Abstract
Enterohemorrhagic E. coli (EHEC) is a foodborne pathogen that causes watery diarrhea and hemorrhagic colitis. In this study, we identified StcE, a secreted zinc metalloprotease that contributes to intimate adherence of EHEC to host cells, in culture supernatants of atypical Shigella boydii 13 (Shigella B13) strains. Further examination of the Shigella B13 strains revealed that this cluster of pathogens does not invade but forms pedestals on HEp-2 cells similar to EHEC and enteropathogenic E. coli. This study also demonstrates that atypical Shigella B13 strains are more closely related to attaching and effacing E. coli and that their evolution recapitulates the progression from ancestral E. coli to EHEC.
Keywords: StcE, E. coli O157:H7, attaching and effacing lesions, Shigella boydii 13
Introduction
Enterohemorrhagic Escherichia coli (EHEC) cause diarrheal disease that ranges from watery diarrhea to hemorrhagic colitis. Virulence factors of EHEC include the chromosomally-encoded Shiga toxin and the locus of enterocyte effacement (LEE). LEE is a 35 kb pathogenicity island that confers the attaching and effacing phenotype to both EHEC and enteropathogenic E. coli (EPEC), wherein intimate adherence of the bacteria to host cells induces formation of actin-rich pedestals beneath the bacteria. The majority of the clinical EHEC disease in United States is caused by serotype O157:H7 (Manning et al., 2007), which carries a 92 kb virulence plasmid, pO157, that encodes many potential virulence factors, including stcE (Burland et al., 1998).
The stcE gene is encoded on the large virulence plasmids of E. coli O157:H7, O157:H-, ON:H7, and O55:H7 (Lathem et al., 2003). In all cases, stcE is found linked to etpD, which encodes the subunit of the type II secretion apparatus responsible for the secretion of StcE protein (Lathem et al., 2002). StcE is a 96 kDa zinc metalloprotease that cleaves specific O-linked glycoproteins and contributes to the intimate adherence of E. coli O157:H7 to HEp-2 cell surfaces (Grys et al., 2005). Evidence supports a role for StcE in EHEC disease in clearing the mucus that forms a protective barrier over the colonic epithelium. Following colonization, intimate adherence, and pedestal formation by EHEC, the clinical syndrome progresses from watery diarrhea to hemorrhagic colitis. At this stage, StcE plays an anti-inflammatory role by localizing the human complement regulator, C1 esterase inhibitor (C1-INH), to cell surfaces, decreasing the complement-mediated lysis of both bacteria and host cells (Lathem et al., 2004; Grys et al., 2006).
Shigella, another enteropathogen, is indistinguishable from E. coli by DNA-DNA hybridization techniques, with the exception of Shigella boydii 13 (Shigella B13) (Pupo et al., 2000). Shigella B13 is more closely related to Escherichia albertii than the E. coli-Shigella group, and lacks the large virulence plasmid, (pINV), that confers the invasion phenotype in all other Shigella. Hyma et al. demonstrated that Shigella B13 and E. albertii strains carry eae, a marker for LEE (Hyma et al., 2005). A small subset of analyzed Shigella B13 strains encoding eae were more related to the E. coli-Shigella group and labeled atypical Shigella B13. Many of these strains also carried markers for the pO157 plasmid, such as ehxA and toxB, suggesting that atypical Shigella B13 may be similar to EHEC and, thus, may encode stcE. This study describes the identification of stcE in atypical Shigella B13 strains and the genetic and phenotypic profile of this unique cluster of Shigella.
