Abstract
The electrophilic metabolites of the polyaromatic hydrocarbon naphthalene have been shown to bind covalently to proteins and covalent adduct formation correlates with the cytotoxic effects of the chemical in the respiratory system. Although 1,2-naphthalene epoxide, naphthalene diol epoxide, 1,2-naphthoquinone, and 1,4-napthoquinone have been identified as reactive metabolites of interest, the role of each metabolite in total covalent protein adduction and subsequent cytotoxicity remains to be established. To better understand the target residues associated with the reaction of these metabolites with proteins, mass spectrometry was used to identify adducted residues following 1) incubation of metabolites with actin and protein disulfide isomerase (PDI), and 2) activation of naphthalene in microsomal incubations containing supplemental actin or PDI. All four reactive metabolites bound to Cys, Lys or His residues in actin and PDI. Cys17 of actin was the only residue adducted by all metabolites; there was substantial metabolite selectivity for the majority of adducted residues. Modifications of actin and PDI, following microsomal incubations containing 14C-naphthalene, were detected readily by 2D gel electrophoresis and phosphor imaging. However, target modifications on tryptic peptides from these isolated proteins could not be readily detected by MALDI/TOF/TOF and only three modified peptides were detected using high resolution – selective ion monitoring (HR-SIM). All the reactive metabolites investigated have the potential to modify several residues in a single protein, but even in tissues with very high rates of naphthalene activation, the extent of modification was too low to allow unambiguous identification of a significant number of modified residues in the isolated proteins.
Keywords: Adducts, mass spectrometry, naphthalene, reactive metabolites, model proteins, microsomal incubations
1. Introduction
Although there is a reasonably well-populated database of proteins which have been shown to be covalently modified by a range of metabolically activated toxicants [1], as well as amino acids in proteins identified as targets of numerous reactive metabolites [2, 3], there still remains a general lack of appreciation for the precise mechanisms by which these modifications subsequently lead to cytotoxic events in a cell. There have been several studies showing modifications of specific amino acids in a protein following direct addition of reactive metabolites to solutions of protein [2–4], however specific residues modified by reactive metabolites generated in a whole cell or an in vivo system have only been identified in a few instances [5–7]. This limited knowledge of reactive metabolite adduction onto proteins in cellular systems is, in part, due to mixture complexity as well as the generally small degree of modification resulting from the reactive metabolites. For example, previous in vivo studies with 1-nitronaphthalene, where the extent of adduction on proteins was detected by a combination of fluorescent protein staining and storage phosphor analysis showed that, with the vast majority of identified, adducted proteins, less than 1% of the protein was modified [8]. Thus, identification of critical sites on proteins for metabolically activated chemicals remains a challenge.
Much of the current insight regarding the importance of protein adduction in cytotoxicity comes from work on compounds where a single reactive metabolite is generated. In cases where multiple reactive metabolites are produced, the understanding of these processes is far more limited, as with naphthalene. Although 1,2-naphthalene epoxide (NPO), naphthalene diol epoxide (NDO), 1,2-naphthoquinone (1,2-NPQ), and 1,4-napthoquinone (1,4-NPQ) have been identified as reactive metabolites of interest, the role of each in covalent protein adduction and subsequent cytotoxicity remains to be established. If the differences in these metabolites’ reactivities with residues translate into disparate effects on whether a particular adduct alters the functional properties of a protein critical to cellular homeostasis, then the overall metabolic disposition of this chemical and inter-individual differences in the enzymes responsible for controlling the formation of these metabolites could substantially influence susceptibility to exposure to compounds like naphthalene.
To determine whether there were differences in residues adducted by the four reactive metabolites of naphthalene, the metabolites were added directly to target proteins, actin and protein disulfide isomerase (PDI), at pH 7.4 (hereafter referred to as synthetic incubations). Actin and PDI have previously been shown to be adducted by naphthalene metabolites in several systems [9, 10] and both are abundant in cysteine, lysine and histidine residues. In addition, both proteins are well-known targets for other metabolically activated chemicals [5, 11–16]. To gain additional insight into the biological distribution of metabolites bound to the amino acid residues, adducts were also generated by the activation of naphthalene using microsomes from target and non-target tissues.
The amino acid targets from both the synthetic and microsomal incubations were analyzed by mass spectrometry following trypsin digestion. Fourteen adducted peptides were observed on actin and twelve on PDI following incubation with naphthalene metabolites. All four metabolites produced adducts with cysteine and lysine, but histidine was shown to be targeted only by 1,2-NPQ and NDO. Adducts on actin and PDI were detected as radioactive 14C-naphthalene spots on 2-DE (two-dimensional gel electrophoresis) of extracts from the microsomal incubations, however, only three specific protein modifications were detected following microsomal incubations. This underscores the challenges of measuring specific modifications of proteins when reactive metabolites are generated biologically rather than synthetically. The two methods investigated in this study were designed to better understand the dynamics between naphthalene metabolites and target amino acids. The first method allowed manipulation of the incubation conditions for a better understanding of the species chemistry; the second method was used to validate model protein adductions in known target tissues for biological applications.
2. Materials & Methods
2.1 Reagents
1,2-Naphthoquinone, 1,4-naphthoquinone, PDI (from bovine liver), iodoacetamide, and Orange G-II dye were purchased from Sigma-Aldrich (St. Louis, MO, USA). Actin was purchased from Cytoskeleton (Denver, CO). Bradford Protein Assay solution and PVDF membranes were purchased from Bio-Rad (Hercules, CA). Protease inhibitor cocktail III was purchased from Calbiochem (La Jolla, CA). Immobilized pH gradient (IPG) buffers and immobiline dry strips (18cm) were purchased from GE Health Care (Piscataway, NJ). Trypsin was purchased from Promega (Madison, WI). [1–14C] Naphthalene was purchased from Moravak Biochemicals (Brea, CA, USA); radiochemical purity was > 98% by HPLC.
2.2 Animals
Animal studies were conducted under protocols approved by the University of California – Davis Animal Use and Care Committee. Male Swiss-Webster mice (25–30g) and Sprague-Dawley rats (150–200g) were purchased from Harlan (San Diego, CA). Animals were housed in a HEPA filtered isolator rack in an AAALAC-accredited facility for one week before use. Food and water were provided ad libitum.
2.3 Synthetic Incubations
2.3.1 Synthesis of Naphthalene Epoxides
Naphthalene 1,2-epoxide and naphthalene 1,2-dihydro-1,2-dihydroxy-3,4-epoxide (NDO) were synthesized using methods published by Yagi and Jerina [17] and Horning et al. [18], respectively. Both products yielded white crystals upon recrystallization. Naphthalene oxide was shown to be free of contamination with 1-naphthol (the primary degradation product) by the absence of a UV peak at 309 nm. NMR of naphthalene diol epoxide in CD3CN yielded signals identical to those reported previously [18]. Products were dissolved in 99.5% ethanol/0.5% triethylamine (epoxide) or ethanol (diol epoxide) and stored at −80° C.
