Abstract
The recent development and application of molecular genetics to the symbionts of invertebrate animal species have advanced our knowledge of the biochemical communication that occurs between the host and its bacterial symbionts. In particular, the ability to manipulate these associations experimentally by introducing genetic variants of the symbionts into naive hosts has allowed the discovery of novel colonization mechanisms and factors. In addition, the role of the symbionts in inducing normal host development has been revealed, and its molecular basis described. In this Review, I discuss many of these developments, focusing on what has been discovered in five well-understood model systems.
This is an exciting time for biologists, and for microbiologists in particular. We are at the convergence of two breakthroughs that are advancing our understanding of how animals and plants live with their microbiota. The first of these advances is the development of methods for dissecting the genetic mechanisms by which organisms signal and respond to each other. The second advance is conceptual: the recognition that higher organisms create a shared living space with a specific set of beneficial microorganisms. Together, these two developments have made it possible to begin to understand how animals and plants communicate with the many bacterial species that live in and on their tissues. Describing the genetic basis of this symbiotic conversation has become a new frontier of biology.
Recent research is expanding to identify and embrace the diversity of microbial symbioses in living systems (FIG. 1). This diversity includes associations in which bacteria perform conserved functions that are common to the needs of many host species (for example, digestive activities1), as well as those in which they perform unusual functions (for example, bioluminescence2) that have specifically evolved in a few host species. Included in this Focus issue are contributions that address a range of symbioses in depth and describe specific phylogenetic groups and metabolic processes. For example, Parniske3 discusses plant-microorganism associations, whereas Werren and colleagues4 concentrate on obligate intracellular partnerships and Ley and colleagues5 discuss complex microbial consortia in vertebrates. By contrast, this Review focuses on a set of experimentally accessible, genetically developed systems that consist of a natural association between an invertebrate host and one or a few bacterial symbionts. These associations illustrate how the application of molecular genetics and genomics to a number of biologically diverse symbioses is revealing the nature of the conversation by which an animal and its microbiota initiate and maintain a shared existence.
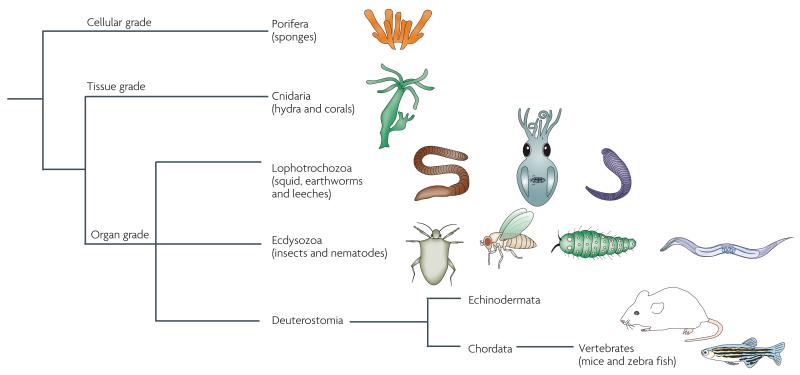
Figure 1. Microbial symbioses occur throughout the phylogeny of animals.
Experimentally accessible associations, including several that are described in this Review, occur in all the main phylogenetic groups. These associations span the breadth of animal diversity, and are represented in cellular-grade, tissue-grade and organ-grade levels of developmental and morphological complexity.
The value of natural experimental models
An emerging awareness of the role of beneficial microorganisms in human health has led to a recent increase in interest in symbioses6,7. For example, there has been success in using gnotobiotic animals that were inoculated with specific combinations of microbial species to dissect the processes that underlie the complex enteric consortia of vertebrates8. These studies, which use constructed systems (FIG. 2) that are artificially simplified to focus on a specific set of events, have already revealed the role of specific bacteria in modulating such diverse and important health issues as obesity and immune dysfunction9-12. As a result of these discoveries, there has been a widespread re-evaluation of the extent to which microorganisms may influence other, as yet unrecognized, aspects of host physiology9.
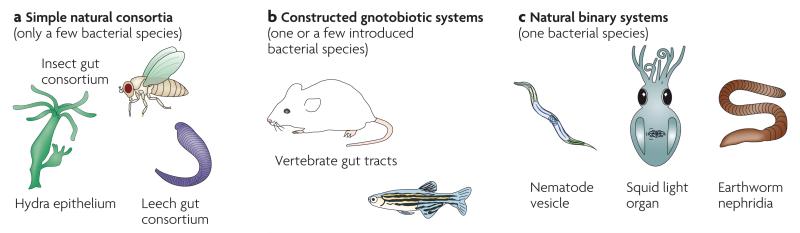
Figure 2. Classes of symbiosis models.
Experimental models of microbial symbioses can be characterized into three types. Gnotobiotic systems (a) have been useful for examining the interactions within the complex consortia that are normally present in vertebrate enteric tracts. In these systems, germ-free host animals are produced, and one or a few bacterial species are introduced to allow an examination of a simplified relationship. An alternative approach is to investigate consortia of invertebrates (b), which are often simpler in species composition. Finally, there are several natural animal models (c) in which only a single bacterial species is present.
A second kind of model that is used in biology, natural systems, allows us to exam how interactions function in the context in which they evolved. In contrast to the genetically modified or inbred host lines that are developed for constructed systems, natural models purposely use a population of genetically heterogeneous hosts to provide insight into the natural range of responses that characterize normal animal populations. However, although these systems may be natural, they are most useful when they have experimentally valuable characteristics, including culturable partners that can be maintained separately from each other. For some symbioses that are established through horizontal transfer, the newly hatched host can survive without its symbiont (aposymbiotic) and the symbiont (or symbionts) can be grown under axenic conditions. such conditions present the opportunity to experimentally study the initiation of the association. Alternatively, in those cases in which mature hosts can be cured of their microbial symbionts and artificially reinfected with novel strains, the persistence of the association can be followed. In any event, if the association is binary (one host and one symbiont species) or consists of a simple bacterial consortium13, it can be easier to focus on a specific set of relationships or events.
Numerous other attributes that allow for technological applications (BOX 1) have been exploited in a range of natural symbioses, but one that has recently begun to open many questions to experimental evaluation is the ability to genetically manipulate one or more of the symbiotic partners and subsequently reconstitute the association with these variants. This genetic level of manipulation has usually been developed in the bacterium, although there are exceptions14. However, in all cases, the desired result is to be able to build and interrogate testable models of the nature of the interaction.
Box 1. Valuable characteristics in a genetic model of symbiosis.
An inexpensive and easily collected or bred host.
A small host with a simple morphology.
Easily imaged symbiotic structures.
Known nutritional characteristics.
A range of ecologically or evolutionarily distinct associations.
An association that is economically important.
An association that is representative of a general biological principle (or principles).
A host and bacterium that can be cultured separately (grown in the laboratory).
A host and bacterium for which genome sequences are available.
An association in which one or both partners are amenable to genetic manipulation.
Over the past 20 years a number of new systems have been developed to study animal-bacteria symbioses15. But why should we promote the development of so many different models? In addition to the ability of an individual model to reveal evolutionary novelties or clearly conserved mechanisms, each model allows a distinct set of difficult questions to be addressed, which when combined with other models provides opportunities that would not be available from any one system6,16. The importance of using a range of different models was illustrated by a study of animal development, in which more than a dozen species were assessed to investigate how animals control the mechanisms by which an individual is built from a single cell. The number of different models that are available has been crucial to the advance of developmental biology: at least seven of these models have directly led to discoveries that were recognized by nobel Prizes (TABLE 1). It is clear that each of these different animal models has provided a unique opportunity to better describe a fundamental process in developmental biology.
Table 1. Examples of Nobel Prize awards in developmental biology*.
| Year‡ | Recipient (or recipients) | Discovery | Model organism§ |
|---|---|---|---|
| 1935 | H. Spemann | The ‘organizer center’ concept∥ | Newt and frog |
| 1995 | E. Lewis, C. Nusslein–Volhard and E. Wieschaus | Homeobox genetic organization | Fruit fly and zebra fish |
| 2001 | L. Hartwell, T. Hunt and P. Nurse | Cyclin regulation of the cell cycle | Sea urchin and frog |
| 2002 | S. Brenner, H. Horvitz and J. Sulston | Programmed cell death | Roundworm |
| 2006 | A. Fire and C. Mello | RNA interference | Roundworm |
| 2007 | M. Capecchi, M. Evans and O. Smithies | Embryonic stem-cell development | Mouse |
Information obtained from Nobelprize.org (see Further information).
