Abstract
Salmonella enterica is a frequent contaminant of minimally-processed fresh produce linked to major foodborne disease outbreaks. The molecular mechanisms underlying the association of this enteric pathogen with fresh produce remain largely unexplored. In our recent study, we showed that the expression of a putative stress regulatory gene, ycfR, was significantly induced in S. enterica upon exposure to chlorine treatment, a common industrial practice for washing and decontaminating fresh produce during minimal processing. Two additional genes, sirA involved in S. enterica biofilm formation and yigG of unknown function, were also found to be differentially regulated under chlorine stress. To further characterize the roles of ycfR, sirA, and yigG in S. enterica attachment and survival on fresh produce, we constructed in-frame deletions of all three genes in two different S. enterica serovars, Typhimurium and Saintpaul, which have been implicated in previous disease outbreaks linked to fresh produce. Bacterial attachment to glass and polystyrene microtiter plates, cell aggregation and hydrophobicity, chlorine resistance, and surface attachment to intact spinach leaf and grape tomato were compared among wild-type strains, single-gene deletion mutants, and their respective complementation mutants. The results showed that deletions of ycfR, sirA, and yigG reduced bacterial attachment to glass and polystyrene as well as fresh produce surface with or without chlorine treatment in both Typhimurium and Saintpaul. Deletion of ycfR in Typhimurium significantly reduced bacterial chlorine resistance and the attachment to the plant surfaces after chlorinated water washes. Deletions of ycfR in Typhimurium and yigG in Saintpaul resulted in significant increase in cell aggregation. Our findings suggest that ycfR, sirA, and yigG collectively contribute to S. enterica surface attachment and survival during post-harvest minimal processing of fresh produce.
Introduction
Salmonellosis, a human infectious disease caused by Salmonella enterica, is the leading cause of bacterial foodborne illnesses (17.4 out of 100,000 persons), hospitalizations (54%) and deaths (43%) in the U. S. [1], [2]. Infections of salmonellosis have mainly been traced back to consumption of products of animal origin; however, an increasing concern is directed to the S. enterica outbreaks associated with fresh produce [3] which poses a significant threat to food safety and public health due to the growing consumption of minimally processed fruits and vegetables as part of a healthy diet [4].
S. enterica can rapidly adapt to environmental stresses and survive for long periods of time in various non-host habitats including agricultural fields and the surface of fresh produce [5]. S. enterica has been implicated in several recent multistate outbreaks linked to contaminated fruits and vegetables including lettuce (S. Typhimurium and S. Braenderup), alfalfa sprouts (S. Enteritidis and S. Saintpaul), jalapeño peppers (S. Saintpaul), tomatoes (S. Saintpaul and S. Typhimurium), papaya (S. Agona) and cantaloupe (S. Saintpaul and S. Panama) [1], [2], [5]–[12].
Among many different S. enterica serovars, we selected to analyze Typhimurium and Saintpaul in this study because these two serovars have been implicated in major human foodborne outbreaks of salmonellosis linked to fresh produce and related products such as tomatoes, alfalfa sprouts, and orange juice [2], [5], [6], [9], [13], [14]. According to the U. S. Centers for Disease Control and Prevention, S. Typhimurium and S. Saintpaul accounted for 17% and 2.2% of all produce related human salmonellosis in 2009, respectively [1], [2].
Current industrial practice to decontaminate fresh produce harvested directly from the field involves the use of wash water supplemented with low concentrations of sodium hypochlorite (or chlorine). Chlorine is an oxidative agent that is used to reduce the overall bacterial load on produce surface and prevent cross-contamination in the wash water during minimal processing. However, studies have shown that S. enterica is capable of attaching to the produce surface and forming protective layers of biofilms [15], making it difficult to completely inactivate the attached pathogens by chlorinated water washes alone.
The genome of S. enterica contains over 4,000 protein-coding genes; some of which have been shown to play roles in bacterial stress resistance, motility, biofilm production, and virulence [16]. Among these genes, ycfR (258-bp) is a highly conserved gene in many Gram-negative bacterial species which is known to play a role in stress resistance and biofilm production in E. coli by altering cell surface hydrophobicity [17]. Our recent studies of ycfR in E. coli O157:H7, S. Typhimurium, and S. Enteritidis showed that this gene was mostly up-regulated under chlorine stress, suggesting a potential role in bacterial chlorine resistance [18]–[20]. Two additional genes, sirA (111-bp) and yigG (459-bp), were selected for analysis in this study because these two genes were also shown to be differentially regulated under chlorine stress and potentially involved in bacterial biofilm production, gene regulation, and virulence in S. enterica [20].
The major objective of this current study was to investigate the functional roles of ycfR, sirA, and yigG in the attachment and survival of S. enterica on fresh produce during procedures related to post-harvest minimal processing. We constructed in-frame deletion and complementation mutants for all three genes in both S. Typhimurium and S. Saintpaul as listed in Table 1. We compared the relative abilities of these mutants with their respective wild-type strains for chlorine resistance, biofilm production, cell aggregation and hydrophobicity, as well as surface attachment to fresh spinach leaves and grape tomatoes. Results from this study may improve our basic understanding of the molecular mechanisms that enable S. enterica to attach to produce and survive post-harvest decontamination processes.
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Designation | Reference |
| S. Typhimurium LT2 | Wild-type | (22) |
| S. Saintpaul | 99A3746 | CSDHa |
| S. Typhimurium LT2 ΔycfR::cat | This study | |
| S. Typhimurium LT2 ΔyigG::cat | This study | |
| S. Typhimurium LT2 ΔsirA::cat | This study | |
| S. Saintpaul ΔycfR::cat | This study | |
| S. Saintpaul ΔyigG::cat | This study | |
| S. Saintpaul ΔsirA::cat | This study | |
| S. Typhimurium LT2 ΔycfR::cat (pJS-8) | This study | |
| S. Typhimurium LT2 ΔyigG::cat (pJS-16) | This study | |
| S. Typhimurium LT2 ΔsirA::cat (pJS-18) | This study | |
| S. Saintpaul ΔycfR::cat (pJS-10) | This study | |
| S. Saintpaul ΔyigG::cat (pJS-16) | This study | |
| S. Saintpaul ΔsirA::cat (pJS-18) | This study | |
| E. coli BW25113 (pKD46) | (12) | |
| E. coli BW25141 (pKD3) | (12) | |
| pACYC177 | NEBb |
California State Department of Health.
