Abstract
Objective
We sought to identify differentially expressed genes in the athero-prone coronary artery and athero-resistant internal mammary arteries.
Methods and Results
Using suppressive subtraction hybridization, we generated reciprocal cDNA collections of representative mRNAs specific to porcine coronary arteries versus porcine mammary arteries. We screened 1000 suppressive subtraction hybridization cDNA clones by dot blot array and sequenced 600 of those showing the most marked expression differences. Northern blot, in situ hybridization, and immunostaining confirmed the differential gene expression patterns identified by the dot blot arrays. Genes associated with mammary arteries included claudin-10 and h-cadherin, which are genes associated with tight junctions and intermediate junctions. In contrast, genes associated with proatherosclerotic processes, such as lipid retention and metabolism, inflammation, and cell growth, were preferentially expressed in coronary arteries.
Conclusions
Normal coronary arteries have gene expression program that is significantly different than internal mammary arteries. These differences may partly explain the resistance of coronary arteries and internal mammary arteries to atherosclerosis.
Keywords: suppression subtractive hybridization, coronary artery, mammary artery, gene expression, arterial phenotype
One of the striking aspects of human atherosclerotic disease is that some arteries, such as the internal mammary and radial arteries, are highly resistant to spontaneous lesion formation even in a strongly proatherogenic environment while at the same time coronary and carotid arteries develop atherosclerosis. Because of these territorial differences in the propensity of various arterial beds to atherosclerosis, the internal mammary artery was the first vessel to be used as a graft to bypass obstructed coronary artery.1–3 The favorable effects on mortality and morbidity are observed irrespective of age, sex, race, or left ventricular function.4 A 15-year survival analysis of all patients in the Coronary Artery Surgery Study registry who had undergone bypass grafting showed that >90% of the internal mammary grafts remain patent.5 A group of patients with left anterior descending coronary artery occlusion who received internal mammary grafts showed >90% patency rate 20 years after surgery.6 Even when atherosclerosis does develop in mammary grafts,7,8 intimal hyperplasia is found primarily at the anastomosis,9 suggesting that it developed in response to surgical trauma. Because of the dramatic benefits afforded by the internal mammary grafts as a conduit, current recommendations are that its use for bypass grafting should be preferred in all but a few specific situations.4
The biological mechanism(s) underlying the athero-resistant property of arteries is completely unknown. The structure and cellular compositions of athero-prone arteries are different from that of internal mammary artery. One major difference between the coronary artery and mammary artery is in intima. Comparison of intimal thickness of coronary, carotid, and mammary arteries have revealed that intimal thickening of left anterior descending coronary artery and carotid artery bulb are more marked, occur earlier, and can be demonstrated as early as 6 months of age.10–21 In contrast, mammary arteries have thin intima and the thickening was not found before 21 years of age.11,21–23 Although not a lesion itself, the tick intima is believed to be a prerequisite for lesion formation.24–26
Despite the obvious importance of the composition of the blood vessels, there has not been a comprehensive analysis to define the differences in the makeup of different arteries. Most studies investigating the regulation of pathological conditions at the level of gene expression have traditionally used a candidate gene approach. However, this strategy is limited in scope because of the sampling of only limited number of a predetermined single gene in isolation from the entire repertoire of mRNA transcripts. We hypothesize that the phenotype of a blood vessel, in part, determines its ability to resist or develop atherosclerosis. Because the phenotypes of tissues within an organism are determined by their pattern of gene expression, we determined differential gene expression pattern between coronary and mammary arteries.
Various approaches to study differential gene expression are applied to vascular tissues.27–34 The compromise between focusing on only the important genes in certain cellular processes and achieving a complete picture is critical for the selection of strategy. We used suppressive subtraction hybridization (SSH) to determine differential gene expression because it provides an approximately 1000-fold enrichment of low copy number (≈10 copies/cell) genes related to defined phenotypes.35–37 We have generated reciprocal cDNA collections representing mRNA specific to porcine coronary versus porcine mammary arteries. We screened 1000 SSH cDNA clones by dot blot array and sequenced 600 of these showing the most marked differences in expression. Northern blot, in situ hybridization, and immunohistochemical staining confirmed the differential gene expression pattern identified by the dot blot arrays. The existence of distinctive sets of genes that mark these two vessels will aid us to understand why the arteries differ in their propensity to develop vascular diseases.
Methods
The porcine mammary artery and proximal segment of left coronary artery from 3 swine (6 months, 40 to 50 lbs) were surgically removed and immediately transferred to a tube containing cold RNA stabilizing solution (RNAlater, Ambion). Histological examination of the segments showed that the segments were normal. The arteries were stripped of the endothelium and periadventitial fatty tissues in the stabilizing solution at 4°C, snap frozen in liquid nitrogen, and stored at −80°C for subsequent RNA extraction. To minimize possible variation between the individual swine, we pooled total RNA extracted from 3 matched coronary and mammary arteries.
Northern blots and in situ hybridizations were performed according to standard procedure. SSH libraries were made using polymerase chain reaction (PCR)-select cDNA subtraction essentially as described by Clonetech using excess (8-fold) mammary and coronary cDNAs as drivers to make coronary-enriched and mammary-enriched libraries. After establishing the efficiency of subtraction, cDNA fragments from the secondary PCR were ligated into a pT-Adv vector using a T/A cloning kit (Clontech) to generate subtracted cDNA libraries. Using colony hybridization, positive clones were identified by differentially screening the library using both forward-subtracted and reverse-subtracted probes (derived by switching between tester and driver populations for another subtractive hybridization). Positive clones, identified by differential screening, were sequenced at the core sequencing facility of Cedars-Sinai Medical Center. The sequences were examined for the presence of vector contamination using vector contamination screening program, VecScreen. The resulting nucleotide sequence was blasted for similarity against all nonredundant and EST databases of the basic local alignment and search tools (BLAST) at the National Center for Biotechnology Information (NIH, Bethesda, MD). The resulting nucleotide sequence was analyzed for similarity to the nonredundant database sequences using the BLAST. Some clones displayed high levels of homology (>90%) to known mRNA present in the nonredundant nucleotide sequence database. However, there were clones that showed low level of homology to the known sequences. These clones were further examined for the presence of any known domain to determine whether they belong to any family of proteins. To classify these genes, we considered 25% identity over a stretch of at least 80 amino acid as significant because it represented the same basic fold.38–41
Results
The aim of this study was to compare the gene expression in porcine coronary and mammary arteries. We used SSH over standard differential or subtractive hybridization methods because it is fully compatible with a PCR-based preamplification method allowing for the synthesis of high-quality double-stranded cDNA from small amounts of total RNA. This is important in vascular tissues because they are hypo-cellular. Starting with 1 μg of total RNA, we generated high-yields of double-stranded porcine coronary and mammary cDNAs suitable for subtracted probe generation. To determine whether the original complexity of the mRNA population was maintained during cDNA synthesis and amplification, we used reverse Northern blot to determine the size range of the amplified products. Because PCR is more efficient at amplifying short sequences, we decided to investigate the presence of several representative large messages, such as fibronectin, laminin, and tenascin-X, in the amplified pool of cDNAs. Northern blot analysis of the amplified cDNAs showed that these large messages were indeed represented in the amplified cDNA made from coronary and mammary arteries (not shown), suggesting that the SMART PCR method faithfully amplified the original cDNA.