Of the past 13 Nobel Prizes awarded in Physiology or Medicine, 5 were from the area of developmental biology.
In several cases, results from more than one model system were specifically recognized.
The first Nobel Prize to be awarded in developmental mechanics (developmental biology).
Examples of beneficial symbioses
With only a few known exceptions (for example, light-organ symbioses of marine fish2), beneficial associations between vertebrates and bacteria exist as complex consortia of tens to hundreds of species6. By contrast, most beneficial symbioses in invertebrates are monospecific or constitute simple (<10 species) consortia, perhaps owing to the limited ability of their hosts to carry out immunological surveillance17. Although most beneficial symbionts of insects are obligately intracellular and are passed vertically through the maternal line18, many bacteria that are associated with these and other invertebrates are passed horizontally, and therefore must be able to live in the external environment. It is therefore not surprising that most of the currently recognized, genetically tractable symbionts that can be readily cultured are horizontally passed, heterotrophic bacteria. Described below are five beneficial symbioses of invertebrate hosts that have been examined by genetically manipulating at least one bacterial species. Three of these systems are normally found as binary associations of a monospecific symbiont population that grows within a specialized tissue of its host, whereas two consist of simple consortia that are found in an animal’s enteric tract (FIG. 3).
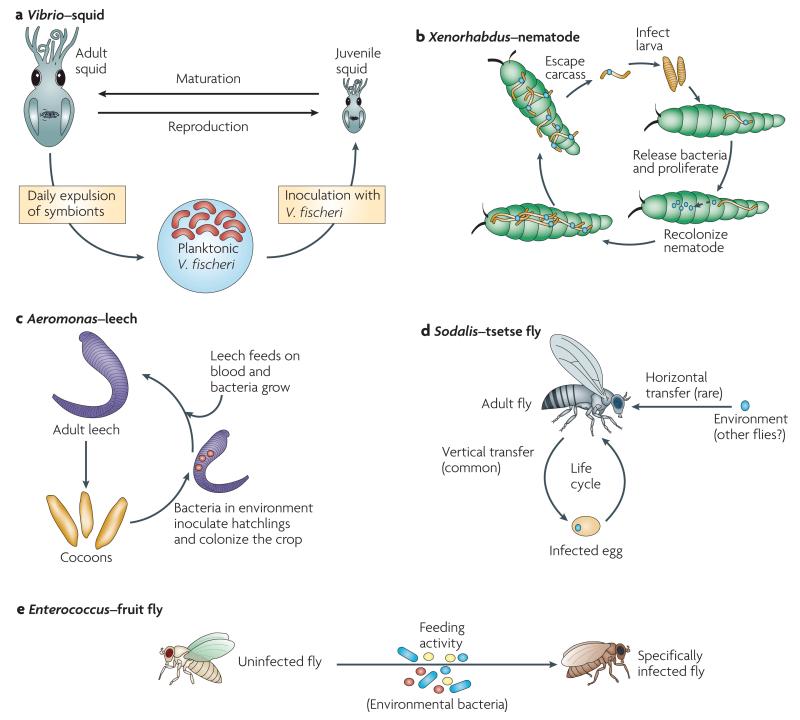
Figure 3. Simplified life cycles of five symbioses.
In each of the symbioses shown, the animal obtains a specific symbiont (or symbionts), which colonizes the host in a particular location. a | The squid obtains its symbionts from sea-water populations, which colonize the nascent light organ. b | The nematode brings its symbiont into the insect host, where both proliferate. The bacteria then recolonize the nematodes, which escape from the carcass. c | Juvenile leeches obtain symbionts after hatching from their cocoon (perhaps from the cocoon itself). They then take up residence in the crop, where they digest their blood meal. d | The tsetse fly can either pass the symbionts maternally to the eggs or pick up new strains from the environment. e | Specific symbionts on the food of the fruit fly colonize and persist in the enteric tract.
Vibrio and sepiolid squid
Many species of marine animals are bioluminescent, and approximately half of these generate their light by developing associations with luminous bacteria in the genera Vibrio or Photobacterium2,19. The best studied of the light-emitting symbioses is that between Vibrio fischeri and the sepiolid squid Euprymna scolopes, although other species of sepiolids that are symbiotic with V. fischeri and/or the related Vibrio logei have also been described20,21. In these symbioses, the host is thought to use the luminescence in a behaviour called counterillumination22. The E. scolopes association is horizontally passed between generations of hosts23, which must individually obtain their bacterial symbionts from environmental populations that live in the ambient sea water24. Thus, the newly hatched juvenile squid is aposymbiotic and, because bioluminescence is not a nutritional product, the host can be maintained in a free-living form in the laboratory for generations25. similarly, V. fischeri cells are easily grown in culture, and can be genetically engineered using recently developed techniques (for example, REFS 26,27). In addition, the completion of two V. fischeri genomes28,29 and associated microarrays30,31, as well as a library of expressed sequence tags32 and a microarray chip for the host squid33, have ushered in an era of genomic-level analysis in both partners of this symbiosis.
Xenorhabdus-Photorhabdus and nematode worms
One of the best-developed systems for the study of beneficial symbioses is a series of specific associations that have evolved between two genera of nematode worms and one of two genera from the Enterobacteriaceae16. These associations share a common biological function: they allow the partnership to infect, kill and grow within insect larvae. As many of the infected insect species are crop pests, an understanding of the basis of bacteria-nematode symbiosis and its potential in biological control is of considerable interest in agriculture. Two pairs of bacteria-host genera are known: Xenorhabdus-Steinernema34 and Photorhabdus–Heterorhabditis35. Although these interactions have distinct features, they share many developmental and genetic characteristics16. Interestingly, in each interaction, the bacterium is highly specific for its particular nematode species, whereas the insect larvae from both taxa are susceptible to both species of nematode.
The nematodes exist in soil in a resting form, and carry dozens of symbionts in a region of their upper enteric tract. In Xenorhabdus, this region consists of a specialized structure called the vesicle. Although initially dormant, this vesicle migrates into the blood system and begins to ingest blood when the nematode invades a larval insect. The nematode then expels its bacterial symbionts, which produce extracellular toxins and degradative enzymes, and proliferate, providing a food source that supports the reproduction of an increasing population of worms. When the insect carcass is depleted of nutrients, the worms revert back into the resting form, and the nematode vesicle becomes colonized by a few cells of the bacterial population36. The nematodes then escape into the soil, where they await the next insect host. Knowledge of the process by which the bacteria colonize their nematode host, and in particular the genetic basis of specificity and development in the bacterial symbionts, has increased markedly in recent years37-39. Although there is considerable interest in the genetic basis of the insect-parasitism stage of the bacteria-nematode life cycle, which has been aided by the development of a tripartite model system that targets larvae of the genetically facile fruit fly Drosophila melanogaster35, the emphasis of this Review is on the beneficial association between the worm and its bacterial symbiont. nevertheless, it should be noted that there is some overlap between the genetic requirements for pathogenic and beneficial symbiosis39.
Sodalis glossinidius and the tsetse fly
Many, if not most, insect species maintain intracellular bacteria that provide essential metabolic and developmental activities for their hosts40,41. These organisms are classified as either primary symbionts (for example, species of Buchnera and Wigglesworthia), which are uniformly present in the host, but are not culturable in their free-living form, or secondary symbionts, which occur more sporadically and in several cases have been grown outside of the host40,42. Primary symbionts are always found in the specialized tissue called the bacteriome, whereas secondary symbionts can occur in several different tissues of the host. As for primary symbionts, secondary symbionts are generally inherited maternally. However, secondary symbionts retain their ability to pass to a new host from either the environment or another host40. Sodalis glossinidius, which is thought to provide a beneficial effect to its specific host, the tsetse fly, is of considerable epidemiological interest as a potential vector that can be engineered by paratransgenetics to artificially produce anti-trypanosomal proteins43. This symbiont is closely related to well-studied enteric species, and microarray-based analyses have shown that its genome is surprisingly similar to that of Escherichia coli44. In addition, because S. glossinidius is not essential to its host, it is possible to eliminate the native bacteria within a fly by antibiotic treatment and reinfect it with genetically modified strains43.