New England BioLabs.
Results
Gene Annotation and Phylogenetic Analysis
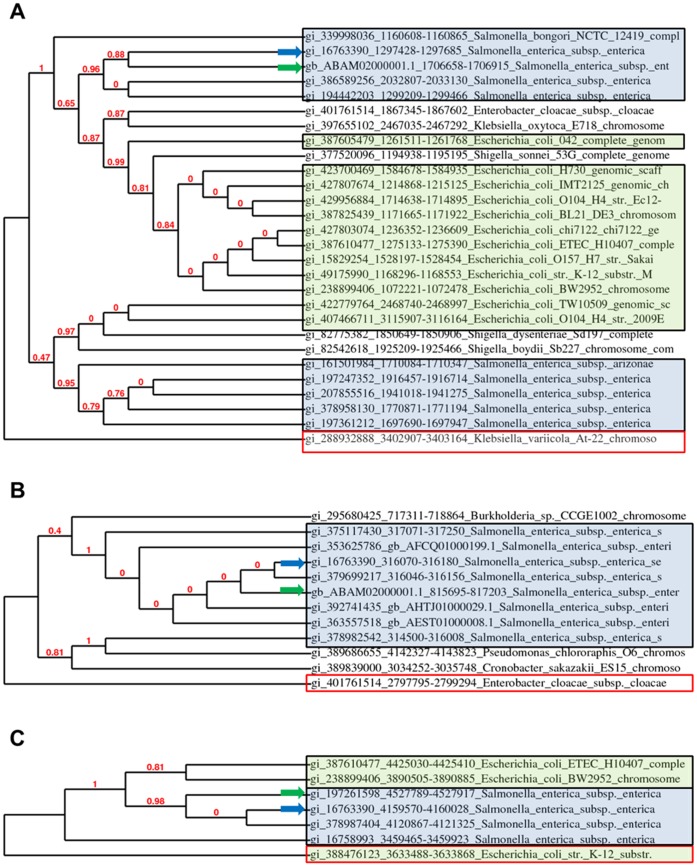
YcfR is a multiple stress resistance protein and biofilm regulator in E. coli K-12, which also plays a role in the chlorine resistance of E. coli O157:H7 [18]. However, the function of ycfR in S. enterica has not been previously reported. BLAST search and sequence alignment of ycfR in S. enterica and its homologs in other Gram-negative bacteria showed that the DNA sequences of this gene display some polymorphisms; however, the amino acid sequences are identical among various S. enterica serovars including Typhi, Saintpaul, Newport, and Montevideo (Fig. S1A). Gene ycfR is also completely conserved in the fully sequenced E. coli K-12 and O157:H7 genomes. BhsA, a multiple stress resistance protein involved in biofilm formation and hydrophobicity, is a homolog of YcfR in Shigella dysenteriae and Klebsiella oxytoca [17], [21]. Fig. 1A is a cladogram that shows the phylogenetic relatedness of ycfR in S. enterica and its homologs in other bacterial species or subspecies including S. bongori, E. coli, S. dysenteriae and K. oxytoca [22]–[28].
Figure 1. Phylogenetic relatedness of ycfR, sirA, and yigG in S. enterica and their homologs in related bacterial species.
Cladograms for ycfR (A), sirA (B), and yigG (C) with branch support values displayed at nodes were reconstructed using PhyML 3.0 aLRT and TreeDyn 198.3. (http://www.phylogeny.fr/) (22–28). Salmonella sp. and E. coli sp. are indicated by blue and green boxes, respectively. An outgroup is indicated with a red box. Blue and green arrows indicate locations of S. Typhimurium and S. Saintpaul, respectively, in the cladograms.
Gene sirA encodes a response regulator Invasol SirA [16]. A BLASTp search showed that SirA in S. Typhimurium shares 100% amino acid sequence identity with its homologs in other S. enterica serovars including Minnesota, Newport, Dublin, and Saintpaul. SirA homologs in other bacterial species include EvpB in Burkholderia sp., ImpC in Enterobacter cloacae, TssC1 in Pseudomonas chlororaphis, and EvpB in Cronobacter sakazakii [29], [30]. Gene yigG encodes a putative inner membrane protein with unknown function and is 100% identical between Typhimurium, Saintpaul, and Typhi. A homolog with 82% amino acid sequence identity was also found in E. coli K-12 and BW2952 genomes. Cladograms of sirA and yigG are shown in Fig. 1B and 1C, respectively [22]–[28]. Similar to ycfR, these two genes also display some sequence polymorphisms among different Gram-negative bacteria (Fig. S1B and S1C). Interestingly, both sirA and yigG are absent in S. Enteritidis, a prevalent S. enterica serotype that causes most foodborne salmonellosis in the U. S. [20].
Chlorine Resistance
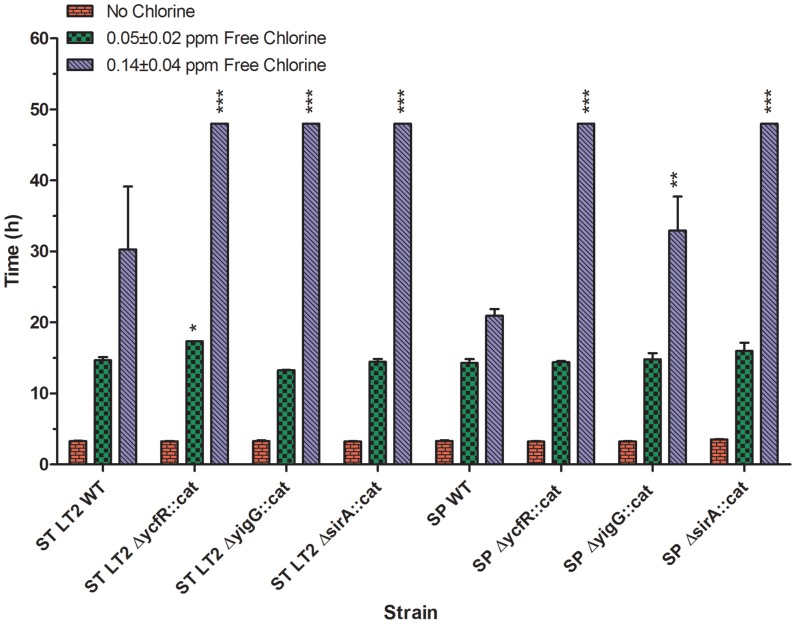
ycfR deletion mutants in S. Typhimurium and S. Saintpaul were compared for their relative resistance to sublethal chlorine stress (i.e. 0.05±0.02 ppm sodium hypochlorite in BHI broth) for 48 h at 37°C in an automated Bioscreen C system. Chlorine resistance was determined based on the extended lag phase, which was defined as the time period (h) between initial inoculation and the time when bacterial OD600 reached 0.2 as previously described [18]. Figure 2 shows that all S. Typhimurium and S. Saintpaul wild-types and mutants had an approximate 3 h lag phase in BHI broth without chlorine. When subjected to 0.05±0.02 ppm free chlorine, ΔycfR::cat in S. Typhimurium displayed the longest lag phase (approximately 17.4 h), significantly different from other mutants (P<0.05). ΔycfR::cat in S. Saintpaul showed a similar lag phase as its parent strain. This suggests that deletion of ycfR led to greater chlorine sensitivity in S. Typhimurium but not in S. Saintpaul. When the free chlorine concentration was increased to 0.14±0.04 ppm, S. Typhimurium and S. Saintpaul wild-types were still able to grow but with significantly extended lag phases at P<0.001 and P<0.0001, respectively. However, both ΔycfR::cat mutants did not grow after 48 h of incubation.
