Abstract
Objective
To evaluate the association of risk and age at onset (AAO) of Alzheimer disease (AD) with single-nucleotide polymorphisms (SNPs) in the chromosome 19 region including apolipoprotein E (APOE) and a repeat-length polymorphism in TOMM40 (poly-T, rs10524523).
Design
Conditional logistic regression models and survival analysis.
Setting
Fifteen genome-wide association study data sets assembled by the Alzheimer's Disease Genetics Consortium.
Participants
Eleven thousand eight hundred forty AD cases and 10 931 cognitively normal elderly controls.
Main Outcome Measures
Association of AD risk and AAO with genotyped and imputed SNPs located in an 800-Mb region including APOE in the entire Alzheimer's Disease Genetics Consortium data set and with the TOMM40 poly-T marker genotyped in a subset of 1256 cases and 1605 controls.
Results
In models adjusting for APOE ε4, no SNPs in the entire region were significantly associated with AAO at P<.001. Rs10524523 was not significantly associated with AD or AAO in models adjusting for APOE genotype or within the subset of ε3/ε3 subjects.
Conclusions
APOE alleles ε2, ε3, and ε4 account for essentially all the inherited risk of AD associated with this region. Other variants including a poly-T track in TOMM40 are not independent risk or AAO loci.
The association of the apolipoprotein E (APOE) polymorphism with late-onset Alzheimer disease (AD) is one of the strongest and most robust genetic risk factors for a common disease. Compared with the common APOE ε3 allele, ε4 increases the risk and lowers the age at onset (AAO) of AD in a dose-dependent fashion whereas the ε2 allele confers a protective benefit.1,2 Although the frequency of ε4 varies among different ethnic groups, the ε4/AD association is evident in diverse populations,3 with a few notable exceptions.4–6 The strength of the association is greatly influenced by age and sex.3 Recent genome-wide association studies (GWAS) have repeatedly reported association signals in APOE and genes in its vicinity,7–9 but the evidence favoring additional AD risk variants in this region is much weaker after accounting for the strong linkage disequilibrium that extends over 3 Mb including these other proposed AD loci.8 Nonetheless, interest in this region remains high because several of these genes have a plausible role in AD pathogenesis.
Roses et al10 reported an association between a variable length poly-T polymorphism (“poly-T”) at rs10524523 in the gene encoding the channel-forming subunit of the translocase of the mitochondrial outer membrane (TOMM40) and risk for and AAO of AD. These investigators used an evolutionary network approach to build phylogenies that provided evidence of selection for variable lengths of the poly-T repeats between cases and controls. The number of poly-T repeats at the rs10524523 locus were grouped into 3 alleles consisting of short (s) (<21), long (l) (21–29), and very long (v) (≥30). Phylogenetic tree analysis indicated that the APOE ε4 allele tracks with the l allele, whereas the APOE ε3 allele tracks with the s and v alleles. The l allele was associated with a 7-year earlier AAO of AD in a small sample (N=34) of APOE ε3/ε4 subjects. Support for an independent role of TOMM40 in AD was obtained from a study showing association of the v/v genotype with lower performance on learning and lower gray matter volume among 117 APOE ε3/ε3 adults.11 A more recent study of this polymorphism in a much larger sample failed to confirm the original findings after adjusting for the effect of APOE ε4.12
In this study, we conducted a comprehensive association study of AD with markers in the APOE region using data from nearly 23 000 subjects assembled by the Alzheimer's Disease Genetics Consortium (ADGC) for a GWAS that identified several new AD risk loci.8 We also evaluated association with the TOMM40 poly-T polymorphism by direct genotyping of 1256 AD cases and 1605 controls and by analysis in the entire GWAS data set of several poly-T proxy single-nucleotide polymorphisms (SNPs).
METHODS
STUDY POPULATION
The primary sample used was 15 GWAS data sets assembled by the ADGC. Details of ascertainment and diagnostic procedures for each data set have been extensively described elsewhere.8 Data from a total of 11 840 AD cases and 10 931 cognitively normal elderly controls were available for this study. All subjects were recruited under protocols approved by the appropriate institutional review boards.
GENOTYPING
GWAS Genotyping
Genotyping for the 15 ADGC cohorts was performed using various genotyping arrays containing between approximately 310 000 and 1.5 million SNPs for each data set.8
APOE Genotyping
APOE genotypes in the Adult Changes in Thought (ACT) Study, the National Institute on Aging (NIA) Alzheimer's Disease Centers (ADCs), the Multi-Site Collaborative Study for Genotype-Phenotype Associations in Alzheimer's Disease Study, the Mayo Clinic, the NIA Late-Onset Alzheimer's Disease Study, and the University of Miami/Vanderbilt University/Mt. Sinai School of Medicine data sets were determined based on allelic combinations of SNPs rs7412 and rs429358. APOE genotyping was performed in the Multi-Institutional Research on Alzheimer's Disease Genetic Epidemiology Study cohort using the Roche Diagnostics LightCycler 480 instrument (Roche Diagnostics) LightMix Kit ApoE C112R R158 (catalog number 40-0445-16) from TIB MOLBIOL.13APOE genotypes in the Translational Genomics Research Institute series 2, the Alzheimer's Disease Neuroimaging Initiative (ADNI) Study, the University of Pittsburgh, and Washington University cohorts were obtained by pyrosequencing14 or restriction fragment length polymorphism analysis.15,16APOE genotypes in the Rush University Religious Orders Study/Memory and Aging Project data set were determined using high-throughput sequencing of codon 112 (position 3937) and codon 158 (position 4075) of exon 4 of the APOE gene on chromosome 19.
Poly-T Genotyping
Three ADGC cohorts were genotyped for poly-T: ACT (290 AD cases, 1271 controls), ADC (831 AD cases, 282 controls), and ADNI (137 AD cases, 162 controls). Poly-T genotypes were determined using a modified short tandem repeat genotyping assay. This assay used a polymerase chain reaction primer set (Ch19_50094815-F: VIC-GCTGACCTCAAGCTGTCCTC that labeled with VIC fluorescent dye and Ch19_50095061-R: GGAGGGACAGGGAAAGAAAA) to amplify a 247–base pair fragment from each subject's genomic DNA. For each polymerase chain reaction, 100 ng of genomic DNA, 12μM primers, 3.75 μL of Qiagen HotStarTaq Master Mix (Qiagen), and 1mM magnesium chloride were mixed together with a final volume of 7.5 μL. Polymerase chain reaction was carried out with a profile of 95°C for 15 minutes and then 30 cycles at 95°C for 30 seconds, 55°C for 30 seconds, and 64°C for 30 seconds. Precise length of the amplified fragments was acquired through an ABI 3130xl Genetic Analyzer and processed with ABI Gene-Mapper version 4.0 software (Applied Biosystems). To increase calling accuracy of poly-T counts of each fragment, we also cloned the same genomic fragments of 4 control poly-T variants (ie, 13xT, 16xT, 22xT, and 35xT) into a DNA vector (pBluescript; Thermo Fisher Scientific) and used them as internal controls to create bins for fragment size standards. Integrity of the bins was further validated by genotyping poly-T inserts from plasmid combinations (eg, 16 plus 22, 16 plus 35, and 22 plus 35). Spacing of the bins was then fine-tuned accordingly. Typically, each allele was associated with a series of peaks and the highest peak in the series was assigned as the allele of interest. Thus, homozygous and heterozygous individuals will have either 1 or 2 alleles, respectively. The final calling of poly-T counts was then determined via manual inspection and cross-checking of the electropherograms.
