Abstract
Notch1 activation by Delta-like (DL) Notch ligands is essential to induce T cell commitment and to suppress B cell development from thymus-seeding progenitors. Thymus-seeding progenitor competition for DL4 is critically regulated by Lunatic Fringe (Lfng), which glycosylates epidermal growth factor repeats in the Notch1 extracellular domain to enhance binding avidity for DL ligands. Notch1 activation is also essential for the process of β-selection, which drives TCRβ+ CD4/CD8 double-negative 3 (DN3) precursors to proliferate and generate a large pool of CD4/CD8 double-positive thymocytes. We have used several genetic approaches to determine the importance of Lfng–Notch1 interactions in regulating competition of preselection and postselection DN3 thymocytes for DL ligands in vivo. Surprisingly, although Lfng overexpression enhanced DL4 binding by preselection DN3a thymocytes, it did not confer them with a competitive advantage in mixed chimeras. In contrast, Lfng overexpression enhanced competition of post–β-selection DN3b precursors for DL ligands. Lfng modification of O-fucose in the Notch1 ligand-binding domain contributed to but was not solely responsible for the developmental effects of Lfng overexpression. Although previous studies have suggested that pre–TCR-deficient DN3 thymocytes compete poorly for DL ligands, Lfng overexpression did not fully restore double-positive thymocyte pools from DN3b cells with pre-TCR signaling defects. Thus, pre-TCR and Notch signaling have largely nonoverlapping functions in β-selection. Collectively, our data reveal that Lfng enhances DN3b precursor competition for intrathymic DL ligands to maximize Notch-induced clonal expansion during the earliest stage of β-selection.
Notch1 signaling induces T lineage commitment and suppresses several alternative cell fates from thymus-seeding progenitors (TSPs) that enter through post-capillary venules near the corticomedullary junction (CMJ) of the thymus. Although all five Notch ligands are expressed in the thymus, Delta-like (DL) 4 expressed by thymic epithelial cells nonredundantly activates Notch1 to suppress B cell development and induce T cell development from TSPs (reviewed in Ref. 1). If TSPs are prevented from activating Notch1, they generate B cells, rather than CD4/CD8 double-negative (DN) CD44+CD117+CD25− early thymic progenitors (ETPs) with robust T lineage potential (2–5). ETPs and their DN2 (CD44+CD117+CD25+) progeny lack robust B cell potential (4–7), but Notch signaling is also required to suppress myeloid potential of these subsets (8–11), ensuring that commitment to the T cell lineage is complete by the DN3 (CD44loCD117loCD25+) stage of T cell development. DN3 thymocytes that express the pre-TCR are induced to survive, proliferate, and differentiate to the Notch1-independent CD4 and CD8 double-positive (DP) stage. Notch1 is essential to efficiently induce this β-selection process in vivo (12, 13). However, the role of Notch signaling at this developmental checkpoint is poorly understood.
Most DN3 thymocytes are resting CD27lo CD5lo (DN3a) cells undergoing V(D)J recombination at Tcrg, Tcrd, and Tcrb loci. Successful Tcrb rearrangement allows preselection DN3a thymocytes to express pre-TCR complexes consisting of Ptcra, TCRβ, and CD3 proteins (14). The pre-TCR signals in a ligand-independent fashion (15) through the Lck and Fyn cytoplasmic Src family tyrosine kinases (16, 17) to induce maturation of CD27lo DN3a cells into CD27hi DN3b blasts (18, 19). DN3b cells then generate CD44−CD117−CD25− DN4 thymocytes that proliferate extensively prior to differentiating to the DP stage.
Interestingly, TCR complexes lacking Ptcra can also promote the DN3-DP transition in a Notch-dependent fashion in vitro (20), but only small numbers of DP thymocytes develop in these cases. Even fewer DP thymocytes develop from Ptcra-deficient DN3 progenitors when they compete with wild-type (WT) DN3 progenitors in vivo (21), revealing that the pre-TCR endows DN3 progenitors with a more robust ability to compete for intrathymic resources. Prockop and Petrie (22) have also shown that DN3 thymocyte competition for limiting stromal niches controls the number of DP thymocytes that can be generated. The poor competitive fitness of Ptcra-deficient DN3 progenitors has been suggested, based on in vitro studies, to reflect inefficient competition for Notch ligands (20). However, the notion that the pre-TCR enhances competition for Notch ligands during the DN3-DP transition in vivo has not been tested experimentally.
Fringe proteins are highly conserved Golgi-localized glycosyl-transferases that critically influence Notch receptor–ligand interactions. They typically enhance Notch activation by ligands belonging to the DL family and diminish Notch activation by Jagged family ligands (reviewed in Ref. 23). Although the Notch ligand(s) needed to induce β-selection in vivo have not been identified, DL1 or DL4 but not Jagged1 can drive β-selection in vitro (24–27). Thus, Fringes represent excellent candidates for regulators of DN3 thymocyte competition for DL ligands in vivo. In mammals, Lunatic Fringe (Lfng) and its homologs Manic Fringe and Radical Fringe proteins add N-acetylglucosamine to O-fucose–modified epidermal growth factor (EGF) repeats of Notch extracellular domains (28, 29). Fucose addition to Ser or Thr residues in Notch receptor EGF repeats is carried out by a protein O-fucosyl-transferase known as Pofut1 (30). On the basis of the amino acid consensus sequence recognized by Pofut1 (31), 21 of 36 EGF repeats in Notch1 are potentially O-fucosylated. However, one of the most conserved O-fucose sites in all Notch receptors is EGF repeat 12, which lies within the ligand-binding domain defined in Drosophila Notch as comprising EGF repeats 11 and 12 (32). Mutation of this site to Ala precludes O-fucose addition and removes the substrate for Fringe in the Notch1 ligand-binding domain. Mice homozygous for this Notch112f mutation have defective T cell development (33), demonstrating that O-fucosylation of EGF12 is important for Notch1 activation in vivo. However, it has not been determined how Fringe modification of O-fucosylated EGF12 affects Notch1 function.
Lfng mRNA is highly expressed from the ETP to DN4 thymocyte stages but drops to undetectable levels by the DP stage. Lfng−/− TSPs generate intrathymic B cells but few T cell progenitors in competitive assays (34), revealing that cell-autonomous Lfng–Notch1 interactions augment DL-dependent Notch1 activation in TSPs, enhancing their ability to compete for limiting CMJ niches. Transgenic overexpression of Lfng using the Lck proximal promoter cell autonomously enhances binding of DL-Fc–soluble Notch ligands by DN3 and DP thymocytes, converting them into “supercompetitors” that dominantly occupy intrathymic niches to inhibit DP thymocyte development from WT DN3 progenitors (34). However, misexpression of Lfng in DN3 and DP thymocytes also causes them to block (cell nonautonomously) DL-dependent Notch1 activation in TSPs, promoting intrathymic B cell development at the expense of T cell development (2, 34).
In this study, we used our Lfng transgenic mouse model to elucidate how Lfng impacts Notch1-dependent β-selection of DN3 thymocytes in vivo. In particular, we designed experiments to determine whether Lfng overexpression improves the competitive fitness of preselection DN3a or postselection DN3b thymocytes. Finally, we assessed the impact of Lfng overexpression on β-selection of WT versus Lck- or Ptcra-deficient DN3 thymocytes to elucidate the functions of pre-TCR versus Notch signaling in the DN3-DP transition. Our studies reveal that Lfng modification of EGF12 in the Notch1 ligand-binding domain contributed to, but was not solely responsible for, the cell-nonautonomous inhibition of T cell development caused by transgenic Lfng expression in DP thymocytes. Furthermore, we show that cell-autonomous Lfng–Notch interactions enhance DN3b (but not DN3a) precursor competition for intrathymic DL ligands to maximize Notch-induced clonal expansion beginning at the earliest stage of β-selection in vivo. Finally, our study argues against the notion that DP thymocyte production from Ptcra-deficient DN3 progenitors reflects inefficient competition for Notch ligands. Rather, our data indicate that pre-TCR and Notch signaling have largely nonoverlapping functions in β-selection.
Materials and Methods
Mice
B6.SJL-Ptprca (B6.CD45.1), C57BL/6NTac-Ptprcb (B6.CD45.2), and B6.129S6-Rag2tm1Fwa N12 (B6.Rag2−/−) mice were purchased (Taconic Farms, Germantown, NY). Ptrca−/− (B6.CD45.2) mice (35) were obtained from Dr. H. von Boehmer (Dana-Farber Cancer Institute, Boston, MA). Lck−/− (B6.CD45.2) and Lfng transgenic (Tg+) mice were described previously (2, 16). Lfng Tg+Ptcra−/− mice were generated by intercrossing Lfng Tg+ B6.CD45.2 and Ptcra−/− B6.CD45.2 mice. Lfng Tg+Lck−/− mice were generated by intercrossing Lfng Tg+ B6.CD45.2 and Lck−/− B6. CD45.2 mice. Lfng Tg+Rag2−/− mice were generated by intercrossing B6. Rag2−/− with Lfng Tg+ B6.CD45.2 mice. Notch112f/+ and Notch112f/12f mice (33) were intercrossed with Lfng Tg+ B6.CD45.2 mice. Mouse genotypes were determined by PCR amplification of tail DNA as described previously. Mice used in this study were bred in the specific pathogen-free facility (Hospital for Sick Children, Toronto, Ontario, Canada) or at the Albert Einstein College of Medicine (Bronx, NY). Animal protocols were approved by the Animal Care Committees of each institution.
