Abstract
Treponema denticola is a consensus periodontal pathogen that has recently been associated with endodontic pathology. In this study, the effect of mono-infection of the dental pulp with T. denticola and with polymicrobial “red-complex” organisms (RC) (Porphyromonas gingivalis, Tannerella forsythia, and T. denticola) in inducing disseminating infections in wild-type (WT) and severe-combined-immunodeficiency (SCID) mice was analyzed. After 21 days, a high incidence (5/10) of orofacial abscesses was observed in SCID mice mono-infected with T. denticola, whereas abscesses were rare in SCID mice infected with the red-complex organisms or in wild-type mice. Splenomegaly was present in all groups, but only mono-infected SCID mice had weight loss. T. denticola DNA was detected in the spleen, heart, and brain of mono-infected SCID mice and in the spleen from mono-infected wild-type mice, which also had more periapical bone resorption. The results indicate that T. denticola has high pathogenicity, including dissemination to distant organs, further substantiating its potential importance in oral and linked systemic conditions.
Keywords: periapical lesion, Tannerella forsythia, Porphyromonas gingivalis, disseminating infection, micro-computed tomography
INTRODUCTION
The microbial etiology of endodontic disease is well-established in animal models and humans (Kakehashi et al., 1965; Sundqvist, 1994). The host immune response plays an important role in localizing these infections to the root canal space. A deficiency in antibody formation by B-lymphocytes (Teles et al., 1997; Hou et al., 2000) or in phagocytic leukocytes (Kawashima et al., 1999) results in more severe disease, including systemic dissemination with sepsis and increased mortality. Thus, the pathogenicity of endodontic bacteria is dependent upon their innate virulence as well as the host’s immune status.
Among oral pathogens, Treponema denticola has been associated with the severity of human periodontal diseases (Socransky et al., 1998; Yoshida et al., 2004), in association with Porphyromonas gingivalis and Tannerella forsythia, forming the so-called “red-complex” organisms (Socransky et al., 1998), and is furthermore linked to severe manifestations in immunodeficient patients (Sela, 2001). T. denticola has been linked with endodontic disease, given its association with orofacial abscesses and periapical radiolucencies (Baumgartner et al., 2003; Foschi et al., 2005; Siqueira and Rocas, 2004). Despite these relationships, the etiological role of T. denticola in endodontic disease, alone and as part of the “red complex” (Socransky et al., 1998; Rocas et al., 2001), has not yet been directly demonstrated in vivo.
The goal of the present study was therefore to determine the role of T. denticola as a mono-infection and as part of “red complex” polymicrobial infection in the etiology of endodontic disease in immunocompetent and severe combined immunodeficient (SCID) mice. The ability of each organism to disseminate in the host, and to stimulate bone resorption—strong indicators of pathogenicity in the endodontic milieu—was also evaluated.
MATERIALS & METHODS
Animals
Six- to eight-week-old SCID (C57BL/6J-Rag1tm1Mom, background C57BL/6J) and wild-type (WT) C57BL/6J male mice were obtained from Jackson Laboratory (Bar Harbor, ME, USA). Animals were maintained, according to IACUC guidelines, in laminar flow isolators in the Forsyth Institute animal facility under pathogen-free conditions.
Periapical Lesion Induction by Microbial Infection
Endodontic pathogens were grown under anaerobic conditions. P. gingivalis (ATCC 33277) was plated on hemin/menadione solid medium, T. forsythia (ATCC 43037) was plated on N-acetyl muramic acid solid medium, and T. denticola (ATCC 35405) was grown in liquid New Oral Spirochete medium (NOS) (Izard et al., 2001). On day 0, animals were anesthetized with 62.5 mg/kg ketamine HCl and 10 mg/kg xylazine in sterile PBS delivered intraperitoneally, and pulp exposures were made in both mandibular first molars with a no. 4 round bur under a surgical microscope, as previously described (Balto et al., 2000).
