Abstract
Background
The increasing incidence of community acquired infection due to Extended-Spectrum Beta-Lactamase (ESBL) -Producing Enterobacteriaceae represent a great concern because there are few therapeutic alternatives. The fecal flora of children in the community can represent a reservoir for ESBLs genes which are located on highly transmissible plasmids and the spread of these genes among bacterial pathogens is concerning. Because intestinal carriage is a key factor in the epidemiology of ESBL-producing Enterobacteriaceae, the study of the prevalence of these resistant bacteria and risk factors in young children is of particular interest.
Methods
We assessed the prevalence and risk factors of community-acquired faecal carriage of extended-spectrum-β-lactamase (ESBL)-producing Enterobacteriaceae in children aged from 6 to 24 months, by means of rectal swabbing in community pediatric practices. Child’s lifestyle and risk factors for carriage of resistant bacteria were noted.
Results
Among the 411 children enrolled, 4.6% carried ESBL-producing Enterobacteriaceae. CTX-M-1, CTX-M-15 and CTX-M-14 were the predominant ESBLs. The 18 E. coli isolates were genetically heterogeneous. Recent third-generation oral-cephalosporin exposure was associated with a higher risk of ESBL carriage (AOR=3.52, 95% CI[1.06-11.66], p=0.04).
Conclusions
The carriage rate of ESBL-producing Enterobacteriacae in young children in the French community setting is noteworthy, underlining the importance of this population as a reservoir. Exposure to third-generation oral cephalosporins was associated with a significant risk of ESBL carriage in our study. Because of the significant public health implications including the treatment of community-acquired urinary tract infections, the spread of organisms producing ESBLs in the community merits close monitoring with enhanced efforts for surveillance.
Keywords: Community carriage, Extended-Spectrum Beta-Lactamase, Enterobacteriaceae, Children
Background
In the past decade there has been an alarming upsurge in antibiotic-resistant Enterobacteriaceae producing extended-spectrum β-lactamases (ESBL), due at least partly to overuse of broad-spectrum cephalosporins [1]. The spread of mobile ESBL genes among bacterial pathogens is of great concern, not only because these enzymes confer resistance to oxy-imino-cephalosporins and other β-lactam antibiotics, but also because they are located on plasmids that confer resistance to other antimicrobial agents, leaving very few treatment options. CTXM–type ESBLs have now supplanted TEM and SHV-type ESBLs both in nosocomial and community settings [1] and Escherichia coli has replaced Klebsiella spp. as the predominant species of ESBL-producing Enterobacteriaceae[1].
Until recently, most infections caused by ESBL-producing E. coli were hospital-acquired. blaCTX-M genes originate from environmental bacteria but have migrated to highly transmissible plasmids, which have been linked to ESBL circulation in the community. The community can thus represent a reservoir for ESBLs not yet detected in clinical isolates [2]. Colonisation in the intestinal compartment by ESBL-producing isolates has been associated with a high risk for developing infection due to ESBL producers [3]. Prevalence of ESBL-carriage in children varies depending on studies and geographical areas. It ranges from 0,1% in Bolivie and Perou [4] to 31% in Niger [5]. Because intestinal carriage is a key factor in the epidemiology of ESBL-producing E. coli infection [6], we investigated the prevalence of community-acquired faecal carriage of ESBL-producing Enterobacteriaceae in children aged between 6 and 24 months presenting to community paediatricians.
Methods
This work was an ancillary study of a nasopharyngeal carriage study conducted in France following implementation of the 7-valent pneumococcal conjugate vaccine [7]. Between October 2010 and March 2011, 18 French pediatricians located in three regions (Ile de France, Lorraine, and Provence-Alpes-Côte d’Azur) took part in this prospective study.
A rectal sample was obtained from children aged 6 to 24 months, either during routine check-ups with normal findings, or when they presented with acute otitis media (AOM). The exclusion criteria were antibiotic treatment whatever the antibiotic was within 7 days before enrolment and severe underlying disease. Once their written informed consent had been granted, we queried the parents or guardians on the child’s demographics, risk factors for carriage of resistant bacteria including use of any antibiotics (between 7 days to 3 months before enrolment), daycare modalities (daycare center, home or child minder), previous hospitalization (during the previous 6 months), and immunization history (pneumococcal conjugate vaccine). The study was approved by the Saint Germain en Laye Hospital Ethics Committee.