Flow cytometry
Single-cell suspensions were stained with fluorochrome-conjugated Abs and secondary reagents, and the immunofluorescence was analyzed on FACSCalibur or LSR-II (BD Biosciences, San Jose, CA) flow cytometers using standard techniques. Data files were analyzed using FlowJo (Tree Star, Ashland, OR). Dead cells and debris have been excluded from all data displayed by propidium iodide and forward scatter gating. Abs used in this study have been described in previous publications (5, 34), and staining mixtures used in individual experiments are available on request. Abfluorochrome conjugates and avidin or anti-IgG second-stage reagents were purchased from BD Pharmingen (San Diego, CA), eBioscience (San Diego, CA), Cedarlane Laboratories (Hornby, Ontario, Canada), or Molecular Probes (Eugene, OR) or produced in our laboratory using standard techniques. Statistical significance for all population comparisons was calculated using the randomization test (two-tailed probability) except where specified otherwise.
Adoptive transfer experiments
Lin− bone marrow cells (enriched for hematopoietic stem cells and progenitors) were prepared by immunomagnetic depletion of cells expressing CD4, CD8, CD3ε, TER-119, GR-1, B220, and CD11b as described previously (5). Mixed or single bone marrow chimeras were generated by injecting 105 Lin− bone marrow cells from Lfng Tg+Rag2−/− (B6.CD45.2) and Rag2−/− (B6.CD45.1) donors into lethally irradiated (1000 cGy with a 137Cs gamma irradiator) WT (B6.CD45.1; CD45.2) hosts (8 wk old) through the tail vein. Thymocytes from individual lobes were analyzed 6 wk post-i.v. injection. For intrathymic injections, equal numbers of Lin− bone marrow progenitors from WT (B6.CD45.1;CD45.2) and Lfng Tg+ (B6.CD45.2), Lfng Tg+Lck−/− (B6.CD45.2), or Lfng Tg+Ptcra−/− (B6. CD45.2) donors were injected (105 cells from each donor type/lobe) into anesthetized 6- to 8-wk-old WT (B6.CD45.1) host mice that had been sublethally irradiated (650 cGy) up to 4 h prior to injection. Thymocytes from individual lobes were analyzed 3 wk postintrathymic injection.
DL4-Fc binding and OP9-DL4 coculture assays
The Fc-pIRES2-EGFP expression construct was described previously (34). The pCR3-Dll4huFc expression construct was provided by F. Radtke (Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland). Fc constructs were linearized and transfected into 293T cells using standard techniques. Stable transfectants were selected by culturing in 500 µg/ml G418 (Invitrogen, Carlsbad, CA). Serum-free supernatant containing Fc or DL4-Fc fusion protein was concentrated using Centricon-70 centrifugal units (Millipore, Bedford, MA) and purified using Montage PROSEP-A spin columns (Millipore) as per manufacturer’s instructions. Fc proteins were buffer exchanged into PBS-containing calcium and magnesium (Wisent Bioproducts, St. Bruno, Quebec, Canada) using Amicon centrifugal units (Millipore). Puritywas verified by SYPRORuby staining (Bio-Rad,Hercules, CA) and Western blot analysis and quantitated by a standard Bradford assay (Bio-Rad). For binding assays, 2 × 106 thymocytes from Rag2−/− or Lfng Tg+ Rag2−/− mice were stained with Abs, followed by saturating amounts of control Fc or DL4-Fc protein (10 µg/ml) and goat anti-human IgG Fc-specific and then analyzed on a FACSCalibur or LSR-II as described previously (34).
The generation of OP9-DL4 cells will be described in detail elsewhere (J.S. Yuan, J.B. Tan, I. Visan, I.R. Matei, P. Urbanellis, K. Xu, J. Danska, S.E. Egan, and C.J. Guidos, submitted for publication). Sorted DN3 or DN4 thymocytes from Lfng Tg+Lck−/−, Lck−/−, Lfng Tg+Ptcra−/−, and Ptcra−/− mice were seeded (2000 cells/well) onto 80% confluent OP9-DL4 monolayers and cultured for 7 or 10 d as previously described for OP9-DL1 (34).
Results
The competitive advantage of the Lfng transgene depends on V(D)J recombination
We have previously shown that Lfng overexpression in DN3 thymocytes enhances their production of DP thymocytes relative to that from WT DN3 cells in the same thymus (34), suggesting that Lfng may critically enhance DN3 thymocyte competition for DL ligands during β-selection. However, Notch signaling also enhances metabolism and survival of RAG-deficient DN3a thymocytes in vitro (36). Thus, we could not rule out the possibility that the competitive advantage of Lfng Tg+ DN3 thymocytes reflected enhanced proliferation or survival of preselection DN3a thymocytes, rather than a direct effect on β-selection. Therefore, we compared the competitive fitness of Rag2−/− (B6 CD45.1) versus Lfng Tg+Rag2−/− (B6 CD45.2) thymocytes to determine whether Lfng overexpression can improve the competitive fitness of preselection DN3a thymocytes. Equal numbers of Lin− bone marrow stem and progenitor cells isolated from each strain were injected together (mixed chimeras) or separately (single chimeras) into lethally irradiated WT (B6 CD45.1;CD45.2) hosts.
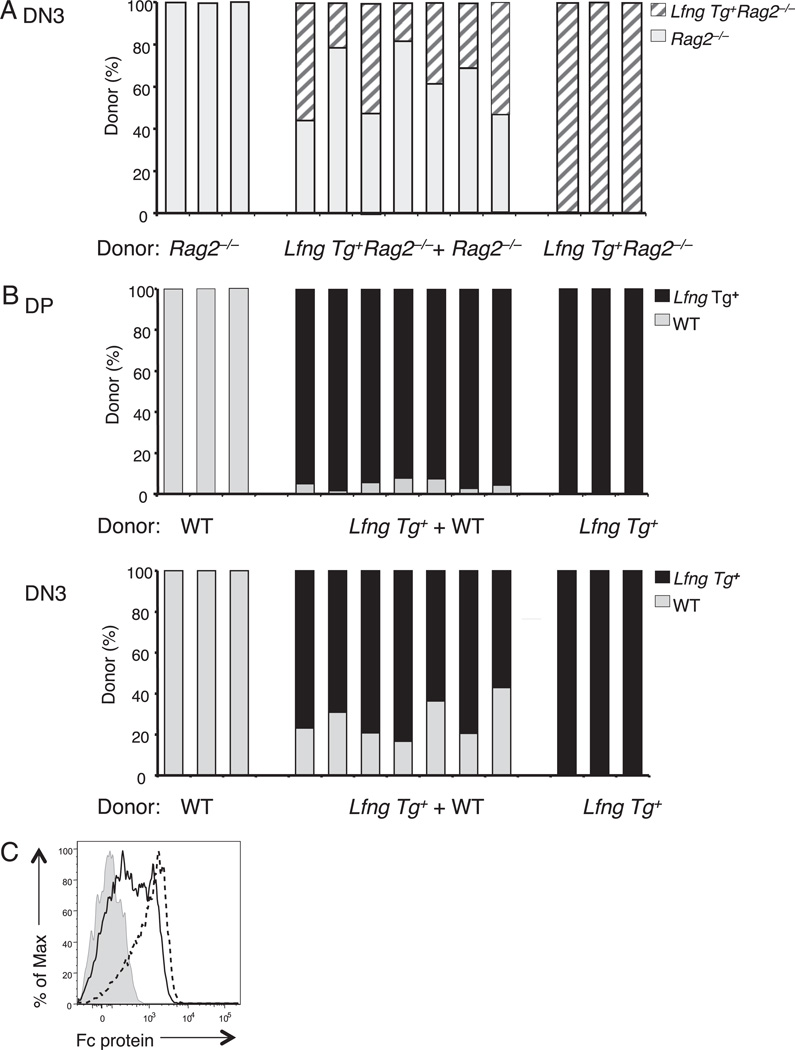
As expected, loss of Rag2 function prevented DN3 thymocytes from Lfng Tg+ and Lfng Tg+ donors from progressing to the DP stage (data not shown). The relative contributions of Lfng Tg+Rag2−/− versus Rag2−/− donors to the thymocyte pool in mixed chimeras was somewhat variable in individual mice (Fig. 1A). On average, Rag2−/− donors contributed 60 ± 14% compared with 36 ± 14% for Lfng Tg+Rag2−/− donors. However, this slight difference did not reach statistical significance when assessed by a paired two-tailed t test (p = 0.07), so we concluded that the two donors have similar competitive fitness. In contrast, the DP pool in control chimeras (generated with an equal mix of Lfng Tg+ and WT B6 donors) was heavily dominated by Lfng Tg+ progeny (Fig. 1B), consistent with our previous study (34). The inability of Lfng Tg+Rag2−/− to outcompete Rag2−/− DN3a thymocytes was not explained by their failure to express the transgene at this developmental stage, because they express at least 10-fold more transgenic Lfng mRNA than endogenous Lfng mRNA (34). Furthermore, Lfng Tg+Rag2−/− DN3a thymocytes bound higher amounts of DL4-Fc fusion protein than Lfng Tg−Rag2−/− DN3a thymocytes (Fig. 1C). Collectively, these data reveal that the competitive advantage afforded by Lfng overexpression is dependent on V(D)J recombination in DN3 thymocytes.
