Abstract
To resolve the controversy about messengers regulating KCNQ ion channels during phospholipase C–mediated suppression of current, we designed translocatable enzymes that quickly alter the phosphoinositide composition of the plasma membrane after application of a chemical cue. The KCNQ current falls rapidly to zero when phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2 or PI(4,5)P2] is depleted without changing Ca2+, diacylglycerol, or inositol 1,4,5-trisphosphate. Current rises by 30% when PI(4,5)P2 is overproduced and does not change when phosphatidylinositol 3,4,5-trisphosphate is raised. Hence, the depletion of PI(4,5)P2 suffices to suppress current fully, and other second messengers are not needed. Our approach is ideally suited to study biological signaling networks involving membrane phosphoinositides.
Phosphoinositide phospholipids are major signaling molecules of cell membranes. Many cellular proteins are inhibited when phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] is hydrolyzed by phospholipase C (PLC) and reactivated when phosphatidylinositol 4-phosphate [PI(4)P] 5-kinase restores PI(4,5)P2 (1, 2). For example, KCNQ K+ channels are closed by muscarinic-receptor–triggered PLC activation (3, 4). Closure of KCNQ2/KCNQ3 channels increases the excitability of central and peripheral neurons, and hypomorphic mutations in KCNQ subunits underlie familial epilepsies, deafness, and arrhythmias (5). Whether the depletion of PI(4,5)P2 suffices to close KCNQ channels upon receptor activation remains controversial because much of the supporting experimental evidence is indirect. Can we rule out that increases of the many signaling molecules downstream of PLC or other changes in phosphoinositides are essential for closing channels instead of or together with the loss of PI(4,5)P2? We used a new method to deplete plasma membrane–associated PI(4,5)P2 in living cells within seconds without activating PLC. Upon addition of a dimerizing drug, PI(4,5)P2 was selectively depleted in living cells without the production of diacylglycerol (DAG), inositol 1,4,5-trisphosphate (IP3), or calcium signals.
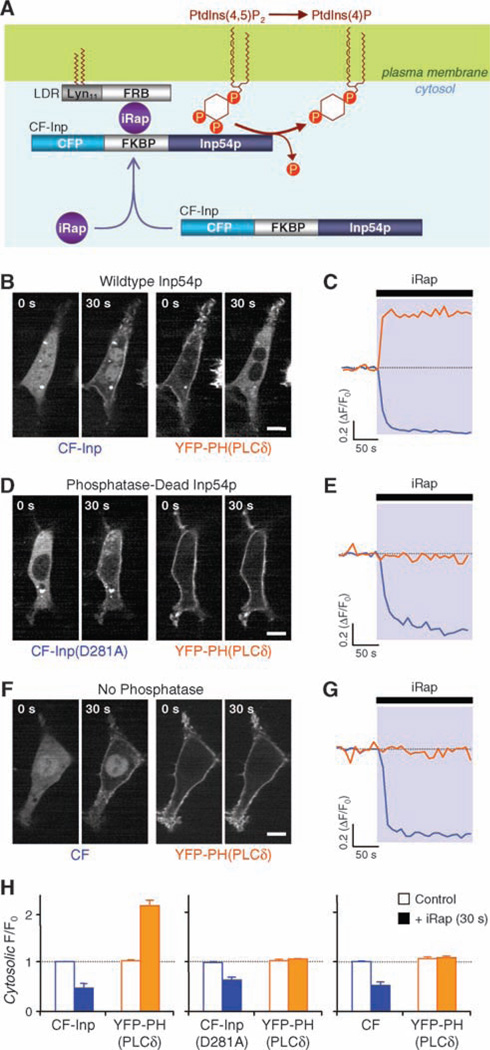
The chemical dimerizer strategy uses heterodimerization of protein domains from FK506 binding protein (FKBP) and from mTOR (FRB) by the immunosuppressant rapamycin (6) or an analog called iRap (7). This approach has previously permitted us to develop a high-speed membrane translocation of Rho guanosine triphosphatases (GTPases) (7). Now we depleted PI(4,5)P2 by inducible membrane translocation of Inp54p, a yeast inositol polyphosphate 5-phosphatase that specifically cleaves the phosphate at the 5 position of PI(4,5)P2 (8) (Fig. 1A). Constitutively membrane-targeted Inp54p has been used to assess