Abstract
Cellular responses to inputs that vary both temporally and spatially are determined by complex relationships between the components of cell signaling networks. Analysis of these relationships requires access to a wide range of experimental reagents and techniques, including the ability to express the protein components of the model cells in a variety of contexts. As part of the Alliance for Cellular Signaling, we developed a robust method for cloning large numbers of signaling ORFs into Gateway® entry vectors, and we created a wide range of compatible expression platforms for proteomics applications. To date, we have generated over 3000 plasmids that are available to the scientific community via the American Type Culture Collection. We have established a website at www.signaling-gateway.org/data/plasmid/ that allows users to browse, search, and blast Alliance for Cellular Signaling plasmids. The collection primarily contains murine signaling ORFs with an emphasis on kinases and G protein signaling genes. Here we describe the cloning, databasing, and application of this proteomics resource for large scale subcellular localization screens in mammalian cell lines.
Analysis of cross-talk between signaling pathways when mammalian cells are challenged with multiple ligand stimuli and the development of molecular models that describe signal integration and processing provide key insight to cellular signaling mechanisms and the regulation of cellular function (1, 2). Two strategic requirements in projects of this nature are the identification of the protein components in a given model system (the so-called “parts list”) and the ability to modulate the expression and function of these protein components. At the outset of the Alliance for Cellular Signaling (AfCS)1 project (1), reagent availability was a central issue, and in the case of cDNA clones, there was no publicly accessible repository of validated mouse sequences. Moreover the nature of the project demanded the establishment of standardized methodology for the isolation of cDNAs and their expression in a variety of contexts. The emergence of recombination-based cloning technologies was timely and allowed the development of a robust cloning platform. We adopted Invitrogen’s Gateway® system that allows facile transfer of DNA segments into multiple expression platforms while maintaining orientation and reading frame register (3–5).
Several reports have described efforts to generate genome-wide collections of cDNAs (6, 7) and the production of “OR-Feome” sets of sequence-validated open reading frames (8–10). The generation of “ORF-only” clones in these latter efforts is key for downstream proteomics applications that require expression of proteins with N- or C-terminal fusion tags (11–14). The AfCS cloning effort has focused on genes involved in cell signaling and therefore does not approach the scale of these genome-wide projects. However, as the described OR-Feome cloning projects have concentrated on model organisms other than mouse, the AfCS-generated plasmids remain, to our knowledge, the largest collection of mouse ORF-only clones available through a non-commercial source.
Using a set of mouse serine-threonine kinase (STK) genes as an example, we describe the development of protocols and vectors contained in the AfCS plasmid database. This repository has grown into a significant resource of several thousand plasmids that have been made available to the scientific community through the ATCC. Certain gene families with key roles in cell signaling such as kinases and heterotrimeric G proteins are highly represented in the database. We have created expression vectors permitting facile recombination-based generation of fluorescent protein fusions for imaging applications and affinity tag fusions for immunoprecipitation of protein complexes. Using the former expression platform, we have carried out large scale subcellular localization studies in murine cell lines to create the AfCS image database. We present selected subcellular localization data to demonstrate the functional utility of this resource.
EXPERIMENTAL PROCEDURES
Detailed protocols developed by the AfCS are available on line at www.signaling-gateway.org/data/ProtocolLinks.html. Brief summaries of key procedures are described below.
Design of Scripts
Cloning Primers
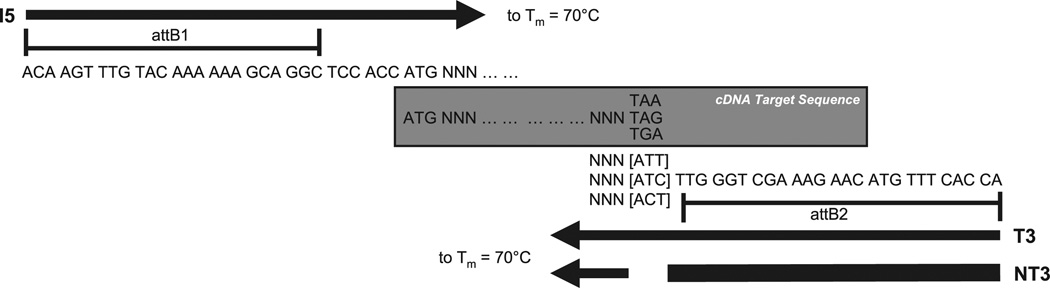
Cloning primers of a defined minimum length, starting at the termini of each ORF and with a melting point above a minimum desired temperature (Tm = 70 °C), were designed using an in-house Perl script. The calculations in the script were performed as described previously (15, 16) based on 50 mM salt and 50 nM DNA concentrations. The attB Gateway recombination sites were added to the primer sequence after the Tm calculations (Fig. 1 and Supplemental Table 1).
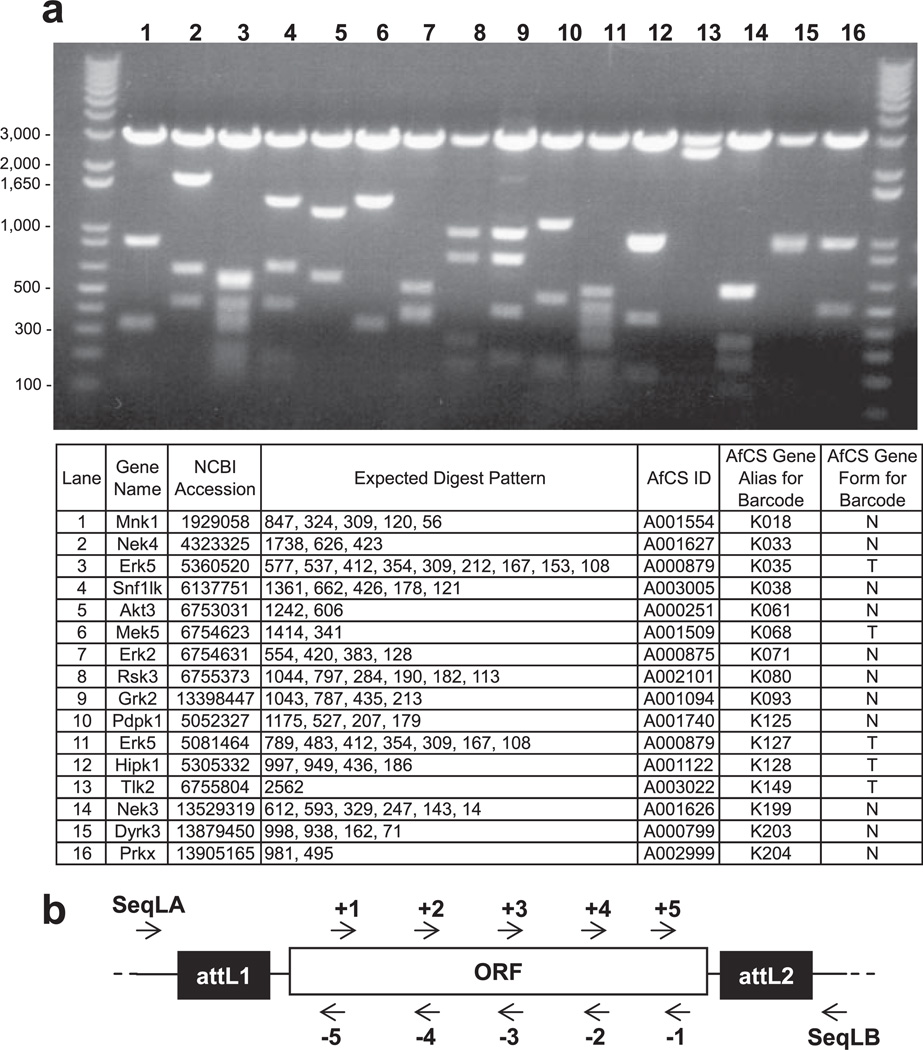
Fig. 3. Entry clone diagnostics and sequencing.