Animals were divided into four experimental groups: A, SCID mice (n = 10) infected with T. denticola, 2 μL of 108 cells; B, SCID mice (n = 9) infected with “red complex” organisms (RC) (P. gingivalis, T. forsythia, and T. denticola), equal numbers in 2 μL, total of 108 cells; C, wild-type mice (n = 8), infected with 2 μL of 108 cells of T. denticola; and D, wild-type mice (n = 10) infected with red-complex organisms, same numbers of cells as group B. Two C57BL/6J wild-type mice served as sham controls. Bacterial concentration was determined with the use of a Petroff-Hausser chamber and a Labophot-2 microscope (Nikon, Tokyo, Japan). After endodontic infection, access cavities were sealed with composite resin (Assure, Reliance Orthodontic Products, Itasca, IL, USA) to prevent superinfection from the oral cavity.
To assess bacterial survival, we extracted infected teeth, pulverized them in a sterile mortar in pre-reduced media, and assessed growth in the selective media used for growth of the organisms. The medium was supplemented with 40 μg/mL of gentamycin for the selection of P. gingivalis and T. forsythia.
Orofacial Abscess-scoring and Body/Splenic Weight Measurements
Animals were monitored daily for orofacial abscess development. Body weights were measured on day 0 and on day 21. Spleen weight was determined at the time of death. Five C57BL/6J wild-type mice with no pulpal exposures served as controls for spleen weight.
PCR Assays
After the animals’ death, the brain, heart, and spleen were removed aseptically, and homogenized in 1 mL of sterile 0.9% saline. DNA was extracted with the use of the DNeasy kit (Qiagen, Valencia, CA, USA). The presence of bacterial DNA (P. gingivalis, T. forsythia, T. denticola) was analyzed with 16S rRNA-specific primers (Invitrogen, Carlsbad, CA, USA) (P. gingivalis, 5′-AATCGTAACGGGCGACACAC-3′ and 5′-GGGTTGCTCCTTCATCACAC-3′; T. denticola, 5′-TAATACCGAATGTGCTCATTTACAT-3′ and 5′-TCAAAGAAGCATTCCCTCTTCTTCTTA-3′; T. forsythia, 5′-AAAACAGGGGTTCCGCATGG-3′ and 5′-TTCACCGCGGACTTAACAGC-3′) (Matto et al., 1998; Tran and Rudney, 1999; Siqueira et al., 2000). Positive controls of DNA from bacterial cultures and negative controls without DNA template were included. The master mix was prepared with Taq PCR Core Kit (Qiagen). The PCR reaction was carried out in a PTC-200 thermalcycler (MJ Research, Waltham, MA, USA) with the following programs (P. gingivalis, 30 cycles, 60″ @ 94°C/60″ @ 70°C/1′ @ 72°C; T. denticola, 36 cycles, 30″ @ 95°C/1′ @ 60°C/1′ @ 72°C; T. forsythia, 35 cycles, 30″ @ 95°C/60″ @ 60°C/60″ @ 72°C), preceded by a 2′ denaturation at 95°C and followed by a 2′ extension at 72°C (Foschi et al., 2005).
Amplification products were analyzed by electrophoresis on 1.5% agarose gels, pre-stained with ethidium bromide (0.5 μg/mL). The PCR products were visualized with a UV transilluminator (FBTI-88, Fischer Biotech, Pittsburgh, PA, USA). The expected size of each band was compared with a 100-bp DNA ladder (Invitrogen). Each positive PCR product was purified (QIAquick Gel Extraction Kit, Qiagen), sequenced, and analyzed with GeneQuest (DNASTAR, Inc., Madison, WI, USA) for confirmation of PCR results.
Micro-CT Analysis
Hemimandibles were removed, fixed, and analyzed with a compact microtomograph (μCT 20, Scanco Medical AG, Bassersdorf, Switzerland) as previously described (Balto et al., 2000). The area of periapical radiolucency was measured with the use of a standard template superimposed on the periapical region of the distal root, and the lesion was quantified with ImageJ software (http://rsb.info.nih.gov/ij, National Institutes of Health, USA).
Statistical Analysis
Data were analyzed with StatgraphicsPlus software (Manugistics, Rockville, MD, USA). A paired Student’s t test, unpaired Student’s t test, and ANOVA Kruskal-Wallis were used as indicated. The estimate of variability (T) was calculated with SPSS v13 (SPSS, Inc., Chicago, IL, USA).