On inclusion, rectal samples were taken with a flexible, sterile, soft rayon swab tip. After sampling, the swabs were immediately inoculated in transport medium (Copan, Brescia, Italy) at room temperature and sent within 48 hours to the National E. coli and Shigella Reference Center-associated laboratory at Robert Debré Hospital, Paris. The rectal swabs were spread on ChromID ESBL screening medium to screen the stool flora for cefpodoxime resistance (bioMérieux, La Balme-les-Grottes, France). ESBL detection was performed using the double-disk synergy test between clavulanic acid and extended spectrum-cephalosporins (ceftazidime and cefotaxime) [8]. Bacterial identification was performed with the API20E system (bioMérieux, Marcy l’Etoile, France).
Multiplex PCR was used to characterize ß-lactamase genes (including blaCTX-M, blaSHV, blaTEMand blaOXA-1), with previously described methods and primers. Amplicons were then sequenced [9,10]. Clonal relationships among E. coli isolates were identified by semi-automated rep-PCR (Diversilab, bioMérieux, France) [11]. The strains were assigned to one of the four main E. coli phylogenetic groups (A, B1, B2 and D), using a previously described multiplex PCR-based method [12].
Data were double-entered using 4D software (version 6.4), and analyzed using Stata SE 9.1 (Stata Corp., College Station, TX, USA) for univariate analysis and multivariate logistic regression (odds ratios [ORs] and 95% confidence intervals [CI]). The Pearson Chi-square test was used in univariate analysis to identify factors related (p<0.10) to ESBL-producing Enterobacteriaceae carriage. Variables identified by univariate analysis were age (continuous variable after testing linearity), the study group (healthy controls vs children with AOM) and recent antibiotic treatment (within 3 months before enrolment).
Results
ESBL carriage was assessed in 411 children none of whom were born preterm. Mean age of the children was 13.3±6.1 months, the sex ratio was 1.07 (M/F). 37.2% of the children attended daycare centers, others were cared for at home, 54.5% had siblings, and 34.1% had received antibiotics within 7 days to 3 months before enrolment. Third-generation oral cephalosporins (O3GC) represented 25% of the antibiotics prescribed and especially cefpodoxime proxetil. ESBL-producing Enterobacteriaceae were found in 19 patients (4.6%): 1 patient with E. coli and Citrobacter freundii, 1 with K. pneumoniae, and 17 with E. coli. None of these children had been hospitalized in the previous 6 months. ESBL-producing Enterobacteriaceae carriage was not related to daycare modalities. The rate of ESBL-producing Enterobacteriaceae carriage was higher in children with recent O3GC exposure (11.1%) than in those without recent antibiotic exposure (4.4%) or with recent aminopenicillin exposure (3.3%) (univariate analysis: OR=3.00, 95% CI [0.94–9.58]). The risk of ESBL-producing Enterobacteriaceae carriage was also higher among children over 1 year old than in younger children (6.5% versus 2.5%, respectively; OR=2.69, 95% CI [0.95–7.61]).