a example of a diagnostic BanII restriction digest profile of selected serine-threonine kinases. The common 3-kb band in lanes 1–16 represents the entry vector backbone. The table lists the expected digest pattern in addition to further gene details. b , schematic demonstrating primer design for ORF sequencing. Gene-specific primers are generated along both strands of each target gene, spaced ~500 bases apart. Two entry vector-specific flanking primers (SeqLA, TCGCGTTAACGCTAGCATGGATCTC; SeqLB, GTAACATCAGAGATTTTGAGACACGGG) provide end sequence.
Sequence Primers
We utilized Primer3 (17) to generate sequencing primers at approximately equal intervals along a sequence (Fig. 3b and Supplemental Table 2). Assuming we need n pairs of primers based on clone length L and interval length l e.g. 500 bp), we then ran Primer3 n + 1 times. Generally we targeted the interval between the coordinates we wanted for the m th primer on the positive strand (minus a window, e.g. 50 bp) and for the (n − m)th primer on the negative strand (plus a window, e.g. 50 bp). The coordinates refer to the positive strand position (i.e. position −y on the reverse strand is actually L – y on the forward strand). For the first reverse primer, we defined one excluded area up to the end of the clone and ran Primer3 to design a pair of primers. We then took only the reverse primer. Primer3 accepts different values for product size, primer GC content, primer size, and primer melting point. For each m > 1 (up to the last one), we used those coordinates to define two excluded zones (from those coordinates to the closest clone ends) and generated a pair of primers using the product size parameter. For the last forward primer, we proceeded similarly to the first reverse primer, with one excluded zone, but only kept the forward primer.
RNA Source and cDNA Preparation
1 µg of mouse brain, testis, or spleen poly(A)+ RNA (Clontech) was used for first strand synthesis using the cDNA Synthesis System (Invitrogen), subaliquoted, and stored at −20 °C.
Amplification of attB PCR Products for Target Genes
Primers were purchased resuspended to 100 µM (Invitrogen). 38 ng of brain cDNA and 38 ng of testis cDNA were combined for amplification of target genes with ProofStart DNA polymerase (Qiagen) including Q-solution in a total reaction volume of 50 µl. Products were amplified using the following cycling program: 95 °C for 5 min, 94 °C for 30 s, 72 °C for 6 min (five cycles), 94 °C for 30 s, 70 °C for 6 min (five cycles), 94 °C for 30 s, 68 °C for 6 min (25 cycles), 4 °C hold. Amplification products were visualized on a 1% agarose, ethidium bromide gel. The correct bands were excised and purified using the QIAquick Gel Extraction kit (Qiagen) and eluted in 30 µl EB buffer (Qiagen).
BP Reactions and Entry Clone Diagnostics
150 ng of pDONR207 was combined with 50–150 ng of purified attB PCR product, 2 µl of BP Clonase enzyme, and 2 µl of BP Reaction Buffer (Invitrogen), and the volume was adjusted to 10 µl/reaction using Milli-Q purified water. After incubation at room temperature for 1 h, 1 µl of Proteinase K was added, and reactions were incubated at 37 °C for 10 min. 5 µl of the reaction mixture was used to transform TOP10 cells (Invitrogen), and recombinants were selected on gentamicin plates (10 µg/ml). Entry clone candidates were identified by digestion with the BanII restriction enzyme (New England Biolabs).
Sequence Reactions, Assembly, and Analysis
Sequence reactions were submitted to Macrogen Inc. (Seoul, South Korea) or Elim Biopharmaceuticals, Inc. (Hayward, CA) in 96-well plate format. 5 µl of miniprep DNA and 5 µl of sequencing primer at 2 µM (Invitrogen) were dispensed per well in separate plates. Sequencing output traces were input into Paracel Genome Assembler (PGA) to generate one contig per sequenced ORF. Contigs were imported into OMIGA 2.0 (Oxford Molecular) for alignment against target ORFs and for further analysis.
LR Reactions and Expression Clone Diagnostics
100 ng of pEN was combined with 100 ng of pDS, 2 µl of LR Clonase enzyme, and 2 µl of LR Reaction Buffer (Invitrogen), and the volume was adjusted to 10 µl/reaction using Milli-Q purified water. After reactions were incubated at room temperature for 1 h, 1 µl of Proteinase K was added, and reactions were incubated at 37 °C for 10 min. 5 µl of the reaction mixture was transformed in TOP10 cells (Invitrogen), and recombinants were selected on kanamycin (10 µg/ ml) or ampicillin (50 µg/ml) plates. Expression clone candidates were validated by restriction digest usually with the NcoI restriction enzyme (New England Biolabs).
Plasmid Database
The AfCS plasmid database structure is broadly composed of three parts. The first part is the vector information that consists of the “parent_vector,” “target_gene,” “cloned_gene,” “construct,” and “misc_plasmid” tables used for storing sequences, features, and related information about vectors. Plasmid map diagrams are generated from this information using CGView (18) and cached in the corresponding “map” tables. For AfCS users, certain functions have been implemented for generating collections of records, e.g. a parent vector may be selected followed by a search for cloned genes to create a set of construct records in batch format. In this case, the construct sequences are generated from the parent vector template and the cloned gene sequence, whereas the construct feature offsets are adjusted from the parent vector template features. The second part is laboratory storage information that uses the “plasmid_prep” and “prep_storage_list” tables to track samples that have been created in the laboratory in terms of their location (freezer, box, and position). The third part is the batch system that allows collections of parent_vector, cloned_gene, construct, and plasmid_prep records to be grouped together. Various functions have been created to operate on batches to simplify routine processing for AfCS laboratory personnel. For example, storage location records may be generated for samples by automatically or manually assigning the next sequentially available box and position; barcode files for label printing may be generated from the batch records; data files may be created for exporting construct information to ATCC. This flexible approach allows additional batch-oriented functions to be easily added to the programs.