Aeromonas, Rikenella and the leech
The diversity and specificity of invertebrate enteric microbiota have begun to be examined using culture-independent techniques, and, with the exception of specialized cellulosic species, such as termites45, these communities seem to be generally composed of simple consortia of fewer than ten species17,46. one such invertebrate is the medicinal leech Hirudo verbana, the diet of which is restricted to vertebrate blood. Because of the initial activity of complement in the ingested blood47, the ingestion of susceptible bacteria by leech haemocytes48 and possibly other leech- or symbiont-produced antimicrobial compounds49, only two species of bacteria (from the genera Aeromonas and Rikenella) are long-term inhabitants of vertebrate blood50. A higher number of species can be detected downstream of the crop in the intestinum, where the blood is digested, but even there Aeromonas and Rikenella remain the most abundant microbial species13.
The role (or roles) of these symbionts in the leech remains unclear, but they might foster the nutrition and/or development of the host. Aeromonas spp. are easily cultured and produce many exoenzymes, whereas Rikenella spp. are fastidiously obligate anaerobes. Interestingly, when in the crop, cells of the two species associate tightly with each other, forming mixed microcolonies that are embedded in a polysaccharide matrix. In addition, the growth of one species is enhanced by the presence of the other50. Taken together, these observations suggest a synergistic interaction between the bacteria partners in this symbiosis.
Enterococcus and the fruit fly
Considerable interest has developed in determining the nature of the microorganisms that inhabit the enteric tracts of insects, and particularly the high number of species that are pests of environmentally and agriculturally important plants51. surprisingly, it seems that despite the high number of different host species that have been examined species of the genus Enterococcus are typically among the natural members of the microbiota. This pattern is apparent even when specimens of D. melanogaster from laboratory-maintained stocks are compared with those from wild-caught populations14. When the anatomical distribution of these bacteria was determined in fruit flies, they were found to be restricted to portions of the foregut, midgut and hindgut. As for Aeromonas spp., native Enterococcus symbionts can be cured, and the consortium re-established using other genetically modified strains. using this approach, cox and Gilmore14 recently showed that an Enterococcus faecalis strain which was engineered to produce a non-native haemolysin was lethal to its host.
Application of molecular genetics
What key mechanisms underlie the development of a beneficial symbiosis? specifically, how have these mechanisms evolved to permit a host and its microbiota to effectively communicate during the initiation, accommodation and subsequent persistence of their symbiotic association (FIG. 3)? This is a dialogue in which the words and phrases are biochemical. However, the conversation as a whole is organized at the genetic and even genomic level. Approaches that apply molecular genetics to help us dissect and define the steps during a pathogenic infection have had a long history of success52, and more recently have begun to provide a useful paradigm for studies of beneficial symbioses. For example, the application of signature-tagged mutagenesis has allowed us to identify symbiont genes that are involved in different steps of an infection, which has begun to reveal new colonization determinants49,53 (FIG. 4).
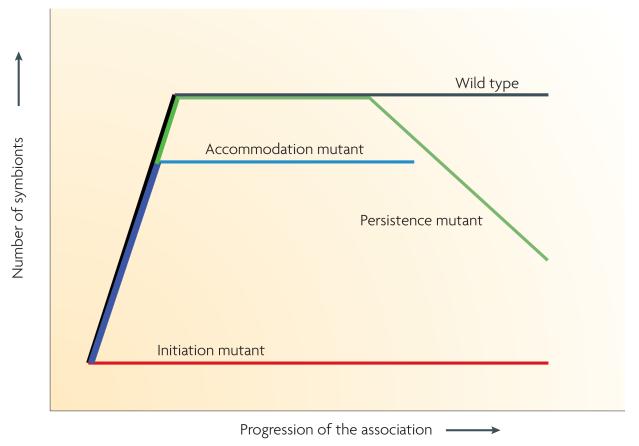
Figure 4. categories of colonization mutants.
Microbial symbionts that are passed horizontally must negotiate several stages of the colonization process. Studies of genetically engineered mutant strains have revealed defects that can be placed in one of several classes. In this example, inoculation with a wild-type strain from the environment allows a few symbionts to colonize, which grow to a specific population size that is then stably maintained over time. Three broad classes of defects have been discovered in several symbiotic systems: initiation mutants, which are unable to inoculate the host; accommodation mutants, which fail to reach the usual population size; and persistence mutants, which at first colonize normally, but are unable to maintain themselves.
Bacterial genetics has been most effectively applied to characterize the well-studied association between V. fischeri and its squid host. The application of bacterial genetics to this association is discussed below, together with the similar and contrasting mechanisms of interaction that have been discovered in the other four experimental genetic associations. It is clear that each of the five systems described above have particular strengths owing both to the specific biology of the symbiosis and the current range of research activities (TABLE 2).
Table 2. Genetic tools and resources for certain bacterial symbionts*.
| Partners | Year initiated‡ |
Clonality | Mutagenesis | Complementation | Screens | Sequenced genome |
Microarray |
|---|---|---|---|---|---|---|---|
| Vibrio–sepiolid squid | 1989 | Yes | Yes | Yes | Yes | Yes | Yes |
|
Xenorhabdus or Photorhabdus–nematode |
1989 | Yes | Yes | Yes | Yes | Yes | No |
| Aeromonas–medicinal leech | 1999 | Yes | Yes | Yes | Yes | No | No |
| Sodalis–tsetse fly | 1995 | Yes | Yes | No | Yes | Yes | No |
| Burkholderia–stink-bug | 2005 | Yes | Yes § | No | No | Yes § | No |
| Enterococcus–fruit fly | 2007 | Yes | Yes § | No | No | Yes § | Yes § |
| Acidovorax–earthworm | 2006 | Yes | No | No | No | Yes | No |
| Bacillus–gypsy moth | 2006 | Yes | No | No | No | Yes § | No |
| Endosymbionts– hydra | 2007 | No | No | No | No | No | No |
This list is rapidly becoming out of date, as these tools are being adapted on an ongoing basis in many of the systems.
Approximate year in which the association was first experimentally described.
Only found in strains from a non-symbiosis source so far.
Surface structures and specificity of the association
A hallmark of all the symbioses considered in this Review is their exclusivity, and therefore the mechanisms that underlie this species specificity are of interest. not surprisingly, surface-associated activities of both the host and the symbiont may be required. For example, mucus that is produced from the surface of the nascent light organ allows the juvenile squid to entrap planktonic cells of V. fischeri in aggregates. These aggregates form near surface pores that lead into the crypt spaces deep within the organ, where the bacteria grow and reside54. The ability of V. fischeri to aggregate in this mucus is dependent on signalling by the Rscs two-component regulation system55. Rscs controls production of an extracellular polysaccharide through its regulation of the syp operon56, and strains with null mutations in either rscS or key syp genes become defective in colonization57,58. similarly, genetic modification of V. fischeri lipopolysaccharide (LPs), either by deletion of pgm, which generates precursors for LPs glycosylation59, or htrB1, which acylates the lipid A component of LPs60, leads to a reduction in light-organ colonization. LPs modification also seems to be important to species of Photorhabdus: pgbE1, a homologue of a gene in the pmr locus of serovars of Salmonella enterica61, is also thought to modify lipid A, and a pgbE1 mutant loses the ability to colonize its nematode host62. In related bacteria, such modifications have also been linked to resistance to host cationic antimicrobial peptides63. similarly, a normal LPs is important for Aeromonas spp. to overcome the activity of host complement and successfully colonize the leech64.
Not surprisingly, other surface factors have also been shown to facilitate a bacterial interaction with host tissue during initiation of the association. In V. fischeri, mutation of either ompU, which encodes an outer membrane protein65, or pilA66, which encodes one of the ten type-Iv pili of this bacterium29, has only a small effect on colonization competence. such defects become more apparent at a low inoculum that might more realistically mimic natural conditions67,68. more impressive effects follow loss of the nil genes of Xenorhabdus species. A nilA mutant strain is attenuated during colonization of the nematode host, whereas deletion of either nilB or nilC, which seem to encode membrane components, completely eliminates symbiosis competency53. The basis of this requirement is as yet unknown, but the similarity of nilB to homologues in mucosal pathogens has suggested it might be involved in an interaction with mucus in the nematode vesicle34. The Xenorhabdus mrxA-encoded pilin protein is another surface structure that has been proposed to function in colonization of the nematode host69.