Materials and Methods
Bacterial culture, DNA extraction and PCR amplification
The S. boydii 7 and 13 and E. albertii strains used in this study are listed in table 2 and were provided by Thomas Whittam. E. coli O157:H7 EDL933 and E. coli O127:H6 E2348/69 were provided by Alison O’Brien. E. coli K12 MG1655 and S. flexneri 5a M90T were provided from Fred Blattner. Internal fragments of Shigella (Venkatesan et al., 2001) and E. coli (Burland et al., 1998) genes were amplified using the primers shown in table 1. Strains stored at −80°C in Luria-Bertani (LB) medium with 50% glycerol were directly inoculated into PCR reactions with GoTaq polymerase (Promega). The stcE gene was sequenced from PCR products amplified with primers IR ApaI 5′ 1 and etpD 3′ 1803 (table 1) and TripleMaster polymerase (Eppendorf) from plasmid DNA extracted from the atypical Shigella B13 strains using a Maxi Prep Kit (Qiagen). The nucleotide sequence for the stcE gene from the atypical Shigella B13 strains 3556–77, 3557–77, 3052–94, and 3053–94 have been submitted to GenBank under accession numbers EU159265, EU159266, EU159267, EU159268, respectively. For Southern blot analysis, plasmid DNA isolated from the atypical Shigella B13 strains was electrophoresed on a 0.6% agarose gel. Gel and stcE probe preparation and hybridization were performed as previously described (Lathem et al., 2003).
Table 2.
Prevalence of E. coli and Shigella specific genes detected by PCR and secreted StcE antigen and C1 cleavage activity detected by immunoblot.
| Straina | Species | E. coli O157:H7 | S. flexneri 5a | StcE secretion | StcE activity | ||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| plasmid
|
chromosome
|
plasmid
|
|||||||
| stcE/etpD | katP | ler/espA | cadA | ipaB/ipaD/icsA/mxiM | virK/ipaH7.8 | ||||
| EDL933 | E. coli O157:H7 | + | + | + | + | − | − | + | + |
| M90T | S. flexneri 5a | − | − | − | − | + | + | − | − |
| 5216–70 | Atypical S. boydii 13 | − | − | − | − | − | + | − | − |
| 3556–77 | Atypical S. boydii 13 | + | − | + | − | − | − | + | + |
| 3557–77 | Atypical S. boydii 13 | + | − | + | − | − | − | − | − |
| 3052–94 | Atypical S. boydii 13 | + | − | + | − | − | − | + | + |
| 3053–94 | Atypical S. boydii 13 | + | − | + | − | − | − | + | + |
stcE was not detected in the following additional strains tested: S. boydii 13 3103–99, S. boydii 13 ATCC
12032, S. boydii 13 2045–54, S. boydii 13 K-694, S. boydii 7 K-1, E. albertii 9194, E. albertii 10790,
E. albertii 10457, E. albertii 12502, E. albertii 19982.
Table 1.
Primers used in this study.
| Primer Name | Sequence |
|---|---|
| stcE 5′ 693 | 5′-CCGCTCCGGTGAACTGGAGAATA-3′ |
| stcE 3′ 1841 | 5′-CCTTATCTGCGGAGGCTGTAGGG-3′ |
| etpD SacI 5′ 1 | 5′-CCGAGCTCCGTGTTCACTACAGTAATTTTG-3′ |
| etpD XbaI 3′ 1929 | 5′-CCTCTAGATTACATCTCCTGCGCATAAA-3′ |
| katP 5′ 16 | 5′-CTTCCTGTTCTGATTCTTCTGG-3′ |