Subcellular Localization in RAW264.7 Cells
A short summary of the steps taken is presented along with the AfCS protocol identification number referenced in parentheses. RAW 264.7 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% heat inactivated fetal bovine serum at 37 °C with 5% CO2 (PP00000159). Cells were transiently transfected with 0.5 µg of each DNA and 1 µl of Lipofectamine 2000 (Invitrogen). The protocol was modified from the manufacturer’s usual method so that the cells were simultaneously transfected and plated onto an 8-well coverglass chamber (PP00000182). Live cell confocal images were collected 20–30 h after transfection. Images were collected on an automated Zeiss Axiovert with 100× oil objective using a Nipkow spinning disk. The automated (PP00000143) microscope and camera (Photometrics CoolSnap HQ) were controlled using MetaMorph software (Molecular Devices Corp.).
RESULTS
Primer Design and Amplification of attB PCR Products
To develop a methodological platform for efficient cloning of large numbers of signaling ORFs for the AfCS research effort, we chose the STK gene family (19, 20) as a pilot project as it was well represented in the public sequence databases at the outset of the AfCS program. We queried the simple modular architecture research tool (SMART) database (21, 22) to generate a comprehensive, yet unique, list of ORFs and identified 217 distinct murine STK ORFs based on the information publicly available in 2001. As different applications might require the ability to express either N- or C-terminal fusions of the cloned cDNAs, we generated two forms of each ORF: T and N. The former includes the stop codon allowing for N-terminal tagging, whereas the latter lacks this codon to permit C-terminal fusions. To allow facile subcloning of cDNAs to multiple expression platforms, we used the Gateway recombination- based cloning technology (4).
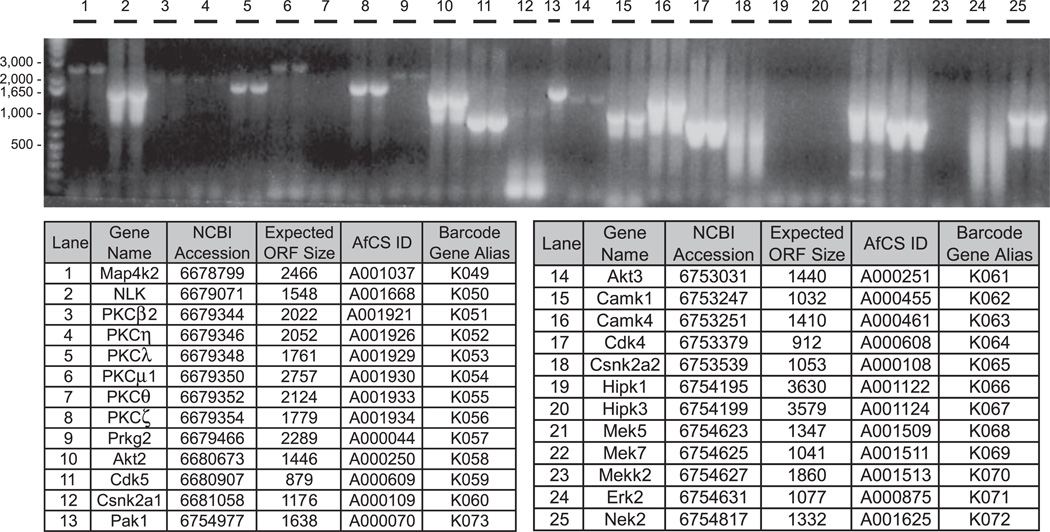
It has been demonstrated that PCR isolation of cDNAs from complex mixtures can be facilitated by amplification protocols that use combined annealing/extension steps that decrease stepwise from high temperatures (so-called “touchdown PCR” (23)). Because these protocols require high initial primer annealing temperatures, we designed a Perl script (see “Experimental Procedures”) that processes input ORF sequences (in FASTA format) to generate cloning primers with a Tm of 70 °C (Supplemental Table 1). The schematic in Fig. 1 shows the design and nomenclature of the cloning primers for amplification of the T and N forms of each ORF. We initially tested several mRNA sources and found a mixture of mouse brain and testes to be optimal for amplification of most signaling target mRNAs, whereas hematopoietic-specific target mRNAs were more often isolated from a mouse spleen source (data not shown). We used the hot start-activated ProofStart DNA polymerase enzyme for amplification as its proprietary chemical modification ensures that primers remain intact during setup to prevent mispriming, and it offers efficient exonuclease activity for high fidelity (10 times greater than Taq DNA polymerase). Fig. 2 shows a typical example of attB PCR amplification of a subset of our targeted STKs from brain/testes cDNA. Of the 434 PCRs attempted for the initial STK target set (T and N forms of 217 ORFs), we amplified 375 products based on expected size (data not shown).
Fig. 1. Design of attB gene-specific PCR amplification primers.
Forward primer (I5) contains attB1 sequence, and reverse primers (T3 and NT3) contain attB2 sequence. I5 and T3 primer pair amplifies the T form of target gene, whereas I5 and NT3 primer pair amplifies the N form. Note that the bracketed sequence complementary to the stop codon is included in the T3 primer but is omitted from NT3 primer. All primers extend to a Tm of 70 °C exclusive of the attB sequence.
Fig. 2. attB PCR amplification of target cDNAs.
An example of attempted attB PCR of 25 murine kinases ranging in size from 0.9 to 3.6 kb is shown. Each reaction is loaded over two lanes except lane 13. 20 of the 25 reactions (80%) produce an amplification product of the expected ORF size.
Recombinational Cloning and Sequence Analysis
Purified PCR products were recombined with pDONR207 in a BP reaction to generate entry vectors for each target mRNA (4). pDONR207 was favored over other commercially available donor vectors as it produces gentamicin-resistant entry clones compatible for recombination to either ampicillin- or kanamycin-based expression vectors.
In moderate to high throughput cloning protocols, it is helpful to have a simple diagnostic method to reliably identify clone candidates prior to sequencing. The redundancy in the BanII recognition site (GPuGCPyC where Pu is purine and Py is pyrimidine) leads to a higher likelihood of internal restriction sites being present in most target genes, increasing variation in expected banding patterns from ORFs of similar length and allowing for more reliable identification of correct clones. Furthermore the entry vector backbone derived from pDONR207 contains only two BanII sites that conveniently flank the ORF. Thus, the parent vector backbone contributes a single 3-kb band in screens for pEN candidates thereby minimizing interference when analyzing sizes of gene-specific bands. We developed a simple Perl script to output expected BanII digest patterns for any number of ORFs input in FASTA format. An example of a diagnostic digest of 16 entry clones is shown in Fig. 3a. Using this screening method, we identified sequence candidates for 95% of the STK ORFs that went through BP recombination in our initial clone set.
