Abstract
PRSS23 and PRSS35 are homologous proteases originally identified in mouse ovaries. In the periimplantation mouse uterus, Prss23 was highly expressed in the preimplantation gestation day 3.5 (D3.5) uterine luminal epithelium (LE). It disappeared from the postimplantation LE and reappeared in the stromal compartment next to the myometrium on D6.5. It was undetectable in the embryo from D4.5 to D6.5 but highly expressed in the embryo on D7.5. Prss35 became detectable in the uterine stromal compartment surrounding the embryo on D4.5 and shifted towards the mesometrial side of the stromal compartment next to the embryo from D5.5 to D7.5. In the ovariectomized uterus, Prss23 was moderately and Prss35 was dramatically downregulated by progesterone and 17β-estradiol. Based on the expression of Prss35 in granulosa cells and corpus luteum of the ovary and the early pregnant uterus, we hypothesized that PRSS35 might play a role in female reproduction, especially in oocyte development, ovulation, implantation, and decidualization. This hypothesis was tested in Prss35(−/−) mice, which proved otherwise. Between wild type (WT) and Prss35(−/−) mice, superovulation of immature females produced comparable numbers of cumulus-oocyte complexes; there were comparable numbers of implantation sites detected on D4.5 and D7.5; there were no obvious differences in the expression of implantation and decidualization marker genes in D4.5 or D7.5 uteri. Comparable mRNA expression levels of a few known protease-related genes in the WT and Prss35(−/−) D4.5 uteri indicated no compensatory upregulation. Comparable litter sizes from WT × WT and Prss35 (−/−)× Prss35 (−/−) crosses suggested that Prss35 gene was unessential for fertility and embryo development. Prss35 gene has been linked to cleft lip/palate in humans. However, no obvious such defects were observed in Prss35(−/−) mice. This study demonstrates the distinct expression of Prss23 and Prss35 in the periimplantation uterus and the dispensable role of Prss35 in fertility and embryo development.
Introduction
Proteases (>600) are categorized into five main groups: metallo, serine, cysteine, aspartic/glutamate, and threonine based on the nature of their active-site catalytic residue. Serine proteases (PRSS) are grouped into 13 classes and 40 families characterized by the presence of serine (Ser) as the nucleophilic amino acid at the enzyme’s active site [1]–[3]. PRSS23 and PRSS35 belong to the trypsin class of serine proteases [4].
PRSS35 was originally identified as a novel mouse ovary-selective gene using suppression subtractive hybridization and PRSS23 was subsequently identified as a homologous protease of PRSS35 via BLAST search [4]. Both genes have two exons with the coding region in exon 2. Although their proteolytic activities have not yet been characterized, both possess general features of serine proteases. However, the canonical Ser is replaced by a threonine (Thr) in PRSS35 [4].
Despite their high sequence homology, Prss35 and Prss23 have their unique expression and regulation patterns in the mouse ovary [4], [5]. Prss35 mRNA is localized in the theca layers of developing follicles, granulosa cells of preovulatory and ovulatory follicles, and the forming and regressing corpus luteum. Prss23 mRNA is highly detected in the granulosa cells of the secondary/early antral follicles, it is also expressed in the ovarian stroma and theca tissues just before ovulation. Prss35 mRNA is upregulated around the time of ovulation and remains elevated in the developing corpus luteum; while Prss23 mRNA is transiently downregulated after ovulation induction and again in the postovulatory period. Prss35 expression is progesterone-dependent prior to follicle rupture and upregulated by gonadotropins; while Prss23 expression is independent of progesterone and downregulated by gonadotropins.
Dot blots of adult mouse tissues indicate that Prss35 is detectable in the ovary only, and Prss23 is detectable in a wide range of tissues, including a high level in the uterus [4]. A recent study shows that Prss23 is localized in heifer’s uterine luminal epithelium (LE) and is upregulated in the heifer uterus from gestation day 7 (D7) to D13 [6], suggesting its uterine regulation during early pregnancy in heifers.
We have been studying the molecular mechanisms for the establishment of uterine receptivity using a mouse model deficient of the third lysophosphatidic acid (LPA) receptor (Lpar3 (−/−)). Lpar3 is mainly detected in the preimplantation D3.5 LE in wild type (WT) mice and Lpar3 (−/−) females have delayed uterine receptivity for embryo implantation [7]. Microarray analysis indicated that Prss23 was the most differentially expressed Prss gene between D4.5 WT and Lpar3 (−/−) LE cells. We analyzed the expression of both Prss23 and its homologous protease gene Prss35 in the periimplantation mouse uterus and found their distinct spatiotemporal expression patterns in the LE and the stromal compartment, respectively.
Proteases play an important role in proteolysis that is essential for tissue remodeling and functions of the ovary and the uterus, which go through extensive tissue remodeling during estrous cycle and pregnancy [8]–[10]. The spatiotemporal expression of Prss23 and Prss35 in the ovary and the uterus led us to hypothesize that PRSS23 and PRSS35 may be involved in ovarian and uterine functions [4], [5]. In this study, we tested the hypothesis on PRSS35 in Prss35 (−/−) mice.
Materials and Methods
Animals and Genotyping
WT and Lpar3 (−/−) mice (C57BL6/129svj mixed background) were from a colony at the University of Georgia, which was originally derived from a colony at The Scripps Research Institute [7]. They were genotyped as previously described [7]. Prss35 (−/−) mice were derived from the mouse strain B6/129S5-Prss35tm1Lex/Mmucd (identification number 032535-UCD) purchased from the Mutant Mouse Regional Resource Center (MMRRC) at UC Davis, a NCRR-NIH funded strain repository. Prss35 (−/−) mice were genotyped using tail genomic DNA and four primers: PRSS35 DNA419.18, PRSS35 DNA419.33, PRSS35 DNA419.34 and PRSS35 GT-IRES (Table 1) in PCR reactions. The PCR cycles were set as follows: 10 cycles of 94°C for 15 s, 65°C for 30 s (decreasing 1°C/cycle), and 72°C for 40 s; and 30 cycles of 94°C for 15 s, 55°C for 30 s and 72°C for 40 s. The expected PCR product sizes for WT (PRSS35 DNA419.18 and PRSS35 DNA419.33) and targeted alleles (PRSS35 DNA419.34 and PRSS35 GT-IRES) were 355 bp and 574 bp, respectively. All mice were housed in polypropylene cages with free access to regular food and water from water sip tubes in a reverse osmosis system. The animal facility is on a 12-hour light/dark cycle (7∶00 AM to 7∶00 PM) at 23±1°C with 30–50% relative humidity. All methods used in this study were approved by the University of Georgia Institutional Animal Care and Use Committee (IACUC) and conform to National Institutes of Health guidelines and public law.