| katP 3′ 2141 | 5′-AACTTATTTCTCGCATCATCC-3′ |
| ler 5′ 38 | 5′-CACATACAACAAGTCCATACATTCAGC-3′ |
| ler 3′ 378 | 5′-CAGCGGTATTATTTCTTCTTCAGTGTCC-3′ |
| espA 5′ 81 | 5′-GTCGAAGGATGAGGTGGTTAAGCTA-3′ |
| espA 3′ 535 | 5′-ATTGCACATCAGAACGTGCACTCG-3′ |
| cadA 5′ 455 | 5′-ACATGGGTGGTACTGCATTCCAGA-3′ |
| cadA 3′ 1603 | 5′-ACAGCAGGTTATACGGACCGGTTT-3′ |
| ipaB 5′ 1 | 5′-ATGCATAATGTAAGCACCACAACC-3′ |
| ipaB 3′ 1743 | 5′-TCAAGCAGTAGTTTGTTGCAAAAT-3′ |
| ipaD 5′ 1 | 5′-ATGAATATAACAACTCTGACTAAT-3′ |
| ipaD 3′ 999 | 5′-TCAGAAATGGAGAAAAAGTTTATC-3′ |
| virK 5′ 188 | 5′-TTCTGGCAATACAACCCACGTTGC-3′ |
| virK 3′ 915 | 5′-TGCATCCAAAGAGCGGATAGCAGT-3′ |
| icsA 5′ 348 | 5′-AGGTCATGGTGGTGCTGGTGATAA-3′ |
| icsA 3′ 2002 | 5′-CTGCAATTTCCAGCCGGTCAGTTT-3′ |
| ipaH7.8 5′ 500 | 5′-ACAGGCTGACAACATTACCCGACT-3′ |
| ipaH7.8 3′ 1624 | 5′-TCTGCTGTTCAGTCTCACGCATCA-3′ |
| mxiM 5′ 27 | 5′-TGCTCTGCAGCAAAGATTAAATAGTGAAGA-3′ |
| mxiM 3′ 407 | 5′-TACCATGTCGAATCATCTGCCTCTCTC-3′ |
| traC 5′ 318 | 5′-TGGTGACAGGATTGAATACGGGCT-3′ |
| traC 3′ 1732 | 5′-GCAACAGCAGACCTTCATGCACTT-3′ |
| IR ApaI 5′ 1 | 5′-AAGGGCCCCTCTGAGGTGTCTGTTAAACCCGTGG-3′ |
| etpD 3′ 1803 | 5′-CGACTGCACCTGTTCCTGATTA-3′ |
StcE activity assay
To examine secretion of StcE, strains were grown in 25 ml Lennox L broth overnight at 37°C with aeration and cells removed by centrifugation. Three mls of culture supernatant were precipitated with 10% trichloroacetic acid on ice for ≥1 hour. To measure StcE activity, 12 ml of culture supernatant were incubated with 0.5 μg C1-INH protein (CompTech) overnight at room temperature prior to TCA precipitation. Precipitated protein was separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with polyclonal anti-rStcE’ antisera (Grys et al., 2005) or anti-C1-INH IgG (Cedarlane Laboratories).
Invasion assay
The gentamicin protection assay was used to determine the invasion phenotypes of the atypical Shigella B13 strains (Elsinghorst et al., 1994). A colony of each strain grown overnight on LB agar was inoculated into 2 ml of LB broth and incubated statically overnight at 37°C. Overnight culture (40 μl) was diluted into a total volume of 1 ml of HEp-2 media (EMEM, 1 mM sodium pyruvate, 10% FBS) prior to the addition to a monolayer of HEp-2 cells in a 24-well tissue culture plate (MOI of 14–95) and incubated at 37°C in 5% CO2 for 2 hours. Monolayers were washed with Dulbecco’s PBS (D-PBS) and fresh media containing 100 μg/ml gentamicin added for an additional 2 hours. The monolayers were washed with D-PBS and lysed with 1 ml 0.1% Triton X-100 per well. Suspensions were serially diluted and plated onto LB agar. Results are presented as the average percent of inoculum recovered after gentamicin treatment and are representative of duplicate samples in three independent experiments. Statistical analysis was preformed using a one-way ANOVA with a Tukey’s post hoc test.