the roles of PI(4,5)P2 in cytoskeleton–plasma membrane adhesion (8). We fused a truncated Inp54p to FKBP already tagged with cyan fluorescent protein (CFP)–FKBP (CF) (9, 10). The CFP label of the resulting fusion protein CF-Inp54p (CF-Inp) exhibited good cytosolic localization in NIH3T3 cells. CF-Inp was then cotransfected along with Lyn11-FRB (LDR), a membrane-anchored FRB (7), and YFP-PH(PLC-δ), a yellow fluorescent protein (YFP)–tagged pleckstrin homology (PH) domain from phospholipase C–δ1 (PLC-δ1) serving as a PI(4,5)P2/IP3 biosensor (11, 12). The addition of iRap led to the translocation of the fluorescent CF-Inp from the cytosol to the plasma membrane and a reciprocal translocation of YFP-PH(PLC-δ) from the plasma membrane to the cytosol (Fig. 1, B and H), demonstrating inducible accumulation of the Inp54p enzyme at the plasma membrane and in situ PI(4,5)P2 depletion. To analyze the kinetics, we measured the fluorescence intensities of CF-Inp and YFP-PH(PLC-δ) in a cytosolic region (Fig. 1C), which revealed quick translocation of both probes [half-time (t1/2) = 12.3 ± 1.2 s and 14.7 ± 2.5 s; n = 12 cells from four experiments]. Control experiments with either a phosphatase-dead mutant of Inp54p [CF-Inp Asp281→Ala281 (D281A)] (13) or a fusion protein lacking the phosphatase (CF) showed no translocation of YFP-PH(PLC-δ), although iRap did induce translocation of the two CFP constructs to the plasma membrane (Fig. 1, D to H).
Fig. 1.
In situ PI(4,5)P2 hydrolysis induced by iRap heterodimerization in NIH3T3 cells. (A) FRB is anchored to plasma membrane via myristoylation and palmitoylation modification sequence Lyn11. Inp54p is recruited from the cytosol upon addition of iRap, forming the tripartite complex FRB-iRap-FKBP. Membrane recruitment of Inp54p rapidly induces specific dephosphorylation at the 5 position of PI(4,5)P2 [PtdIns(4,5)P2]. PtdIns(4)P, PI(4)P. (B, D and F) Time-series confocal fluorescent images of cells expressing LDR, YFP-PH(PLC-δ) and either CF-Inp (B), CF-Inp(D281A) (D), or CF (F). Images before and after the 30-s addition of iRap (5 µM). Scale bar, 10 µm. (C, E and G) Cytosolic fluorescence intensities of CFP (blue) and YFP (yellow) for the cells shown in B (C), D (E), or F (G). Normalized fluorescence intensities are shown as a function of time. ΔF/F0, fluorescence change divided by initial fluorescence. (H) Normalized fluorescence intensity of CFP and YFP in the cytosol from more than four independent experiments with the same conditions as [(B) to (G)] before (white bars) and after (colored bars) 30-s iRap treatment. The black box refers to both the solid orange and solid blue bars, and the white box refers to the white bars, whether outlined with orange or blue. Mean values are shown and error bars indicate SEM.