2.3.2 Chemical Preparation of Metabolite-Protein Adducts
Approximately 10 µmoles of each metabolite, in ethanol (epoxides) or DMSO (quinones), were added to a solution of protein (1 mg/ml) in 0.1M sodium phosphate buffer (pH 7.4, argon sparged) and the mixture was incubated in a sealed vial. The total volume of each actin incubation was 1 mL. Metabolites were in 500-fold molar excess to actin based on amount of protein and approximately 14-fold molar excess based on adduction sites (C, K, and H). The total volume of each PDI incubation was 0.5mL. Metabolites were in 500-fold molar excess to PDI based on amount of protein and approximately 7-fold molar excess based on potential sites. Following metabolite addition to the vial, the sample was stirred on a vortex mixer for 15 s and then mixed by continuous inversion for 1 h at room temperature. pH was measured at the beginning and the end of the incubation and remained constant. For control incubations, the metabolite was omitted. Reactions were stopped by adding 10 µl formic acid (FA).
2.3.3 In-solution Digest
Protein samples were denatured in 4 mM DTT at 65°C prior to trypsin digest at 37°C overnight in a Gene Amp PCR System 2400 (Perkin Elmer, MA). Digests were quenched by adding 0.1% FA. Samples were dried in a vacuum centrifuge, reconstituted in 0.1% FA, and stored at −20°C prior to MS analysis.
2.3.4 Protein Identification by LC-MS/MS
Tryptic digests from the synthetic incubations were analyzed with a LTQ Orbitrap XL (Thermo Fisher, San Jose, CA) operated in positive mode with the IonMax electrospray ionization source and N2 collision gas. Compounds were separated by HPLC using a 1 × 50 mm Waters Symmetry C18 column. Mobile phase A was 0.1% FA/H2O and mobile phase B was 0.1% FA/acetonitrile (ACN); initial gradient conditions were 95% A and 5% B at flow rate of 200 µL/min. The gradient was increased linearly from 5% B to 50% B over 25 min, increased to 100% B over the next 3 min, then returned to initial conditions the last 3 min. The HPLC effluent was monitored in FT mode in m/z 200 to 2000 range with a spray voltage of 5.00 kV, capillary temperature of 250°C, and capillary voltage of 49 V. Collision induced dissociation was used to fragment [M+H]+ ions with 35% normalized collision energy. The resolving power was 30,000 for full scan MS and MS/MS events. Calibration was performed with a Cal mix of caffeine, MRFA, and Ultramark, and yielded mass accuracies of 5 ppm or better.
2.4 Microsomal Incubations
2.4.1 Collection of Mouse and Rat tissue
Rodents were killed with an overdose of pentobarbital administered i.p. Mouse livers were removed and rinsed in ice-cold buffer. For rat nasal compartments, the head of each rodent was removed from the carcass, the lower jaw and skin were removed, the head was split along the medial suture, and the nasal olfactory epithelium was removed by careful dissection [19].
2.4.2 Microsomal Preparation and Incubation
Nasal tissue and livers were homogenized in four volumes of pH 7.4 homogenization buffer consisting of 20 mM Tris, 150 mM KCl, 0.2 mM sodium EDTA, 0.5 mM dithiothreitol, and 15% glycerol at 4 °C with glass homogenizers. Washed microsomal pellets were prepared by differential ultracentrifugation as described previously [20]. Final microsomal pellets were resuspended in 0.1M sodium phosphate buffer (pH 7.4). Protein concentration was determined using the Bradford method with BSA as the standard.
All microsomal incubations were performed in triplicate (i.e., three radiolabeled and three nonlabeled). Incubations were prepared on ice using 0.1 M sodium phosphate buffer (pH 7.4) and consisted of: either 0.25 mM unlabeled naphthalene or 0.25 mM [14C] – naphthalene (106,000 DPM/nmole), microsomes (400µg), NADPH generating system (15 mM MgCl2, 60 mM glucose-6-phosphate, 2 mM NADP, and 15 iu/mL glucose-6-phosphate dehydrogenase). 100 µg of excess actin or PDI was added to the model systems; controls contained an equivalent volume of phosphate buffer. Capped vials were incubated for 30 min in a 37°C shaking water bath then placed on ice, transferred to centrifuge tubes, and centrifuged at 14,000g for 60 min at 4°C. The supernatant was separated from the pellet and both were stored at −80°C.
2.4.3 Separation and identification of adducted proteins
Methods for separation, localization, and identification of adducted proteins were described in detail previously [21]. Briefly, pelleted samples were not pooled and separate gels were run for each with 14C-labeled (for imaging) and unlabeled (for identification) naphthalene. Pelleted samples were solubilized and proteins were resolved by 2-DE on 18-cm IPG strips, pI 4–7. Following 2-DE, gels from incubations with non-labeled naphthalene were fixed (10% acetic acid, 40% methanol, and 50% H2O); gels from radiolabeled incubations were electroblotted to PVDF membranes. Blots were washed twice with H2O and dried overnight in a vacuum desiccator. Dried blots were placed against storage phosphor screens (GE) in a lead-shielded container to assess the location of radioactive bands. Phosphor screens were exposed for 15 days and scanned on a Typhoon 8600 Variable Mode Scanner (GE) for visualization. Phosphor images were registered by overlay with corresponding non-labeled gels to guide gel spot selection for excision.
Gels containing non-labeled samples were silver stained and imaged on a 16-bit gray scale scanner. From silver-stained gels, protein spots of interest were excised, destained, washed, dried, and rehydrated according to established protocols [22]. Proteins were digested with sequencing grade trypsin (25 ng/mg of protein target) at 37°C for 24 h. Peptides from the tryptic gel digests were desalted, mixed with matrix solution (α-cyano-4-hydroxycinnamic acid in 0.1% TFA in 70% ACN), applied to the target plate, and analyzed with an ABI 4700 MALDI TOF/TOF (time of flight mass spectrometer) (Applied Biosystems, Foster City, CA) equipped with a Nd:YAG laser (337 nm). Mass spectra were acquired in the reflectron mode. Internal mass calibration performed with two trypsin autodigestion fragments (842.5 and 2211.1 Da), typically resulted in mass accuracies of 50 ppm or better.
2.5 Identification of Metabolite Modifications on Tryptic Digests of Model Proteins
MS/MS data (from both the LTQ-Orbitrap and 4700 MALDI TOF/TOF) were inputted into the Mascot server (Matrix Sciences, Boston, MA, http://www.matrixscience.com) to search for matching adducted peptides for actin and PDI from the UniProtKB/Swiss-Prot mouse database (http://www.uniprot.org). Databases were searched with a precursor tolerance of 150 ppm, MS/MS fragment tolerance of 0.8Da, and a maximum of two missed cleavages. Custom modifiers were created for each metabolite with preferences for the target amino acids (C, K, and H): NPO (C10H8O, m/z 144.0575 Da) for the naphthalene monoepoxide; NDO (C10H10O3, m/z 178.0629) for the naphthalene diol epoxide; NPQ (C10H6O2, m/z 158.0368) for the 1,2-naphthoquinone and the 1,4-naphthoquinone. Samples were searched against each metabolite modifier separately as fixed custom modifications. Variable modifications such as oxidation and carbamidomethylation were included in searches.