Table 1. Primers used for realtime PCR, making probes for in situ hybridization, and genotyping of Prss35 (−/−) mice.
| Primer | Sequence | GenBankAccession no. | Productsize (bp) |
| Prss8 | F: AGAGAACACAGCAGGGAAG | NM_133351.1 | 399 |
| R: CATGTCCTGCTGGATAGTGT | |||
| Prss12 | F: AGCTGCTCAGGAAAAGAAGT | NM_008939 | 351 |
| R: TGTGCACTTCACATTATCCA | |||
| Prss23 | F: GAAAGGGACACAGAAACTCC | NM_029614.3 | 372 |
| R: TGCTGGTAGAGAAGGTCGTA | |||
| Prss35 | F: GGACGGGAGGATACAGTAAG | NM_178738 | 347 |
| R: CTTTCACCCTGGTTAAGGTT | |||
| Isp1 | F: GCTGGCAGTGTCTGTACAAT | XM_001477507 | 371 |
| R: TGAGTCACCATAGCAGGAAT | |||
| Isp2 | F: ACCCTCAGTGGATTCTGACT | NM_053259 | 280 |
| R: ACAGCTCTTGTTGTCCTGAA | |||
| SerpinA3N | F: CCTACTTCCGAGATGAGGAG | NM_009252.2 | 390 |
| R: GGACAAATTTGACTCCAGTG | |||
| SerpinG1 | F: CACTAAAGGGCTTTTCATCC | NM_009776.2 | 396 |
| R: CCTTCAAAGTATGGTCATCG | |||
| Spink3 | F: AGTTCTTCTGGCTTTTGCAC | NM_009258 | 340 |
| R: TTATTCAACGAACCCACTTG | |||
| Hprt1 | F: CAATCAAGACATTCTTTCCAGT | NM_013556 | 172 |
| R: GCTGACCTGCTGGATTACAT | |||
| Cox-2 | F: CTACATCCTGACCCACTTCA | NM_011198.3 | 387 |
| R: GGTCCTCGCTTATGATCTGT | |||
| Gja1 | F: CCTACATCATCAGCATCCTC | NM_010288.2 | 397 |
| R: TACCCAGGAGGAGACATAGG | |||
| Gjb2 | F: CAGCATTGGAAAGATCTGG | NM_008125.2 | 397 |
| R: GAAGACGGCTTCAAAGATG | |||
| Abp1 | F: TACCCTAATGGTGTGATGGA | NM_029638.1 | 398 |
| R: TCAGCCATAGAGTGGATCTG | |||
| Dtprp | F: GCTCAGATCCCCTTGTGAT | NM_010088.1 | 396 |
| R: GGTCATCATGGATTTCTCTG | |||
| Prlpi | F: TATGTGACTGCCACACGATA | NM_013766.1 | 379 |
| R: CCCTCCAGAACGACTTTATT | |||
| Gapdh | F: GCCGAGAATGGGAAGCTTGTCAT | XM_001476707 | 230 |
| R: GTGGTTCACACCCATCACAAACAT | |||
| PRSS35 DNA419.18 | GCATCGAATGTCAGGAAGAG | 355 | |
| PRSS35 DNA419.33 | CTGCCTTTGACATAGTCCTTC | ||
| PRSS35 DNA419.34 | TGGAGCAGTGAACACGCAGAG | 574 | |
| PRSS35 GT-IRES | CCCTAGGAATGCTCGTCAAGA |
F: forward primer; R: reverse primer. 5′→3′.
In situ Hybridization
Gestation day 3.5 (D3.5) ∼ D7.5 WT uterus, D4.5 WT ovary, D3.5 and D4.5 Lpar3 (−/−) uterus, D4.5 Prss35 (−/−) ovary, D4.5 and D7.5 Prss35 (−/−) uterus were snap-frozen and in situ hybridization was performed as previously descried [11]–[13]. Sense and antisense probes for Prss23, Prss35, Gjb2 (gap junction protein, beta 2), Gja1 (gap junction protein, alpha 1), Cox-2 (cyclooxygenase 2), Dtprp (decidual/trophoblast prolactin-related protein), Prlpi (prolactin-like protein I), and Abp1 (amiloride binding protein 1) were synthesized from cDNA fragment amplified with their respective gene specific primer pairs (Table1).
Hormonal Treatment
Hormonal treatment on ovariectomized WT mice was done as previously described [13], [14]. Briefly, in the vehicle-treated group and the progesterone (P4)-treated group, the ovariectomized virgin C57BL6 mice (recovered for 2 weeks after surgery) were injected with 0.1 ml vehicle (oil) or P4 (2 mg in 0.1 ml oil) three times on 0 h, 24 h, and 48 h, respectively. In the 17β-estradiol (E2)-treated group, the ovariectomized mice were injected with 0.1 ml oil on 0 h and 24 h, then 100 ng E2 (in 0.1 ml oil) on 48 h. In P4+ E2-treated group, the mice were treated the same as the P4-treated group except an additional injection of 100 ng E2 on 48 h. All the mice were dissected 6 hours after the last injection. The total treatment time for P4 was 54 hours and that for E2 was 6 hours. The uteri were dissected and snap-frozen for realtime PCR.
Realtime PCR
Realtime PCR was used to quantify the expression levels of Prss23 and Prss35 in the ovariectomized WT uterus upon hormonal treatment and the mRNA expression levels of a few protease-related genes in the D4.5 WT uterus and the D4.5 Prss35 (−/−) uterus. The protease-related genes examined in the D4.5 uterus included Prss8, Prss12, Prss23, Prss35, ISP1 (implantation serine proteinase 1), ISP2, Spink3 (serine peptidase inhibitor, Kazal type 3), SerpinA3N (serine (or cysteine) proteinase inhibitor, clade A, member 3), and SerpinG1 (serpin peptidase inhibitor, clade G (C1 inhibitor), member 1). The gene specific primers were listed in Table 1. Realtime PCR was done as previously described [11]–[13]. The mRNA expression levels were normalized by the expression of GAPDH (glyceraldehyde-3-phosphate dehydrogenase). HPRT1 (hypoxanthine phosphoribosyltransferase 1) served as the second house-keeping gene (Table 1).
Vaginal Opening, Superovulation, Embryo Implantation, Gestation Period, and Litter Size
The vaginas of WT, Prss35 (+/−), and Prss35 (−/−) females were checked daily from postnatal day 22 until vaginal opening was detected, which was recorded as the age of vaginal opening. Superovulation was done on immature 21 days old WT and Prss35 (−/−) females. They were injected intraperitoneally with 5 IU eCG (equine chorionic gonadotropin, Sigma-Aldrich) and 48 hours later with 5 IU hCG (human chorionic gonadotropin, Sigma-Aldrich). The cumulus-oocyte complexes were collected from the oviduct 16 hours after hCG injection and counted. Embryo implantation was detected on D4.5 and D7.5 using blue dye reaction as previously described [7]. Gestation period and litter size were recorded as previously described [7].
Statistical Analysis
Statistical analyses were done using two-tail, unequal variance Student’s t test. The significant level was set at p<0.05.
Access to online data about Prss35tm1Lex can be found at: http://www.informatics.jax.org/javawi2/servlet/WIFetch?page=alleleDetail&key=665880. From this site, click on “data” at the row of “Notes”, it will lead to the detailed information about the knockout mice. For example, “Fertility” data are under “Diagnostics”, http://mmrrc.mousebiology.org/phenotype/Genentech/PRT215N1/Diagnostics/Fertility/Level_I/PRT215N1-Diagnostics-Fertility-Level_I.html, and RT-PCR results are under “Expression” then “WT Panel”, http://mmrrc.mousebiology.org/phenotype/Genentech/PRT215N1/Expression/WT_Panel/Level_I/popups/PRT215N1-Expression-WT_Panel-imageViewer-3349.html.