FIGURE 1.
The competitive advantage of the Lfng transgene depends on V(D)J recombination. A, Lin− bone marrow progenitors from Lfng Tg+ Rag2−/− (B6.CD45.2) and Rag2−/− (B6.CD45.1) donors were injected i.v. in equal ratios into lethally irradiated WT hosts (B6.CD45.1;CD45.2). The relative contribution of Lfng Tg+Rag2−/− and Rag2−/− donors to the DN3 thymocyte pool was assessed 3 wk later by staining total thymocytes with Abs against CD4, CD8, CD25, CD117, CD45.1, and CD45.2. B, Lin− bone marrow progenitors from Lfng Tg+ and WT donors were intrathymically injected separately or together as a 50:50 mix into sublethally irradiated WT hosts. The relative contribution of Lfng Tg+ and WT donors to the DP and DN3 thymocyte pools was assessed 3 wk later by staining total thymocytes with Abs against CD4, CD8, CD25, CD117, CD45.1, and CD45.2. C, Lfng overexpression enhances DL4-Fc binding by Rag2−/− DN3a thymocytes. Flow cytometry was used to measure binding of DL4-Fc protein by DN3a (CD117−CD25+) thymocytes from Lfng Tg+Rag2−/− (dashed line) compared with Rag2−/− (solid line) mice. Binding of control Fc protein to Lfng Tg+ Rag2−/− DN3a thymocytes is shown for comparison (shaded line). Binding of control Fc protein to Rag2−/− DN3a thymocytes had a pattern similar to Lfng Tg+Rag2−/− DN3 thymocytes (data not shown). Each experiment was performed at least three times with similar results.
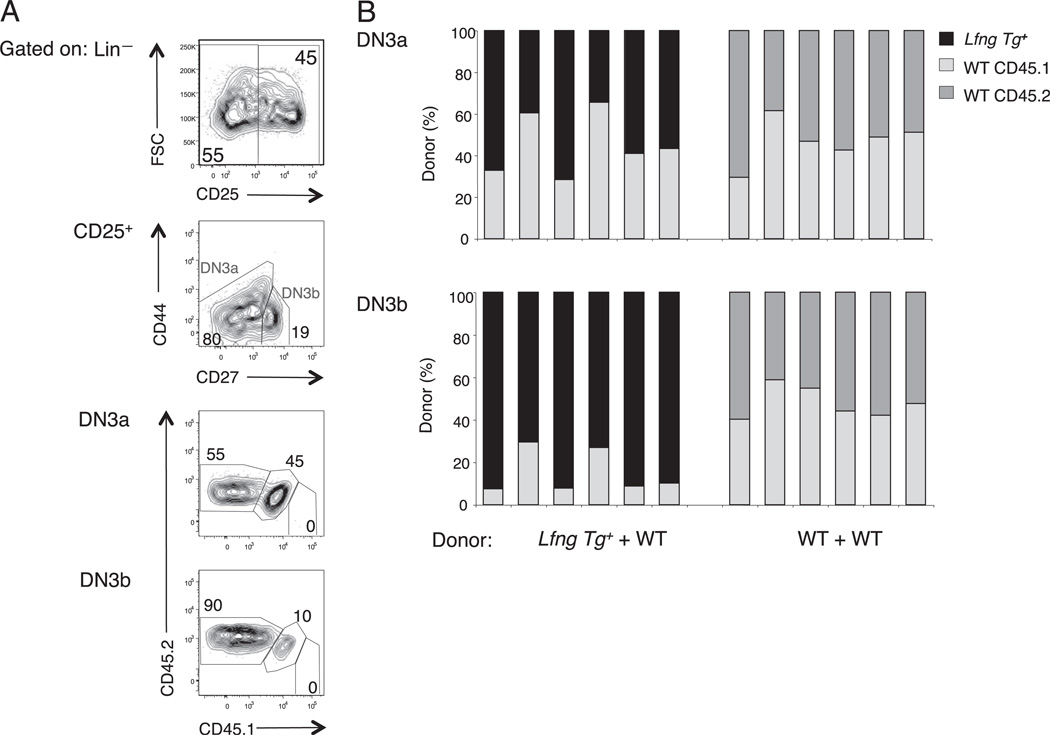
Our observation that transgenic Lfng confers a competitive advantage to WT but not Rag2−/− DN3 thymocytes suggested that it specifically improves the competitive fitness of DN3b thymocytes. To directly assess this possibility, we devised an immunophenotyping strategy that included CD44 and CD27 to distinguish DN3a from DN3b thymocytes and to quantify their relative abundance from Lfng Tg+ versus Tg− B6 donor cells in mixed chimera experiments (Fig. 2A). This classification scheme was originally devised by Taghon et al. (19) and does not exclude DN2 thymocytes from the DN3a pool. We observed that Lfng Tg+ and Tg− donors made roughly equal contributions to the DN3a thymocyte subset of mixed chimeras (Fig. 2B), consistent with the results obtained in mixed chimeras made from Lfng Tg+Rag2−/− and Rag2−/− donors (Fig. 1A). In contrast, Lfng Tg+ progeny profoundly dominated the DN3b subset (Fig. 2B) as well as the DP thymocyte pool (data not shown). Because DN2s account for only 1–5% of total CD25+ DN thymocytes, excluding DN2s from the DN3a subset would not alter this conclusion. These data show that transgenic Lfng confers a competitive advantage specifically to DN3b thymocytes at the earliest stage of β-selection in vivo.
FIGURE 2.
Transgenic Lfng competitive advantage is restricted to DN3b thymocytes undergoing β-selection. Equal numbers of Lin− bone marrow progenitors from Lfng Tg+ B6.CD45.2 and WT B6.CD45.1;CD45.2 mice were intrathymically injected into WT B6.CD45.1 hosts. Abs against Lineage markers (CD4, CD8, CD3ε, B220, Mac-1, GR-1, and NK1.1), CD25, CD44, CD27, CD45.1, and CD45.2 were used to assess the relative contribution of Lfng Tg+ and WT cells to the DN3a and DN3b subsets of individual thymic lobes 18 d later. A, Gating strategy used to assess donor chimerism in the DN3a and DN3b thymocyte pools. B, Relative contribution of Lfng Tg+ and WT donor cells to DN3a and DN3b thymocytes subsets in Lfng Tg+ + WT chimeras and control WT + WT chimeras. Each experiment was performed at least three times with similar results.