RESULTS
Effects of Pathogens on Orofacial Abscess Development
The pathogenicity of mono-infection with T. denticola (108 organisms) or infection with T. denticola in combination with the other members of the “red complex” (P. gingivalis, T. forsythia, 108 total organisms) was determined in a well-established model of pulpal infection (Balto et al., 2000) in WT and SCID mice.
T. denticola mono-infection in SCID animals was characterized by the highest incidence of severe orofacial infections, with 5/10 animals exhibiting odontogenic abscesses. Abscesses developed on days 5 to 14 after infection. Infection of SCID mice with a mixture of red-complex organisms showed a surprisingly low frequency of abscesses (1/9), possibly due to the lower numbers of T. denticola (3.3 × 107) in the mixture. T. denticola or red-complex infection in WT mice resulted in abscesses in 1/8 and 1/10 animals, respectively. Mice with abscesses presented with malaise and general weakness. One animal in each of Groups B and C died before the end of the experiment.
Effects of Pathogens on Body Weight and Splenomegaly
Weight loss and cachexia were indicators of disseminating endodontic infection and sepsis in this model (Teles et al., 1997; Hou et al., 2000). There was a significant decrease in body weight of approximately 10% from day 0 to day 21 in SCID mice mono-infected with T. denticola (Group A) (Fig. 1). Animals with abscesses had more severe weight loss, suggesting that the development of orofacial abscesses was correlated with sepsis and cachexia (p < 0.01) (Fig. 1). Although not significant, SCID animals with red complex infection (Group B) also had modest weight loss, whereas WT mice gained weight during the course of the infection (Fig. 1).
Figure 1.
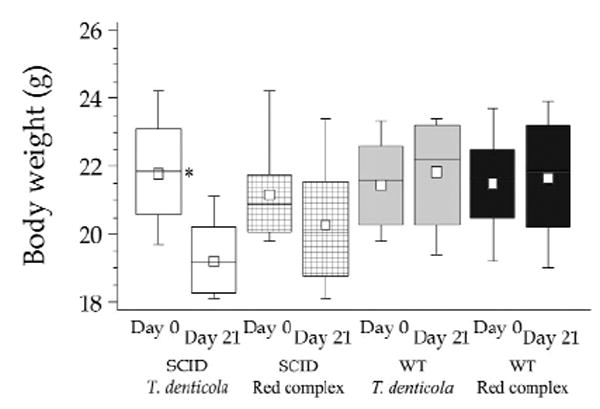
Effect of endodontic infection on body weight in immunocompetent and immunodeficient mice on day 0 and day 21 after infection. SCID mice infected with T. denticola showed a significant decrease of body weight from days 0 to 21. *p < 0.01. White boxes, SCID mice with T. denticola infection (n = 10; T = 8.03; 21.78 ± 1.52; 19.20 ± 1.04) (n, number of animals; T, variability, mean ± standard deviation @ day 0; mean ± standard deviation @ day 21); checkered boxes, SCID mice with red complex (RC) infection (n = 9; T = 3; 21.03 ± 1.41; 19.92 ± 2.05); gray boxes, WT mice with T. denticola infection (n = 8; T = 0.36; 21.18 ± 1.44; 21.82 ± 1.56); black boxes, WT mice with RC infection (n = 10; T = 0.74; 21.50 ± 1.46; 21.66 ± 1.80).
Splenomegaly was also monitored as an indicator of systemic infection. All experimental groups had similar levels of splenomegaly, compared with uninfected controls (p < 0.05), suggesting dissemination of micro-organisms to this organ (Table 1).
Table 1.
Mean Spleen Weight following Pulpal Infection with Pathogens
| Experimental Group | Mean Spleen Weight (g) ± SD |
|---|---|
| A (SCID/T. denticola) | 0.13 ± 0.02* |
| B (SCID/red complex) | 0.10 ± 0.02* |
| C (WT/T. denticola) | 0.13 ± 0.02* |
| D (WT/red complex) | 0.12 ± 0.02* |
| Control (WT, no infection) | 0.06 ± 0.01 |
SD, standard deviation
p < 0.05 vs. non-infected controls. There were no significant differences among the infected groups.