The Primer3 PCR primer program (17) was customized to autogenerate sequence primers along both DNA strands of each target ORF spaced ~500 bases apart (see “Experimental Procedures” and Supplemental Table 2). Two entry vector-specific flanking primers were also used for each ORF to provide extensive coverage of both strands of each sequenced clone (Fig. 3b). We used PGA software to assemble the output from each set of sequencing reactions into a single contig. PGA utilizes an algorithm that allows comparison of base calls in the raw sequencing data to evaluate and select the most accurate reading. We found that the combination of generous overlap between individual contigs from the 500-nucleotide spacing of primers and the efficient analysis of the PGA software provided a robust protocol for generation of high quality sequence data. More importantly, the generation of N and T forms for each ORF, from separate amplification reactions, allowed us to better differentiate between PCR-generated mutations and genuine variants due to splice differences or polymorphisms. We found many cases where our amplified sequence did not precisely match the reference sequence in GenBank™. This disagreement could be caused by randomly generated PCR-based mutations. However, if the same “mutation” occurred in the independently amplified T and N forms of a given gene, the chance that this difference was introduced randomly by amplification was considered negligible, and the clones in question were databased as valid. Details of any such differences between our target and cloned cDNAs (target ORF versus AfCS ORF) were recorded in the AfCS plasmid database (described below).
Design, Structure, and Content of the AfCS Plasmid Database
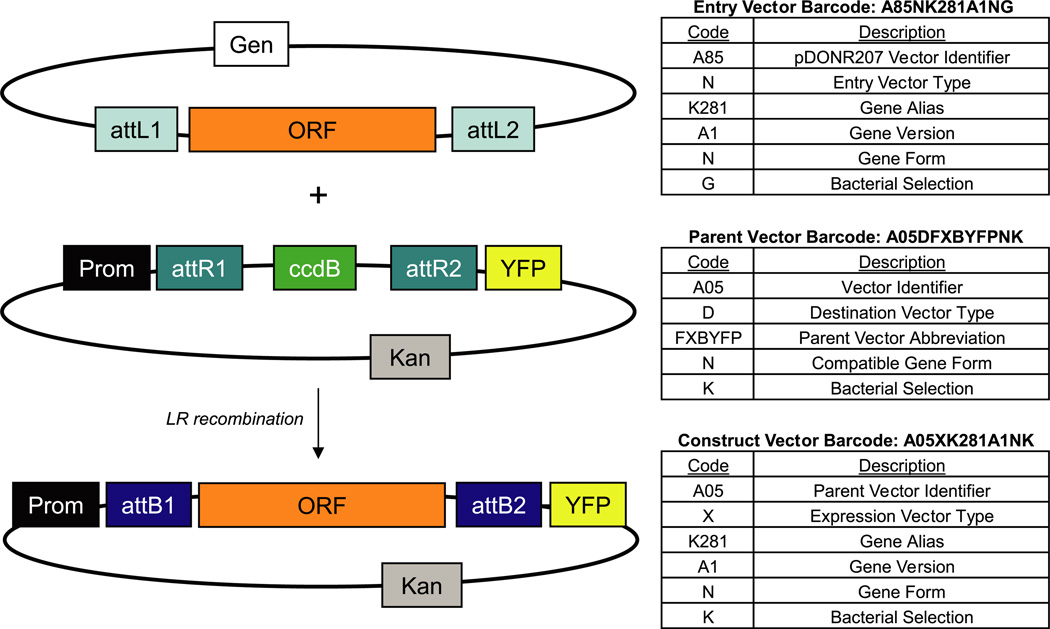
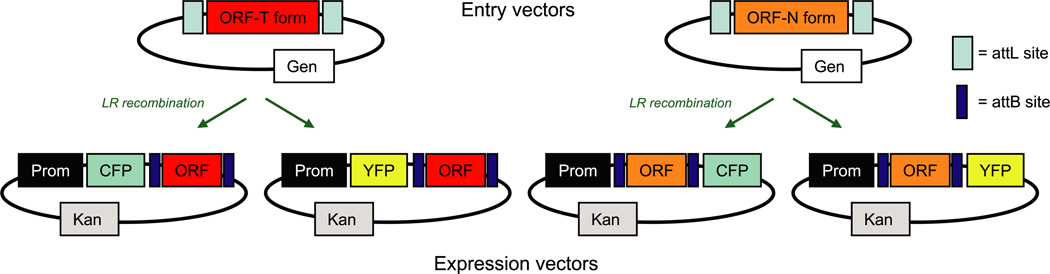
One advantage of the Gateway cloning platform is that it consists of standard backbones for each parent vector into which cDNAs are incorporated at invariant recombination sites. In this context, the union of a “parent” vector backbone and a sequence-validated “cloned gene” generates a construct. With regard to standard Gateway nomenclature in this system, both entry vectors (pEN) and expression vectors (pEX) containing a cloned cDNA are constructs, whereas destination vectors (pDS) are parents (Fig. 4). We designed the AfCS plasmid database to build and curate these various vector types. This database combines cloned gene and parent vector template information, and it uses custom Perl scripts to automatically generate detailed construct records and maps (Supplemental Fig. 1, a and b). To facilitate tracking of these vectors, we created a barcode system for ready identification of relevant components and properties (Fig. 4). This database structure supports the generation of detailed records for the large number of constructs generated by the AfCS consortium. The details for a given cloned cDNA only require one initial entry to the database; all subsequent construct records created using that sequence are autogenerated. We found this particularly valuable for 96-well-based applications where a plate of 96 entry clones could be sub-cloned by LR recombination to a set of four CFP and YFP fusion expression vectors for microscopy studies (Fig. 5). The database was designed to permit effortless combination of the 96 “cloned cDNAs” with each of the four parent vector sequence “templates” to create 384 construct records containing full sequence and recommended diagnostic digests to validate recombinants. The plasmid database software is a standard web-based database application that consists of a collection of Perl common gateway interface (CGI) programs for display and data entry and uses an Oracle database for persistence (Supplemental Fig. 2 and “Experimental Procedures”).
Fig. 4. AfCS plasmid database vector types and barcodes.
The ORF from the entry vector recombines with the parent vector backbone to generate an expression construct. In the AfCS plasmid database, entry vectors and expression vectors, which both contain a cloned ORF, are classified as constructs, whereas parent vectors remain in a separate category. The tables to the right describe the components of the AfCS barcode system for each vector type.
Fig. 5. Schematics of ORF transfer by recombinational cloning to generate expression clones for subcellular localization studies.
ORF-T form contains the stop codon, whereas ORF-N form lacks the stop codon. ORF sequence is transferred to expression vectors via LR recombination. The T form is CFP- or YFP-tagged at the N terminus; the N form is CFP- or YFP-tagged at the C terminus.
The publicly accessible version of the database contains constructs that have been made available to the research community through the ATCC (see their molecular genomics clone search on line). Users can view, browse, and search the list of available vectors at www.signaling-gateway.org/data/plasmid/ where barcodes link to the detailed construct records including full sequence and plasmid maps (Supplemental Fig. 1, a and b). The database also contains details on a number of available AfCS-developed parent vectors that can be used to create expression constructs for a range of applications from tagging of cDNAs with GFP derivatives to affinity tags for proteomics applications (Supplemental Table 3). Although the majority of AfCS “expression-ready” constructs available from the ATCC are CFP or YFP fusions (1898 of a total 3076 constructs containing cDNA), we have also deposited all of the cloned ORFs as entry vectors (1178 of the 3076) that can be used to move the ORFs to any of the parent vectors described in Supplemental Table 3 or to any other Gateway-ready vector.