Lfng acts in part through the O-fucose site in the Notch1 ligand-binding domain
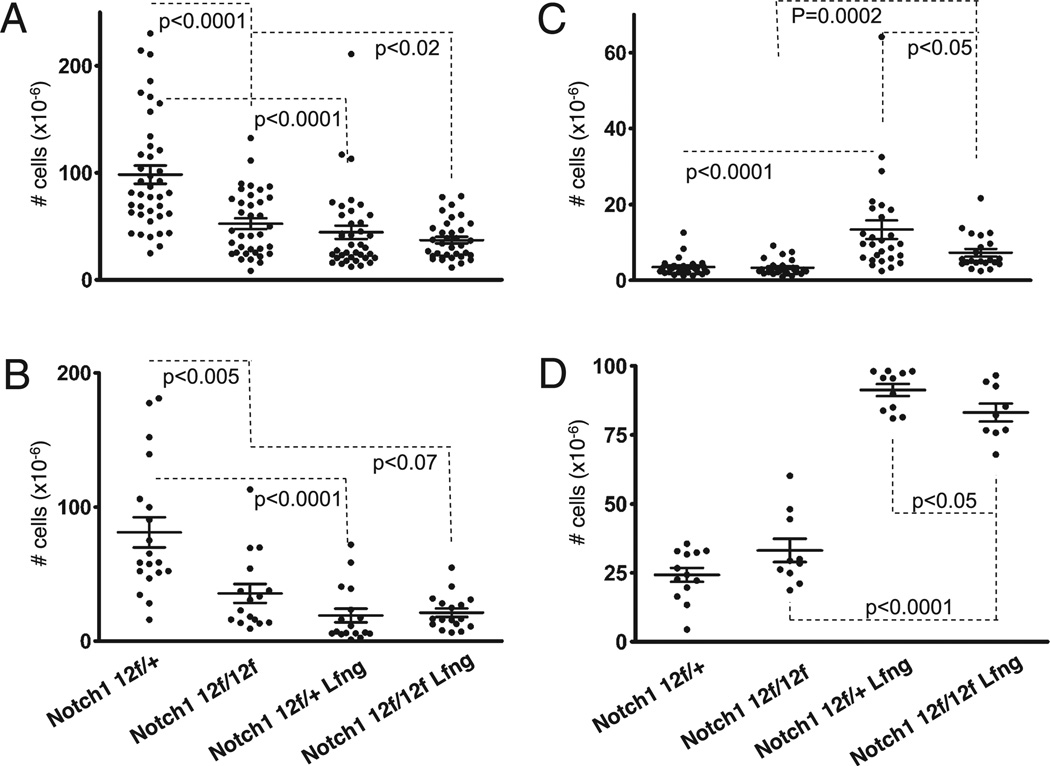
Previously, we have shown that misexpression of Lfng in DP thymocytes causes them to block (cell nonautonomously) DL-dependent Notch1 activation in TSPs, promoting intrathymic B cell development at the expense of T cell development (2, 5, 34). To determine whether the O-fucose site in the Notch1 ligandbinding domain contributes to these developmental effects of Lfng overexpression, mice expressing the Lfng transgene and homozygous or heterozygous for the Notch112f mutation in EGF12 were generated and their thymi evaluated. Control mice were from the same crosses but were not transgenic. As expected from our previous studies of Lfng Tg+Notch1+/+ mice, we observed that Lfng Tg+Notch112f/+ mice had significantly fewer total and DP thymocytes (p < 0.0001) (Fig. 3A, 3B) relative to their Tg−Notch112f/+ littermates. They also had enlarged DN thymocyte pools (p < 0.0001) (Fig. 3C), consisting predominantly of B cells (Fig. 3D), indicating ectopic B cell development in the thymus.
FIGURE 3.
The O-fucose site in the Notch1 ligand binding domain is partly responsible for the effects of Lfng overexpression. Absolute numbers of total thymocytes (A), DP thymocytes (B), DN thymocytes (C), and B220+ B cells (D) were evaluated in Lfng Tg+ Notch112f/+ and Lfng Tg+Notch112f/12f mice (7–8 wk old) relative to their Lfng Tg− littermates. Errors bars represent SE and p values that were derived from unpaired two-tailed Student t test.
As reported previously, Notch112f/f thymi had markedly reduced numbers of DP thymocytes relative to Notch112f/+ mice (p < 0.0001), because of defective β-selection, but both strains had normal-sized intrathymic B cell pools (Fig. 3 and data not shown) (33). Lfng overexpression only marginally reduced the already very small DP thymocyte pools in Notch112f/f mice (p < 0.07) (Fig. 3B). However, Lfng Tg+ Notch112f/f mice had significantly larger DN thymocytes pools (p = 0.0002) that contained a much higher frequency of B cells (p < 0.0001) than seen in Notch112f/f mice (Fig. 3C, 3D). This relative B cell increase within the DN pool reflected a significant increase in absolute numbers of intrathymic B cells (p = 0.0001). These data demonstrate that preventing O-fucose modification of EGF12 does not abrogate the intrathymic B cell development caused by Lfng overexpression. Thus, Lfng can target O-fucose–modified EGF repeats outside the Notch ligand-binding domain. However, Lfng Tg+ Notch112f/f mice had slightly smaller DN thymocyte pools (p < 0.05) with fewer B cells (p < 0.05) than Lfng Tg+ Notch112f/+ mice. Thus, Lfng overexpression had less pronounced developmental effects in Notch112f/f relative to Notch112f/+ mice, indicating that Lfng modification of O-fucose on EGF12 contributes to the developmental impact of Lfng overexpression in this system. Collectively, these data show that Lfng modification of EGF12 contributed to, but was not solely responsible for, the developmental effects of Lfng over- and misexpression in this system.
Lfng enhances DP thymocyte generation from DN3b progenitors with pre-TCR signaling defects
It has been suggested that inefficient DP thymocyte generation by Ptcra-deficient DN3 thymocytes, particularly under competitive conditions, reflects a poor ability to compete for Notch ligands (20). This model would predict that Lfng overexpression should greatly improve DP thymocyte production from Ptcra-deficient DN3s, because Lfng enhances DN3b thymocyte competition for intrathymic DL ligands. Therefore, we assessed the capacity of transgenic Lfng to improve DP thymocyte generation from DN3 thymocytes lacking Ptcra or Lck. Lck-deficient DN3 progenitors can only transduce pre-TCR signals using the less effective Fyn Src family kinase, leading to a 10- to 50-fold reduction in the size of the DP thymocyte pool (16, 17). Thus, DN3b thymocytes from Lck−/− mice have an intrinsic defect in signaling through Ptcra-containing pre-TCR complexes, whereas those from Ptcra−/− mice express Ptcra-deficient TCR complexes that promote the DN3b to DP transition inefficiently.
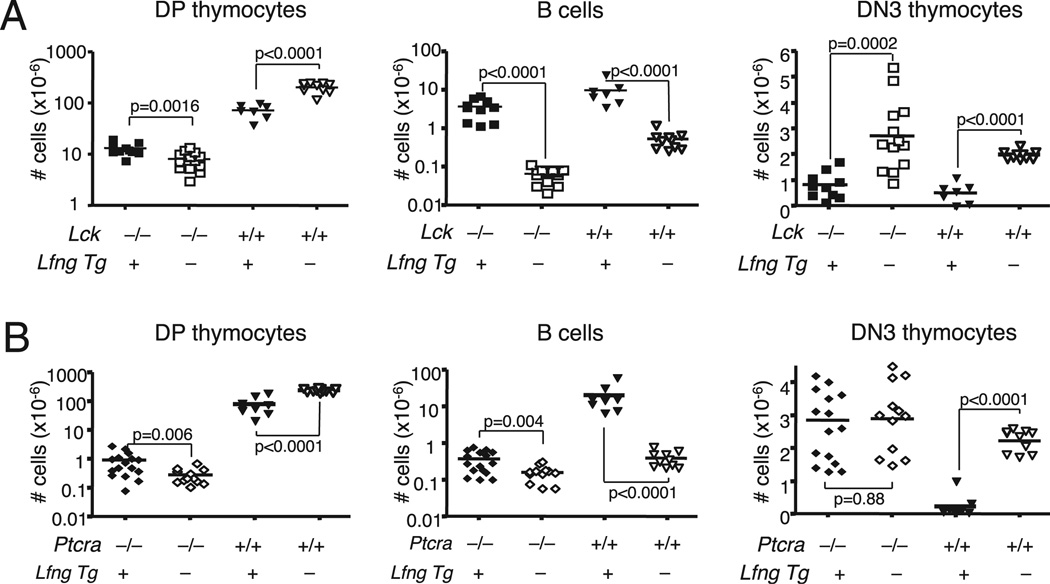
To determine whether Lfng could improve DP thymocyte generation from DN3 thymocytes with defects in pre-TCR expression (Ptcra−/−) or signaling (Lck−/−), we generated Lfng Tg+ Lck−/− and Lfng Tg+Ptcra−/− mice and first assessed the impact of the Lfng transgene on the numbers of different thymocyte subsets at steady state. We found that transgenic Lfng induced a small but statistically significant increase in the number of DP thymocytes in Lfng Tg+ WT, Lfng Tg+Lck−/−, and Lfng Tg+ Ptcra−/− mice (Fig. 4A, 4B, Table I). However, as we noted previously, transgenic Lfng decreased the size of DN3 thymocyte pools in WT mice because Lfng Tg+ DP thymocytes dominantly block CMJ niches and divert many TSPs to the B cell fate (5, 34). Similarly, Lfng Tg+Lck−/− mice also had abnormally large intrathymic B cell pools and small DN3 thymocyte pools relative to their Tg−Lck−/− littermates (Fig. 4A). Thus, although loss of Lck reduced the size of the DP thymocyte pool by ~90%, this small number of Lfng Tg+ DP progenitors could still block Notch activation in TSPs. In contrast, Lfng overexpression caused a very minor increase in intrathymic B cells in Ptcra−/− mice, and their DN3 pool size was not decreased relative to Lfng Tg−Ptcra−/− mice (Fig. 4B).
