Abstract
The mitochondrial genome of metazoan animal typically encodes 22 tRNAs. Nematode mt-tRNAs normally lack the T-stem and instead feature a replacement loop. In the class Enoplea, putative mt-tRNAs that are even further reduced have been predicted to lack both the T- and the D-arm. Here we investigate these tRNA candidates in detail. Three lines of computational evidence support that they are indeed minimal functional mt-tRNAs: (1) the high level of conservation of both sequence and secondary structure, (2) the perfect preservation of the anticodons, and (3) the persistence of these sequence elements throughout several genome rearrangements that place them between different flanking genes.
Keywords: D-loop, Enoplea, Mermithidae, T-loop, mitochondrial tRNA
Introduction
TRNAs (tRNAs) are present in all types of cells and are nearly ubiquitously encoded in organelle genomes. Animal mitogenomes usually contain 22 mt-tRNA genes.1 In contrast to their nuclear counterparts, mt-tRNA not only evolve rapidly at sequence level but also exhibit a variety of deviations from the common clover-leaf structure.2-4 An example of a complete, canonical cloverleaf structure is given in Figure 1. “Bizarre” mt-tRNAs have long been known in particular in the Nematoda,5 where they appear to be the rule rather than an exception, see Figure 1 for a typical structure. Throughout the Chromadorea (one of the two major nematode clades) 20 of the 22 mt-tRNA are T-armless, and the two serine tRNAs lack the D-arm.6-8 It was shown early-on that these tRNAs with deletions are functional.9 The evolution of the deletions inmt-tRNAs is intimately linked to that of the EF-Tu protein.10,11 Following the duplication of the EF-Tu gene early in ecdysozoan evolution its paralogs acquired differential binding abilities to tRNAs with deleted domains.
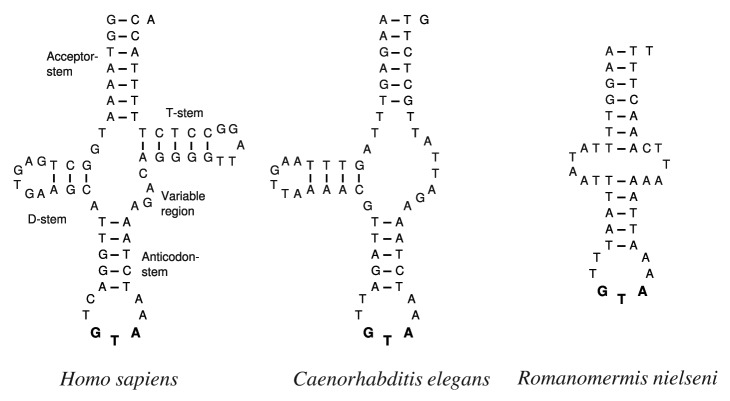
Figure 1. Human mitochondrial tRNATyr has a canonical clover-leaf structure forming four stems. Its ortholog in Caenorhabditis elegans lacks the T-arm. In Romanomermis nielseni both arms are missing. The anticodons are highlighted with bold characters.
Besides the well studied Chromadorea, the phylum Nematoda includes the Enoplea with, among others, the orders Dorylaimida, Mermithida, and Trichocephalida.12 Within these three, much less well-studied orders, the situation is more heterogeneous than in Chromadorea. In Trichinella (Trichocephalida) there are still eight tRNAs with a canonical clover-leaf structure, 12 already lack the T-arm, and the two serine tRNAs are D-armless as usual.13,14 The tRNA complement of Thaumamermis cosgrovei (Mermithida) consists of 24 tRNAs (two of which are recent duplicates) with structures matching those in the Chromadorea.15 Mitochondrial genome structure can vary dramatically within Enoplea. Romanomermis culicivorax (Mermithida) has one of the largest metazoan mitochondrial genomes(26,000 bp) featuring long repeats.16 The mitogenome of Xiphinema americanum (Dorylaimida), on the other hand, is among the smallest in Metazoa. Most genes, including the tRNAs, are somewhat smaller than usual and several gene overlaps have been observed.17 The genome appears to lack tRNACys, tRNAAsn, and tRNASer(UGA).17 Surprisingly, three of the 19 known mt-tRNAs are D-armless. An annotation of the canonical set of 22 tRNAs in Romanomermis iyengari reports tRNAAla and tRNACys as superimposed reverse complements at the same genomic location.18
In a recent survey of animal mt-tRNAs19 we detected a large number of reduced tRNAs. While the majority conforms to the well-described models of T-armless or D-armless structures, we also found several candidates for minimal tRNA structures that simultaneously lack the D- and T-stems. In particular, we recovered candidates for all 22 mt-tRNAs in Xiphinema americanum without the need to postulate nearly completely overlapping tRNAs on opposite strands, as currently annotated in several Enoplea genomes.18 Furthermore, for several tRNA families, including tRNAAla, tRNAHis, and tRNATyr, only extremely truncated sequences comprising acceptor- and anticodon stems were detectable for Mermithida. The high level of conservation of the acceptor stem, which defines the 3′/5′ ends of tRNA genes, suggested that these tRNA candidates are indeed correct annotations.19
In the present contribution we investigate these minimal tRNA candidates in more detail and provide compelling computational evidence that they are indeed functional tRNAs. To this end we constructed nematode-specific covariance models (CMs),20 used these to complete the mt-tRNA annotation, and proceeded to studying their evolution throughout the Enoplea.