Dissemination of Endodontic Pathogens
We used PCR detection of the 16S rRNA gene of the 3 pathogens to determine if the infecting organisms had disseminated to distant sites, including the brain, heart, and spleen. T. denticola was present in a total of 9 of 117 examined organs, while P. gingivalis and T. forsythia were not detected in any of the samples (Table 2). SCID animals infected with T. denticola had the highest frequency of detection, with 4/10 animals positive for T. denticola.
Table 2.
Detection of T. denticola Genomic DNA in Organs of Mice with Concomitant Presence of Orofacial Abscesses
| Sample | |||||
|---|---|---|---|---|---|
| Group | Animal # | Brain | Heart | Spleen | Abscess |
| A (SCID/T. denticola) | 917 | + | + | + | |
| 918 | + | + | + | + | |
| 919 | + | + | |||
| 920 | + | ||||
| C (WT/T. denticola) | 986 | + | |||
| 987 | + | + | |||
Detected by PCR.
Effect on Periapical Bone Destruction
The effect of pulpal infection with T. denticola and the red complex on periapical bone loss was assessed by micro-computed tomography (Fig. 2A). All infected groups had increased periapical bone destruction compared with sham-infected controls (p < 0.01) (Fig. 2B). Analysis of differences among infected groups showed that animals mono-infected with T. denticola exhibited greater periapical bone resorption compared with SCID and WT animals infected with the red complex (p < 0.05), but had resorption similar to that of WT mice infected with T. denticola. Mono-infection with T. denticola thus caused significant bone destruction following pulpal infection.
Figure 2.
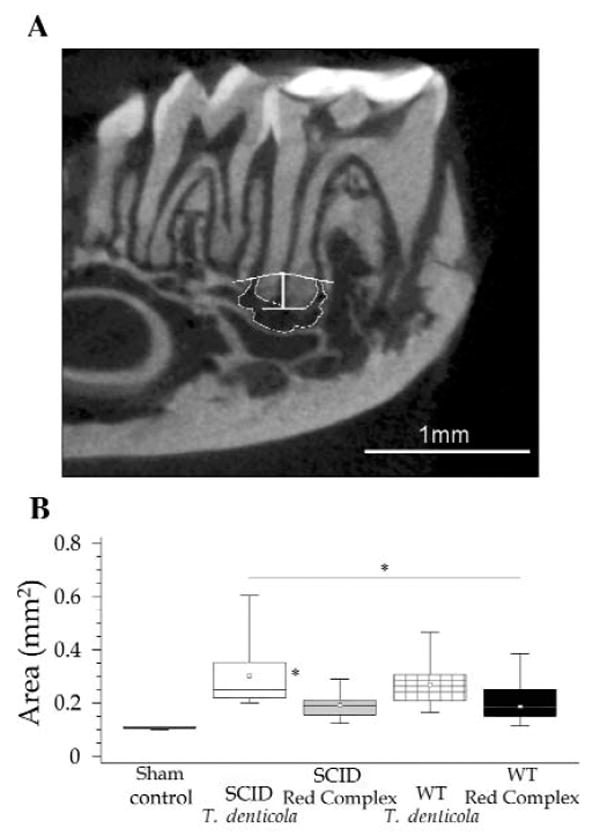
Periapical bone resorption caused by endodontic infection. (A) Micro CT image showing the pivot section of a mandibular first left molar and the area of resorption (outlined in white). (B) Comparison of areas (expressed in mm2) of periapical bone resorption in the different experimental groups. SCID animals infected with T. denticola showed periapical lesions of greater size compared with those in SCID and WT animals infected with red complex (p < 0.05). Sham controls presented significantly smaller periapical lesions compared with all other groups (p < 0.01). *p < 0.05. White boxes, SCID mice with T. denticola infection (n = 10; T = 4.555; 0.307 ± 0.132) (n, number of animals; T, variability; mean ± standard deviation). Grey boxes, SCID mice with red complex (RC) infection (n = 9; T = 8.492; 0.203 ± 0.073); checkered boxes, WT mice with T. denticola infection (n = 8; T = 9.58; 0.253 ± 0.091); black boxes, WT mice with RC infection (n = 10; T = 13.67; 0.192 ± 0.045). Sham control (n = 2; T = 7248; 0.115 ± 0.002).