Pedestal formation assay
To determine the ability of atypical Shigella B13 strains to form pedestals, HEp-2 cells were seeded onto 8-well microscope slides (Nalge Nunc International) 48 hours prior to infection so that cells would reach 50–80% confluency. Overnight bacterial cultures (10 μl of 2.5×108 to 9.0×108 CFUs/ml) grown as for the invasion assay were diluted into a total volume of 250 μl with HEp-2 media and added to each well of washed HEp-2 cells. The mixtures were incubated at 37°C in 5% CO2 for a total of 6–7 hours with a media exchange after 3 hours. Wells were washed with D-PBS and the cells fixed with 3% paraformaldehyde and permeabilized with 0.1% Triton X-100. Bacterial cells were stained with 1:200 goat anti-lipid A (Abcam), followed by 1:200 anti-goat-Alexa 488 and HEp-2 cells stained with 1:100 phalloidin-Alexa 594 (Invitrogen). Preparations were mounted with Prolong Gold (Invitrogen) and analyzed by epifluorescence microscopy (Carl Zeiss MicroImaging Inc.).
Results
PCR screen for presence of stcE in atypical S. boydii and E. albertii strains
We set out to identify stcE in other bacterial species recently found to carry eae, the gene that encodes the bacterial adhesin (intimin) required for pedestal formation. A PCR screen of numerous S. boydii and E. albertii strains showed that an internal fragment of stcE can be PCR amplified from only a subset of the S. boydii strains known as atypical S. boydii 13 (table 2). Atypical Shigella B13 strains 3557–77, 3556–77, 3052–94, and 3053–94, which form a distinct phylogenetic cluster, were all positive for the stcE gene. Atypical Shigella B13 strain 5216–70, which is phylogenetically clustered with enteroinvasive E. coli strains, was negative for the stcE gene. The presence or absence of the stcE gene in all strains was confirmed by Southern blot (data not shown). Analysis of isolated plasmid DNA by Southern blot demonstrated that stcE was encoded on the large plasmid of the four atypical Shigella B13 strains (data not shown). Sequence analysis of the 2.7 kb stcE gene showed only three synonymous substitutions shared among the atypical Shigella B13 strains and a Q727L substitution in strain 3556–77 compared to the EHEC EDL933 allele (data not shown). Six substitutions within 220 nucleotides of the intergenic region upstream of the predicted stcE promoter are present in the plasmids of all four atypical Shigella B13 strains compared to pO157.
Activity of StcE in atypical S. boydii B13 strains
To determine if the StcE protein was expressed and secreted by the atypical Shigella B13 strains, TCA-precipitated supernatants of overnight cultures were analyzed by immunoblot. StcE protein was identified in supernatants from strains 3556–77, 3052–94, and 3053–94, but not from 3557–77 or 5216–70 (table 2). StcE activity in culture supernatants was assayed for C1-INH proteolysis by immunoblots, and detected with all atypical Shigella B13 strains except 3557–77 and 5216–70 (Fig. 1, table 2).
Figure 1. StcE activity of atypical Shigella B13 strains.
Immunoblot of overnight culture supernatants of atypical Shigella B13 strains 5216–70, 3556–77, 3557–77, 3052–94, and 3053–94 incubated overnight at room temperature with purified C1-INH and probed with anti-C1-INH IgG. E. coli O157:H7 EDL933 (EHEC) and S. flexneri 5a M90T were used as positive and negative controls for C1-INH cleavage, respectively.
Atypical Shigella B13 strains encode other EHEC virulence factors
To determine if the atypical Shigella B13 plasmid encoding stcE is similar to the large invasion plasmid of Shigella (pINV), several pINV-encoded virulence factors were sought by PCR amplification (table 2). None of the pINV-encoded virulence factors could be amplified from the atypical Shigella B13 strains. PCR analysis using primers specific for pO157-encoded genes resulted in amplification of etpD, but not katP. The gene, traC, which is an F plasmid gene that is also encoded on the large virulence plasmid of E. coli O157:H-, pSFO157, did not PCR amplify from any of the atypical Shigella B13 strains tested.