Results
All Chromadorea tRNA sequences show the same structural patterns. With the exception of the two D-armless tRNASer1 and tRNASer2 they all lack the T-arm. Since each mitochondrial tRNA of the complete complement has lost exactly one of its arm, all 22 mt-tRNAs feature a replacement loop instead. These observations confirm previous reports.11
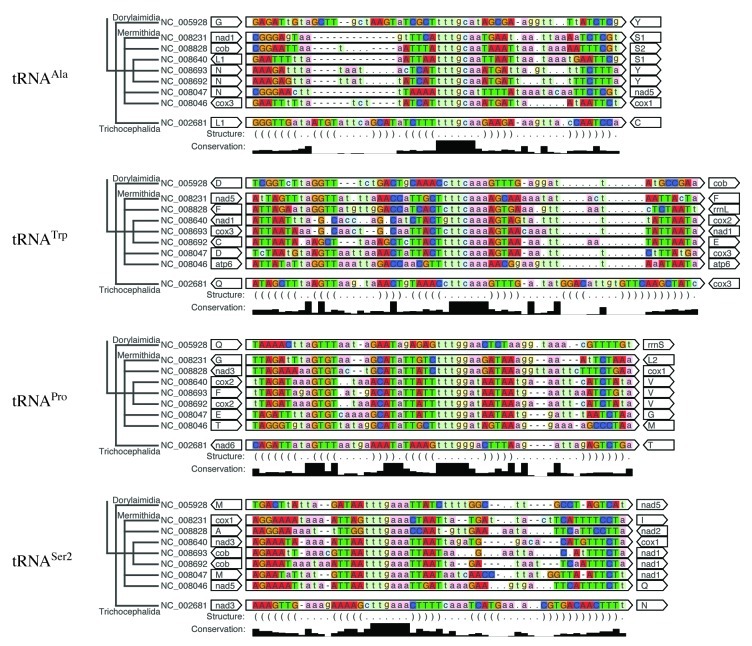
Our primary aim was to annotate the Enoplea mitochondrial tRNA genes as completely as possible. As mentioned previously19 this is a difficult task. A complete tRNA complement was found only for Trichinella spiralis (Trichocephalida), while several mt-tRNA remained missing in Xiphinema americanum (Dorylaimida). In this initial search a given tRNA family was either consistently found or consistently missed in the available Mermithida genomes. In addition, we had identified several weak candidate sequences that lack both arms, among them tRNACys of Thaumamermis cosgrovei, tRNAPhe of Romanomermis culicivorax and tRNAArg of Hexamermis agrotis. Using the nematode-specific models, however, we found that these truncated structures appear systematically in Mermithida. The iterative inclusion of newly identified armless mt-tRNAs eventually lead to a complete annotation of mt-tRNAs in Enoplea, summarized in Table 2. All Enoplea with a sequenced mitogenome appear to have a (nearly) complete complement of mt-tRNAs. All retrieved Dorylaimida and Mermithida tRNAs appear as abnormal, missing either the D-arm (9%), the T-arm (56%), or both (35%). Figure 2 shows alignments for selected examples (see Fig. S1 for the complete set of sequences). Intact clover-leaf structures were described for Trichinella spiralis.13 Since we used the T-armless nematode CM model for homology search, the initial alignments required some manual editing to recover the published clover-leaf structures.