To estimate the efficiency of subtraction, the abundance of a known cDNA was compared before and after subtraction. The populations of unsubtracted and subtracted cDNAs were analyzed by PCR with primers specific for the commonly expressed β-actin gene. In nonsubtracted libraries, a 500-bp β-actin-specific PCR product is visible by 28 cycles of amplification and becomes saturated at 33 cycles (not shown). In contrast, the β-actin PCR product was not detected in the subtracted libraries. We conclude that the subtractions were effective in markedly reducing the abundance of genes shared by both arteries.
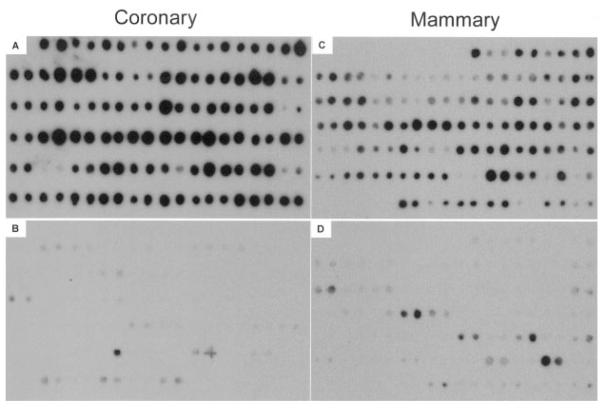
After establishing that the subtractions were effective, the subtracted cDNAs were cloned and 1000 clones were randomly selected for screening. Two approaches have been used to screen the differentially expressed libraries: screening with the unsubtracted cDNA probes and screening with the subtracted probes. We used primarily the second approach because subtracted cDNA probes allow the identification of low copy number genes.42 In addition, some clones were screened by unsubtracted probes. Coronary-enriched clones that hybridized with mammary-subtracted probes but not with coronary-subtracted probes were considered differentially expressed coronary genes. Similarly, mammary clones that hybridized with coronary-subtracted cDNAs but not mammary-subtracted probes were considered differentially expressed mammary genes. More than 90% of the coronary-enriched clones showed a strong positive signal with the mammary-subtracted probe (Figure 1A) and failed to hybridize, or gave a very low signal, with the mammary-subtracted cDNAs (Figure 1B). Likewise, more than 80% of the mammary-enriched clones strongly hybridized with the coronary-subtracted probe (Figure 1C) and generally failed to hybridize with the mammary-subtracted cDNAs (Figure 1D). Those coronary clones that hybridized with the mammary probe (Figure 2) and those mammary clones that hybridized with the coronary probe (Figure 2) were considered false-positives. The positive clones that were identified by the subtracted probes were considered to contain differentially expressed high and low copy number genes. A large number of distinct gene fragments were present in the resulting subtraction products, as determined by sequencing 600 clones with at least 3-fold difference in the intensity of the spots. Tables 1 and 2 show the genes with known functions. We considered these genes as candidate genes that mark coronary and mammary phenotypes.
Figure 1.
Representative dot blot array of differentially expressed gene clones. Replicate dot blot arrays from coronary- (A and B) and mammary- (C and D) enriched clones were hybridized with both coronary-enriched (A and B) and mammary-enriched (C and D) cDNA probes. Coronary clones hybridized with coronary probe (A) and mammary clones hybridized with mammary probe (C) were considered differentially expressed genes that were used for sequencing. Coronary clones that hybridized with mammary probe (B) and mammary clones that hybridized with coronary probe (D) were considered nonspecific.
Figure 2.
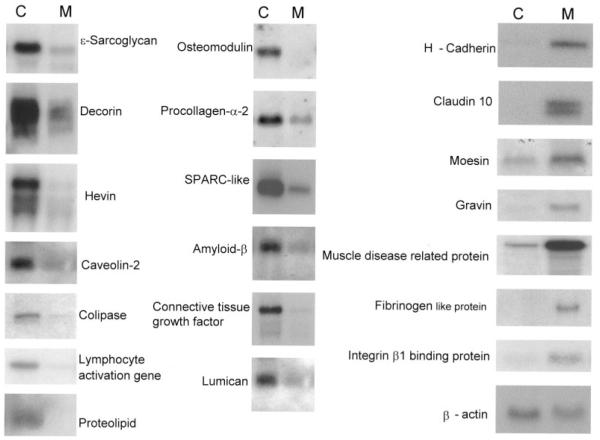
Representative Northern blot analysis of a selected differentially expressed coronary and mammary genes: Total RNA isolated from matched porcine coronary (C, left) and mammary (M, right) arteries (10 μg/lane) were electrophoresed on a 1% agarose gel, transferred to nylon filters, and hybridized under high-stringency conditions with indicated 32P-labled cDNA probes. Each lane represents a pool of total RNA from 3 porcine arteries (n=12). The blots were stripped and reused 3 times. The β-actin is a representative of the loaded RNA, indicating that RNA loading was uniform.
TABLE 1.