DISCUSSION
T. denticola has been implicated as a key pathogen in periodontal diseases (Sela, 2001) and, more recently, has been associated with severe endodontic disease (Baumgartner et al., 2003; Foschi et al., 2005), although a direct demonstration of its role in disease pathogenesis has been lacking. The present study investigated whether T. denticola alone, or as a part of the red complex that includes P. gingivalis and T. forsythia, has a role in the etiology of endodontic pathology. Our results demonstrate that T. denticola was highly pathogenic as a mono-infection in SCID mice, with 50% of animals demonstrating orofacial abscesses, splenomegaly, cachexia, and increased periapical bone destruction. T. denticola mono-infection also resulted in increased bone loss in WT mice. All 3 organisms were recovered by culture from teeth for up to 21 days post-infection (data not shown). However, only T. denticola, but not P. gingivalis or T. forsythia, was present in distant organs, including the spleen, heart, and brain, indicating its potential for dissemination from the root canal. Analysis of these data strongly indicated that T. denticola is an important endodontic pathogen that has the potential to cause systemic manifestations, particularly in immunocompromised hosts.
T. denticola dissemination may be promoted by its ability to penetrate tight junctions of epithelial monolayers in vitro (Peters et al., 1999; Lux et al., 2001). The unique spiral shape, the periplasmic location of the flagellar filaments, and the presence of surface protease (Lux et al., 2001; Sela, 2001) are associated with its invasive potential.
T. denticola mono-infection induced significant bone resorption, which could be mediated through induction of cytokine expression, or via stimulation by T. denticola lipo-oligosaccharide. The lipo-oligosaccharide is a glycolipid that has been shown to up-regulate RANKL and, conversely, to down-regulate osteoprotegerin (Choi et al., 2003). Other pathogenic factors of T. denticola that may contribute to pathology include an immunosuppressive protein that arrests human lymphocyte proliferation at the G1 phase (Lee et al., 2004), hemolysins, and extracellular matrix protein-binding capabilities (Holt and Ebersole, 2005).
Our findings indicated that mono-infection with T. denticola was more pathogenic alone, rather than as a constituent of a polybacterial red complex infection. This most likely reflects the absolute numbers of T. denticola in the inoculum (108 in mono-infection vs. 3.3 × 107 in polymicrobial red complex infection). Previous studies in a subcutaneous injection model demonstrated increased abscess formation infection with combinations of T. denticola and P. gingivalis, although the bacterial challenge was 2-3 logs higher than in the present study (Kesavalu et al., 1998). The lack of synergism in the present study might also reflect the different ecological niches of the root canal space vs. the periodontal pocket or subcutaneous sites. In this regard, samples from infected human pulps revealed that the incidence of the red complex (8%) is significantly lower than the single constituents of the consortium (T. denticola, 44%; P. gingivalis, 30%; T. forsythia, 26%) (Rocas et al., 2001). Taken together with our data, these findings suggest that the red complex consortium plays a different role in the etiology of endodontic vs. periodontal disease.
In humans, organ dissemination of Treponema has been described to the heart (Cavrini et al., 2005), at sites of esophageal cancer (Narikiyo et al., 2004), and to the brain in Alzheimer’s patients (Riviere et al., 2002). Systemic conditions, including coronary heart disease (CHD) and low birthweight, have been linked to periodontal diseases (Loesche, 1999), and red complex bacteria have been detected in atherosclerotic plaques from coronary, carotid, and aortic arteries (Ishihara et al., 2004; Cavrini et al., 2005). A recent study indicated that the most significant association is with the titer of antibodies against oral bacteria, rather than with clinical periodontal status itself, suggesting a link to recent infectious challenge (Beck et al., 2005). In endodontics, a retrospective study found a correlation between the number of root-filled teeth and CHD, although there was a lack of correlation with periapical radiolucencies, suggesting that endodontic status may not be a risk factor for CHD (Frisk et al., 2003). Analysis of these data, taken together, suggests that root canal infection with T. denticola and other oral spirochetes may lead to dissemination to important target organs, particularly in immunosuppressed hosts.
Acknowledgments
This work was supported by grant DE-11664 (PS) from the NIDCR and by the John W. Hein Fellowship from The Forsyth Institute (JI).