As a check on genotyping accuracy, we genotyped 352 samples from the NIA Late-Onset Alzheimer's Disease Study included in a previous study of the poly-T polymorphism.12 There were no discrepancies between the 2 laboratories in calling the s, l, and v alleles. In addition, there was complete agreement in the genotypes for 90 ADNI subjects included in this and the Cruchaga et al12 studies. One genotype was discordant with the genotype publically available from the ADNI website. Finally, genotypes from 16 subjects were confirmed by genomic DNA cloning and Sanger capillary sequencing independently at the University of Washington and the University of Pennsylvania.
GENOTYPE IMPUTATION AND QUALITY CONTROL
The APOE region was defined as SNPs located between map positions 45 000 000 and 45 800 000 base pairs according to the University of California, Santa Cruz Genome browser (hg19, GRCh37). This region encompasses CEACAM22P and EXOC3L2, which contained previously identified significant association signals (P<10−4) without adjustment for APOE genotype.8 Genotypes for all SNPs in this region were imputed with the Markov chain haplotyping software17 using reference haplotypes for white subjects in the HapMap phase 2 (release 22) database. This procedure also filled in missing data for the genotyped SNPs. Individuals with high genotyping call rates (>95%) and SNPs with 95% call rates or better were used as seeds for the imputation procedure. We excluded SNPs with low minor allele frequency (<2%), SNPs not in Hardy-Weinberg equilibrium (P<10−6), and SNPs with potential for undetected strand flips (C/G and A/T coding) to ensure consistency of allele frequencies between the test and reference haplotypes and to improve the quality of imputation. Imputation quality was determined as R2, which estimates the squared correlation between imputed and true genotypes. We applied stringent criteria for quality control assessment of imputed SNPs (R2≥0.8 in each data set), since inclusion of SNPs with lower-quality imputation may lead to spurious associations.18 After filtering, 367 SNPs in the APOE region were available for this study.
ASSESSMENT OF POPULATION SUBSTRUCTURE
We examined population substructure in each data set by analyzing tagging SNPs from the genome-wide panels using the smartpca module from EIGENSTRAT software19 in a manner described previously.8 The strength of association of the top 10 principal components was tested with the outcome (presence of AD and AAO of AD) and also with the rs10524523 genotype. The top 3 principal components were included in association models to adjust for hidden substructure, though none of the principal components were associated with either presence or AAO of AD at P<10−3.
GENETIC ASSOCIATION ANALYSES
Poly-T genotypes were determined in the ACT, ADNI, and ADC data sets as described previously.10,11 Association of AD risk with poly-T was evaluated using logistic regression models including a term for poly-T defined as dosage for one of the alleles. We also tested genotype models assigning v/v as the reference genotype. Linear regression was used to test association of poly-T with AAO in the case sample. Models for AD risk included covariates for population substructure within data sets, age (AAO or age at death if deceased and AAO unknown in cases; age at last examination or death in controls), and sex. Population substructure and sex were included in models for AAO. The influence of APOE on the associations with poly-T was evaluated in 2 ways. In the first approach, an additive model with a term for the number of APOE ε4 alleles (0, 1, or 2) was added to the models. Significant SNPs were further evaluated in models including APOE genotype as a covariate and random-effects models allowing for heterogeneity of the association among data sets. In the second approach, models were evaluated in APOE genotype subgroups; conversely, we assessed the effect of the APOE ε4 allele within the poly-T subgroups. To capture information about association with poly-T in other ADGC data sets, we tested association with genotyped SNPs that were in high linkage disequilibrium (LD) (r2≥0.8) with rs10524523. All regression analyses were conducted using the R statistical package in each data set separately, and the results were meta-analyzed using an inverse-variance method as implemented in the package METAL.20 The respective influences of the APOE and poly-T loci on AAO were also evaluated by comparing Kaplan-Meier survival curves derived using R for subgroups of AD cases defined by APOE and poly-T genotypes. Association of all other genotyped and imputed SNPs from the APOE region with AD risk and AAO was evaluated in all ADGC data sets using the strategy described earlier.
RESULTS
ASSOCIATION OF POLY-T WITH AD RISK AND AAO
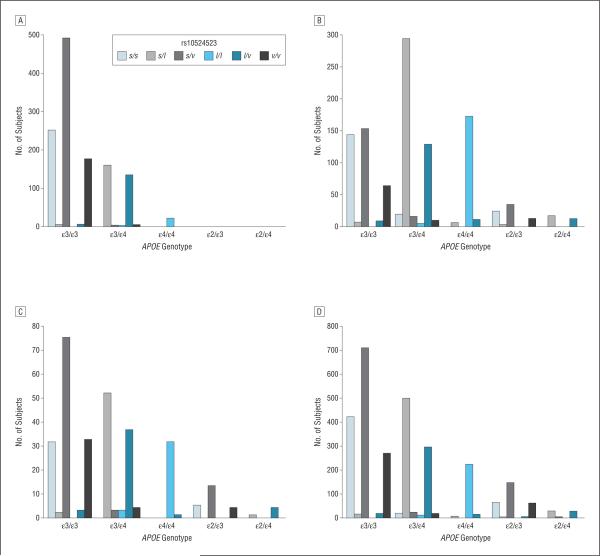
To determine if poly-T genotypes at rs10524523 confer risk for AD or affect AAO for AD, we genotyped 1256 AD cases and 1605 controls from the ACT, ADC, and ADNI cohorts (Table 1). The mean AAO in the ACT cohort was about 12 years higher (83.8 years) than that in the ADC (71.2 years) and ADNI (71.7 years) cohorts. The distribution of the poly-T lengths within each APOE genotype subgroup was comparable with the corresponding distributions reported in the original study,10 and these patterns were similar across data sets (eFigure 1, http://www.archneurol.com). Nearly all subjects with the s/s or s/v genotypes had APOE genotypes ε3/ε3 or ε2/ε3 (eTable 1). Similarly, there was a very high correlation between heterozygosity for the ε4 and l alleles, and nearly all l homozygotes were homozygous for ε4 (Figure 1).
Table 1.