Figure 2. Extended lag phase of S. enterica wild-types and mutants in BIH broth with and without chlorine stress.
Standard deviations represent three independent experiments. Significant differences in comparison to the parent strains under the same experimental condition are shown as *(P<0.5), **(P<0.001), and ***(P<0.0001).
To determine if sirA and yigG also contribute to the chlorine resistance of S. enterica, isogenic ΔsirA and ΔyigG mutants in S. Typhimurium and S. Saintpaul were constructed and subjected to low sublethal chlorine stress (0.05±0.02 ppm free chlorine). All ΔsirA::cat and ΔyigG::cat mutants in S. Typhimurium and S. Saintpaul displayed similar lag phases to those of wild types (Fig. 2). However, when subjected to a higher concentration of chlorine (0.14±0.04 ppm free chlorine), all mutants except for ΔyigG::cat in S. Saintpaul were unable to grow, indicating that both sirA and yigG are required at least for S. Typhimurium to grow under chlorine stress.
Cell Aggregation and Hydrophobicity
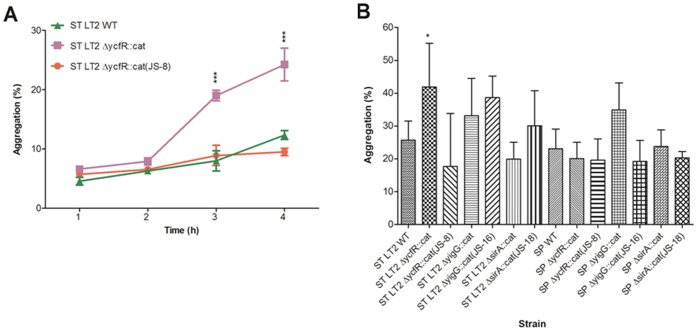
To test whether deletion of ycfR in S. enterica may cause changes to cell surface properties such as aggregation and hydrophobicity as previously reported in its close relative E. coli O157:H7 [18], S. enterica wild-type strains, ΔycfR::cat and complementation mutants were compared for these phenotypes. In the cell aggregation assay, S. Typhimurium wild-type and the ycfR complement mutant settled moderately whereas the ΔycfR::cat in S. Typhimurium aggregated quickly with significant differences (P<0.0001) at 3 h and 4 h (Fig. 3A). After 24 h, significant differences (P<0.05) were detected between ΔycfR::cat in S. Typhimurium and its parent strain (Fig. 3B). Interestingly, no change in aggregation was seen with ΔycfR::cat in S. Saintpaul, indicating that deletion of ycfR leads to greater aggregation in S. Typhimurium but not in S. Saintpaul. Assays were also conducted to test the hydrophobicity of the outer membrane of the bacterium. No remarkable difference in hydrophobicity was noted among the wild-types and mutants (data not shown), suggesting that, unlike in E. coli, ycfR may not be involved in hydrophobicity in S. enterica. In addition, hydrophobicity and aggregation (Fig. 3B) of the ΔsirA::cat and ΔyigG::cat mutants were assayed. No significant change of hydrophobicity and aggregation was evident for the mutants.
Figure 3. S. enterica aggregation assays.
A) Aggregation of S. Typhimurium LT2 WT, ΔycfR::cat mutant, and ΔycfR::cat (JS-8) complement over 4 h. B) Aggregation of all S. enterica strains after 24 h incubation at room temperature. Standard deviations represent three independent experiments. Significant differences in comparison to the parent strains under the same condition are shown as *(P<0.5), **(P<0.001), and ***(P<0.0001).