Bacterial behaviour and gene regulation
For horizontally acquired symbionts, which may exist for long periods in sea water or soil, a rapid and effective adaptation to the specific conditions inside their host can be crucial. In pathogenic associations, the expression of suites of genes that encode colonization factors is often triggered by the activities of specific transcriptional modulators, such as quorum sensing systems or two-component regulators70,71. Thus, many studies have targeted such transcription factors for mutagenesis as a way to affect their downstream regulons. In V. fischeri, there are two sets of quorum-sensing regulators, Ains–AinR and LuxI–LuxR, that work sequentially to induce distinct but partially overlapping regulons30,31. mutations in either system lead to symbiosis defects. For example, an ainS mutant colonizes the squid light organ more slowly than the wild type, and when it does finally infect, it is unable to persist at normal levels72. Global expression analyses of V. fischeri revealed more than 280 genes that are differentially regulated in the mutant (e.G.R., unpublished observations), including those that encode the flagellar apparatus31 and the central metabolic-switch enzyme acetyl-coA synthetase (Acs)73,74. These microarray results led to construction of a V. fischeri acs mutant, which was found to be defective in colonization, indicating an important role in acetate metabolism for the symbiont population.
V. fischeri LuxIR positively regulates the luminescence (or lux) genes, as well as a few other loci75, in a cell-density-dependent manner70. mutation of the luxIR genes or a gene that encodes luciferase (luxA), results in a symbiosis persistence defect that could relate to the inability of these mutants to produce normal light levels in the host33,76. However, in a separate microarray-based study, 18 new genes were found to be regulated by the LuxIR system, and several of these were outer-membrane proteins or secreted proteases30. Thus, it remains possible that LuxIR, which fully induces its regulon only under the high cell densities that are achieved in the light organ77, controls a number of different symbiosis factors, the activities of which are necessary for a long-term, stable colonization of the host. In any case, quorum signalling is a crucial activity in the squid–Vibrio association, as well as in certain plant-microorganism interactions78; the role of cell-density sensing in other beneficial bacteria-animal symbioses is less well defined.
Host-induced bacterial-gene expression is another way by which symbionts adjust to life in the tissue of an animal. not surprisingly, two-component systems that detect specific environmental cues and modulate transcription output are important in this adjustment. In a pioneering effort, Hussa et al.55 used a bioinformatics approach to identify all the putative response regulators that are encoded in the V. fischeri genome. Individual mutations were created in 35 of 40 of these genes, and the resulting strains were compared with the wild-type parent for colonization competence. of these 35 genes, 12 had a symbiosis defect, although further examination of these individual regulators is needed to understand the basis of their effects. For example, in nematode symbioses, HexA, a LysR-like repressor that has been studied in species of both Photorhabdus and Xenorhabdus, seems to control nematode colonization in Photorhabdus but not Xenorhabdus16. specifically, among the >100 genes that are repressed by HexA in Photorhabdus spp. are cipA and cipB, which encode small, crystal-forming proteins that promote nematode development when expressed in trans in E. coli79. mutation of hexA results in a strain that supports the growth of a higher number of symbionts80. It seems that the primary function of HexA is to enhance bacterial virulence in its insect host, which indicates that there is a reciprocal relationship between those Photorhabdus genes that are required for pathogenesis and those that lead to beneficial colonization39.
The importance of global changes in symbiont gene expression has also been revealed by mutating genes that encode sigma factors in V. fischeri (rpoN) and Xenorhabdus spp. (rpoS and rpoE), which leads to defects in flagellar motility81 and resistance to oxidative stress53, respectively, thereby indicating that these activities are required for symbiotic colonization. However, the fact that these transcription factors control the proper expression of dozens of genes makes it difficult to identify the individual contributions of the many downstream targets.
In addition to targeted mutagenesis of suspected symbiosis determinants, phenotypic screens of mutant libraries have provided a powerful tool for identifying both predicted and unexpected colonization factors and activities. For example, V. fischeri mutants that were created by random transposon insertion have been screened for strains that are defective in phenotypes such as flagellar motility and chemotaxis82,83, siderophore production84 and luminescence85. subsequent colonization studies have revealed that these phenotypes are all required symbiosis factors. By contrast, unbiased screening of entire libraries in largescale colonization studies has revealed the importance of new factors (for example, REFS 48,53,86,87). Finally, the transfer of genes that encode fluorescent proteins into different strains of bacteria has been applied not only to tag and localize different strains of symbionts within host tissues, but also to track their transcriptional activity26,88.
Adaptation to host defences
Because all host animals must protect themselves against colonization by inappropriate or pathogenic microorganisms, a central theme in beneficial bacteria-host interactions is that the symbiont either avoids damage by the defences of the host or communicates with host cells to modulate them89. The ability to modulate host defences might therefore also contribute to symbiont specificity. Two ways in which invertebrates confront bacteria that enter their tissues are to produce oxidative or nitrosative stress molecules, or to initiate phagocytosis. evidence for the avoidance of oxidative or nitrosative stress molecules by the symbiont came from the colonization defects of mutant strains that were unable to produce catalase90 or autoinducer 2 (REF. 91). By contrast, Aeromonas spp. do not seem to require a functional catalase to colonize their leech host92. V. fischeri cells effectively avoid phagocytic haemocytes that are present in the light organ. studies of these haemocytes in primary culture suggest that prior exposure to symbionts decreases the ability of the host cell to attach and engulf V. fischeri, but not other bacteria (e.G.R., unpublished observations). mutation of ompU results in an increased susceptibility to phagocytosis, although the mechanism that underlies this phenomenon is unknown. In a related case, in which sensitivity to an immunity factor was encountered in the symbiosis, an Aeromonas LPs mutant was less able to escape killing by complement factors that were present in the blood meal of the leech64.
A type III secretion system (T3ss) which exports effector proteins that moderate host defences by interfering with the antimicrobial function of the host is emerging as a common theme in several beneficial symbioses. Leech-associated Aeromonas spp. are the first extracellular animal symbionts to be shown to use a T3ss to avoid host phagocytosis48. specifically, mutation and complementation analyses of ascU, a homologue of inner-membrane components in other T3sss, have linked this gene to Aeromonas survival in the leech crop. Interestingly, wild-type cells did not rescue the T3ss mutant in co-infection experiments, indicating that the positive effect of the T3ss is restricted to those bacteria that can express it. In a similar manner, mutation of the inv (also known as spa) gene clusters that encode a Sodalis spp. T3ss eliminated the ability of this bacterium either to invade cells of the tsetse fly or even colonize cultured cells after introduction by microinjection86. subsequent analysis revealed the presence of two separate T3sss in the Sodalis genome that have sequential functions during host colonization93. Photorhabdus spp. also produce a functional T3ss that secretes a homologue of YopT, a protein effector that inhibits phagocytosis of Yersinia pestis by host phagocytes94. However, it has not been definitively shown that this T3ss is important to Photorhabdus spp. in host colonization.
Induction of host development
One of the most exciting outcomes of recent studies of bacteria-host interactions is the recognition that symbionts can play a crucial part in triggering the development of their host. often these effects lead to dramatic morphological changes that are required for the proper functioning of the association (for example, REFS 35,95,96). The bacteria in the V. fischeri–E. scolopes symbiosis have a remarkably important role in light-organ development95. several of these developmental events have been linked to specific bacterial gene products through bacterial mutant analysis: infection by lux mutants fails to induce the oedemic swelling that is characteristic of the epithelial cells that line colonized crypts76, which suggests that the host tissue normally responds to symbiont bioluminescence or to the hypoxia that is associated with luciferase activity97. experiments that were designed to determine which of these factors triggers this developmental response will allow us to better understand the mechanisms that underlie this interaction.
The striking discovery that developmental regression of the ciliated surface of the light organ of the juvenile squid is induced by a bacterial cell-wall monomer98, which was previously described as tracheal cytotoxin (TcT), shows how a normal bacteria-host signal can also serve a pathogenic function. Identification in the V. fischeri genome of homologues of genes that encode the activities which are necessary for TcT secretion has allowed these activities to be genetically linked to the developmental biology of this symbiosis. The role of GacA, a common bacterial transcriptional regulator, in squid development has also recently been reported99. specifically, mucus secretion and ciliated-surface apoptosis are not induced in squid that are colonized by a gacA mutant. Because GacA affects the LPs structure of the bacterium, it has been suggested that the developmental defects are linked to outer-membrane modifications100.