Table 2. Secondary structures of predicted mt-tRNAs in Enoplea.
| Organism | A | C | D | E | F | G | H | I | K | L1 | L2 | M | N | P | Q | R | S1 | S2 | T | V | W | Y |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Dorylaimida |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Xiphinema americanum |
⊣ |
| |
⊣ |
⊣ |
⊣ |
⊣ |
⊣ |
| |
⊣ |
⊣ |
⊣ |
⊣ |
⊣ |
⊣ |
⊣ |
⊣ |
⊢ |
⊢ |
⊣ |
⊣ |
⊣ |
| |
|
Mermithida |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Agamermis sp BH-2006 |
| |
⊣ |
⊣ |
⊣ |
| |
⊣ |
| |
| |
⊣ |
⊣ |
⊣ |
⊣ |
| |
⊣ |
⊣ |
| |
⊢ |
⊢ |
⊣ |
| |
⊣ |
| |
|
Hexamermis agrotis |
| |
| |
⊣ |
⊣ |
| |
⊣ |
| |
| |
⊣ |
⊣ |
⊣ |
⊣ |
| |
⊣ |
⊣ |
| |
⊢ |
⊢ |
⊣ |
⊣ |
⊣ |
| |
|
Romanomermis culicivorax |
| |
| |
⊣ |
⊣ |
| |
⊣ |
| |
| |
⊣ |
⊣ |
⊣ |
⊣ |
| |
⊣ |
⊣ |
| |
⊢ |
⊢ |
| |
⊣ |
⊣ |
| |
|
Romanomermis iyengari |
| |
| |
⊣ |
⊣ |
| |
⊣ |
| |
| |
⊣ |
⊣ |
⊣ |
⊣ |
| |
⊣ |
⊣ |
| |
⊢ |
⊢ |
| |
⊣ |
⊣ |
| |
|
Romanomermis nielseni |
| |
| |
⊣ |
⊣ |
| |
⊣ |
| |
| |
⊣ |
⊣ |
⊣ |
⊣ |
| |
⊣ |
⊣ |
| |
⊢ |
⊢ |
| |
⊣ |
⊣ |
| |
|
Strelkovimermis spiculatus |
| |
| |
⊣ |
⊣ |
| |
⊣ |
| |
| |
⊣ |
⊣ |
⊣ |
⊣ |
| |
⊣ |
⊣ |
| |
⊢ |
⊢ |
⊣ |
⊣ |
⊣ |
| |
|
Thaumamermis cosgrovei |
| |
| |
⊣ |
⊣ |
| |
⊣ |
| |
| |
⊣ |
⊣ |
⊣ |
⊣ |
| |
⊣ |
⊣ |
⊣ |
⊢ |
⊢ |
⊣ |
| |
⊣ |
| |
|
Trichocephalida |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Trichinella spiralis | ⊣ | ⊣ | + | ⊣ | ⊣ | ⊣ | ⊣ | + | + | + | + | + | ⊣ | ⊣ | ⊣ | + | ⊢ | ⊢ | ⊣ | ⊣ | + | | |
The first row enumerates all 20 amino acids. As mitochondrial genomes encode two distinct tRNALeu and tRNASer genes, both are listed twice as L1/L2 and S1/S2, respectively. Typical nematode tRNAs with a D-arm but no T-arm are indicated by (⊣). The two serine tRNAs have retained their T-arm and lack the D-arm (⊢). Structures lacking both the T-arm and the D-arm are denoted by (|). The intact clover-leaf structures in Trichinella spiralis are shown as (+).
Figure 2. Alignments and genomic context of Enoplea mt-tRNAs. The square of the ClustalX conservation is shown to enhance the contrast between conserved and non-conserved parts of the sequences. tRNAAla lost the D-stem in all Mermithida while the other two stems are highly conserved. In contrast, the D-stem of tRNATrp is preserved in most species, but it is reduced already in two Romanomermis sequences (NC_008640 and NC_008693). The situation is similar for the T-stem of tRNASer2, again in Romanomermis. In contrast, tRNAPro has maintained a highly conserved D-stems. The base pairs of the (partially) retained T-stem in Trichinella spiralis mt-tRNA are not shown explicitly here. Base pairs are highlighted by saturated colors and capital letters, loop regions are shown in light colors and small letters; colors emphasize the sequence.