Subcellular Localization of cDNAs in RAW264.7
As described above, a large proportion of the vectors generated for AfCS studies were CFP and YFP fusion constructs for microscopy studies. In an effort to generate a database describing the subcellular localization of signaling genes in AfCS model cell types, we utilized multiple constructs for each cloned gene. We carried out a large scale assessment of subcellular localization of the STKs in both the RAW264.7 murine macrophage cell line and the WEHI231 murine B cell line. This data set is part of the freely available image database found on the AfCS data center (www.signaling-gateway.org/data/Data.html). In this analysis, we co-expressed the same protein N- or C-terminally fused with C/YFP in the following fashion: protein-CFP + YFP-protein and CFP-protein + protein-YFP. We visualized the expression patterns of the STKs using live cell confocal microscopy, allowing us to make a subjective determination of the subcellular localization of the STKs. Importantly, screening for localization by this method allowed us to determine whether the N- and C-terminally tagged versions co-localized and if there were differences in localization patterns across different cells.
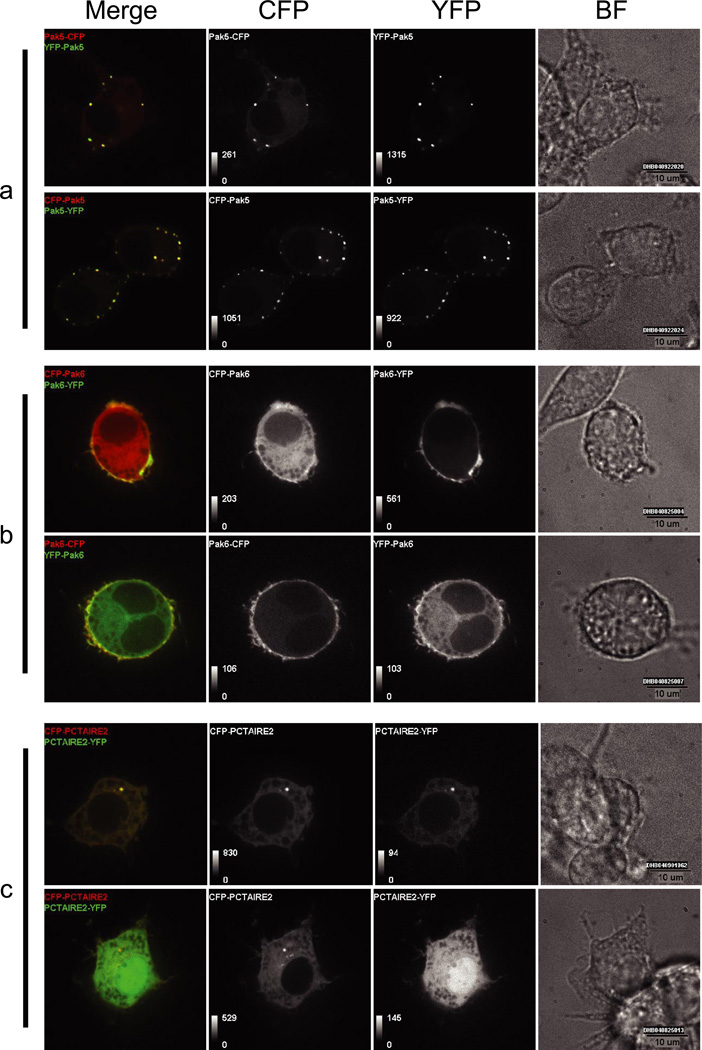
Here we highlight three examples in RAW264.7 cells to emphasize the relevance and utility of a large scale cloning and subcellular localization screen. The p21-activated kinases (Paks) bind to and may be stimulated by activated forms of the small GTPases Cdc42 and Rac. The Pak5 and Pak6 isoforms belong to the recently recognized Group II Paks (24). In the RAW264.7 line, we observed Pak5 localization as a punctate pattern along the cellular membrane (Fig. 6 a), which differs from previously published reports showing association with mitochondria (25). The same pattern of Pak5 localization is observed with both N and C-terminal CFP/YFP fusions. We noted differential Pak6 localization depending on the terminus at which CFP/YFP is expressed. With N-terminal tagging, we observed the disperse cytoplasmic localization of Pak6 described previously in CV-1 cells (26). However, with C-terminal CFP or YFP, we detected a more distinct localization of Pak6 to the plasma membrane (Fig. 6b). This finding may provide insight into the cellular mechanisms that regulate Pak6 localization.
Fig. 6. Subcellular localization of murine kinases in RAW264.7 cells.
Shown are live cell confocal images of RAW 264.7 cells expressing C/YFP-tagged Pak5 (a), Pak6 (b), and Pctaire2 (c). Brightfield (BF), CFP, YFP, and CFP/YFP merged images are shown. Pak5 localizes to punctate structures near the plasma membrane, Pak6 shows differential tag-dependent localization, and Pctaire2 localizes to the centrosome.
Cyclin-dependent kinases (Cdks) have been shown to be key players in the control of cell cycle progression. Their activity is regulated by interaction with specific subunits known as cyclins, phosphorylation by other protein kinases, and dephosphorylation by phosphatases (27). Based on sequence homology, Pctaire2 is a Cdk-related gene and is likely to play a role in cell cycle progression. Here we show the first example of punctate localization of Pctaire2 potentially at the centrosome (Fig. 6c). In most cells, N- or C-terminal tagging does not seem to disrupt this localization (see Fig. 6c upper panels). However, in some cells, we find that C-terminal tagging does disrupt Pctaire2 localization from the centrosome in the same cell in which an N-terminally tagged version still localizes to this region (see Fig. 6c lower panels). Overall these data demonstrate that large scale subcellular localization studies cannot only confirm previous findings but also reveal alternative or novel data that provide the basis for new hypotheses about protein function.