FIGURE 4.
Effect of transgenic Lfng on intrathymic B cell, DN3, and DP pool sizes in Lck−/− and Ptcra−/− mice. A, Absolute numbers of DP, DN3 thymocytes, and B cells in thymi of 8- to 10 wk-old Lfng Tg+Lck−/− (n = 10), Lck−/− (n = 13), Lfng Tg+ (n = 7), and WT (n = 10) mice. B, Absolute numbers of DP, DN3 thymocytes, and B cells in thymi of 8- to 10 wk-old Lfng Tg+Ptcra−/− (n = 16), Ptcra−/− (n = 12), Lfng Tg+ (n = 9), and WT (n = 10) mice. Horizontal bars indicate means.
Table I.
Mean and SD values from experiments in Fig. 4
| No. of DP Thymocytes (×10−6) | No. of B Cells (×10−6) | No. of DN3 Thymocytes (×10−6) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fig. 4Aa | ||||||||||||
| Lck | −/− | −/− | +/+ | +/+ | −/− | −/− | +/+ | +/+ | −/− | −/− | +/+ | +/+ |
| Lfng Tg | + | − | + | − | + | − | + | − | + | − | + | − |
| Mean | 13.12 | 8.11 | 72.60 | 202.56 | 3.45 | 0.06 | 9.44 | 0.53 | 0.82 | 2.73 | 0.57 | 1.98 |
| SD | 3.55 | 3.20 | 21.60 | 42.51 | 1.92 | 0.02 | 6.97 | 0.28 | 0.50 | 1.36 | 0.38 | 0.17 |
| Fig. 4Bb | ||||||||||||
| Ptcra | −/− | −/− | +/+ | +/+ | −/− | −/− | +/+ | +/+ | −/− | −/− | +/+ | +/+ |
| Lfng Tg | + | − | + | − | + | − | + | − | + | − | + | − |
| Mean | 0.85 | 0.26 | 81.13 | 246.51 | 0.39 | 0.16 | 20.1 | 0.35 | 2.82 | 2.87 | 0.22 | 2.17 |
| SD | 0.74 | 0.15 | 55.57 | 40.77 | 0.24 | 0.08 | 16.8 | 0.17 | 1.27 | 1.05 | 0.30 | 0.34 |
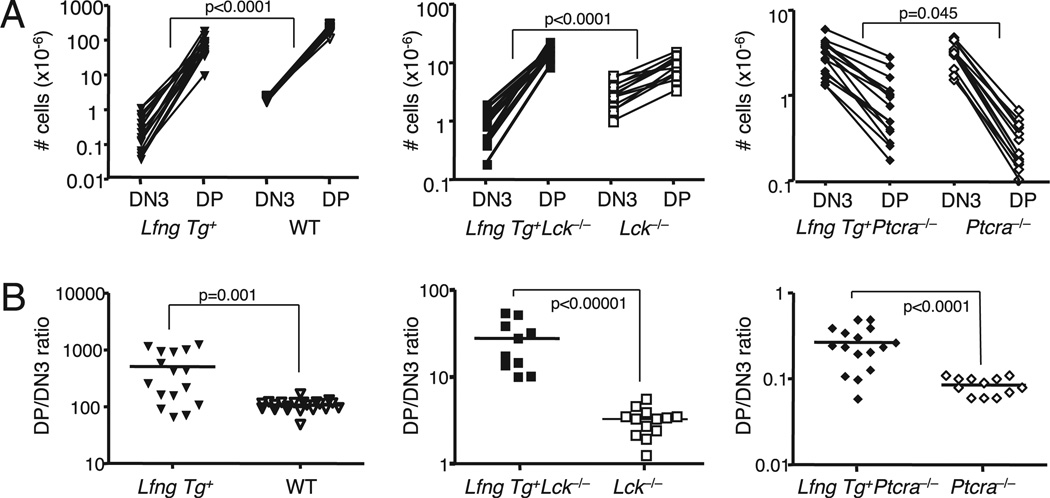
The variable ability of Lfng overexpression to divert TSPs to the B cell fate and away from the DN3 fate in the different strains made it difficult to directly assess the impact of transgenic Lfng on the DN3 to DP transition. Therefore, we compared the DP thymocyte pool size relative to the DN3 pool size in individual mice of each genotype at steady state. The slope of the lines connecting the number of DN3 precursors to the number of DP progeny thus gives a reasonable measure of survival and proliferative expansion during DN3-DP transition in each strain. The results show that transgenic Lfng significantly improved DP thymocyte production during β-selection in WT as well as in Lck−/− mice (Fig. 5A; p < 0.0001 for both comparisons). Interestingly, the lines had negative slopes in both Lfng Tg+Ptcra−/− and Ptcra−/− strains (Fig. 5B), suggesting a profound survival and/or proliferation defect during the DN3 to DP transition in this strain, as has been noted previously (37). However, Lfng overexpression allowed generation of ~3-fold more DP thymocytes from Ptcra-deficient DN3 cells (Fig. 4B, Table I), thus slightly decreasing the slope of the DN3 to DP decline in Ptcra−/− thymi (Fig. 5A). However, Lfng overexpression did not reverse the survival defect in this strain.
FIGURE 5.
Transgenic Lfng enhances DP thymocyte generation from DN3 thymocytes in Lck−/− and Ptcra−/− mice. A, Absolute numbers of DN3 and DP thymocytes, with lines connecting values from individual mice for every given genotype. B, Ratios of the number of DP and DN3 thymocytes in each thymus by genotype. Lfng Tg+ (n = 16), WT (n = 20), Lfng Tg+Lck−/− (n = 10), Lck−/− (n = 13), Lfng Tg+Ptcra−/− (n = 16), and Ptcra−/− (n = 12).
In all comparisons, the DP/DN3 ratios were also increased by expression of Lfng (Fig. 5B). For example, the average DP/DN3 ratio in WT B6 mice was 112, whereas this average was 480 for Lfng Tg+ WT mice. By comparison, transgenic Lfng improved the average DP/DN3 ratio from 3.3 to 22.4 in Lck−/− mice but only from 0.09 to 0.26 in Ptcra−/− mice. These improvements were highly significant in all three comparisons, demonstrating that Lfng increases the number of DP progeny that can be generated from each DN3b thymocyte regardless of the type of TCR or the Src family kinase through which TCR signals are transduced.
Lfng improves the competitive fitness of Lck-deficient and Ptcra-deficient DN3b thymocytes
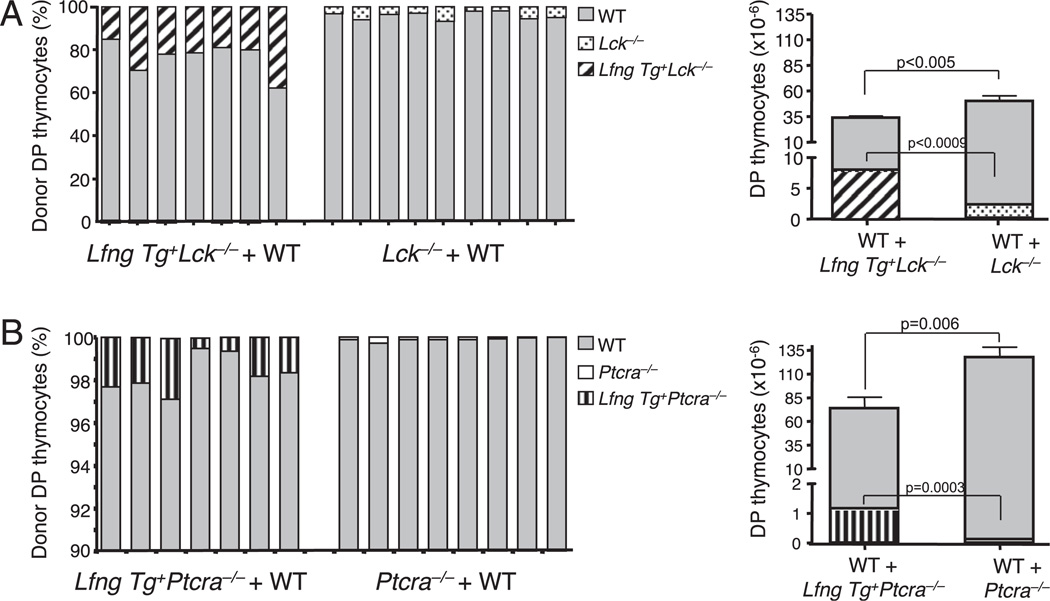
To formally test whether transgenic Lfng could improve the ability of DN3 thymocytes with defective pre-TCR expression or signaling to compete for Notch ligands in vivo, we carried out competitive repopulation experiments. We injected equal mixes of Lin− bone marrow progenitors from Lfng Tg+Lck−/− (B6 CD45.2) and WT (B6CD45.1/CD45.2) mice into thymi of sublethally irradiated B6 CD45.1 hosts. Control chimeras received equal mixtures of Lin− bone marrow cells from Lck−/− (CD45.2) and WT (B6 CD45.1/CD45.2) mice. Interestingly, Lck-deficient cells were profoundly outcompeted by WT progenitors and generated even fewer DP thymocytes in this competitive situation (1.9 ± 1 × 106) (Fig. 6A) than they did at steady state (8.1 ± 3.2 × 106) (Table I). DL ligands likely represents at least one limiting resource, because Lfng Tg+Lck−/− progeny achieved higher degrees of chimerism in the DP pool than Lck−/− progeny (12–30 versus 1–10%) (Fig. 6A, left panel). Furthermore, four times more DP thymocytes (on average) were generated from Lfng Tg+Lck−/− progenitors (8.2 × 106 cells) compared with Lck−/− progenitors (1.9 × 106 cells) in control mixed chimeras (Fig. 6A, right panel). Thus, transgenic Lfng significantly improved clonal expansion of Lck-deficient DN3b thymocytes under competitive conditions.