References
- Balto K, Muller R, Carrington DC, Dobeck J, Stashenko P. Quantification of periapical bone destruction in mice by micro-computed tomography. J Dent Res. 2000;79:35–40. doi: 10.1177/00220345000790010401. [DOI] [PubMed] [Google Scholar]
- Baumgartner JC, Khemaleelakul SU, Xia T. Identification of spirochetes (treponemes) in endodontic infections. J Endod. 2003;29:794–797. doi: 10.1097/00004770-200312000-00002. [DOI] [PubMed] [Google Scholar]
- Beck JD, Eke P, Heiss G, Madianos P, Couper D, Lin D, et al. Periodontal disease and coronary heart disease: a reappraisal of the exposure. Circulation. 2005;112:19–24. doi: 10.1161/CIRCULATIONAHA.104.511998. [DOI] [PubMed] [Google Scholar]
- Cavrini F, Sambri V, Moter A, Servidio D, Marangoni A, Montebugnoli L, et al. Molecular detection of Treponema denticola and Porphyromonas gingivalis in carotid and aortic atheromatous plaques by FISH: report of two cases. J Med Microbiol. 2005;54:93–96. doi: 10.1099/jmm.0.45845-0. [DOI] [PubMed] [Google Scholar]
- Choi BK, Lee HJ, Kang JH, Jeong GJ, Min CK, Yoo YJ. Induction of osteoclastogenesis and matrix metalloproteinase expression by the lipooligosaccharide of Treponema denticola. Infect Immun. 2003;71:226–233. doi: 10.1128/IAI.71.1.226-233.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foschi F, Cavrini F, Montebugnoli L, Stashenko P, Sambri V, Prati C. Detection of bacteria in endodontic samples by polymerase chain reaction assays and association with defined clinical signs in Italian patients. Oral Microbiol Immunol. 2005;20:289–295. doi: 10.1111/j.1399-302X.2005.00227.x. [DOI] [PubMed] [Google Scholar]
- Frisk F, Hakeberg M, Ahlqwist M, Bengtsson C. Endodontic variables and coronary heart disease. Acta Odontol Scand. 2003;61:257–262. doi: 10.1080/00016350310005510. [DOI] [PubMed] [Google Scholar]
- Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005;38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- Hou L, Sasaki H, Stashenko P. B-cell deficiency predisposes mice to disseminating anaerobic infections: protection by passive antibody transfer. Infect Immun. 2000;68:5645–5651. doi: 10.1128/iai.68.10.5645-5651.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara K, Nabuchi A, Ito R, Miyachi K, Kuramitsu HK, Okuda K. Correlation between detection rates of periodontopathic bacterial DNA in coronary stenotic artery plaque [corrected] and in dental plaque samples. J Clin Microbiol. 2004;42:1313–1315. doi: 10.1128/JCM.42.3.1313-1315.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izard J, Samsonoff WA, Limberger RJ. Cytoplasmic filament-deficient mutant of Treponema denticola has pleiotropic defects. J Bacteriol. 2001;183:1078–1084. doi: 10.1128/JB.183.3.1078-1084.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakehashi S, Stanley HR, Fitzgerald RJ. The effects of surgical exposures of dental pulps in germ-free and conventional laboratory rats. Oral Surg Oral Med Oral Pathol. 1965;20:340–349. doi: 10.1016/0030-4220(65)90166-0. [DOI] [PubMed] [Google Scholar]
- Kawashima N, Niederman R, Hynes RO, Ullmann-Cullere M, Stashenko P. Infection-stimulated infraosseus inflammation and bone destruction is increased in P-/E-selectin knockout mice. Immunology. 1999;97:117–123. doi: 10.1046/j.1365-2567.1999.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesavalu L, Holt SC, Ebersole JL. Virulence of a polymicrobic complex, Treponema denticola and Porphyromonas gingivalis, in a murine model. Oral Microbiol Immunol. 1998;13:373–377. doi: 10.1111/j.1399-302x.1998.tb00694.x. [DOI] [PubMed] [Google Scholar]
- Lee W, Pankoski L, Zekavat A, Shenker BJ. Treponema denticola immunoinhibitory protein induces irreversible G1 arrest in activated human lymphocytes. Oral Microbiol Immunol. 2004;19:144–149. doi: 10.1111/j.0902-0055.2004.00129.x. [DOI] [PubMed] [Google Scholar]