Biofilm Production
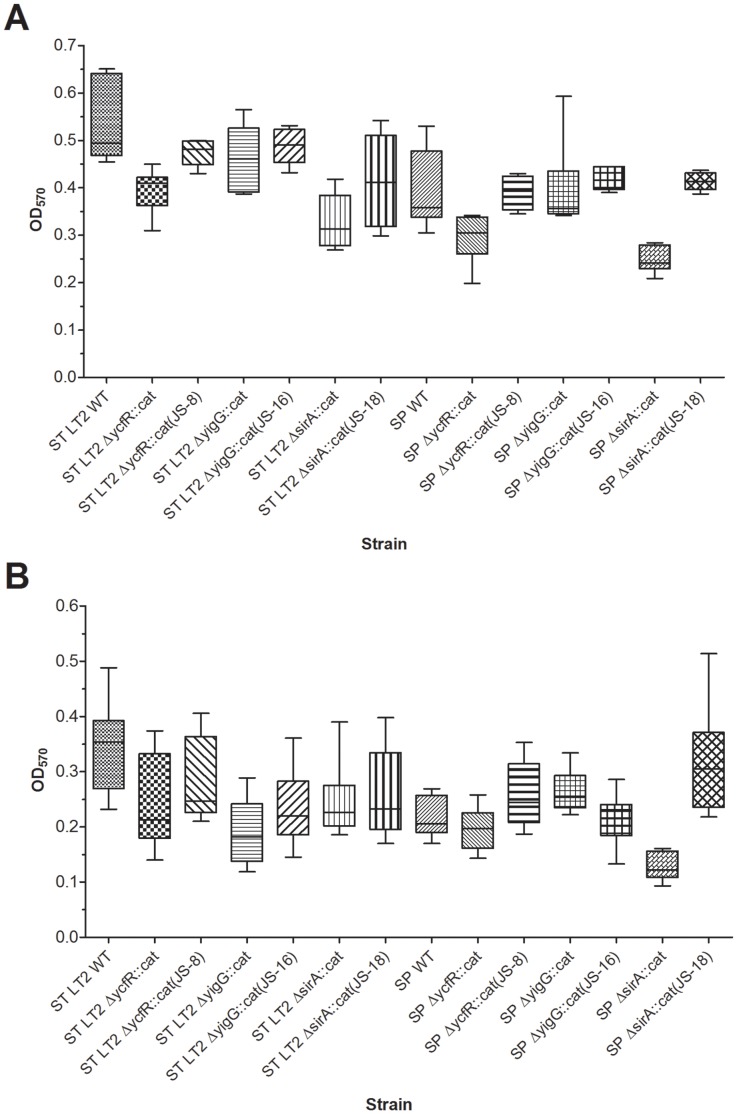
Crystal violet assays were performed to determine if ycfR contributes to biofilm formation in S. enterica, as previously reported in E. coli O157:H7 [18]. All the tested strains and mutants attached better on glass (Fig. 4A) than on polystyrene surfaces (Fig. 4B) when incubated in BHI broth at 37°C. S. Typhimurium showed a significant difference in attachment to glass surface between the wild-type and ΔycfR::cat mutant (P = 0.006) whereas the differences between the S. Saintpaul wild-type and ΔycfR::cat mutant was less notable (P = 0.035, see Table 2).
Figure 4. S. enterica crystal violet attachment assays.
The bacterial attachment is quantified by OD570 readings. A) Quantification of bacterial attachment in glass test tubes. B) Quantification of bacterial attachment in polystyrene 96-well plates. All assays were conducted for 24 h at 37°C. Middle horizontal line in each box represents the median of the entire data set; the upper and lower horizontal lines represent the upper quadrant median and the lower quadrant median, respectively. Detailed data analysis is given in Table 2.
Table 2. Statistical analysis of S. enterica crystal violet attachment assays on glass and polystyrene surfaces.
| Strain | ST LT2 WTa | SP WTb | ||
| Glass | Polystyrene | Glass | Polystyrene | |
| mutants | ||||
| S. Typhimurium LT2 ΔycfR::cat | 0.0063c | 0.021 | ||
| S. Typhimurium LT2 ΔyigG::cat | NS | 0.0003 | ||
| S. Typhimurium LT2 ΔsirA::cat | 0.0007 | 0.009 | ||
| complements | ||||
| S. Typhimurium LT2 ΔycfR::cat (pJS-8) | 0.023 | NS | ||
| S. Typhimurium LT2 ΔyigG::cat (pJS-16) | NS | 0.006 | ||
| S. Typhimurium LT2 ΔsirA::cat (pJS-18) | NS | 0.040 | ||
| mutants | ||||
| S. Saintpaul ΔycfR::cat | 0.035 | NS | ||
| S. Saintpaul ΔyigG::cat | NS | 0.020 | ||
| S. Saintpaul ΔsirA::cat | 0.0025 | <0.0001 | ||
| complements | ||||
| S. Saintpaul ΔycfR::cat (pJS-10) | NS | NS | ||
| S. Saintpaul ΔyigG::cat (pJS-16) | NS | NS | ||
| S. Saintpaul ΔsirA::cat (pJS-18) | NS | 0.010 | ||
ST LT2 wild type strain under the same condition is used as a reference in t-test analysis.
SP wild type strain under the same condition is used as a reference in t-test analysis.
All values are given as probability (P). P>0.05 indicates not significant (NS).
In contrast, ΔsirA::cat in S. Saintpaul displayed significantly decreased attachment on both glass (Fig. 4A) and polystyrene surfaces (Fig. 4B) (P = 0.0025 and P<0.0001, respectively, see Table 2) whereas the complementation mutant restored its attachment phenotype similar to that of the wild-type, indicating that sirA is necessary for biofilm production in S. Saintpaul. Interestingly, deletion of sirA in S. Typhimurium caused a less considerable but still significant reduction in attachment to both surfaces; in-trans complementation of sirA did not restore the attachment phenotype as that of the wild-type S. Typhimurium. In addition, ΔyigG::cat in S. Typhimurium showed a significant reduction in attachment in polystyrene plates (P = 0.0003) but not in glass test tubes. A slight difference was also detected for the ΔyigG::cat in S. Saintpaul on polystyrene surface (significant at P = 0.02).
