Abstract
Our recent finding that the Candida albicans RNase III enzyme CaDcr1 is an unusual, multifunctional RNase III coupled with data on the RNase III enzymes from other fungal species prompted us to seek a model that explained the evolution of RNase III’s in modern budding yeast species. CaDcr1 has both dicer function (generates small RNA molecules from dsRNA precursors) and Rnt1 function, (catalyzes the maturation of 35S rRNA and U4 snRNA). Some budding yeast species have two distinct genes that encode these functions, a Dicer and RNT1, whereas others have only an RNT1 and no Dicer. As none of the budding yeast species has the canonical Dicer found in many other fungal lineages and most eukaryotes, the extant species must have evolved from an ancestor that lost the canonical Dicer, and evolved a novel Dicer from the essential RNT1 gene. No single, simple model could explain the evolution of RNase III enzymes from this ancestor because existing sequence data are consistent with two equally plausible models. The models share an architecture for RNase III evolution that involves gene duplication, loss, subfunctionalization, and neofunctionalization. This commentary explains our reasoning, and offers the prospect that further genomic data could further resolve the dilemma surrounding the budding yeast RNase III’s evolution.
Keywords: Candida, DCR1, RNT1, RNase III, bifunctional dicer, evolution, yeast
The RNase III Family of Proteins
RNase III enzymes are a family of double-strand RNases conserved from bacteria to higher eukaryotes.1,2 RNase III family members participate in a wide variety of RNA-processing reactions including the generation of small interfering RNAs (siRNAs)3 capable of eliciting gene silencing and the processing of rRNA, snoRNA, or snRNA precursor molecules.4-7 The hallmark of the RNase III family of proteins is the RNase III domain. This domain is distinguished by the presence of both a dsRNA-binding domain (dsRBD); and an RNase III signature motif. The RNase III signature motif contains a number of highly conserved amino acids important for coordination of 2 Mg2+ ions and RNase III catalytic activity8-11 while the dsRBD binds 2’-OH of the ribose moieties and the phosphate backbone of dsRNA. Furthermore, some RNase IIIs contain multiple dsRBDs that can act collectively to identify and bind specific substrates.12 As their diverse biological functions suggest, RNase III enzymes may contain accessory domains such as PAZ, helicase, or additional dsRBD domains, which play roles in targeting the enzyme to a specific RNA substrate or facilitating specific biochemical tasks once present at the target RNA.13
In S. cerevisiae, the only protein predicted to contain an RNase III domain is Rnt1 (SceRnt1). The RNT1 gene encodes a protein that is important for the initial steps of 35S rRNA processing, and for processing snoRNA and snRNA.4-7 However, the SceRnt1 lacks dicer function, and S. cerevisiae does not encode an Argonaute, another key component of RNAi silencing systems. By contrast the Saccharomyces castellii genome encodes not only a RNT1 ortholog, but a second RNase III gene. This second RNase III, ScaDcr1, is a noncanonical Dicer enzyme. Although ScaDcr1 generates siRNA capable of mediating gene silencing, it encodes only a single RNase III domain, and lacks PAZ and helicase domains typically encoded by canonical Dicers.14 Furthermore, ScaDcr1 is distinguished from Rnt1 enzymes by the presence of a second dsRBD at its C-terminus.