The only mitogenome among the Enoplea that may not have a complete complement of mt-tRNAs is that of Hexamermis agrotis. Ofthe two (weak) candidates for the tRNATyr, one overlaps the annotated Cox2 protein, the other one is located anti-sense to tRNAVal. Similar anti-sense arrangements recently have been proposed as a rather general phenomenon.18 For all previously proposed examples of this type, however, we found credible, alternative candidates in “empty” parts of the respective mitogenomes. Nevertheless, some of the annotated tRNA candidates have moderate overlaps with adjacent genes. This phenomenon is not uncommon in metazoan mitogenomes.26 The largest overlap among tRNA genes was found between tRNALeu1 and tRNAHis of Thaumamermis cosgrovei with 14 nt (Fig. S2). In all other cases the predicted overlap of a tRNA with another tRNA or an adjacent protein-coding gene is less than 10 nt. Larger overlaps were found with rRNA genes in Agamermis sp BH-2006, Romanomermis culicivorax, Romanomermis nielseni, and Thaumamermis cosgrovei. These can probably be attributed to the notorious difficulties of determining the exact boundaries of mitochondrial rRNA genes.21
Many of the predicted tRNAs conform to the expected structure with an intact D-stem. Structural features are largely conserved within each of these tRNA families. If a D-stem is present, then the stem region is highly conserved throughout the Enoplea. These tRNAs were, with a few exceptions, already detected unambiguously in the first iteration of the search with the CMs built from the Chromadorea sequences.
We find that tRNAs lacking both D- and T-arms have evolved in ten mt-tRNA families: tRNAAla, tRNACys, tRNAPhe, tRNAHis, tRNAIle, tRNAAsn, tRNAArg, tRNAThr, tRNAVal, and tRNATyr. As an example, Figure 1 compares the armless tRNATyr of Romanomermis nielseni with its T-armless counterparts in Caenorhabditis elegans and the canonical clover-leaf structure in Homo sapiens. The tRNA complement of all armless mitochondrial tRNAs of a typical mermithid (Romanomermis nielseni) are presented in Figure. S3. At least the three Romanomermis species lost the D-stem in all these mt-tRNAs except tRNAVal. While Agamermis sp BH-2006 and Xiphinema americanum have only a few D-armless mt-tRNAs, the other Mermithida share mostly the Romanomermis pattern.
Several lines of computational evidence can be invoked to argue that the minimal mt-tRNA candidates are indeed functional tRNAs. The first arguments concerns the high level of sequence conservation. From a comparison of 12S rRNA we infer that the evolutionary distances within Enoplea are comparable to those within vertebrates. The divergence between the Romanomermis species, for instance, matches that of the major gnathostome lineages, and the distance between Trichocephalida and Mermithida is nearly the same as between mammals and Enoplea. Since DNA sequences become unalignable in the absence of stabilizing selection already between mammalian orders,27 we can safely conclude that the sequences of all our mt-tRNA candidates are under very strong stabilizing selection. Furthermore, a comparison of the evolutionary distances of the acceptor stem sequences between the tRNAs that lack only the T-arm and the armless candidates does not reveal a substantial acceleration in the armless group (Fig. 3).This indicates that the selective constraints are comparable between typical nematode mt-tRNAs and the newly identified armless candidates. In addition, the sequence of the entire anticodon loop, and in particular the anticodon itself is nearly absolutely conserved throughout the Enoplea for all 22 mt-tRNA families.
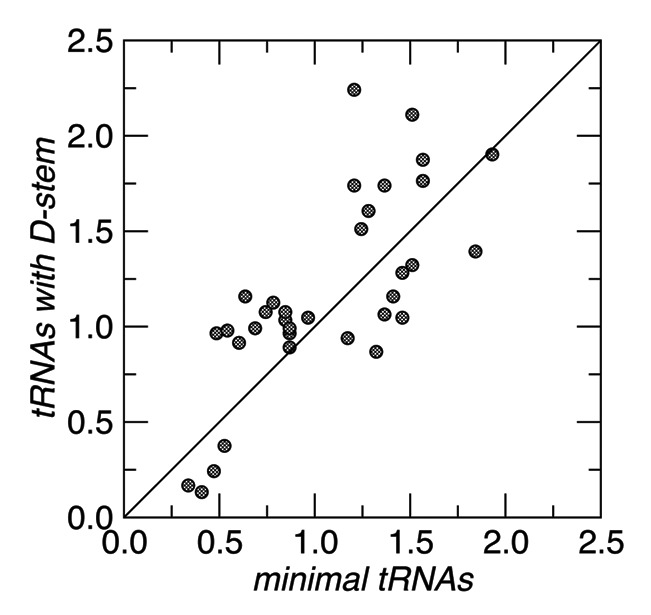
Figure 3. Comparison of the Jukes-Cantor distances between Enoplea species estimated from the acceptor stems of the mt-tRNAs with and without D- and T-stems, respectively. The two tRNASer families are not included.
