Abstract
Prokaryotes possess various defense mechanisms against invading DNA. Adaptive defense by CRISPR/Cas relies on incorporation of invader DNA sequences in the host genome. In Escherichia coli, processed transcripts of these incorporated sequences (crRNAs) guide Cascade-mediated invader DNA recognition.1–4 Cascade is a multisubunit ribonucleoprotein complex, consisting of one crRNA and five proteins: Cse1, Cse2, Cas7, Cas5 and Cas6e.1, 2 Cascade-mediated DNA recognition requires a conserved sequence adjacent to the target (protospacer adjacent motif, PAM) and a negatively supercoiled DNA topology.3, 4 While Cse1 carries out PAM recognition,5 the Cascade structure suggests that Cse2 may interact with target DNA in the PAM-distal end of the protospacer.6 Using Electrophoretic Mobility Shift Assays, we here describe the function of the Cse1 and Cse2 subunits in the context of protospacer recognition on negatively supercoiled DNA. While Cse1 is required for nonspecific DNA binding, Cse2 appears to be important for specific binding, presumably by mediating stabilizing interactions with the displaced strand, the R-loop, or both. Furthermore, we performed Scanning Force Microscopy using linearized DNA molecules, which facilitates accurate and reliable measurements of Cascade-mediated bending. This analysis reveals that Cascade binding induces flexibility in the DNA target, most likely due to single stranded DNA regions flanking the R-loop.
Keywords: CRISPR, DNA, RNA, bacteria, defense, immune, nucleic acids, phage, prokaryotes, ribonucleoprotein complex
Bacteria and archaea defend themselves with the recently discovered CRISPR/Cas system against invading DNA such as phages and conjugative plasmids (reviewed in refs. 7–17). Nowadays ten different subtypes of CRISPR/Cas systems are recognized, that belong to three main types.18 The E. coli K12 Type I-E is one of the most extensively studied systems and consists of a CRISPR locus (Clustered Regularly Interspaced Short Palindromic Repeats) with type 2 repeat sequences19 and 8 CRISPR-associated (cas) genes (cas3, cse1, cse2, cas7, cas5, cas6e, cas1, cas2). The CRISPR locus is characterized by repetitive sequences of 29 nt, that are separated by 32 nt of invader-derived sequences, known as spacers. During the adaptation stage, new spacer sequences can be incorporated in the existing array through as yet unknown mechanisms.20, 21 During the subsequent expression stage, the CRISPR locus is transcribed into a precursor CRISPR RNA (pre-crRNA) molecule, which is then cleaved by a dedicated Cas protein.1 This protein, Cas6e, cleaves the pre-crRNA in the repeat sequence to generate 61 nt mature crRNA.2 These mature crRNA molecules form a key component of Cascade, a ribonucleoprotein complex consisting of crRNA and 5 different Cas proteins: Cse1, Cse2, Cas7, Cas5 and Cas6e.1, 2 During the interference stage, invader DNA with a sequence complementary to the crRNA is recognized by the Cascade complex through base pairing between the crRNA and the target DNA sequence (protospacer).2, 4 Following recognition, Cascade as well as the target DNA undergo conformational changes. 2, 4, 6 Most notably, the positions and orientations of the Cse1, Cse2 and Cas6e subunits alter upon nucleic acid binding,6 and the target DNA is strongly bent at the site of Cascade binding.4 Finally, Cas3 is recruited and the invader DNA is degraded by the joint nuclease and helicase activities of the Cas3 HD-nuclease domain and the Cas3 SF2 helicase domain.4