Bacterial Attachment to Plant Surfaces
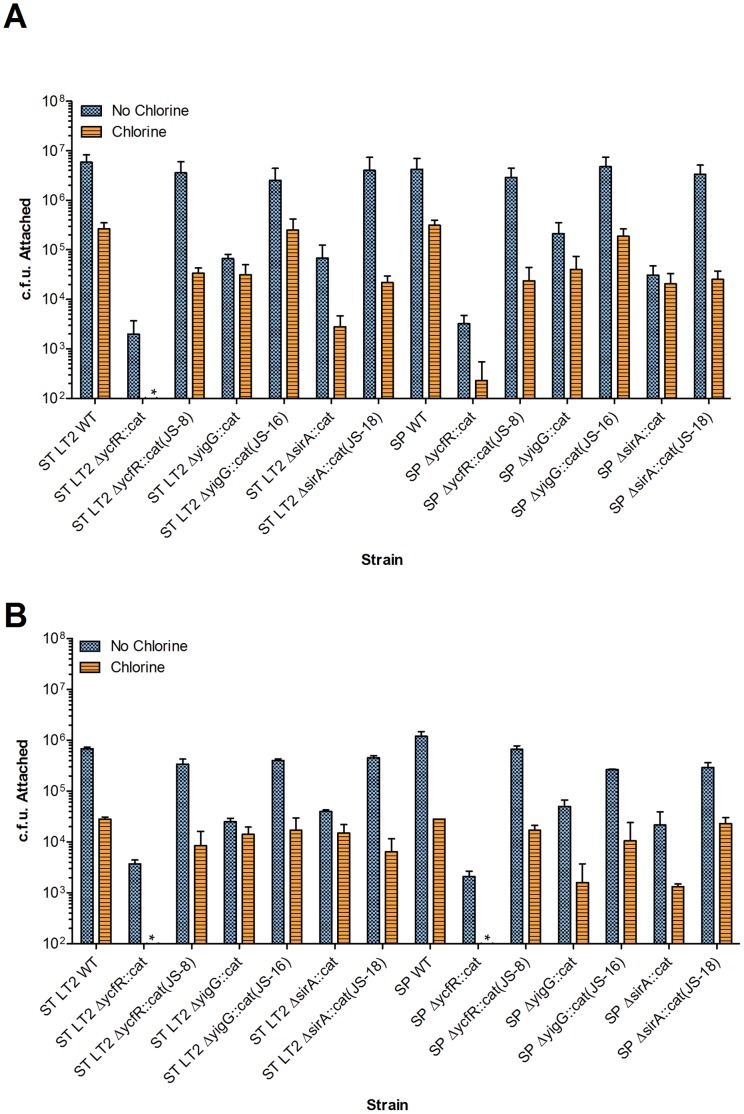
In our recent studies, we showed that deletion of ycfR in E. coli O157:H7 decreased the attachment efficiency of the bacterium to baby spinach leaves under oxidative chlorine stress [18]. To determine if ycfR is involved in attachment of S. Typhimurium and S. Saintpaul to plant surfaces, we conducted attachment assays using both intact spinach leaves (Fig. 5A) and grape tomatoes (Fig. 5B). Deletion of ycfR in both serovars led to a decrease of 102 to 103 c.f.u. in attachment to spinach leaves and grape tomatoes after chlorine treatment. Interestingly, the ΔycfR::cat in S. Typhimurium was completely unable to attach to spinach leaves and tomatoes after chlorine treatment (P<0.0001, see Table 3). In addition, the attachment of ΔycfR::cat in S. Saintpaul to spinach leaves decreased more than 103 c.f.u. after chlorine treatment (significant at P<0.0001). This mutant did not attach at all to tomatoes under the same stress conditions (significant at P<0.0001). In most cases, the complementation strains were able to restore the wild-type level of attachment after chlorine treatment. In the cases where the complement was not able to restore the wild-type attachment, we speculated that the plasmid used for complementation might interfere with how S. enterica responds to chlorine treatment, although this was not further tested in the current study. All the above collectively suggest that ycfR plays a critical role in bacterial plant surface attachment and subsequent chlorine resistance in both S. Typhimurium and Saintpaul.
Figure 5. S. enterica attachment to fresh produce surface.
A) Bacterial attachment to intact spinach leaves. B) Bacterial attachment to grape tomatoes. Attached bacteria were enumerated on XLD agar. Standard deviations represent three or four independent experiments. * indicates data is pΔresent but below the threshold for the graph. Detailed data analysis is given in Table 3.
Table 3. Statistical analysis of S. enterica spinach and tomato attachment assays with and without chlorine treatment.
| Strain | ST LT2 WTa | SP WTb | ||||||
| Spinach | Tomato | Spinach | Tomato | |||||
| −c | +d | − | + | − | + | − | + | |
| mutants | ||||||||
| S. Typhimurium LT2 ΔycfR::cat | <0.0001e | <0.0001 | <0.0001 | |||||
| S. Typhimurium LT2ΔyigG::cat | <0.0001 | <0.0001 | <0.0001 | 0.025 | ||||
| S. Typhimurium LT2 ΔsirA::cat | <0.0001 | <0.0001 | <0.0001 | 0.027 | ||||
| complements | ||||||||
| S. Typhimurium LT2ΔycfR::cat (pJS-8) | NS | <0.0001 | 0.040 | 0.040 | ||||
| S. Typhimurium LT2ΔyigG::cat (pJS-16) | 0.010 | NS | 0.021 | NS | ||||
| S. Typhimurium LT2ΔsirA::cat (pJS-18) | NS | <0.0001 | 0.044 | 0.030 | ||||
| mutants | ||||||||
| S. Saintpaul ΔycfR::cat | <0.0001 | <0.0001 | 0.003 | <0.0001 | ||||
| S. Saintpaul ΔyigG::cat | 0.0005 | <0.0001 | 0.003 | <0.0001 | ||||
| S. Saintpaul ΔsirA::cat | <0.0001 | <0.0001 | 0.002 | <0.0001 | ||||
| complements | ||||||||
| S. Saintpaul ΔycfR::cat (pJS-10) | NS | <0.0001 | NS | 0.044 | ||||
| S. SainΔtpaul ΔyigG::cat (pJS-16) | NS | NS | NS | NS | ||||
| S. Saintpaul ΔsirA::cat (pJS-18) | NS | <0.0001 | NS | NS | ||||
ST LT2 wild type strain under the same condition is used as a reference in t-test analysis.
SP wild type strain under the same condition is used as a reference in t-test analysis.
(−) and d(+) represent without and with chlorine treatment, respectively.
All values are given as probability (P). P>0.05 indicates not significant (NS).
Attachment assays to intact spinach leaves (Fig. 5A) and grape tomatoes (Fig. 5B) were also performed with ΔsirA::cat and ΔyigG::cat mutants in S. Typhimurium and Saintpaul. Compared with their respective wild-type strains, sirA and yigG deletion mutants in both serovars showed decreased attachment to spinach leaves and grape tomatoes without chlorine treatment. The most noticeable reduction in the attachment to the plant surfaces after chlorine treatment was observed in ΔsirA::cat in S. Typhimurium and Saintpaul (significant at P<0.0001, see Table 3).