FIGURE 6.
Lfng improves the competitive fitness of Lck-deficient and Ptcra-deficient DN3b thymocytes. A, DP thymocyte production in Lfng Tg+ Lck−/− + WT versus Lck−/− + WT mixed chimeras. Left panel, Relative contribution of Lfng Tg+Lck−/−, Lck−/−, and WT donor cells to the DP thymocyte pool in individual thymic lobes. Right panel, Absolute numbers of DP thymocytes derived from Lfng Tg+Lck−/− (diagonal lines), Lck−/− (dots), and WT (gray) donors in individual thymic lobes. B, DP thymocyte production in Lfng Tg+Ptcra−/− + WT versus Ptcra−/− + WT mixed chimeras. Left panel, Relative contribution of Lfng Tg+Ptcra−/−, Ptcra−/−, and WT donor cells to the DP thymocyte pool in individual thymic lobes. Right panel, Absolute numbers of DP thymocytes derived from Lfng Tg+Ptcra−/− (vertical lines), Ptcra−/− (white), and WT (gray) in individual thymic lobes. Chimeras were generated and analyzed after 3 wk as described for Fig. 2.
In similar experiments, Ptcra−/− donor cells competed very poorly against WT cells and contributed only 0.01–0.3% to the DP pool, in line with previous observations made by another group (21). However, Lfng Tg+Ptcra−/− progeny achieved higher degrees of chimerism (0.5–3%) against their WT competitors (Fig. 6B, left panel) and generated on average 10 times more DP thymocytes than did Ptcra−/− donors (1.1 × 106 cells compared with 0.1 × 106 cells) (Fig. 6B, right panel). These observations demonstrate that Lfng significantly improves the ability of Ptcra-deficient and Lck-deficient DN3b thymocytes to compete for intrathymic DL Notch ligands. However, these mutant DN3b progenitors still generated very small numbers of DP thymocytes, demonstrating that defective pre-TCR signaling cannot be bypassed by enhancing competition for Notch ligands.
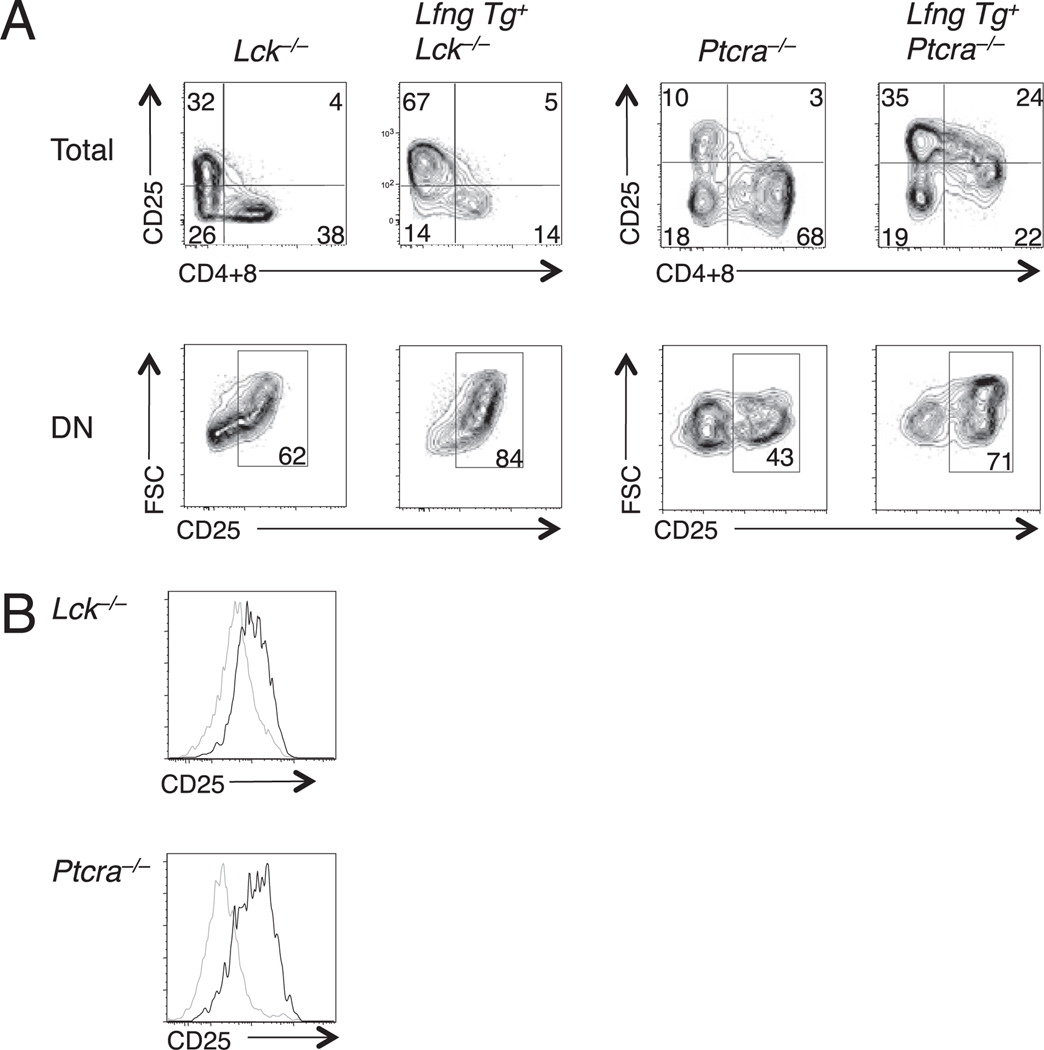
We have found that transgenic Lfng sustains DL4-induced Notch signaling for a longer time during the DN3 to DP transition of WT thymocytes, prolonging proliferation of CD25+ DN3 progenitors while delaying their differentiation to the DP stage (J.S. Yuan et al., submitted for publication). To determine whether transgenic Lfng can similarly delay differentiation and enhance self-renewal of Lck−/− or Ptcra−/− DN3 thymocytes, we cultured Lfng Tg+ and nontransgenic DN3 thymocytes from each strain on OP9-DL4 stromal cells. Examination of their progeny 7–10 d later showed that there were more CD25+ DN thymocytes and fewer CD4- and/or CD8-expressing thymocytes in cultures of Lfng Tg+Lck−/− or Lfng Tg+Ptcra−/− DN3 thymocytes relative to their nontransgenic counterparts (Fig. 7A, top panels). Furthermore, a higher proportion of DN progeny were CD25+ blasts in cultures initiated with Lfng Tg+ DN3 thymocytes (Fig. 7A, bottom panels). Finally, DP progeny of Lfng Tg+ DN3 progenitors expressed higher levels of CD25 than the DP progeny of nontransgenic DN3 progenitors (Fig. 7B). We have recently shown that DP thymocytes that develop within 1–2 d after DN4 progenitors are cocultured with OP9-DL4 that express CD25, whereas those that develop on OP9 are CD25− (J.S. Yuan et al., submitted for publication). Thus, CD25 expression in DP thymocytes is reflective of active Notch signaling, in keeping with the demonstration that CD25 is a direct Notch1 target (13). Therefore, these findings demonstrate that Lfng overexpression enhances DL4-induced Notch activation in DP thymocytes. Furthermore, these data reveal that transgenic Lfng enhances DL4-induced self-renewal of Lck−/− and Ptcra−/− DN3b thymocytes and provide a potential mechanism to explain how transgenic Lfng improves the competitive fitness of Lck−/− and Ptcra−/− DN3b thymocytes during β-selection in vivo.
FIGURE 7.
