD, Pin1 is oxidatively modified, down-regulated, and has decreased activity as compared to brains from normal elderly controls.35 In cancer, Pin1-targeted therapies have been suggested as possible chemotherapeutic agents. EGCG, a green tea polyphenol with potent anticancer activity, has recently been identified as a Pin1 inhibitor.14 EGCG interacts with both the WW and the PPIase domains of Pin1, interfering with Pin1-substrate binding and catalytic activity. Furthermore, Pin1 knockout mouse embryonic fibroblasts (MEFs) were protected from EGCG-induced cell death as compared to Pin1-expressing (WT) MEFs.14 We observed that Pin1 siRNA partially protected MDA-MB-231 cells against HNE toxicity as compared to scrambled control siRNA cells. The magnitude of the increase in the IC50 of Pin1 siRNA cells treated with HNE was less than the increase observed with EGCG (Figure 6b,c). This is likely due to the fact that HNE only modifies the catalytic cysteine, whereas EGCG binds to both the active site and the WW domain and to the fact that HNE does not modify all of the Pin1 in the MDA-MB-231 cells at the concentrations used for the toxicity experiment. In addition, HNE modifies many protein targets so that Pin1 is not the sole determinant of toxicity.
Unlike the pharmacological agents EGCG, PiB, and juglone, HNE is produced endogenously by cells and is elevated in conditions of oxidative stress. The results presented demonstrate the modification of the catalytic cysteine of Pin1 by an endogenously produced species that may have potential implications in disease where oxidative stress and deregulation of Pin1 coexist.
Glossary
Abbreviations
- Pin1
peptidylprolyl cis/trans-isomerase A1
- HNE
4-hydroxynonenal
- AD
Alzheimer’s disease
- EGCG
epigallocatechin gallate
- PPIase
peptidyl-prolyl cis/trans-isomerase
- ATCC
American Type Culture Collection
- aHNE
alkynyl-4-hydroxynonenal
- BCA
bicinchoninic acid assay
- RT
room temperature
- TBST
tris-buffered saline plus tween-20
- CHCA
α-cyano-4-hydroxycinnamic acid
- MEF
mouse embryonic fibroblast
Supporting Information Available
Figures of MALDI-TOF/TOF fragmentation of spectra of peaks and base peak and extracted ion chromatograms. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported by NIH Grants 5P01ES013125 to L.J.M. and 1F32GM100726-01A1 to C.D.A.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Markesbery W. R. (1999) The role of oxidative stress in Alzheimer disease. Arch. Neurol. 56, 1449–1452. [DOI] [PubMed] [Google Scholar]
- Butterfield D. A.; Lauderback C. M. (2002) Lipid peroxidation and protein oxidation in Alzheimer’s disease brain: Potential causes and consequences involving amyloid beta-peptide-associated free radical oxidative stress. Free Radical Biol. Med. 32, 1050–1060. [DOI] [PubMed] [Google Scholar]
- Marnett L. J. (2000) Oxyradicals and DNA damage. Carcinogenesis 21, 361–370. [DOI] [PubMed] [Google Scholar]
- Schneider C.; Porter N. A.; Brash A. R. (2008) Routes to 4-hydroxynonenal: Fundamental issues in the mechanisms of lipid peroxidation. J. Biol. Chem. 283, 15539–15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markesbery W. R.; Lovell M. A. (1998) Four-hydroxynonenal, a product of lipid peroxidation, is increased in the brain in Alzheimer’s disease. Neurobiol. Aging 19, 33–36. [DOI] [PubMed] [Google Scholar]
- Butterfield D. A.; Reed T.; Perluigi M.; De Marco C.; Coccia R.; Cini C.; Sultana R. (2006) Elevated protein-bound levels of the lipid peroxidation product, 4-hydroxy-2-nonenal, in brain from persons with mild cognitive impairment. Neurosci. Lett. 397, 170–173. [DOI] [PubMed] [Google Scholar]
- Rahman I.; van Schadewijk A. A.; Crowther A. J.; Hiemstra P. S.; Stolk J.; MacNee W.; De Boer W. I. (2002) 4-Hydroxy-2-nonenal, a specific lipid peroxidation product, is elevated in lungs of patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 166, 490–495. [DOI] [PubMed] [Google Scholar]