Interestingly, research over the last few years has revealed that Cascade DNA recognition is constrained by several features of the target DNA. First, the protospacer needs to be flanked by a conserved motif, known as the protospacer adjacent motif (PAM).3, 22 At least four different PAM sequences are allowed,4 which are being recognized by a loop structure of the Cse1 subunit of the Cascade complex.5 Second, although mismatches between the crRNA and the target DNA are allowed at some positions, the 3′ end of the protospacer needs to be fully complementary at the so-called seed region, corresponding to positions 1 to 5 and positions 7 to 8 of the protospacer.3 Third, the target DNA needs to be negatively supercoiled (nSC).4 The nSC topology contributes half of the energy required for strand separation of the double stranded target DNA during base pairing between the crRNA, contained by Cascade, and the protospacer.4
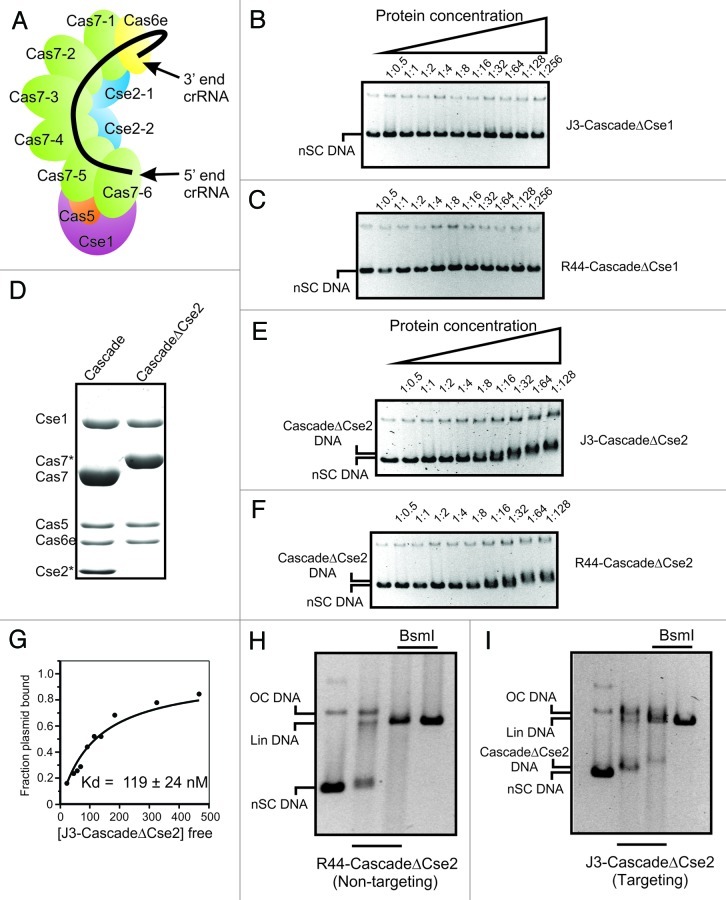
The prime task of the Cascade surveillance complex is to locate protospacer sequences in alien DNA and to form an R-loop at this target site through base pairing between the crRNA and the target DNA strand. The complex architecture of Cascade, which has recently been resolved by cryo-EM,6 reflects well the complexity of this task (see Fig. 1A for a schematic model of Cascade). Each of the subunits plays a crucial role in this process, as seen from the lack of in vivo resistance when any single Cascade subunit is absent.2 The function of some of the 5 different protein subunits has been (partially) revealed during recent studies. The Cas6e subunit cleaves the pre-crRNA and binds the 3′ end of crRNA to incorporate it in the Cascade complex.1, 23, 24 The Cas7 subunit plays a structural role,2 forming the backbone of the complex.6 Interestingly, Cas7 appears to make very close contacts with the crRNA and causes disruption of the R-loop to yield 5 helical segments instead of a continuous crRNA-DNA heteroduplex.6 The Cse1 subunit has been shown to be involved in PAM recognition.5 It has been possible to generate Cascade subcomplexes lacking either Cse1 or both Cse1 and Cse2, which enables the effects of the absence of these proteins to be assessed.2 Electrophoretic Mobility Shift Assays using short linear dsDNA of 85 bp show that CascadeΔCse1 loses its nonspecific DNA binding activity,2 in agreement with a role for Cse1 in PAM recognition.5 This result is further corroborated in the context of negatively supercoiled DNA by the finding that J3-CascadeΔCse1 (CascadeΔCse1 loaded with the previously described J3 crRNA)25 does not bind to pUC-λ, a 3 kb negatively supercoiled plasmid containing the J3 protospacer sequence (Fig. 1B). Moreover, CascadeΔCse1 loaded with an unrelated non-targeting R44-crRNA (R44-CascadeΔCse1) also does not show any nonspecific interaction with pUC-λ (Fig. 1C).