Protein identifications with a probability-based MOWSE protein score (> 52) for both of the proteins and ion score (> 30) for the peptides obtained through MASCOT were considered statistically significant (p < 0.05). Spectra from significant peptide PTM hits were analyzed with Xcalibur Qual Browser software (Thermo Fisher) and validated against predicted fragment ions using Protein Prospector (http://prospector.ucsf.edu).
2.6 Selective Ion Monitoring by ESI-MS
Samples obtained following microsomal incubation, pellet separation, 2-DE protein isolation, and digestion with trypsin were also injected and scanned with the LTQ Orbitrap XL using SIM (selective ion monitoring) for enhanced detection of the adducted ions based on the adducted parent masses identified from the synthetic incubations. Gradient conditions for the SIM were the same as the full scan for the synthetic incubations. The HPLC effluent was monitored in FT mode centered on the target m/z ion with a width of 10Da. Monitoring was performed with positive ionization, a spray voltage of 5.00 kV, a capillary temperature of 250°C, and a capillary voltage of 49 V. The resolving power was 60,000 for the SIM-MS.
3. Results
3.1 Adduction Sites of Naphthalene Reactive Metabolites on Actin
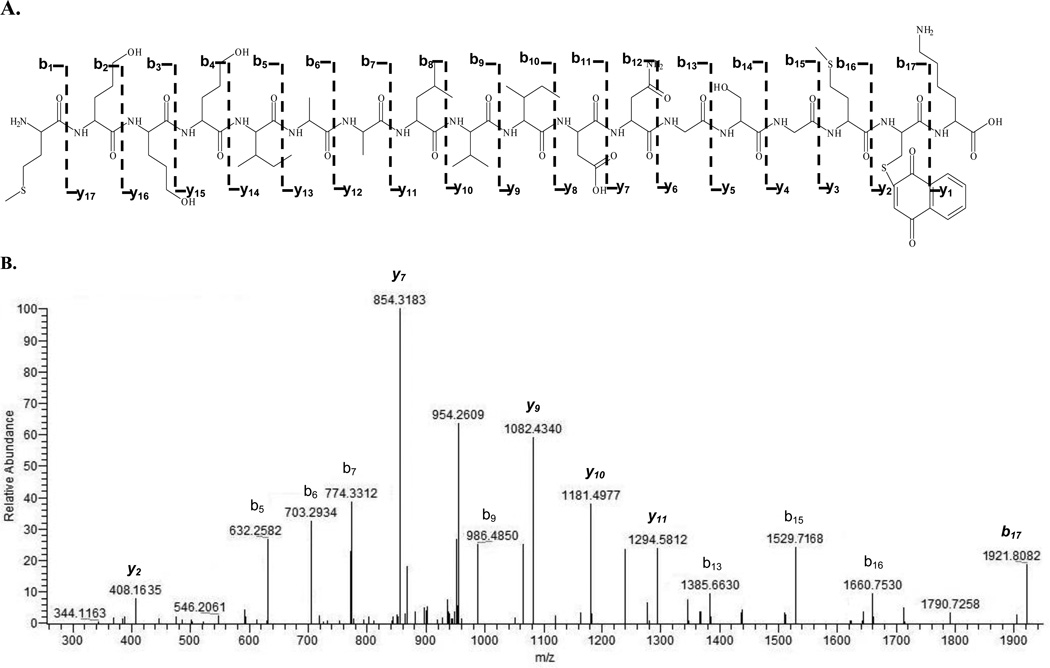
Actin was reacted in vitro with four naphthalene metabolites: NPO, NDO, 1,2-NPQ, and 1,4-NPQ. A mapping scheme of observed actin adducts is shown in Figure 1 and the adducted actin peptides are summarized in Table 1. After each LC/MS run, MS/MS spectra were analyzed with MASCOT for automated detection of metabolite modification. Matches were then manually validated by examining the MS/MS. For example, Figure 2 shows the CID spectrum of a [M+H]2+ at 1033 representing an actin tryptic peptide (−) MEEEIAALVIDNGSGMCK adducted on Cys17 by 1,4-NPQ. The presence of a singly charged modified b17 (m/z 1921.8082) and singly charged modified y ions (y2, y7, and y9 – y11) in the CID spectrum confirmed cysteine adduction. It should be noted there were four cases (* in Table 1) where an adducted peptide was observed with appropriate ion scores (individual ions scores > 30 indicate identity or extensive homology (p<0.05)), but there was not sufficient manual validation of the MS/MS to confirm adduction.
Figure 1.
Mapping of actin adduction sites by reactive metabolites of naphthalene. Observed binding sites are underlined and in bold. Amino acid sites of trypsin cleavage are in red.
Table 1.
Adducted sequences on actin observed with metabolites of interest.
| Actin Observed Adducted Peptides by Reactive Metabolites of Naphthalene | |||||
|---|---|---|---|---|---|
| Peptide Order |
Sequence | Predicted Adducted Mass |
Charge | Observed Mass |
Modifier |
| 1 – 18 | MEEEIAALVIDNGSGMCK | 2053.9304 | 2 | 1026.9643 | NPO |
| 1 – 18 | MEEEIAALVIDNGSGMCK* | 2087.9332 | 2 | 1043.9679 | NDO |
| 1 – 18 | MEEEIAALVIDNGSGMCK | 2067.9070 | 2 | 1033.9498 | 1,2NPQ |
| 1 – 18 | MEEEIAALVIDNGSGMCK | 2067.9070 | 2 | 1033.9521 | 1,4NPQ |
| 40 – 50 | HQGVMVGMGQK | 1349.6240 | 2 | 674.8164 | NDO |
| 40 – 50 | HQGVMVGMGQK | 1329.5978 | 2 | 664.7988 | 1,2NPQ |
| 40 – 50 | HQGVMVGMGQK | 1329.5978 | 2 | 664.7953 | 1,4NPQ |
| 63 – 68 | GILTLK | 802.4608 | 1 | 802.4601 | 1,4NPQ |
| 85 – 95 | IWHHTFYNELR | 1673.7758 | 2 | 836.8874 | 1,2NPQ |
| 96 – 113 | VAPEEHPVLLTEAPLNPK | 2132.1170 | 2 | 1066.0590 | NDO |
| 182 – 196 | DLTDYLMKILTER* | 1754.8935 | 2 | 877.4465 | NPO |
| 210 – 214 | DIKEK | 790.3878 | 1 | 790.3892 | 1,4NPQ |
| 257 – 283 | CPEALFQPSFLGMESCGIHETTFNSIMK* | 3275.4458 | 3 | 1091.8143 | 1,2NPQ |
| 328 – 335 | IKIIAPPER* | 1180.6985 | 2 | 590.3488 | NPO |
The diadduct is highlighted in gray.