- Zhang B.; Shi Z.; Duncan D. T.; Prodduturi N.; Marnett L. J.; Liebler D. C. (2011) Relating protein adduction to gene expression changes: a systems approach. Mol. Biosyst. 7, 2118–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs A. T.; Marnett L. J. (2010) Systems analysis of protein modification and cellular responses induced by electrophile stress. Acc. Chem. Res. 43, 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila A.; Tallman K. A.; Jacobs A. T.; Liebler D. C.; Porter N. A.; Marnett L. J. (2008) Identification of protein targets of 4-hydroxynonenal using click chemistry for ex vivo biotinylation of azido and alkynyl derivatives. Chem. Res. Toxicol. 21, 432–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West J. D.; Marnett L. J. (2005) Alterations in gene expression induced by the lipid peroxidation product, 4-hydroxy-2-nonenal. Chem. Res. Toxicol. 18, 1642–1653. [DOI] [PubMed] [Google Scholar]
- Yaffe M. B.; Schutkowski M.; Shen M.; Zhou X. Z.; Stukenberg P. T.; Rahfeld J. U.; Xu J.; Kuang J.; Kirschner M. W.; Fischer G.; Cantley L. C.; Lu K. P. (1997) Sequence-specific and phosphorylation-dependent proline isomerization: A potential mitotic regulatory mechanism. Science 278, 1957–1960. [DOI] [PubMed] [Google Scholar]
- Zhou X. Z.; Kops O.; Werner A.; Lu P. J.; Shen M.; Stoller G.; Kullertz G.; Stark M.; Fischer G.; Lu K. P. (2000) Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol. Cell 6, 873–883. [DOI] [PubMed] [Google Scholar]
- Urusova D. V.; Shim J. H.; Kim D. J.; Jung S. K.; Zykova T. A.; Carper A.; Bode A. M.; Dong Z. (2011) Epigallocatechin-gallate suppresses tumorigenesis by directly targeting Pin1. Cancer Prev. Res. 4, 1366–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida T.; Takamiya M.; Takahashi M.; Miyashita H.; Ikeda H.; Terada T.; Matsuo Y.; Shirouzu M.; Yokoyama S.; Fujimori F.; Hunter T. (2003) Pin1 and Par14 peptidyl prolyl isomerase inhibitors block cell proliferation. Chem. Biol. 10, 15–24. [DOI] [PubMed] [Google Scholar]
- Jacobs A. T.; Marnett L. J. (2007) Heat shock factor 1 attenuates 4-Hydroxynonenal-mediated apoptosis: critical role for heat shock protein 70 induction and stabilization of Bcl-XL. J. Biol. Chem. 282, 33412–33420. [DOI] [PubMed] [Google Scholar]
- Tang X.; Sayre L. M.; Tochtrop G. P. (2011) A mass spectrometric analysis of 4-hydroxy-2-(E)-nonenal modification of cytochrome c. J. Mass Spectrom. 46, 290–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislinger T.; Humeny A.; Peich C. C.; Becker C. M.; Pischetsrieder M. (2005) Analysis of protein glycation products by MALDI-TOF/MS. Ann. N.Y. Acad. Sci. 1043, 249–259. [DOI] [PubMed] [Google Scholar]
- Codreanu S. G.; Zhang B.; Sobecki S. M.; Billheimer D. D.; Liebler D. C. (2009) Global analysis of protein damage by the lipid electrophile 4-hydroxy-2-nonenal. Mol. Cell Proteomics 8, 670–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. Y.; Tallman K. A.; Liebler D. C.; Porter N. A. (2009) An azido-biotin reagent for use in the isolation of protein adducts of lipid-derived electrophiles by streptavidin catch and photorelease. Mol. Cell Proteomics 8, 2080–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustighi A.; Tiberi L.; Soldano A.; Napoli M.; Nuciforo P.; Rosato A.; Kaplan F.; Capobianco A.; Pece S.; Di Fiore P. P.; Del Sal G. (2009) The prolyl-isomerase Pin1 is a Notch1 target that enhances Notch1 activation in cancer. Nat. Cell Biol. 11, 133–142. [DOI] [PubMed] [Google Scholar]
- Connor R. E.; Marnett L. J.; Liebler D. C. (2011) Protein-selective capture to analyze electrophile adduction of hsp90 by 4-hydroxynonenal. Chem. Res. Toxicol. 24, 1275–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart D. E.; Sarkar A.; Wampler J. E. (1990) Occurrence and role of cis peptide bonds in protein structures. J. Mol. Biol. 214, 253–260. [DOI] [PubMed] [Google Scholar]
- Pal D.; Chakrabarti P. (1999) Cis peptide bonds in proteins: residues involved, their conformations, interactions and locations. J. Mol. Biol. 294, 271–288. [DOI] [PubMed] [Google Scholar]