Results and Discussion
Expression of Prss23 and Prss35 in Periimplantation Mouse Uterus
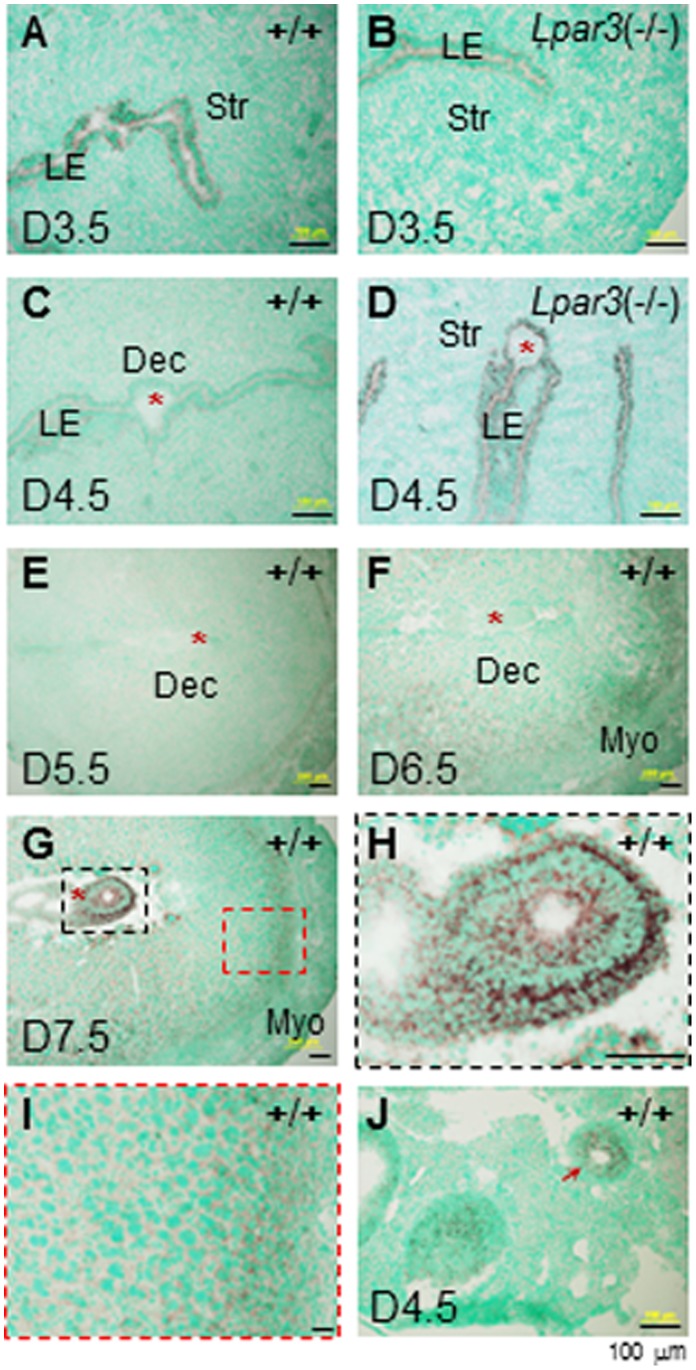
In situ hybridization showed expression of Prss23 in the uterine luminal epithelium (LE) but no detectable expression in other uterine compartments of the D3.5 WT uterus (Fig. 1A). The expression level of Prss23 was greatly decreased from LE in the D4.5 WT uterus and it was undetectable in other uterine compartments (Fig. 1C). It was detected in the LE of the D3.5 and the D4.5 Lpar3 (−/−) uteri (Figs. 1B, 1D), which had delayed uterine receptivity for embryo implantation [7]. These data demonstrated that Prss23 was downregulated in the LE upon embryo implantation, which normally initiates around D4.0 in WT mice [11], [15]. No significant levels of Prss23 were detectable in the D5.5 WT uterus (Fig. 1E). It reappeared at a relatively low level in the stromal compartment next to the myometrium in the D6.5 WT uterus (Fig. 1F). It remained there in the D7.5 WT uterus and interestingly, it was highly expressed in the D7.5 embryo (Figs. 1G∼1I). Positive control indicated Prss23 expression in the granulosa cells of the D4.5 WT ovary (Fig. 1J) [4], [5].
Figure 1. Expression of Prss23 in the periimplantation uterus by in situ hybridization.
A. Gestation day 3.5 (D3.5), wild-type (WT, +/+) uterus. B. D3.5 Lpar3 (−/−) uterus. C. D4.5 WT uterus. D. D4.5 Lpar3 (−/−) uterus. E. D5.5 WT uterus. F. D6.5 WT uterus. G. D7.5 WT uterus. H. D7.5 WT embryo enlarged from the black rectangle in G. I. D7.5 WT uterus enlarged from the red rectangle in G. J. D4.5 WT ovary as a positive control, red arrow indicating granulosa cells. Prss23 sense probe was used as a negative control, no specific signal was detected (data not shown). Red star, embryo; LE, luminal epithelium; Str, stroma; Dec, decidual zone; Myo, myometrium; scale bar, 100 µm.
Prss35 was undetectable in the D3.5 WT uterus by in situ hybridization (Fig. 2A). It was detected in the D4.5 WT stromal compartment surrounding the embryo (Fig. 2C). It was undetectable in the D3.5 and the D4.5 Lpar3 (−/−) uteri (Figs. 2B, 2D), in which embryo implantation had not occurred yet [7], and it became detectable in the stromal compartment in the D5.5 Lpar3 (−/−) uterus (Figs. 2F, 2H). Strong staining shifted to the stromal compartment on the mesometrial side of the D5.5 WT uterus (Figs. 2E, 2G). Such shifting continued on D6.5 although the intensity of expression decreased (Figs. 2I, 2J). Prss35 remained detectable at a lower expression level in the stromal compartment on the mesometrial side of the D7.5 WT uterus (Figs. 2K, 2L), but it was undetectable in the embryo from D4.5 to D7.5 (Figs. 2C, 2D, 2E, 2F, 2I, 2K). The D4.5 WT ovary was used as a positive control for Prss35 (Fig. 2M).
Figure 2. Expression of Prss35 in the periimplantation uterus by in situ hybridization.