Gene Fragments Preferentially Expressed in the Porcine Coronary Arteries
| Species | Artery | Expressed Genes |
|---|---|---|
| H sapiens | Coronary | SPARC-like 1 (mast9, hevin) (SPARCL1) |
| S scrofa | Coronary | Decorin |
| H sapiens | Coronary | Procollagen alpha 2 (COL1A2) |
| H sapiens | Coronary | Platelet-derived growth factor receptor, alpha (PDGFRA) mRNA |
| H sapiens | Coronary | Collagen type I alpha 2 (COL1A2) mRNA |
| H sapiens | Coronary | Tissue inhibitor of metalloproteinase 3 (TIMP3), mRNA |
| H sapiens | Coronary | Lumican (LUM) mRNA |
| H sapiens | Coronary | Angiogenin, ribonuclease, Rnase family, 5 precursor (ANG) |
| S scrofa | Coronary | Tenascin mRNA |
| S scrofa | Coronary | Pleiotropic factor beta |
| H sapiens | Coronary | Tumor necrosis factor type 1 receptor associated protein |
| H sapiens | Coronary | Lipoma-preferred partner |
| H sapiens | Coronary | Sarcoglycan, epsilon (SGCE), mRNA |
| S scrofa | Coronary | Proteolipid protein (plp) gene |
| H sapiens | Coronary | NICE-5 protein |
| H sapiens | Coronary | Amyloid beta A4 precursor-like protein 2 |
| H sapiens | Coronary | MDM2 gene intron 9 exon 10, partial sequence |
| H sapiens | Coronary | T-cell activation protein (PGR1) gene, complete cds |
| H sapiens | Coronary | Osteomodulin (OMD), mRNA |
| H sapiens | Coronary | Caveolin 2 (CAV2) mRNA |
| H sapiens | Coronary | Lymphocyte-activation gene 1 |
| H sapiens | Coronary | Connective tissue growth factor mRNA |
| H sapiens | Coronary | Death associated protein 3 (DAP3) mRNA |
| H sapiens | Coronary | Kruppel-like factor 5 (KLF5) mRNA, complete cds |
| H sapiens | Coronary | Caveolin 1 (CAV1) gene, exon 3 and partial cds |
| S scrofa | Coronary | Colipase gene, partial sequence |
| H sapiens | Coronary | Peanut-like 2 (PNUTL2) mRNA |
| H sapiens | Coronary | Uncharacterized hematopoietic stem/progenitor cells |
| H sapiens | Coronary | MO25 protein (LOC51719), mRNA |
| H sapiens | Coronary | HSPC297 mRNA, partial cds |
| H sapiens | Coronary | BNIP3H (BNIP3H) gene, complete cds |
| H sapiens | Coronary | Palladin (KIAA0992) mRNA |
| H sapiens | Coronary | KIAA0630 protein mRNA, partial cds |
| H sapiens | Coronary | KIAA0092 gene mRNA, complete cds |
| H sapiens | Coronary | KIAA1411 protein, partial cds |
| H sapiens | Coronary | KIAA0965 protein, partial cds |
| H sapiens | Coronary | KIAA1275 protein, partial cds |
| H sapiens | Coronary | cDNA FLJ21653 fis similar to Human protein kinase mRNA |
| H sapiens | Coronary | Chromosome 8 clone 36G5 map 8p23.1, complete sequence |
| H sapiens | Coronary | Chromosome 19-specific cosmid F14150 complete genomic |
| H sapiens | Coronary | Chromosome 14 map 14q24.3, complete sequence |
| H sapiens | Coronary | Chromosome 14 map 14q24.3, complete sequence |
| H sapiens | Coronary | PRO0989 mRNA, complete cds |
| H sapiens | Coronary | PRO1873 mRNA, complete cds |
| H sapiens | Coronary | Hypothetical protein (HSU79253), mRNA |
| H sapiens | Coronary | Sequence from clone RP4–540A13 on chromosome Xq22.1–22.3 |
| H sapiens | Coronary | Sequence from clone RP5–1018E9 on chromosome 20… |
| H sapiens | Coronary | Sequence from clone RP11–18I14 on chromosome 10… |
| H sapiens | Coronary | CGI-75 protein mRNA, complete cds |
| H sapiens | Coronary | CGI-81 protein mRNA, complete cds |
| H sapiens | Coronary | CGI-54 protein mRNA, complete cds |
| H sapiens | Coronary | CGI-07 protein mRNA, complete cds |
TABLE 2.
Gene Fragments Preferentially Expressed in the Porcine Internal Mammary Arteries
| Species | Artery | Expressed Genes |
|---|---|---|
| H sapiens | Mammary | Alpha-2-macroglobulin (A2 mol/L) |
| H sapiens | Mammary | Cadherin 13, H-cadherin (CDH13) |
| H sapiens | Mammary | Integrin beta 1 binding protein (melusin) 2 (ITGB1BP2) |
| H sapiens | Mammary | FLICE-like inhibitory protein short form |
| H sapiens | Mammary | Caspase 3, apoptosis-related cystein protease (CASP3) |
| H sapiens | Mammary | Abl-interactor protein 1 long (Abl1) |
| H sapiens | Mammary | Fibrinogen-like protein 2 |
| H sapiens | Mammary | RBP1-like protein (LOC51742) |
| H sapiens | Mammary | A kinase (PRKA) anchor protein (gravin) 12 (AKAP12) |
| H sapiens | Mammary | Leucyl/cystinyl aminopeptidase (LNPEP) |
| H sapiens | Mammary | Claudin-10 (CLDN10) |
| S scrofa | Mammary | SINE sequence SSPRE DNA |
| H sapiens | Mammary | Sema domain (semaphorin) 3E (SEMA3E) |
| H sapiens | Mammary | Muscle disease related protein (LOC51725) |
| H sapiens | Mammary | Antizyme inhibitor |
| H sapiens | Mammary | MSTP032 |
| H sapiens | Mammary | Hypothetical protein FLJ20585 |
| H sapiens | Mammary | Hypothetical protein FLJ13350 |
| H sapiens | Mammary | Hypothetical protein (HSPC194) |
| H sapiens | Mammary | Hypothetical protein PR02975 |
| H sapiens | Mammary | Hypothetical protein, expressed in osteoblast |
| H sapiens | Mammary | Hypothetical protein (LOC54675) |
| H sapiens | Mammary | Hypothetical protein FLJ10830 |
| H sapiens | Mammary | Hypothetical protein PR01489 |
We confirmed differential gene expression of selected genes by Northern blot and in situ hybridization. Total RNA isolated from two matched coronary and mammary arteries were pooled and used for Northern blot. We randomly selected 21 genes that were identified using the subtracted probes for Northern blot analysis (Figure 2). Among these genes, 13 were hybridized primarily with the mammary-subtracted cDNA probes and presumably represent coronary-enriched genes. Northern blot analyses confirmed differential expression of 11 genes. These included ∊-sarcoglycan, caveolin-2, colipase, lymphocyte activating factor, proteolipid protein, osteomodulin, procollagen-α-2, SPARC-like protein, amyloid beta A4 precursor-like protein 2, lumican, and connective tissue growth factor. In addition, we selected 8 genes that were hybridized with coronary-subtracted cDNA probes, but not mammary-subtracted probes, which presumably represent mammary-enriched genes. Northern blot analysis confirmed differential expression of 7 genes. These included h-cadherin, claudin-10, moesin, gravin, muscle disease–related protein, fibrinogen-like protein-2, and integrin β1 binding protein 2. A representative of loading control, β-actin, is also shown. In addition to these genes that were identified by subtracted probes, we confirmed differential gene expression of 3 selected genes that were identified by unsubtracted coronary (hevin and decorin) and mammary (h-cadherin) cDNA probes.