- Pahlke D.; Freund C.; Leitner D.; Labudde D. (2005) Statistically significant dependence of the Xaa-Pro peptide bond conformation on secondary structure and amino acid sequence. BMC Struct. Biol. 5, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahlke D.; Leitner D.; Wiedemann U.; Labudde D. (2005) COPS--cis/trans peptide bond conformation prediction of amino acids on the basis of secondary structure information. Bioinformatics 21, 685–686. [DOI] [PubMed] [Google Scholar]
- Lu K. P.; Finn G.; Lee T. H.; Nicholson L. K. (2007) Prolyl cis-trans isomerization as a molecular timer. Nat. Chem. Biol. 3, 619–629. [DOI] [PubMed] [Google Scholar]
- Butterfield D. A.; Bader Lange M. L.; Sultana R. (2010) Involvements of the lipid peroxidation product, HNE, in the pathogenesis and progression of Alzheimer’s disease. Biochim. Biophys. Acta 1801, 924–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair J.; Barbin A.; Velic I.; Bartsch H. (1999) Etheno DNA-base adducts from endogenous reactive species. Mutat. Res. 424, 59–69. [DOI] [PubMed] [Google Scholar]
- Mattson M. P. (2009) Roles of the lipid peroxidation product 4-hydroxynonenal in obesity, the metabolic syndrome, and associated vascular and neurodegenerative disorders. Exp. Gerontol. 44, 625–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverso N.; Menini S.; Odetti P.; Pronzato M. A.; Cottalasso D.; Marinari U. M. (2002) Diabetes impairs the enzymatic disposal of 4-hydroxynonenal in rat liver. Free Radical Biol. Med. 32, 350–359. [DOI] [PubMed] [Google Scholar]
- Esterbauer H.; Schaur R. J.; Zollner H. (1991) Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radical Biol. Med. 11, 81–128. [DOI] [PubMed] [Google Scholar]
- Siems W.; Grune T. (2003) Intracellular metabolism of 4-hydroxynonenal. Mol. Aspects Med. 24, 167–175. [DOI] [PubMed] [Google Scholar]
- Lovell M. A.; Ehmann W. D.; Mattson M. P.; Markesbery W. R. (1997) Elevated 4-hydroxynonenal in ventricular fluid in Alzheimer’s disease. Neurobiol. Aging 18, 457–461. [DOI] [PubMed] [Google Scholar]
- Sultana R.; Boyd-Kimball D.; Poon H. F.; Cai J.; Pierce W. M.; Klein J. B.; Markesbery W. R.; Zhou X. Z.; Lu K. P.; Butterfield D. A. (2006) Oxidative modification and down-regulation of Pin1 in Alzheimer’s disease hippocampus: A redox proteomics analysis. Neurobiol. Aging 27, 918–925. [DOI] [PubMed] [Google Scholar]
- Butterfield D. A.; Poon H. F.; Clair D. St; Keller J. N.; Pierce W. M.; Klein J. B.; Markesbery W. R. (2006) Redox proteomics identification of oxidatively modified hippocampal proteins in mild cognitive impairment: insights into the development of Alzheimer’s disease. Neurobiol. Dis 22, 223–232. [DOI] [PubMed] [Google Scholar]
- Fang J.; Holmgren A. (2006) Inhibition of thioredoxin and thioredoxin reductase by 4-hydroxy-2-nonenal in vitro and in vivo. J. Am. Chem. Soc. 128, 1879–1885. [DOI] [PubMed] [Google Scholar]
- Ranganathan R.; Lu K. P.; Hunter T.; Noel J. P. (1997) Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell 89, 875–886. [DOI] [PubMed] [Google Scholar]
- Behrsin C. D.; Bailey M. L.; Bateman K. S.; Hamilton K. S.; Wahl L. M.; Brandl C. J.; Shilton B. H.; Litchfield D. W. (2007) Functionally important residues in the peptidyl-prolyl isomerase Pin1 revealed by unigenic evolution. J. Mol. Biol. 365, 1143–1162. [DOI] [PubMed] [Google Scholar]
- Hennig L.; Christner C.; Kipping M.; Schelbert B.; Rucknagel K. P.; Grabley S.; Kullertz G.; Fischer G. (1998) Selective inactivation of parvulin-like peptidyl-prolyl cis/trans isomerases by juglone. Biochemistry 37, 5953–5960. [DOI] [PubMed] [Google Scholar]
- Wakita C.; Maeshima T.; Yamazaki A.; Shibata T.; Ito S.; Akagawa M.; Ojika M.; Yodoi J.; Uchida K. (2009) Stereochemical configuration of 4-hydroxy-2-nonenal-cysteine adducts and their stereoselective formation in a redox-regulated protein. J. Biol. Chem. 284, 28810–28822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balogh L. M.; Roberts A. G.; Shireman L. M.; Greene R. J.; Atkins W. M. (2008) The stereochemical course of 4-hydroxy-2-nonenal metabolism by glutathione S-transferases. J. Biol. Chem. 283, 16702–16710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.