DISCUSSION
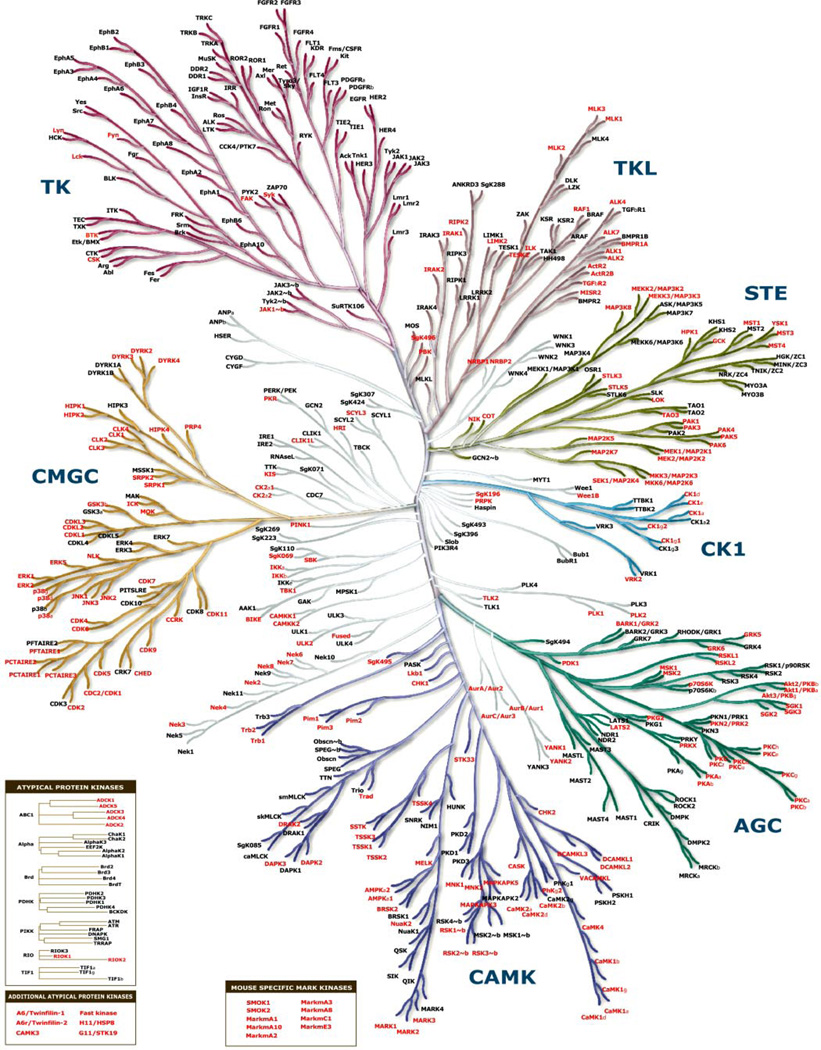
The cloning of large numbers of cDNA ORFs is a challenging task that requires capable software systems and robust technical methodology as well as efficient, facile, and accessible data curation. We describe the generation of a publicly available cDNA collection by the AfCS consortium with emphasis on the STK gene family. Although reference is made to an initial set of 217 STK target genes (of which 124 were successfully cloned and databased), this group was limited by the lack of a publicly available mouse genome sequence at the outset of the project. Our scope was expanded with the completion of the mouse genome (28), and the entire set of murine kinase genes (the mouse “kinome”) is now proposed to number 540 genes (19) of which the AfCS has cloned and distributed >260. Fig. 7 shows a phylogenetic tree derived from the human kinome (20) that highlights the murine protein kinase orthologs cloned and distributed as part of the AfCS effort. In addition to wild type kinase cDNAs, we also created mutant cDNAs encoding kinases where the essential lysine residue in the kinase domain was mutated to arginine. This “Lys to Arg” mutation creates a potential dominant negative for almost 200 kinases. These mutants have also been distributed to the ATCC.
Fig. 7. Protein kinase phylogenetic tree.
Relationships between members of the protein kinase superfamily (kinome dendrogram reprinted with permission from Cell Signaling Technology, Inc. and Manning et al. 20)) are shown. Group names are: AGC containing cAMP-dependent protein kinase (PKA), cGMP-dependent protein kinase (PKG), and protein kinase C (PKC); CAMK calcium/calmodulin-dependent protein kinase; CK1 casein kinase 1; CMGC containing CDK, mitogen-activated protein kinase (MAPK), glycogen synthase kinase 3 (GSK3), and CDC-like kinase (CLK) families; STE homologs of the yeast Sterile kinases; TK tyrosine kinase; TKL tyrosine kinase-like. Insets highlight several atypical and mouse-specific kinases in the AfCS collection. The murine orthologs of all kinases highlighted red are represented in the AfCS plasmid database and are available through the ATCC (see Supplemental Table 4 for clone IDs).
We managed the curation of our cloned cDNAs by creating the AfCS plasmid database (www.signaling-gateway.org/data/plasmid/). This database is a dynamic and flexible resource that represents a long term and ongoing repository of not only STK-containing but all publicly available AfCS vectors. Users can also perform blast searches of the AfCS collection at this site.
In addition to the kinase cDNA set, many additional genes with key roles in cell signaling were cloned as part of the AfCS effort. Other gene families strongly represented in this group include heterotrimeric G proteins, G protein-coupled receptors, small G proteins, guanine nucleotide exchange factors, and phospholipases. A comprehensive set of pleckstrin homology domains were also cloned and distributed. The total number of unique full-length ORFs cloned and fully sequenced in Gateway entry vectors, and currently available at the ATCC, is 449 (Supplemental Table 4). The majority of these are available in two forms (±a termination codon) to permit expression as either N- or C-terminal fusions with various tags (see Supplemental Table 3 for list of expression options). Although Supplemental Table 4 shows only one representative clone for each unique gene, splice variants were cloned for several genes. For example, a keyword search with the Mek7 gene name at www.signaling-gateway.org/data/plasmid/ finds 44 constructs containing six different variants of Mek7.
A primary application for these cDNAs in the AfCS program was the creation of a subcellular localization database generated through expression of cDNAs as CFP and YFP fusion proteins in the RAW264.7 macrophage cell line and the WEHI231 B cell line (www.signaling-gateway.org/data/Data.html) (29). Thus, the majority of “ready-to-use” expression constructs available through the ATCC are CFP or YFP fusion constructs for mammalian expression. An additional resource that was created as part of the imaging project was a comprehensive set of localization markers for various organelles (34). These have been made available by the ATCC in a 96-well plate to provide the entire 64 plasmid set in an accessible format (ATCC ID MBA-91 and Supplemental Table 5).
We describe localization data in RAW264.7 macrophages for the kinases Pak5, Pak6, and Pctaire2. Pak5 and Pak6 are highly homologous and belong to the recently recognized Group II Paks (Paks 4, 5, and 6) that retain 40–50% identity to the kinase domains of the Group I Paks (Paks 1, 2, and 3) (24, 30). We observed a distinct punctate pattern for Pak5 localization along the cellular membrane that does not seem to be disrupted by N- or C-terminal tagging (Fig. 6a). Because previous studies observed Pak5 localization in mitochondria of Chinese hamster ovary cells, it is possible that Pak5 localization is dynamic and varies between cell types. Expression of the N-terminal fusion of Pak6 exhibits the previously observed disperse cytoplasmic localization (26). However, the C-terminal fusion reveals a more distinct plasma membrane location (Fig. 6b). Pak6 contains a PAK box domain (PBD), characteristic of the Pak kinases (31). It is possible that C-terminal tagging disrupts the function of the PBD domain and, by extension, the cellular compartmentalization of Pak6. On the other hand, the specific membrane localization of the C-terminal CFP/YFP fusion may provide insight into how engagement of the PBD domain could dynamically regulate Pak6 localization.