Furthermore, we confirmed differential expression of 4 selected genes by in situ hybridization of which 3 were identified with mammary-subtracted probe (caveolin-1, pleiotrophin, lipoma-preferred partner) and 1 was identified by coronary-subtracted probe (α-2 macroglobulin). Figure 3 shows differential expression of caveolin-1 mRNA. Caveolin-1 was strongly expressed in the coronary artery. Its distribution was heterogeneous and concentrated primarily in the intima. The sense strand probe for caveolin-1 did not stain the coronary. Caveolin-1 mRNA was substantially lower in the mammary artery than coronary artery and similar to the sense strand. This pattern of expression was observed in 10 of 12 porcine coronary arteries. We did not detect caveolin-1 mRNA in two other porcine coronary arteries. In addition to caveolin-1, we validated differential expression of pleiotrophin and lipoma-preferred partner by in situ hybridization. Our data showed that these genes are preferentially expressed in the coronary arteries (not shown). Further, we confirmed preferential expression of α-2 macroglobulin mRNA in the porcine mammary arteries (not shown).
Figure 3.
In situ hybridization of caveolin-1 in the porcine coronary and mammary arteries. In situ hybridization was performed essentially as described70,71 using caveolin-1 digoxigenin-labeled sense and antisense riboprobes.
To further demonstrate the blood vessel selectivity of candidate gene expression, we examined expression of tenascin and angiogenin in porcine coronary, mammary, and radial arteries. These genes were identified by screening the coronary-enriched library with unsubtracted coronary and mammary cDNA probes and, therefore, these genes are enriched in the coronary arteries. Northern blot analysis of porcine coronary, mammary, and radial arteries revealed that the two isoforms of tenascin are predominantly expressed in the coronary arteries (Figure 4). In addition, we found preferential expression of angiogenin in the coronary arteries but not mammary or radial arteries. These results further support other data suggesting that the coronary artery has a specific gene expression program that is different from either mammary or radial arteries.
Figure 4.

Northern blot analysis of tenascin and angiogenin expression in different arteries. Total RNA isolated from porcine coronary (C), mammary (M), and radial (R) arteries were analyzed by Northern blot using tenascin and angiogenin as described in the legend to Figure 3. RNA loading was examined by reprobing the blot with β-actin.
Discussion
We used SSH to determine differential gene expression between coronary and mammary arteries. To work with a relatively homogeneous cellular population, we removed endothelium and adventitia; therefore, our data primarily represent differential gene expression of medial and intimal smooth muscle cells. To screen the libraries, we used subtracted probes to identify rare messages.42 From the subtracted libraries, we randomly selected 27 genes that were identified with the subtracted probes for further analysis by Northern blot, in situ hybridization, and immunostaining. The results confirmed differential expression of 24 of the genes in coronary and mammary arteries. These results indicate that the false-positive rate was approximately 10%. This false-positive rate could be the result of an inherent feature of this methodology,36 similarities between mRNA patterns of coronary and mammary arteries,42 or a combination thereof. If we assume that this low rate of false-positive can be extended to other genes listed in Tables 1 and 2, then our data suggest there are substantial differences in the gene expression patterns of coronary and mammary arteries.
Although the vascular function of majority of these genes has not been defined, extrapolation from other systems would suggest that signals generated locally by these genes might either alone or in combination with invading inflammatory cells result in initiation and development of lesion in the coronary arteries. Northern blot analysis of porcine coronary and mammary arteries showed that extracellular matrix proteins are consistently overexpressed in 12 coronary and mammary arteries examined. For example, decorin, lumican, procollagen-α, tenascin, secreted protein acidic and rich in cystein (SPARC)-like molecule, hevin, ∊-sarcoglycan, and osteomodulin are preferentially expressed in the coronary arteries. Although vascular expression and activity of some of these genes are unknown, they may play an important role in processes that are critical to the pathogenesis of atherosclerosis. Decorin promotes binding of LDL to collagen,43 which increases the capacity of coronary arteries to retain lipids. This interaction increases the ability of LDL to oxidize,44 which could lead to formation of foam cells.45 Tenascin and SPARC-like/hevin are antiadhesive extracellular molecules preferentially expressed in the coronary arteries. We have previously shown that tenascin prevents adhesion of rat vascular smooth muscle cells to fibronectin46 and promotes migration of these cells.47 Others reported that tenascin promotes proliferation of smooth muscle cells.48,49 SPARC-like/hevin is another antiadhesive molecule that we have found to be differentially expressed in the porcine coronary arteries. We have previously reported that SPARC-like/hevin is primarily expressed in the aorta and vena cava and is primarily a medial marker.50 Taken together, these data suggest that the medial gene expression profile is dependent on arterial phenotype.