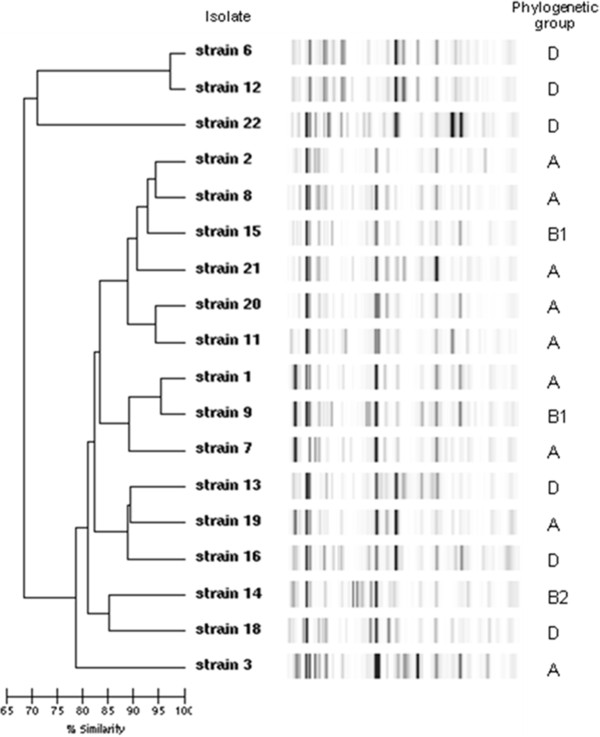
After adjustment in a logistic regression model, recent O3GC use was associated with a significant risk of ESBL carriage (Adjusted Odds Ratio (AOR) =3.52, 95% CI [1.06-11.66], p=0.04). Table 1 shows the bacterial species, the E. coli phylogenetic groups, the ESBLs produced, other ß-lactamase genes, and associated resistance patterns. Phylogenetic group A (9/18) predominated, and the most frequent ESBL was CTX-M-1 (9/20). 11/16 of the CTX-M-producing E. coli carried one other β-lactamase-encoding gene (mainly TEM-1) and 14/16 were resistant to at least one other antibiotic family. Figure 1 shows the genetic heterogeneity of the ESBL-producing E. coli; 17 different profiles were found among the 18 isolates with a cut-off for genetic relation of 97%.
Table 1.
Bacterial species, phylogenetic group of E. coli, ESBL produced, others ß-lactamase genes and associated resistance
| Isolate number | Strains | Phylogenetic group | ESBL type | Other beta-lactamase type | Associated resistance patterns* |
|---|---|---|---|---|---|
| 1 |
E. coli |
A |
CTX-M-1 |
TEM-135 |
NONE |
| 2 |
E. coli |
A |
CTX-M-1 |
|
TSU,NA |
| 7 |
E. coli |
A |
CTX-M-1 |
TEM-1 |
TSU, GT |
| 19 |
E. coli |
A |
CTX-M-1 |
TEM-1 |
TSU, NA, CIP |
| 20 |
E. coli |
A |
CTX-M-1 |
TEM-1 |
TSU, NA, KA |
| 8 |
E. coli |
A |
CTX-M-15 |
TEM-1 |
TSU, NA, GT,TM |
| 11 |
E. coli |
A |
CTX-M-15 |
OXA-1 |
TSU, NA, CIP, K, GT, TM |
| 3 |
E. coli |
A |
SHV-12 |
|
NA |
| 21 |
E. coli |
A |
TEM-52 |
|
TSU, NA |
| 9 |
E. coli |
B1 |
CTX-M-1 |
TEM-1 |
TSU |
| 15 |
E. coli |
B1 |
CTX-M-1 |
|
TSU |
| 14 |
E. coli |
B2 |
CTX-M-15 |
|
TSU, NA, CIP, K, GT, TM |
| 13 |
E. coli |
D |
CTX-M-1 |
|
TSU, NA, CIP |
| 22 |
E. coli |
D |
CTX-M-1 |
TEM-1 |
TSU, NA |
| 6 |
E. coli |
D |
CTX-M-14 |
|
NA |
| 12 |
E. coli |
D |
CTX-M-14 |
TEM-1 |
NA |
| 18 |
E. coli |
D |
CTX-M-27 |
TEM-1 |
TSU, NA, CIP |
| 16 |
E. coli** |
D |
CTX-M-32 |
TEM-1 |
NONE |
| |
C. freundii** |
|
CTX-M-32 |
|
NONE |
| K. pneumoniae | CTX-M-14 | SHV-1 | TSU |
*TSU, trimethoprim-sulfamethoxazole; NA, nalidixic acid; CIP, ciprofloxacin; GT,gentamicin; KA, kanamycin; TM, tobramycin.
**same patient.
Figure 1.
Genetic diversity of ESBL-producing E. coli. Dendrogram showing the genetic heterogeneity of ESBL-producing E. coli. 17 different profiles were found among the 18 isolates with a cut-off for genetic relation of 97%.