Second, both the acceptor stem and the anticodon stem appear as nearly perfectly conserved secondary structure elements. A large number of compensatory point mutation as well as compensation of the structure by “moving” the entire stem by one nucleotide show that not only the sequence but also the secondary structure is under strong stabilizing selection.
A third, independent line of evidence is provided by the frequent rearrangements of the mitogenomic gene order within the Enoplea. We observe that the genomic context of a particular tRNA changes frequently even within the Mermithida. For instance tRNAAla is found between eight different pairs of neighbors, see Figure 2. The different neighbors on both sides show that the mitochondrial gene order was broken multiple times between tRNAAla and its neighbors. We can conclude, therefore, that the armless tRNAAla is propagated as a unit. The same argument can be made for all other mt-tRNA genes, independent of whether they are armless, have a D-arm, or a T-arm only (see Fig. S1).
Discussion
Mitochondrial tRNAs in Nematodes exhibit well-known special features, in particular the almost ubiquitous loss of either the T-arm or the D-arm. A careful re-annotation of the nematode mitogenomes with iteratively improved nematode-specific CMs showed that all nematode mitogenomes sequenced to-date harbor the complete complement of the 22 mt-RNA families typical for animal mitochondria. Losses of tRNAs suggested in the literature are the consequence of insufficient sensitivity of the homology search methods employed in previous annotation efforts. This is explained by the “bizarre” structure of many of the mt-tRNAs in particular in two of the three major clades of the Enoplea. Both Mermithida and Dorylaimida feature tRNAs that have simultaneously lost both their T-arm and their D-arm. Several lines of computational evidence converge to the conclusion that the minimal, armless mt-tRNAs are indeed functional: (1) the strong conservation of sequence and in particular of the anticodon, (2) the large number of compensatory substitutions in the secondary structure, and (3) the undisturbed integrity of sequence elements through multiple genome rearrangements. As we found armless tRNAs only in Enoplea but not in other nematodes it is likely that their appearance represents an independent evolutionary event within this clade.
It remains an open question how these minimal mt-tRNAs function. Although we do not have experimental support for the minimal worm mt-tRNAs, it has been shown previously that artificial sequences without D- and T-arms are recognized by synthetases and are aminoacylated.28,29 Since the evolution of the reduced nematode mt-tRNAs is intimately related to the two paralogous elongation factors EF-Tu1 and EF-Tu2,11 it is tempting to hypothesize that the further reduction of the mt-tRNA structures in Dorylaimida and Mermithida are compensated by corresponding changes in the EF-Tu proteins. In the same vein, it would also be of interest to analyze the corresponding aminoacyl tRNA synthetases for structural adaptations to armless tRNAs. At present, however, no pertinent nuclear sequence data are available for these clades that would allows us to pursue this topic. However, this situation will soon change when the genome of Romanomermis culicivorax becomes available.
The presence of minimal, armless mt-tRNAs in Enoplea begs the question whether other animal groups, in which only incomplete complements of mt-tRNAs have been found, might also harbor yet unidentified structurally reduced or otherwise aberrant tRNAs. This concerns some arthropod mitogenomes, in particular Cecidomyiidae30 and Arachnida.31 The Onychophoran Opisthopatus cinctipes32 and the Chaetognatha33 (depending on their actual phylogenetic position) might also share the EF-Tu duplication that is common to the arthropods and nematodes.11 Independent types of “bizarre” mt-tRNAs might be present in the Rotifera,19,34 a small lophotrochozoan phylum. The pseudogenization and loss of tRNALys in marsupials, on the other hand, is compensated by the import of the corresponding nuclear tRNA.35 This mechanism is also at work in many diploblasts with reduced mt-tRNA complement.36,37 The minimal mt-tRNAs in the Enoplea, finally, might also shed new light on the minimal requirements for tRNA function, in particular on the specificity of recognition by aminoacyl-tRNA synthetase.