The Candida albicans Dicer, CaDcr1, is a multifunctional RNase III
Inspection of budding yeast genome sequences revealed that non-canonical Dicer enzymes could be found in a number of species including the human pathogen C. albicans.14 Since small RNAs with the structure similar to those of Argonaute-bound guide RNAs had previously been identified in C. albicans, we hypothesized that this C. albicans RNase III (CaDCR1) was responsible for siRNA generation. C. albicans strains homozygous for a deletion of the resident CaDCR1, but encoding an inducible copy at an ectopic site were very slow growing when the exogenous copy was not expressed, suggesting that in contrast to ScaDCR1, CaDCR1 was important for WT growth.14 However, knockout of a C. albicans Argonaute (CaAGO1) had no obvious growth defect, which suggested that the Cadcr1/Cadcr1 growth defect was unlikely to be a consequence of diminished silencing.15 These results raised the question; does CaDcr1 function as a Dicer? Subsequent experiments showed that the small-RNA levels observed in wild type were decreased in the C. albicans CaDCR1 knockout and that Candida DCR1 complemented a S. castellii dcr1 in a GFP silencing assay. Moreover, in vitro CaDcr1 could process a dsRNA molecule into sRNA fragments, and the size of these fragments was consistent with the sRNA fragments identified in C. albicans cell extracts. These data suggested CaDcr1 was acting as a Dicer in vivo, but failed to explain why CaDcr1 was essential.15
Further inspection of the Candida genome revealed that the only other RNase III enzyme, Candida Dicer Like1 (CDL1), was likely to be catalytically inactive since a number of residues known to coordinate Mg2+ in RNase III active sites had been altered.15 As CaDcr1 was the only active RNase III enzyme in C. albicans, we hypothesized that CaDCR1 might carry out functions similar to those of S. cerevisiae RNT1, Loss of Rnt1 function in S. cerevisiae led to defects in rRNA and snRNA processing, and indeed, Northern analysis showed that knock out of CaDCR1 similarly lead to an increased accumulation of rRNA and U4 precursor molecules.4,15,16
Our analysis showed that members of the budding-yeast lineage contain at least three distinct types of RNase III enzymes. S. cerevisiae contains only a single RNase III enzyme, SceRnt1, which carries out rRNA, snoRNA, and snRNA processing reactions. S. castellii contains two functional RNase III enzymes, one an ortholog of SceRnt1 that is likely to function similarly, and a noncanonical Dicer, that generates siRNA capable of gene silencing. Members of the Candida clade appear to encode two proteins with RNase III motifs: an active enzyme, CaDcr1, that functions both as a non-canonical Dicer and Rnt1 in vivo (Table 1), and Candida Dicer Like 1 (Cdl1), that lacks essential residues for RNase III activity.
Table 1.
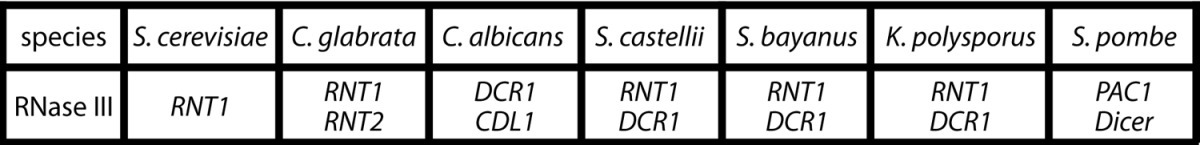
List of the RNase III domain containing proteins found in budding yeast mentioned throughout this commentary and S. pombe (fission yeast).
Evolution of the Budding Yeast RNase III
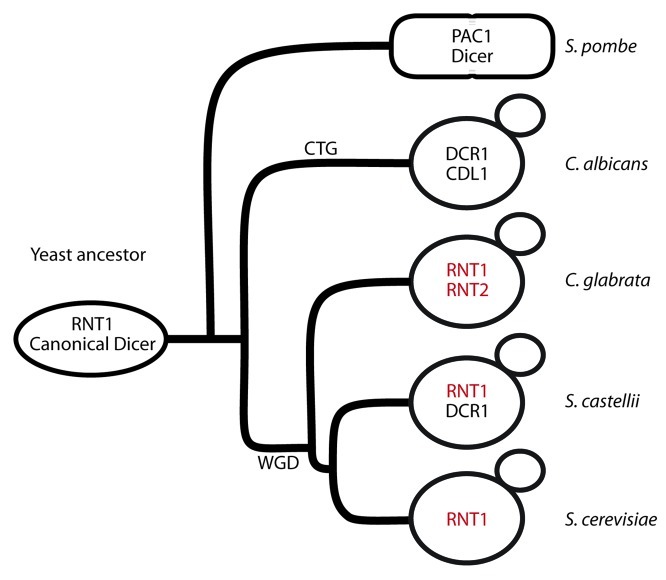
The discovery that extant members of the budding-yeast lineage encode such a wide variety of RNase III enzymes prompted our attempt to reconstruct an evolutionary history that could engender such diversity. Our aim was to identify a linear progression from a presumptive ancestral RNase III complement to that found in the current species of budding yeasts. This route should be compatible with the known genome sequence relationships, assume no lateral transfer of genes, or multiple independent and simultaneous evolution of the noncanonical Dicer (DCR) and RNT1 genes. Based on these considerations, we made the underlying assumption (suggested previously)15 that both the budding yeast and non-budding yeast (e.g., S. pombe) lineages emerged from an ancestor that had both a canonical Dicer enzyme (DICER) as well as an RNT1 ortholog (Fig. 1) (Table 1), and that DICER was lost from the budding-yeast lineage.