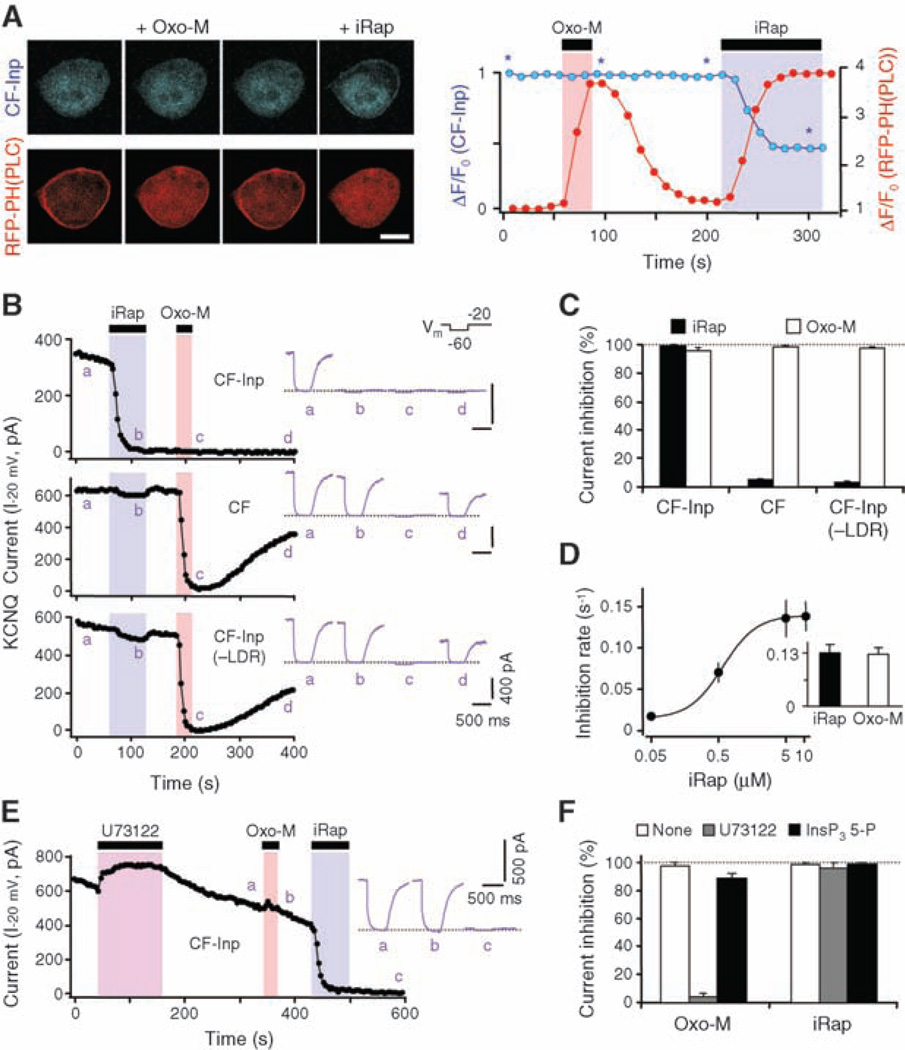
We then studied whether PI(4,5)P2 depletion suppresses currents in KCNQ K+ channels in human embryonic kidney tsA-201 (tsA) cells. As controls, iRap alone had only minor, reversible effects on current amplitude and no effect on the voltage dependence of activation of KCNQ current (iRap, −24.2 ± 1.2 mV; controls, −23.9 ± 0.7 mV; n = 6 cells). Furthermore, in cells expressing the full Inp54p translocation system together with the M1 subtype of muscarinic receptors (M1R) and the red fluorescent protein (RFP)–PH(PLC-δ) probe, only iRap and not the muscarinic agonist oxotremorine-M (Oxo-M) translocated the CF-Inp enzyme to the plasma membrane (Fig. 2A). The fast, irreversible translocation of CF-Inp reached half-maximal rate at 1.2 ± 0.1 µM iRap (n = 5 to 7 cells) (fig. S1). Both Oxo-M and iRap caused translocation of RFP-PH(PLC-δ), reflecting rapid depletion of PI(4,5)P2, by PLC in one case and by the phosphatase in the other (Fig. 2A). Only the effect of Oxo-M was reversible. We were now ready to test our hypothesis. The application of iRap to cells expressing channels and the full Inp54p translocation system suppressed KCNQ current rapidly, fully, and irreversibly (Fig. 2, B and C). Suppression was not seen when the Inp54p enzyme or the membrane anchor was omitted, although Oxo-M acting via transfected M1Rs was still effective at suppressing current reversibly. The saturated rate constant for suppression of KCNQ current by iRap was the same as for Oxo-M, equivalent to a 5-s t1/2, and reached half-maximal at 0.6 ± 0.1 µM iRap (n = 5 to 9 cells) (Fig. 2D).
Fig. 2.