Lfng overexpression prolongs DN3 thymocyte responsiveness to DL4 in vitro. DN3 thymocytes from Lfng Tg+Lck−/−, Lck−/−, Lfng Tg+Ptcra−/−, and Ptcra−/− mice were seeded onto OP9-DL4 stromal cells and cultured for 7 and 10 d. A, Delayed differentiation of Lfng Tg+ versus Lfng Tg− thymocytes. Representative plots of CD4+CD8 versus CD25 expression profiles are shown for Lfng Tg+Ptcra−/− and Ptcra−/− DN3 progeny after 7 d of culture and after 10 d for for Lfng Tg+Lck−/− and Lck−/− DN3 progeny. B, Lfng overexpression prolongs Notch signaling in DP thymocyte progeny of Lck−/− and Ptcra−/− DN3 thymocytes. Plots show CD25 expression after 7 d of culture gated on DP progeny of Lfng Tg− (gray line) and Lfng Tg+ (black line) DN3 thymocytes from each genotype.
Discussion
In this study, we demonstrate that Lfng overexpression greatly enhances thymocyte competitive fitness specifically at the DN3b stage, dramatically enhancing DN3b contribution to the DP thymocyte pool. Lfng modification of O-fucose on EGF12 within the Notch1 ligand-binding domain contributed to, but was not solely responsible for, the developmental effects of Lfng overexpression. Previous studies have shown that Notch1 plays an important role in regulating glucose metabolism and viability of preselection DN3a thymocytes (36). Although Lfng overexpression enhanced the ability of preselection Rag2−/− DN3a thymocytes to bind DL4, it did not provide these cells with a competitive advantage in vivo. Similarly, Lfng overexpression did not provide WT DN3a thymocytes with a competitive advantage in mixed chimeras. These findings suggest that preselection DN3a thymocytes do not compete for DL4 to maintain their glucose metabolism and viability in vivo. Collectively, these data highlight DN3b thymocytes as critical targets of Notch-mediated regulation during β-selection in vivo and demonstrate that Lfng maximizes Notch-induced clonal expansion of these early products of β-selection. This notion was independently confirmed by our recent demonstration that DN3 thymocytes conditionally lacking Lfng generate very small numbers of DP thymocytes in vivo (J.S. Yuan et al., submitted for publication).
Ectopic expression of Lfng in WT DP thymocytes causes inappropriate binding of DL4 ligands and blocks TSP from DL4-induced Notch1 activation in the CMJ (34). Therefore, Lfng Tg+ thymi are characterized by a massive accumulation of B cells, decreased ETP generation, and a reduction of the DN3 thymocyte pool. A similar phenotype was observed in Lfng Tg+Lck−/− mice but not in Lfng Tg+Ptcra−/− mice. This suggests that the pool of ~107 DP thymocytes in Lfng Tg+Lck−/− thymi can saturate DL4 in the CMJ niche, whereas the smaller pool of ~106 DP thymocytes in Lfng Tg+Ptcra−/− thymi are not sufficient to saturate DL4 in CMJ niches. Thus, it takes only ~10% of the normal number of DP thymocytes expressing Lfng to block TSP interactions with DL4 in CMJ niches. These data highlight the importance of Lfng downregulation in DP thymocytes.
The reliance of DN3b but not DN3a thymocytes on Lfng–Notch1 interactions to enhance their competitive fitness has several possible explanations. DN3a thymocytes may reside in a thymic niche that provides ample amounts of DL4, whereas DN3b thymocytes may reside in a distinct intrathymic niche characterized by low abundance of DL4. However, because most CD25+ DN thymocytes reside in the subcapsular zone of the thymic cortex (38), this explanation seems unlikely. Another possibility is that weak or low-threshold Notch1 signals may be sufficient to maintain glucose metabolism in resting DN3a thymocytes, whereas much stronger, Lfng-enhanced Notch1 signals may be required to induce proliferation of DN3b thymocytes at the onset of β-selection. Alternatively or in addition, Lfng may be particularly critical at the DN3b stage because these progenitors express slightly lower levels of Notch1 than DN3a thymocytes (19, 39).
A previous study suggested that Notch independence emerges by the DN3b thymocyte stage because these progenitors generated DP thymocytes in the absence of Notch ligands in vitro, whereas DN3a progenitors did not (19). However, DN3b and DN4 thymocytes remain highly responsive to DL Notch ligands, and many more DP thymocytes are generated when DN3b or DN4 thymocytes are cultured on OP9-DL1 or OP9-DL4 than when they are cultured on OP9 stromal cells (20) (J.S. Yuan, submitted for publication). Furthermore, overexpression of Lfng in DN3 and DN4 thymocytes prolongs their proliferation in response to DL ligands in vitro (J.S. Yuan et al., submitted for publication). Finally, we show in this study that Lfng overexpression greatly increases the competitive fitness of DN3b thymocytes, suggesting that they are highly Notch1 dependent in vivo.
If Lfng-modified Notch1 signaling is a major driver of proliferation during β-selection, why does defective pre-TCR signaling also result in very small numbers of DP thymoctes? It has been suggested that this reflects a reduced ability of pre–TCR-deficient DN3b progenitors to compete for Notch ligands (20). Indeed, Lfng overexpression increased the competitive fitness of Lck−/− and Ptcra−/− DN3 thymocytes, demonstrating that Lfng can enhance competition for DL ligands during the DN3-DP transition when pre-TCR expression or signaling is defective. However, the effect of Lfng overexpression was minor, because Lfng Tg+Ptcra−/− thymi still contained many fewer DP thymocytes than DN3 progenitors. Thus, improving the capacity of Ptcra-deficient DN3s to bind and compete for DL ligands compensated only slightly for the intrinsically poor efficiency with which Ptcra-deficient TCRs promote the DN3-DP transition. These data argue against the notion that DP thymocyte production from Ptcra-deficient DN3 progenitors reflects inefficient competition for Notch ligands. Rather, they indicate that pre-TCR and Notch signaling have largely nonoverlapping functions in β-selection. The pre-TCR appears to have unique functions in promoting differentiation during the DN3b to DP transition, whereas Lfng enhances Notch signaling to promote robust proliferation of pre-TCR+ thymocytes before they differentiate to the DP stage.
Acknowledgments
We thank Freddy Radtke for the DL4-Fc construct, Juan-Carlos Zuniga-Pflucker for OP9-DL1 cells, and Wen Dong for excellent technical assistance.
This work was supported by operating Grant FRN 11530 from the Canadian Institutes of Health Research (to C.J.G.) and the U.S. National Institutes of Health/National Cancer Institute Grant R01 95022 (to P.S.). Partial support to P. S. was also provided by National Cancer Institute Grant P01 13330 to the Albert Einstein Cancer Center. J.S.Y. and I.V. were supported by the Research Training Competition fund from the Hospital for Sick Children Research Institute. I.V. was also supported by a postdoctoral fellowship award from the Canadian Institutes of Health Research.