Figure 1. Cartoon depicting where WGD and CTG codon usage changes are estimated to have occurred in the budding yeast lineage. Genes colored in red are syntenic with one another.
Simple models require an unlikely sequence of events
A simple model that could account for the diversity of RNase IIIs in budding yeast is that this lineage traces its origin back to an ancestor that had lost DICER and had only a single RNT1 ortholog. After the whole-genome duplication event (WGD), there were two copies of this ancestral RNT1. In S. castellii, this duplication was followed by neo-functionalization of one copy to create ScaRNT1 and ScaDCR1. In the Candida clade one of the RNT1 ohnologs evolved to gain an additional Dicer activity and the second, CDL1, was inactivated. This model is unsatisfactory because it requires that the WGD occur prior to the formation of the Candida clade. However, evidence suggests the Candida clade split from the Saccharomyces clade prior to the WGD, making the WGD an improbable explanation for the duplication of an ancestral RNase III gene in the Candida clade.17 Furthermore, regions of the Candida genome surrounding CDL1 and DCR1 are not syntenic, thus the two Candida albicans RNase III paralogs are likely to be a product of a more restricted duplication event. A comparison of the genes neighboring ScaDCR1 with those adjacent to ScaRNT1 also failed to reveal synteny consistent with a WGD derivation. By contrast, C. glabrata RNT1 paralogs show synteny in the genes flanking each copy suggesting that the C. glabrata RNT1’s are possible products of the WGD (Fig. 1). So, while there is support for duplication of RNT1 by WGD in the budding yeast lineage, the WGD event is unlikely to be responsible for the RNase III genes in S. castellii (ScaDCR1and ScaRNT1) or the Candida clade (CaDCR1 and CDL1) .
A second model posits that the evolution of the second RNase III protein present in S. castellii and the Candida clade involved rare independent events caused by the duplication of RNT1 and its subsequent evolution to obtain Dicer function. This sequence of events seems an unlikely scenario as antecedent events for S. castellii for several reasons. First, if the ScaDCR1 RNase III were the result of a recent autonomous duplication event of ScaRNT1, then ScaDcr1 should be more related to the ScaRnt1 than to other budding yeast Dcr1s. However, the RNase III domain sequences of ScaDcr1 are more closely related to the RNase III domains of Saccharomyces bayanus and Kluyveromyces polysporus, than they are to that of ScaRnt1 (Table 2).18 S. bayanus and K. polysporus, are two yeasts from disparate branches of the Saccharomycete tree that like S. castellii encode two RNase IIIs, one homologous to ScaRNT1 and one homologous to ScaDCR1. Not only are the RNase III domains of these DCR1’s more closely related to each other than to the RNT1 gene family, they possess a distinct domain structure containing an additional C-terminal double stranded binding domain not found in Rnt1s. Although, the sequence of CaDcr1 RNase III domain is slightly more similar to the SceRnt1 and ScaRnt1 than the budding yeast Dcr1s RNase III domains, Candida Dcr1 possesses the distinct domain structure of S. bayanus, K. polysporus, and S. castellii Dcr1s (Table 2). These data suggest S. bayanus, K. polysporus, S. castellii, and C. albicans DCR1 did not independently evolve via recent duplications of RNT1. Instead, it is likely they originated from a common ancestral enzyme present before these lineages split.