Binding sites on the peptides are underlined and in bold.
represents observed adducted sequences that did not have strong MS/MS fragmentation patterns.
Spectra are shown in supplemental data.
Figure 2.
Actin peptide MEEEIAALVIDNGSGMCK (1–18) adducted by 1,4-naphthoquinone with a MASCOT ion score of 60. A is the fragmentation pattern of depicted b- and y-ion series for the adducted peptide. B is the CID spectrum of [M+H]2+ at m/z 1033 (modified peptide); 1,4-NPQ is bound to the Cys17. Italicized ions y2, y7, and y9 – y11 are modified y-ions in the presence of the NPQ. Italicized b17 (m/z 1921.8082) represents the modified N-terminal fragment peptide. The peptide was doubly charged.
For actin, there were nine adducted tryptic peptides and 14 modified sites detected. Two of the 6 cysteines (33%) in actin had detectable modifications: Cys17 was the only amino acid observed to be adducted by all four metabolites; Cys272 modification was only detected with 1,2-NPQ. Two adjacent histidines (out of 9; 22.2%) and six lysines (out of 19, 31.8%) were adducted, with Lys50 adducted by both quinones and the diol epoxide. Lys68 and Lys191 formed adducts with naphthalene epoxide, but not with diol epoxide. Some selectivity was also observed with the 1,2- and 1,4-NPQ adducts: they did not inclusively adduct the same sequences. In addition, 1,2-NPQ formed the only diadduct (when a metabolite adducts onto the same peptide twice, regardless of the modification site) on adjacent histidines on IWHHTFYNELR.
3.2 Adduction Sites of Reactive Naphthalene Metabolites on PDI
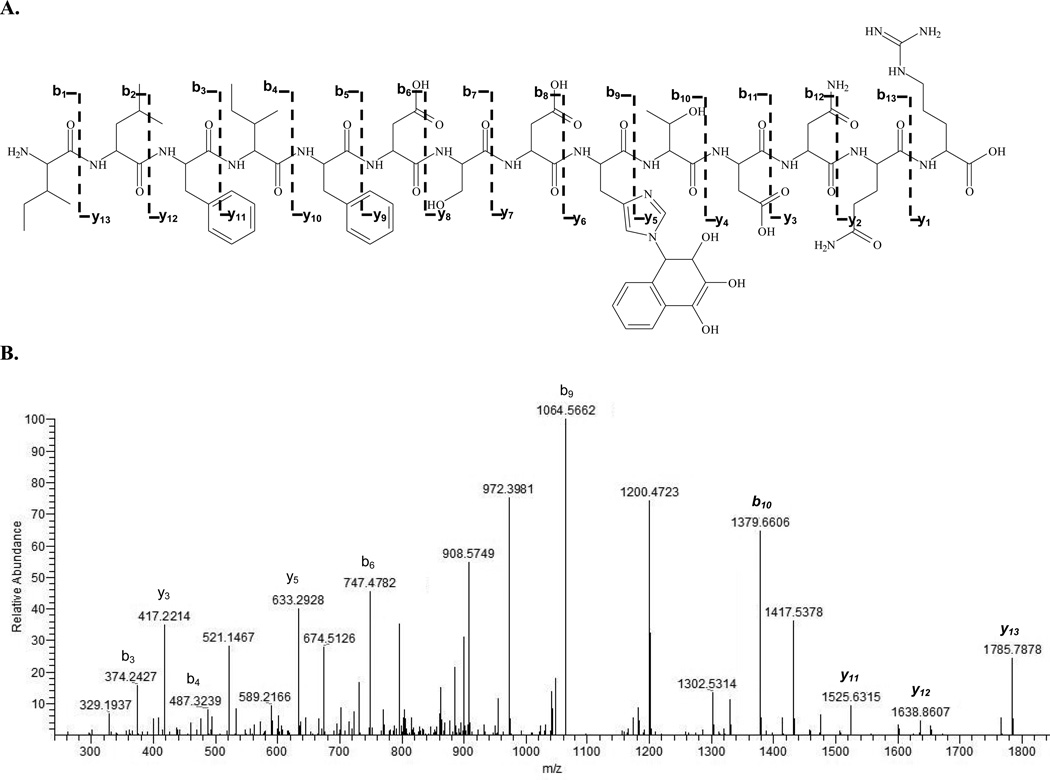
Six cysteines, 10 histidines and 49 lysines were potential targets for reactive naphthalene metabolites on PDI. Incubations of PDI with each of the metabolites in vitro yielded nine distinct adducted tryptic peptides (Table 2, Figure 3) and 12 observed adductions. The CID spectrum of a tryptic PDI peptide at [M+H]+2 of m/z 1006 (ILFIFIDSDHTDNQR) adducted by NDO is shown in Figure 4. Like the modified actin sequence, site modification of the PDI peptide was confirmed from the y- and b-ion series in the MS/MS spectra. Two adducts (* in Table 2) could not be manually validated by MS/MS because the ions observed did not show inclusive fragmentation patterns for the metabolite modifier on the appropriate y and b ions. NDO formed adducts on Cys55, Lys164 (also adducted by NPO), and His258 (also adducted by 1,2-NPQ). With the exception of Cys345, 1,2-NPQ and 1,4-NPQ did not adduct any of the same sites. Adduct formation appeared to be selective with no clear relationship between the metabolites.
Table 2.
Adducted sequences on PDI observed with metabolites of interest.
| PDI Observed Adducted Peptides by Reactive Metabolites of Naphthalene | |||||
|---|---|---|---|---|---|
| Peptide Order |
Sequence | Predicted Adducted Mass |
Charge | Observed Mass |
Modifier |
| 4 – 16 | MLSRALLCLALAWAAR | 1903.0445 | 2 | 951.5223 | NPO |
| 45 – 59 | YLLVEFYAPWCGHCK* | 2007.9252 | 2 | 1003.9616 | NDO |
| 123 – 132 | EADDIVNWLK | 1360.6412 | 1 | 1360.6425 | 1,2NPQ |
| 135 – 164 | TGPAATTLSDTAAAESLVDSSEVTVIGFFK | 3143.5307 | 2 | 1571.7659 | 1,2NPQ |
| 135 – 164 | TGPAATTLSDTAAAESLVDSSEVTVIGFFK | 3163.5569 | 2 | 1581.7780 | NDO |
| 165 – 172 | DVESDSAK* | 994.4364 | 1 | 994.4378 | NPO |
| 257 – 265 | THILLFLPK | 1259.7400 | 1 | 1259.7401 | NDO |
| 257 – 265 | THILLFLPK | 1397.7568 | 1 | 1397.7574 | 1,2NPQ |
| 280 – 285 | AAEGFK | 780.3563 | 1 | 780.3569 | 1,4NPQ |
| 288 – 302 | ILFIFIDSDHTDNQR | 2012.9832 | 2 | 1006.4910 | NDO |
| 341 – 347 | ITEFCHR | 1063.4666 | 1 | 1063.4670 | 1,2NPQ |
| 341 – 347 | ITEFCHR | 1063.4666 | 1 | 1063.4661 | 1,4NPQ |
The diadduct is highlighted in gray.