Based on sequence homology, Pctaire2 is a Cdk-related gene and likely plays a role in cell cycle progression (27, 32). Previous localization studies of Pctaire1 show abundant cytoplasmic localization (32). Here we show the first example of distinct localization of Pctaire2 that, in most cells, appears to be at the centrosome with both N- or C-terminal tagging (Fig. 6c). It is noteworthy that we find a small proportion of cells where C-terminally tagged Pctaire2 is delocalized from the centrosome while the N-terminally tagged form remains localized (Fig. 6c lower panels). This observation may provide insight to a dynamic role for the Pctaire2 C terminus during certain stages of the cell cycle. These data demonstrate how expression constructs from the AfCS collection can provide informative data on the localization of signaling genes that may lead to important insights into protein function. In addition, the CFP/YFP fluorescent tags were chosen for their compatibility in fluorescence resonance energy transfer studies, which can provide data on the real time interaction kinetics of dynamically associated proteins (e.g. heterotrimeric G protein subunits). Indeed all of the constructs available in the AfCS plasmid database can provide the foundation for further analyses working toward a better understanding of complex cellular signaling networks.
In general, the cDNA cloning protocol we describe proved to be robust for the isolation of a large number of murine signaling genes. As a standard source material, a mixture of brain and testes cDNA appears to provide the most comprehensive coverage of genes involved in the major cell signaling pathways. Analysis of the genes that we failed to amplify shows a bias toward large cDNAs. However, we were still able to clone 17 ORFs of >3 kb using the ProofStart polymerase. We found that this enzyme provided the best balance of robustness of amplification versus fidelity. Furthermore we could readily identify randomly generated PCR mutations by running separate PCRs for cDNAs with and without the termination codon (Fig. 1). In most cases, we expect that the differences we observed between our cloned sequences and the GenBank reference were caused by either splice variation between the originally sequenced ORF and ours, genuine polymorphisms, or sequence inaccuracies from more dated sequencing technologies. For example, we cloned the heterotrimeric G protein Gαi2 from the sequence in GI: 6680036 and observed five differences in the nucleotide sequence. Comparison with the current RefSeq record for Gαi2 (GI: 41054805) shows a perfect match between the AfCS and RefSeq sequences at these five locations. Interestingly there are three presumed polymorphisms between the AfCS and RefSeq sequences at other locations, but these three nucleotide differences are “silent” with respect to amino acid sequence (data not shown).
Our adoption of the Gateway cloning technology at the outset of this effort greatly facilitated the cloning process (4). The resource described here is specific to cDNA cloning, but we have also used Gateway extensively in the cloning of short hairpin RNAs for RNA interference, initially as RNA polymerase III promoter-driven stem loops (www.signaling-gateway.org/data/plasmid/RNAi_vector_guide.pdf) and more recently as conditionally expressed microRNA-like-short hairpin RNAs (33). The recombination reactions used for both cloning PCR products and subcloning to expression platforms for functional analysis have proven to be highly robust and scalable to 96-well plate formats. Moreover our development of custom software scripts allowed us to automate the generation of plasmid database records, an approach that was particularly useful for batch calculations of diagnostic restriction maps for validating constructs after recombination.
In summary, the constructs generated and distributed to the ATCC by the AfCS consortium represent a valuable experimental resource for the cell signaling research community. We present only a small subset of the localization data available at the AfCS image database (www.signaling-gateway.org/data/Data.html), so this data resource can provide significantly more insight and experimental leads than what we are able to highlight in this report. Moreover these localization screens are only one of many applications for this Gateway-compatible clone set. The AfCS ORFs described here are also available from the ATCC in Gateway entry vectors. Thus, subcloning to other Gateway-compatible expression platforms (such as those detailed in Supplemental Table 3 and others available commercially) would allow many additional proteomics applications to further investigate the molecular function of the signaling genes in the AfCS collection.
Supplementary Material
Acknowledgments
We thank Linda Holloway, Judy Kantor, Barry Westfall, and Jennifer Zinck at the ATCC for assistance in facilitating the distribution of AfCS clones and Mike Brasch, Joel Jessee, Ray Harris, Virginia Heatwole, and Graziella Piras at Invitrogen for technical advice on the Gateway system and provision of plasmids prior to commercial release. We thank Gerard Manning for early access to the human and mouse kinome sequences; these sequence data were invaluable in helping to curate kinase ORFs prior to cloning. We are grateful to colleagues in the AfCS for insightful discussions on a wide range of plasmid vector development projects.
Footnotes
This work was supported by contributions from public and private sources, including the NIGMS, National Institutes of Health, Glue Grant Initiative U54 GM062114. A complete listing of the AfCS sponsors can be found at www.signaling-gateway.org/aboutus/sponsors.html. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement ” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The abbreviations used are: AfCS, Alliance for Cellular Signaling; ATCC, American Type Culture Collection; STK, serine-threonine kinase; PGA, Paracel Genome Assembler; CFP, cyan fluorescent protein; YFP, yellow fluorescent protein; Pak, p21-activated kinase; Cdk, cyclin-dependent kinase; PBD, PAK box domain.
REFERENCES
- 1.Gilman AG, Simon MI, Bourne HR, Harris BA, Long R, Ross EM, Stull JT, Taussig R, Bourne HR, Arkin AP, Cobb MH, Cyster JG, Devreotes PN, Ferrell JE, Fruman D, Gold M, Weiss A, Stull JT, Berridge MJ, Cantley LC, Catterall WA, Coughlin SR, Olson EN, Smith TF, Brugge JS, Botstein D, Dixon JE, Hunter T, Lefkowitz RJ, Pawson AJ, Sternberg PW, Varmus H, Subramaniam S, Sinkovits RS, Li J, Mock D, Ning Y, Saunders B, Sternweis PC, Hilgemann D, Scheuermann RH, DeCamp D, Hsueh R, Lin KM, Ni Y, Seaman WE, Simpson PC, O’Connell TD, Roach T, Simon MI, Choi S, Eversole-Cire P, Fraser I, Mumby MC, Zhao Y, Brekken D, Shu H, Meyer T, Chandy G, Heo WD, Liou J, O’Rourke N, Verghese M, Mumby SM, Han H, Brown HA, Forrester JS, Ivanova P, Milne SB, Casey PJ, Harden TK, Arkin AP, Doyle J, Gray ML, Meyer T, Michnick S, Schmidt MA, Toner M, Tsien RY, Natarajan M, Ranganathan R, Sambrano GR. Overview of the Alliance for Cellular Signaling. Nature. 2002;420:703–706. doi: 10.1038/nature01304. [DOI] [PubMed] [Google Scholar]