Figure 1. Cse1 and Cse2 subunits of Cascade are involved in nonspecific and specific binding to nSC plasmid DNA, respectively. (A) Schematic model of Cascade, indicating the relative position of each of the 11 different Cascade subunits. (B) Gel-shift of nSC plasmid DNA with J3-CascadeΔCse1, containing a targeting (J3) crRNA. pUC-λ was mixed with 2-fold increasing amounts of J3-CascadeΔCse1, from a pUC-λ: Cascade molar ratio of 1: 0.5 up to a 1: 256. The first lane contains only pUC-λ. (C) Gel-shift as in (B) with R44-CascadeΔCse1 containing a non-targeting (R44) crRNA. (D) SDS PAGE of J3-Cascade (lane 1) and J3-CascadeΔCse2 (lane 2). Asterisks indicate proteins containing an N-terminal StrepII purification tag. (E) Gel-shift as in (B) with J3-CascadeΔCse2, with a pUC-λ: Cascade molar ratio of 1: 0.5 up to a 1: 128 (F) Gel-shift as in (E) with R44-CascadeΔCse2 containing a non-targeting (R44) crRNA. (G) Kd determination of J3-CascadeΔCse2 DNA binding using a y = x/(a+x) fit with y = fraction bound plasmid, x = free Cascade concentration and a = Kd (H) Specific binding of R44-CascadeΔCse2 to the protospacer monitored by BsmI footprinting at a pUC-λ: CascadeΔCse2 molar ratio of 32:1. Lane 1 contains only pUC- λ. Lane 2 contain pUC- λ mixed with CascadeΔCse2. Lane 3 contains pUC- λ mixed with CascadeΔCse2 and subsequent BsmI addition. Lane 4 contains pUC- λ mixed with BsmI. Lin indicates linear plasmid. OC indicates plasmids with a relaxed open circular topology. (I) Specific binding of J3-CascadeΔCse2 to the protospacer monitored as in (H).
The Cse2 subunit has basic patches and may be involved in Cascade nucleic acid contacts.26 Based on the cryo-EM structure, such interactions probably take place in the non-seed area of the target DNA.6 To understand the role of the Cse2 subunit in more detail, we made a subcomplex of Cascade lacking only the Cse2 subunit. This subcomplex has the same apparent stoichiometry as the entire Cascade, as estimated by SDS-PAGE (Fig. 1D). Interestingly, J3-CascadeΔCse2 is able to bind the pUC-λ target plasmid (Fig. 1E), albeit with an almost 10-fold lower affinity (Kd = 119 ± 24 nM) than J3-Cascade (Kd = 13 ± 1.4 nM) (Fig. 1G and ref. 4). In addition, R44-CascadeΔCse2 is also able to interact nonspecifically with negatively supercoiled plasmid DNA (Fig. 1F) with roughly the same affinity (203 ± 36 nM) as Cascade (429 ± 152 nM), indicating that the nonspecific interaction is not affected by the absence of Cse2 and hence may be primarily mediated by the Cse1 subunit. The lowered specific binding affinity of CascadeΔCse2 strongly suggests that Cse2 plays an important role during R-loop formation. The previously described basic patches on the Cse2 surface,26 together with the position of Cse2 near the non-seed region of the crRNA,6 make it tempting to speculate that Cse2 plays a role in either stabilizing the base pairing between the crRNA and the target DNA strand in the non-seed region, or in stabilizing or positioning the displaced DNA strand, or in both. As reported before, nearly half of the energy for strand separation is derived from the negatively supercoiled topology of the target DNA.4 The other half of the energy may be derived from the base pairing between crRNA and the target DNA and from stabilizing interactions of Cascade components with the R-loop, such as Cse2-mediated stabilization of the displaced strand or the non-seed base pairing region. The BsmI site, which is located within the J3 protospacer,4 is, as expected, not protected by R44-CascadeΔCse2 after binding to pUC-λ (Fig. 1H). Intriguingly, and in contrast to J3-Cascade,4 J3-CascadeΔCse2 also does not protect the BsmI site (Fig. 1I). This further suggests that Cse2 plays an important role in stabilization of the R-loop structure.