Materials and Methods
We analyzed 32 nematode mitochondrial genomes derived from the RefSeq database.12 This compilation includes genomes of 9 Enoplea species listed in Table 1. For comparison, the 23 Chromadorea genomes listed in the Supplemental Material were analyzed.
Table 1. Enoplea mitogenomes used in this study.
| Taxonomy | Organism | Accession |
|---|---|---|
| Dorylaimida |
Xiphinema americanum |
NC_005928 |
| Mermithida |
Agamermis sp BH-2006 |
NC_008231 |
| |
Hexamermis agrotis |
NC_008828 |
| |
Romanomermis culicivorax |
NC_008640 |
| |
Romanomermis iyengari |
NC_008693 |
| |
Romanomermis nielseni |
NC_008692 |
| |
Strelkovimermis spiculatus |
NC_008047 |
| |
Thaumamermis cosgrovei |
NC_008046 |
| Trichocephalida | Trichinella spiralis | NC_002681 |
All genomes were re-annotated using the Mitos pipeline.21 In brief, proteins are annotated with blast using a large library of queries, rRNA (rRNA) annotation uses a general metazoan covariance models (CM) for 12S and 18S rRNA, resp. The pipeline also invokes the MiTFi script to identify tRNA genes.19 It uses rules to ensure the consistency of the annotation and accounts for inaccuracies in the predicted gene boundaries. We identified nearly all mt-tRNAs in Chromadorea using the default Metazoa-specific CMs used in the MiTFi script. Exceptions occurred in Spirurida and Tylenchida. Here we predicted only 85% and 50% genes of the complete set. All the tRNAs genes missed by the automatic pipeline were then identified by homology searches using infernal.20
The complete set of Chromadorea mt-tRNAs was then used to construct a nematode-specific CM for each of the 22 families with cmbuild from the infernal 1.0 package.20 These CMs were used to re-annotate the Enoplea mitogenomes listed in Table 1 with cmsearch in semi-global alignment mode. In addition, we used the MiTFi pipeline to obtain tRNA candidates. Newly identified tRNA genes were added to the alignments after manual inspection, which were then used to compute revised CMs. Alignments of mt-tRNAs were edited using the emacs editor using the ralee mode.22 After several iterations we obtained a stable complete set of 22 mt-tRNA candidates for each of the Enoplea species.
Evolutionary distances among the Enoplea, and for comparison among a collection of vertebrates, were estimated from 12S rRNAs as annotated by the Mitos pipeline. Alignments were constructed with infernal using the CMs included in the Mitos system. They were manually inspected and poorly aligned parts at the ends were removed. Pairwise distances were computed with DNADIST.23,24 In order to estimate evolutionary distances between Enoplea mt-tRNAs we used the concatenated sequence information of the acceptor stems. The 10 tRNA families that lack both loops and the 10 families with a T-loop only were considered separately. The two serine tRNAs were excluded because of their exceptional structure.
Figures of alignments and conservation scores were constructed by ClustalX.25 The NCBI taxonomy was used throughout.