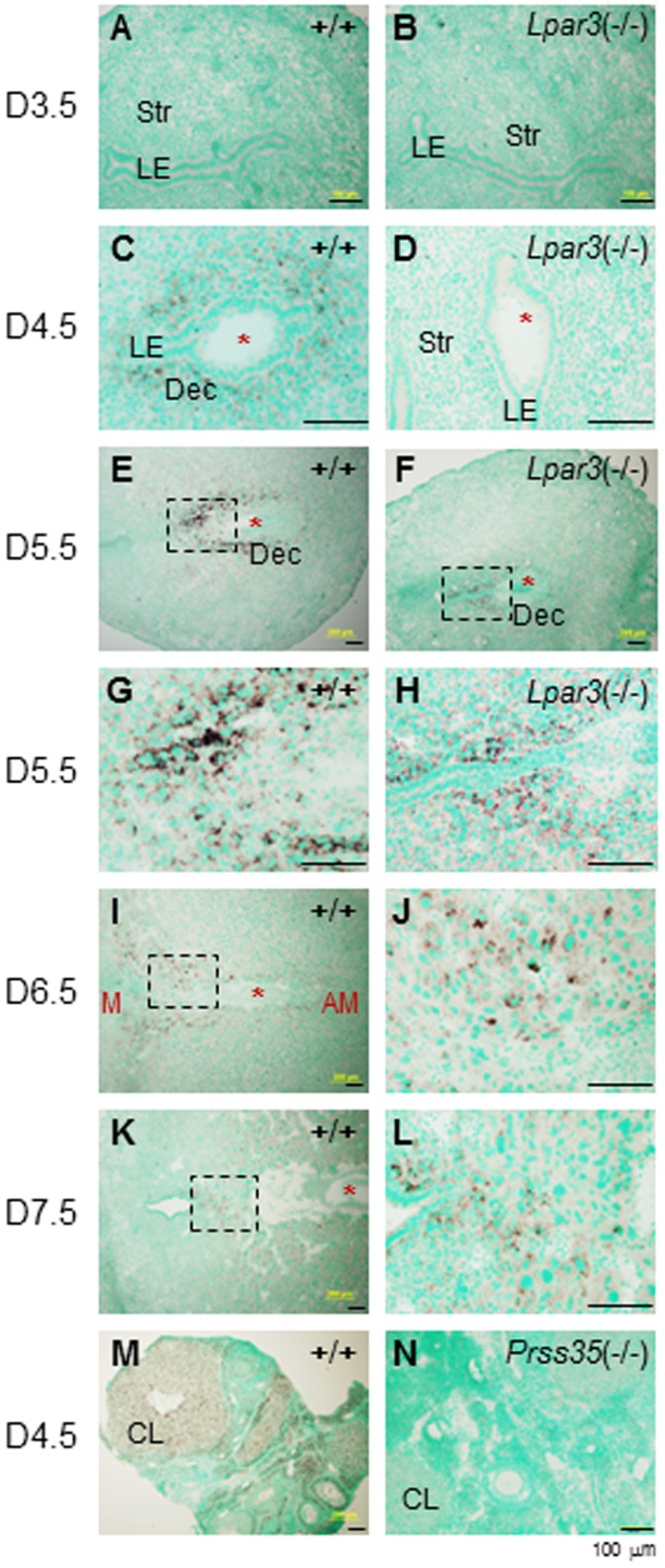
A. Gestation day 3.5 (D3.5), wild-type (WT, +/+) uterus. B. D3.5 Lpar3 (−/−) uterus. C. D4.5 WT uterus. D. D4.5 Lpar3 (−/−) uterus. E. D5.5 WT uterus. F. D5.5 Lpar3 (−/−) uterus. G. D5.5 WT uterus enlarged from the black rectangle in E. H. D5.5 Lpar3 (−/−) uterus enlarged from the black rectangle in F. I. D6.5 WT uterus. M, mesometrial side; AM, antimesometrial side. J. D6.5 WT uterus enlarged from the black rectangle in I. K. D7.5 WT uterus. L. D7.5 WT uterus enlarged from the black rectangle in K. M. D4.5 WT ovary as a positive control. Prss35 sense probe was used as a negative control, no specific signal was detected (data not shown). N. D4.5 Prss35 (−/−) ovary. Red star, embryo; LE, luminal epithelium; Str, stroma; Dec, decidual zone; CL, corpus luteum; scale bars, 100 µm.
In the original study that identified PRSS35 [4], Prss35 was only detectable in the mouse ovary but not other adult mouse tissues, including the uterus examined using dot blot. Our study indicated that Prss35 expression was only detected in the stromal compartment upon implantation during pregnancy, which may explain why dot blot couldn’t detect Prss35 in the non-pregnant adult mouse uterus [4]. RT-PCR data from MMRRC website (http://mmrrc.mousebiology.org/phenotype/Genentech/PRT215N1/Expression/WT_Panel/Level_I/popups/PRT215N1-Expression-WT_Panel-imageViewer-3349.html) indicate expression of Prss35 in most tissues (no uterus) examined, including several tissues that had undetectable levels of Prss35 by dot blot [4].
Despite their high sequence homology [4], Prss23 and Prss35 had no obvious overlapping spatiotemporal expression in the periimplantation mouse uterus (Figs. 1, 2), suggesting that they have potentially different roles in uterine remodeling during early pregnancy. Since Prss23 is expressed in the LE before embryo implantation, it is possible that it could be involved in the LE preparation for the initial implantation processes, such as embryo attachment. On the other hand, Prss35 is only detected in the stromal compartment (Fig. 2). Interestingly, the main localization of Prss35 is on the mesometrial side of the embryo (Figs. 2E, 2G).
Hormonal Regulation of Prss23 and Prss35 in Ovariectomized Mouse Uterus
In the ovariectomized WT uterus, Prss23 was moderately but significantly downregulated (<1-fold) upon P4 or E2 treatments. There was no significant difference in Prss23 expression levels between vehicle and P4+ E2-treat groups (Fig. 3). Prss35 was dramatically downregulated (>5-fold) upon P4 or E2 treatments. It was also significantly downregulated by P4+ E2 treatment (Fig. 3). The house-keeping gene HPRT1 was not regulated by these hormonal treatments (Fig. 3).
Figure 3. Regulation of Prss23 and Prss35 in ovariectomized wild-type mouse uterus upon treatments of progesterone (P4), 17β-estradiol (E2), or P4+E2.
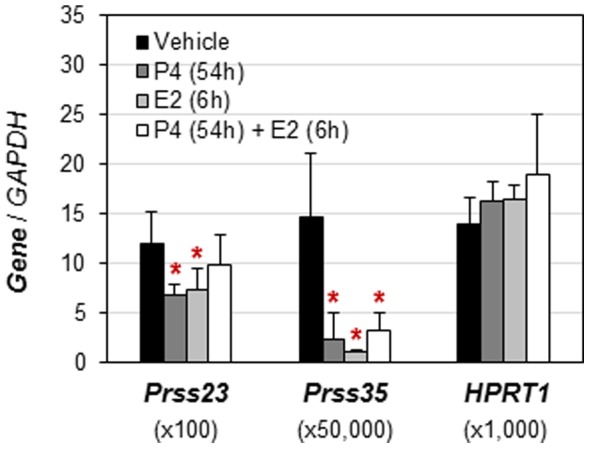
The mRNA expression levels were normalized by the expression of GAPDH (glyceraldehyde-3-phosphate dehydrogenase). HPRT1 (hypoxanthine phosphoribosyltransferase 1) served as the second house-keeping gene. N = 6. Error bars, standard deviation. *p<0.05, compared to vehicle-treated group.
Since the magnitude of uterine Prss23 upregulation during early pregnancy is decreased in heifers with low P4 [6], it suggests that P4 might upregulate Prss23 in the early pregnant heifer uterus. Prss23 mRNA can be induced by E2-activated ERα in MCF-7 breast cancer cells [16]. However, Prss23 is moderately downregulated by P4 and E2 in the ovariectomized mouse uterus under the treatment regimen (Fig. 3). These observations suggest that Prss23 could potentially be differentially regulated by P4 in the uterus in different species and/or under different experimental settings, such as natural pregnancy [6] and ovariectomy (Fig. 3), as well as that Prss23 could be differentially regulated by E2 under different experimental settings, such as breast cancer cells [16] and the ovariectomized mouse uterus (Fig. 3).