The proper development of nematode-infective juveniles has also been linked to the functions of different genes in species of Xenorhabdus and Photorhabdus. The Xenorhabdus global regulator Lrp is required for normal colonization and subsequent host maturation through both its repression (with nilR) of the nil genes and its induction of nematode development16. microarray analyses of the Lrp regulon will help us identify genes that trigger nematode development. By contrast, the ability of Photorhabdus spp. to promote nematode growth and development seems to be largely due to the HexA repressor protein (discussed above). nematodes that are infected by a Photorhabdus ngrA mutant are unable to develop from the infective juvenile stage to the self-fertile hermaphrodite stage101. Because ngrA is a homologue of a non-ribosomal peptide-synthesis complex, it has been suggested that the symbiont produces a peptide signal that induces nematode development39. Finally, although the Photorhabdus crystalforming proteins cipA and cipB promote nematode development through an unexplained mechanism, it is not yet clear whether the analogous Xenorhabdus crystal protein that is encoded by pixA has an important role in the biology of the host16.
Nutritional and metabolic accommodation
In a mutually successful symbiosis, both partners must obtain sufficient organic and inorganic nutrients to grow and sustain the relationship. not surprisingly, virtually all symbiotic relationships include a transfer of such materials between the host and the bacterium, in at least one direction. sometimes, the symbionts must be able to synthesize missing nutrients owing to the nature of the food, such as blood, that is provided by the host. The details of this nutritional dependency can often be revealed by following the colonization of genetic mutants that are defective either in specific biosynthetic or metabolic pathways1, or in the ability to adjust to changing nutrient levels102.
A central question in the Vibrio–squid symbiosis is: what does the host feed its bacterial symbionts and does this change over the course of the relationship? mutant library screening for auxotrophs of V. fischeri yielded a collection of strains that were defective in the biosynthesis of specific amino acids103. The colonization levels that were achieved by these mutants provided a relative measure of the extent to which the host provided each of these amino acids. The resulting pattern suggested that peptides of a typical protein composition were available to the symbiont population103. By contrast, a similar study of nematode colonization by Xenorhabdus auxotrophs indicated that two specific amino acids, methionine and threonine, were of limited availability in the vesicle88. supporting this conclusion, the Xenorhabdus crystal protein PixA is methionine rich, and a pixA mutant out-competes its parent for growth in the vesicle, which suggests that excess methionine synthesis is a metabolic drain on the symbiont104. Because colonization by Xenorhabdus spp. requires some or all of the isc-hsc-fdx locus, which encodes activities that synthesize [Fe-s] centres, it is possible that this bacterium depends on some non-haem redox activity, such as anaerobic respiration, when in the host105. Interestingly, studies of the squid–Vibrio association also suggest that the generation of acetate73 and metabolic pathways which require anaerobic respiration106 are important to V. fischeri in the light organ.
As is typical for pathogenic infections107, free iron seems to be the main inorganic nutrient that limits growth in beneficial animal symbioses. several studies have focused on identifying the genetic determinants of iron acquisition that allow symbiotic bacteria to colonize their hosts. In V. fischeri, a mutation in the glnD gene results in pleiotropic phenotypes, including reduced production of at least one siderophore in culture84. When assayed for colonization, the glnD mutant failed to persist, but this defect was reversed when ferric chloride was added to the assay. similarly, the addition of iron rescued colonization by a putative siderophore-uptake mutant (exbD) of a Photorhabdus species108. A putative iron-regulated promoter element has also been detected upstream of ngrA39, a gene that could function to synthesize either a peptide signal or siderophore109, which further indicates that iron levels are important to Photorhabdus species. Thus, the availability of iron can have both nutritional and regulatory implications.
Conclusions
The molecular genetics of symbiosis is a rapidly expanding field, and new systems are continually adding to its breadth and impact. It is important to continue to develop systems such as the earthworm nephridia-Acidovorax symbiosis110,111, the caterpillar gut-Enterococcus association51, the as-yet-uncultured epithelial symbionts of hydra112 and the recently described louse fly-Arsenophonus relationship113. one particularly promising system is that between a Burkholderia species and its host, the broad-headed stink-bug114. In this association the bacterium is environmentally acquired at each generation, provides a clear benefit to the host and is extracellularly located115. A successful history of molecular studies of other Burkholderia species makes it highly likely that it will not be long before the symbiont will be amenable to molecular genetics.
The development of experimental strategies and technological approaches will be important to the continued growth of the field of symbiosis. creating a community of researchers that can develop genomic (for example, genomes and microarrays) and molecular genetic sequencing resources (for example, mutants, vectors and methodology), either within a single species, or more generally across the field, will quicken the pace of advance in all systems. similarly, in addition to increasing the number of bacterial symbionts with sequenced genomes (TABLE 2), efforts to obtain genome sequences for different strains of the same symbiont species have proven valuable for comparative studies28.
In most symbioses described here14, the application of molecular genetics to the animal partner lags behind efforts with the bacterial symbiont. nevertheless, in the past year there have been promising results in the host using a reverse-genetic RnA interference approach to silence specific genes116 and with the application of microarray technology to determine the differential effect of colonization by mutant symbionts on host gene expression33. Future studies should focus on developing interactive genomics to track patterns of gene expression simultaneously in both the host and the symbiont. In this way we will not only be able to interrogate this conversation, but perhaps learn how to manipulate it.
Acknowledgements
The author thanks M. McFall-Ngai for helpful discussions and ideas, and H. Goodrich-Blair, J. Graf and M. Mandel for reading parts of the manuscript. Support was provided by grants from the National Institutes of Health (grant number RR-12294) and the National Science Foundation (grant number IOB-0517007).
Glossary
- Bioluminescence
The process by which some bacteria and other organisms produce light as the result of a chemical reaction. During symbiosis, this light can be used in behaviours such as counterillumination, in which the bioluminescence is used to eliminate the shadow of the host’s silhouette
- Gnotobiotic
An animal that is born under aseptic conditions and is exposed only to experimentally introduced microorganisms. Gnotobiotic animals are used to investigate the symbiotic relationship between an animal and one or more of the consortia of interacting microbial species that normally inhabit its body
- Horizontal transfer
The process by which an animal or plant obtains its natural microbial constituents from the environment at each generation. By contrast, vertical transfer occurs when a young organism receives its microbiota from its parent, usually in or on the egg
- Expressed sequence tag (eST)
One of a series of short nucleotide sequences which represent a pool of mRNAs that are expressed under a certain environmental or developmental condition. libraries of eSTs can be used to identify gene transcripts in global expression studies
- Two-component regulation system
A stimulus-response coupling mechanism that allows an organism to sense and respond to various changes in environmental conditions
- Lipopolysaccharide
A major component of the outer membrane of Gram-negative bacteria. The immune systems of animals generally sense and react to the presence of lipopolysaccharide
- Quorum sensing
A system by which bacteria respond to increased population density by coordinately controlling expression of a specific set of genes. By sensing the concentration of one of several continuously secreted signal molecules, including acyl homoserine lactones, peptides and autoinducer 2, the population can recognize when it reaches a ‘quorum’.
- Auxotroph
An organism, or mutant derivative, which is unable to synthesize a particular compound (for example, an amino acid) that is required as a building block for its growth.