Table 2.

Pair wise comparison of CaDcr1, CaCdl1, SceRnt1, ScaDcr1, ScaRnt1, SbDcr1, KpDcr1, CgRnt1, CgRnt2, SpDcr1, SpDcr2, and SpRnt1 RNase III domains. Higher scores indicate greater homology.
In addition, our second model does not satisfactorily explain the origin of the two RNase IIIs found in C. albicans. The Candida clade is distinct from other budding yeast because they do not encode a Rnt1 homolog and one of their RNase III homologs, Cdl1, appears to have been inactivated through precise alteration of several key catalytic residues.11,15 Since multiple highly conserved residues in RNase III domains have been altered in Cdl1, it is not surprising that Cdl1’s RNase III domain is less homologous to active RNase III domains.18 Nonetheless, Cdl1’s RNase III domain is most highly homologous to CaDcr1’s RNase III domain, and the Cdl1 domain structure is consistent with it being more closely related to a noncanonical dicer than a Rnt1 enzyme14(Table 2). In view of these considerations, it is likely that Cdl1 is a product of gene duplication and subsequent evolution of CaDcr1. This duplication occurred after the noncanonical Dicer ancestor had been established in the budding-yeast lineage. We cannot rule out that the duplication creating Cdl1 occurred prior to the split of the C. albicans, S. bayanus, K. polysporus, and S. castellii lineages, and the duplicated gene was subsequently lost from all except the Candida clade, where it was instead inactivated.
We considered a third model in which duplication of an ancestral canonical Dicer followed by subfunctionalization and neofunctionalization of one of the copies resulted in the generation of a noncanonical dicer-like enzyme. Loss of canonical dicer in S. castellii would then account for the present day allocation of its RNase III enzymes. According to this model, loss of canonical Dicer as well as the noncanonical Dicer would leave only RNT1 as is presently observed in S. cerevisiae. Duplication of noncanonical Dicer as well as apparent inactivation of one copy accompanied by RNT1 and loss of the canonical Dicer would result in the complement observed in the Candida clade. We found these scenarios to be unlikely because the RNase III domain sequence from CaDcr1 and ScaDcr1 are more similar to one another, and to SceRnt1 or ScaRnt1 than they are to modern-day canonical Dicer RNase III domains18 (Table 2). This suggests that the noncanonical Dicers likely evolved from an ancestral Rnt1-like enzyme and not from a canonical Dicer.
Two Models that Explain the Evolution of Budding Yeast DCR1 and RNT1
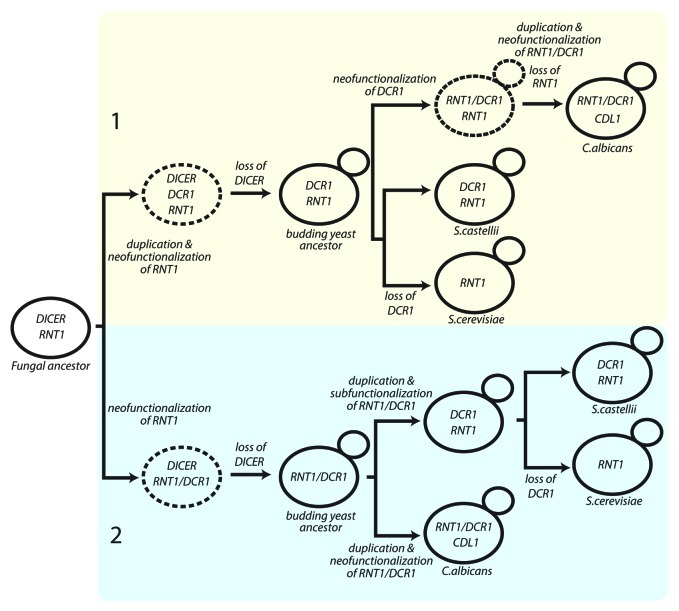
The failure of these models to explain the diversity of contemporary fungal RNase IIIs led us to propose that both the budding-yeast DCR1s evolved from an ancestral Rnt1 enzyme (Fig. 2).
