Abstract
Single-molecule fluorescence imaging has provided unprecedented access to the dynamics of ribosome function, revealing transient intermediate states that are critical to ribosome activity. Imaging platforms have now been developed that are capable of probing many hundreds of molecules simultaneously at temporal and spatial resolutions approaching the sub-millisecond time and the sub-nanometer scales. These advances enable both steady- and pre-steady state measurements of individual steps in the translation process as well as processive reactions. The data generated using these methods have yielded new, quantitative structural and kinetic insights into ribosomal activity. They have also shed light on the mechanisms of antibiotics targeting the translation apparatus, revealing features of the structure-function relationship that would be difficult to obtain by other means. This review provides an overview of the types of information that can be obtained using such imaging platforms and a blueprint for using the technique to assess how small-molecule antibiotics alter macromolecular functions.
Keywords: antibiotics, ribosome, smFRET, translation, tRNA
The origins of our understanding of translation
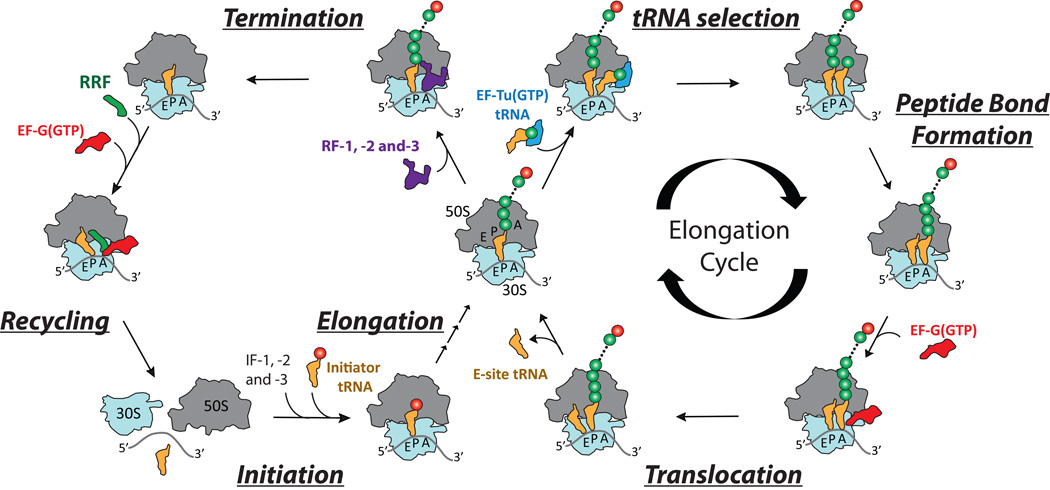
Since the ribosome was identified as the site of protein synthesis (1–3), technological advances have enabled many of the key insights into the mechanism of translation. The components required for ribosome-catalyzed protein synthesis were first identified through the advent of cell-free translation assays together with the development of biochemical and biophysical methods enabling the physical isolation of the essential protein and RNA cofactors. Subsequent biochemical and biophysical investigations ultimately led to a global framework for understanding how the ribosome “decodes” messenger RNA (mRNA) to produce a linear polypeptide. Key steps in developing this framework were the identification of the ribosome’s adaptor substrate molecules, highly structured and aminoacylated “transfer” RNAs (tRNAs), as well as the protein factors that interact with the ribosome to coordinate the four principal phases of translation: initiation, elongation, termination and recycling (Fig. 1).
Figure 1.
Schematic representation of the biochemical and structural features of the translating ribosome. The four principal stages of protein synthesis catalyzed by the ribosome—Initiation, Elongation, Termination and Recycling—are shown in the cycle on the left. A more detailed description of the steps of the elongation cycle, including tRNA selection, peptide bond formation and translocation, are shown on the right.
In bacteria, initiation factors (IF-1, IF-2 and IF-3) facilitate the assembly of the 70S ribosome at the site on the mRNA where protein synthesis starts (often an AUG codon). Elongation factors Tu (EF-Tu) and G (EF-G) catalyze the synthesis phase of translation, in which aminoacylated, “elongator” tRNAs enter and transit the ribosome, adding amino acids to the growing polypeptide. Release factors (RF) -1, -2 and -3 promote release of this peptide and the termination of protein synthesis at “nonsense” codons (UAA, UGA and UAG). Finally, ribosome recycling factor (RRF), in conjunction with EF-G, enables the disassembly of the complex, releasing the subunits, mRNA, and tRNA. The recognition that IF-2, EF-Tu, EF-G, and RF-3 bind GTP and hydrolyze it while bound to the ribosome led to speculation that the energy of hydrolysis directly powers the translation mechanism.
Our understanding of the ribosome itself, enabled principally by rigorous biochemical methods, has been greatly expanded by the development of new genetic, chemical and biophysical tools (4–10). These advances afforded perspective on the architectural features of the 2.4 MDa bacterial ribosome and further insights into the nature of tRNA interactions with the small and large ribosomal subunits (30S and 50S in bacteria; 40S and 60S in eukaryotic organisms). Critically, these experiments revealed how the 25 kDa, L-shaped tRNA molecules physically bridge the distinct activities of the ribosome: mRNA decoding within the small subunit and peptide bond formation within the large subunit. They also shed light on the spatial orientation of tRNA molecules as they transition between binding sites located within a solvent channel that traverses the interface between the two subunits. Ultimately, three physically distinct tRNA binding sites were identified and characterized: the aminoacyl (A), peptidyl (P), and exit (E) sites. The anticodons of tRNA in the A- and P-sites base pair with single-stranded mRNA displayed at the interface of the head, platform, and body domains of the small subunit. Within the A and P sites, the CCA sequence at the 3′ end of the tRNA forms Watson-Crick base pairs with highly conserved nucleotides within structured RNA elements of the large subunit’s peptidyl transferase center (PTC). In the E site, the terminal adenosine of tRNA stacks within an RNA helix of the large subunit to form a non-canonical base pair with C2394 that includes direct contacts with the vicinal hydroxyl groups of its ribose moiety. The nature of this interaction in the E site correspondingly precludes aminoacyl- or peptidyl-tRNA binding at this site.
The tools developed during this era also generated important insights into the nature of tRNA movements through the ribosome during protein synthesis. For instance, in line with early hypotheses regarding the substrate translocation mechanism (11, 12), A- and P-site tRNAs were shown to spontaneously adopt “hybrid” positions with respect to the ribosomal subunits (13). During this process, the 3’-CCA end of both tRNAs move with respect to the large subunit in the direction of translocation (towards the E site) before the mRNA and tRNA anticodons move relative to the small subunit. Large-scale movements of this kind were shown to be favored by peptide bond formation (13), suggesting that energy liberated by catalysis could also provide a driving force for directional tRNA movements on the ribosome.
Landmark achievements in structural biology have ushered in the most recent era of discovery. This work was also enabled by technological advances, including critical developments in cryogenic electron microscopy (cryo-EM) during the 1990’s. These breakthroughs revealed the ribosome’s structure in unprecedented detail, and provided new insights into the nature of its interactions with tRNAs and external factors. For instance, Frank and colleagues generated a low-resolution structure of the ribosome complexed with elongation factor-G, which captured the P-site tRNA in its hybrid (P/E) configuration (14). These findings provided the first clues that large-scale conformational events at the subunit interface—described as a “ratchet-like” rotation of the small subunit with respect to the large—are critical to hybrid state formation and the process of translocation.
The foundations provided by cryo-EM, together with improvements of cryogenic crystallography techniques (15, 16), paved the way for the first atomic resolution views of the protein synthesis apparatus. Advances in generating heavy metal derivatives enabled the generation of nearly complete structures of the individual small and large subunits, reported in August 2000. (17–19) These important milestones were quickly extended of the elucidation of ribosome structures bound to antibiotics, tRNA mimics, mRNAs, and external factors. (20–24) In late 2005, these efforts culminated in atomic resolution structures of the complete bacterial ribosome (a 70S particle). (25) A diverse range of functional ribosomal complexes trapped in intermediate states of the elongation cycle, have since been reported. Breakthroughs on this front continue at a rapid pace, and are likely to continue to enhance our knowledge of the translation process.