Modulation of KCNQ current induced by CF-Inp and iRap in tsA cells. (A) Translocation of CF-Inp (blue) and RFP-PH(PLC-δ) (red) upon addition of Oxo-M (10 µM) or iRap (5 µM) to cells cotransfected with M1R. Scale bar, 10 µm. Asterisks indicate times of the four images shown. (B) Channel modulation by iRap (5 µM) or Oxo-M (10 µM) in cells expressing CF constructs with LDR or CF-Inp alone. Insets show the current waveforms at indicated times a to d; the dotted line indicates zero current. Vm, membrane voltage protocol; I−20 mV, KCNQ current recorded at −20 mV. (C) Current inhibition by iRap and Oxo-M in cells expressing combinations of LDR and CF constructs. (D) Rate constants of current inhibition with various iRap concentrations. Inset shows the rate constants (s−1) for inhibition by iRap (5 µM) or Oxo-M (10 µM) in CF-Inp–expressing cells (n = 3 to 10 cells). (E) Effect of Oxo-M and iRap on KCNQ channel in a cell treated with the PLC inhibitor U73122 (3 µM). (F) Current inhibition by Oxo-M and iRap in cells treated with U73122 or overexpressing IP3 5-phosphatase (n = 3 to 7 cells). Error bars in (C), (D), and (F) indicate SEM.
The goal of our study was to use an enzyme distinct from PLC to determine whether depletion of PI(4,5)P2 suffices to turn off KCNQ channels. Therefore, we had to rule out the alternative possibility that some essential downstream products of PLC, such as IP3, DAG, or calcium transients, might also be generated by membrane translocation of Inp54p. In tsA cells where PLC was blocked with the inhibitor U73122 or where IP3 accumulation was prevented by overexpressing IP3 5-phosphatase (14), the iRap-induced translocation of CF-Inp, the depletion of PI(4,5)P2 (fig. S2, A and B), and the suppression of KCNQ current were unaltered (Fig. 2, E and F). Further, in tsA cells expressing the YFP-labeled C1 domain of protein kinase C–γ, a DAG indicator (15), iRap induced no translocation of the indicator, but Oxo-M did (fig. S2C). Hence, CF-Inp/iRap does not induce DAG production. Finally, we measured intracellular calcium using fura-2 as a calcium indicator in tsA cells expressing M1Rs. Application of Oxo-M led to a transient calcium rise that was abolished by overexpressing IP3 5-phosphatase (fig. S2D). On the other hand, the application of iRap led to no calcium elevation. Analogous experiments in NIH3T3 cells also showed neither DAG production nor calcium elevation with CF-Inp/iRap.