The presence of additional E. coli-specific chromosomally-encoded genes was determined by colony PCR (table 2). The LEE-encoded regulator (Ler) is a global virulence regulator that has been shown to positively regulate the expression of LEE (Mellies et al., 1999), stcE, and the etp operon in E. coli O157:H7 (Lathem et al., 2002). PCR analysis of the atypical Shigella B13 strains identified the ler gene in the four atypical Shigella B13 strains encoding eae and stcE. An additional LEE-encoded gene, espA, encodes a subunit of the type III secretion system unique to EPEC and EHEC and is encoded by the atypical Shigella B13 strains encoding eae and stcE. PCR analysis of cadA, which encodes lysine decarboxylase and is universally absent in Shigella but present in most E. coli strains (Day et al., 2001), revealed that none of the atypical Shigella B13 strains encoded cadA.
Some atypical Shigella B13 strains invade and form pedestals on HEp-2 cells like E. coli
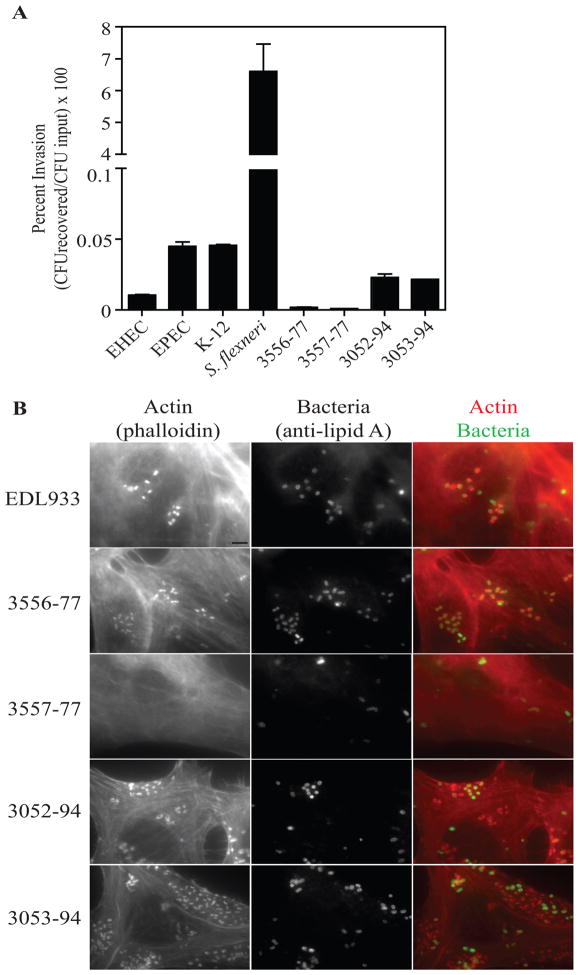
The abilities of the atypical Shigella B13 strains to invade HEp-2 cells were determined. Strains 3556–77 and 3557–77 showed invasion levels below the level of detection, whereas strains 3052–94 and 3053–94 showed relative levels of invasion more similar to E. coli than S. flexneri strains (Fig. 2A). The presence of the LEE operon and stcE suggested that the atypical Shigella B13 strains might form pedestals on host cells. We tested this hypothesis by infecting HEp-2 cells and observing for co-localization of bacteria with actin bundles on the surface of cells. Pedestal formation on HEp-2 cells could be detected for atypical Shigella B13 strains 3556–77, 3052–94, and 3053–94, but not 3557–77 (Fig. 2B).
Figure 2. Invasion and pedestal formation of atypical Shigella B13 strains.
A, Percent HEp-2 invasion by E. coli O157:H7 EDL933 (EHEC), E. coli O127:H6 E2348/69 (EPEC), E. coli K12 MG1655, S. flexneri 5a M90T, and the atypical Shigella B13 strains 3556–77, 3557–77, 3052–94, and 3053–94 as determined by the gentamicin protection assay with duplicate samples in three independent experiments. Statistical analysis was preformed using a one-way ANOVA with a Tukey’s post hoc test. S. flexneri invasion is significantly different from all other strains (P<0.001). Invasion was not significantly different among all other strains (P>0.05). B, Pedestal formation of the atypical Shigella B13 strains and E. coli O157:H7 EDL933. Infected HEp-2 cells were fixed and permeabilized and were stained with anti-lipid A antibody, followed by anti-goat-Alexa 488 and phalloidin-Alexa 594 antibodies. The scale bar located in the lower right corner of the EDL933 actin field represents 5 μm and is applicable to all micrographs.