Small-molecule inhibitors of the translation apparatus
The use of translation-targeting antibiotics in clinical medicine began in 1948, when the antibiotic streptomycin, derived from soil bacteria, was introduced to combat the spread of tuberculosis in the United States. (26, 27) Today, almost every step of the translation cycle can be targeted by small-molecule antibiotics (reviewed in Ref. (26)). Hundreds of chemically distinct ribosomal inhibitors are described in the literature; some block translation universally, while others target translation in a species-specific manner. However, as for many anti-infective agents, drug-resistant organisms have invariably emerged for each translation inhibitor class. (26, 28, 29) Target modification, including post-transcriptional or post-translational modifications and/or point mutations within the drug binding site, serves as an important drug resistance mechanism. For example, streptomycin resistance can be acquired through any of a number of single point mutations in ribosomal protein S12, which resides within the small subunit decoding region. (30–33) These mutations in S12 are generally not deleterious to ribosome function and one or more of these mutations is found in most contemporary tuberculosis isolates. (34)
Therapies targeting the translation apparatus remain in widespread use, and new strategies for targeting the components of translation continue to be pursued. A key reason for this is that protein synthesis plays a ubiquitous and central role in all forms of life and the inhibition of translation is a proven strategy for broad spectrum bacterial growth inhibition. Anti-infective agents directly targeting functional centers in rRNA are particularly sought after as most bacteria possess multiple rRNA operons in their genome, making it unlikely that resistance mutations can occur in all rRNA operons simultaneously. As the functional centers in the ribosome are highly conserved, resistance mutations that do arise are often associated with defects in translation that are deleterious to cell growth. This propensity is exacerbated by the fact that actively expressed mRNAs are typically translated by many ribosomes simultaneously. In this context, even small deficiencies in ribosome function can result in perturbations to an organism’s capacity to effectively control gene expression.
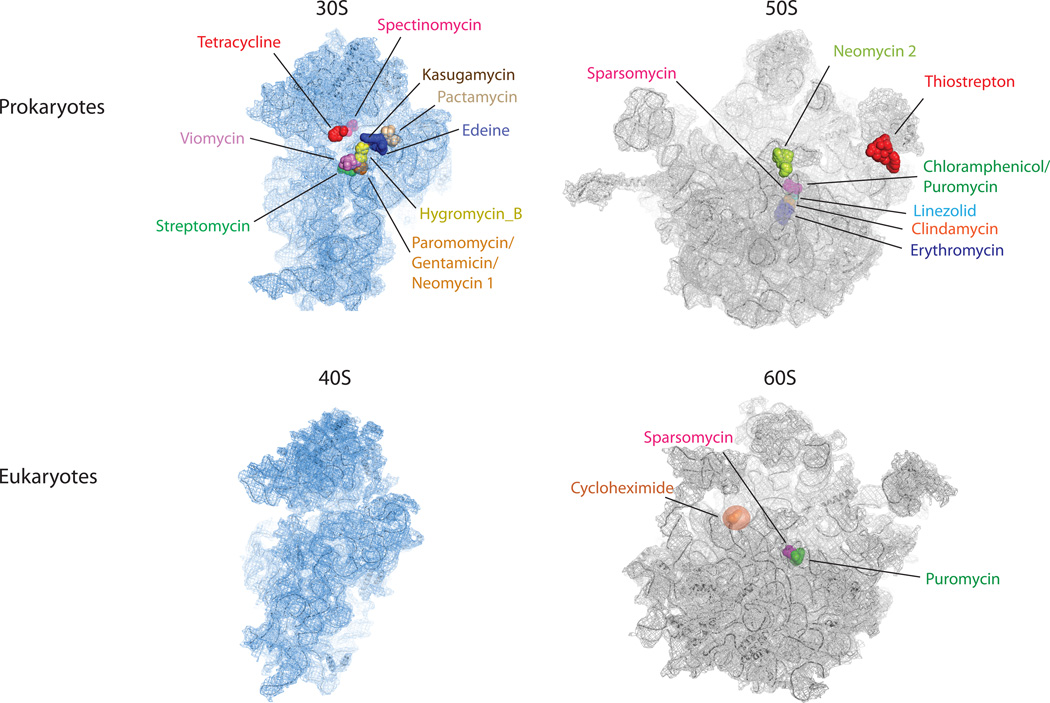
Elegant biochemical, genetic and structural investigations (reviewed in Ref. (26)) have revealed that the majority of known ribosome-targeting antibiotics bind functional centers within rRNA. These sites are often near the core regions of the particle, typically within the large subunit PTC or the small subunit decoding region (Fig. 2). Despite this understanding, there are major limitations to our ability to rationally target ribosome function. The mechanisms of only a subset of existing ribosome-targeting molecules are presently understood to an extent that enables rational design initiatives. This situation is unlikely to change rapidly as there are significant challenges associated with biophysical investigations of the translation apparatus and a paucity of tools that provide quantitative information regarding mechanisms of action.
Figure 2.
Antibiotic binding sites on the large and small subunits of the ribosome identified by crystallographic means. Small subunits (30S and 40S) are shown in blue and large subunits (50S and 60S) are shown in grey. The two most common drug targets are located in the small subunit decoding site (paromomycin, gentamicin and neomycin) and in the large subunit peptidyl transferase center (sparsomycin, chloramphenicol, puromycin, linezolid, clindamycin and erythromycin). The PDB accession codes for the structures are: 3R8O 3R8T, 3O30 and 3O5H.
The role of dynamics in ribosome function
Despite the steadily growing number of atomic-resolution descriptions of the translation machinery and the nature of ribosome-ligand interactions, such insights on their own fail to fully report on the mechanism of protein synthesis. These data only provide static snapshots of ribosomal activity and even when organized into a plausible time series—they lack the information required to reveal the structural and kinetic features of the ground and excited states of the system. (35, 36) Such efforts paint an incomplete picture by suggesting that all molecular processes related to ribosome functions are deterministic in nature. (37) A first principle consideration of thermal forces and solvent-ligand interactions argues that biological systems are constantly fluctuating between many distinct conformations and that the processes they govern may be largely stochastic in nature.
The dynamic nature of the translation process has long been appreciated (11, 12) and efforts to probe it have been pursued for more than three decades. Progress on this front has yielded insights into time-dependent changes in the movements of tRNA ligands and/or ribosome components with respect to each other upon thermal activation and peptide bond formation as well as during the process of tRNA selection and EF-G-mediated translocation. (38–47) However, such perspectives have largely been restricted to investigations of populations of ribosomes. These bulk methods suffer from technical challenges, such as the need to account for biochemically inactive components present in complex systems and incomplete reactivities or labeling (static heterogeneity). Bulk methods are also challenged by the dynamic nature of “ground state” ribosome configurations, which may differ before and after a given reaction, and the inherently asynchronous nature of events that occur during multistep processes (dynamic heterogeneity).
Imaging at the single-molecule scale
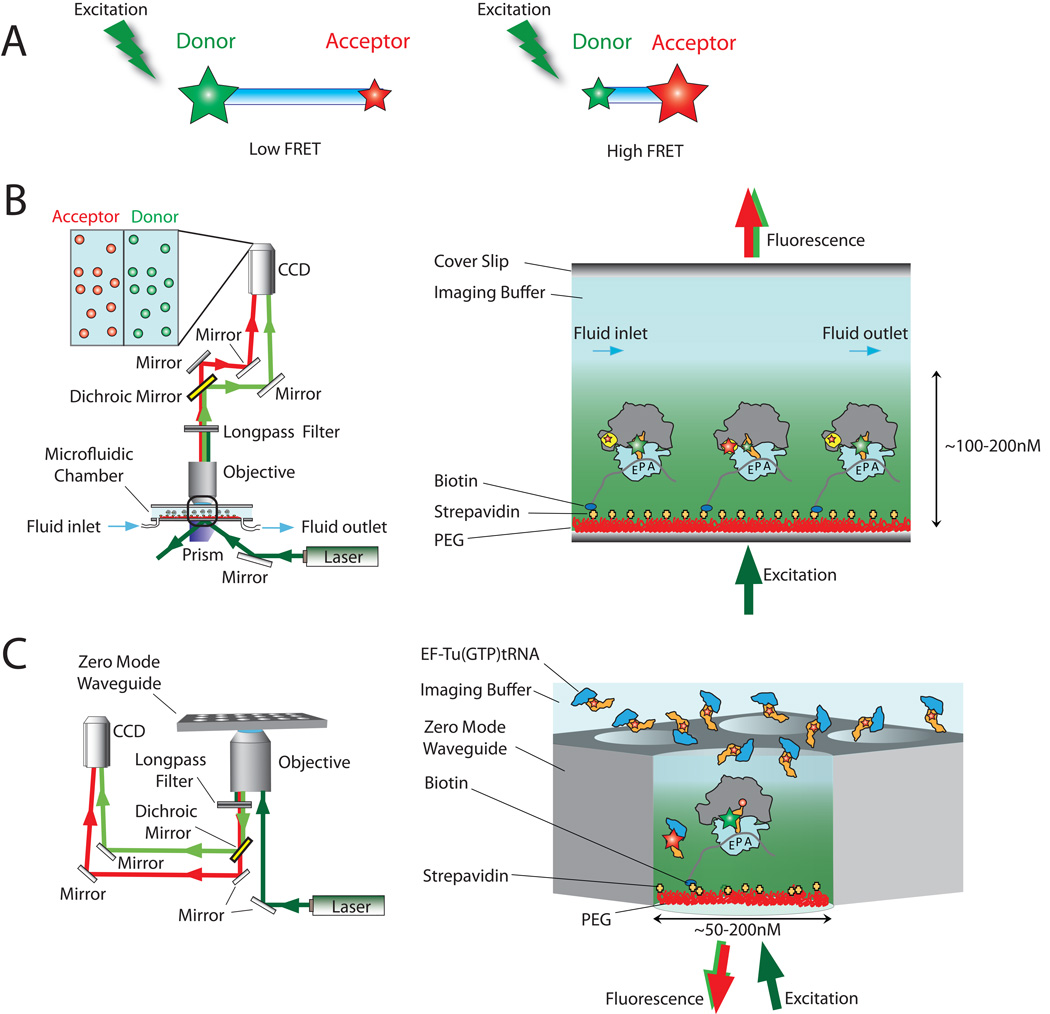
Single-molecule fluorescence imaging obviates many issues regarding dynamic heterogeneities present within a population of molecules, enabling direct measurements of asynchronous events critical to the translation mechanism across the full gamut of relevant time scales (ranging from 10 ms for individual steps to minutes for a complete protein to be synthesized). It also bypasses many of the complications arising from functional or physical heterogeneities, eliminating the need for large quantities of homogeneous components that are uniquely and uniformly tagged. Fluorescence and fluorescence resonance energy transfer (FRET) remain one of the most powerful tools in the scientific arsenal for detecting biomolecular localization, structure, and function. (48–51) Fluorescence from an individual point emitter—such as an organic dye molecule, a fluorescent protein or a quantum dot—can be resolved with nanometer resolution. (52–54) FRET, a process where an excited fluorophore returns to the ground state by transferring its energy through space to an acceptor fluorophore, is even more sensitive and enables the measurement of intra- and inter-molecular distances on the molecular scale (2–10 nm) with sub-nanometer accuracy. (50, 51, 55) The sensitivity of FRET-based measurements is a consequence of the dipole-dipole nature of the interaction, where the efficiency of energy transfer, E, scales inversely with the sixth-power of the distance between the two interacting dyes, R: E=1/[1+(R/R0)6]. Here, R0 is a normalization factor that reports on the spectral properties of the fluorophore pairs and the relative orientations of their dipole moments (Fig. 3A). (51)
Figure 3.