Poly-T (rs10524523) Genotype Frequencies in All Subjects and in APOE ε3/ε3 Subjects
| All Subgroups |
ε3/ε3 Subgroup |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases |
Controls |
Cases |
Controls |
|||||||||
| Study | No. | Freq | AAO,y | No. | Freq | AAE,y | No. | Freq | AAO,y | No. | Freq | AAE,y |
|
| ||||||||||||
| ACT | ||||||||||||
| s/s | 60 | 0.208 | 84.85 | 225 | 0.193 | 82.41 | 56 | 0.357 | 84.91 | 193 | 0.253 | 82.5 |
| s/l | 50 | 0.174 | 81.98 | 124 | 0.107 | 81.77 | 1 | 0.006 | 85 | 5 | 0.007 | 83.4 |
| s/v | 82 | 0.285 | 84.39 | 507 | 0.436 | 81.52 | 68 | 0.433 | 84.56 | 416 | 0.546 | 81.85 |
| l/l | 11 | 0.038 | 82.91 | 15 | 0.013 | 80.00 | 0 | 0 | NA | 0 | 0 | NA |
| l/v | 43 | 0.149 | 82.56 | 112 | 0.096 | 80.69 | 0 | 0 | NA | 6 | 0.008 | 81 |
| v/v | 42 | 0.146 | 84.38 | 181 | 0.155 | 81.47 | 32 | 0.204 | 84.28 | 142 | 0.186 | 81.69 |
| ADC | ||||||||||||
| s/s | 97 | 0.117 | 72.51 | 85 | 0.304 | 78.08 | 80 | 0.369 | 72.48 | 61 | 0.407 | 77.98 |
| s/l | 274 | 0.330 | 72.26 | 44 | 0.157 | 73.72 | 4 | 0.018 | 76 | 2 | 0.013 | 63.5 |
| s/v | 117 | 0.141 | 72.22 | 82 | 0.293 | 79.34 | 94 | 0.433 | 71.82 | 56 | 0.373 | 79.05 |
| l/l | 164 | 0.197 | 68.36 | 9 | 0.032 | 67.00 | 0 | 0 | NA | 0 | 0 | NA |
| l/v | 133 | 0.160 | 71.35 | 23 | 0.082 | 76.87 | 3 | 0.014 | 69 | 5 | 0.033 | 76.4 |
| v/v | 46 | 0.055 | 70.41 | 37 | 0.132 | 79.76 | 36 | 0.166 | 70.22 | 26 | 0.173 | 81.11 |
| ADNI | ||||||||||||
| s/s | 13 | 0.095 | 74.85 | 23 | 0.143 | 78.48 | 11 | 0.256 | 74.45 | 20 | 0.202 | 78.7 |
| s/l | 34 | 0.248 | 71.06 | 20 | 0.124 | 79.40 | 1 | 0.023 | 81 | 1 | 0.01 | 75 |
| s/v | 21 | 0.153 | 73.09 | 69 | 0.429 | 78.43 | 19 | 0.442 | 72.89 | 55 | 0.556 | 78.8 |
| l/l | 29 | 0.212 | 68.34 | 5 | 0.031 | 76.80 | 0 | 0 | NA | 0 | 0 | NA |
| l/v | 26 | 0.190 | 71.69 | 18 | 0.112 | 78.55 | 1 | 0.023 | 72 | 2 | 0.02 | 75 |
| v/v | 14 | 0.102 | 75.43 | 26 | 0.161 | 78.69 | 11 | 0.256 | 77.27 | 21 | 0.212 | 78.71 |
| Combined | ||||||||||||
| s/s | 170 | 0.135 | 77.04 | 333 | 0.207 | 81.03 | 147 | 0.353 | 77.28 | 274 | 0.271 | 79.73 |
| s/l | 358 | 0.285 | 73.50 | 188 | 0.117 | 79.63 | 6 | 0.014 | 80.67 | 8 | 0.008 | 73.97 |
| s/v | 220 | 0.175 | 76.84 | 658 | 0.410 | 80.92 | 181 | 0.434 | 76.42 | 527 | 0.521 | 79.9 |
| l/l | 204 | 0.162 | 69.14 | 29 | 0.018 | 75.41 | 0 | 0 | NA | 0 | 0 | NA |
| l/v | 202 | 0.161 | 73.78 | 153 | 0.095 | 79.86 | 4 | 0.01 | 70.5 | 13 | 0.013 | 77.47 |
| v/v | 102 | 0.081 | 76.85 | 244 | 0.152 | 80.91 | 79 | 0.189 | 77.26 | 189 | 0.187 | 80.5 |
Abbreviations: ACT, Adult Changes in Thought Study; AAE, mean age at examination; AAO, mean age at onset; ADC, National Institute on Aging Alzheimer's Disease Centers; ADNI, Alzheimer's Disease Neuroimaging Initiative Study; APOE, apolipoprotein E; Freq, frequency; l, long allele; NA, not applicable; No., total sample size; s, short allele; v, very long allele.
Figure 1.
Distribution of TOMM40 rs10524523 genotypes (derived from combinations of the short [s], long [l], and very long [v] alleles) according to apolipoprotein E (APOE) genotype in the Adult Changes in Thought Study (A), National Institute on Aging Alzheimer's Disease Centers (B), Alzheimer's Disease Neuroimaging Initiative Study (C), and the combined (D) data sets.
Without adjustment for APOE ε4, the poly-T l allele was significantly associated with increased AD risk (meta-analysis P value [meta-P] = 3.9 × 10−33), whereas the other alleles were protective (meta-P value: s = 5.9 × 10−8 and v = 1.9 × 10−8) (Table 2 and eTable 2). The dosage of the l allele was associated with an increased risk of AD (odds ratio [OR], 2.83; 95% CI, 2.39–3.36), while those of the s and v alleles were protective (s: OR, 0.69; 95% CI, 0.61–0.79; v: OR, 0.68; 95% CI, 0.59–0.78). However, the effect of the l allele on AD risk was greatly diminished after adjustment for the APOE ε4 allele (meta-P = .02; OR, 1.70; 95% CI, 1.09–2.65) and not significant in the ε3/ε3 subgroup (meta-P = .45), suggesting that risk of AD is influenced directly and specifically by APOE genotype and not the poly-T genotype. The apparent lack of association of ε4 with AD risk in the l-negative subgroup and l with AD in the ε4-negative subgroup is explained by the observation that virtually all persons with the ε4 allele also had the l allele. Thus, because very few AD cases and controls had ε4 but not the l allele, these particular association tests have very little power.
Table 2.
Association of the rs10524523 /allele With AD Risk and Age at Onset
| Basic Modela |
Conditional on ε4 Dosageb |
ε3/ε3 Subgroupa |
||||
|---|---|---|---|---|---|---|
| Study | OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value |
|
| ||||||
| AD | ||||||
| ACT | 2.08 (1.62–2.68) | 9.1 × 10−9 | 0.91 (0.38–2.16) | .83 | 0.49 (0.06–3.85) | .50 |
| ADC | 3.86 (2.94–5.06) | 1.1 × 10−22 | 2.05 (1.14–3.7) | .016 | 0.57 (0.18–1.83) | .35 |
| ADNI | 3.22 (2.06–5.04) | 2.9 × 10−7 | 2.38 (0.8–7.08) | .12 | 1.68 (0.25–11.25) | .59 |
| Meta-analysis | 2.83 (2.39–3.36) | 3.9 × 10−33 | 1.70 (1.09–2.65) | .020 | 0.71 (0.29–1.73) | .45 |
| Age at onset | β(SE) | PValue | β(SE) | PValue | β(SE) | PValue |
| ACT | −1.73 (0.49) | 5.3 × 10−4 | 1.15 (1.59) | .47 | −0.91 (4.57) | .84 |
| ADC | −1.62 (0.45) | 3.3 × 10−4 | 1.75 (1.37) | .20 | 2.04 (4.63) | .66 |
| ADNI | −2.77 (0.94) | .0037 | 1.42 (2.66) | .59 | 1.67 (6.36) | .79 |
| Meta-analysis | −1.79 (0.31) | 1.0 × 10−8 | 1.48 (0.97) | .12 | 0.78 (2.90) | .79 |
Abbreviations: ACT, Adult Changes in Thought Study; AD, Alzheimer disease; ADC, National Institute on Aging Alzheimer's Disease Centers; ADNI, Alzheimer's Disease Neuroimaging Initiative Study; APOE, apolipoprotein E; l, long allele; OR, odds ratio.
Adjusted for population substructure, age, and sex for AD risk and population substructure and sex for age at onset.
Adjusted for population substructure, age, sex, and number of APOE ε4 alleles for AD risk and population substructure, sex, and number of APOE ε4 alleles for age at onset.
Analogously, there was evidence of significant association of the l allele with AAO in the combined sample (meta-P = 1.0 × 10−8) and within each data set without accounting for the number of APOE ε4 alleles (Table 2 and eTable 3). These data show that each dose of the l allele is associated with a 2-year earlier onset of AD symptoms. However, this association was no longer significant after conditioning on the number of APOE ε4 alleles (meta-P = .12). Specificity of the association of AAO with APOE was supported by the lack of association with the l allele in the subgroup lacking ε4 (meta-P = .87) and evidence for a moderate association with the ε4 allele in the subgroup lacking the l allele (meta-P = .022) (eTable 2 and eTable 3). These results suggest that APOE ε4 has an effect on AAO independent of the TOMM40 poly-T l allele, whereas the association of the poly-T polymorphism is more likely due to confounding with APOE.