Figure 2. Two potential models to explain the evolution of budding yeast DCR1 and RNT1. DICER, canonical Dicer as found in S. pombe; DCR1, budding yeast Dicer as found in S. castellii, K. polysporus, and S. bayanus; RNT1, Ribonuclease III as in S. cerevisiae; RNT1/DCR1, multifunctional Dicer found in the Candida clade; CDL1 Candida Dicer-like from the Candida clade. Proposed transitional species are shown in dashed lines. (This figure is nearly identical to the original model figure published in ref. 15).
Model 1 proposes duplication of RNT1 with neofunctionalization of one copy to generate a non-canonical Dicer gene (DCR) in a transitional species. The loss of DICER left both RNT1 and DCR1, as in present-day S. castellii. Loss of DCR1 and the rest of the RNAi pathway in many budding-yeast lineages left these lineages with only RNT1, as observed in present-day S. cerevisiae and other members of the Saccharomyces complex that lack Argonaute and Dicer homologs. Meanwhile, neofunctionalization of DCR1 in the S. castellii-like ancestor of the Candida clade led to the multifunctional enzyme Rnt1/Dcr1 (CaDcr1). RNT1 loss (and CDL1 gain through RNT1/DCR1 duplication with neofunctionalization/inactivation) generated the RNase III genes of C. albicans.15
Model 2 posits early neofunctionalization of an ancestral RNT1 to generate a transitional species with the multifunctional RNT1/DCR1. Subsequent canonical Dicer loss then generated the Candida-like budding yeast ancestor, which gained CDL1 through RNT1/DCR1 duplication with neofunctionalization of CDL1 in the Candida lineage. Meanwhile, RNT1/DCR1 duplication with subfunctionalization of each copy generated the two RNase III enzymes present in S. castellii.15
Model 1 predicts that the S. castellii Dcr1 RNase III domain should be more similar to the RNase III domain of CaDcr1 (Rnt1/Dcr1) than that of S. castellii Rnt1 whereas Model 2 predicts that the S. castellii Dcr1 RNase III domain should be more similar to the RNase III domain of S. castellii Rnt1 than that of CaDcr1, however, the S. castellii Dcr1 RNase III domain is not significantly more similar to the RNase III domains of ScaRnt1 or CaDcr1 (Rnt1/Dcr1) (Table 2). We are thus unable to distinguish between the two models.
Substrates affect RNase III evolution
What are the selective pressures that shaped the evolution of these RNase IIIs, their loss, and the emergence of novel functions over time? The answers to these questions require consideration of not only the RNases, but also the interplay between these proteins and their substrates.
dsRNA viruses shaped the fungal genome
Drinnenberg et al. observed that loss of dicing activity in yeast correlates with the acquisition of killer virus. While dicing activity is hypothesized to play a protective role limiting the expression of transposons,19 acquisition of killer virus provides a competitive advantage over yeast lacking the virus as these killer virus strains secrete a toxin that kills neighbors not infected with the virus.20 Consequently, the budding yeast lineage has been faced with opposing selections: Retain the advantage of Dicer and RNAi at the risk of elimination by a strain lacking Dicer but harboring the killer virus. The advantage gained from the killer virus could explain why many budding yeast lack either a canonical or noncanonical Dicer. The dichotomy between species with RNAi and no killer and those with killer and no RNAi is a striking example of how the fungal genome has been molded by a dsRNA substrate.20
RRNA evolution could affect RNase III evolution
Slight changes in the sequence of the ribosome substrate of Rnt1 could have led to the loss of the canonical RNT1, and the emergence of the multifunctional Candida CaDcr1. For example, mutation of a substrate in an essential reaction, such as the rRNA 3′ETS, might have permitted the eventual loss of canonical RNT1 if Dcr1 acquired or possessed the ability to make this cleavage. Both in vitro and in vivo analysis have affirmed that the dsRBD binding of the AGNN tetra loop is required for SceRnt1 to cleave the double stranded 35S rRNA 3′ETS,21-23 whereas preliminary analysis of 35S rRNA 3′ETS from the Candida clade suggests that a conserved AGNN tetra loop is not required for efficient cleavage.24 Such an adaptation could explain our observation that S. cerevisiae RNT1 is unable to complement a CaDCR1 knockout. In the ancestor of modern C. albicans, mutation in the rRNA 3′ETS could have allowed CaDcr1 to process this altered substrate more efficiently than Rnt1. This change coupled with an ensuing period of Rnt1 decay could have permitted changes to additional Rnt1 substrates to be cemented in the Candida lineage and CaDcr1 to take over the role/s of an ancestral Rnt1.