Another group of molecules that we found to be expressed preferentially in the coronary arteries is related to lipid binding and metabolizing molecules, such as caveolin-1 and 2, proteolipid protein, colipase, and lipoma-preferred partner. Caveolins are integral membrane proteins that form the framework of caveolae, which are 50- to 100-nm plasma membrane invaginations. Caveolin-1 binds to cholesterol and 7-keto cholesterol and mediates their transport, suggesting that caveolin-1 is important in regulating cellular cholesterol homeostasis during atherosclerosis.51 Caveolin-2 also appears to be involved in trafficking of lipid molecules mediated by caveolins.52 Colipases are involved in the absorption, transport, storage, and mobilization of lipids.53 Proteolipid protein is another coronary gene that codes for the most abundant protein in the central nervous system, however, its vascular function is unknown.54
Lipoproteins, once retained and metabolized, can induce inflammatory reactions.55 Inflammation is initiated by rolling adhesion of monocytes and lymphocytes to the endothelial surface, penetration of endothelial junction, and migration to the subendothelium. Monocytes are converted to macrophages, whereupon they ingest lipids and become foam cells leading to formation of fatty streaks. We have found that the coronary arteries are enriched in genes that are involved in every one of these events. For example, tenascin and hevin are reported to support the tethering and rolling of lymphocytes and lymphoblastic cell lines under flow conditions.56,57 Compared with rolling of the same cell type on E-selectin, rolling on tenascin was found to be smoother at all shear stresses tested, suggesting that cells formed a larger number of bonds on tenascin substrate than on E-selectin substrate.56 We also found preferential expression of T-cell activation protein and lymphocyte-activation gene in the coronary arteries, which promote monocyte chemotaxis58 and in combination with macrophage-colony stimulating factor or granulocyte macrophage-colony stimulating factor, stimulate proliferation of mature tissue macrophages.59
In contrast with these coronary artery genes, internal mammary arteries are enriched in genes involved in cell–cell interaction. For example, we found a strong expression of h-cadherin and claudin-10. Cadherins are transmembrane proteins that mediate homophilic cell–cell interaction. In addition, cadherins modulate β1 integrin activity thus regulating cell–matrix interaction.60 Further, cadherins, interact with growth factor signaling, thus influencing cell growth and migration.61 H-cadherin is a newly characterized cadherin molecule whose expression is decreased in a variety of human carcinoma cells, especially in high-grade invasive epithelial ovarian62 and gastric cancers.63 Transfection of h-cadherin cDNA into two breast tumor cell lines in which there was no basal h-cadherin expression resulted in diminished tumor cell growth and a significant change from invasive morphology to a normal cell-like morphology.64 Injection of h-cadherin–transfected cells into mammary fat pads of nude mice produced a marked inhibition of tumor growth and modified the morphology of tumor cells.65 In addition to anti-invasive activity, h-cadherin inhibits neovascularization.66 Claudin-10 is a member of tight junction proteins.67 The function of claudins are unknown; however, in kidney tubules, claudins appear to regulate extracellular or paracellular permeability, through resorption of Mg2+ and Ca2+.68
The differential gene expression between coronary and mammary arteries suggests that these blood vessels have different phenotypes and, therefore, their response to a proatherosclerotic environment would be different. Therefore, although of comparable diameter and in close physical proximity to coronary arteries for 50 or more years in humans, mammary arteries do not develop atheroma in a proatherosclerotic environment. This freedom from atherosclerosis continues for decades thereafter when these vessels are used for coronary revascularization. In addition, the response of mammary arteries to mechanical injury is different from that of coronary arteries. Human coronary arteries respond to balloon angioplasty by promoting cell migration and proliferation, leading to the formation of neointima and restenosis in approximately 40% of cases. However, unlike coronary arteries and saphenous vein grafts, restenosis was not found in mammary artery grafts after percutaneous transluminal angioplasty.69 The resistance of internal mammary artery grafts to restenosis and atherosclerosis combined with our data regarding the differential gene expression between normal coronary and mammary arteries suggest that these two blood vessels have inherently different properties that may explain their divergent pathologies.
In summary, we have been able to detect differential expression of several genes between coronary and mammary arteries. The most striking aspect of our findings is the extensive differences in RNA expression in the internal mammary artery and coronary artery smooth muscle cells. The differential expression of porcine genes associated with lipid retention and metabolism, inflammation, neovascularization, apoptosis, cell growth, and anchoring junction should not be regarded simply as a list of genes and their associated expression patterns, but rather it is a representation of the state of the coronary and mammary arteries (ie, atheropermissive genes are expressed in the coronary artery and athero-resistant genes are expressed in the mammary artery). Future development will yield an even more comprehensive view of the gene expression pattern between various arterial phenotypes.
Acknowledgments
This work was supported by National Institute of Health grant HL50566, Established Investigator Award 0040201N from American Heart Association, and the Eisner Foundation.
Contributor Information
Minghui Qin, Atherosclerosis Research Center, Division of Cardiology, and Burns and Allen Research Institute, Cedars-Sinai Medical Center, Los Angeles, Calif.