Discussion
ESBL-producing Enterobacteriaceae are recognized as important nosocomial pathogens in children, and are often associated with outbreaks [13]. Screening for intestinal carriage is crucial to predict the risk of ESBL infection, as the colon serves as a reservoir for extra-intestinal pathogenic E. coli[14-16]. ESBL-producing E. coli have previously been described as community-acquired in adult patients [17]. The rate of ESBL-producing- E. coli carriage in healthy subjects were 3.7% and 5.8% in a Spanish [2] and Swiss study [18], respectively. Detection of ESBL in fecal isolates from healthy children has also been previously reported. Pallecchi et al. detected ESBL-producing Enterobacteriaceae in 4 (0.1%) of 3208 children, and molecular characterization revealed the presence of CTX-M type β-lactamase genes in the isolates from all 4 children [4]. Recently, Guimaraes et al. studied fecal carriage of ESBL-producing Enterobacteriaceae in 112 healthy children and found that 3 (2.6%) harbored ESBL-producing E. coli (CTX-M-1, TEM-52 and SHV-12) [19]. These isolates belonged to phylogenetic groups A, B1 and D, respectively [19]. In our study, 4.6% (19/411) of children aged from 6 to 24 months carried ESBL-producing Enterobacteriaceae. The prevalence of carriage of ESBL-producing Enterobacteriaceae is probably underestimated in this study because children with antibiotic treatment within 7 days before enrolment and severe underlying disease were excluded from the study. However, the percentage of children carrying ESBL-producing Enterobacteriaceae strains in our population of children is comparable to the percentage of ESBL-producing-E. coli isolated from urinary tract infection in pediatric patients found in France (data on file). Most of the isolates of our study (18/20, 90%) produced a CTX-M type ESBL, and were resistant to multiple antibiotic classes (Table 1). Virulent extra-intestinal strains of E. coli belong to phylogenetic group B2 and, to a lesser extent, group D, whereas commensal strains mainly belong to group A or B1 [20]. Most of our ESBL-producing E. coli isolates belonged to group A/B1, possibly because of greater antibiotic exposure of group A/B1 strains belonging to the fecal flora [21]. Interestingly, one group A E. coli strain isolated here carried a TEM-52 ESBL. Neonatal meningitis caused by TEM-52-producing group A E. coli was recently described [13].
Rep-PCR showed a high level of genomic diversity among the 18 E. coli isolates, illustrating the high capacity for spread among different genetic backgrounds found in the community, via mobile conjugative elements. Exposure to third-generation oral cephalosporins was associated with a significant risk of ESBL carriage in our study (AOR=3.52, 95% CI [1.06-11.66], p=0.04). These results support efforts to reduce prescribing of cephalosporins for control of resistant Enterobacteriaceae not only for the treatment of urinary tract infection (UTIs) but also upper respiratory infection.
Conclusion
The carriage rate of ESBL-producing Enterobacteriaceae in young children in the French community setting (4.6%) is noteworthy, underlining the importance of this population as a reservoir [22].
Exposure to third-generation oral cephalosporins was associated with a significant risk of ESBL carriage in our study. Because of the significant public health implications including the treatment of community-acquired UTIs, the spread of organisms producing ESBLs in the community merits close monitoring with enhanced efforts for surveillance.
Competing interests
All authors: no conflicts.
Authors' contribution
AB: carried out the molecular genetic studies of bacterial strains and drafted a part of the manuscript. RC: have been involved in designing the study, collecting samples and informations, analysing data, revising critically the article and drafting a part of the manuscript. Have given final approval of the version to be published. CL: have been involved in designing the study, collecting samples and informations, analysing data, revising critically the article and drafting a part of the manuscript. Have given final approval of the version to be published. PB: have been involved in the molecular genetic studies, in the sequence alignement and in interpretation of data. CC: participated in the molecular genetic studies and in collection of samples and data. MB: was one of the main investigators of the study and have been involved in designing the study. FT: was one of the main investigators of the study and have been involved in designing the study. EB: have been involved in designing the study, analysing data, drafting a part of the manuscript and revising critically the article. Have given final approval of the version to be published. All authors read and approved the final manuscript.