Prss35 is regulated by P4 in the mouse ovary [4]. Treatment of eCG-primed mice with steroid synthesis inhibitor trilostane (TRL) reduces ovarian P4 and Prss35 levels and the synthetic progestin R5020 could reverse the inhibitory effect of TRL on ovarian Prss35 expression [4], indirectly indicating that P4 could upregulate Prss35 in the mouse ovary. Prss35 is downregulated by P4 in the ovariectomized mouse uterus (Fig. 3). These observations suggest tissue-specific regulation of Prss35 by P4 in mice.
General Characterization of Prss35 (−/−) Mice
The unique spatiotemporal expression patterns of Prss35 in the ovary [4], [5] and in the periimplantation uterus (Fig. 2) implied that PRSS35 might play roles in female reproduction. This implication was seemingly reinforced when the limited fertility data from MMRRC database indicated reduced litter sizes of 3 (normal ∼7–8) each from two Prss35 (−/−) females mated with WT males (http://mmrrc.mousebiology.org/phenotype/Genentech/PRT215N1/Diagnostics/Fertility/Level_I/PRT215N1-Diagnostics-Fertility-Level_I.html). Therefore, we decided to study fertility in Prss35 (−/−) females.
The deletion of Prss35 alleles was confirmed by genotyping (data not shown) and in situ hybridization. Prss35 mRNA was undetectable in the D4.5 Prss35 (−/−) ovary (Fig. 2N) where Prss35 was normally expressed (Fig. 2M). Prss35 (−/−) mice were indistinguishable from their Prss35 (+/−) or WT littermates in body weight or appearance (data not shown). The mating activities of Prss35 (−/−) females were indistinguishable from that of the WT females (data not shown). PRSS35 has been associated with cleft lip/palate in humans [17]. However, there was no such defect observed in the Prss35 (−/−) mice (data not shown).
Normal Ovarian and Uterine Functions during Puberty and Pregnancy in Prss35 (−/−) Females
The ovary is involved in puberty [18], [19]. Vaginal opening is an indication of puberty onset in mice [20]. The average ages of vaginal opening for WT, Prss35 (+/−), and Prss35 (−/−) females were comparable, around postnatal 31 days (Fig. 4A). The ovary is also involved in oocyte development and ovulation. Comparable numbers of cumulus-oocyte complexes were ovulated from superovulated immature WT and Prss35 (−/−) females (Fig. 4B), indicating no obvious defect in oocyte development and ovulation in Prss35 (−/−) females.
Figure 4. Deletion of Prss35 on the age of vaginal opening (A) and the number of superovulated cumulus-oocyte complexes (COCs) from each ovary (B).
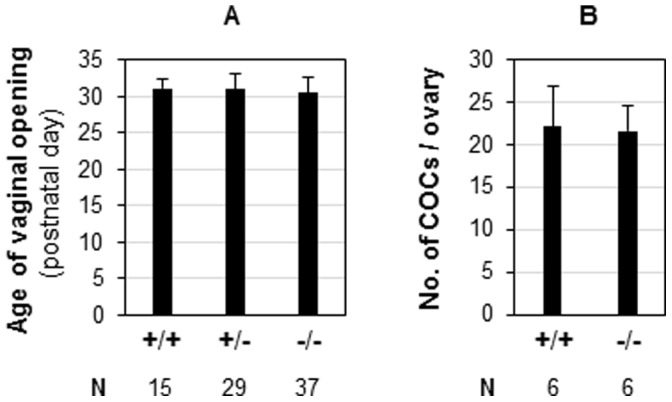
+/+, wild-type; +/−, Prss35 (+/−); −/−, Prss35 (−/−); N, the number of female mice (A) or the number of ovaries (B) in each group; error bars, standard deviation.
The corpus luteum, in which Prss35 is highly expressed (Fig. 2M) [4], [5], secretes P4 that is essential for establishing a receptive uterus for embryo implantation and for stimulating decidualization in the uterine stromal compartment upon embryo implantation [21]. The following data demonstrated that the corpus luteum in Prss35 (−/−) females had normal function. First, embryo implantation, including the timing of implantation, which was based on the appearance of the blue bands that indicated the implantation sites, the spacing among the implantation sites along the uterine horns, and the average number of implantation sites detected on D4.5, was comparable between WT and age-matched Prss35 (−/−) females (Figs. 5A∼5C). Normal embryo implantation was also confirmed at the molecular level. In situ hybridization confirmed the localization of Prss35 in the D4.5 WT uterus (Fig. 5D) and the absence of Prss35 in the D4.5 Prss35 (−/−) uterus (Fig. 5E). It detected comparable expression of implantation markers, such as Gjb2 (Figs. 5F, 5G) [22], Gja1 (Figs. 5H, 5I) [22], and Cox-2 (Figs. 5J, 5K) [23], between the D4.5 WT uterus and the D4.5 Prss35 (−/−) uterus. Second, decidualization was well developed in the D7.5 Prss35 (−/−) uterus, which had comparable sizes and numbers of implantation sites compared to those in the D7.5 WT uterus (Figs. 6A∼6C). Normal decidualization in the D7.5 Prss35 (−/−) uterus was also confirmed by the comparable expression of decidualization markers, such as Dtprp (Figs. 6D, 6E) [24], Prlpi (Figs. 6F, 6G) [24], and Abp1 (Figs. 6H, 6I) [25]. Interestingly, these decidualization markers were mainly localized in the antimesometrial side (Figs. 6D–6I), while Prss35 was mainly localized in the mesometrial side (Figs. 2E, 2I, 2K). These data also demonstrated that the Prss35 (−/−) uterus functioned normally during early pregnancy. In addition, all the females, regardless of genotypes, had gestation periods of ∼19.5 days. Therefore, PRSS35 is not essential for ovarian and uterine functions.
Figure 5. Deletion of Prss35 on embryo implantation and the expression of implantation markers in D4.5 uterus.