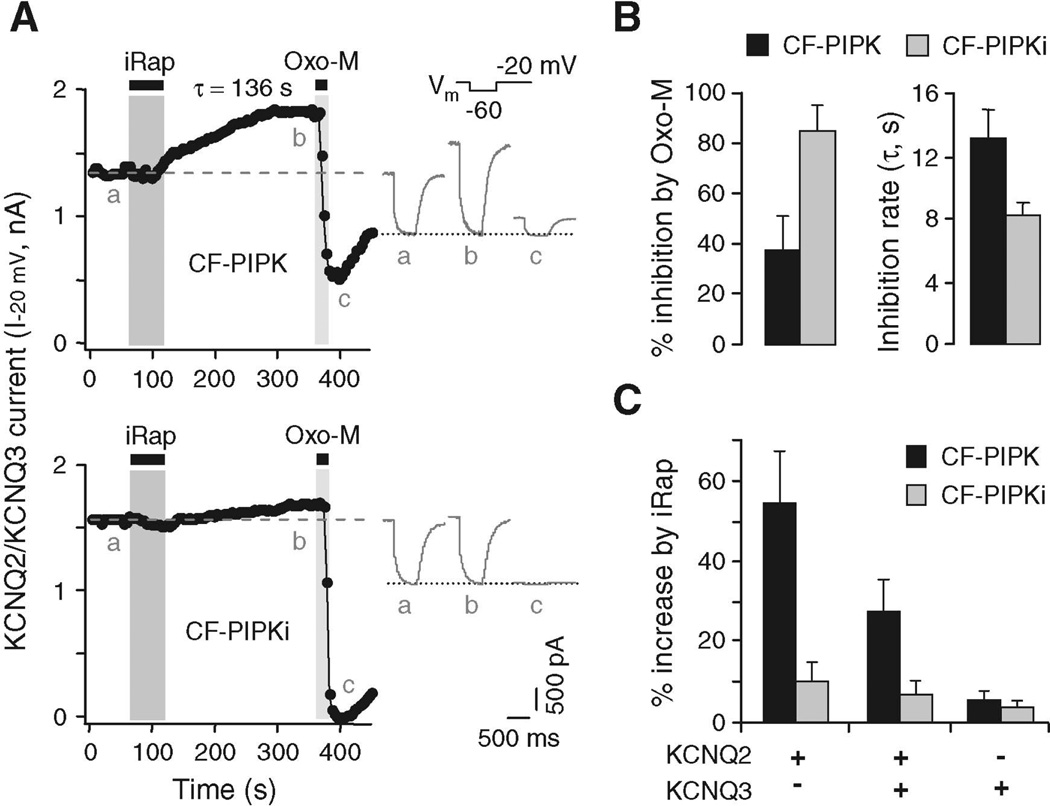
As a further test of the phosphoinositide hypothesis, we used a similar strategy to increase PI(4,5)P2 in the plasma membrane with a lipid kinase. We made a translocatable construct CF-PIPK by combining most of the enzyme PI(4)P 5-kinase type I–γ (PIPKI-γ) (16) with CFP-FKBP. In NIH3T3 cells and tsA cells, CF-PIPK fluorescence was largely in the cytosol at rest and moved to the plasma membrane irreversibly upon addition of iRap (fig. S3). In tsA cells also expressing KCNQ subunits, the KCNQ current was slowly augmented after the addition of iRap (exponential time constant τ = 138 ± 23 s; n = 4 cells), as might be predicted if there is a slow increase of membrane PI(4,5)P2 (Fig. 3A). The midpoint of channel activation did not change (iRap, −28.5 ± 2.0 mV; controls, −27.3 ± 1.8 mV; n = 5 cells). However, current suppression by subsequent addition of Oxo-M was greatly slowed and incomplete after only 20 s of application (Fig. 3B), as if accumulation and speeded synthesis of PI(4,5)P2 decreased the depletion by PLC. The effects of the translocated CF-PIPK enzyme were paralleled in cells overexpressing the nontranslocating full-length original enzyme PIPKI-γ (fig. S4). Again, in those cells, the current suppression by Oxo-M was slowed and only partial [compare to similar experiments with PIPKI-β (17)]; current suppression by the coexpressed CF-Inp/iRap system was also slowed but remained nearly complete. A kinase-dead version of the kinase construct (Asp253→Ala253, CF-PIPKi) (18) increased KCNQ current little and had almost no effect on subsequent suppression by muscarinic agonist (Fig. 3, A and B).
Fig. 3.
Increase of KCNQ current by activation of PI(4)P 5-kinase. (A) Modulation of KCNQ2/KCNQ3 channels in tsA cells expressing CF-PIPK (top) or CF-PIPKi (bottom) constructs by application of iRap (5 µM) for 1 min. The dashed line indicates current amplitude before iRap. Insets show the current waveforms at indicated times a to c. (B) Percent inhibition by Oxo-M and time constant of inhibition after previous iRap translocation of CF-PIPK or CF-PIPKi. (C) Effects of CF-PIPK or CF-PIPKi on iRap-induced augmentation of current in homomeric KCNQ2, heteromeric KCNQ2/KCNQ3, or homomeric KCNQ3 channels (n = 4–10 cells). Error bars in (B) and (C) indicate SEM.