Footnotes
DATABASES
Entrez Gene: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene
acs | ainS | gacA | glnD | htrB1 | luxA | ompU | pgm | rpoN | rscS
Entrez Genome Project: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=genomeprj
Drosophila melanogaster | Enterococcus faecalis | Escherichia coli | Sodalis glossinidius | Vibrio fischeri |Yersinia pestis
FURTHER INFORMATION
Edward G. Ruby’s homepage: http://www.medmicro.wisc.edu/department/faculty/ruby.html
Nobelprize.org (All Nobel Laureates): http://nobelprize.org/nobel_prizes/lists/all/
References
- 1.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 2.Haygood MG. Light organ symbioses in fishes. Crit. Rev. Microbiol. 1993;19:191–216. doi: 10.3109/10408419309113529. [DOI] [PubMed] [Google Scholar]
- 3.Parniske Nature Rev. Microbiol. 2008;10 doi: 10.1038/nrmicro1987. xxx-xxx. [DOI] [PubMed] [Google Scholar]
- 4.Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nature Rev. Microbiol. 2008;10 doi: 10.1038/nrmicro1969. xxx-xxx. [DOI] [PubMed] [Google Scholar]
- 5.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nature Rev. Microbiol. 2008;10 doi: 10.1038/nrmicro1978. xxx-xxx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 8.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host–archaeal–bacterial mutualism. Proc. Natl Acad. Sci. USA. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personal heath care. Nature Rev. Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 10.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 12.Vaishnava S, Behrendt CL, Hooper LV. Innate immune responses to commensal bacteria in the gut epithelium. J. Pediatr. Gastroenterol. Nutr. 2008;46(Suppl. 1):E10–E11. doi: 10.1097/01.mpg.0000313823.93841.65. [DOI] [PubMed] [Google Scholar]
- 13.Graf J, Kikuchi Y, Rio RV. Leeches and their microbiota: naturally simple symbiosis models. Trends Microbiol. 2006;14:365–371. doi: 10.1016/j.tim.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 14.Cox CR, Gilmore MS. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect. Immun. 2007;75:1565–1576. doi: 10.1128/IAI.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McFall-Ngai MJ, Gordon JI. In: Evolution of Microbial Virulence. Seifert H, DiRita VJ, editors. ASM; Washington DC: 2006. Au:please provide page numbers? [Google Scholar]
- 16.Goodrich-Blair H, Clarke DJ. Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: two roads to the same destination. Mol. Microbiol. 2007;64:260–268. doi: 10.1111/j.1365-2958.2007.05671.x. [DOI] [PubMed] [Google Scholar]
- 17.McFall-Ngai M. Adaptive immunity: care for the community. Nature. 2007;445:153. doi: 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- 18.Moran NA. Bacterial menageries inside insects. Proc. Natl Acad. Sci. USA. 2001;98:1338–1340. doi: 10.1073/pnas.98.4.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nealson KH, Hastings JW. In: The Prokaryotes, a Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Applications. 2nd edn Balows A, Truper HG, Dworkin M, Harder W, Schleifer KH, editors. Springer; Berlin: 1991. pp. 1332–1345. [Google Scholar]
- 20.Fidopiastis PM, von Boletzky S, Ruby EG. A new niche for Vibrio logei, the predominant light organ symbiont of squids in the genus Sepiola. J. Bacteriol. 1998;180:59–64. doi: 10.1128/jb.180.1.59-64.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishiguchi MK, Nair VS. Evolution of symbiosis in the Vibrionaceae: a combined approach using molecules and physiology. Int. J. Syst. Evol. Microbiol. 2003;53:2019–2026. doi: 10.1099/ijs.0.02792-0. [DOI] [PubMed] [Google Scholar]
- 22.Jones BW, Nishiguchi MK. Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca:Cephalopoda) Mar. Biol. 2004;144:1151–1155. [Google Scholar]
- 23.Wei SL, Young RE. Development of symbiotic bacterial luminescence in a nearshore cephalopod, Euprymna scolopes. Mar. Biol. 1989;103:541–546. [Google Scholar]
- 24.Ruby EG, Lee KH. The Vibrio fischeri-Euprymna scolopes light organ association: current ecological paradigms. Appl. Environ. Microbiol. 1998;64:805–812. doi: 10.1128/aem.64.3.805-812.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Claes MF, Dunlap PV. Aposymbiotic culture of the sepiolid squid Euprymna scolopes: role of the symbiotic bacterium Vibrio fischeri in host animal growth, development, and light organ morphogenesis. J. Exp. Zool. 2000;286:280–296. [PubMed] [Google Scholar]
- 26.Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol. 2006;72:802–810. doi: 10.1128/AEM.72.1.802-810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stabb EV, Ruby EG. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 2002;358:413–426. doi: 10.1016/s0076-6879(02)58106-4. [DOI] [PubMed] [Google Scholar]
- 28.Mandel MJ, Stabb EV, Ruby EG. Comparative genomics-based investigation of resequencing targets in Vibrio fischeri: focus on point miscalls and artefactual expansions. BMC Genomics. 2008;9:138. doi: 10.1186/1471-2164-9-138. Introduced novel technological approaches to apply a comparative genomics approach to two strains of a beneficial bacterial symbiont.
- 29.Ruby EG, et al. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl Acad. Sci. USA. 2005;102:3004–3009. doi: 10.1073/pnas.0409900102. Sequenced the V. fischeri genome, which allowed molecular genetics to be applied to the squid-Vibrio system and opened up new approaches of genetic analysis.
- 30.Antunes LC, et al. Transcriptome analysis of the Vibrio fischeri LuxR–LuxI regulon. J. Bacteriol. 2007;189:8387–8391. doi: 10.1128/JB.00736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lupp C, Ruby EG. Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. J. Bacteriol. 2005;187:3620–3629. doi: 10.1128/JB.187.11.3620-3629.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chun CK, et al. An annotated cDNA library of juvenile Euprymna scolopes with and without colonization by the symbiont Vibrio fischeri. BMC Genomics. 2006;7:154. doi: 10.1186/1471-2164-7-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chun C, et al. Effects of colonization, luminescence, and autoinducer on host transcription during development of the squid-vibrio association. Proc. Natl Acad. Sci. USA. 2008 Aug 5; doi: 10.1073/pnas.0802369105. doi:10.1073/pnas.0802369105. First large-scale transcriptional analysis of the host response to colonization by bacteria that possess mutations in their symbiosis genes.
- 34.Herbert EE, Goodrich-Blair H. Friend and foe: the two faces of Xenorhabdus nematophila. Nature Rev. Microbiol. 2007;5:634–646. doi: 10.1038/nrmicro1706. [DOI] [PubMed] [Google Scholar]
- 35.Hallem EA, Rengarajan M, Ciche TA, Sternberg PW. Nematodes, bacteria, and flies: a tripartite model for nematode parasitism. Curr. Biol. 2007;17:898–904. doi: 10.1016/j.cub.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 36.Ciche TA, Kim KS, Kaufmann-Daszczuk B, Nguyen KC, Hall DH. Cell invasion and matricide during Photorhabdus luminescens transmission by Heterorhabditis bacteriophora nematodes. Appl. Environ. Microbiol. 2008;74:2275–2287. doi: 10.1128/AEM.02646-07. The first study to focus on development of the host during an association with a nematode.
- 37.Cowles CE, Goodrich-Blair H. The Xenorhabdus nematophila nilABC genes confer the ability of Xenorhabdus spp. to colonize Steinernema carpocapsae nematodes. J. Bacteriol. 2008;190:4121–4128. doi: 10.1128/JB.00123-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goodrich-Blair H. They’ve got a ticket to ride: Xenorhabdus nematophila-Steinernema carpocapsae symbiosis. Curr. Opin. Microbiol. 2007;10:225–230. doi: 10.1016/j.mib.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 39.Joyce SA, Watson RJ, Clarke DJ. The regulation of pathogenicity and mutualism in Photorhabdus. Curr. Opin. Microbiol. 2006;9:127–132. doi: 10.1016/j.mib.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 40.Dale C, Moran NA. Molecular interactions between bacterial symbionts and their hosts. Cell. 2006;126:453–465. doi: 10.1016/j.cell.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 41.Moran NA. Symbiosis. Curr. Biol. 2006;16:R866–R871. doi: 10.1016/j.cub.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 42.Toh H, et al. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 2006;16:149–156. doi: 10.1101/gr.4106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss BL, et al. Interspecific transfer of bacterial endosymbionts between tsetse fly species: infection establishment and effect on host fitness. Appl. Environ. Microbiol. 2006;72:7013–7021. doi: 10.1128/AEM.01507-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rio RV, Lefevre C, Heddi A, Aksoy S. Comparative genomics of insect-symbiotic bacteria: influence of host environment on microbial genome composition. Appl. Environ. Microbiol. 2003;69:6825–6832. doi: 10.1128/AEM.69.11.6825-6832.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warnecke F, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 46.Broderick NA, Raffa KF, Goodman RM, Handelsman J. Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl. Environ. Microbiol. 2004;70:293–300. doi: 10.1128/AEM.70.1.293-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Indergand S, Graf J. Ingested blood contributes to the specificity of the symbiosis of Aeromonas veronii biovar sobria and Hirudo medicinalis, the medicinal leech. Appl. Environ. Microbiol. 2000;66:4735–4741. doi: 10.1128/aem.66.11.4735-4741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silver AC, et al. Interaction between innate immune cells and a bacterial type III secretion system in mutualistic and pathogenic associations. Proc. Natl Acad. Sci. USA. 2007;104:9481–9486. doi: 10.1073/pnas.0700286104. Discovered a T3SS in a beneficial bacteria-animal symbiosis, and described one of its functions.