After binding of Cascade to negatively supercoiled targets, Cascade is predominantly located at the apex of a supercoiled loop, as demonstrated by scanning force microscopy.4 This indicates that Cascade introduces strong bending or possibly wrapping of the target DNA. To analyze this in more detail, J3-Cascade binding to pUC-λ was followed by the addition of a probe complementary to the displaced strand, which serves to stabilize the R-loop (as described before in ref. 4). Unlike the previous study in which we investigated the structure of Cascade bound to supercoiled plasmid DNA using scanning force microscopy, we now linearized these complexes using NdeI (which cleaves ~400 bp upstream of the protospacer) before visualization by scanning force microscopy (as described in ref. 4). The linearization of the Cascade-bound plasmid DNA facilitates reliable measurements of the length of the DNA and the angles in the DNA at the site where Cascade is bound, providing insight in the degree of bending and/or wrapping by Cascade.
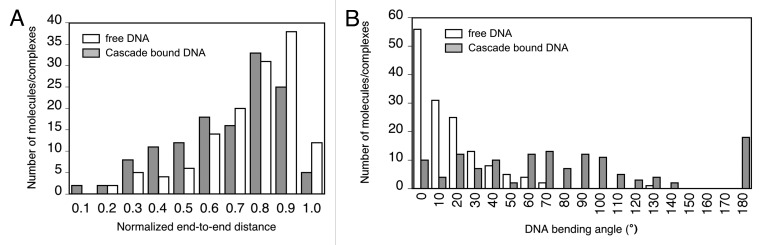
A total number of 136 Cascade-DNA complexes and 185 bare DNA molecules were imaged (Fig. 2). In the Cascade-DNA complexes Cascade was consistently bound at the same position: 123 ± 9 nm (or 367 ± 27 bp) from one of the DNA extremities, indicating specific binding (the center of the protospacer is at a distance of 403 bp (or 134 nm) from the extremity). Extensive wrapping of DNA around Cascade would reduce the contour length of Cascade-DNA complexes compared with that of bare DNA molecules. Similar measurements have previously revealed DNA wrapping by E. coli RNA polymerase and UvrB proteins.27, 28 We measured the contour length of both bare DNA molecules (960 ± 24 nm) and Cascade-DNA complexes (957 ± 26 nm). As these values are not significantly different, these experiments yield no direct evidence for extensive wrapping of DNA around Cascade. Subsequently, we investigated the bending at the position of the bound Cascade using two complementary approaches. First, we used an approach that relies on measuring changes in distance between two defined points on a DNA molecule (usually the two extremities).29 This distance (generally referred to as the end-to-end distance, EED) is a measure of DNA bending. As the linear DNA molecules used in these studies are relatively long (about 3 kb), we adapted this approach to measure the distance between the extremity closest to the bound Cascade and a second point on the other DNA arm at the same distance from the bound protein. The EED distribution of Cascade-DNA complexes is distinctly different from that of bare DNA molecules: it is shifted toward shorter distances, indicating that Cascade bends DNA (Fig. 3A). Next, we directly measured (as described in