The ability of translocated CF-PIPK to augment KCNQ current suggests that KCNQ channels are not fully saturated by resting levels of plasma membrane–associated PI(4,5)P2. We tested this further by taking advantage of a reported difference in PI(4,5)P2 affinities for different KCNQ subunit isoforms: Increasing concentrations of short-chain PI(4,5)P2 are reportedly needed to activate channels expressed from KCNQ3 alone, KCNQ2 and KCNQ3 together, and KCNQ2 alone, respectively (19). When we compared these channel types for responses to CF-PIPK/iRap, KCNQ2 current was augmented by 54 ± 12% (n = 5 cells), KCNQ2/KCNQ3 current by 27 ± 8% (n = 10 cells), and KCNQ3 channels by only 5 ± 2% (n = 5 cells) (Fig. 3C). Augmentation of current after iRap addition must be due to accumulation of extra PI(4,5)P2. The experiments of Fig. 3C with KCNQ2/KCNQ3 heteromers suggest, therefore, that the degree of saturation by resting PI(4,5)P2 is about 70%, which is in good agreement with preliminary published estimates of 72% (20) and 81% (17). KCNQ2 channels are even less saturated, and KCNQ3 channels are more saturated.
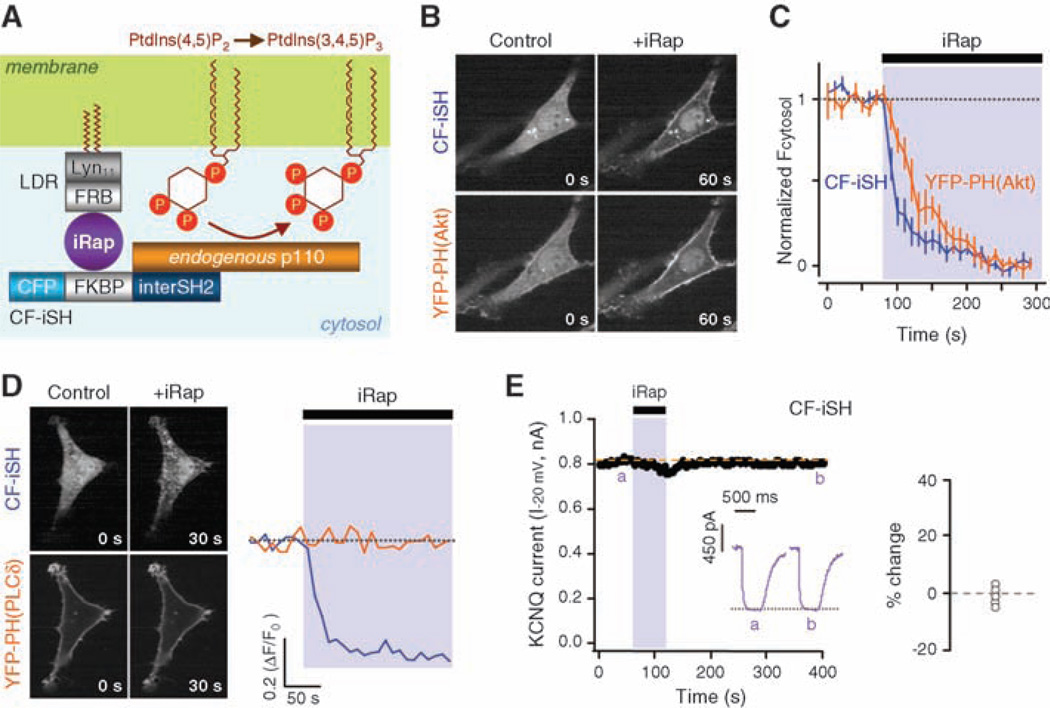
Triply phosphorylated PI(3,4,5)P3 is a potent second messenger for growth factor signaling (21). Roles in ion channel regulation are less studied. Applied PI(3,4,5)P3 restores the function of G protein–activated, inward rectifier Kir3.1/3.4 K+ channels (but not constitutively active Kir2.1 channels) about as effectively as PI(4,5)P2 in excised membrane patches (22). Therefore, we asked whether PI(3,4,5)P3 affects KCNQ currents in intact cells. TsA cells grown in normal serum-containing medium have some plasma membrane–associated PI(3,4,5)P3 as reported by the PI(3,4,5)P3 indicator YFP-PH(Akt) (23), which is derived from the PH domain of Akt (fig. S5A). This resting PI(3,4,5)P3 disappears if phosphatidylinositol 3-kinase (PI 3-kinase) activity is lowered by growth in serum-free conditions, by treatment with 0.5 to 1 µM of the inhibitor wortmannin, or by overexpression of the dominant-negative PI 3-kinase regulatory subunit Δp85 (fig. S5, A to C) (24). Still, the current and its suppression by Oxo-M remained normal (fig. S5, B and D) (3). To raise PI(3,4,5)P3, we made CF-iSH, a translocatable construct combining CFP-FKBP with the inter–Src homology 2 (iSH2) domain from p85, which complexes in cells with endogenous PI 3-kinase p110 (Fig. 4A) (25). In NIH3T3 cells and tsA cells, the CF-iSH construct translocated rapidly to the plasma membrane (t1/2 = 14.4 ± 3.6 s, n = 8 cells) upon addition of iRap and induced membrane translocation of YFP-PH(Akt) (t1/2 = 36.5 ± 7.6 s) (Fig. 4, B and C). Nevertheless, the PI(4,5)P2 indicator YFP-PH(PLC-δ) was not translocated (Fig. 4D), indicating that CF-iSH initiates synthesis of PI(3,4,5)P3 at the plasma membrane with little depletion of PI(4,5)P2. This agrees with reports that the amount of PI(3,4,5)P3 made by receptor activation of endogenous 3-kinases is only a few percent of the available PI(4,5)P2 (26). Adding iRap to tsA cells expressing CF-iSH and grown in serum-free conditions had little effect on the amplitude of KCNQ2/KCNQ3 current (Fig. 4E). It did reduce the subsequent suppression by Oxo-M slightly so that suppression was not complete in 20 s (fig. S6).