Discussion
S. Typhimurium is a frequent contaminant of leafy vegetables and other fresh produce items [1], [6], [31]. The mechanisms that enable the survival of this pathogen during post-harvest processing, in particular with chlorinated water washes, have not been completely understood. In our studies of E. coli O157:H7, we demonstrated that ycfR was involved in bacterial chlorine resistance and survival on the surface of spinach leaves [18]. The results of this current study provided additional evidence to show that ycfR, which is the most up-regulated gene in S. enterica under chlorine stress [20], is also important for the chlorine resistance and attachment of S. enterica on plant surfaces.
ycfR encodes a putative membrane protein in E. coli K-12 and regulates biofilm production via a process that involves changing surface properties of the bacterial cell [17]. This gene also helps E. coli cope with multiple environmental stresses such as fluctuating temperature and pH [18], [20], [21]. In E. coli O157:H7, ycfR plays a critical role in the bacterial stress response and was most up-regulated under oxidative stress [20], [32]. Unlike in K-12, ycfR in O157:H7 does not alter the cell surface properties or biofilm formation [18]. Similar to E. coli, some functional differences of ycfR were observed between S. enterica serovars Typhimurium and Saintpaul. For instance, deletion of ycfR reduced attachment of S. Saintpaul to grape tomato to a greater extent than S. Typhimurium. S. Saintpaul has been predominantly associated with foodborne outbreaks linked to produce items such as tomatoes, jalapeño peppers, and alfalfa sprouts [9] but not leafy greens. It remains to be determined whether ycfR also plays a role in the attachment of other S. enterica serovars to various fruit and vegetable surfaces.
In our previous study on the global gene expression of S. Typhimurium under chlorine stress, sirA and yigG were up- and down-regulated, respectively [20]. sirA encodes Invasol SirA, a Type VI secretion protein of the EvpB family [16], [33]. This family of proteins consists of putative cytoplasmic, periplasmic, and outer membrane localizing proteins that are commonly found in Gram-negative organisms which are associated with eukaryotic cells in either a pathogenic or symbiotic manner [32]–[34]. The precise role and mode of action of this secretion system has not been thoroughly studied. Here we showed that sirA is involved in biofilm formation of S. enterica on both glass and polystyrene. Although the crystal violet staining method used here more readily quantifies attachment of bacteria and not necessarily biofilm formation, this technique was used as a generalized experiment to estimate bacterial biofilm productivity under different experimental conditions. In addition, we showed that both sirA and yigG, which encodes a putative inner membrane protein [35], are also involved in S. enterica attachment to spinach leaves and grape tomatoes. Admittedly, it remained unknown whether ycfR, sirA and yigG are involved in a single or multiple regulatory circuits in S. enterica or other pathogenic bacteria that control the oxidative stress response and plant surface attachment. More in-depth investigations may shed new light on the specific roles of these genes in the interaction of S. enterica with plant surfaces.
It should be noted that the findings from this study were based on laboratory-scale experiments of two S. enterica strains on baby spinach leaves and grape tomatoes. Future studies should include analysis of strains representing other S. enterica serotypes and strains, additional fresh produce items, and ideally scaled-up experimental approaches using actual processing lines to ascertain the roles of specific genes on S. enterica surface attachment in order to develop more effective and targeted strategies to minimize the contamination of this pathogen in fruit and vegetables, as well as on food contact surfaces during post-harvest minimal processing of fresh produce. We based this study on chlorine which is the most commonly used sanitizer in the current fresh produce industry. Use of other sanitizing agents or combined methods has been recommended to improve the killing of bacterial pathogens on fresh produce and minimize environmental and health hazards. Functional roles of S. enterica genes in response to these and other preventive control measures await to be further investigated in the future.
Materials and Methods
Bacterial Strains and Culture Conditions
Bacterial strains and mutants used in this study are listed in Table 1. S. Typhimurium strain LT2 (ATCC 19585) was obtained from the American Type Culture Collection (ATCC, Manassas, VA) and S. Saintpaul strain 99A3746 was kindly provided by the California State Department of Health. E. coli strains BW25113 (pKD46) and BW25141 (pKD3) were kindly provided by the E. coli Genetic Stock Center at Yale University. Vector pACYC177 was purchased from New England BioLabs, Beverly, MA. All mutants were derived from laboratory stocks in this study. Bacteria were grown in brain heart infusion (BHI) (Becton, Dickinson and Co., Franklin Lakes, NJ) supplemented with chloramphenicol (30 µg ml−1), ampicillin (50 µg ml−1), or kanamycin (30 µg ml−1) when necessary.
Construction of in-frame Deletion and Complementation Mutants
In-frame gene deletions of ycfR, sirA, and yigG in S. Typhimurium and Saintpaul were generated using the lambda red recombinase method as described by Datsenko and Wanner [36]. The primer pairs used here are listed in Table S1. Briefly, 60-bp upstream and downstream regions of each target gene were fused to a chloramphenicol cartridge (cat) from pKD3 and transformed into competent S. enterica cells containing lambda recombinase plasmid pKD46 induced by 10 mM L-arabinose. Colonies of transformants with a deletion marked with the cat cassette were selected for on BHI agar plates containing chloramphenicol and verified by colony PCR and subsequent DNA sequencing.
Complementation mutants were constructed by cloning a fragment containing the original gene along with its promoter-region into the vector pACYC177. The BamHI restriction site was used to obtain complementation vectors pJS-8, pJS-16, and pJS-18, which harbored gene ycfR, yigG and sirA, respectively. The vector was subsequently transformed into competent S. enterica deletion mutants. Colonies were selected on BHI agar plates containing kanamycin and verified by colony PCR and DNA sequencing.
Measurement of Total and Free Chorine in BHI Broth
The total and free chlorine concentrations were measured at room temperature using ChloroSense (Palintest Limited, Tyne & Wear, England) according to the manufacturer’s instructions. Seventy ml BHI broth was supplemented with 2.7, or 3.0 µl per ml of a 13% fresh stock sodium hypochlorite solution, resulting in final free chlorine concentrations of 0.05±0.02, and 0.14±0.04 ppm in the broth, respectively.
Bacterial Growth Under Chlorine Stress
Growth curves of S. enterica at 37°C were performed using a Bioscreen C automatic growth curve system (Growth Curves, Piscataway, NJ). Overnight cultures of S. enterica in BHI broth were normalized to an OD600 of 0.8. A 1∶10,000 dilution was aliquoted into BHI broth supplemented with 0.05±0.02 and 0.14±0.04 ppm free chlorine. A 200-µl aliquot was loaded in triplicate into a 100-well honeycomb plate for analysis. Uninoculated BHI broth was used as a negative control. Bacterial growth was monitored by recording the cell turbidity every 5 min, after a 10 s shaking period, over a period of 48 h. All experiments were repeated at least three times using independent cultures for statistical analysis.