refs. 29 and 30) the Cascade-induced bend and compared the bending angle distribution of Cascade-DNA complexes with that of bare DNA (Fig. 3B). As expected, the bare DNA has a bending angle distribution centered around 0°. The bending angle distribution of Cascade–DNA complexes deviates in width and in position from the distribution found for bare DNA. It exhibits no preferential angle and the broad distribution of bending angles found suggests that the flexibility of Cascade-DNA complexes is significantly higher than that of bare double stranded DNA. Similar observations have been reported for HU and HMG-box proteins, which bend DNA and enhance its flexibility.30,31 We attribute the enhanced flexibility in the Cascade-DNA complexes to helix unwinding or DNA melting at the DNA ‘entry/exit’ points into Cascade. This explanation is in agreement with a nuclease P1 footprint on Cascade-bound double stranded target DNA, which shows that ~5 nucleotides located at the 3′ end of the PAM are highly sensitive to cleavage by this single stranded DNA specific endonuclease.2
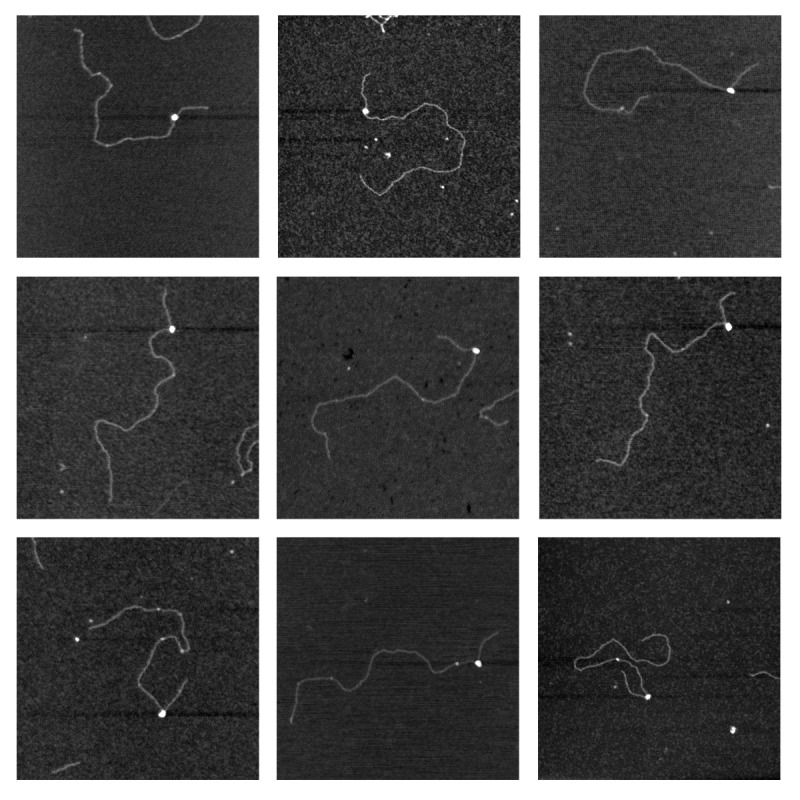
Figure 2. Cascade is bound specifically at the position of the protospacer. Scanning force microscopy images of pUC- λ (linearized by digestion with NdeI) with J3-Cascade containing a targeting (J3) crRNA. pUC-λ was mixed with J3-Cascade at a pUC-λ:Cascade ratio of 1:2. Each image shows a 750 × 750 nm surface area. White dots along the DNA correspond to Cascade.
Figure 3. Cascade bound specifically at the position of the protospacer bends DNA. (A) End-to-end distributions (normalized to DNA contour length) of bare DNA molecules (white) and J3-Cascade-DNA complexes (gray). The reduced end-to-end distance for Cascade-DNA complexes indicates DNA bending. (B) Bending angle distributions of bare DNA molecules (white) and J3-Cascade-DNA complexes (gray). The broad distribution of bending angles suggests enhanced DNA flexibility at the position of the bound Cascade.
Altogether, the data presented here corroborate, in the context of negatively supercoiled DNA, that Cse1 is required for nonspecific binding activity of Cascade, in line with its recently described role in PAM recognition.5 In addition, Cse2 appears to be important for R-loop stabilization, possibly by interacting with the displaced DNA strand of the double stranded target, or by interacting with the non-seed region of the crRNA-DNA heteroduplex, or both. Finally, we show that Cascade induces flexibility in the DNA target, most likely due to unwinding of the double stranded DNA adjacent to the base pairing region. This enhanced flexibility explains why plasmid-bound Cascade is predominantly found at the apex of a supercoiled loop,4 since it facilitates the strong bending of the DNA at this position.