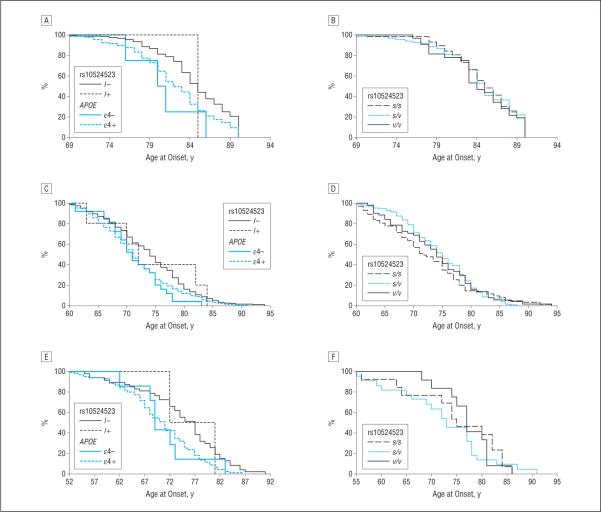
The effect of poly-T on AAO was further examined by survival analysis in each data set (Figure 2). Among subjects with AD in the l-negative subgroup, the ε4 allele showed a trend of association with earlier onset, but the effect of the l allele among subjects lacking ε4 was inconclusive because of a small sample size (Figure 2A, C, and E). There were no distinguishable differences in AAO according to poly-T genotype among ε3/ε3 subjects with AD, which is not surprising because few of these individuals had an l allele (Figure 2B, D, and F).
Figure 2.
Survival analysis curves for age at onset of Alzheimer disease in the Adult Changes in Thought Study (A and B), National Institute on Aging Alzheimer's Disease Centers (C and D), and Alzheimer's Disease Neuroimaging Initiative Study (E and F) data sets. The effect of the presence or absence of the TOMM40 long (l) allele at rs10524523 and of the apolipoprotein E (APOE) ε4 allele on age at onset is shown in all subjects (A, C, and E) and in the APOE ε3/ε3 subgroup (B, D, and E). s Indicates short allele and v, very long allele.
Evaluation of the LD structure in this region revealed that in each data set rs10524523 was strongly correlated only with SNPs in the interval including TOMM40 and APOE (eFigure 2). We identified 5 SNPs (rs157580, rs2075650, rs8106922, rs405509, and rs439401) in high LD with rs10524523 (eFigure 2) and thus considered these SNPs as proxies for poly-T in analyses in the other ADGC data sets, which were not genotyped for rs10524523. None of these SNPs was significantly associated with AD or AAO after adjustment for APOE ε4 (Table 3).
Table 3.
Association of SNPs Tagging rs10524523 With AD Risk and AAO in all ADGC Data Sets
| Average r2 With rs10524523a |
L0ADb |
AAOc |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | BP | Near Gene | RA | RAF | s-v | s-l | v-l | OR (95% CI) | P | β(SE) | P |
|
| |||||||||||
| rs157580 | 45395266 | TOMM40 | A | 0.671 | 0.80 | 0.00 | 0.72 | 1.05 (0.98–1.12) | .16 | −0.34 (0.12) | .005 |
| rs2075650 | 45395619 | TOMM40 | A | 0.768 | 0.00 | 0.55 | 0.41 | 0.89 (0.73–1.05) | .16 | 0.15 (0.15) | .31 |
| rs8106922 | 45401666 | TOMM40 | A | 0.646 | 0.90 | 0.87 | 0.00 | 0.97 (0.9–1.04) | .36 | 0.15 (0.12) | .23 |
| rs405509 | 45408836 | APOE | T | 0.529 | 0.67 | 0.55 | 0.03 | 0.97 (0.9–1.04) | .43 | 0.07 (0.14) | .62 |
| rs439401 | 45414451 | Intergenic | T | 0.326 | 0.37 | 0.00 | 0.44 | 0.95 (0.88–1.02) | .16 | 0.2 (0.13) | .11 |
Abbreviations: AAO, age at onset; AD, Alzheimer disease; ADGC, Alzheimer's Disease Genetics Consortium; APOE, apolipoprotein E; BP, chromosome position in base pairs; l, long allele; LOAD, National Institute on Aging Late-Onset Alzheimer's Disease Study; OR, odds ratio; P, meta-analysis P value; RA, reference allele; RAF, reference allele frequency; s, short allele; SNP, single-nucleotide polymorphism; v, very long allele.
Rs10524523 genotypes were categorized in 3 ways: s-v(s/s, s/v, v/v, and others as missing), s-l(s/s, s/l, l/l, and others as missing), and v-l(v/v, v/l, l/l, and others as missing). The pairwise linkage disequilibrium coefficients for SNPs in the APOE region with rs10524523 genotypes were computed separately within and then averaged across the Adult Changes in Thought Study, National Institute on Aging Alzheimer's Disease Centers, and Alzheimer's Disease Neuroimaging Initiative Study data sets.
Adjusted for population substructure, age, sex, and number of APOE ε4 alleles.
Adjusted for population substructure, sex, and number of APOE ε4 alleles.
ASSOCIATION OF AD WITH SNPs THROUGHOUT THE APOE REGION
To evaluate the hypothesis that multiple loci in the APOE region influence risk or AAO of AD, we tested association using the entire ADGC sample (eTable 4) with all SNPs spanning the 800-kb region surrounding APOE that encompasses previously reported genome-wide significant findings in several genes.21 Eight SNPs spanning the entire region were significantly associated with AD risk at P < .001 in models adjusting for the number of APOE ε4 alleles, and one of these results (rs445925 located between APOE and APOC1) was genome-wide significant (P = 4.1 × 10−11). However, significance of these results was greatly diminished after taking into account heterogeneity across data sets and APOE genotypes including the ε2 allele (Table 4). In the model including all APOE genotypes, nominal significance was observed for 3 SNPs (rs29651, P = .04; rs37451, P = .0063; and rs20756, P = .01), but none of these results remained significant after correcting for the number of tests. No SNPs were significantly associated with AAO at P < .001 in models adjusting for dose of ε4 (eTable 5).
Table 4.
Top-Ranked Results (P < .001) for Association of AD Risk in the Conditional Model Including Dose of ε4
| Conditional on ε4 Dosagea |
Conditional on APOE Genotypeb |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | BP | Near Gene | RA | RAF | OR (95% CI) | P | REM-P | OR (95% CI) | P | REM-P |
|
| ||||||||||
| rs2965109 | 45225345 | CEACAM16/BCL3 | T | 0.376 | 0.92 (0.89–0.94) | .0027 | .0027 | 0.94 (0.92–0.97) | .0420 | .04 |
| rs7254776 | 45227742 | CEACAM16/BCL3 | T | 0.636 | 1.08 (1.05–1.10) | .0036 | .0036 | 1.04 (1.01–1.07) | .12 | .12 |
| rs2965101 | 45237812 | CEACAM16/BCL3 | T | 0.686 | 1.07 (1.05–1.10) | .0055 | .0055 | 1.03 (1.01–1.06) | .20 | .20 |
| rs3745150 | 45385759 | PVRL2 | C | 0.392 | 1.11 (1.07–1.14) | .0036 | .0050 | 1.03 (1.00–1.07) | .38 | .42 |
| rs6857 | 45392254 | PVRL2 | T | 0.253 | 1.23 (1.17–1.29) | 3.2 × 10−5 | .0026 | 1.22 (1.16–1.28) | 6.4 × 10−5 | .0063 |
| rs2075650 | 45395619 | T0MM40 | A | 0.767 | 0.84 (0.80–0.88) | 6.4 × 10−5 | .0034 | 0.85 (0.81–0.88) | 1.6 × 10−4 | .01 |
| rs445925 | 45415640 | APOE/APOC1 | A | 0.115 | 0.74 (0.71–0.78) | 4.1 × 10−11 | 7.9 × 10−4 | 0.93 (0.87–0.99) | .25 | .47 |
Abbreviations: AD, Alzheimer disease; APOE, apolipoprotein E; BP, chromosome position in base pairs; OR, odds ratio under a fixed-effects model; P, meta-analysis P value under a fixed-effects model; RA, reference allele; RAF, reference allele frequency; REM-P, meta-analysis P value under a random-effects model; SNP, single-nucleotide polymorphism; 95% CI, 95% confidence interval under a fixed-effects model.