Experimental measurements of fluorescence resonance energy transfer. (A) Cartoon illustrating the principle of FRET: the amount of energy transfer depends on the distance between an excited donor fluorophore (green star) and acceptor fluorophore (red star). (B) Basic set-up of a prism-based single-molecule FRET microscope. (C) Basic set-up of a zero-mode waveguide (ZMW)-based fluorescent microscope. (88, 96, 97, 100)
Early foundations of single-molecule imaging
The first direct imaging of FRET in individual biomolecules in aqueous environments at ambient temperature was reported in 1996 (49). This study showed that distances between the ends of double helical DNA could be measured using single-molecule FRET (smFRET) by physically tethering the fluorescently labeled molecules near an optically transparent surface. The surface-immobilization approach was a significant breakthrough, as it enabled dramatic increases in the signal-to-noise ratio of imaging over more conventional illumination strategies. It also allowed one molecule to be continuously monitored over extended periods and subjected to fluid flow. Pulse-chase type experiments were important to the subsequent era in which such technologies were applied to a variety of pure RNA systems (56–60) and the eventual adaptation of analogous methods for investigations of ribosome function at the single-molecule scale. Significant modifications to the surface chemistries were needed to transition from imaging relatively simple nucleic acid species to more complex RNA-protein assemblies as early manufacturing procedures rendered quartz surfaces highly prone to non-specific binding, producing a physically heterogeneous population of molecules to be imaged.
Efforts to perform single-molecule investigations of ribosome function began as early as 1998. The first experiments documenting that activities arising from individual ribosome molecules could be observed was reported in 2003. (61) Single-molecule fluorescence experiments establishing that surface-immobilized ribosomes were fully active in basal translation reactions were reported in 2004. (62, 63) The implementation of these methods to probe ribosome function relied heavily on the foundations provided by the previous decades of bulk biochemical investigations. These studies guided efforts to fluorescently label components of the translation apparatus in a way that did not affect function (see (63) and references therein) and provided the experimental contexts for interpreting the findings obtained. The successful implementation of this approach also drew on the era of ribosome structure determination.
Today, microfluidic devices and instrument configurations exist that allow translation reactions to be robustly tracked at the single-molecule scale at high spatial and temporal resolution (Fig. 3B and 3C). Platforms of this kind enable the simultaneous imaging of fluorescence and FRET from several hundred to several thousand molecules under both steady state and pre-steady state conditions. The ability to study ribosome function at the single-molecule scale can reveal new information about the mechanisms of antibiotic action on the translation machinery. In this review, we briefly highlight some of the remarkable progress that has already been afforded using this approach, although we refer the reader elsewhere for more detailed discussions of the many specific findings obtained by these novel techniques. (36, 64, 65–67) It is our present view that continued investigations into known translation-targeting antibiotics are warranted and that deeper insight into antibiotic action requires a detailed understanding of the dynamic energy landscape of the ribosome. The need for additional knowledge in this area is particularly acute in the case of small molecules allosterically altering ribosome function. Studies of this kind are in their infancy, and we hope that the present work will serve as a useful guide for those interested in this line of research.
Steady-state measurements of antibiotic activity
The observation that ribosomes spontaneously transit between distinct native-state conformations led to the proposal that ribosomal functions are governed by the complex’s metastable energy landscape. (35, 36) Here, multiple basins on the energy landscape, each separated by activation barriers, represent globally similar ensembles of ribosome conformations that are likely to be significantly populated. The depths of these basins and heights of the activation barriers determine the stability of each ensemble and the transition rates between them. By providing quantitative measures of the occupancy within each basin and transition probabilities between distinct configurations, smFRET provides direct access to features of the underlying energy landscape.
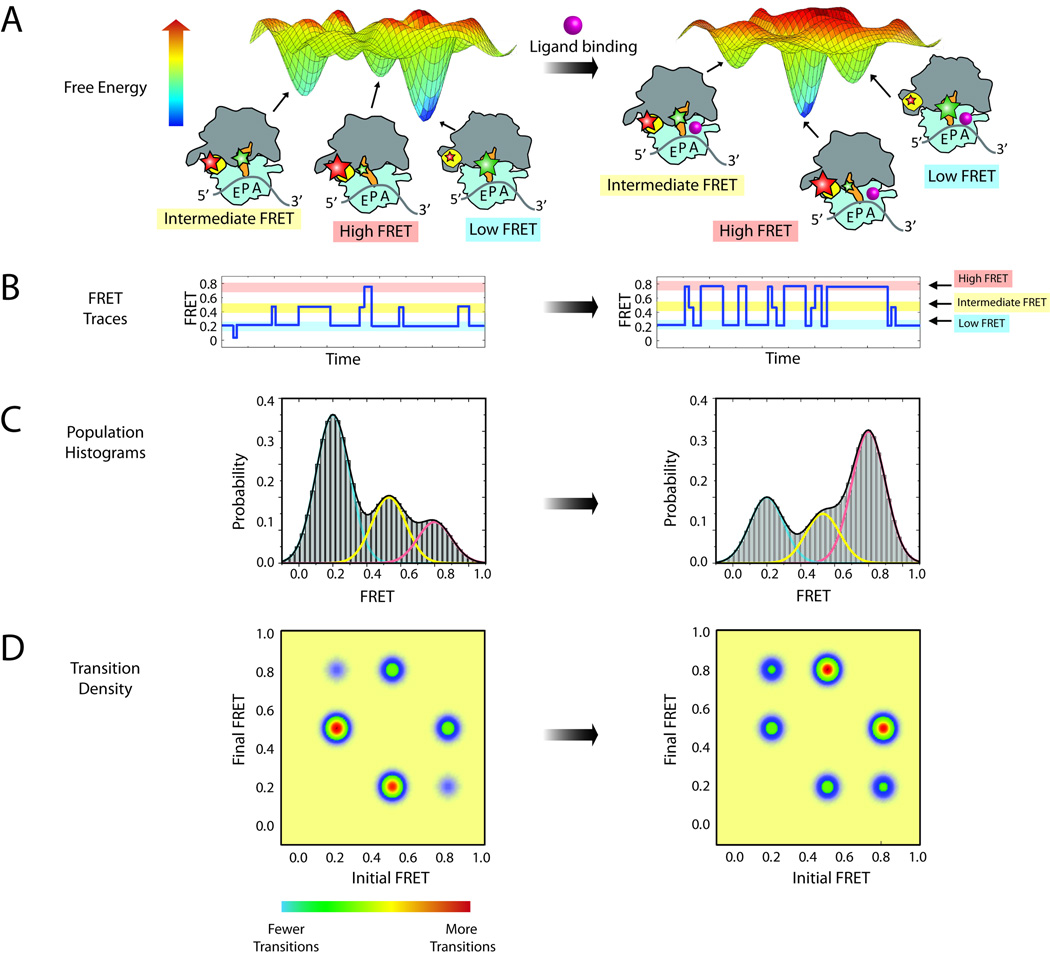
The energy landscape framework stipulates that intrinsic dynamics are essential to function and that ligands and translation factors regulate ribosome function by remodeling the ground state energies of distinct configurations of the system and/or the height of the energy barriers separating them. This framework enables the data obtained via smFRET to shed important new light on the mode of action of ribosome-targeting antibiotics, as it can reveal how these chemicals alter ribosome conformational dynamics and how such modifications perturb the ribosome’s interaction with exogenous translation components. Figure 4 illustrates how smFRET can be applied to explore the features of a hypothetical energy landscape.