- 2.Natarajan M, Lin KM, Hsueh RC, Sternweis PC, Ranganathan R. A global analysis of cross-talk in a mammalian cellular signalling network. Nat. Cell Biol. 2006;8:571–580. doi: 10.1038/ncb1418. [DOI] [PubMed] [Google Scholar]
- 3.Brasch MA, Hartley JL, Vidal M. ORFeome cloning and systems biology: standardized mass production of the parts from the parts-list. Genome Res. 2004;14:2001–2009. doi: 10.1101/gr.2769804. [DOI] [PubMed] [Google Scholar]
- 4.Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walhout AJ, Temple GF, Brasch MA, Hartley JL, Lorson MA, van den Heuvel S, Vidal M. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- 6.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, Yamanaka I, Kiyosawa H, Yagi K, Tomaru Y, Hasegawa Y, Nogami A, Schonbach C, Gojobori T, Baldarelli R, Hill DP, Bult C, Hume DA, Quackenbush J, Schriml LM, Kanapin A, Matsuda H, Batalov S, Beisel KW, Blake JA, Bradt D, Brusic V, Chothia C, Corbani LE, Cousins S, Dalla E, Dragani TA, Fletcher CF, Forrest A, Frazer KS, Gaasterland T, Gariboldi M, Gissi C, Godzik A, Gough J, Grimmond S, Gustincich S, Hirokawa N, Jackson IJ, Jarvis ED, Kanai A, Kawaji H, Kawasawa Y, Kedzierski RM, King BL, Konagaya A, Kurochkin IV, Lee Y, Lenhard B, Lyons PA, Maglott DR, Maltais L, Marchionni L, McKenzie L, Miki H, Nagashima T, Numata K, Okido T, Pavan WJ, Pertea G, Pesole G, Petrovsky N, Pillai R, Pontius JU, Qi D, Ramachandran S, Ravasi T, Reed JC, Reed DJ, Reid J, B. Ring Z, Ringwald M, Sandelin A, Schneider C, Semple CA, Setou M, Shimada K, Sultana R, Takenaka Y, Taylor MS, Teasdale RD, Tomita M, Verardo R, Wagner L, Wahlestedt C, Wang Y, Watanabe Y, Wells C, Wilming LG, Wynshaw-Boris A, Yanagisawa M, Yang I, Yang L, Yuan Z, Zavolan M, Zhu Y, Zimmer A, Carninci P, Hayatsu N, Hirozane-Kishikawa T, Konno H, Nakamura M, Sakazume N, Sato K, Shiraki T, Waki K, Kawai J, Aizawa K, Arakawa T, Fukuda S, Hara A, Hashizume W, Imotani K, Ishii Y, Itoh M, Kagawa I, Miyazaki A, Sakai K, Sasaki D, Shibata K, Shinagawa A, Yasunishi A, Yoshino M, Waterston R, Lander ES, Rogers J, Birney E, Hayashizaki Y. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 7.Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Madan A, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. SciUSA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marsischky G, LaBaer J. Many paths to many clones: a comparative look at high-throughput cloning methods. Genome Res. 2004;14:2020–2028. doi: 10.1101/gr.2528804. [DOI] [PubMed] [Google Scholar]
- 9.Rual JF, Hirozane-Kishikawa T, Hao T, Bertin N, Li S, Dricot A, Li N, Rosenberg J, Lamesch P, Vidalain PO, Clingingsmith TR, Hartley JL, Esposito D, Cheo D, Moore T, Simmons B, Sequerra R, Bosak S, Doucette-Stamm L, Le Peuch C, Vandenhaute J, Cusick ME, Albala JS, Hill DE, Vidal M. Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res. 2004;14:2128–2135. doi: 10.1101/gr.2973604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiemann S, Arlt D, Huber W, Wellenreuther R, Schleeger S, Mehrle A, Bechtel S, Sauermann M, Korf U, Pepperkok R, Sultmann H, Poustka A. From ORFeome to biology: a functional genomics pipeline. Genome Res. 2004;14:2136–2144. doi: 10.1101/gr.2576704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rual JF, Hill DE, Vidal M. ORFeome projects: gateway between genomics and omics. Curr. Opin. Chem. Biol. 2004;8:20–25. doi: 10.1016/j.cbpa.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 12.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 13.Simpson JC, Wellenreuther R, Poustka A, Pepperkok R, Wiemann S. Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep. 2000;1:287–292. doi: 10.1093/embo-reports/kvd058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiemann S, Bechtel S, Bannasch D, Pepperkok R, Poustka A. The German cDNA network: cDNAs, functional genomics and proteomics. J. Struct. Funct. Genomics. 2003;4:87–96. doi: 10.1023/a:1026148428520. [DOI] [PubMed] [Google Scholar]
- 15.Breslauer KJ, Frank R, Blocker H, Marky LA. Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. SciUSA. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rychlik W, Spencer WJ, Rhoads RE. Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res. 1990;18:6409–6412. doi: 10.1093/nar/18.21.6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 18.Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- 19.Caenepeel S, Charydczak G, Sudarsanam S, Hunter T, Manning G. The mouse kinome: discovery and comparative genomics of all mouse protein kinases. Proc. Natl. Acad. SciUSA. 2004;101:11707–11712. doi: 10.1073/pnas.0306880101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 21.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaffer ZM, Chernoff J. p21-activated kinases: three more join the Pak. IntJBiochem. Cell Biol. 2002;34:713–717. doi: 10.1016/s1357-2725(01)00158-3. [DOI] [PubMed] [Google Scholar]
- 25.Cotteret S, Jaffer ZM, Beeser A, Chernoff J. p21-activated kinase 5 (Pak5) localizes to mitochondria and inhibits apoptosis by phosphorylating BAD. Mol. Cell. Biol. 2003;23:5526–5539. doi: 10.1128/MCB.23.16.5526-5539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang F, Li X, Sharma M, Zarnegar M, Lim B, Sun Z. Androgen receptor specifically interacts with a novel p21-activated kinase, PAK6. J. Biol. Chem. 2001;276:15345–15353. doi: 10.1074/jbc.M010311200. [DOI] [PubMed] [Google Scholar]
- 27.Solomon MJ. Activation of the various cyclin/cdc2 protein kinases. Curr. Opin. Cell Biol. 1993;5:180–186. doi: 10.1016/0955-0674(93)90100-5. [DOI] [PubMed] [Google Scholar]
- 28.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O’Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 29.O’Rourke NA, Meyer T, Chandy G. Protein localization studies in the age of ‘Omics’. Curr. Opin. Chem. Biol. 2005;9:82–87. doi: 10.1016/j.cbpa.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 30.Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- 31.Osada S, Izawa M, Koyama T, Hirai S, Ohno S. A domain containing the Cdc42/Rac interactive binding (CRIB) region of p65PAK inhibits transcriptional activation and cell transformation mediated by the Ras-Rac pathway. FEBS Lett. 1997;404:227–233. doi: 10.1016/s0014-5793(97)00139-7. [DOI] [PubMed] [Google Scholar]
- 32.Besset V, Rhee K, Wolgemuth DJ. The cellular distribution and kinase activity of the Cdk family member Pctaire1 in the adult mouse brain and testis suggest functions in differentiation. Cell Growth Differ. 1999;10:173–181. [PubMed] [Google Scholar]
- 33.Shin KJ, Wall EA, Zavzavadjian JR, Santat LA, Liu J, Hwang JI, Rebres R, Roach T, Seaman W, Simon MI, Fraser ID. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc. Natl. Acad. SciUSA. 2006;103:13759–13764. doi: 10.1073/pnas.0606179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandy G, Mukai T, Mi Q, Zavzavadjian J, Gehrig E, Verghese M, Fung E, Couture S, Park WS, O’Rourke N, Fraser I. Building an atlas of subcellular localization markers in WEHI-231 cells. AfCS Brief Communications. 2003 http://www.afcs.org/reports/v1/DA0002/ DA0002.pdf. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.