Binding sites on the peptides are underlined and in bold.
represents observed adducted sequences that did not have strong MS/MS fragmentation patterns.
Spectra are shown in supplemental data.
Figure 3.
Mapping of PDI adduction sites by naphthalene reactive metabolites. Observed binding sites are underlined and in bold. Amino acid sites of trypsin cleavage are in red.
Figure 4.
PDI peptide ILFIFIDSDHTDNQR (288–302) adducted by naphthalene diol epoxide with a MASCOT ion score of 96. A is the fragmentation pattern with depicted b- and y-series ions for the adducted peptide. B is the CID spectrum of [M+H]2+ at m/z 1006 of the adducted peptide; NDO is bound to His297. Italicized ions y11 – y13 and b10 are modified y-ions in the presence of the NDO. Italicized b10 (m/z 1379.8098) represents the modified N-terminal fragment with histidine. The peptide was doubly charged.
Identified modified amino acid sites in actin and PDI are listed in Table 3.
Table 3.
Residues identified as binding sites on model proteins after incubation with synthetic naphthalene metabolites at pH 7.4.
| Identified Binding Sites | |
|---|---|
| Actin | PDI |
| Cys17 | Cys8 |
| Lys50 | Cys55 |
| Lys68 | Lys132 |
| His87 | Lys163 |
| His88 | Lys172 |
| Lys112 | His258 |
| Lys191 | Lys265 |
| Lys212 | Lys285 |
| Cys272 | His297 |
| Lys328 | Cys345 |
3.3 Microsomally Generated Metabolites, Trapping with Model Proteins
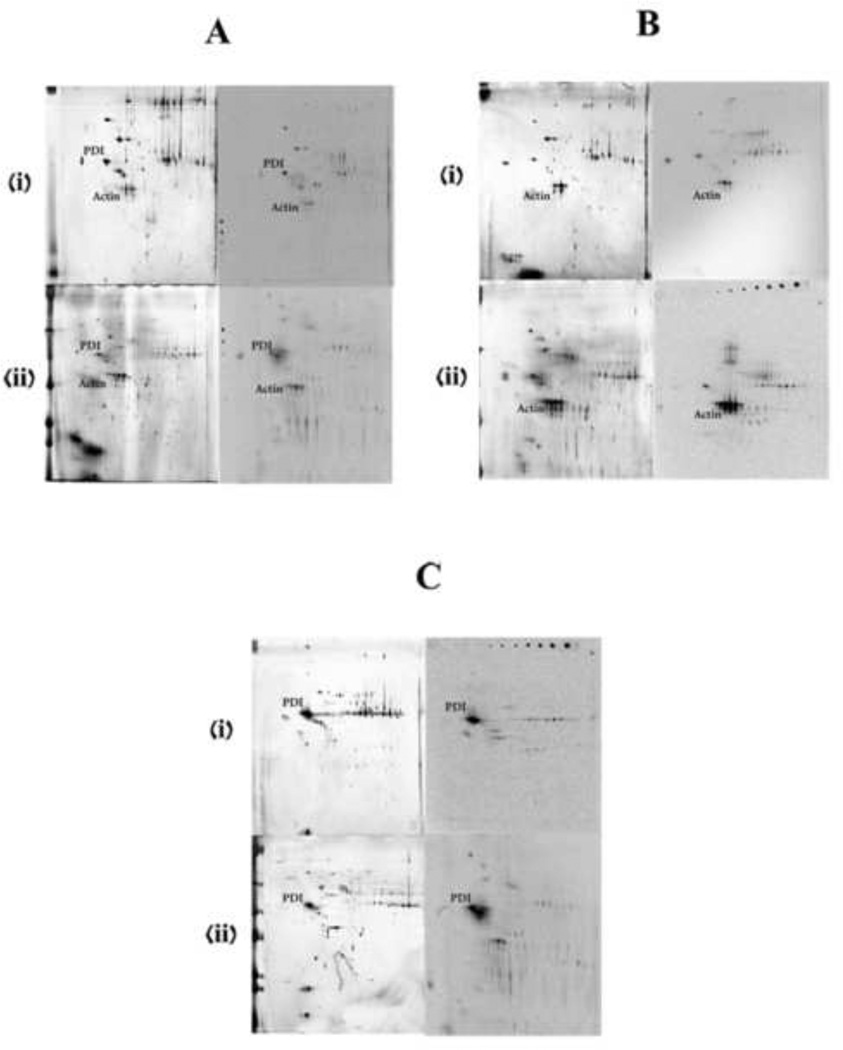
To determine whether the spectrum of metabolites bound covalently to residues in actin and PDI was dependent on the source of metabolites, incubations of model protein and microsomes from target (rat nose) and nontarget tissues (mouse liver) were conducted. Half of the incubations contained 14C-naphthalene to assist in localizing adducted proteins of interest on storage phosphor screens. There was minimal inter-gel or inter-blot variation (Figure 5). Actin and PDI adducts were readily detected in control microsomal incubations without addition of exogenous actin or PDI (Fig 5A, storage phosphor images). Furthermore, the addition of actin (Fig 5B) or PDI (Fig 5C) markedly increased the amounts of adducted proteins detected by storage phosphor analysis, especially in incubations with the rat olfactory epithelial microsomes.
Figure 5.
Silver-stained gels (left) and storage phosphor screens (right) of the proteins isolated from the pellet of microsomal incubations with actin or PDI. (i) and (ii) are for liver microsomal and nasal microsomal incubations respectively. A is the control incubations without the addition of any external protein. B is with added actin and C is with added PDI. Target proteins are labeled on both the silver-stained gels and phosphor screen images.
Actin or PDI recovered from 2D gels of extracts of microsomal incubations were digested with trypsin and samples were applied to MALDI plates for detection of modified peptides. However, MS and MS/MS data from MALDI-TOF/TOF failed to yield signals consistent with those observed with the synthetic incubations. Peptide signals appropriate for unmodified actin peptide ions were prominent in the MS of the microsomal incubations as compared to the synthetic incubations, where the relative ratios of modified versus unmodified for actin peptides like MEEEIAALVIDNGSGMCK or HQGVMVGMGQK were approximately 4:1 or 3:1, respectively (not shown). Signals for metabolite-modified actin peptides were not detected. Similar observations were made with PDI; modified versus unmodified peptide ions ranged from 5:1 to 3:1 in the synthetic incubations for the observed adducts, but modified ions could not be detected in the microsomal incubations. Although specific modified binding sites from the microsomal incubations could not be detected with MALDI TOF/TOF, the studies confirmed previous observations that actin and PDI are targets of naphthalene metabolites and contain numerous sites for covalent binding.