Figure 4.
Steady state measurements of dynamic processes within the ribosome. (A) A hypothetical energy landscape of a ribosome complex with a P-site tRNA. Three dominant populations of ribosome conformations correspond to the three largest energy basins on the energy landscape. Donor and acceptor fluorophores (green and red stars, respectively) can be attached to the ribosome to report on the lifetime of these states. The addition of ligand (pink sphere) may alter the energy landscape, thus changing the dynamic equilibrium of the complex. (B) Single-molecule traces of the ribosome complex before (left) and after (right) addition of ligand. Three FRET states can be observed: high FRET (red), intermediate FRET (yellow) and low FRET (blue). (C) Population histograms from summation of single-molecule traces shown in (B). Each population can be fitted with three Gaussians, corresponding to each of the three conformations. (D) Transition density plots showing the relative number of transitions between each state.
In this example of the energy landscape (Fig. 4A, left panel), there are three deep energy basins, corresponding to three metastable conformations of a ribosome complex bearing tRNA in the P site. By placing fluorophores at strategic locations on this complex, FRET can report on the relative populations of three structural states and the rates of transition between them. A hypothetical smFRET trace to illustrate this case is shown in Figure 4B, left panel. Here, the low-FRET ribosome configuration is most stable and the high-FRET configuration is least stable and discrete transitions between these states are observed as a step-wise change in FRET efficiency. In such experiments, a large number of smFRET traces are summed together to yield a population histogram as shown in Figure 4C, left panel. These data, which are a direct reflection of the relative free energies of the structural configurations of the system, can then be fit to Gaussian distributions corresponding to the states observed in the individual traces and the population FRET histogram. Here, the widths of these Gaussian fits reflect both the intrinsic experimental noise of the measurement and fast dynamics within the conformational basin observed. Transition density plots, as shown in Figure 4D, display the relative number of transitions between pairs of states and correspondingly reflect the free energy barriers between them. By determining the rates of transitions between each state, the Gibbs free energy of activation (ΔG‡) can be calculated using the equation ΔG‡=−RT*ln[h*k/(kB*T)], where k is the reaction (transition) rate, h is Planck’s constant and kB is the Boltzmann constant.
Ligand binding could alter the dynamic equilibrium between states by changing the activation energy of one or more distinct transitions or the energy of one or more of the metastable states. (Fig. 4A, right panel). The smFRET trace in Figure 4B, right panel reflects a situation where a ligand has lowered the energy of the highest energy FRET state, while increasing the energy of the remaining states and changing the activation energies between states. More transitions to the high-FRET configuration are observed as well as shorter lifetimes in the low-FRET configuration. Changes in the dynamic equilibrium between states is also illustrated at the population level in the FRET histogram (Fig. 4C, right panel), where the predominant population shifts from low to high FRET upon ligand binding, and in the transition density plot (Fig. 4D, right panel), which displays a greater number of transitions into the high FRET state. These kinetic data can be used to calculate the ΔG‡ before and after the ligand binding (ΔΔG‡), information that reports on how an energetic barrier is altered for a particular transition.
Pre-steady state measurements of antibiotic activity
Dynamic processes that regulate ribosome activity can also be investigated through pre-steady state, non-equilibrium experiments. These studies can complement steady state observations and are typically required to gain insights into processes that use energy in the form of ATP or GTP hydrolysis to drive translation reactions. Pre-steady state measurements can be particularly advantageous when measuring the rates of factor and/or antibiotic binding, or when conformational dynamics drive a non-equilibrium process. Collecting sufficient data from single-molecule studies of transient processes remains a challenge for these experiments. In part, this is due to the finite photon collection efficiencies of current imaging platforms, and the finite lifespan of fluorophores prior to photobleaching. Using wide-field imaging methods, relatively modest levels of shot noise in the detected signal can now be achieved at temporal resolutions >10 ms (100 frames/s).
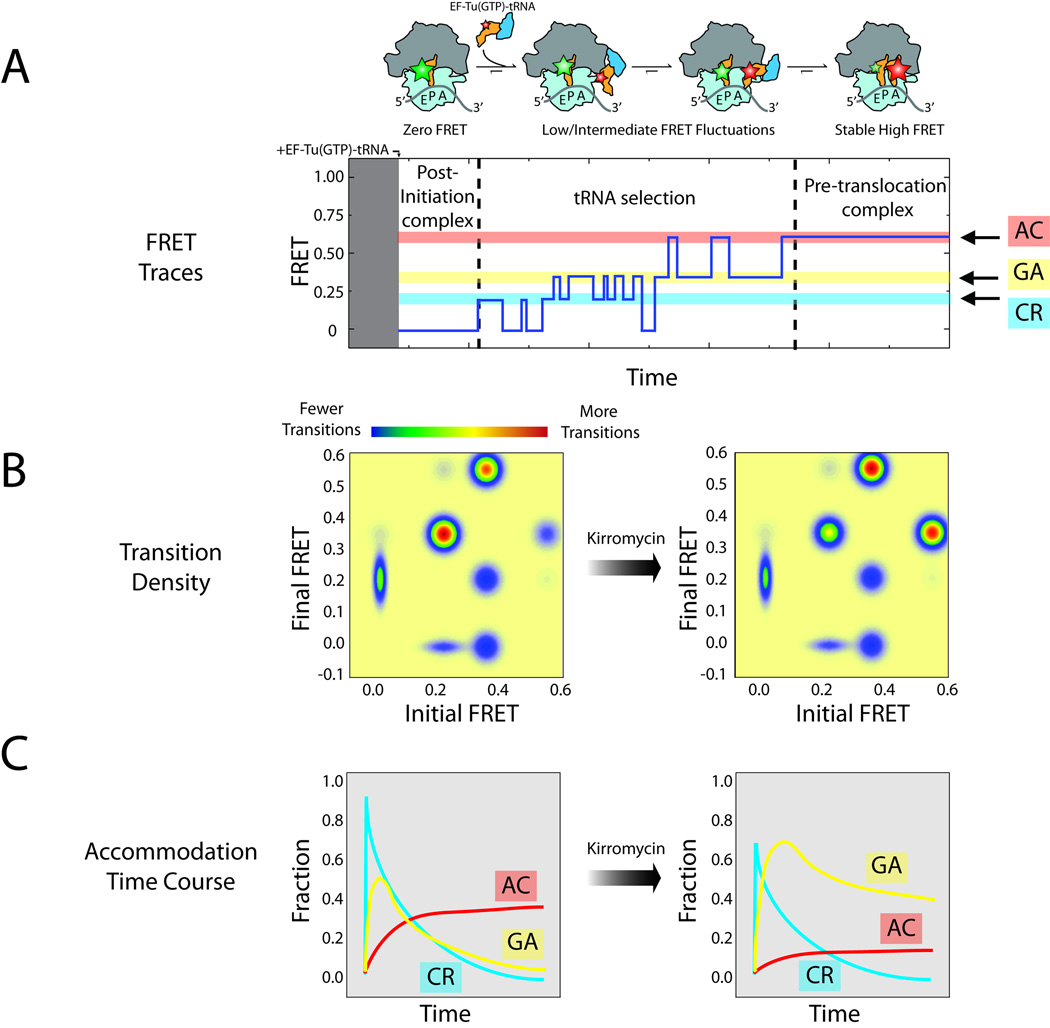
Figure 5 highlights the findings of a recent exploration of structural changes that occur on the ribosome during the process of EF-Tu-catalyzed tRNA selection (68). In this study, smFRET was employed to monitor the movements involved in this multistep process, tracking a Cy5-labeled Phe-tRNAPhe as it enters the A site by following its position with respect to Cy3-labeled tRNAfMet within the P site. The post-initiation ribosome complex starts with only a Cy3 fluorophore on P-site tRNA (Fig. 5A), and the initial FRET value observed is zero. Upon the introduction of Cy5-labeled Phe-tRNAPhe in a ternary complex with EF-Tu and GTP, transient, low-FRET events are observed, which correspond to sampling of the mRNA codon in the A site by Cy5-labeled Phe-tRNAPhe. For cognate tRNA (correctly paired to the mRNA codon within the A site), these early codon recognition (CR) events are followed by reversible excursions to higher-FRET states. These events ultimately lead to the tRNA achieving a fully accommodated (AC), high-FRET position on the ribosome, in which peptide bond formation occurs. This final state is termed the pre-translocation complex, as it is the substrate for EF-G-catalyzed translocation. (36)
Figure 5.