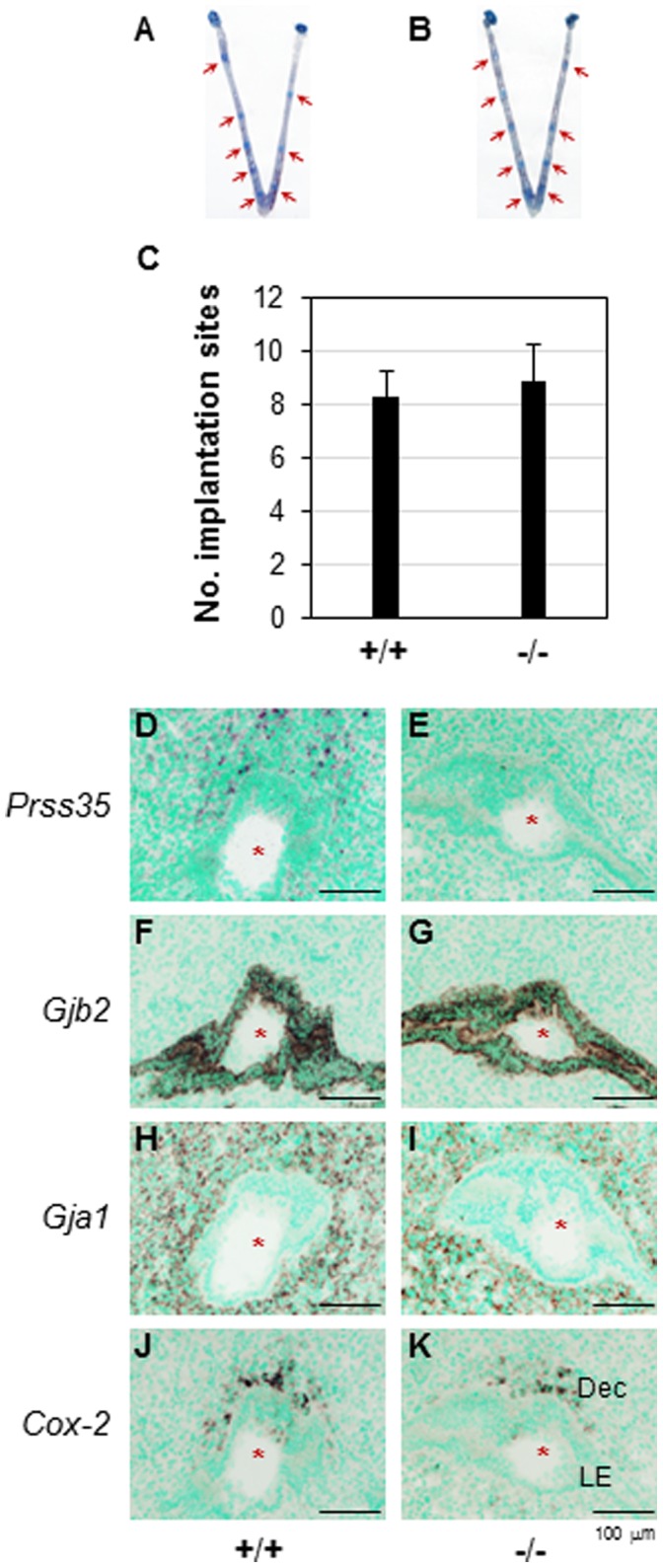
A. A representative uterus from D4.5 WT mice. B. A representative uterus from D4.5 Prss35 (−/−) mice. A & B. Red arrows, implantation sites. C. The average number of implantation sites per mouse in the WT (+/+, N = 8) or Prss35 (−/−) (−/−, N = 8) females. All the mating males were Prss35 (−/−) for both groups except 2 WT males mated with 2 females in the WT group. Error bars, standard deviation. D∼K. Gene expression in D4.5 uterus by in situ hybridization. D. Prss35 in WT uterus. E. Prss35 in Prss35 (−/−) uterus. F. Gjb2 in WT uterus. G. Gjb2 in Prss35 (−/−) uterus. H. Gja1 in WT uterus. I. Gja1 in Prss35 (−/−) uterus. J. Cox-2 in WT uterus. K. Cox-2 in Prss35 (−/−) uterus. Red star, embryo; LE, luminal epithelium; Dec, decidual zone; scale bars, 100 µm.
Figure 6. Deletion of Prss35 on embryo implantation and the expression of decidualization markers in D7.5 uterus.
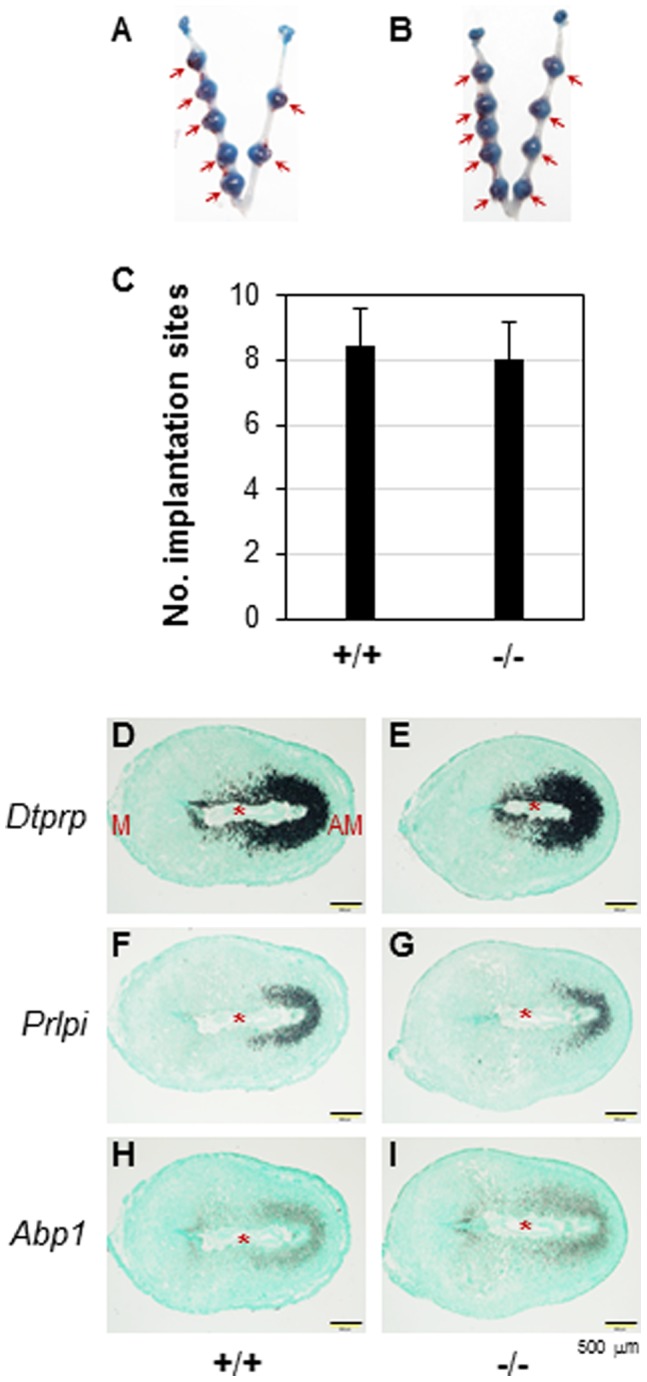
A. A representative uterus from D7.5 WT mice. B. A representative uterus from D7.5 Prss35 (−/−) mice. A & B. Red arrows, implantation sites. C. The average number of implantation sites per mouse in the WT (+/+, N = 7) or Prss35 (−/−) (−/−, N = 7) females. All the mating males were Prss35 (−/−) for both groups except one WT male mated with one female in the WT group. Error bars, standard deviation. D∼I. Expression of decidualization markers in D7.5 uterus by in situ hybridization. D. Dtprp in WT uterus. E. Dtprp in Prss35 (−/−) uterus. F. Prlpi in WT uterus. G. Prlpi in Prss35 (−/−) uterus. H. Abp1 in WT uterus. I. Abp1 in Prss35 (−/−) uterus. Red star, embryo; M, mesometrial side; AM, antimesometrial side; scale bars, 500 µm.