Actin and PDI recovered from microsomal incubations were also analyzed by SIM to probe the formation of the 26 previously detected adducted sites in a biological setting. HPLC/MS parameters similar to the synthetic incubations were used. Two modified tryptic peptides were detected in both actin microsomal incubations: IKIIAPPER (NPO) and VAPEEHPVLLTEAPLNPK (NDO) and confirmed by MS/MS. A single NPO adduct on MLSRALLCLALAWAAR was observed from the hepatic microsomal incubations with PDI; however, modified PDI peptides were not detected in the olfactory epithelial incubations.
4. Discussion
4.1 Key factors in the mechanism of reactive metabolite-mediated toxicity
A thorough understanding of the mechanisms by which reactive metabolites generated in target cells lead to a loss of cellular homeostasis depends upon an appreciation of which metabolite modifications lead to altered protein structure/function and in instances where multiple reactive metabolites may be involved, insight on the roles of each in the overall modification of critical sites of key proteins. One of the first steps in assessing which electrophilic protein interactions are critical to events leading to cellular necrosis involves understanding which amino acids are targets, which metabolites are involved, and whether such interactions result in a decrement of function. The present studies used a straightforward approach to evaluate such covalent modification by investigating the conjugation of actin and PDI with naphthalene metabolites.
4.2 Functional importance of adduct formation with actin and PDI
One of the first signs of injury related to naphthalene exposure involves swelling and vacuolization of Clara cells in the airway epithelium [23]. This is consistent with changes noted in actin fiber morphology in naphthalene-treated mice where Alexa-labeled phalloidin was used to visualize actin filaments; irregular fiber structure and the appearance of frayed actin filaments were observed by confocal microscopy 3 or 6 hrs following naphthalene treatment [24]. Posttranslational modification of actin has been suggested as a key event in membrane blebbing associated with redox cycling quinones; however, the key amino acids involved in controlling the polymerization/depolymerization of this cytoskeletal protein were not identified [25]. Later work showed Cys374 as a binding site for aldehyde decomposition products such as 4-hydroxynonenal and acrolein [26, 27], but modification of this site by either aldehyde did not appear to influence actin polymerization. Rather, at very high aldehyde to actin ratios (20:1, 50:1), additional amino acids are modified including His87 which appeared to be an important factor in slowing actin polymerization. In the current studies, His87 was modified by 1,2-NPQ but modification by other reactive metabolites was not detected.
Likewise, Cys57 in rat PDI (analogous to Cys55 in the current work) is adducted by 4-hydroxynonenal and, at high aldehyde to protein ratios, this modification significantly decreases the catalytic activity of PDI [28]. This cysteine is one of two in PDI involved in the thioredoxin-like catalytic sites (-CGHC-), one located near the N- (Cys55) and the other near the C-terminus (Cys399). These catalytic sites form the active sites of the oxidoreductase and when the cysteines form a disulfide-bond, PDI can function as an oxidase for protein thiols emerging in the ER lumen [29, 30]. Studies with rat recombinant PDI showed that, in incubations with the glutathione conjugate of 1,2-dichloroethane, a decrease in protein activity was reportedly due to modifications of either Cys57 and Cys381 [31]. Furthermore, in yeast PDI, it was shown that the N-terminal Cys of the two active site sequences of yeast PDI were found to be required for cell viability, but mutation of the C-terminal Cys was not lethal [32]. Thus, modification of this N-terminal active cysteine by the reactive metabolites of naphthalene may play a significant role in the functions of PDI as an oxidant of protein thiols.
4.3 Relative activities of naphthalene metabolites
In the synthetic incubations, NPO adducted fewer sites on either model protein than NDO or the quinones. These results were consistent with previous work showing fewer adducts when NPO was incubated with model peptides in comparison with either NDO or NPQ (Pham et al., submitted). Most sites adducted by NPO in both actin and PDI were adjacent to neutral, nonpolar amino acids with little electron withdrawing effect: Lys191 and Lys328 of actin are flanked by methionine and isoleucine, and two isoleucines, respectively; Cys8 of PDI is flanked between two leucines. With little influence from neighboring amino acids, it seemed that these sites were more susceptible to adduct formation with the naphthalene epoxide. A few sequences modified by 1,2-NPQ were also adducted by 1,4-NPQ, consistent with their similar electrophilic properties. The only diadducts observed in both proteins were formed with 1,2-NPQ. Although many amino acid sites adducted by the NPQs were surrounded by acidic or basic residues, the stronger electrophilicity of the naphthoquinones, in comparison to the epoxide, appeared to be sufficient to overcome the large energy transition necessary to adduct these residues.
In addition to metabolite reactivity, other factors like the microenvironment as well as protein structure are important in assessing adduct formation. For instance, only Cys17 of actin was adducted by all four metabolites. Furthermore, although adduction was interspersed across the actin sequence, adduction in the C-terminal region of PDI was noticeably absent, possibly attributable to the highly acidic C-terminal domain [13]. PDI also did not have adducted sites between positions 173 – 255 supporting speculation that adduction not only depends on the metabolite, but also on factors such as site nucleophilicity and/or protein conformation. NDO adduction on the Cys55 of PDI is particularly interesting because the cysteine is located in an active site and further investigation into the role of this modification is necessary.
There are several sites on the PDI sequence where lysines were adjacent to each other (132 and 133, 209 and 210, 310 and 311, 388 and 389, and 469 and 470), but only one adducted lysine of those pairs was detected: Lys132. It was possible other Lys residues were adducted, but that the extent of modification on those residues was not sufficient for detection or the adducted residues were not ionized in the MS source. Overall, adducted sites were only identified preceding a trypsin cleavage site or in the middle of a peptide sequence suggesting that 1) a covalent bond following a trypsin site may be labile enough to break during digest or that 2) the presence of an adduct may block the trypsin cleavage. We recognize the in vitro studies conducted here do not necessarily accurately mimic in vivo metabolite-protein interactions, but the studies do demonstrate reasonable target selectivity which likely reflects the effects of pH and neighboring amino acids on the nucleophilicity of the residue [33–35].
4.4 Microsomal incubation and adduct formation with actin and PDI
Use of radiolabeled naphthalene in rat nasal and hepatic microsomal incubations confirmed the generation of adducts with both actin and PDI as indicated by co-localization of radioactive spots with both proteins on 2D gels (Figure 5). However, the specific modified site and metabolite bound to tryptic peptides of actin or PDI recovered from the complementary 2D gels with unlabeled naphthalene could not be determined with MALDI/TOF/TOF. It is likely the inability to detect protein modifications in the microsomal incubations is related to the relatively smaller amounts of reactive metabolite generated as well as the partial microsomal metabolism of the reactive species to less reactive products. In such instances, only a very small amount of the same modified peptides may be present, resulting in a failure to observe the modified peptide, even if adduction occurs [36]. This is consistent with results where we reanalyzed the microsomal samples using SIM to specifically detect modified peptides from the tryptic digests based on [M+H]+ identified from our synthetic incubations. These experiments detected only three epoxide-modified peptides in the two model proteins. Although we were only able to detect a few selected ions, the results are consistent with earlier work from other investigators in which they were either unable to detect modified peptides in vivo [37] or could do so only with agents generating very high levels of protein adduct in vivo [5].