Pre-steady state measurements of dynamic processes within the ribosome. (A) Cartoon illustrating the principal steps in tRNA selection. Donor and acceptor fluorophores can be attached to tRNAs to monitor this process using single-molecule FRET. Low, intermediate and high FRET states are observed, corresponding to codon recognition (CR, blue), GTPase activation (GA, yellow) and tRNA accommodation (AC, red). (B) A transition density plot can be calculated to illustrate changes in the number of transitions between each state before (left) and after (right) the addition of kirromycin, an antibiotic inhibiting the release of inorganic phosphate from EF-Tu after GTP hydrolysis. (C) Changes in AC, GA and CR populations over time.
Here, the period of fast, reversible fluctuations was shown to reflect the fidelity process in tRNA selection, as indicated by the fact that the time scales and directionalities of these events depend on the nature of the mRNA codon and GTP hydrolysis. The intermediate-FRET state was assigned as the configuration where GTP hydrolysis occurs (the GTPase-activated [GA] state) through investigations using non-hydrolyzable forms of GTP. This conclusion was reinforced through experiments performed in the presence of kirromycin, an antibiotic that binds directly to EF-Tu to block conformational changes in the protein required for the release of tRNA immediately after GTP hydrolysis (69). Here, kirromycin was observed to increase the lifetime of the intermediate-FRET state. Quantitative analysis of these data, schematized in Figure 5B and 5C, were employed to reveal the order, timing and amplitude of the reversible excursions observed during the selection process, critical structural and kinetic features of the fidelity mechanism. Treatment of the data in this way showed that each configuration observed during the selection process was transiently and reversibly sampled, and that cognate tRNA can enter the A site only after sequentially passing through CR and GA states on path to its fully accommodated position. Imaging experiments of the same process from a distinct structural perspective showed that the observed FRET transitions arose principally from motions of Cy5-labeled Phe-tRNAPhe in the A site (68, 70). Here, the effects of antibiotics on the selection process are quantified by changes in the transition types, transition frequencies and the efficiency with which the pre-translocation complex is formed. This quantitative framework for exploring antibiotic action on the selection mechanism is at an early, nascent stage that has yet to be leveraged to its full potential.
Using smFRET to establish structure-function relationships and the mechanisms of antibiotic action
Because it provides direct access to the functional, structural, and kinetic features of a biomolecular system, smFRET can also be an important tool for guiding structure-determination efforts. (71– 74) Here we highlight one recent example, where smFRET, together with crystallographic efforts, was used to show that the 2-deoxystreptamine (2-DOS) aminoglycoside antibiotic neomycin allosterically inhibits global aspects of the translation mechanism through a previously uncharacterized binding site. (74) Single-molecule investigations, which predated this investigation, provided orthogonal structural perspectives that directly supported these results. (75)
2-DOS aminoglycosides are broad-spectrum antibiotics that are effective against Gram-negative bacterial infections. In vivo, they are known to target the tRNA selection process by inducing local rearrangements in rRNA within helix 44 (h44), which resides in the decoding site of the small subunit. (76) This rearrangement promotes the inappropriate incorporation of both near-cognate aa-tRNAs (one mismatch with the mRNA codon) and non-cognate aa-tRNAs (two or three mismatches with the mRNA codon) into the A site. However, in vitro experiments have also shown that 2-DOS aminoglycosides inhibit a range of distinct steps in the translation process, including initiation, translocation and recycling (see (74) and references therein). These diverse effects suggest that 2-DOS aminoglycosides may also inhibit features of the ribosomal machinery that are shared by these processes, such as rearrangement at the subunit interface.
To test if 2-DOS aminoglycosides affect global conformational processes in the ribosome, a new smFRET approach was developed for detecting intersubunit rotation and the process by which the ribosome interconverts between structurally distinct “locked” and “unlocked” configurations (corresponding to unrotated/classical and rotated/hybrid configurations, respectively). This method involved introducing fluorophores site-specifically within ribosomal proteins of the large and small subunit (Fig. 6A). With this approach, ribosome complexes were observed to exchange between two predominant configurations—low- (~0.19) and high- (~0.55) FRET states—corresponding to “locked” and “unlocked” configurations. (74)
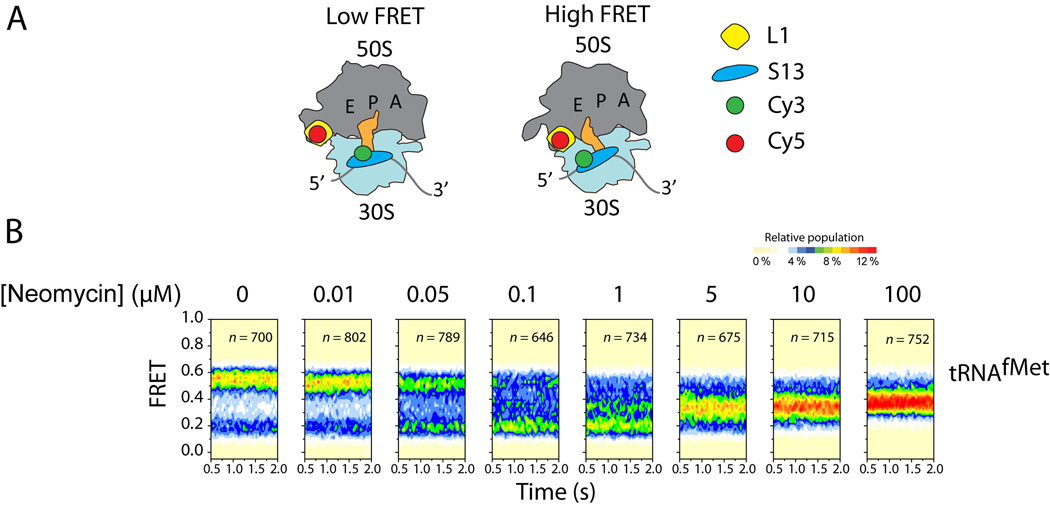
Figure 6.
Neomycin stabilizes a new ribosome rotation state. (A) Cartoon illustrating the labeling strategy used to probe subunit rotation. The low FRET state reports on the unrotated, locked conformation; the high FRET state reports on the rotated, unlocked conformation. (B) Population FRET histograms across a range of neomycin concentrations. At neomycin concentrations <1 µM, the low FRET state is stabilized. At neomycin concentrations >1 µM, a new FRET state emerges and is stabilized, reporting on a previously uncharacterized intermediate state of subunit rotation. Figure adapted from. (74)
When neomycin, a 2-DOS aminoglycoside, was added to ribosomes labeled in this fashion, a bimodal effect on subunit rotation dynamics was observed (Fig. 6B). At neomycin concentrations <0.1 µM, the low-FRET, locked ribosome configuration was stabilized, while an intermediate-FRET (~0.37) configuration predominated at neomycin concentrations >0.1 µM. This suggests that neomycin could stabilize a ribosome configuration that is intermediate between “locked” and “unlocked” states. Pre-steady state smFRET measurements showed that the stabilization of this configuration correlated with substantial reductions in a number of translation activities, including tRNA selection, translocation and ribosome recycling. These functional data suggest that the neomycin-stabilized ribosome conformation is somehow incompatible with the basic mechanics of translation. Notably, the dynamic properties of the ribosome were strongly suppressed in the neomycin-stabilized ribosome, suggesting that such complexes may be amenable to structure determination techniques.