Non-essential Role of PRSS35 in Embryo Development
When Prss35 (−/−) males were mated with Prss35 (+/−) or Prss35 (−/−) females, the average litter sizes were comparable to that from WT × WT crosses (Fig. 7), indicating that PRSS35 did not play a critical role in embryo development.
Figure 7. Deletion of Prss35 on litter size. +/+, wild-type; +/−, Prss35 (+/−); −/−, Prss35 (−/−); N, the number of female mice in each group.
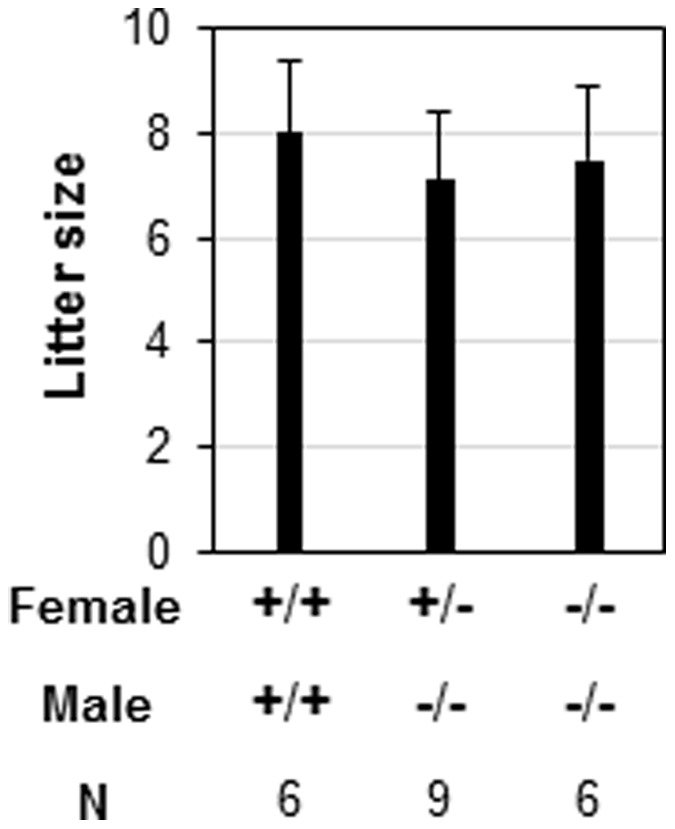
Error bars, standard deviation.
No Compensatory Upregulation of the mRNA Levels of a Few Other Protease-related Genes in the D4.5 Prss35 (−/−) Uterus
Compensatory upregulation of other proteases has been reported in the uterus upon deletion of one protease, such as in the case of upregulation of matrix metalloproteinase-3 (MMP-3) and MMP-10 in the MMP-7-deficient uterus [26]. To determine if deletion of Prss35 could lead to compensatory upregulation of other proteases in the uterus, several known uterine expressed protease-related genes were examined in D4.5 WT and Prss35 (−/−) uteri. These genes included Prss8 [27], Prss12 [28], Prss23 [4], ISP1 [29], ISP2 [30], Spink3 [31], SerpinA3N [32], and SerpinG1 [33]. Since Prss35 was upregulated in the D4.5 WT uterus upon implantation (Fig. 2C), the expression levels of these genes were examined in the D4.5 WT and Prss35 (−/−) uteri. Comparable mRNA expression levels of these protease-related genes between D4.5 WT and Prss35 (−/−) uteri indicated no compensatory upregulation of these genes in the Prss35 (−/−) uterus (Fig. 8). However, it could not rule out the possible compensatory upregulation of other protease-related genes in the uterus or the possible compensatory upregulation of protease-related genes in other tissues.
Figure 8. Deletion of Prss35 on the expression of a few protease-related genes in D4.5 uterus.
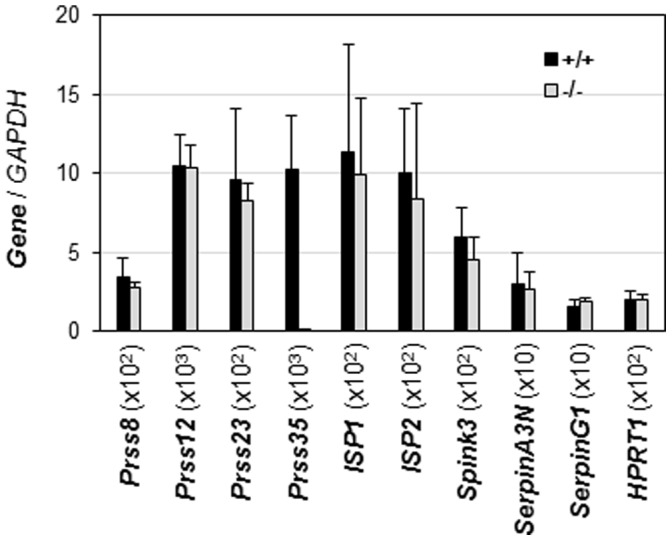
+/+, wild-type; −/−, Prss35 (−/−). X-axis indicated the names of different genes. Y-axis showed the normalized mRNA expression levels by GAPDH x the number in the parentheses following each gene on X-axis in order to present the relative expression levels of these genes in the same figure. Prss35 was included to confirm that Prss35 was deleted in the Prss35 (−/−) uterus. N = 6 (+/+) and N = 4 (−/−). Error bars, standard deviation.
In summary, this study demonstrates the distinct spatiotemporal expression patterns of Prss23 and Prss35 in the periimplantation mouse uterus as well as the non-essential role of Prss35 in ovarian function, uterine function, and embryo development using the Prss35 (−/−) mouse model.
Acknowledgments
The authors thank Dr. James N. Moore and Dr. Zhen Fu in the College of Veterinary Medicine, University of Georgia for access to the ABI 7900-Real-Time PCR machine and the imaging system, respectively. We thank Mrs. Kali King for proofreading the manuscript.