Adjusted for population substructure, age, sex, and number of APOE ε4 alleles.
Adjusted for population substructure, age, sex, and APOE genotype.
COMMENT
Our study of nearly 12 000 AD cases and 11 000 cognitively normal controls was unable to confirm association of disease risk or variation of AAO of AD symptoms with SNPs in any gene in the APOE region other than APOE. Although we observed genome-wide significance with many SNPs in several genes in this region, the residual effect of these variants dissipated dramatically in models adjusting for APOE genotype.
We also considered the possibility of an independent effect of the TOMM40 variable repeat length polymorphism (rs10524523), which has been reported as a modifier of AAO,10 by genotyping and evaluating this association in a subset of 1256 AD cases and 1605 controls. We were unable to replicate the original finding in models adjusting for APOE genotype or in subgroups stratified by APOE genotype, even though we used a much larger data set than others published to date. This result is consistent with negative findings in several other recent studies.12,22–25 Moreover, association findings were also negative for 5 SNPs in high LD with rs10524523 evaluated in the entire ADGC GWAS sample. Although Cruchaga et al12 found a significantly lower frequency of the rs10524523 v allele in cases compared with controls among APOE ε3 homozygotes in a large case-control series, the effect was in the opposite direction as reported in the original study.10 In our study, there was no effect in either direction for s/s homozygotes with an APOE ε3/ε3 genotype. In a subset of 733 subjects from the Cruchaga et al study, there was no evidence of association of rs10524523 with cerebrospinal fluid tau or β-amyloid 42 levels.12 Johnson et al11 reported an association of rs10524523 with lower performance on learning tests and with decreasing gray matter volume in a brain region affected early in AD development in a small sample of APOE ε3/ε3 adult children of subjects with AD, but a study of a larger community-based cohort between the ages of 79 and 87 years was unable to disentangle the confounding effects of the rs10524523 l allele and APOE ε4 on poorer performance of verbal memory and abstract reasoning.26
Since the association of AD with APOE was established nearly 2 decades ago,1,2 numerous studies have reported significant associations with other genes in the region surrounding APOE,27–29 whereas other studies concluded that these findings are not true independent contributors to AD risk.30,31 Attempts to resolve this controversy have been complicated by very strong LD in this region, which contains many biologically plausible candidate genes.25,29 However, further insight regarding multiple independent association signals can be obtained from analyses in other populations (eg, those of black African descent) having a narrower LD structure in the APOE region. Tycko et al32 excluded independent influence of APOE or APOC1 promoter polymorphisms on risk of AD in samples of African American and Caribbean Hispanic individuals. Logue et al33 identified highly significant associations of AD with 3 markers within 25 kb of APOE including PVRL2 SNP rs6859 (P = 5.39×10−7) and TOMM40 SNPs rs157582 (P = 3.26×10−6) and rs10119 (P = 5.95×10−7) in a sample of 513 well-characterized African American AD cases and 504 ethnically matched cognitively normal controls. However, only rs6859 remained nominally significant (P = .008) after adjustment for APOE genotype, which was very strongly associated with AD (P = 9.69×10−23).
Our study has several strengths that lead to more conclusive findings than previous association studies of genes in the APOE region. First, genotypes for the APOE iso-forms in all ADGC data sets were determined directly using robust methods,8 rather than by inference using imputed genotypes for the 2 SNPs that determine APOE genotype. The genotype for 1 of the APOE SNPs (rs429538) imputed in the ADGC data sets using the 1000 Genomes reference panel (October 2011; release ICHG2011) was only modestly correlated (r2 about 0.5) with the actual APOE genotype (data not presented). Second, our sample size is several-fold larger than those in any previous study of this issue and had sufficient power to detect associations with ORs of 1.2 or greater.21 Thus, even if there were other loci in this region independent of APOE that influenced AD risk or AAO, we would have detected a signal whereas smaller studies probably could not. Third, we conducted a comprehensive examination of all markers in the region, including the poly-T repeat in TOMM40, and tested multiple models to address confounding with APOE.
Although there is some evidence from gene expression, cell biology, and immunohistochemistry studies supporting a connection of AD to the immediate neighbors of APOE including PVRL2, TOMM40 and APOC1,31,34–36 results of our study weigh heavily against the hypothesis of inherited susceptibility to AD due to common variation in genes in the APOE region other than APOE.
Acknowledgments
Additional Contributions: We thank D. Stephen Snyder, PhD, and Marilyn Miller, PhD, from NIA who are ex officio ADGC members. We are grateful to contributors, including the Alzheimer's Disease Centers who collected samples used in this study, as well as patients and their families, whose help and participation made this work possible.