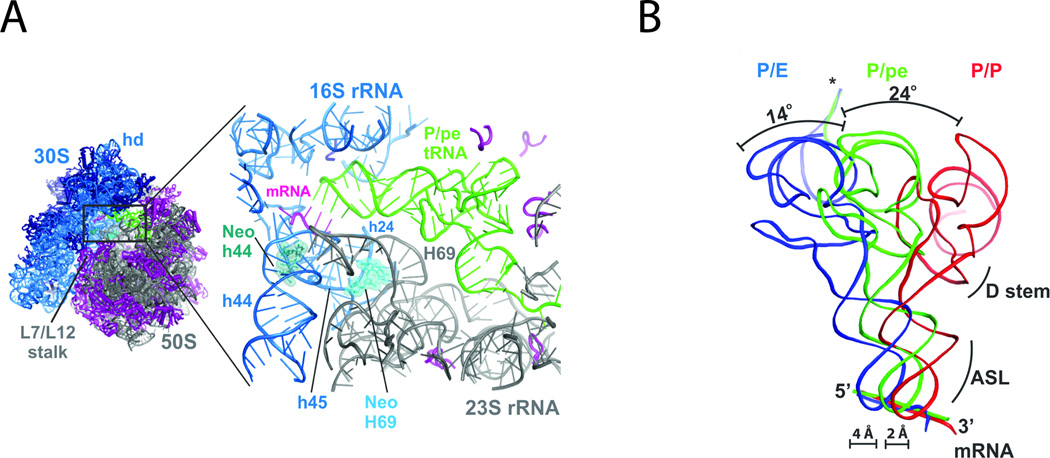
To gain further insights into this intermediate configuration, structures of neomycin-bound ribosome complexes bearing P-site tRNAPhe were solved using X-ray crystallography. As expected, positive electron density for neomycin was found in its canonical H44 decoding site. However, additional neomycin density was also found near the base of H69 in the large subunit (Fig. 7A). Strikingly, for the ribosome in the asymmetric unit that normally adopted a rotated configuration (73), the overall extent of small subunit rotation was markedly attenuated with neomycin bound to H69. P-site tRNAPhe was also observed to adopt a position intermediate between its classical (P/P) and hybrid (P/E) configurations (Fig. 7B). These findings, together with direct measurements of ribosome-factor interactions, provided a grounded physical framework within which the observed inhibition profiles caused by neomycin could be rationalized. Specifically, the data suggested that neomycin exhibits pleiotropic effects on the translation mechanism as a direct result of its binding to H69 of the large subunit, where its presence inhibits relative motions of the small and large subunits required for function. This example highlights the power of smFRET to serve as a tool to guide structure-determining efforts that yield important insights into the nature of structure-function relationships. It also exemplifies the utility of the approach to provide fundamentally new insights into the mechanisms of antibiotic action.
Figure 7.
The neomycin-bound ribosome. (A) Neomycin binds the small subunit (blue) in h44 (green) and the large subunit (grey) in H69 (light blue). (B) Ribosomes with neomycin bound at H69 display an intermediate position of P-site tRNA (green), between the hybrid (blue) and classical (red) configurations. Figure adapted from. (74)
Probing the effects of antibiotics on processive translation
In addition to detailed studies of single steps in the translation elongation cycle, smFRET has recently been applied to the investigation of processive translation, where it was used to observe multiple rounds of amino acid incorporation and translocation by individual ribosomes. By using Cy3-labeled 30S subunits and Cy5-labeled 50S subunits, Aitken and Puglisi tracked the elongation cycle of translation where ribosomes traverse mRNA while synthesizing protein. (77) In their system, ribosomes cycle between two FRET states (a low FRET state and a high FRET state) during each round of elongation, transitions that were assigned as reporting on peptide bond formation (unlocking) and translocation (locking). The authors also demonstrated the power of this system to resolve the effects of elongation factors and ribosome-targeting antibiotics on processive translation.
As expected, antibiotics such as fusidic acid, viomycin, and spectinomycin inhibited translation. However, significant reductions in protein synthesis were only observed following several cycles of elongation. Fusidic acid lengthened the lifetime of the high-FRET (locked) state, but did not affect the low-FRET (unlocked) state, and this effect was apparent only after the first elongation cycle. These observations are consistent with the known mechanism of fusidic acid translation inhibition, (78–81) in which the drug prevents the dissociation of EF-G(GDP) from the ribosome, thereby blocking ternary complex entry to the A site. In contrast, spectinomycin lengthened the lifetime of the low-FRET state without impacting the high-FRET state lifetime. This finding is consistent with structural data suggesting that spectinomycin “traps” the 30S head domain in a conformation that blocks translocation. (80, 82–86) Viomycin, one of the most potent translocation inhibitors known, lengthened the lifetime of both the high- and low-FRET states, suggesting that it may raise the energy barrier between locked and unlocked ribosome configurations. Notably, the codon-resolved impacts of the antibiotics investigated highlight how subtle effects that may not have significant impact on single rounds of elongation can be amplified across several elongation cycles.
Essential information about the mechanism of translation can be learned through direct observations of ligand-ribosome interactions (tRNAs, EF-G and/or EF-Tu) that drive protein synthesis reactions. However, imaging processive translation using smFRET can be hampered when physiological concentrations (ca. µM) of fluorescently-labeled ligands are employed. This is due to the high background signal arising from fluorescent molecules free in solution. While translation reactions proceed at lower than physiological concentrations (albeit at reduced rates) zero-mode waveguides (ZMWs) have been developed that circumvent this limitation by providing a way to drastically reduce the excitation volume. ZMWs are effectively zeptolitre (10−21 L) reaction chambers that restrict the penetration of light into the imaged area by virtue of being smaller than the wavelength of light. Their size also limits the number of fluorescently labeled molecules that are present in the chamber and illuminated volume simultaneously. By adapting ZMWs to the study of translation, as illustrated in Figure 3C, Uemura et al. were able to observe multiple rounds of elongation using fluorescently labeled tRNAs at near micromolar concentrations. (87, 88) This imaging platform allowed the authors to use three distinct tRNA species, labeled with Cy2, Cy3, and Cy5 respectively, to monitor the incorporation of each as it arrived at surface-immobilized ribosome complexes. Addition of erythromycin, an antibiotic that binds to the exit tunnel and traps the nascent polypeptide chain, halted tRNA incorporation after six to eight amino acids. This imaging strategy has many potential applications for the study of dynamic processes on the ribosome during processive translation reactions and its potential for exploring antibiotic action in this context is only beginning to be tapped.
Perspectives on the future
Current and future single-molecule fluorescence and FRET investigations hold great promise for providing deeper insights into the mechanisms of antibiotic action on the ribosome. However, progress in this area requires advancements on a number of critical fronts. First, improvements in instrument sensitivity and design are needed to broaden the range of time scales and motions that are accessible to study. Such efforts would be aided by parallel pursuits of enhancements in fluorophore performance (89, 90) and new methods by which to incorporate these probes into specific sites in native biological systems. (91, 92) Continued pursuit of a greater range of highly stable fluorophores and compatible detectors to enable photon detection at increasing speeds will also be paramount. The robustness and scalability of single-molecule imaging approaches will also need to be improved by implementing novel means to increase experimental and analytical throughput. Scaling efforts of this kind will require progress in the area of patterned microfluidic devices and automated systems, along with an expanded repertoire of robust methods for biomolecule immobilization. Computational tools for rapid, quantitative analysis will also need to scale accordingly.
While the power of smFRET to provide structural and functional insights into ribosome function has now been demonstrated in the context of both traditional structural biology approaches (71–74, 93) and molecular dynamics simulations (94), this relatively new approach remains in an era of validation; its full potential in the study of ribosome function has yet to be reached. Milestones that highlight both the promise and limitations of the field are likely to be encountered through the continued study of biological systems from a range of structural perspectives (95) as well as computational efforts aiming to understand the precise relationship between the dynamics observed by smFRET and complete atomistic representations of the structural changes they represent. As the field of single-molecule imaging achieves greater time resolution and computational methods extend to longer time scales, overlap between these two methods for understanding dynamic processes may soon be achieved.
Future single-molecule ribosome imaging pursuits must also probe the relationship between single-step ribosome reactions and processive translation (87, 88, 96, 97)—particularly in the context of polysomes. The continued pursuit of such investigations using the more complex and heterogeneous translation machinery present in eukaryotic systems represents an essential, and relatively uncharted, frontier. (72, 98) These efforts would be greatly aided by the advent of technologies enabling the simultaneous detection of irreversible chemical steps during each translation cycle (e.g. GTP hydrolysis and peptide bond formation) in order to provide fiducial markers for discrete steps in the protein synthesis mechanism. The capacity to work with ever smaller amounts of material will continue to be a critical asset for advancing the field. While incredibly difficult to imagine just a few years ago, the prospect of single-molecule imaging of biological systems inside of living cells (99) is likely to serve as an important beacon for future technology development, one that will entice the imaginations of biologists and biophysicists alike. Ultimately, efforts made towards that goal will be critical to fully understanding the physiological effects of translation inhibitors and their mechanisms of action.
Acknowledgements
We would like to thank Daniel S. Terry and Angelica Ferguson for critical feedback on the manuscript. This work was supported by the US National Institutes of Health (GM079238 and GM098859) and the National Science Foundation (0747230). M.B.F. is a trainee in the Weill Cornell/Rockefeller University/Sloan-Kettering Tri-Institutional MD/PhD Program supported by US National Institutes of Health Medical Scientist Training Program grant GM07739. M.R.W. is supported by US National Institutes of Health National Research Service Award fellowship 5F31DC012026-02.