Funding Statement
This work was supported by the grants from the National Institutes of Health (NIH R15HD066301 and NIH R01HD065939 to X.Y.), and funding from the Office of the Vice President for Research, the Graduate School, the Department of Physiology & Pharmacology, and the Interdisciplinary Toxicology Program at University of Georgia, Athens, Georgia, USA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Madala PK, Tyndall JD, Nall T, Fairlie DP (2010) Update 1 of: Proteases universally recognize beta strands in their active sites. Chem Rev 110: PR1–31. [DOI] [PubMed] [Google Scholar]
- 2. Di Cera E (2009) Serine proteases. IUBMB Life 61: 510–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hedstrom L (2002) Serine protease mechanism and specificity. Chem Rev 102: 4501–4524. [DOI] [PubMed] [Google Scholar]
- 4. Miyakoshi K, Murphy MJ, Yeoman RR, Mitra S, Dubay CJ, et al. (2006) The identification of novel ovarian proteases through the use of genomic and bioinformatic methodologies. Biol Reprod 75: 823–835. [DOI] [PubMed] [Google Scholar]
- 5. Wahlberg P, Nylander A, Ahlskog N, Liu K, Ny T (2008) Expression and localization of the serine proteases high-temperature requirement factor A1, serine protease 23, and serine protease 35 in the mouse ovary. Endocrinology 149: 5070–5077. [DOI] [PubMed] [Google Scholar]
- 6.Forde N, Mehta JP, Minten M, Crowe MA, Roche JF, et al.. (2012) Effects of Low Progesterone on the Endometrial Transcriptome in Cattle. Biol Reprod. [DOI] [PubMed]
- 7. Ye X, Hama K, Contos JJ, Anliker B, Inoue A, et al. (2005) LPA3-mediated lysophosphatidic acid signalling in embryo implantation and spacing. Nature 435: 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Salamonsen LA, Nie G (2002) Proteases at the endometrial-trophoblast interface: their role in implantation. Rev Endocr Metab Disord 3: 133–143. [DOI] [PubMed] [Google Scholar]
- 9. Curry TE Jr, Osteen KG (2003) The matrix metalloproteinase system: changes, regulation, and impact throughout the ovarian and uterine reproductive cycle. Endocr Rev 24: 428–465. [DOI] [PubMed] [Google Scholar]
- 10.Salamonsen LA, Dimitriadis E, Jones RL, Nie G (2003) Complex regulation of decidualization: a role for cytokines and proteases–a review. Placenta 24 Suppl A: S76–85. [DOI] [PubMed]
- 11. Diao H, Paria BC, Xiao S, Ye X (2011) Temporal expression pattern of progesterone receptor in the uterine luminal epithelium suggests its requirement during early events of implantation. Fertil Steril 95: 2087–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diao H, Xiao S, Zhao F, Ye X (2010) Uterine luminal epithelium-specific proline-rich acidic protein 1 (PRAP1) as a marker for successful embryo implantation. Fertil Steril 94: 2808–2811 e2801. [DOI] [PubMed]
- 13.Ye X, Herr DR, Diao H, Rivera R, Chun J (2011) Unique uterine localization and regulation may differentiate LPA3 from other lysophospholipid receptors for its role in embryo implantation. Fertil Steril 95: 2107–2113 e2104. [DOI] [PMC free article] [PubMed]
- 14. Brown N, Deb K, Paria BC, Das SK, Reese J (2004) Embryo-uterine interactions via the neuregulin family of growth factors during implantation in the mouse. Biol Reprod 71: 2003–2011. [DOI] [PubMed] [Google Scholar]
- 15. Finn CA, McLaren A (1967) A study of the early stages of implantation in mice. J Reprod Fertil 13: 259–267. [DOI] [PubMed] [Google Scholar]
- 16. Chan HS, Chang SJ, Wang TY, Ko HJ, Lin YC, et al. (2012) Serine protease PRSS23 is upregulated by estrogen receptor alpha and associated with proliferation of breast cancer cells. PLoS One 7: e30397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Letra A, Menezes R, Fonseca RF, Govil M, McHenry T, et al. (2010) Novel cleft susceptibility genes in chromosome 6q. J Dent Res 89: 927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hamm ML, Bhat GK, Thompson WE, Mann DR (2004) Folliculogenesis is impaired and granulosa cell apoptosis is increased in leptin-deficient mice. Biol Reprod 71: 66–72. [DOI] [PubMed] [Google Scholar]
- 19.Kinder JE, Bergfeld EG, Wehrman ME, Peters KE, Kojima FN (1995) Endocrine basis for puberty in heifers and ewes. J Reprod Fertil Suppl 49: 393–407. [PubMed]
- 20. Safranski TJ, Lamberson WR, Keisler DH (1993) Correlations among three measures of puberty in mice and relationships with estradiol concentration and ovulation. Biol Reprod 48: 669–673. [DOI] [PubMed] [Google Scholar]
- 21. Liu HC, Pyrgiotis E, Davis O, Rosenwaks Z (1995) Active corpus luteum function at pre-, peri- and postimplantation is essential for a viable pregnancy. Early Pregnancy 1: 281–287. [PubMed] [Google Scholar]
- 22. Grummer R, Reuss B, Winterhager E (1996) Expression pattern of different gap junction connexins is related to embryo implantation. Int J Dev Biol 40: 361–367. [PubMed] [Google Scholar]
- 23. Lim H, Paria BC, Das SK, Dinchuk JE, Langenbach R, et al. (1997) Multiple female reproductive failures in cyclooxygenase 2-deficient mice. Cell 91: 197–208. [DOI] [PubMed] [Google Scholar]
- 24. Bany BM, Cross JC (2006) Post-implantation mouse conceptuses produce paracrine signals that regulate the uterine endometrium undergoing decidualization. Dev Biol 294: 445–456. [DOI] [PubMed] [Google Scholar]
- 25. Liang XH, Zhao ZA, Deng WB, Tian Z, Lei W, et al. (2010) Estrogen regulates amiloride-binding protein 1 through CCAAT/enhancer-binding protein-beta in mouse uterus during embryo implantation and decidualization. Endocrinology 151: 5007–5016. [DOI] [PubMed] [Google Scholar]
- 26. Rudolph-Owen LA, Hulboy DL, Wilson CL, Mudgett J, Matrisian LM (1997) Coordinate expression of matrix metalloproteinase family members in the uterus of normal, matrilysin-deficient, and stromelysin-1-deficient mice. Endocrinology 138: 4902–4911. [DOI] [PubMed] [Google Scholar]
- 27. List K, Hobson JP, Molinolo A, Bugge TH (2007) Co-localization of the channel activating protease prostasin/(CAP1/PRSS8) with its candidate activator, matriptase. J Cell Physiol 213: 237–245. [DOI] [PubMed] [Google Scholar]
- 28. Chen Y, Ni H, Ma XH, Hu SJ, Luan LM, et al. (2006) Global analysis of differential luminal epithelial gene expression at mouse implantation sites. J Mol Endocrinol 37: 147–161. [DOI] [PubMed] [Google Scholar]
- 29. O’Sullivan CM, Liu SY, Karpinka JB, Rancourt DE (2002) Embryonic hatching enzyme strypsin/ISP1 is expressed with ISP2 in endometrial glands during implantation. Mol Reprod Dev 62: 328–334. [DOI] [PubMed] [Google Scholar]
- 30. O’Sullivan CM, Liu SY, Rancourt SL, Rancourt DE (2001) Regulation of the strypsin-related proteinase ISP2 by progesterone in endometrial gland epithelium during implantation in mice. Reproduction 122: 235–244. [DOI] [PubMed] [Google Scholar]
- 31. Chen W, Han BC, Wang RC, Xiong GF, Peng JP (2010) Role of secretory protease inhibitor SPINK3 in mouse uterus during early pregnancy. Cell Tissue Res 341: 441–451. [DOI] [PubMed] [Google Scholar]
- 32.Pelch KE, Schroder AL, Kimball PA, Sharpe-Timms KL, Davis JW, et al.. (2010) Aberrant gene expression profile in a mouse model of endometriosis mirrors that observed in women. Fertil Steril 93: 1615–1627 e1618. [DOI] [PMC free article] [PubMed]
- 33. Mitko K, Ulbrich SE, Wenigerkind H, Sinowatz F, Blum H, et al. (2008) Dynamic changes in messenger RNA profiles of bovine endometrium during the oestrous cycle. Reproduction 135: 225–240. [DOI] [PubMed] [Google Scholar]