Aggregation Assay
Aggregation assays were conducted based on the method by Yonezawa et al. [37]. Briefly, bacteria were grown overnight at 37°C with shaking, centrifuged at 4,000 rpm for 20 min, washed with 5 ml PBS, and finally resuspended in 5 ml PBS. The bacteria were then incubated at room temperature without shaking and the OD600 was measured and recorded at 1, 2, 3, 4, and 24 h, respectively. The percent aggregation was calculated as (OD600 before incubation - OD600 after incubation)/OD600 before incubation × 100. Experiments were repeated at least three times with triplicate samples for statistical analysis.
Cell Surface Hydrophobicity Assay
Cell adherence to hexane was measured as previously described by Deng et al. [18]. Briefly, overnight cultures incubated at 37°C were centrifuged and resuspended in PBS containing differing amounts of hexane. After incubation at room temperature for 1 h, the hexane phase was removed and OD600 of the remaining aqueous suspension was measured. Hydrophobicity is represented as the calculated percentage of bacteria remaining in the aqueous phase. Experiments were repeated at least three times with triplicate samples for statistical analysis.
Crystal Violet Attachment Assay
Crystal violet attachment assay was performed as previously described by Deng et al. [18]. Briefly, individual bacterial cultures were grown overnight at 37°C in BHI broth, normalized, and then grown overnight without shaking in glass test tubes or polystyrene 96-well microtiter plates. After overnight growth, the liquid was removed, and attached bacteria remaining in the test tubes or wells were washed three times with PBS and incubated with 3 ml (for test tubes) or 200 µl (for microtiter plates) of 1% crystal violet for 15 min. The test tubes and plates were then washed three times with PBS and incubated with 95% ethanol for 20 min. The OD570 readings, which reflect the amount of attached bacteria, were measured. All experiments were performed at least three times with triplicate samples for statistical analysis.
Bacterial Attachment to Spinach Leaf and Grape Tomato
Attachment assays were conducted for each wild-type strain and its corresponding deletion and complementation mutants, based on the method by Deng et al. [18] with minor modifications. Baby spinach and grape tomatoes were purchased from a local retail grocer. S. enterica cultures were grown overnight in BHI at 37°C with shaking. All cultures were normalized to 1×108 c.f.u./ml and 1 ml was added into a 50-ml conical tube containing 45 ml PBS. For each experiment, six pieces of intact spinach leaves (approximately 1 g each) were placed into the conical tubes and incubated with S. enterica culture for 10 min. After incubation, leaves were pulled out and air dried in a biohazard cabinet in sterile petri dishes for 1 h. Three leaves was washed three times with PBS as the untreated control; the other three leaves was immersed in a 50 ppm aqueous chlorine solution (made from a 13% sodium hypochlorite stock solution) for 10 s and the reaction was stopped by adding 4.5 ml of 1 M sodium thiosulfate. The chlorine-treated leaves were then washed three times with PBS. To recover attached bacteria, five sterile 6-mm glass beads were added to the leaves in 50-ml conical tubes containing 10 ml PBS and vortexed vigorously for 1 min. Serial dilutions of the eluted bacteria were plated on XLD agar (Becton Dicksinson and Co.) in duplicate. The plates were incubated at 37°C for 24 h before c.f.u. were enumerated. Attachment assays with tomatoes were performed similarly using six intact grape tomatoes (approximately 8 g each) for each experiment. All attachment experiments were conducted independently for at least three times.
Phylogenetic Analysis
Gene sequences of ycfR, sirA, and yigG in S. enterica and their homologs in other bacterial genomes were retrieved from GenBank under accession numbers as shown in Fig. 1. Multiple gene alignments were performed using MUSCLE 3.7 and Gblocks 0.91 b modules of Phylogeny.fr. Phylogenetic analysis was performed using PhyML 3.0 aLRT and cladograms were generated using TreeDyn 198.3 module of Phylogeny.fr. All the above software and modules are freely available at http://www.phylogeny.fr/ [23].
Statistical Analysis
Student’s t-test analysis was performed using GraphPad Prism software package (GraphPad Software, Version 5).
Supporting Information
Gene sequence alignment of ycfR , sirA , and yigG in S. enterica and their homologs in related bacterial species. Multiple gene sequence alignments of ycfR (A), sirA (B), and yigG (C).
(DOCX)
Primers used in this study. A list of all primers used in the study along with sequences.
(DOCX)
Acknowledgments
We are grateful to Zhang and Tortorello lab members for discussion and technical support in this study.
Funding Statement
This work was supported by the United States Department of Agriculture National Institute of Food and Agriculture Grant no. 2011-67017-30016 under the Agriculture and Food Research Initiative Foundational Program Priority Area of Foodborne Pathogen-Plant Interactions (Program Area Code - A1301). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Centers for Disease Control and Prevention (2011) National Salmonella surveillance annual data summary, 2009. U.S. Department of Health and Human Services, Atlanta, Georgia.
- 2. Centers for Disease Control and Prevention CfDCaP (2011) Vital signs: incidence and trends of infection with pathogens transmitted commonly through food- foodborne diseases active surveillance network, 10 U.S. sites, 1996–2010. MMWR Morb Mortal Wkly Rep 60: 749–755. [PubMed] [Google Scholar]
- 3. Berger CN, Sodha SV, Shaw RK, Griffin PM, Pink D, et al. (2010) Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ Microbiol 12: 2385–2397. [DOI] [PubMed] [Google Scholar]
- 4. Lynch MF, Tauxe RV, Hedberg CW (2009) The growing burden of foodborne outbreaks due to contaminated fresh produce: risks and opportunities. Epidemiol Infect 137: 307–315. [DOI] [PubMed] [Google Scholar]
- 5. Humphrey T (2004) Salmonella, stress responses and food safety. Nat Rev Microbiol 2: 504–509. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention (2006) Salmonellosis- outbreak investigation, October 2006. U. S. Department of Health and Human Services, Atlanta, Georgia.