References
- 1.Siekevitz P, Palade GE. Distribution of newly synthesized amylase in microsomal subfractions of guinea pigs pancreas. The Journal of cell biology. 1966 Sep;30:519. doi: 10.1083/jcb.30.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palade GE. A small particulate component of the cytoplasm. The Journal of biophysical and biochemical cytology. 1955 Jan;1:59. doi: 10.1083/jcb.1.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siekevitz P, Zamecnik PC. Ribosomes and protein synthesis. The Journal of cell biology. 1981 Dec;91:53s. doi: 10.1083/jcb.91.3.53s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green R, Noller HF. Ribosomes and translation. Annual review of biochemistry. 1997;66:679. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 5.Moore PB. The three-dimensional structure of the ribosome and its components. Annual review of biophysics and biomolecular structure. 1998;27:35. doi: 10.1146/annurev.biophys.27.1.35. [DOI] [PubMed] [Google Scholar]
- 6.Scheinman A, et al. Mapping the three-dimensional locations of ribosomal RNA and proteins. Biochimie. 1992 Apr;74:307. doi: 10.1016/0300-9084(92)90108-q. [DOI] [PubMed] [Google Scholar]
- 7.Jaynes EN, Jr, Grant PG, Giangrande G, Wieder R, Cooperman BS. Photoinduced affinity labeling of the Escherichia coli ribosome puromycin site. Biochemistry. 1978 Feb 21;17:561. doi: 10.1021/bi00597a001. [DOI] [PubMed] [Google Scholar]
- 8.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989 Nov 9;342:142. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 9.Nomura M. Early days of ribosome research. Trends in biochemical sciences. 1990 Jun;15:244. doi: 10.1016/0968-0004(90)90038-d. [DOI] [PubMed] [Google Scholar]
- 10.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000 Jul 20;406:318. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 11.Bretscher MS. Translocation in protein synthesis: a hybrid structure model. Nature. 1968 May 18;218:675. doi: 10.1038/218675a0. [DOI] [PubMed] [Google Scholar]
- 12.Spirin AS. A model of the functioning ribosome: locking and unlocking of the ribosome subparticles. Cold Spring Harbor symposia on quantitative biology. 1969;34:197. doi: 10.1101/sqb.1969.034.01.026. [DOI] [PubMed] [Google Scholar]
- 13.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 14.Frank J, Agrawal R. Ratchet-like movements between the two ribosomal subunits: Their implications in elongation factor recognition and tRNA translocation. Cold Spring Harbor symposia on quantitative biology. 2001;66:67. doi: 10.1101/sqb.2001.66.67. [DOI] [PubMed] [Google Scholar]
- 15.Yonath A. Approaching atomic resolution in crystallography of ribosomes. Annual review of biophysics and biomolecular structure. 1992;21:77. doi: 10.1146/annurev.bb.21.060192.000453. [DOI] [PubMed] [Google Scholar]
- 16.Yonath A, Mussig J, Wittmann HG. Parameters for crystal growth of ribosomal subunits. J Cell Biochem. 1982;19:145. doi: 10.1002/jcb.240190205. [DOI] [PubMed] [Google Scholar]
- 17.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 18.Wimberly BT, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 19.Schluenzen F, et al. Structure of functionally activated small ribosomal subunit at 3.3 angstrom resolution. Cell. 2000;102:615. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 20.Ogle JM, Carter AP, Ramakrishnan V. Insights into the decoding mechanism from recent ribosome structures. Trends in biochemical sciences. 2003 May;28:259. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- 21.Hansen JL, et al. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Molecular cell. 2002 Jul;10:117. doi: 10.1016/s1097-2765(02)00570-1. [DOI] [PubMed] [Google Scholar]
- 22.Harms JM, Bartels H, Schlunzen F, Yonath A. Antibiotics acting on the translational machinery. J Cell Sci. 2003 Apr 15;116:1391. doi: 10.1242/jcs.00365. [DOI] [PubMed] [Google Scholar]
- 23.Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002 Feb 22;108:557. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 24.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 angstrom resolution. Science. 2001;292:883. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 25.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005 Nov 4;310:827. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 26.Blanchard SC, Cooperman BS, Wilson DN. Probing translation with small-molecule inhibitors. Chemistry & biology. 2010 Jun 25;17:633. doi: 10.1016/j.chembiol.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schatz A, Bugie E, Waksman SA. Streptomycin, a substance exhibiting antibiotic activity against gram-positive and gram-negative bacteria. 1944. Clinical orthopaedics and related research. 2005 Aug;:3. doi: 10.1097/01.blo.0000175887.98112.fe. [DOI] [PubMed] [Google Scholar]
- 28.Tenson T, Mankin A. Antibiotics and the ribosome. Molecular microbiology. 2006 Mar;59:1664. doi: 10.1111/j.1365-2958.2006.05063.x. [DOI] [PubMed] [Google Scholar]
- 29.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009 Aug 28;325:1089. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurland CG. Translational accuracy and the fitness of bacteria. Annual review of genetics. 1992;26:29. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- 31.Carr JF, Hamburg DM, Gregory ST, Limbach PA, Dahlberg AE. Effects of streptomycin resistance mutations on posttranslational modification of ribosomal protein S12. Journal of bacteriology. 2006 Mar;188:2020. doi: 10.1128/JB.188.5.2020-2023.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bilgin N, Claesens F, Pahverk H, Ehrenberg M. Kinetic properties of Escherichia coli ribosomes with altered forms of S12. Journal of molecular biology. 1992 Apr 20;224:1011. doi: 10.1016/0022-2836(92)90466-w. [DOI] [PubMed] [Google Scholar]
- 33.Kurland CG, Ehrenberg M. Growth-optimizing accuracy of gene expression. Annual review of biophysics and biophysical chemistry. 1987;16:291. doi: 10.1146/annurev.bb.16.060187.001451. [DOI] [PubMed] [Google Scholar]
- 34.Gregory ST, Cate JHD, Dahlberg AE. Streptomycin-resistant and streptomycin-dependent mutants of the extreme thermophile Thermus thermophilus. Journal of molecular biology. 2001;309:333. doi: 10.1006/jmbi.2001.4676. [DOI] [PubMed] [Google Scholar]
- 35.Munro JB, Sanbonmatsu KY, Blanchard SC. A new view of protein synthesis: Mapping the free energy landscape of the ribosome using single-molecule FRET. Biopolymers. 2008 Jul;89:565. doi: 10.1002/bip.20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munro JB, Sanbonmatsu KY, Spahn CM, Blanchard SC. Navigating the ribosome's metastable energy landscape. Trends in biochemical sciences. 2009 Aug;34:390. doi: 10.1016/j.tibs.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore PB. How should we think about the ribosome? Annual review of biophysics. 2012;41:1. doi: 10.1146/annurev-biophys-050511-102314. [DOI] [PubMed] [Google Scholar]
- 38.Rodnina MV, Fricke R, Wintermeyer W. Transient conformational states of aminoacyl-tRNA during ribosome binding catalyzed by elongation factor Tu. Biochemistry. 1994;33:12267. doi: 10.1021/bi00206a033. [DOI] [PubMed] [Google Scholar]
- 39.Borowski C, Rodnina MV, Wintermeyer W. Truncated elongation factor G lacking the G domain promotes translocation of the 3' end but not of the anticodon domain of peptidyl-tRNA. Proc. Nat. Acad. Sci. USA. 1996;93:4202. doi: 10.1073/pnas.93.9.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 41.Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E-coli ribosome. Embo Journal. 1998;17:7490. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cochella L, Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005 May 20;308:1178. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seo HS, et al. Kinetics and thermodynamics of RRF, EF-G, and thiostrepton interaction on the Escherichia coli ribosome. Biochemistry. 2004 Oct 12;43:12728. doi: 10.1021/bi048927p. [DOI] [PubMed] [Google Scholar]
- 44.Pan D, Kirillov S, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Molecular cell. 2007 Feb 23;25:519. doi: 10.1016/j.molcel.2007.01.014. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robertson JM, Wintermeyer W. Effect of translocation on topology and conformation of anticodon and D loops of tRNAPhe. Journal of molecular biology. 1981;151:57. doi: 10.1016/0022-2836(81)90221-7. [DOI] [PubMed] [Google Scholar]
- 46.Odom OW, Stoffler G, Hardesty B. Movement of the 3'-end of 16 S RNA towards S21 during activation of 30 S ribosomal subunits. FEBS Lett. 1984;173:155. doi: 10.1016/0014-5793(84)81037-6. [DOI] [PubMed] [Google Scholar]
- 47.Odom OW, Picking WD, Hardesty B. Movement of tRNA but not the nascent peptide during peptide bond formation on ribosomes. Biochemistry. 1990;29:10734. doi: 10.1021/bi00500a004. [DOI] [PubMed] [Google Scholar]
- 48.Stryer L, Haugland RP. Energy transfer: a spectroscopic ruler. Proc. Natl. Acad. Sci., USA. 1967;58:719. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ha T, et al. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc Natl Acad Sci U S A. 1996;93:6264. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selvin PR. The renaissance of fluorescence resonance energy transfer. Nat Struct Biol. 2000;7:730. doi: 10.1038/78948. [DOI] [PubMed] [Google Scholar]