Discussion
In this study, we discovered the stcE gene in the atypical Shigella B13 cluster. The relatively low incidence of three nucleotide substitutions within the 2.7 kb stcE gene compared to the six nucleotide substitutions within 220 nucleotides of the upstream intergenic region suggests selection for the preservation of StcE function. The acquisition of the large plasmid carrying stcE and the etp operon, in combination with the LEE element encoded on the chromosome, may provide a selective advantage by increasing the level of intimate adherence to host cells. A role of StcE in intimate adherence is further supported by the observation that a lack of extracellular StcE coincides with absence of pedestal formation by strain 3557–77.
The current model of Shigella evolution proposes that multiple ancestral E. coli clones acquired the pINV Shigella invasion plasmid, leading to selection for the loss of traits such as motility and lysine decarboxylation (Pupo et al., 2000). In contrast, the atypical Shigella B13 strains show loss of E. coli traits in the apparent absence of pINV selective forces. Furthermore, strains 3556–77 and 3557–77 display metabolic phenotypes intermediate between Shigella and E. coli, and atypical Shigella B13 DNA is more similar to E.coli than other Shigella B13 strains based on DNA-DNA hybridization assays (Brenner et al., 1982). These atypical Shigella B13 strains also form a distinct phylogenetic cluster and possess intermediate chromosomal genotypes between E. coli and Shigella groups (Hyma et al., 2005). As was previously suggested by Hyma et al., these data indicate that the atypical Shigella B13 strains were misclassified as Shigella and that they actually represent a lineage that evolved from ancestral forms of Shigella and attaching and effacing E. coli. The data presented here strengthen this argument by showing the acquisition of LEE and a pO157-like plasmid encoding stcE which we suggest recapitulates the model of EHEC evolution, described as the step-wise acquisition of the LEE element, followed by pO157 and then the Shiga toxin phage (Reid et al., 2000). We therefore propose to reclassify the atypical Shigella B13 strains as an E. coli group that, through convergent evolution or horizontal transfer of virulence genes on an ancestral background that shared both E. coli and Shigella characteristics, has evolved to closely resemble pathotypes of E. coli that form attaching and effacing lesions.
Sequelogs of stcE, historically named tagA, have been functionally characterized in other pathogens, including Vibrio cholerae and Aeromonas hydrophila (Szabady et al., 2010 and Pillai et al., 2006). Like StcE, V. cholerae TagA is a secreted mucinase and contributes to colonization of the intestinal epithelium (Szabady et al., 2010). The A. hydrophilia TagA exhibits an additional StcE function by cleaving and localizing C1-INH the surface of bacterium, increasing the serum resistance of the bacterium in vitro. An isogenic deletion mutant of tagA decreased the mortality of mice compared to wild-type A. hydrophila in a mouse model of peritonitis (Pillai et al., 2006). Thus, StcE-like metalloproteases play a role in the virulence phenotypes of A. hydrophila, V. cholerae and E. coli O157:H7. In this study, we identified stcE as a possible virulence factor in atypical Shigella B13 strains, and further characterized this unique cluster of attaching and effacing pathogens.
Acknowledgments
We would like to thank Thomas Whittam, Alison O’Brien, and Fred Blattner for bacterial strains, Nancy Strockbine for information regarding the atypical Shigella B13 strains, Jay Bangs for use of his epifluorescence microscope, and Rose Szabady and Becca Moritz for insightful discussions regarding the project and critical reading of the manuscript. This work was supported by NIH grant RO1 AI051735.