- 49.Silver AC, Rabinowitz NM, Kuffer S, Graf J. Identification of Aeromonas veronii genes required for colonization of the medicinal leech, Hirudo verbana. J. Bacteriol. 2007;189:6763–6772. doi: 10.1128/JB.00685-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kikuchi Y, Graf J. Spatial and temporal population dynamics of a naturally occurring two-species microbial community inside the digestive tract of the medicinal leech. Appl. Environ. Microbiol. 2007;73:1984–1991. doi: 10.1128/AEM.01833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Broderick NA, Raffa KF, Handelsman J. Midgut bacteria required for Bacillus thuringiensis insecticidal activity. Proc. Natl Acad. Sci. USA. 2006;103:15196–15199. doi: 10.1073/pnas.0604865103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Falkow S. Molecular Koch’s postulates applied to bacterial pathogenicity — a personal recollection 15 years later. Nature Rev. Microbiol. 2004;2:67–72. doi: 10.1038/nrmicro799. [DOI] [PubMed] [Google Scholar]
- 53.Heungens K, Cowles CE, Goodrich-Blair H. Identification of Xenorhabdus nematophila genes required for mutualistic colonization of Steinernema carpocapsae nematodes. Mol. Microbiol. 2002;45:1337–1353. doi: 10.1046/j.1365-2958.2002.03100.x. One of the first applications of an advanced genetic screen for bacterial colonization factors in an animal symbiont.
- 54.Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. Establishment of an animal–bacterial association: recruiting symbiotic vibrios from the environment. Proc. Natl Acad. Sci. USA. 2000;97:10231–10235. doi: 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hussa EA, O’Shea TM, Darnell CL, Ruby EG, Visick KL. Two-component response regulators of Vibrio fischeri: identification, mutagenesis, and characterization. J. Bacteriol. 2007;189:5825–5838. doi: 10.1128/JB.00242-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yip ES, Geszvain K, Deloney-Marino CR, Visick KL. The symbiosis regulator RscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol. Microbiol. 2006;52:1586–1600. doi: 10.1111/j.1365-2958.2006.05475.x. Provided a breakthrough in our understanding of the regulation of genes that are involved in symbiosis initiation in the squid–Vibrio association.
- 57.Darnell CL, Hussa EA, Visick KL. The putative hybrid sensor kinase SypF coordinates biofilm formation in Vibrio fischeri by acting upstream of two response regulators, SypG and VpsR. J. Bacteriol. 2008;190:4941–4950. doi: 10.1128/JB.00197-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Geszvain K, Visick KL. Roles of bacterial regulators in the symbiosis between Vibrio fischeri and Euprymna scolopes. Prog. Mol. Subcell. Biol. 2006;41:277–290. doi: 10.1007/3-540-28221-1_13. [DOI] [PubMed] [Google Scholar]
- 59.DeLoney CR, Bartley TM, Visick KL. Role for phosphoglucomutase in Vibrio fischeri–Euprymna scolopes symbiosis. J. Bacteriol. 2002;184:5121–5129. doi: 10.1128/JB.184.18.5121-5129.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adin DM, et al. Characterization of htrB and msbB mutants of the light organ symbiont Vibrio fischeri. Appl. Environ. Microbiol. 2007;74:633–644. doi: 10.1128/AEM.02138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gunn JS, Ryan SS, Van Velkinburgh JC, Ernst RK, Miller SI. Genetic and functional analysis of a PmrA–PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 2000;68:6139–6146. doi: 10.1128/iai.68.11.6139-6146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bennett HP, Clarke DJ. The pbgPE operon in Photorhabdus luminescens is required for pathogenicity and symbiosis. J. Bacteriol. 2005;187:77–84. doi: 10.1128/JB.187.1.77-84.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gunn JS, et al. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 64.Braschler TR, Merino S, Tomas JM, Graf J. Complement resistance is essential for colonization of the digestive tract of Hirudo medicinalis by Aeromonas strains. Appl. Environ. Microbiol. 2003;69:4268–4271. doi: 10.1128/AEM.69.7.4268-4271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aeckersberg F, Lupp C, Feliciano B, Ruby EG. Vibrio fischeri outer membrane protein OmpU plays a role in normal symbiotic colonization. J. Bacteriol. 2001;183:6590–6597. doi: 10.1128/JB.183.22.6590-6597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stabb EV, Ruby EG. Contribution of pilA to competitive colonization of the squid Euprymna scolopes by Vibrio fischeri. Appl. Environ. Microbiol. 2003;69:820–826. doi: 10.1128/AEM.69.2.820-826.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee K-H, Ruby EG. Detection of the light organ symbiont, Vibrio fischeri, in Hawaiian seawater by using lux gene probes. Appl. Environ. Microbiol. 1992;58:942–947. doi: 10.1128/aem.58.3.942-947.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee K-H, Ruby EG. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl. Environ. Microbiol. 1994;60:1565–1571. doi: 10.1128/aem.60.5.1565-1571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.He H, Snyder HA, Forst S. Unique organization and regulation of the mrx fimbrial operon in Xenorhabdus nematophila. Microbiology. 2004;150:1439–1446. doi: 10.1099/mic.0.26853-0. [DOI] [PubMed] [Google Scholar]
- 70.Fuqua C, Greenberg EP. Listening in on bacteria: acyl-homoserine lactone signalling. Nature Rev. Mol. Cell Biol. 2002;3:685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 71.Alegado RA, Campbell MC, Chen WC, Slutz SS, Tan MW. Characterization of mediators of microbial virulence and innate immunity using the Caenorhabditis elegans host-pathogen model. Cell Microbiol. 2003;5:435–444. doi: 10.1046/j.1462-5822.2003.00287.x. [DOI] [PubMed] [Google Scholar]
- 72.Lupp C, Ruby EG. Vibrio fischeri LuxS and AinS: comparative study of two signal synthases. J. Bacteriol. 2004;186:3873–3881. doi: 10.1128/JB.186.12.3873-3881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Studer SV, Mandel MJ, Ruby EG. AinS quorum sensing regulates the Vibrio fischeri acetate switch. J. Bacteriol. 2008 doi: 10.1128/JB.00148-08. doi:10.1128/JB.00148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wolfe AJ. The acetate switch. Microbiol. Mol. Biol. Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Callahan SM, Dunlap PV. LuxR- and acyl-homoserine-lactone-controlled non-lux genes define a quorum-sensing regulon in Vibrio fischeri. J. Bacteriol. 2000;182:2811–2822. doi: 10.1128/jb.182.10.2811-2822.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Visick KL, Foster J, Doino J, McFall-Ngai M, Ruby EG. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 2000;182:4578–4586. doi: 10.1128/jb.182.16.4578-4586.2000. A key paper that links the production of luminescence to both the induction of normal host development and the capacity for persistent colonization.