Abbreviations used in this paper
- CMJ
corticomedullary junction
- DL
Delta-like
- DN
double-negative
- DP
double-positive
- EGF
epidermal growth factor
- ETP
early thymic progenitor
- Lfng
Lunatic Fringe
- TSP
thymus-seeding progenitor
- WT
wild-type
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Yuan JS, Kousis PC, Suliman S, Visan I, Guidos CJ. Functions of notch signaling in the immune system: consensus and controversies. Annu. Rev. Immunol. 2010;28:343–365. doi: 10.1146/annurev.immunol.021908.132719. [DOI] [PubMed] [Google Scholar]
- 2.Koch U, Lacombe TA, Holland D, Bowman JL, Cohen BL, Egan SE, Guidos CJ. Subversion of the T/B lineage decision in the thymus by lunatic fringe-mediated inhibition of Notch-1. Immunity. 2001;15:225–236. doi: 10.1016/s1074-7613(01)00189-3. [DOI] [PubMed] [Google Scholar]
- 3.Wilson A, MacDonald HR, Radtke F. Notch 1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J. Exp. Med. 2001;194:1003–1012. doi: 10.1084/jem.194.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sambandam A, Maillard I, Zediak VP, Xu L, Gerstein RM, Aster JC, Pear WS, Bhandoola A. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat. Immunol. 2005;6:663–670. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- 5.Tan JB, Visan I, Yuan JS, Guidos CJ. Requirement for Notch1 signals at sequential early stages of intrathymic T cell development. Nat. Immunol. 2005;6:671–679. doi: 10.1038/ni1217. [DOI] [PubMed] [Google Scholar]
- 6.Balciunaite G, Ceredig R, Rolink AG. The earliest subpopulation of mouse thymocytes contains potent T, significant macrophage, and natural killer cell but no B-lymphocyte potential. Blood. 2005;105:1930–1936. doi: 10.1182/blood-2004-08-3087. [DOI] [PubMed] [Google Scholar]
- 7.Benz C, Bleul CC. A multipotent precursor in the thymus maps to the branching point of the T versus B lineage decision. J. Exp. Med. 2005;202:21–31. doi: 10.1084/jem.20050146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taghon T, Yui MA, Rothenberg EV. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat. Immunol. 2007;8:845–855. doi: 10.1038/ni1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 10.Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, Kawamoto H. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- 11.Feyerabend TB, Terszowski G, Tietz A, Blum C, Luche H, Gossler A, Gale NW, Radtke F, Fehling HJ, Rodewald HR. Deletion of Notch1 converts pro-T cells to dendritic cells and promotes thymic B cells by cell-extrinsic and cell-intrinsic mechanisms. Immunity. 2009;30:67–79. doi: 10.1016/j.immuni.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Wolfer A, Wilson A, Nemir M, MacDonald HR, Radtke F. Inactivation of Notch1 impairs VDJbeta rearrangement and allows pre-TCR-independent survival of early alpha beta Lineage Thymocytes. Immunity. 2002;16:869–879. doi: 10.1016/s1074-7613(02)00330-8. [DOI] [PubMed] [Google Scholar]
- 13.Maillard I, Tu L, Sambandam A, Yashiro-Ohtani Y, Millholland J, Keeshan K, Shestova O, Xu L, Bhandoola A, Pear WS. The requirement for Notch signaling at the beta-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J. Exp. Med. 2006;203:2239–2245. doi: 10.1084/jem.20061020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Boehmer H. Unique features of the pre-T-cell receptor alpha-chain: not just a surrogate. Nat. Rev. Immunol. 2005;5:571–577. doi: 10.1038/nri1636. [DOI] [PubMed] [Google Scholar]
- 15.Yamasaki S, Ishikawa E, Sakuma M, Ogata K, Sakata-Sogawa K, Hiroshima M, Wiest DL, Tokunaga M, Saito T. Mechanistic basis of pre-T cell receptor-mediated autonomous signaling critical for thymocyte development. Nat. Immunol. 2006;7:67–75. doi: 10.1038/ni1290. [DOI] [PubMed] [Google Scholar]
- 16.Groves T, Smiley P, Cooke MP, Forbush K, Perlmutter RM, Guidos CJ. Fyn can partially substitute for Lck in T lymphocyte development. Immunity. 1996;5:417–428. doi: 10.1016/s1074-7613(00)80498-7. [DOI] [PubMed] [Google Scholar]
- 17.van Oers NS, Lowin-Kropf B, Finlay D, Connolly K, Weiss A. alpha beta T cell development is abolished in mice lacking both Lck and Fyn protein tyrosine kinases. Immunity. 1996;5:429–436. doi: 10.1016/s1074-7613(00)80499-9. [DOI] [PubMed] [Google Scholar]
- 18.Hoffman ES, Passoni L, Crompton T, Leu TM, Schatz DG, Koff A, Owen MJ, Hayday AC. Productive T-cell receptor beta-chain gene rearrangement: coincident regulation of cell cycle and clonality during development in vivo. Genes Dev. 1996;10:948–962. doi: 10.1101/gad.10.8.948. [DOI] [PubMed] [Google Scholar]
- 19.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging beta- and gammadelta-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 20.Garbe AI, Krueger A, Gounari F, Zúñiga-Pflücker JC, von Boehmer H. Differential synergy of Notch and T cell receptor signaling determines alphabeta versus gammadelta lineage fate. J. Exp. Med. 2006;203:1579–1590. doi: 10.1084/jem.20060474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borowski C, Li X, Aifantis I, Gounari F, von Boehmer H. Pre-TCRalpha and TCRalpha are not interchangeable partners of TCRbeta during T lymphocyte development. J. Exp. Med. 2004;199:607–615. doi: 10.1084/jem.20031973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prockop SE, Petrie HT. Regulation of thymus size by competition for stromal niches among early T cell progenitors. J. Immunol. 2004;173:1604–1611. doi: 10.4049/jimmunol.173.3.1604. [DOI] [PubMed] [Google Scholar]
- 23.Stanley P. Regulation of Notch signaling by glycosylation. Curr. Opin. Struct. Biol. 2007;17:530–535. doi: 10.1016/j.sbi.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohtashami M, Zúñiga-Pflücker JC. Three-dimensional architecture of the thymus is required to maintain Delta-like expression necessary for inducing T cell development. J. Immunol. 2006;176:730–734. doi: 10.4049/jimmunol.176.2.730. [DOI] [PubMed] [Google Scholar]
- 25.Lehar SM, Dooley J, Farr AG, Bevan MJ. Notch ligands Delta1 and Jagged1 transmit distinct signals to T cell precursors. Blood. 2005;105:1440–1447. doi: 10.1182/blood-2004-08-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciofani M, Schmitt TM, Ciofani A, Michie AM, Cuburu N, Aublin A, Maryanski JL, Zúñiga-Pflücker JC. Obligatory role for cooperative signaling by pre-TCR and Notch during thymocyte differentiation. J. Immunol. 2004;172:5230–5239. doi: 10.4049/jimmunol.172.9.5230. [DOI] [PubMed] [Google Scholar]
- 27.Huang EY, Gallegos AM, Richards SM, Lehar SM, Bevan MJ. Surface expression of Notch1 on thymocytes: correlation with the double-negative to double-positive transition. J. Immunol. 2003;171:2296–2304. doi: 10.4049/jimmunol.171.5.2296. [DOI] [PubMed] [Google Scholar]
- 28.Moloney DJ, Shair LH, Lu FM, Xia J, Locke R, Matta KL, Haltiwanger RS. Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. J. Biol. Chem. 2000;275:9604–9611. doi: 10.1074/jbc.275.13.9604. [DOI] [PubMed] [Google Scholar]
- 29.Rampal R, Li AS, Moloney DJ, Georgiou SA, Luther KB, Nita-Lazar A, Haltiwanger RS. Lunatic fringe, manic fringe, and radical fringe recognize similar specificity determinants in O-fucosylated epidermal growth factor-like repeats. J. Biol. Chem. 2005;280:42454–42463. doi: 10.1074/jbc.M509552200. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Shao L, Shi S, Harris RJ, Spellman MW, Stanley P, Haltiwanger RS. Modification of epidermal growth factor-like repeats with O-fucose: molecular cloning and expression of a novel GDP-fucose protein O-fucosyltransferase. J. Biol. Chem. 2001;276:40338–40345. doi: 10.1074/jbc.M107849200. [DOI] [PubMed] [Google Scholar]
- 31.Panin VM, Shao L, Lei L, Moloney DJ, Irvine KD, Haltiwanger RS. Notch ligands are substrates for protein O-fucosyltransferase-1 and Fringe. J. Biol. Chem. 2002;277:29945–29952. doi: 10.1074/jbc.M204445200. [DOI] [PubMed] [Google Scholar]
- 32.Haines N, Irvine KD. Glycosylation regulates Notch signalling. Nat. Rev. Mol. Cell Biol. 2003;4:786–797. doi: 10.1038/nrm1228. [DOI] [PubMed] [Google Scholar]
- 33.Ge C, Stanley P. The O-fucose glycan in the ligand-binding domain of Notch1 regulates embryogenesis and T cell development. Proc. Natl. Acad. Sci. USA. 2008;105:1539–1544. doi: 10.1073/pnas.0702846105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Visan I, Tan JB, Yuan JS, Harper JA, Koch U, Guidos CJ. Regulation of T lymphopoiesis by Notch1 and Lunatic fringe-mediated competition for intrathymic niches. Nat. Immunol. 2006;7:634–643. doi: 10.1038/ni1345. [DOI] [PubMed] [Google Scholar]
- 35.Fehling HJ, Krotkova A, Saint-Ruf C, von Boehmer H. Crucial role of the pre-T-cell receptor α gene in development of αβ but not γδ T cells. Nature. 1995;375:795–798. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- 36.Ciofani M, Zúñiga-Pflücker JC. Notch promotes survival of pre-T cells at the β-selection checkpoint by regulating cellular metabolism. Nat. Immunol. 2005;6:881–888. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- 37.Aifantis I, Mandal M, Sawai K, Ferrando A, Vilimas T. Regulation of T-cell progenitor survival and cell-cycle entry by the pre-T-cell receptor. Immunol. Rev. 2006;209:159–169. doi: 10.1111/j.0105-2896.2006.00343.x. [DOI] [PubMed] [Google Scholar]
- 38.Porritt HE, Gordon K, Petrie HT. Kinetics of steady-state differentiation and mapping of intrathymic-signaling environments by stem cell transplantation in nonirradiated mice. J. Exp. Med. 2003;198:957–962. doi: 10.1084/jem.20030837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yashiro-Ohtani Y, He Y, Ohtani T, Jones ME, Shestova O, Xu L, Fang TC, Chiang MY, Intlekofer AM, Blacklow SC, et al. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 2009;23:1665–1676. doi: 10.1101/gad.1793709. [DOI] [PMC free article] [PubMed] [Google Scholar]