To the Editor:
Amelogenesis imperfecta (AI) is a collection of non-syndromic inherited diseases featuring a variety of abnormal enamel phenotypes, patterns of inheritance, and causative genes. The term is also used to indicate the presence of an enamel phenotype in syndromes. Dental enamel is the most highly mineralized tissue in the body, lacks collagen, and is the product of specialized epithelial cells (ameloblasts). During the formation of enamel, ameloblasts rely upon many genes that function in other tissues, like laminin-332 and type XVII collagen. Mutations in such genes typically cause syndromes that include enamel malformations as a phenotype. Non-syndromic AI is caused by defects in genes that are more functionally specialized for dental enamel formation. Mutations in AMELX, ENAM, FAM83H, WDR72, KLK4 and MMP20 cause non-syndromic AI, and account for about half of all AI cases (1). The other causative genes are unknown. To date, no non-dental phenotype or history of an associated non-dental phenotype has been reported in any person with an AI-causing defect in any of these genes.
Increasingly, AI is classified according to the pattern of inheritance and genetic etiology. Most cases of autosomal dominant amelogenesis imperfecta are caused by mutations in FAM83H (family with sequence similarity, member H; 8q24.3) or ENAM (enamelin; 4q21), the rest are of unknown etiology. FAM83H mutations cause autosomal dominant hypocalcified AI (OMIM: #130900), which has a distinctive enamel phenotype. At eruption, the enamel has a pale-cream color and is poorly mineralized. It is soft and undergoes rapid attrition and staining so that only the cervical enamel near the gingiva may remain. In contrast, ENAM mutations are dose dependent and cause local hypoplastic AI (OMIM: #104500), featuring regions of abnormally thin enamel or a nearly complete lack of enamel when both alleles are defective.
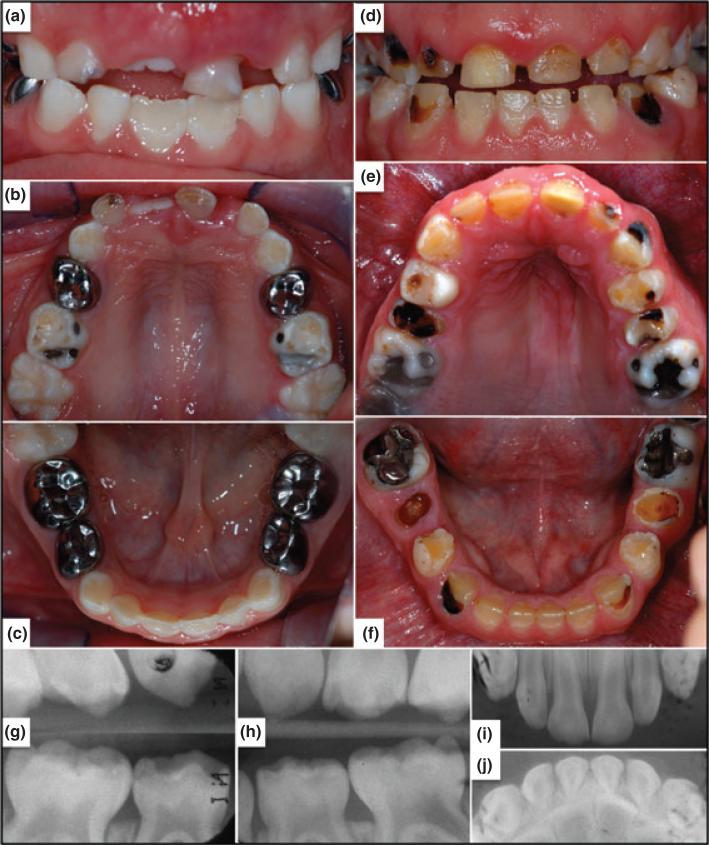
Recently, we recruited a Caucasian family with autosomal dominant hypoplastic AI that falls within the range of phenotypes characteristic of ENAM mutations. The proband was a 6.5 years old Caucasian male. His mixed dentition displayed thin enamel that was only slightly more radiopaque than dentin and was secondarily affected with dental caries (Fig. 1a–c). The incisal edges of the primary anterior teeth were chipped. The permanent mandibular central incisors had recently erupted and exhibited particularly thin enamel over the incisal two thirds of the crown. This was not due to attrition, but was how the teeth formed. The proband's father had similar enamel defects (Fig. 1d–f) and reported that his teeth came in small, just like his baby teeth. His dentition was characterized as having small, well-separated teeth with extensive carious lesions and attrition. The remaining enamel surfaces were rough and pitted.
Fig. 1.