- 77.Boettcher KJ, Ruby EG. Detection and quantification of Vibrio fischeri autoinducer from symbiotic squid light organs. J. Bacteriol. 1995;177:1053–1058. doi: 10.1128/jb.177.4.1053-1058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sanchez-Contreras M, Bauer WD, Gao M, Robinson JB, Allan Downie J. Quorum-sensing regulation in rhizobia and its role in symbiotic interactions with legumes. Philos. Trans. R. Soc. Lond. B. 2007;362:1149–1163. doi: 10.1098/rstb.2007.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.You J, Liang S, Cao L, Liu X, Han R. Nutritive significance of crystalline inclusion proteins of Photorhabdus luminescens in Steinernema nematodes. FEMS Microbiol. Ecol. 2006;55:178–185. doi: 10.1111/j.1574-6941.2005.00015.x. [DOI] [PubMed] [Google Scholar]
- 80.Joyce SA, Clarke DJ. A hexA homologue from Photorhabdus regulates pathogenicity, symbiosis and phenotypic variation. Mol. Microbiol. 2003;47:1445–1457. doi: 10.1046/j.1365-2958.2003.03389.x. [DOI] [PubMed] [Google Scholar]
- 81.Wolfe AJ, Millikan DS, Campbell JM, Visick KL. Vibrio fischeri σ54 controls motility, biofilm formation, luminescence, and colonization. Appl. Environ. Microbiol. 2004;70:2520–2524. doi: 10.1128/AEM.70.4.2520-2524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.DeLoney-Marino CR, Wolfe AJ, Visick KL. Chemoattraction of Vibrio fischeri to serine, nucleosides, and N-acetylneuraminic acid, a component of squid light-organ mucus. Appl. Environ. Microbiol. 2003;69:7527–7530. doi: 10.1128/AEM.69.12.7527-7530.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Graf J, Dunlap PV, Ruby EG. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J. Bacteriol. 1994;176:6986–6991. doi: 10.1128/jb.176.22.6986-6991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Graf J, Ruby EG. Novel effects of a transposon insertion in the Vibrio fischeri glnD gene: defects in iron uptake and symbiotic persistence, as well as nitrogen utilization. Mol. Microbiol. 2000;37:168–179. doi: 10.1046/j.1365-2958.2000.01984.x. [DOI] [PubMed] [Google Scholar]
- 85.Bose JL, et al. Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol. Microbiol. 2007;65:538–553. doi: 10.1111/j.1365-2958.2007.05809.x. [DOI] [PubMed] [Google Scholar]
- 86.Dale C, Young SA, Haydon DT, Welburn SC. The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc. Natl Acad. Sci. USA. 2001;98:1883–1888. doi: 10.1073/pnas.021450998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Visick KL, Ruby EG. TnluxAB insertion mutants of Vibrio fischeri with symbiosis-regulated phenotypes. Nova Acta Leopoldina. 2003;333:93–100. [Google Scholar]
- 88.Martens EC, Russell FM, Goodrich-Blair H. Analysis of Xenorhabdus nematophila metabolic mutants yields insight into stages of Steinernema carpocapsae nematode intestinal colonization. Mol. Microbiol. 2005;58:28–45. doi: 10.1111/j.1365-2958.2005.04742.x. [DOI] [PubMed] [Google Scholar]
- 89.Davidson SK, Koropatnick TA, Kossmehl R, Sycuro L, McFall-Ngai MJ. NO means ‘yes’ in the squid–vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell. Microbiol. 2004;6:1139–1151. doi: 10.1111/j.1462-5822.2004.00429.x. [DOI] [PubMed] [Google Scholar]
- 90.Visick KL, Ruby EG. The periplasmic, group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and approach to stationary phase. J. Bacteriol. 1998;180:2087–2092. doi: 10.1128/jb.180.8.2087-2092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Krin E, et al. Pleiotropic role of quorum-sensing autoinducer 2 in Photorhabdus luminescens. Appl. Environ. Microbiol. 2006;72:6439–6451. doi: 10.1128/AEM.00398-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rio RV, Anderegg M, Graf J. Characterization of a catalase gene from Aeromonas veronii, the digestivetract symbiont of the medicinal leech. Microbiology. 2007;153:1897–1906. doi: 10.1099/mic.0.2006/003020-0. [DOI] [PubMed] [Google Scholar]
- 93.Dale C, Jones T, Pontes M. Degenerative evolution and functional diversification of type-III secretion systems in the insect endosymbiont Sodalis glossinidius. Mol. Biol. Evol. 2005;22:758–766. doi: 10.1093/molbev/msi061. [DOI] [PubMed] [Google Scholar]
- 94.Brugirard-Ricaud K, et al. Site-specific antiphagocytic function of the Photorhabdus luminescens type III secretion system during insect colonization. Cell. Microbiol. 2005;7:363–371. doi: 10.1111/j.1462-5822.2004.00466.x. [DOI] [PubMed] [Google Scholar]
- 95.Nyholm SV, McFall-Ngai MJ. The winnowing: establishing the squid-Vibrio symbiosis. Nature Rev. Microbiol. 2004;2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 96.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl Acad. Sci. USA. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ruby EG, McFall-Ngai MJ. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends Microbiol. 1999;7:414–420. doi: 10.1016/s0966-842x(99)01588-7. [DOI] [PubMed] [Google Scholar]
- 98.Koropatnick TA, et al. Microbial factor-mediated development in a host-bacterial mutualism. Science. 2004;306:1186–1188. doi: 10.1126/science.1102218. Developed a new paradigm by showing that a bacterial toxin serves as a required developmental signal compound in a beneficial host-microorganism association.
- 99.Whistler CA, Ruby EG. GacA regulates symbiotic colonization traits of Vibrio fischeri and facilitates a beneficial association with an animal host. J. Bacteriol. 2003;185:7202–7212. doi: 10.1128/JB.185.24.7202-7212.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Whistler CA, Koropatnick TA, Pollack A, McFall-Ngai MJ, Ruby EG. The GacA global regulator of Vibrio fischeri is required for normal host tissue responses that limit subsequent bacterial colonization. Cell. Microbiol. 2007;9:766–778. doi: 10.1111/j.1462-5822.2006.00826.x. [DOI] [PubMed] [Google Scholar]
- 101.Faraldo-Gomez JD, Sansom MS. Acquisition of siderophores in Gram-negative bacteria. Nature Rev. Mol. Cell Biol. 2003;4:105–116. doi: 10.1038/nrm1015. [DOI] [PubMed] [Google Scholar]
- 102.Cowles KN, Cowles CE, Richards GR, Martens EC, Goodrich-Blair H. The global regulator Lrp contributes to mutualism, pathogenesis and phenotypic variation in the bacterium Xenorhabdus nematophila. Cell. Microbiol. 2007;9:1311–1323. doi: 10.1111/j.1462-5822.2006.00873.x. [DOI] [PubMed] [Google Scholar]
- 103.Graf J, Ruby EG. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc. Natl Acad. Sci. USA. 1998;95:1818–1822. doi: 10.1073/pnas.95.4.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Goetsch M, Owen H, Goldman B, Forst S. Analysis of the PixA inclusion body protein of Xenorhabdus nematophila. J. Bacteriol. 2006;188:2706–2710. doi: 10.1128/JB.188.7.2706-2710.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Martens EC, et al. Xenorhabdus nematophila requires an intact iscRSUA–hscBA–fdx operon to colonize Steinernema carpocapsae nematodes. J. Bacteriol. 2003;185:3678–3682. doi: 10.1128/JB.185.12.3678-3682.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dunn AK, Stabb EV. The twin arginine translocation system contributes to symbiotic colonization of Euprymna scolopes by Vibrio fischeri. FEMS Microbiol. Lett. 2008;279:251–258. doi: 10.1111/j.1574-6968.2007.01043.x. [DOI] [PubMed] [Google Scholar]
- 107.Schaible UE, Kaufmann SH. Iron and microbial infection. Nature Rev. Microbiol. 2004;2:946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 108.Watson RJ, Joyce SA, Spencer GV, Clarke DJ. The exbD gene of Photorhabdus temperata is required for full virulence in insects and symbiosis with the nematode Heterorhabditis. Mol. Microbiol. 2005;56:763–773. doi: 10.1111/j.1365-2958.2005.04574.x. [DOI] [PubMed] [Google Scholar]
- 109.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Davidson SK, Stahl DA. Transmission of nephridial bacteria of the earthworm Eisenia fetida. Appl. Environ. Microbiol. 2006;72:769–775. doi: 10.1128/AEM.72.1.769-775.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Davidson SK, Stahl DA. Selective recruitment of bacteria during embryogenesis of an earthworm. ISME J. 2008;2:510–518. doi: 10.1038/ismej.2008.16. [DOI] [PubMed] [Google Scholar]
- 112.Fraune S, Bosch TC. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc. Natl Acad. Sci. USA. 2007;104:13146–13151. doi: 10.1073/pnas.0703375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dale C, Beeton M, Harbison C, Jones T, Pontes M. Isolation, pure culture, and characterization of ’Candidatus Arsenophonus arthropodicus,’ an intracellular secondary endosymbiont from the hippoboscid louse fly Pseudolynchia canariensis. Appl. Environ. Microbiol. 2006;72:2997–3004. doi: 10.1128/AEM.72.4.2997-3004.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kikuchi Y, Meng XY, Fukatsu T. Gut symbiotic bacteria of the genus Burkholderia in the broad-headed bugs Riptortus clavatus and Leptocorisa chinensis (Heteroptera: Alydidae) Appl. Environ. Microbiol. 2005;71:4035–4043. doi: 10.1128/AEM.71.7.4035-4043.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kikuchi Y, Hosokawa T, Fukatsu T. Insect-microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl. Environ. Microbiol. 2007;73:4308–4316. doi: 10.1128/AEM.00067-07. Introduced an emerging experimental symbiosis system that had intriguing parallels with the squid-Vibrio association.
- 116.Ciche TA, Sternberg PW. Postembryonic RNAi in Heterorhabditis bacteriophora: a nematode insect parasite and host for insect pathogenic symbionts. BMC Dev. Biol. 2007;7:101. doi: 10.1186/1471-213X-7-101. [DOI] [PMC free article] [PubMed] [Google Scholar]