- Loesche WJ. Anaerobic periodontal infections as risk factors for medical diseases. Curr Infect Dis Rep. 1999;1:33–38. doi: 10.1007/s11908-999-0007-5. [DOI] [PubMed] [Google Scholar]
- Lux R, Miller JN, Park NH, Shi W. Motility and chemotaxis in tissue penetration of oral epithelial cell layers by Treponema denticola. Infect Immun. 2001;69:6276–6283. doi: 10.1128/IAI.69.10.6276-6283.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matto J, Saarela M, Alaluusua S, Oja V, Jousimies-Somer H, Asikainen S. Detection of Porphyromonas gingivalis from saliva by PCR by using a simple sample-processing method. J Clin Microbiol. 1998;36:157–160. doi: 10.1128/jcm.36.1.157-160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narikiyo M, Tanabe C, Yamada Y, Igaki H, Tachimori Y, Kato H, et al. Frequent and preferential infection of Treponema denticola, Streptococcus mitis, and Streptococcus anginosus in esophageal cancers. Cancer Sci. 2004;95:569–574. doi: 10.1111/j.1349-7006.2004.tb02488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters SR, Valdez M, Riviere G, Thomas DD. Adherence to and penetration through endothelial cells by oral treponemes. Oral Microbiol Immunol. 1999;14:379–383. doi: 10.1034/j.1399-302x.1999.140609.x. [DOI] [PubMed] [Google Scholar]
- Riviere GR, Riviere KH, Smith KS. Molecular and immunological evidence of oral Treponema in the human brain and their association with Alzheimer’s disease. Oral Microbiol Immunol. 2002;17:113–118. doi: 10.1046/j.0902-0055.2001.00100.x. [DOI] [PubMed] [Google Scholar]
- Rocas IN, Siqueira JF, Jr, Santos KR, Coelho AM. “Red complex” (Bacteroides forsythus, Porphyromonas gingivalis, and Treponema denticola) in endodontic infections: a molecular approach. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2001;91:468–471. doi: 10.1067/moe.2001.114379. [DOI] [PubMed] [Google Scholar]
- Sela MN. Role of Treponema denticola in periodontal diseases. Crit Rev Oral Biol Med. 2001;12:399–413. doi: 10.1177/10454411010120050301. [DOI] [PubMed] [Google Scholar]
- Siqueira JF, Jr, Rocas IN. Treponema species associated with abscesses of endodontic origin. Oral Microbiol Immunol. 2004;19:336–339. doi: 10.1111/j.1399-302x.2004.00156.x. [DOI] [PubMed] [Google Scholar]
- Siqueira JF, Jr, Rocas IN, Favieri A, Santos KR. Detection of Treponema denticola in endodontic infections by 16S rRNA gene-directed polymerase chain reaction. Oral Microbiol Immunol. 2000;15:335–337. doi: 10.1034/j.1399-302x.2000.150512.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Sundqvist G. Taxonomy, ecology, and pathogenicity of the root canal flora. Oral Surg Oral Med Oral Pathol. 1994;78:522–530. doi: 10.1016/0030-4220(94)90047-7. [DOI] [PubMed] [Google Scholar]
- Teles R, Wang CY, Stashenko P. Increased susceptibility of RAG-2 SCID mice to dissemination of endodontic infections. Infect Immun. 1997;65:3781–3787. doi: 10.1128/iai.65.9.3781-3787.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran SD, Rudney JD. Improved multiplex PCR using conserved and species-specific 16S rRNA gene primers for simultaneous detection of Actinobacillus actinomycetemcomitans, Bacteroides forsythus, and Porphyromonas gingivalis. J Clin Microbiol. 1999;37:3504–3508. doi: 10.1128/jcm.37.11.3504-3508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A, Kawada M, Suzuki N, Nakano Y, Oho T, Saito T, et al. TaqMan real-time polymerase chain reaction assay for the correlation of Treponema denticola numbers with the severity of periodontal disease. Oral Microbiol Immunol. 2004;19:196–200. doi: 10.1111/j.0902-0055.2004.00142.x. [DOI] [PubMed] [Google Scholar]