Nucleophilicity/electrophilicity as well as protein structure and conformation strongly influence whether an adduction occurs. Using mass spectrometric findings from these model systems, we now have a clearer idea of the target preferences by naphthalene reactive intermediates and future work will focus on utilizing these established MS/MS patterns in a method akin to multiple reaction monitoring mass spectrometry (MRM-MS) [38, 39] to investigate precursor ions and product ions for analytes of interest in vivo. In summary, the potential downstream effects following a toxicant exposure may be dependent not only on the metabolites available and the stoichiometric amounts of those intermediates, but also on which proteins and where on the sequence they adduct.
Supplementary Material
Acknowledgements
Mass spectral analysis was performed at the Campus Mass Spectrometry Facilities, University of California Davis. This work was supported by the National Institutes of Environmental Health [Grant ES 04311, ES 04699]. NP was supported by a fellowship from the Floyd and Mary Schwall Foundation.
Abbreviations
- SIM
selective ion monitoring
- HRMS
high resolution mass spectrometry
- CID
collision-induced dissociation
- ESI
electrospray ionization
- ACN
acetonitrile
- DMSO
dimethyl sulfoxide
- FA
formic acid
- NPO
naphthalene epoxide
- NDO
naphthalene diol epoxide
- 1,2–NPQ
1,2–naphthoquinone
- 1,4–NPQ
1,4–naphthoquinone
- NPQ
naphthoquinone
- TFA
trifluoroacetic acid
- 2-DE
two-dimensional gel electrophoresis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare there are no conflicts of interest.
References
- 1.Hanzlik RP, Koen YM, Theertham B, Dong Y, Fang J. The reactive metabolite target protein database (TPDB)--a web-accessible resource. BMC Bioinformatics. 2007;8:95. doi: 10.1186/1471-2105-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones JA, Liebler DC. Tandem MS analysis of model peptide adducts from reactive metabolites of the hepatotoxin 1,1-dichloroethylene. Chem Res Toxicol. 2000;13(12):1302–1312. doi: 10.1021/tx000148w. [DOI] [PubMed] [Google Scholar]
- 3.Hettick JM, Siegel PD. Determination of the toluene diisocyanate binding sites on human serum albumin by tandem mass spectrometry. Anal Biochem. 2011;414(2):232–238. doi: 10.1016/j.ab.2011.03.035. [DOI] [PubMed] [Google Scholar]
- 4.Codreanu SG, Adams DG, Dawson ES, Wadzinski BE, Liebler DC. Inhibition of protein phosphatase 2A activity by selective electrophile alkylation damage. Biochemistry. 2006;45(33):10020–10029. doi: 10.1021/bi060551n. [DOI] [PubMed] [Google Scholar]
- 5.Ikehata K, Duzhak TG, Galeva NA, Ji T, Koen YM, Hanzlik RP. Protein targets of reactive metabolites of thiobenzamide in rat liver in vivo. Chem Res Toxicol. 2008;21(7):1432–1442. doi: 10.1021/tx800093k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koen YM, Williams TD, Hanzlik RP. Identification of three protein targets for reactive metabolites of bromobenzene in rat liver cytosol. Chem Res Toxicol. 2000;13(12):1326–1335. doi: 10.1021/tx000165l. [DOI] [PubMed] [Google Scholar]
- 7.Koen YM, Yue W, Galeva NA, Williams TD, Hanzlik RP. Site-specific arylation of rat glutathione s-transferase A1 and A2 by bromobenzene metabolites in vivo. Chem. Res. Toxicol. 2006;19:1426–1434. doi: 10.1021/tx060142s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheelock AM, Boland BC, Isbell M, Morin D, Wegesser TC, Plopper CG, Buckpitt AR. In vivo effects of ozone exposure on protein adduct formation by 1-nitronaphthalene in rat lung. Am J Respir Cell Mol Biol. 2005;33(2):130–137. doi: 10.1165/rcmb.2005-0047OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Isbell MA, Morin D, Boland B, Buckpitt A, Salemi M, Presley J. Identification of proteins adducted by reactive naphthalene metabolites in vitro. Proteomics. 2005;5(16):4197–4204. doi: 10.1002/pmic.200401278. [DOI] [PubMed] [Google Scholar]
- 10.Lin CY, Boland BC, Lee YJ, Salemi MR, Morin D, Miller LA, Plopper CG, Buckpitt AR. Identification of proteins adducted by reactive metabolites of naphthalene and 1-nitronaphthalene in dissected airways of rhesus macaques. Proteomics. 2006;6(3):972–982. doi: 10.1002/pmic.200500170. [DOI] [PubMed] [Google Scholar]
- 11.Aldini G, Orioli M, Carini M. Alpha,beta-unsaturated aldehydes adducts to actin and albumin as potential biomarkers of carbonylation damage. Redox Rep. 2007;12(1):20–25. doi: 10.1179/135100007X162310. [DOI] [PubMed] [Google Scholar]
- 12.Ghitescu LD, Gugliucci A, Dumas F. Actin and annexins I and II are among the main endothelial plasmalemma-associated proteins forming early glucose adducts in experimental diabetes. Diabetes. 2001;50(7):1666–1674. doi: 10.2337/diabetes.50.7.1666. [DOI] [PubMed] [Google Scholar]
- 13.Hatahet F, Ruddock LW. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal. 2009;11(11):2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- 14.Martin JL, Kenna JG, Martin BM, Thomassen D, Reed GF, Pohl LR. Halothane hepatitis patients have serum antibodies that react with protein disulfide isomerase. Hepatology. 1993;18(4):858–863. doi: 10.1002/hep.1840180417. [DOI] [PubMed] [Google Scholar]
- 15.Hayes MJ, Rescher U, Gerke V, Moss SE. Annexin-actin interactions. Traffic. 2004;5(8):571–576. doi: 10.1111/j.1600-0854.2004.00210.x. [DOI] [PubMed] [Google Scholar]
- 16.Lin TI, Dowben RM. Fluorescence spectroscopic studies of pyrene-actin adducts. Biophys Chem. 1982;15(4):289–298. doi: 10.1016/0301-4622(82)80012-4. [DOI] [PubMed] [Google Scholar]
- 17.Yagi H, Jerina D. A General Synthetic Method for Non-K-Region Arene Oxides. J Am Chem Soc. 1975;97(11) [Google Scholar]
- 18.Tsang W, Griffin G, Horning M, Stillwell W. Chemistry of anti- and syn-1,2, 3,4-naphthalene dioxides and their potential relevance as metabolic intermediates. J Org Chem. 1981;47(27):5339–5353. [Google Scholar]