Supplementary Material
Acknowledgments
This work was funded in part by the Centre National de la Recherche Scientifique (CNRS), Université de Strasbourg, Association Française contre les Myopathies (MNM1 2009), ANR MITOMOT [ANR-09-BLAN-0091–01]; Deutsche Forschungsgemeinschaft [SPP-1174 (“Deep Metazoan Phylogeny”) project STA 850/3–2 and STA 850/2]; French-German PROCOPE program [DAAD D/0628236, EGIDE PHC 14770PJ]; French-German University (DFH-UFA, Cotutelle de thèse CT-08–10); a doctoral fellowship of the German Academic Exchange Service [DAAD D/10/43622]; and a bridge scholarship of the Collège Doctoral Européen (CDE), Université de Strasbourg.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/rnabiology/21630
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/21630
References
- 1.Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–80. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arcari P, Brownlee GG. The nucleotide sequence of a small (3S) seryl-tRNA (anticodon GCU) from beef heart mitochondria. Nucleic Acids Res. 1980;8:5207–12. doi: 10.1093/nar/8.22.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Bruijn MH, Schreier PH, Eperon IC, Barrell BG, Chen EY, Armstrong PW, et al. A mammalian mitochondrial serine transfer RNA lacking the “dihydrouridine” loop and stem. Nucleic Acids Res. 1980;8:5213–22. doi: 10.1093/nar/8.22.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helm M, Brulé H, Friede D, Giegé R, Pütz D, Florentz C. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA. 2000;6:1356–79. doi: 10.1017/S1355838200001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolstenholme DR, Macfarlane JL, Okimoto R, Clary DO, Wahleithner JA. Bizarre tRNAs inferred from DNA sequences of mitochondrial genomes of nematode worms. Proc Natl Acad Sci U S A. 1987;84:1324–8. doi: 10.1073/pnas.84.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu M, Chilton NB, Gasser RB. The mitochondrial genomes of the human hookworms, Ancylostoma duodenale and Necator americanus (Nematoda: Secernentea) Int J Parasitol. 2002;32:145–58. doi: 10.1016/S0020-7519(01)00316-2. [DOI] [PubMed] [Google Scholar]
- 7.Keddie EM, Higazi T, Unnasch TR. The mitochondrial genome of Onchocerca volvulus: sequence, structure and phylogenetic analysis. Mol Biochem Parasitol. 1998;95:111–27. doi: 10.1016/S0166-6851(98)00102-9. [DOI] [PubMed] [Google Scholar]
- 8.Montiel R, Lucena MA, Medeiros J, Simões N. The complete mitochondrial genome of the entomopathogenic nematode Steinernema carpocapsae: insights into nematode mitochondrial DNA evolution and phylogeny. J Mol Evol. 2006;62:211–25. doi: 10.1007/s00239-005-0072-9. [DOI] [PubMed] [Google Scholar]
- 9.Okimoto R, Wolstenholme DR. A set of tRNAs that lack either the T psi C arm or the dihydrouridine arm: towards a minimal tRNA adaptor. EMBO J. 1990;9:3405–11. doi: 10.1002/j.1460-2075.1990.tb07542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suematsu T, Sato A, Sakurai M, Watanabe K, Ohtsuki T. A unique tRNA recognition mechanism of Caenorhabditis elegans mitochondrial EF-Tu2. Nucleic Acids Res. 2005;33:4683–91. doi: 10.1093/nar/gki784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohtsuki T, Watanabe Y. T-armless tRNAs and elongated elongation factor Tu. IUBMB Life. 2007;59:68–75. doi: 10.1080/15216540701218722. [DOI] [PubMed] [Google Scholar]
- 12.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35(Database issue):D61–5. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lavrov DV, Brown WM. Trichinella spiralis mtDNA: a nematode mitochondrial genome that encodes a putative ATP8 and normally structured tRNAS and has a gene arrangement relatable to those of coelomate metazoans. Genetics. 2001;157:621–37. doi: 10.1093/genetics/157.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arita M, Suematsu T, Osanai A, Inaba T, Kamiya H, Kita K, et al. An evolutionary ‘intermediate state’ of mitochondrial translation systems found in Trichinella species of parasitic nematodes: co-evolution of tRNA and EF-Tu. Nucleic Acids Res. 2006;34:5291–9. doi: 10.1093/nar/gkl526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang S, Hyman BC. Mitochondrial genome haplotype hypervariation within the isopod parasitic nematode Thaumamermis cosgrovei. Genetics. 2007;176:1139–50. doi: 10.1534/genetics.106.069518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azevedo JL, Hyman BC. Molecular characterization of lengthy mitochondrial DNA duplications from the parasitic nematode Romanomermis culicivorax. Genetics. 1993;133:933–42. doi: 10.1093/genetics/133.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He Y, Jones J, Armstrong M, Lamberti F, Moens M. The mitochondrial genome of Xiphinema americanum sensu stricto (Nematoda: Enoplea): considerable economization in the length and structural features of encoded genes. J Mol Evol. 2005;61:819–33. doi: 10.1007/s00239-005-0102-7. [DOI] [PubMed] [Google Scholar]