Authors' information
EB is a French microbiologist specialist, scientific director of a research unit (EA3105) and head of the microbiologic department of Hopital Robert-Debré, pediatric hospital. He is also director of the associated laboratory of the national center of reference of shiga toxin-producing E. coli. His main research interests are epidemiologic studies and clinical trials in community and hospital acquired infections including those due to Extended-Spectrum-beta-lactamases-producing Enterobacteriaceae. He has published more than 250 papers in English language in these fields.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
André Birgy, Email: birgy.a@gmail.com.
Robert Cohen, Email: robert.cohen@wanadoo.fr.
Corinne Levy, Email: activ@wanadoo.fr.
Philippe Bidet, Email: philippe.bidet@rdb.aphp.fr.
Céline Courroux, Email: celine.courroux@rdb.aphp.fr.
Mohamed Benani, Email: activ@wanadoo.fr.
Franck Thollot, Email: activ@wanadoo.fr.
Edouard Bingen, Email: edouard.bingen@rdb.aphp.fr.
Acknowledgements
We thank all the practitioners who participated in the study, namely Benani Mohamed, Bensoussan Catherine, Bougle Jacky, Corrard François, Deberdt Patrice, Derks Véronique, D'Ovidio Nadia, Elbez Annie, Koskas Marc, Martin Patrick, Michot Anne-Sylvestre, Romain Olivier, Turberg-Romain Catherine, Rondeau Marie-Charlotte, Schlemmer Catherine, Thollot Franck, and Wollner Alain.
We are grateful to M Boucherat, A Liboz, S Tortorelli, M. Pereira, I. Ramay, D. Menguy, and S. Axelrad for their technical assistance.
Financial support was given by ACTIV.
References
- Canton R, Coque TM. The CTX-M beta-lactamase pandemic. Curr Opin Microbiol. 2006;9(5):466–475. doi: 10.1016/j.mib.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Valverde A, Coque TM, Sanchez-Moreno MP, Rollan A, Baquero F, Canton R. Dramatic increase in prevalence of fecal carriage of extended-spectrum beta-lactamase-producing Enterobacteriaceae during nonoutbreak situations in Spain. J Clin Microbiol. 2004;42(10):4769–4775. doi: 10.1128/JCM.42.10.4769-4775.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P, Malczynski M, Obias A, Reiner S, Jin N, Huang J, Noskin GA, Zembower T. Screening for extended-spectrum beta-lactamase-producing Enterobacteriaceae among high-risk patients and rates of subsequent bacteremia. Clin Infect Dis. 2007;45(7):846–852. doi: 10.1086/521260. [DOI] [PubMed] [Google Scholar]
- Pallecchi L, Malossi M, Mantella A, Gotuzzo E, Trigoso C, Bartoloni A, Paradisi F, Kronvall G, Rossolini GM. Detection of CTX-M-type beta-lactamase genes in fecal Escherichia coli isolates from healthy children in Bolivia and Peru. Antimicrob Agents Chemother. 2004;48(12):4556–4561. doi: 10.1128/AAC.48.12.4556-4561.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woerther PL, Angebault C, Jacquier H, Hugede HC, Janssens AC, Sayadi S, El Mniai A, Armand-Lefevre L, Ruppe E, Barbier F. et al. Massive increase, spread, and exchange of extended spectrum beta-lactamase-encoding genes among intestinal Enterobacteriaceae in hospitalized children with severe acute malnutrition in Niger. Clin Infect Dis. 2011;53(7):677–685. doi: 10.1093/cid/cir522. [DOI] [PubMed] [Google Scholar]
- Valverde A, Grill F, Coque TM, Pintado V, Baquero F, Canton R, Cobo J. High rate of intestinal colonization with extended-spectrum-beta-lactamase-producing organisms in household contacts of infected community patients. J Clin Microbiol. 2008;46(8):2796–2799. doi: 10.1128/JCM.01008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen R, Levy C, Bonnet E, Grondin S, Desvignes V, Lecuyer A, Fritzell B, Varon E. Dynamic of pneumococcal nasopharyngeal carriage in children with acute otitis media following PCV7 introduction in France. Vaccine. 2010;28(37):6114–6121. doi: 10.1016/j.vaccine.2009.05.037. [DOI] [PubMed] [Google Scholar]