Zhaohui Zeng, Atherosclerosis Research Center, Division of Cardiology, and Burns and Allen Research Institute, Cedars-Sinai Medical Center, Los Angeles, Calif.
Jie Zheng, Atherosclerosis Research Center, Division of Cardiology, and Burns and Allen Research Institute, Cedars-Sinai Medical Center, Los Angeles, Calif.
Prediman K. Shah, Atherosclerosis Research Center, Division of Cardiology, and Burns and Allen Research Institute, Cedars-Sinai Medical Center, Los Angeles, Calif
Stephen M. Schwartz, Department of Pathology, University of Washington, Seattle, Wash.
Lawrence D. Adams, Department of Pathology, University of Washington, Seattle, Wash.
Behrooz G. Sharifi, Atherosclerosis Research Center, Division of Cardiology, and Burns and Allen Research Institute, Cedars-Sinai Medical Center, Los Angeles, Calif
References
- 1.Kolessov VI. Mammary artery-coronary artery anastomosis as method of treatment for angina pectoris. J Thorac Cardiovasc Surg. 1967;54:535–544. [PubMed] [Google Scholar]
- 2.Green GE, Stertzer SH, Reppert EH. Coronary arterial bypass grafts. Ann Thorac Surg. 1968;5:443–450. doi: 10.1016/s0003-4975(10)66377-1. [DOI] [PubMed] [Google Scholar]
- 3.Green GE, Paul RS, Wallsh E, Tice DA. Coronary artery bypass grafting. Surg Forum. 1968;19:159–161. [PubMed] [Google Scholar]
- 4.Loop FD. Internal-thoracic-artery grafts. Biologically better coronary arteries. N Engl J Med. 1996;334:263–265. doi: 10.1056/NEJM199601253340411. [DOI] [PubMed] [Google Scholar]
- 5.Cameron A, Davis KB, Green G, Schaff HV. Coronary bypass surgery with internal-thoracic-artery grafts—effects on survival over a 15-year period. N Engl J Med. 1996;334:216–219. doi: 10.1056/NEJM199601253340402. [DOI] [PubMed] [Google Scholar]
- 6.Boylan MJ, Lytle BW, Loop FD, Taylor PC, Borsh JA, Goormastic M, Cosgrove DM. Surgical treatment of isolated left anterior descending coronary stenosis: comparison of left internal mammary artery and venous autograft at 18 to 20 years of follow-up. J Thorac Cardiovasc Surg. 1994;107:657–662. [PubMed] [Google Scholar]
- 7.Sisto T, Yla-Herttuala S, Luoma J, Riekkinen H, Nikkari T. Biochemical composition of human internal mammary artery and saphenous vein. J Vasc Surg. 1990;11:418–422. doi: 10.1067/mva.1990.17248. [DOI] [PubMed] [Google Scholar]
- 8.Kay HR, Korns ME, Flemma RJ, Tector AJ, Lepley D., Jr Atherosclerosis of the internal mammary artery. Ann Thorac Surg. 1976;21:504–507. doi: 10.1016/s0003-4975(10)63917-3. [DOI] [PubMed] [Google Scholar]
- 9.Ojha M, Leask RL, Johnston KW, David TE, Butany J. Histology and morphology of 59 internal thoracic artery grafts and their distal anastomoses. Ann Thorac Surg. 2000;70:1338–1344. doi: 10.1016/s0003-4975(00)01975-5. [DOI] [PubMed] [Google Scholar]
- 10.van Son JA, Smedts F, Vincent JG, van Lier HJ, Kubat K. Comparative anatomic studies of various arterial conduits for myocardial revascularization. J Thorac Cardiovasc Surg. 1990;99:703–707. [PubMed] [Google Scholar]
- 11.Sims FH. A comparison of coronary and internal mammary arteries and implications of the results in the etiology of arteriosclerosis. Am Heart J. 1983;105:560–566. doi: 10.1016/0002-8703(83)90478-7. [DOI] [PubMed] [Google Scholar]
- 12.Sims FH, Koelmeyer TD, Zhang YP, Lambie N, Edgar SG. Primary plexogenic pulmonary hypertension shows imperfect formation of the internal elastic lamina of the pulmonary arteries. Exp Lung Res. 1995;21:367–383. doi: 10.3109/01902149509023714. [DOI] [PubMed] [Google Scholar]
- 13.Sims FH. Discontinuities in the internal elastic lamina: a comparison of coronary and internal mammary arteries. Artery. 1985;13:127–143. [PubMed] [Google Scholar]
- 14.Sims FH. The internal elastic lamina in normal and abnormal human arteries: a barrier to the diffusion of macromolecules from the lumen. Artery. 1989;16:159–173. [PubMed] [Google Scholar]
- 15.Sims FH. A comparison of structural features of the walls of coronary arteries from 10 different species. Pathology. 1989;21:115–124. doi: 10.3109/00313028909059547. [DOI] [PubMed] [Google Scholar]
- 16.Sims FH, Chen X, Gavin JB. The importance of a substantial elastic lamina subjacent to the endothelium in limiting the progression of atherosclerotic changes. Histopathology. 1993;23:307–317. doi: 10.1111/j.1365-2559.1993.tb01213.x. [DOI] [PubMed] [Google Scholar]
- 17.Wendelhag I, Wiklund O, Wikstrand J. Atherosclerotic changes in the femoral and carotid arteries in familial hypercholesterolemia: ultrasonographic assessment of intima-media thickness and plaque occurrence. Arterioscler Thromb. 1993;13:1404–1411. doi: 10.1161/01.atv.13.10.1404. [DOI] [PubMed] [Google Scholar]
- 18.Sramek A, Bosch JG, Reiber JH, Van Oostayen JA, Rosendaal FR. Ultrasound assessment of atherosclerotic vessel wall changes: reproducibility of intima-media thickness measurements in carotid and femoral arteries. Invest Radiol. 2000;35:699–706. doi: 10.1097/00004424-200012000-00001. [DOI] [PubMed] [Google Scholar]
- 19.Lekakis JP, Papamichael CM, Cimponeriu AT, Stamatelopoulos KS, Papaioannou TG, Kanakakis J, Alevizaki MK, Papapanagiotou A, Kalofoutis AT, Stamatelopoulos SF. Atherosclerotic changes of extracoronary arteries are associated with the extent of coronary atherosclerosis. Am J Cardiol. 2000;85:949–952. doi: 10.1016/s0002-9149(99)00907-8. [DOI] [PubMed] [Google Scholar]
- 20.O’Leary DH, Polak JF, Kronmal RA, Manolio TA, Burke GL, Wolfson SK., Jr Carotid-artery intima and media thickness as a risk factor for myocardial infarction and stroke in older adults: Cardiovascular Health Study Collaborative Research Group. N Engl J Med. 1999;340:14–22. doi: 10.1056/NEJM199901073400103. [DOI] [PubMed] [Google Scholar]
- 21.Weninger WJ, Muller GB, Reiter C, Meng S, Rabl SU. Intimal hyperplasia of the infant parasellar carotid artery: a potential developmental factor in atherosclerosis and SIDS. Circ Res. 1999;85:970–975. doi: 10.1161/01.res.85.10.970. [DOI] [PubMed] [Google Scholar]