Of course, we cannot rule out a dramatic change in the Candida albicans DCR1, such as the addition of a C-terminal dsRBD, which allowed it to compete more efficiently for substrates previously cleaved by Rnt1. Such a change could have paved the way for the eventual loss of Rnt1 from the Candida lineage, and the subsequent drift of 35S rRNA 3′ETS sequence. Alternatively, evolution through loss of a dsRBD could have provided an equally significant functional adaptation as is posited in Model 2. Eukaryotic proteins have been found to encode up to five dsRBDs, and even seemingly minor changes in the number of dsRBDs can have profound effects on their biological function.25,26 Furthermore, additional dsRNases could have played compensatory roles during the postulated transitional stages. Although some bacterial RNase III enzymes are essential,27 in other species they are dispensable presumably because other RNases perform essential RNase III functions in their absence.28,29 Similar overlap of functions by ribonucleases with broad specificity could have supported budding yeast RNase III evolution.
Summary and Future Directions
The discovery of the multifunctional CaDcr1 suggested that the RNase IIIs of extant species evolved from a common ancestor by gene duplication events coupled with subfunctionalization, neofunctionalization, and gene loss. The relationship among extant species is consistent with either of two models, but does not permit the choice between them. The ambiguities could be resolved by genome sequences of additional yeast species. For example, the identification of an extant budding yeast with a complement of RNase IIIs labeled as transitional in Figure 2 could pave the way to a single model that satisfactorily traces the route from the ancestral species to modern budding yeast.
Acknowledgments
The authors would like to acknowledge David Bartel and David Weinberg for helpful discussions. This work was funded by A.C.S. Grant PF-09–072–01-MBC to D.A.B., N.I.H. NRSA F32 AI729353 to V.K.V., Herman Sokol Fellowship to V.K.V, and N.I.H. Grant GM040266 (to G.R.F.).
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/21360
References
- 1.Lamontagne B, Larose S, Boulanger J, Elela SA. The RNase III family: a conserved structure and expanding functions in eukaryotic dsRNA metabolism. Curr Issues Mol Biol. 2001;3:71–8. [PubMed] [Google Scholar]
- 2.Apirion D, Miczak A. RNA processing in prokaryotic cells. Bioessays. 1993;15:113–20. doi: 10.1002/bies.950150207. [DOI] [PubMed] [Google Scholar]
- 3.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–9. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 4.Elela SA, Igel H, Ares M., Jr. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–24. doi: 10.1016/S0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 5.Chanfreau G, Elela SA, Ares M, Jr., Guthrie C. Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev. 1997;11:2741–51. doi: 10.1101/gad.11.20.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abou Elela S, Ares M., Jr. Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 1998;17:3738–46. doi: 10.1093/emboj/17.13.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chanfreau G, Legrain P, Jacquier A. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J Mol Biol. 1998;284:975–88. doi: 10.1006/jmbi.1998.2237. [DOI] [PubMed] [Google Scholar]
- 8.Robertson HD, Webster RE, Zinder ND. Purification and properties of ribonuclease III from Escherichia coli. J Biol Chem. 1968;243:82–91. [PubMed] [Google Scholar]
- 9.Chen SM, Takiff HE, Barber AM, Dubois GC, Bardwell JC, Court DL. Expression and characterization of RNase III and Era proteins. Products of the rnc operon of Escherichia coli. J Biol Chem. 1990;265:2888–95. [PubMed] [Google Scholar]
- 10.Blaszczyk J, Tropea JE, Bubunenko M, Routzahn KM, Waugh DS, Court DL, et al. Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure. 2001;9:1225–36. doi: 10.1016/S0969-2126(01)00685-2. [DOI] [PubMed] [Google Scholar]