Acknowledgments
This work was financially supported by an NWO-TOP grant to J.O. (854.10.003), an NWO Veni grant to S.J.J.B. (863.08.014) and an NWO Vidi grant to R.T.D. (864.08.001). E.R.W. was financially supported by the NWO Spinoza award to Willem M. de Vos.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/21410
References
- 1.Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–36. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 3.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A. 2011;108:10098–103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westra ER, van Erp PB, Künne T, Wong SP, Staals RH, Seegers CL, et al. CRISPR Immunity Relies on the Consecutive Binding and Degradation of Negatively Supercoiled Invader DNA by Cascade and Cas3. Mol Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell. 2012;46:606–15. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJ, van der Oost J, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–9. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 2011;45:273–97. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 8.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–8. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 9.Al-Attar S, Westra ER, van der Oost J, Brouns SJ. Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol Chem. 2011;392:277–89. doi: 10.1515/bc.2011.042. [DOI] [PubMed] [Google Scholar]
- 10.Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475–93. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 11.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–70. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 12.Jore MM, Brouns SJ, van der Oost J. RNA in defense: CRISPRs protect prokaryotes against mobile genetic elements. Cold Spring Harb Perspect Biol. 2012;4:1–12. doi: 10.1101/cshperspect.a003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karginov FV, Hannon GJ. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol Cell. 2010;37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–27. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 15.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–90. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol. 2011;14:321–7. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009;34:401–7. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–77. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swarts DC, Mosterd C, van Passel MW, Brouns SJ. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–76. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mojica FJM, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–40. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 23.Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol. 2011;18:680–7. doi: 10.1038/nsmb.2043. [DOI] [PubMed] [Google Scholar]
- 24.Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat Struct Mol Biol. 2011;18:688–92. doi: 10.1038/nsmb.2042. [DOI] [PubMed] [Google Scholar]
- 25.Westra ER, Pul U, Heidrich N, Jore MM, Lundgren M, Stratmann T, et al. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol Microbiol. 2010;77:1380–93. doi: 10.1111/j.1365-2958.2010.07315.x. [DOI] [PubMed] [Google Scholar]
- 26.Agari Y, Yokoyama S, Kuramitsu S, Shinkai A. X-ray crystal structure of a CRISPR-associated protein, Cse2, from Thermus thermophilus HB8. Proteins. 2008;73:1063–7. doi: 10.1002/prot.22224. [DOI] [PubMed] [Google Scholar]
- 27.Verhoeven EE, Wyman C, Moolenaar GF, Hoeijmakers JH, Goosen N. Architecture of nucleotide excision repair complexes: DNA is wrapped by UvrB before and after damage recognition. EMBO J. 2001;20:601–11. doi: 10.1093/emboj/20.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dame RT, Wyman C, Wurm R, Wagner R, Goosen N. Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1. J Biol Chem. 2002;277:2146–50. doi: 10.1074/jbc.C100603200. [DOI] [PubMed] [Google Scholar]
- 29.Dame RT, van Mameren J, Luijsterburg MS, Mysiak ME, Janićijević A, Pazdzior G, et al. Analysis of scanning force microscopy images of protein-induced DNA bending using simulations. Nucleic Acids Res. 2005;33:e68. doi: 10.1093/nar/gni073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT. Dual architectural roles of HU: formation of flexible hinges and rigid filaments. Proc Natl Acad Sci U S A. 2004;101:6969–74. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, McCauley MJ, Maher LJ, 3rd, Williams MC, Israeloff NE. Mechanism of DNA flexibility enhancement by HMGB proteins. Nucleic Acids Res. 2009;37:1107–14. doi: 10.1093/nar/gkn1011. [DOI] [PMC free article] [PubMed] [Google Scholar]