- 22.Svendsen E, Dregelid E, Eide GE. Internal elastic membrane in the internal mammary and left anterior descending coronary arteries and its relationship to intimal thickening. Atherosclerosis. 1990;83:239–248. doi: 10.1016/0021-9150(90)90169-j. [DOI] [PubMed] [Google Scholar]
- 23.Ferro M, Conti M, Novero D, Botto Micca F, Palestro G. Incidence of atherosclerosis of the internal mammary artery compared with the coronary artery: a study of 22 non-selected autopsy cases. Minerva Cardioangiol. 1990;38:325–330. [PubMed] [Google Scholar]
- 24.Velican C, Velican D. The precursors of coronary atherosclerotic plaques in subjects up to 40 years old. Atherosclerosis. 1980;37:33–46. doi: 10.1016/0021-9150(80)90091-x. [DOI] [PubMed] [Google Scholar]
- 25.Velican D, Velican C. Atherosclerotic involvement of the coronary arteries of adolescents and young adults. Atherosclerosis. 1980;36:449–460. doi: 10.1016/0021-9150(80)90238-5. [DOI] [PubMed] [Google Scholar]
- 26.Schwartz SM, deBlois D, O’Brien ER. The intima: soil for atherosclerosis and restenosis. Circ Res. 1995;77:445–465. doi: 10.1161/01.res.77.3.445. Review. [DOI] [PubMed] [Google Scholar]
- 27.Armstrong PJ, Johanning JM, Calton WC, Jr, Delatore JR, Franklin DP, Han DC, Carey DJ, Elmore JR. Differential gene expression in human abdominal aorta: aneurysmal versus occlusive disease. J Vasc Surg. 2002;35:346–355. doi: 10.1067/mva.2002.121071. [DOI] [PubMed] [Google Scholar]
- 28.Hiltunen MO, Tuomisto TT, Niemi M, Brasen JH, Rissanen TT, Toronen P, Vajanto I, Yla-Herttuala S. Changes in gene expression in atherosclerotic plaques analyzed using DNA array. Atherosclerosis. 2002;165:23–32. doi: 10.1016/s0021-9150(02)00187-9. [DOI] [PubMed] [Google Scholar]
- 29.Tung WS, Lee JK, Thompson RW. Simultaneous analysis of 1176 gene products in normal human aorta and abdominal aortic aneurysms using a membrane-based complementary DNA expression array. J Vasc Surg. 2001;34:143–150. doi: 10.1067/mva.2001.113310. [DOI] [PubMed] [Google Scholar]
- 30.Shiffman D, Porter JG. Gene expression profiling of cardiovascular disease models. Curr Opin Biotechnol. 2000;11:598–601. doi: 10.1016/s0958-1669(00)00150-6. [DOI] [PubMed] [Google Scholar]
- 31.Tai JT, Brooks EE, Liang S, Somogyi R, Rosete JD, Lawn RM, Shiffman D. Determination of temporal expression patterns for multiple genes in the rat carotid artery injury model. Arterioscler Thromb Vasc Biol. 2000;20:2184–2191. doi: 10.1161/01.atv.20.10.2184. [DOI] [PubMed] [Google Scholar]
- 32.Shiffman D, Mikita T, Tai JT, Wade DP, Porter JG, Seilhamer JJ, Somogyi R, Liang S, Lawn RM. Large scale gene expression analysis of cholesterol-loaded macrophages. J Biol Chem. 2000;275:37324–37332. doi: 10.1074/jbc.M004732200. [DOI] [PubMed] [Google Scholar]
- 33.Tyson KL, Weissberg PL, Shanahan CM. Heterogeneity of gene expression in human atheroma unmasked using cDNA representational difference analysis. Physiol Genomics. 2002;9:121–130. doi: 10.1152/physiolgenomics.00116.2001. [DOI] [PubMed] [Google Scholar]
- 34.Wuttge DM, Sirsjo A, Eriksson P, Stemme S. Gene expression in atherosclerotic lesion of ApoE deficient mice. Mol Med. 2001;7:383–392. [PMC free article] [PubMed] [Google Scholar]
- 35.Diachenko LB, Ledesma J, Chenchik AA, Siebert PD. Combining the technique of RNA fingerprinting and differential display to obtain differentially expressed mRNA. Biochem Biophys Res Commun. 1996;219:824–828. doi: 10.1006/bbrc.1996.0317. [DOI] [PubMed] [Google Scholar]
- 36.Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci U S A. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gurskaya NG, Diatchenko L, Chenchik A, Siebert PD, Khaspekov GL, Lukyanov KA, Vagner LL, Ermolaeva OD, Lukyanov SA, Sverdlov ED. Equalizing cDNA subtraction based on selective suppression of polymerase chain reaction: cloning of Jurkat cell transcripts induced by phytohemaglutinin and phorbol 12-myristate 13-acetate. Anal Biochem. 1996;240:90–97. doi: 10.1006/abio.1996.0334. [DOI] [PubMed] [Google Scholar]
- 38.Altschul SF, Boguski MS, Gish W, Wootton JC. Issues in searching molecular sequence databases. Nat Genet. 1994;6:119–129. doi: 10.1038/ng0294-119. [DOI] [PubMed] [Google Scholar]
- 39.Tatusov RL, Altschul SF, Koonin EV. Detection of conserved segments in proteins: iterative scanning of sequence databases with alignment blocks. Proc Natl Acad Sci U S A. 1994;91:12091–12095. doi: 10.1073/pnas.91.25.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Argos P, Vingron M, Vogt G. Protein sequence comparison: methods and significance. Protein Eng. 1991;4:375–383. doi: 10.1093/protein/4.4.375. [DOI] [PubMed] [Google Scholar]
- 41.Vingron M, Argos P. Motif recognition and alignment for many sequences by comparison of dot- matrices. J Mol Biol. 1991;218:33–43. doi: 10.1016/0022-2836(91)90871-3. [DOI] [PubMed] [Google Scholar]
- 42.Diatchenko L, Lukyanov S, Lau YF, Siebert PD. Suppression subtractive hybridization: a versatile method for identifying differentially expressed genes. Methods Enzymol. 1999;303:349–380. doi: 10.1016/s0076-6879(99)03022-0. [DOI] [PubMed] [Google Scholar]
- 43.Pentikainen MO, Oorni K, Lassila R, Kovanen PT. The proteoglycan decorin links low density lipoproteins with collagen type I. J Biol Chem. 1997;272:7633–7638. doi: 10.1074/jbc.272.12.7633. [DOI] [PubMed] [Google Scholar]
- 44.Hurt-Camejo E, Camejo G, Rosengren B, Lopez F, Ahlstrom C, Fager G, Bondjers G. Effect of arterial proteoglycans and glycosaminoglycans on low density lipoprotein oxidation and its uptake by human macrophages and arterial smooth muscle cells. Arterioscler Thromb. 1992;12:569–583. doi: 10.1161/01.atv.12.5.569. [DOI] [PubMed] [Google Scholar]
- 45.Steinberg D. Atherogenesis in perspective: hypercholesterolemia and inflammation as partners in crime. Nat Med. 2002;8:1211–1217. doi: 10.1038/nm1102-1211. [DOI] [PubMed] [Google Scholar]
- 46.LaFleur DW, Fagin JA, Forrester JS, Rubin SA, Sharifi BG. Cloning and characterization of alternatively spliced isoforms of rat tenascin: platelet-derived growth factor-BB markedly stimulates expression of spliced variants of tenascin mRNA in arterial smooth muscle cells. J Biol Chem. 1994;269:20757–20763. [PubMed] [Google Scholar]