- 18.Seligmann H. Undetected antisense tRNAs in mitochondrial genomes? Biol Direct. 2010;5:39. doi: 10.1186/1745-6150-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jühling F, Pütz J, Bernt M, Donath A, Middendorf M, Florentz C, et al. Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res. 2012;40:2833–45. doi: 10.1093/nar/gkr1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–7. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernt M, et al. Mitos: Improved de novo metazoan mitochondrial genome annotation. Mol Phylog Evol 2012. In revision; see http://www.bioinf.uni-leipzig.de/Publications/PREPRINTS/09-028.pdf for a preprint. [DOI] [PubMed]
- 22.Griffiths-Jones S. RALEE—RNA alignment editor in Emacs. Bioinformatics. 2005;21:257–9. doi: 10.1093/bioinformatics/bth489. [DOI] [PubMed] [Google Scholar]
- 23.Felsenstein J. PHYLIP – phylogeny inference package (version 3.2) Cladistics. 1989;5:164–6. [Google Scholar]
- 24.Jukes TH, Cantor CR. Evolution of protein molecules. In HN Munro, editor, Mammalian Protein Metabolism Academic Press, New York, 1969, 21–132. [Google Scholar]
- 25.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 26.Reichert A, Rothbauer U, Mörl M. Processing and editing of overlapping tRNAs in human mitochondria. J Biol Chem. 1998;273:31977–84. doi: 10.1074/jbc.273.48.31977. [DOI] [PubMed] [Google Scholar]
- 27.Ogurtsov AY, Sunyaev S, Kondrashov AS. Indel-based evolutionary distance and mouse-human divergence. Genome Res. 2004;14:1610–6. doi: 10.1101/gr.2450504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolfson AD, Khvorova AM, Sauter C, Florentz C, Giegé R. Mimics of yeast tRNAAsp and their recognition by aspartyl-tRNA synthetase. Biochemistry. 1999;38:11926–32. doi: 10.1021/bi9908383. [DOI] [PubMed] [Google Scholar]
- 29.Giegé R, Jühling F, Pütz J, Stadler P, Sauter C, Florentz C. Structure of transfer RNAs: similarity and variability. Wiley Interdiscip Rev RNA. 2012;3:37–61. doi: 10.1002/wrna.103. [DOI] [PubMed] [Google Scholar]
- 30.Beckenbach AT, Joy JB. Evolution of the mitochondrial genomes of gall midges (Diptera: Cecidomyiidae): rearrangement and severe truncation of tRNA genes. Genome Biol Evol. 2009;1:278–87. doi: 10.1093/gbe/evp027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masta SE, Boore JL. The complete mitochondrial genome sequence of the spider Habronattus oregonensis reveals rearranged and extremely truncated tRNAs. Mol Biol Evol. 2004;21:893–902. doi: 10.1093/molbev/msh096. [DOI] [PubMed] [Google Scholar]
- 32.Braband A, Cameron SL, Podsiadlowski L, Daniels SR, Mayer G. The mitochondrial genome of the onychophoran Opisthopatus cinctipes (Peripatopsidae) reflects the ancestral mitochondrial gene arrangement of Panarthropoda and Ecdysozoa. Mol Phylogenet Evol. 2010;57:285–92. doi: 10.1016/j.ympev.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 33.Miyamoto H, Machida RJ, Nishida S. Complete mitochondrial genome sequences of the three pelagic chaetognaths Sagitta nagae, Sagitta decipiens and Sagitta enflata. Comp Biochem Physiol Part D Genomics Proteomics. 2010;5:65–72. doi: 10.1016/j.cbd.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 34.Suga K, Mark Welch DB, Tanaka Y, Sakakura Y, Hagiwara A. Two circular chromosomes of unequal copy number make up the mitochondrial genome of the rotifer Brachionus plicatilis. Mol Biol Evol. 2008;25:1129–37. doi: 10.1093/molbev/msn058. [DOI] [PubMed] [Google Scholar]
- 35.Dörner M, Altmann M, Pääbo S, Mörl M. Evidence for import of a lysyl-tRNA into marsupial mitochondria. Mol Biol Cell. 2001;12:2688–98. doi: 10.1091/mbc.12.9.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haen KM, Pett W, Lavrov DV. Parallel loss of nuclear-encoded mitochondrial aminoacyl-tRNA synthetases and mtDNA-encoded tRNAs in Cnidaria. Mol Biol Evol. 2010;27:2216–9. doi: 10.1093/molbev/msq112. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Lavrov DV. Seventeen new complete mtDNA sequences reveal extensive mitochondrial genome evolution within the Demospongiae. PLoS One. 2008;3:e2723. doi: 10.1371/journal.pone.0002723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.