Fig. 4.
Translocation of PI 3-kinase without change of KCNQ current amplitude. (A) The translocatable inter-SH2 domain of p85 and its complexation with endogenous p110 PI 3-kinase. PtdIns(3,4,5)P3, PI(3,4,5)P3. (B) Translocation of CF-iSH and YFP-PH(Akt) in NIH3T3 cells after a 1-min addition of iRap (5 µM). (C) Time course of translocation of CF-iSH and YFP-PH(Akt) by iRap. Error bars indicate SEM. (D) Translocation in cells coexpressing CF-iSH and YFP-PH(PLC-δ). (E) Modulation of KCNQ2/KCNQ3 channel activity by iRap in a tsA cell expressing LDR plus CF-iSH in serum-free conditions. Inset shows the current change after 4 min of iRap; mean, −0.4 ± 2.0% (n = 5 cells).
With intact cells, we have shown that decreases of PI(4,5)P2 quickly turn off current in KCNQ2/KCNQ3 channels by more than 95% in the complete absence of the cascade of IP3, calcium, and DAG signals normally generated by the activation of PLC. Conversely, an increase of PI(4,5)P2 augments the current, whereas synthesis of PI(3,4,5)P3 does not change the amplitude. During the activation of CF-Inp, rapid dephosphorylation of the PI(4,5)P2 pool would generate a bolus of additional PI(4)P. Modeling shows that this dephosphorylation will produce a large transient elevation of PI(4)P, followed by a maintained plateau of extra PI(4)P. Nevertheless, our experiments show that this elevated PI(4)P is not able to sustain KCNQ current when PI(4,5)P2 is depleted. These results give clarity to our hypothesis that the function of KCNQ channels is dependent on plasma membrane–associated PI(4,5)P2. Our study also shows the utility of the iRap translocation strategy for perturbing the lipid composition of the plasma membrane. This method is noninvasive, inducible, rapid, and specific, a combination not found when using RNA interference, antibodies, or pharmacology. We have used this strategy in an accompanying paper studying the plasma membrane targeting of small GTPases (27). The phosphoinositide dependence of many cellular functions is now directly testable.
Supplementary Material
Acknowledgments
Supported by NIH grants NS08174 and AR17803 (B.H.) and MH64801 and GM63702 (T.M.). T.I. is a recipient of a fellowship from the Quantitative Chemical Biology Program. We thank J. York for the Inp54p plasmid and advice on construction of CF-Inp, Alliance for Cellular Signaling for the p85 plasmid, T. Martin and Y. Aikawa (University of Wisconsin) for the PIPKI-γ plasmid and advice on construction of CF-PIPK, T. Wandless and J. Grimley for the iRap compound, J. Duman for help with calcium measurements, and K. Mackie for advice and discussion. Some images used the University of Washington Keck Imaging Center.