Oral photos and dental radiographs. (a–c) Frontal and maxillary and mandibular occlusal photos of the proband at age 6.5 years. Note the chips on the incisal edges of primary anterior teeth and that the enamel is particularly thin on the incisal 2/3 of the recently erupted mandibular central incisors. (d–f) Oral photographs of proband's father. Note the spacing of the maxillary anterior teeth secondary to having thin enamel on the crowns. The mandibular incisors have thicker enamel in the cervical third like the proband. (h–j) Oral radiographs of primary dentition taken at age 3 prior to the placement of dental restorations.
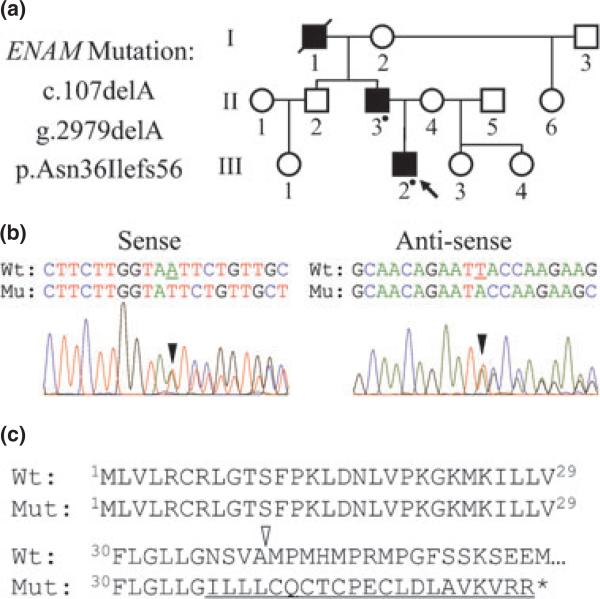
The pedigree indicated an autosomal dominant pattern of inheritance (Fig. 2a). Mutational analyses identified a novel heterozygous frameshift mutation in exon 4 of ENAM (c.107delA; g.2979delA; p.Asn36Ilefs56) in both the proband and his father. The frameshift occurs in the coding region for the signal peptide and would have precluded synthesis of the entire secreted protein (Fig. 2c). This sequence variation is not listed among the known polymorphisms in the current single nucleotide polymorphism database (dbSNP, National Center for Biotechnology Information).
Fig. 2.
Identifying the AI-causing ENAM mutation. (a) Pedigree showing the autosomal dominant pattern of inheritance. A dot indicates individuals who were examined and contributed DNA. (b) DNA sequencing chromatograms showing the single nucleotide deletion (c.107delA; g.2979delA) that shifted the reading frame (black arrowheads) in one ENAM allele. (c) Conceptual translation of the wild-type (WT) and mutant (Mut) alleles. The frameshift (p.Asn36Ilefs56) occurs in codon 36 prior to the signal peptide cleavage site (white arrowhead) and would probably cause the mutant mRNA to be degraded by nonsense mediated decay.
This AI-causing ENAM mutation is the 12th to be reported (Fig. S1). To help clinicians identify autosomal dominant AI patients with probable ENAM mutations based upon their dental phenotypes, previously published oral photographs of patients with defined mutations in a single ENAM allele are provided in Figs S2 and S3. When only one ENAM allele is defective, the enamel malformations vary in severity, but rarely show a complete lack of penetrance. In its mildest form, the enamel defects are well-circumscribed enamel pits, often arranged in horizontal lines. With increasing severity, the enamel defects appear as horizontal grooves, usually in the cervical third of the crown, or as slightly thin enamel that transitions abruptly to very thin enamel with a border that runs horizontally around the crown. When both ENAM alleles are defective, the enamel layer is either completely absent or appears as a very thin mineral layer that only partially covers the crown (Fig. S4).
The International Classification of Diseases lists AI under hereditary disturbances in tooth structure, not elsewhere classified (K00.5) (2). As AI is a diverse collection of diseases caused by more than six genes that can be transmitted in an autosomal dominant, autosomal recessive, or X-linked pattern and can be mistaken for enamel malformations associated with non-dental manifestations, a more definitive diagnosis is warranted when practicable. When the pattern of inheritance and genotype–phenotype correlations implicate a single gene as being the probable cause, mutational analyses should be considered to establish a specific diagnosis.
Supplementary Material
Acknowledgements
We thank the participants in this study for their participation. We thank the publishers who gave permission to reproduce previously published oral photographs online. This work was supported by NIDCR/NIH grants DE015846 and DE011301.
Footnotes
Supporting Information
The following Supporting information is available for this article:
References
- 1.Chan HC, Estrella NM, Milkovich RN, Kim JW, Simmer JP, Hu JC. Target gene analyses of 39 amelogenesis imperfecta kindreds. Eur J Oral Sci. 2011;119(Suppl. 1):311–323. doi: 10.1111/j.1600-0722.2011.00857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . International Statistical Classification of Diseases and Related Health Problems 10th Revision (ICD-10) World Health Organization; 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.