- 51.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nature methods. 2008 Jun;5:507. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gordon MP, Ha T, Selvin PR. Single-molecule high-resolution imaging with photobleaching. Proc Natl Acad Sci U S A. 2004 Apr 27;101:6462. doi: 10.1073/pnas.0401638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nature methods. 2006 Oct;3:793. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hell SW. Far-field optical nanoscopy. Science. 2007 May 25;316:1153. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 55.Sugawa M, Arai Y, Iwane AH, Ishii Y, Yanagida T. Single molecule FRET for the study on structural dynamics of biomolecules. Biosystems. 2007;88:243. doi: 10.1016/j.biosystems.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 56.Ha T, et al. Ligand-induced conformational changes observed in single RNA molecules. Proc Natl Acad Sci U S A. 1999;96:9077. doi: 10.1073/pnas.96.16.9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhuang X, et al. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 58.Zhuang X, et al. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002;296:1473. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 59.Kim HD, et al. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc Natl Acad Sci U S A. 2002;99:4284. doi: 10.1073/pnas.032077799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bartley LE, Zhuang X, Das R, Chu S, Herschlag D. Exploration of the transition state for tertiary structure formation between an RNA helix and a large structured RNA. Journal of molecular biology. 2003 May 16;328:1011. doi: 10.1016/s0022-2836(03)00272-9. [DOI] [PubMed] [Google Scholar]
- 61.Vanzi F, Vladimirov S, Knudsen C, Goldman Y, Cooperman B. Protein synthesis by single ribosomes. RNA. 2003;9:1174. doi: 10.1261/rna.5800303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol. 2004 Oct;11:1008. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 63.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci USA. 2004;101:12893. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marshall RA, Aitken CE, Dorywalska M, Puglisi JD. Translation at the Single-Molecule Level. Annual review of biochemistry. 2008 Jul 7;77:177. doi: 10.1146/annurev.biochem.77.070606.101431. [DOI] [PubMed] [Google Scholar]
- 65.Blanchard SC. Single-molecule observations of ribosome function. Current opinion in structural biology. 2009 Feb;19:103. doi: 10.1016/j.sbi.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frank J, Jr, Gonzalez RL. Structure and dynamics of a processive Brownian motor: the translating ribosome. Annual review of biochemistry. 2010;79:381. doi: 10.1146/annurev-biochem-060408-173330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bustamante C, Cheng W, Mejia YX. Revisiting the central dogma one molecule at a time. Cell. 2011 Feb 18;144:480. doi: 10.1016/j.cell.2011.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Geggier P, et al. Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. Journal of molecular biology. 2010 Jun 18;399:576. doi: 10.1016/j.jmb.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rodnina MV, Gromadski KB, Kothe U, Wieden HJ. Recognition and selection of tRNA in translation. FEBS Lett. 2005 Feb 7;579:938. doi: 10.1016/j.febslet.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 70.Whitford PC, et al. Accommodation of aminoacyl-tRNA into the ribosome involves reversible excursions along multiple pathways. Rna. 2010 Jun 1;16:1196. doi: 10.1261/rna.2035410. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fu J, Munro JB, Blanchard SC, Frank J. Cryoelectron microscopy structures of the ribosome complex in intermediate states during tRNA translocation. Proc Natl Acad Sci U S A. 2011 Mar 22;108:4817. doi: 10.1073/pnas.1101503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Budkevich T, et al. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Molecular cell. 2011 Oct 21;44:214. doi: 10.1016/j.molcel.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011 May 20;332:981. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang L, et al. Allosteric control of the ribosome by small-molecule antibiotics. Nat Struct Mol Biol. 2012 Aug 19; doi: 10.1038/nsmb.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feldman MB, Terry DS, Altman RB, Blanchard SC. Aminoglycoside activity observed on single pre-translocation ribosome complexes. Nature chemical biology. 2010 Jan;6:54. doi: 10.1038/nchembio.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annual review of biochemistry. 2005;74:129. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 77.Aitken CE, Puglisi JD. Following the intersubunit conformation of the ribosome during translation in real time. Nat Struct Mol Biol. 2010 Jul;17:793. doi: 10.1038/nsmb.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bodley JW, Zieve FJ, Lin L, Zieve ST. Formation of the ribosome-G factor-GDP complex in the presence of fusidic acid. Biochemical and biophysical research communications. 1969 Oct 22;37:437. doi: 10.1016/0006-291x(69)90934-6. [DOI] [PubMed] [Google Scholar]
- 79.Gao YG, et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009 Oct 30;326:694. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Munro JB, Wasserman MR, Altman RB, Wang L, Blanchard SC. Correlated conformational events in EF-G and the ribosome regulate translocation. Nat Struct Mol Biol. 2010 Dec;17:1470. doi: 10.1038/nsmb.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Savelsbergh A, Rodnina MV, Wintermeyer W. Distinct functions of elongation factor G in ribosome recycling and translocation. Rna. 2009 May;15:772. doi: 10.1261/rna.1592509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carter AP, et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics [see comments] Nature. 2000;407:340. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 83.Jerinic O, Joseph S. Conformational changes in the ribosome induced by translational miscoding agents. Journal of molecular biology. 2000 Dec 15;304:707. doi: 10.1006/jmbi.2000.4269. [DOI] [PubMed] [Google Scholar]
- 84.Borovinskaya MA, Shoji S, Holton JM, Fredrick K, Cate JHD. A Steric Block in Translation Caused by the Antibiotic Spectinomycin. ACS Chem. Biol. 2007;2:545. doi: 10.1021/cb700100n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. Journal of molecular biology. 2004 Nov 5;343:1183. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 86.Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Molecular cell. 2007 Feb 23;25:519. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Uemura S, et al. Single-molecule imaging of full protein synthesis by immobilized ribosomes. Nucleic acids research. 2008 Jul;36:e70. doi: 10.1093/nar/gkn338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Uemura S, et al. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010 Apr 15;464:1012. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Altman RB, et al. Enhanced photostability of cyanine fluorophores across the visible spectrum. Nature methods. 2012 May;9:428. doi: 10.1038/nmeth.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Altman RB, et al. Cyanine fluorophore derivatives with enhanced photostability. Nature methods. 2012 Jan;9:68. doi: 10.1038/nmeth.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Heal WP, Wright MH, Thinon E, Tate EW. Multifunctional protein labeling via enzymatic N-terminal tagging and elaboration by click chemistry. Nature protocols. 2012 Jan;7:105. doi: 10.1038/nprot.2011.425. [DOI] [PubMed] [Google Scholar]
- 92.Wang H, Chen X. Site-specifically modified fusion proteins for molecular imaging. Frontiers in bioscience : a journal and virtual library. 2008;13:1716. doi: 10.2741/2795. [DOI] [PubMed] [Google Scholar]
- 93.Wang L, Altman RB, Blanchard SC. Insights into the molecular determinants of EF-G catalyzed translocation. Rna. 2011 Dec;17:2189. doi: 10.1261/rna.029033.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Whitford PC, et al. Accommodation of aminoacyl-tRNA into the ribosome involves reversible excursions along multiple pathways. Rna. 2010 Jun;16:1196. doi: 10.1261/rna.2035410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Whitford PC, et al. In: Ribosomes: Structure, Function, and Dynamics. Rodnina MV, Wintermeyer W, Green R, editors. New York: Springer-Verlag/Wien; 2011. pp. 303–319. [Google Scholar]
- 96.Petrov A, et al. Dynamics of the translational machinery. Current opinion in structural biology. 2011 Feb;21:137. doi: 10.1016/j.sbi.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tsai A, et al. Heterogeneous pathways and timing of factor departure during translation initiation. Nature. 2012 Jul 19;487:390. doi: 10.1038/nature11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Petrov A, Puglisi JD. Site-specific labeling of Saccharomyces cerevisiae ribosomes for single-molecule manipulations. Nucleic acids research. 2010 Jul;38:e143. doi: 10.1093/nar/gkq390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xie XS, Choi PJ, Li G-W, Lee NK, Lia G. Single-Molecule Approach to Molecular Biology in Living Bacterial Cells. Annual review of biophysics. 2008;37:417. doi: 10.1146/annurev.biophys.37.092607.174640. [DOI] [PubMed] [Google Scholar]
- 100.Levene MJ, et al. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003 Jan 31;299:682. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]