- 19.Fanucchi MV, Harkema JR, Plopper CG, Hotchkiss JA. In vitro culture of microdissected rat nasal airway tissues. Am J Respir Cell Mol Biol. 1999;20(6):1274–1285. doi: 10.1165/ajrcmb.20.6.3451. [DOI] [PubMed] [Google Scholar]
- 20.Buckpitt A, Buonarati M, Avey LB, Chang AM, Morin D, Plopper CG. Relationship of Cytochrome-P450 activity to Clara cell cytotoxicity. 2. Comparison of stereoselectivity of naphthalene epoxidation in lung and nasal-mucosa of mouse, hamster, rat and Rhesus-monkey. J Pharmacol Exp Ther. 1992;261(1):364–372. [PubMed] [Google Scholar]
- 21.Lin CY, Isbell MA, Morin D, Boland BC, Salemi MR, Jewell WT, Weir AJ, Fanucchi MV, Baker GL, Plopper CG, Buckpittt AR. Characterization of a structurally intact in situ lung model and comparison of naphthalene protein adducts generated in this model vs lung microsomes. Chem Res Tox. 2005;18(5):802–813. doi: 10.1021/tx049746r. [DOI] [PubMed] [Google Scholar]
- 22.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68(5):850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 23.Van Winkle LS, Johnson ZA, Nishio SJ, Bronn CD, Plopper CG. Early events in naphthalene-induced acute Clara cell toxicity - Comparison of membrane permeability and ultrastructure. Am J Resp Cell Mol Biol. 1999;21(1):44–53. doi: 10.1165/ajrcmb.21.1.3630. [DOI] [PubMed] [Google Scholar]
- 24.Phimister AJ, Williams KJ, Van Winkle LS, Plopper CG. Consequences of abrupt glutathione depletion in murine Clara cells: Ultrastructural and biochemical investigations into the role of glutathione loss in naphthalene cytotoxicity. J Pharmacol Exper Therap. 2005;314(2):506–513. doi: 10.1124/jpet.105.084533. [DOI] [PubMed] [Google Scholar]
- 25.Bellomo G, Mirabelli F, Vairetti M, Iosi F, Malorni W. Cytoskeleton as a target in menadione-induced oxidative stress in cultured mammalian cells. I. Biochemical and immunocytochemical features. J Cell Physiol. 1990;143(1):118–128. doi: 10.1002/jcp.1041430116. [DOI] [PubMed] [Google Scholar]
- 26.Dalle-Donne I, Carini M, Vistoli G, Gamberoni L, Giustarini D, Colombo R, Maffei Facino R, Rossi R, Milzani A, Aldini G. Actin Cys374 as a nucleophilic target of alpha,beta-unsaturated aldehydes. Free Radic Biol Med. 2007;42(5):583–598. doi: 10.1016/j.freeradbiomed.2006.11.026. [DOI] [PubMed] [Google Scholar]
- 27.Aldini G, Dalle-Donne I, Vistoli G, Maffei Facino R, Carini M. Covalent modification of actin by 4-hydroxy-trans-2-nonenal (HNE): LC-ESI-MS/MS evidence for Cys374 Michael adduction. J Mass Spectrom. 2005;40(7):946–954. doi: 10.1002/jms.872. [DOI] [PubMed] [Google Scholar]
- 28.Carbone DL, Doorn JA, Kiebler Z, Petersen DR. Cysteine modification by lipid peroxidation products inhibits protein disulfide isomerase. Chem Res Toxicol. 2005;18(8):1324–1331. doi: 10.1021/tx050078z. [DOI] [PubMed] [Google Scholar]
- 29.Mezghrani A, Fassio A, Benham A, Simmen T, Braakman I, Sitia R. Manipulation of oxidative protein folding and PDI redox state in mammalian cells. EMBO J. 2001;20(22):6288–6296. doi: 10.1093/emboj/20.22.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lappi AK, Ruddock LW. Reexamination of the role of interplay between glutathione and protein disulfide isomerase. J Mol Biol. 2011;409(2):238–249. doi: 10.1016/j.jmb.2011.03.024. [DOI] [PubMed] [Google Scholar]
- 31.Kaetzel RS, Stapels MD, Barofsky DF, Reed DJ. Alkylation of protein disulfide isomerase by the episulfonium ion derived from the glutathione conjugate of 1,2-dichloroethane and mass spectrometric characterization of the adducts. Arch Biochem Biophys. 2004;423(1):136–147. doi: 10.1016/j.abb.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 32.Luz JM, Lennarz WJ. The nonactive site cysteine residues of yeast protein disulfide isomerase are not required for cell viability. Biochem Biophys Res Commun. 1998;248(3):621–627. doi: 10.1006/bbrc.1998.8992. [DOI] [PubMed] [Google Scholar]
- 33.Zhang M, Vogel HJ. Determination of the side chain pKa values of the lysine residues in calmodulin. J Biol Chem. 1993;268(30):22420–22428. [PubMed] [Google Scholar]
- 34.Ferrer-Sueta G, Manta B, Botti H, Radi R, Trujillo M, Denicola A. Factors affecting protein thiol reactivity and specificity in peroxide reduction. Chem Res Toxicol. 2011;24(14):434–450. doi: 10.1021/tx100413v. [DOI] [PubMed] [Google Scholar]
- 35.Spiess PC, Deng B, Hondal RJ, Matthews DE, van der Vliet A. Proteomic profiling of acrolein adducts in human lung epithelial cells. J Proteomics. 2011;74(11):2380–2394. doi: 10.1016/j.jprot.2011.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mortz E, O'Connor PB, Roepstorff P, Kelleher NL, Wood TD, McLafferty FW, Mann M. Sequence tag identification of intact proteins by matching tanden mass spectral data against sequence data bases. Proc Natl Acad Sci U S A. 1996;93(16):8264–8267. doi: 10.1073/pnas.93.16.8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stamper BD, Mohar I, Kavanagh TJ, Nelson SD. Proteomic analysis of acetaminophen-induced changes in mitochondrial protein expression using spectral counting. Chem Res Toxicol. 2011 doi: 10.1021/tx1004198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abbatiello SE, Mani DR, Keshishian H, Carr SA. Automated detection of inaccurate and imprecise transitions in peptide quantification by multiple reaction monitoring mass spectrometry. Clin Chem. 2010;56(2):291–305. doi: 10.1373/clinchem.2009.138420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thakur SS, Geiger T, Chatterjee B, Bandilla P, Froehlich F, Cox J, Mann M. Deep and highly sensitive proteome coverage by LC-MS/MS without pre-fractionation. Mol Cell Proteomics. 2011 doi: 10.1074/mcp.M110.003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.