- 47.LaFleur DW, Chiang J, Fagin JA, Schwartz SM, Shah PK, Wallner K, Forrester JS, Sharifi BG. Aortic smooth muscle cells interact with tenascin-C through its fibrinogen-like domain. J Biol Chem. 1997;272:32798–32803. doi: 10.1074/jbc.272.52.32798. [DOI] [PubMed] [Google Scholar]
- 48.Jones PL, Cowan KN, Rabinovitch M. Tenascin-C, proliferation and subendothelial fibronectin in progressive pulmonary vascular disease. Am J Pathol. 1997;150:1349–1360. [PMC free article] [PubMed] [Google Scholar]
- 49.Chung CY, Murphy-Ullrich JE, Erickson HP. Mitogenesis, cell migration, and loss of focal adhesions induced by tenascin-C interacting with its cell surface receptor, annexin II. Mol Biol Cell. 1996;7:883–892. doi: 10.1091/mbc.7.6.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Geary RL, Wong JM, Rossini A, Schwartz SM, Adams LD. Expression profiling identifies 147 genes contributing to a unique primate neointimal smooth muscle cell phenotype. Arterioscler Thromb Vasc Biol. 2002;22:2010–2016. doi: 10.1161/01.atv.0000038147.93527.35. [DOI] [PubMed] [Google Scholar]
- 51.Sleer LS, Brown AJ, Stanley KK. Interaction of caveolin with 7-ketocholesterol. Atherosclerosis. 2001;159:49–55. doi: 10.1016/s0021-9150(01)00486-5. [DOI] [PubMed] [Google Scholar]
- 52.Fujimoto T, Kogo H, Ishiguro K, Tauchi K, Nomura R. Caveolin-2 is targeted to lipid droplets, a new “membrane domain” in the cell. J Cell Biol. 2001;152:1079–1086. doi: 10.1083/jcb.152.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Tilbeurgh H, Bezzine S, Cambillau C, Verger R, Carriere F. Colipase, structure and interaction with pancreatic lipase. Biochim Biophys Acta. 1999;1441:173–184. doi: 10.1016/s1388-1981(99)00149-3. [DOI] [PubMed] [Google Scholar]
- 54.Baumgartner BG, Deppe A, Rettenberger G, Leeb T, Hameister H, Brenig B. Molecular analysis of the porcine proteolipid protein (PLP) gene. Mamm Genome. 1999;10:895–899. doi: 10.1007/s003359901110. [DOI] [PubMed] [Google Scholar]
- 55.Binder CJ, Chang MK, Shaw PX, Miller YI, Hartvigsen K, Dewan A, Witztum JL. Innate and acquired immunity in atherogenesis. Nat Med. 2002;8:1218–1226. doi: 10.1038/nm1102-1218. [DOI] [PubMed] [Google Scholar]
- 56.Clark RA, Erickson HP, Springer TA. Tenascin supports lymphocyte rolling. J Cell Biol. 1997;137:755–765. doi: 10.1083/jcb.137.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Girard JP, Springer TA. Modulation of endothelial cell adhesion by hevin, an acidic protein associated with high endothelial venules. J Biol Chem. 1996;271:4511–4517. doi: 10.1074/jbc.271.8.4511. [DOI] [PubMed] [Google Scholar]
- 58.Uguccioni M, D’Apuzzo M, Loetscher M, Dewald B, Baggiolini M. Actions of the chemotactic cytokines MCP-1, MCP-2, MCP-3, RANTES, MIP-1 alpha and MIP-1 beta on human monocytes. Eur J Immunol. 1995;25:64–68. doi: 10.1002/eji.1830250113. [DOI] [PubMed] [Google Scholar]
- 59.Fahey TJ, 3rd, Tracey KJ, Tekamp-Olson P, Cousens LS, Jones WG, Shires GT, Cerami A, Sherry B. Macrophage inflammatory protein 1 modulates macrophage function. J Immunol. 1992;148:2764–2769. [PubMed] [Google Scholar]
- 60.Arregui C, Pathre P, Lilien J, Balsamo J. The nonreceptor tyrosine kinase fer mediates cross-talk between N-cadherin and beta1-integrins. J Cell Biol. 2000;149:1263–1274. doi: 10.1083/jcb.149.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saffell JL, Williams EJ, Mason IJ, Walsh FS, Doherty P. Expression of a dominant negative FGF receptor inhibits axonal growth and FGF receptor phosphorylation stimulated by CAMs. Neuron. 1997;18:231–242. doi: 10.1016/s0896-6273(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 62.Kawakami M, Staub J, Cliby W, Hartmann L, Smith DI, Shridhar V. Involvement of H-cadherin (CDH13) on 16q in the region of frequent deletion in ovarian cancer. Int J Oncol. 1999;15:715–720. doi: 10.3892/ijo.15.4.715. [DOI] [PubMed] [Google Scholar]
- 63.Mori Y, Matsunaga M, Abe T, Fukushige S, Miura K, Sunamura M, Shiiba K, Sato M, Nukiwa T, Horii A. Chromosome band 16q24 is frequently deleted in human gastric cancer. Br J Cancer. 1999;80:556–562. doi: 10.1038/sj.bjc.6690391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee SW. H-cadherin, a novel cadherin with growth inhibitory functions and diminished expression in human breast cancer. Nat Med. 1996;2:776–782. doi: 10.1038/nm0796-776. [DOI] [PubMed] [Google Scholar]
- 65.Lee SW, Reimer CL, Campbell DB, Cheresh P, Duda RB, Kocher O. H-cadherin expression inhibits in vitro invasiveness and tumor formation in vivo. Carcinogenesis. 1998;19:1157–1159. doi: 10.1093/carcin/19.6.1157. [DOI] [PubMed] [Google Scholar]
- 66.Nagashima H, Okada M, Hidai C, Hosoda S, Kasanuki H, Kawana M. The role of cadherin-catenin-cytoskeleton complex in angiogenesis: antisense oligonucleotide of plakoglobin promotes angiogenesis in vitro, and protein kinase C (PKC) enhances angiogenesis through the plakoglobin signaling pathway. Heart Vessels. 1997;(suppl):110–112. [PubMed] [Google Scholar]
- 67.Tsukita S, Furuse M. Pores in the wall: claudins constitute tight junction strands containing aqueous pores. J Cell Biol. 2000;149:13–16. doi: 10.1083/jcb.149.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wong V, Goodenough DA. Paracellular channels! Science. 1999;285:62. doi: 10.1126/science.285.5424.62. [DOI] [PubMed] [Google Scholar]
- 69.Enomoto S, Horita K, Naito K, Okazaki H, Ueda O, Kohchi K, Koga N, Ooteki H, Itoh T. Percutaneous transluminal angioplasty in patients with prior coronary artery bypass grafting. Kyobu Geka. 1989;42:814–817. [PubMed] [Google Scholar]
- 70.Wallner K, Li C, Shah PK, Fishbein MC, Forrester JS, Kaul S, Sharifi BG. Tenascin-C is expressed in macrophage-rich human coronary atherosclerotic plaque. Circulation. 1999;99:1284–1289. doi: 10.1161/01.cir.99.10.1284. [DOI] [PubMed] [Google Scholar]
- 71.Wallner K, Sharifi BG, Shah PK, Noguchi S, DeLeon H, Wilcox JN. Adventitial remodeling after angioplasty is associated with expression of tenascin mRNA by adventitial myofibroblasts. J Am Coll Cardiol. 2001;37:655–661. doi: 10.1016/s0735-1097(00)01117-7. [DOI] [PubMed] [Google Scholar]