References
- Brenner DJ, Steigerwalt AG, Wathen HG, Gross RJ, Rowe B. Confirmation of aerogenic strains of Shigella boydii 13 and further study of Shigella serotypes by DNA relatedness. J Clin Microbiol. 1982;16:432–36. doi: 10.1128/jcm.16.3.432-436.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burland V, Shao Y, Perna NT, Plunkett G, Sofia HJ, Blattner FR. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 1998;26:4196–204. doi: 10.1093/nar/26.18.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day WA, Fernández RE, Maurelli AT. Pathoadaptive mutations that enhance virulence: genetic organization of the cadA regions of Shigella spp. Infect Immun. 2001;69:7471–80. doi: 10.1128/IAI.69.12.7471-7480.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsinghorst EA. Measurement of invasion by gentamicin resistance. Methods Enzymol. 1994;236:405–20. doi: 10.1016/0076-6879(94)36030-8. [DOI] [PubMed] [Google Scholar]
- Grys TE, Walters LL, Welch RA. Characterization of the StcE protease activity of Escherichia coli O157:H7. J Bacteriol. 2006;188:4646–53. doi: 10.1128/JB.01806-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grys TE, Siegel MB, Lathem WW, Welch RA. The StcE protease contributes to intimate adherence of enterohemorrhagic Escherichia coli O157:H7 to host cells. Infect Immun. 2005;73:1295–303. doi: 10.1128/IAI.73.3.1295-1303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyma KE, Lacher DW, Nelson AM, et al. Evolutionary genetics of a new pathogenic Escherichia species: Escherichia albertii and related Shigella boydii strains. J Bacteriol. 2005;187:619–28. doi: 10.1128/JB.187.2.619-628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathem WW, Bergsbaken T, Welch RA. Potentiation of C1 esterase inhibitor by StcE, a metalloprotease secreted by Escherichia coli O157:H7. J Exp Med. 2004;199:1077–87. doi: 10.1084/jem.20030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathem WW, Bergsbaken T, Witowski SE, Perna NT, Welch RA. Acquisition of stcE, a C1 esterase inhibitor-specific metalloprotease, during the evolution of Escherichia coli O157:H7. J Infect Dis. 2003;187:1907–14. doi: 10.1086/374719. [DOI] [PubMed] [Google Scholar]
- Lathem WW, Grys TE, Witowski SE, et al. StcE, a metalloprotease secreted by Escherichia coli O157:H7, specifically cleaves C1 esterase inhibitor. Mol Microbiol. 2002;45:277–88. doi: 10.1046/j.1365-2958.2002.02997.x. [DOI] [PubMed] [Google Scholar]
- Manning D, Madera T, Schneider, et al. Surveillance for Shiga Toxin-producing Escherichia coli, Michigan, 2001–2005. Emerg Infect Dis. 2007;13:318–321. doi: 10.3201/eid1302.060813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellies JL, Elliott SJ, Sperandio V, Donnenberg MS, Kaper JB. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler) Mol Microbiol. 1999;33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- Pillai L, Sha J, Erova TE, Fadl AA, Khajanchi BK, Chopra AK. Molecular and functional characterization of a ToxR-regulated lipoprotein from a clinical isolate of Aeromonas hydrophila. Infect Immun. 2006;74:3742–55. doi: 10.1128/IAI.00402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pupo GM, Lan R, Reeves PR. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci U S A. 2000;97:10567–72. doi: 10.1073/pnas.180094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SD, Herbelin CJ, Bumbaugh AC, Selander RK, Whittam TS. Parallel evolution of virulence in pathogenic Escherichia coli. Nature. 2000;406:64–7. doi: 10.1038/35017546. [DOI] [PubMed] [Google Scholar]
- Venkatesan MM, Goldberg MB, Rose DJ, Grotbeck EJ, Burland V, Blattner FR. Complete DNA sequence and analysis of the large virulence plasmid of Shigella flexneri. Infect Immun. 2001;69:3271–85. doi: 10.1128/IAI.69.5.3271-3285.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]