- Drieux L, Brossier F, Sougakoff W, Jarlier V. Phenotypic detection of extended-spectrum beta-lactamase production in Enterobacteriaceae: review and bench guide. Clin Microbiol Infect. 2008;14(Suppl 1):90–103. doi: 10.1111/j.1469-0691.2007.01846.x. [DOI] [PubMed] [Google Scholar]
- Birgy A, Doit C, Mariani-Kurkdjian P, Genel N, Faye A, Arlet G, Bingen E. Early detection of colonization by VIM-1-producing Klebsiella pneumoniae and NDM-1-producing Escherichia coli in two children returning to France. J Clin Microbiol. 2011;49(8):3085–3087. doi: 10.1128/JCM.00540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65(3):490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- Bonacorsi S, Bidet P, Mahjoub F, Mariani-Kurkdjian P, Ait-Ifrane S, Courroux C, Bingen E. Semi-automated rep-PCR for rapid differentiation of major clonal groups of Escherichia coli meningitis strains. Int J Med Microbiol. 2009;299(6):402–409. doi: 10.1016/j.ijmm.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66(10):4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissenet D, Salauze B, Clermont O, Bingen E, Arlet G, Denamur E, Merens A, Mitanchez D, Vu-Thien H. Meningitis caused by Escherichia coli producing TEM-52 extended-spectrum beta-lactamase within an extensive outbreak in a neonatal ward: epidemiological investigation and characterization of the strain. J Clin Microbiol. 2010;48(7):2459–2463. doi: 10.1128/JCM.00529-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emori TG, Gaynes RP. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin Microbiol Rev. 1993;6(4):428–442. doi: 10.1128/cmr.6.4.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman SL, Seed PC. Procession to pediatric bacteremia and sepsis: covert operations and failures in diplomacy. Pediatrics. 2010;126(1):137–150. doi: 10.1542/peds.2009-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef M, Al Shurman A, Chachaty E, Bsoul AR, Andremont A. Use of molecular typing to investigate bacterial translocation from the intestinal tract in malnourished children with Gram-negative bacteremia. Clin Microbiol Infect. 1998;4(2):70–74. doi: 10.1111/j.1469-0691.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- Ho PL, Poon WW, Loke SL, Leung MS, Chow KH, Wong RC, Yip KS, Lai EL, Tsang KW. Community emergence of CTX-M type extended-spectrum beta-lactamases among urinary Escherichia coli from women. J Antimicrob Chemother. 2007;60(1):140–144. doi: 10.1093/jac/dkm144. [DOI] [PubMed] [Google Scholar]
- Geser NSR, Korczak BM, Beutin L, Hachler H. Molecular identification of blaESBL genes from Enterobacteriaceae isolated from healthy human carriers in Switzerland. Antimicrob Agents Chemother. 2012;56(3):1609–1612. doi: 10.1128/AAC.05539-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes B, Barreto A, Radhouani H, Figueiredo N, Gaspar E, Rodrigues J, Torres C, Igrejas G, Poeta P. Genetic detection of extended-spectrum beta-lactamase-containing Escherichia coli isolates and vancomycin-resistant enterococci in fecal samples of healthy children. Microb Drug Resist. 2009;15(3):211–216. doi: 10.1089/mdr.2009.0910. [DOI] [PubMed] [Google Scholar]
- Branger C, Zamfir O, Geoffroy S, Laurans G, Arlet G, Thien HV, Gouriou S, Picard B, Denamur E. Genetic background of Escherichia coli and extended-spectrum beta-lactamase type. Emerg Infect Dis. 2005;11(1):54–61. doi: 10.3201/eid1101.040257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houdouin V, Bonacorsi S, Bidet P, Bingen-Bidois M, Barraud D, Bingen E. Phylogenetic background and carriage of pathogenicity island-like domains in relation to antibiotic resistance profiles among Escherichia coli urosepsis isolates. J Antimicrob Chemother. 2006;58(4):748–751. doi: 10.1093/jac/dkl326. [DOI] [PubMed] [Google Scholar]
- Ho PL, Wong RC, Chow KH, Yip K, Wong SS, Que TL. CTX-M type beta-lactamases among fecal Escherichia coli and Klebsiella pneumoniae isolates in non-hospitalized children and adults. J Microbiol Immunol Infect. 2008;41(5):428–432. [PubMed] [Google Scholar]