- 11.Weinberg DE, Nakanishi K, Patel DJ, Bartel DP. The inside-out mechanism of Dicers from budding yeasts. Cell. 2011;146:262–76. doi: 10.1016/j.cell.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saunders LR, Barber GN. The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J. 2003;17:961–83. doi: 10.1096/fj.02-0958rev. [DOI] [PubMed] [Google Scholar]
- 13.MacRae IJ, Doudna JA. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr Opin Struct Biol. 2007;17:138–45. doi: 10.1016/j.sbi.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, et al. RNAi in budding yeast. Science. 2009;326:544–50. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein DA, Vyas VK, Weinberg DE, Drinnenberg IA, Bartel DP, Fink GR. Candida albicans Dicer (CaDcr1) is required for efficient ribosomal and spliceosomal RNA maturation. Proc Natl Acad Sci USA. 2012;109:523–8. doi: 10.1073/pnas.1118859109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol. 2000;20:441–52. doi: 10.1128/MCB.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dujon B. Yeast evolutionary genomics. Nat Rev Genet. 2010;11:512–24. doi: 10.1038/nrg2811. [DOI] [PubMed] [Google Scholar]
- 18.Stothard, P., The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques, 2000. 28(6): p. 1102, 1104. [DOI] [PubMed]
- 19.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–68. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drinnenberg IA, Fink GR, Bartel DP. Compatibility with killer explains the rise of RNAi-deficient fungi. Science. 2011;333:1592. doi: 10.1126/science.1209575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chanfreau G, Buckle M, Jacquier A. Recognition of a conserved class of RNA tetraloops by Saccharomyces cerevisiae RNase III. Proc Natl Acad Sci USA. 2000;97:3142–7. doi: 10.1073/pnas.070043997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaudin C, Ghazal G, Yoshizawa S, Elela SA, Fourmy D. Structure of an AAGU tetraloop and its contribution to substrate selection by yeast RNase III. J Mol Biol. 2006;363:322–31. doi: 10.1016/j.jmb.2006.08.029. [DOI] [PubMed] [Google Scholar]
- 23.Wu H, Yang PK, Butcher SE, Kang S, Chanfreau G, Feigon J. A novel family of RNA tetraloop structure forms the recognition site for Saccharomyces cerevisiae RNase III. EMBO J. 2001;20:7240–9. doi: 10.1093/emboj/20.24.7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chanfreau G. Conservation of RNase III processing pathways and specificity in hemiascomycetes. Eukaryot Cell. 2003;2:901–9. doi: 10.1128/EC.2.5.901-909.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palavicini JP, O’Connell MA, Rosenthal JJ. An extra double-stranded RNA binding domain confers high activity to a squid RNA editing enzyme. RNA. 2009;15:1208–18. doi: 10.1261/rna.1471209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palavicini JP, Correa-Rojas RA, Rosenthal JJ. Extra double-stranded RNA binding domain (dsRBD) in a squid RNA editing enzyme confers resistance to high salt environment. J Biol Chem. 2012;287:17754–64. doi: 10.1074/jbc.M112.366005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herskovitz MA, Bechhofer DH. Endoribonuclease RNase III is essential in Bacillus subtilis. Mol Microbiol. 2000;38:1027–33. doi: 10.1046/j.1365-2958.2000.02185.x. [DOI] [PubMed] [Google Scholar]
- 28.Takiff HE, Chen SM, Court DL. Genetic analysis of the rnc operon of Escherichia coli. J Bacteriol. 1989;171:2581–90. doi: 10.1128/jb.171.5.2581-2590.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Babitzke P, Granger L, Olszewski J, Kushner SR. Analysis of mRNA decay and rRNA processing in Escherichia coli multiple mutants carrying a deletion in RNase III. J Bacteriol. 1993;175:229–39. doi: 10.1128/jb.175.1.229-239.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]