Funding/Support: The National Institutes of Health NIA supported this work through the following grants: ADGC, U01 AG032984 and RC2 AG036528; National Alzheimer's Coordinating Center, U01 AG016976; National Cell Repository for Alzheimer's Disease, U24 AG021886; NIA Late-Onset Alzheimer's Disease Study, U24 AG026395 and U24 AG026390; Banner Sun Health Research Institute, P30 AG019610; Boston University, P30 AG013846, U01 AG10483, R01 CA129769, R01 MH080295, R01 AG017173, R01 AG025259, and R01 AG33193; Columbia University, P50 AG008702 and R37 AG015473; Duke University, P30 AG028377, R01 AG05128, R01 NS39764, and R01 MH60451; Emory University, AG025688; Group Health Research Institute, UO1 AG06781 and UO1 HG004610; Indiana University, P30 AG10133; Johns Hopkins University, P50 AG005146 and R01 AG020688; Massachusetts General Hospital, P50 AG005134; Mayo Clinic, P50 AG016574; Mount Sinai School of Medicine, P50 AG005138 and P01 AG002219; New York University, P30 AG08051, MO1 RR00096, and UL1 RR029893; Northwestern University, P30 AG013854; Oregon Health & Science University, P30 AG008017 and R01 AG026916; Rush University, P30 AG010161, R01 AG019085, R01 AG15819, R01 AG17917, and R01 AG30146; Translational Genomics Research Institute, R01 NS059873 and R01 AG034504; University of Alabama at Birmingham, P50 AG016582 and UL1 RR02777; University of Arizona, R01 AG031581; University of California, Davis, P30 AG010129; University of California, Irvine, P50 AG016573, P50 AG016575, P50 AG016576, and P50 AG016577; University of California, Los Angeles, P50 AG016570; University of California, San Diego, P50 AG005131; University of California, San Francisco, P50 AG023501 and P01 AG019724; University of Kentucky, P30 AG028383 and AG05144; University of Michigan, P50 AG008671; University of Pennsylvania, P30 AG010124; University of Pittsburgh, P50 AG005133 and AG030653; University of Southern California, P50 AG005142; University of Texas Southwestern, P30 AG012300; University of Miami, R01 AG027944, AG010491, AG027944, AG021547, and AG019757; University of Washington, P50 AG005136; Vanderbilt University, R01 AG019085; and Washington University, P50 AG005681 and P01 AG03991. Samples from the National Cell Repository for Alzheimer's Disease, which receives government support under cooperative agreement grant U24 AG21886 awarded by the NIA, were used in this study. The Translational Genomics Research Institute series was also funded by the Banner Alzheimer's Foundation, the Johnnie B. Byrd Sr Alzheimer's Center & Research Institute, the Medical Research Council, and the state of Arizona and also includes samples from the following sites: Newcastle Brain Tissue Resource (funding via the Medical Research Council, local National Health Service trusts, and Newcastle University), MRC London Brain Bank for Neurodegenerative Diseases (funding via the Medical Research Council), South West Dementia Brain Bank (funding via numerous sources including the Higher Education Funding Council for England, Alzheimer's Research Trust, and BRACE as well as North Bristol NHS Trust Research and Innovation Department and DeNDRoN), the Netherlands Brain Bank (funding via numerous sources including Stichting MS Research, BrainNet Europe, Hersenstichting Nederland Breinbrekend Werk, International Parkinson Fonds, and Internationale Stiching Alzheimer Onderzoek), Institut de Neuropatologia, Servei Anatomia Patologica, and Universitat de Barcelona. Funding for ADNI is through the Northern California Institute for Research and Education by grants from the Alzheimer's Association, Alzheimer's Drug Discovery Foundation, the Dana Foundation, and the National Institute of Biomedical Imaging and Bioengineering and NIA grants U01 AG024904, RC2 AG036535, and K01 AG030514. Support was also from Alzheimer's Association grants IIRG-08-89720 (Dr Farrer) and IIRG-05-14147 (Dr Pericak-Vance) and the US Department of Veterans Affairs Administration, Office of Research and Development, Biomedical Laboratory Research Program. Dr St George-Hyslop is supported by Wellcome Trust, Howard Hughes Medical Institute, and the Canadian Institute of Health Research.
Footnotes
Author Contributions: Study concept and design: Jun, Vardarajan, Buros, Baldwin, Beecham, Bowen, Cummings, Hakonarson, Hardy, Naj, Trojanowski, Haines, Lunetta, Pericak-Vance, Schellenberg, and Farrer. Acquisition of data: Yu, Hawk, Dombroski, Crane, Larson, Apostolova, Arnold, Baldwin, Barmada, Beach, Beekly, Bennett, Bigio, Bird, Blacker, Bowen, Boxer, Buxbaum, Cairns, Cantwell, Cao, Carney, Carrasquillo, Carroll, Corneveaux, Cotman, Crocco, Cruchaga, Cummings, De-Carli, DeKosky, Demirci, Diaz-Arrastia, Dick, Dickson, Duara, Ellis, Ertekin-Taner, Evans, Faber, Fallon, Farlow, Ferris, Foroud, Galasko, Ganguli, Gearing, Gesch-wind, Ghetti, Gilbert, Gilman, Giordani, Glass, Goate, Graff-Radford, Green, Hamilton, Harrell, Head, Honig, Huentelman, Hulette, Hyman, Jarvik, Jicha, Jin, Kamboh, Karydas, Kauwe, Kaye, Kim, Kowall, Kramer, Kukull, Lah, Levey, Lieberman, Lopez, Mack, Martiniuk, Mash, McCormick, McCurry, McDavid, McKee, Mesulam, C. A. Miller, J. W. Miller, Montine, Morris, Myers, Naj, Nowotny, Parisi, Peskind, Poon, Quinn, Raj, Rajbhandary, Raskind, Reiman, Reisberg, Reitz, Ringman, Roberson, Rogaeva, Rosenberg, Sano, Saykin, J. A. Schneider, L. S. Schneider, Seeley, Sonnen, Spina, St George-Hyslop, Stern, Trojanowski, Troncoso, Tsuang, Van Deerlin, Vinters, Vonsattel, Wang, Weintraub, Woltjer, Younkin, Mayeux, Pericak-Vance, Schellenberg, and Farrer. Analysis and interpretation of data: Jun, Vardarajan, Buros, Larson, Barmada, Beecham, Boeve, Corneveaux, De Jager, DeCarli, Frosch, Ghetti, Hakonarson, Huentelman, Martin, Masliah, McKee, B. L. Miller, Naj, Petersen, Potter, Rajbhandary, Tanzi, Trojanowski, Valladares, Wright, Haines, Lunetta, Pericak-Vance, Schellenberg, and Farrer. Drafting of the manuscript: Jun, Vardarajan, Buros, Beekly, De Jager, Karydas, Martin, McCurry, B. L. Miller, Tanzi, Trojanowski, Valladares, Pericak-Vance, and Farrer. Critical revision of the manuscript for important intellectual content: Jun, Vardarajan, Yu, Hawk, Dombroski, Crane, Larson, Apostolova, Arnold, Baldwin, Barmada, Beach, Beecham, Bennett, Bigio, Bird, Blacker, Boeve, Bowen, Boxer, Buxbaum, Cairns, Cantwell, Cao, Carney, Carrasquillo, Carroll, Corneveaux, Cotman, Crocco, Cruchaga, Cummings, DeCarli, DeKosky, Demirci, Diaz-Arrastia, Dick, Dickson, Duara, Ellis, Ertekin-Taner, Evans, Faber, Fallon, Farlow, Ferris, Foroud, Frosch, Galasko, Ganguli, Gearing, Geschwind, Ghetti, Gilbert, Gilman, Giordani, Glass, Goate, Graff-Radford, Green, Hakonarson, Hamilton, Hardy, Harrell, Head, Honig, Huentelman, Hulette, Hyman, Jarvik, Jicha, Jin, Kamboh, Kauwe, Kaye, Kim, Kowall, Kramer, Kukull, Lah, Levey, Lieberman, Lopez, Mack, Martiniuk, Mash, Masliah, McCormick, McDavid, McKee, Mesulam, C. A. Miller, J. W. Miller, Montine, Morris, Myers, Naj, Nowotny, Parisi, Peskind, Petersen, Poon, Potter, Quinn, Raj, Rajbhandary, Raskind, Reiman, Reisberg, Reitz, Ringman, Roberson, Rogaeva, Rosenberg, Sano, Saykin, J. A. Schneider, L. S. Schneider, Seeley, Sonnen, Spina, St George-Hyslop, Stern, Trojanowski, Troncoso, Tsuang, Van Deerlin, Vinters, Vonsattel, Wang, Weintraub, Woltjer, Wright, Younkin, Mayeux, Haines, Lunetta, Pericak-Vance, Schellenberg, and Farrer. Statistical analysis: Jun, Vardarajan, Buros, Barmada, Beecham, Hakonarson, Jarvik, Masliah, Naj, Saykin, Trojanowski, Haines, Lunetta, Pericak-Vance, and Farrer. Obtained funding: Crane, Larson, Beach, Bennett, Blacker, DeKosky, Giordani, Goate, Green, Hardy, Harrell, Head, Huentelman, Hulette, Jarvik, Kamboh, Kauwe, Levey, Martin, Montine, Morris, Naj, Poon, Potter, Reiman, Reisberg, Rogaeva, Saykin, Seeley, St George-Hyslop, Younkin, Schellenberg, and Farrer. Administrative, technical, and material support: Yu, Crane, Larson, Arnold, Baldwin, Beach, Beekly, Boxer, Cairns, Cantwell, Cao, Carroll, Corneveaux, Cotman, Cruchaga, Cummings, DeCarli, DeKosky, Demirci, Diaz-Arrastia, Dick, Duara, Ertekin-Taner, Evans, Faber, Farlow, Ferris, Foroud, Frosch, Geschwind, Ghetti, Gilman, Giordani, Green, Hakonarson, Hamilton, Honig, Huentelman, Hyman, Jarvik, Karydas, Kaye, Kowall, Lieberman, Mack, Martiniuk, McCormick, McCurry, Mesulam, B. L. Miller, C. A. Miller, J. W. Miller, Montine, Myers, Nowotny, Peskind, Poon, Potter, Quinn, Raj, Rajbhandary, Raskind, Ringman, Roberson, Rosenberg, Sano, J. A. Schneider, Seeley, Trojanowski, Troncoso, Tsuang, Valladares, Van Deerlin, Vinters, Woltjer, Mayeux, Haines, Pericak-Vance, Schellenberg, and Farrer. Study supervision: Buxbaum, Diaz-Arrastia, Ferris, Gilbert, Harrell, Lopez, Mack, L. S. Schneider, St George-Hyslop, Trojanowski, Van Deerlin, Lunetta, Schellenberg, and Farrer. Alzheimer's Disease Genetics Consortium: Liana G. Apostolova, MD, Steven E. Arnold, MD, Clinton T. Baldwin, PhD, Michael M. Barmada, PhD, Thomas G. Beach, MD, PhD, Gary W. Beecham, PhD, Duane Beekly, BS, David A. Bennett, MD, Eileen H. Bigio, MD, Thomas D. Bird, MD, Deborah Blacker, MD, Bradley F. Boeve, MD, James D. Bowen, MD, Adam Boxer, MD, PhD, Joseph D. Buxbaum, PhD, Nigel J. Cairns, PhD, FRCPath, Laura B. Cantwell, MPH, Chuanhai Cao, PhD, Regina M. Carney, MD, Minerva M. Carrasquillo, PhD, Steven L. Carroll, MD, PhD, Jason Corneveaux, BS, Carl W. Cotman, PhD, Elizabeth A. Crocco, MD, Carlos Cruchaga, PhD, Jeffrey L. Cummings, MD, Philip L. De Jager, MD, PhD, Charles DeCarli, MD, Steven T. DeKosky, MD, F. Yesim Demirci, MD, Ramon Diaz-Arrastia, MD, PhD, Malcolm Dick, PhD, Dennis W. Dickson, MD, Ranjan Duara, MD, William G. Ellis, MD, Nilufer Ertekin-Taner, MD, PhD, Denis Evans, MD, Kelley M. Faber, MS, Kenneth B. Fallon, MD, Martin R. Farlow, MD, Steven Ferris, PhD, Tatiana M. Foroud, PhD, Matthew P. Frosch, MD, PhD, Douglas R. Galasko, MD, Mary Ganguli, MD, Marla Gearing, PhD, Daniel H. Geschwind, MD, PhD, Bernardino Ghetti, MD, John R. Gilbert, PhD, Sid Gilman, MD, FRCP, Bruno Giordani, PhD, Jonathan D. Glass, MD, Alison M. Goate, DPhil, Neill R. Graff-Radford, MD, Robert C. Green, MD, MPH, Hakon Hakonarson, MD, PhD, Ronald L. Hamilton, MD, John Hardy, PhD, Lindy E. Harrell, MD, PhD, Elizabeth Head, PhD, Lawrence S. Honig, MD, PhD, Matthew J. Huentelman, PhD, Christine M. Hulette, MD, Bradley T. Hyman, MD, PhD, Gail P. Jarvik, MD, PhD, Gregory A. Jicha, MD, PhD, Lee-Way Jin, MD, PhD, M. Ilyas Kamboh, PhD, Anna Karydas, BA, John S. K. Kauwe, PhD, Jeffrey A. Kaye, MD, Ronald Kim, MD, Neil W. Kowall, MD, Patricia Kramer, PhD, Walter A. Kukull, PhD, James J. Lah, MD, PhD, Allan I. Levey, MD, PhD, Andrew P. Lieberman, MD, PhD, Oscar L. Lopez, MD, Wendy J. Mack, PhD, Eden R. Martin, PhD, Frank Martiniuk, PhD, Deborah C. Mash, PhD, Eliezer Masliah, MD, Wayne C. McCormick, MD, MPH, Susan M. McCurry, PhD, Andrew N. McDavid, BA, Ann C. McKee, MD, Marsel Mesulam, MD, Bruce L. Miller, MD, Carol A. Miller, MD, Joshua W. Miller, PhD, Thomas J. Montine, MD, PhD, John C. Morris, MD, Amanda J. Myers, PhD, Adam C. Naj, PhD, Petra Nowotny, PhD, Joseph E. Parisi, MD, Elaine Peskind, MD, Ronald C. Petersen, MD, PhD, Wayne W. Poon, PhD, Huntington Potter, PhD, Joseph F. Quinn, MD, Ashok Raj, MD, Ruchita A. Rajbhandary, MPH, Murray Raskind, MD, Eric M. Reiman, MD, Barry Reisberg, MD, Christiane Reitz, MD, PhD, John M. Ringman, MD, Erik D. Roberson, MD, PhD, Ekaterina Rogaeva, PhD, Roger N. Rosenberg, MD, Mary Sano, PhD, Andrew J. Saykin, PsyD, Julie A. Schneider, MD, Lon S. Schneider, MD, William W. Seeley, MD, Joshua A. Sonnen, MD, Salvatore Spina, MD, Peter St George-Hyslop, MD, FRCP, Robert A. Stern, PhD, Rudolph E. Tanzi, PhD, John Q. Trojanowski, MD, PhD, Juan C. Troncoso, MD, Debby W. Tsuang, MD, Otto Valladares, MS, Vivianna M. Van Deerlin, MD, PhD, Harry V. Vinters, MD, Jean Paul Vonsattel, MD, Li-San Wang, PhD, Sandra Weintraub, PhD, Randall L. Woltjer, MD, PhD, Clinton B. Wright, MD, MS, Steven G. Younkin, MD, PhD.
Financial Disclosure: The Kathleen Price Bryan Brain Bank at Duke University Medical Center is funded by GlaxoSmithKline. Genotyping of the Translational Genomics Research Institute series 2 cohort was supported by Kronos Science. Funding for ADNI is through the Northern California Institute for Research and Education by grants from Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson & Johnson, Eli Lilly and Co, Medpace Inc, Merck and Co Inc, Novartis AG, Pfizer Inc, F. Hoffman-La Roche, Schering-Plough, and Synarc Inc.
Online-Only Material: The eFigures and eTables are available at http://www.archneurol.com.