- 7. Centers for Disease Control and Prevention CfDCaP (2006) Multistate outbreak of Salmonella Typhimurium infections associated with eating ground beef- United States, 2004. MMWR Morb Mortal Wkly Rep 55: 180–182. [PubMed] [Google Scholar]
- 8. Centers for Disease Control and Prevention CfDCaP (2008) Outbreak of Salmonella serotype Saintpaul infections associated with multiple raw produce items- United States, 2008. MMWR Morb Mortal Wkly Rep 57: 929–934. [PubMed] [Google Scholar]
- 9. Behravesh CB, Mody RK, Jungk J, Gaul L, Redd JT, et al. (2011) 2008 outbreak of Salmonella Saintpaul infections associated with raw produce. N Engl J Med 364: 918–927. [DOI] [PubMed] [Google Scholar]
- 10. Gajraj R, Pooransingh S, Hawker JI, Olowokure B (2012) Multistate outbreaks of Salmonella Braenderup associated with consumption of iceberg lettuce. Int J Environ Health Res 22: 150–155. [DOI] [PubMed] [Google Scholar]
- 11. Munnoch SA, Ward K, Sheridan S, Fitzsimmons GJ, Shadbolt CT, et al. (2009) A multi-state outbreak of Salmonella Saintpaul in Australia associated with cantaloupe consumption. Epidemiol Infect 137: 367–374. [DOI] [PubMed] [Google Scholar]
- 12. Takkinen J, Nakari UM, Johansson T, Niskanen T, Siitonen A, et al. (2005) A nationwide outbreak of multiresistant Salmonella Typhimurium in Finland due to contaminated lettuce from Spain, May 2005. Euro Surveill 10: E050630.050631. [DOI] [PubMed] [Google Scholar]
- 13. Centers for Disease Control and Prevention (2009) Multistate outbreak of Salmonella infections associated with peanut butter and peanut butter-containing products- United States, 2008–2009. MMWR Morb Mortal Wkly Rep 58: 85–90. [PubMed] [Google Scholar]
- 14. Mody RK, Greene SA, Gaul L, Sever A, Pichette S, et al. (2011) National outbreak of Salmonella serotype Saintpaul infections: importance of Texas restaurant investigations in implicating jalapeño peppers. PLoS ONE 6: e16579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brandl MT (2006) Fitness of human enteric pathogens on plants and implications for food safety. Ann Rev Phytopathol 44: 367–392. [DOI] [PubMed] [Google Scholar]
- 16. McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, et al. (2001) Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413: 852–856. [DOI] [PubMed] [Google Scholar]
- 17. Zhang XS, Garcia-Contreras R, Wood TK (2007) YcfR (BhsA) influences Escherichia coli biofilm formation through stress response and surface hydrophobicity. J Bacteriol 189: 3051–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deng K, Wang S, Rui X, Zhang W, Tortorello ML (2011) Functional analysis of ycfR and ycfQ in Escherichia coli O157: H7 linked to outbreaks of illness associated with fresh produce. Appl Environ Microbiol 77: 3952–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang S, Deng K, Zaremba S, Deng X, Lin C, et al. (2009) Transcriptomic response of Escherichia coli O157: H7 to oxidative stress. Appl Environ Microbiol 75: 6110–6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang S, Phillippy AM, Deng K, Rui X, Li Z, et al. (2010) Transcriptomic responses of Salmonella enterica serovars Enteritidis and Typhimurium to chlorine-based oxidative stress. Appl Environ Microbiol 76: 5013–5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wood TK (2009) Insights on Escherichia coli biofilm formation and inhibition from whole-transcriptome profiling. Environ Microbiol 11: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dereeper A, Audic S, Claverie JM, Blanc G (2010) BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 12;10(8). [DOI] [PMC free article] [PubMed]
- 23.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 1;36 (Web Server Issue): W465–9. [DOI] [PMC free article] [PubMed]
- 24. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 19 32(5): 1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17(4): 540–52. [DOI] [PubMed] [Google Scholar]
- 26. Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52(5): 696–704. [DOI] [PubMed] [Google Scholar]
- 27. Anisimova M, Gascuelo O (2006) Approximate likelihood ratio test for branchs: A fast, accurate and powerful alternative. Syst Biol 55(4): 539–52. [DOI] [PubMed] [Google Scholar]
- 28. Chevenet F, Brun C, Banuls AL, Jacq B (2006) TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 10 7: 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Filloux A, Hachani A, Bleves S (2008) The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiol 154(6): 1570–1583. [DOI] [PubMed] [Google Scholar]
- 30. Aubert D, MacDonald DK, Valvano MA (2010) BcsKc is an essential protein for the type VI secretion system activity in Burkholderia cenocepa that forms an outer membrane complex with BcsLB . J Biol Chem 12 285(46): 35988–35998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Doyle MP, Erickson MC (2008) Summer meeting 2007- the problems with fresh produce: an overview. J Appl Microbiol 105: 317–330. [DOI] [PubMed] [Google Scholar]
- 32. Fink RC, Black EP, Hou Z, Sugawara M, Sadowsky MJ, et al. (2012) Transcriptional responses of Escherichia coli K-12 and O157: H7 associated with lettuce leaves. Appl Environ Microbiol 78: 1752–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fricke WF, Mammel MK, McDermott PF, Tartera C, White DG, et al. (2011) Comparative genomics of 28 Salmonella enterica isolates: evidence for CRISPR-mediated adaptive sublineage evolution. J Bacteriol 193: 3556–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Folkesson A, Lofdahl S, Normark S (2002) The Salmonella enterica subspecies I specific centisome 7 genomic island encodes novel protein families present in bacteria living in close contact with eukaryotic cells. Res Microbiol 153: 537–545. [DOI] [PubMed] [Google Scholar]
- 35. Drew D, Sjostrand D, Nilsson J, Urbig T, Chin CN, et al. (2002) Rapid topology mapping of Escherichia coli inner-membrane proteins by prediction and PhoA/GFP fusion analyss. Proc Natl Acad Sci U S A 99: 2690–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yonezawa H, Osaki T, Kurata S, Zaman C, Hanawa T, et al. (2010) Assessment of in vitro biofilm formation by Heliobacter pylori. . J Gastroenterol Hepatol 25 Suppl: S90–94 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene sequence alignment of ycfR , sirA , and yigG in S. enterica and their homologs in related bacterial species. Multiple gene sequence alignments of ycfR (A), sirA (B), and yigG (C).
(DOCX)
Primers used in this study. A list of all primers used in the study along with sequences.
(DOCX)