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/1131163/DC1
Materials and Methods
Figs. S1 to S6
References
References
- 1.Hilgemann DW, Feng S, Nasuhoglu C. Sci. STKE. 2001;2001:re19. doi: 10.1126/stke.2001.111.re19. [DOI] [PubMed] [Google Scholar]
- 2.Suh BC, Hille B. Curr. Opin. Neurobiol. 2005;15:370. doi: 10.1016/j.conb.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Suh BC, Hille B. Neuron. 2002;35:507. doi: 10.1016/s0896-6273(02)00790-0. [DOI] [PubMed] [Google Scholar]
- 4.Delmas P, Brown DA. Nat. Rev. Neurosci. 2005;6:850. doi: 10.1038/nrn1785. [DOI] [PubMed] [Google Scholar]
- 5.Lerche H, Weber YG, Jurkat-Rott K, Lehmann-Horn F. Curr. Pharm. Des. 2005;11:2737. doi: 10.2174/1381612054546815. [DOI] [PubMed] [Google Scholar]
- 6.Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Science. 1993;262:1019. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 7.Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. Nat. Methods. 2005;2:415. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raucher D, et al. Cell. 2000;100:221. doi: 10.1016/s0092-8674(00)81560-3. [DOI] [PubMed] [Google Scholar]
- 9.Materials and methods are available as supporting material on Science Online.
- 10.Correspondence about the iRap-inducible enzyme systems should be directed to T.I. (jctinoue@stanford.edu).
- 11.Stauffer TP, Ahn S, Meyer T. Curr. Biol. 1998;8:343. doi: 10.1016/s0960-9822(98)70135-6. [DOI] [PubMed] [Google Scholar]
- 12.Hirose K, Kadowaki S, Tanabe M, Takeshima H, Iino M. Science. 1999;284:1527. doi: 10.1126/science.284.5419.1527. [DOI] [PubMed] [Google Scholar]
- 13.Tsujishita Y, Guo S, Stolz LE, York JD, Hurley JH. Cell. 2001;105:379. doi: 10.1016/s0092-8674(01)00326-9. [DOI] [PubMed] [Google Scholar]
- 14.Horowitz LF, et al. J. Gen. Physiol. 2005;126:243. doi: 10.1085/jgp.200509309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oancea E, Meyer T. Cell. 1998;95:307. doi: 10.1016/s0092-8674(00)81763-8. [DOI] [PubMed] [Google Scholar]
- 16.Ishihara H, et al. J. Biol. Chem. 1998;273:8741. doi: 10.1074/jbc.273.15.8741. [DOI] [PubMed] [Google Scholar]
- 17.Winks JS, et al. J. Neurosci. 2005;25:3400. doi: 10.1523/JNEUROSCI.3231-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA. Nature. 2002;420:89. doi: 10.1038/nature01082. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Gamper N, Hilgemann DW, Shapiro MS. J. Neurosci. 2005;25:9825. doi: 10.1523/JNEUROSCI.2597-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suh BC, Horowitz LF, Hirdes W, Mackie K, Hille B. J. Gen. Physiol. 2004;123:663. doi: 10.1085/jgp.200409029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cantley LC. Science. 2002;296:1655. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 22.Rohacs T, Chen J, Prestwich GD, Logothetis DE. J. Biol. Chem. 1999;274:36065. doi: 10.1074/jbc.274.51.36065. [DOI] [PubMed] [Google Scholar]
- 23.Kontos CD, et al. Mol. Cell. Biol. 1998;18:4131. doi: 10.1128/mcb.18.7.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hara K, et al. Proc. Natl. Acad. Sci. U.S.A. 1994;91:7415. doi: 10.1073/pnas.91.16.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Q, Klippel A, Muslin AJ, Fantl WJ, Williams LT. Science. 1995;268:100. doi: 10.1126/science.7701328. [DOI] [PubMed] [Google Scholar]
- 26.Auger KR, Serunian LA, Soltoff SP, Libby P, Cantley LC. Cell. 1989;57:167. doi: 10.1016/0092-8674(89)90182-7. [DOI] [PubMed] [Google Scholar]
- 27.Heo WD, et al. Science. 2006;314:1458. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.