REFERENCES
- 1.Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 2.Corder EH, Saunders AM, Risch NJ, et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet. 1994;7(2):180–184. doi: 10.1038/ng0694-180. [DOI] [PubMed] [Google Scholar]
- 3.Farrer LA, Cupples LA, Haines JL, et al. APOE and Alzheimer Disease Meta Analysis Consortium Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease: a meta-analysis. JAMA. 1997;278(16):1349–1356. [PubMed] [Google Scholar]
- 4.Bowirrat A, Treves TA, Friedland RP, Korczyn AD. Prevalence of Alzheimer's type dementia in an elderly Arab population. Eur J Neurol. 2001;8(2):119–123. doi: 10.1046/j.1468-1331.2001.00183.x. [DOI] [PubMed] [Google Scholar]
- 5.Gureje O, Ogunniyi A, Baiyewu O, et al. APOE epsilon4 is not associated with Alzheimer's disease in elderly Nigerians. Ann Neurol. 2006;59(1):182–185. doi: 10.1002/ana.20694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pericak-Vance MA, Johnson CC, Rimmler JB, et al. Alzheimer's disease and apolipoprotein E-4 allele in an Amish population. Ann Neurol. 1996;39(6):700–704. doi: 10.1002/ana.410390605. [DOI] [PubMed] [Google Scholar]
- 7.Hollingworth P, Harold D, Sims R, et al. Alzheimer's Disease Neuroimaging Initiative; CHARGE consortium; EADI1 consortium Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43(5):429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naj AC, Jun G, Beecham GW, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43(5):436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seshadri S, Fitzpatrick AL, Ikram MA, et al. CHARGE Consortium; GERAD1 Consortium; EADI1 Consortium Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303(18):1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roses AD, Lutz MW, Amrine-Madsen H, et al. A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer's disease. Pharmacogenomics J. 2010;10(5):375–384. doi: 10.1038/tpj.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson SC, La Rue A, Hermann BP, et al. The effect of TOMM40 poly-T length on gray matter volume and cognition in middle-aged persons with APOE ε3/ε3 genotype. Alzheimers Dement. 2011;7(4):456–465. doi: 10.1016/j.jalz.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruchaga C, Nowotny P, Kauwe JS, et al. Alzheimer's Disease Neuroimaging Initiative Association and expression analyses with single-nucleotide polymorphisms in TOMM40 in Alzheimer disease. Arch Neurol. 2011;68(8):1013–1019. doi: 10.1001/archneurol.2011.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wittwer CT, Ririe KM, Andrew RV, David DA, Gundry RA, Balis UJ. The Light-Cycler: a microvolume multisample fluorimeter with rapid temperature control. Biotechniques. 1997;22(1):176–181. doi: 10.2144/97221pf02. [DOI] [PubMed] [Google Scholar]
- 14.Ahmadian A, Gharizadeh B, Gustafsson AC, et al. Single-nucleotide polymorphism analysis by pyrosequencing. Anal Biochem. 2000;280(1):103–110. doi: 10.1006/abio.2000.4493. [DOI] [PubMed] [Google Scholar]
- 15.Hixson JE, Vernier DT. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J Lipid Res. 1990;31(3):545–548. [PubMed] [Google Scholar]
- 16.Lai E, Riley J, Purvis I, Roses A. A 4-Mb high-density single nucleotide polymorphism-based map around human APOE. Genomics. 1998;54(1):31–38. doi: 10.1006/geno.1998.5581. [DOI] [PubMed] [Google Scholar]
- 17.Li M, Boehnke M, Abecasis GR. Efficient study designs for test of genetic association using sibship data and unrelated cases and controls. Am J Hum Genet. 2006;78(5):778–792. doi: 10.1086/503711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinnott JA, Kraft P. Artifact due to differential error when cases and controls are imputed from different platforms. Hum Genet. 2012;131(1):111–119. doi: 10.1007/s00439-011-1054-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 20.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naj AC, Beecham GW, Martin ER, et al. Dementia revealed: novel chromosome 6 locus for late-onset Alzheimer disease provides genetic evidence for folate-pathway abnormalities. PLoS Genet. 2010;6(9):e1001130. doi: 10.1371/journal.pgen.1001130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu SH, Roeder K, Ferrell RE, et al. TOMM40 poly-T repeat lengths, age of onset and psychosis risk in Alzheimer disease. Neurobiol Aging. 2011;32(12):2328–2329. e1–e9. doi: 10.1016/j.neurobiolaging.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maruszak A, Pepłońska B, Safranow K, Chodakowska-ZŻebrowska M, Barcikowska M, Zekanowski C. TOMM40 rs10524523 polymorphism's role in late-onset Alzheimer's disease and in longevity. J Alzheimers Dis. 2012;28(2):309–322. doi: 10.3233/JAD-2011-110743. [DOI] [PubMed] [Google Scholar]
- 24.Pomara N, Bruno D, Nierenberg JJ, et al. TOMM40 poly-T variants and cerebro-spinal fluid amyloid beta levels in the elderly. Neurochem Res. 2011;36(6):1124–1128. doi: 10.1007/s11064-011-0459-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu CE, Seltman H, Peskind ER, et al. Comprehensive analysis of APOE and selected proximate markers for late-onset Alzheimer's disease: patterns of linkage disequilibrium and disease/marker association. Genomics. 2007;89(6):655–665. doi: 10.1016/j.ygeno.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schiepers OJ, Harris SE, Gow AJ, et al. APOE E4 status predicts age-related cognitive decline in the ninth decade: longitudinal follow-up of the Lothian Birth Cohort 1921. Mol Psychiatry. 2012;17(3):315–324. doi: 10.1038/mp.2010.137. [DOI] [PubMed] [Google Scholar]
- 27.Cervantes S, Samaranch L, Vidal-Taboada JM, et al. Genetic variation in APOE cluster region and Alzheimer's disease risk. Neurobiol Aging. 2011;32(11):2107–2117. e7–e17. doi: 10.1016/j.neurobiolaging.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 28.Chartier-Harlin MC, Parfitt M, Legrain S, et al. Apolipoprotein E, epsilon 4 allele as a major risk factor for sporadic early and late-onset forms of Alzheimer's disease: analysis of the 19q13.2 chromosomal region. Hum Mol Genet. 1994;3(4):569–574. doi: 10.1093/hmg/3.4.569. [DOI] [PubMed] [Google Scholar]
- 29.Takei N, Miyashita A, Tsukie T, et al. Japanese Genetic Study Consortium for Alzheimer Disease Genetic association study on in and around the APOE in late-onset Alzheimer disease in Japanese. Genomics. 2009;93(5):441–448. doi: 10.1016/j.ygeno.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 30.Deelen J, Beekman M, Uh HW, et al. Genome-wide association study identifies a single major locus contributing to survival into old age: the APOE locus revisited. Aging Cell. 2011;10(4):686–698. doi: 10.1111/j.1474-9726.2011.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin ER, Lai EH, Gilbert JR, et al. SNPing away at complex diseases: analysis of single-nucleotide polymorphisms around APOE in Alzheimer disease. Am J Hum Genet. 2000;67(2):383–394. doi: 10.1086/303003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tycko B, Lee JH, Ciappa A, et al. APOE and APOC1 promoter polymorphisms and the risk of Alzheimer disease in African American and Caribbean Hispanic individuals. Arch Neurol. 2004;61(9):1434–1439. doi: 10.1001/archneur.61.9.1434. [DOI] [PubMed] [Google Scholar]
- 33.Logue MW, Schu M, Vardarajan BN, et al. Multi-Institutional Research on Alzheimer Genetic Epidemiology (MIRAGE) Study Group A comprehensive genetic association study of Alzheimer disease in African Americans. Arch Neurol. 2011;68(12):1569–1579. doi: 10.1001/archneurol.2011.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paik YK, Chang DJ, Reardon CA, Walker MD, Taxman E, Taylor JM. Identification and characterization of transcriptional regulatory regions associated with expression of the human apolipoprotein E gene. J Biol Chem. 1988;263(26):13340–13349. [PubMed] [Google Scholar]
- 35.Bekris LM, Lutz F, Yu CE. Functional analysis of APOE locus genetic variation implicates regional enhancers in the regulation of both TOMM40 and APOE. J Hum Genet. 2012;57(1):18–25. doi: 10.1038/jhg.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bullido MJ, Artiga MJ, Recuero M, et al. A polymorphism in the regulatory region of APOE associated with risk for Alzheimer's dementia. Nat Genet. 1998;18(1):69–71. doi: 10.1038/ng0198-69. [DOI] [PubMed] [Google Scholar]