Abstract
Recent sequencing efforts have described the mutational landscape of the pediatric brain tumor medulloblastoma. Although MLL2 is among the most frequent somatic single nucleotide variants (SNV), the clinical and biological significance of these mutations remains uncharacterized. Through targeted re-sequencing, we identified mutations of MLL2 in 8 % (14/175) of MBs, the majority of which were loss of function. Notably, we also report mutations affecting the MLL2-binding partner KDM6A, in 4 % (7/175) of tumors. While MLL2 mutations were independent of age, gender, histological subtype, M-stage or molecular subgroup, KDM6A mutations were most commonly identified in Group 4 MBs, and were mutually exclusive with MLL2 mutations. Immunohistochemical staining for H3K4me3 and H3K27me3, the chromatin effectors of MLL2 and KDM6A activity, respectively, demonstrated alterations of the histone code in 24 % (53/220) of MBs across all subgroups. Correlating these MLL2-and KDM6A-driven histone marks with prognosis, we identified populations of MB with improved (K4+/K27−) and dismal (K4−/K27−) outcomes, observed primarily within Group 3 and 4 MBs. Group 3 and 4 MBs demonstrate somatic copy number aberrations, and transcriptional profiles that converge on modifiers of H3K27-methylation (EZH2, KDM6A, KDM6B), leading to silencing of PRC2-target genes. As PRC2-mediated aberrant methylation of H3K27 has recently been targeted for therapy in other diseases, it represents an actionable target for a substantial percentage of medulloblastoma patients with aggressive forms of the disease.
Keywords: MLL2, KDM6A, Histone lysine methylation, Medulloblastoma, PRC2
Introduction
Medulloblastoma (MB) is the most common malignant pediatric brain tumor and an emerging model for the integrative use of clinical and molecular features to improve patient stratification [24]. Although 5-year survival rates for standard risk patients exceed 70 %, many survivors suffer from radiotherapy-related neurocognitive side effects, highlighting the need for targeted therapies [12]. Our contemporary understanding of MB has shifted rather strikingly as large cohort studies have effectively dissected MB from a single disease into four disparate molecular subgroups each with specific transcriptional, genomic and clinical features [25]. While the copy number and transcriptional heterogeneity has been well described, characterization of the mutational landscape has only recently emerged [9, 19, 20].
The first genome-wide MB mutational screen, performed by Parsons et al. [17] identified MLL2 as among the most frequent somatic mutation, occurring in 14 % (12/88) of sequenced tumors. While this finding supported initial reports of copy number-driven alterations of the histone code in the pathogenesis of MB [16], the clinical and molecular significance of the novel mutations were not described. Given the strong prognostic association between CTNNB1 mutations and improved patient outcome [5] and the subgroup-dependent survival effect of TP53 mutations [18], we investigated the role of mutations targeting MLL2 and its binding partner KDM6A across a large cohort of MB patients to determine if there was an association with patient outcome.
MLL2 is the catalytic component of trithorax group (trxG) proteins, that methylate histone H3 tails at the fourth lysine position (H3K4me) and creating a transcriptionally permissive chromatin state [22]. KDM6A, also a trxG protein, binds MLL2, but acts as a H3K27-trimethylation (me3) demethylase [22]. TrxG proteins function in concert to activate gene expression, through the combinatorial accumulation of active H3K4me3 residues and the removal of H3K27me3 repressive marks. The actions of TrxG proteins are opposed by Polycomb (PcG) group proteins, composed of two subunits (PRC1 and PRC2), which function antagonistically to repress gene transcription through accumulation of H3K27me3 marks [23].
Recent studies have demonstrated that the progenitor state is maintained through bivalent chromatin domains, a balancing act of inhibitory (H3K27me) and activating (H3K4me) chromatin marks at promoters of cell determinant factors, including many neural genes [23] (Fig. 5a). Differentiation is induced in part through shifts in the chromatin state of promoters, allowing pro-differentiation genes to be expressed [23]. In tumor cells, the chromatin state is hijacked through up-regulation of a number of PcG genes and loss of TrxG remodeling genes, resulting in inappropriate silencing of pro-differentiation genes [23]. These changes, which include MLL2 and KDM6A mutations, identified in numerous cancers [8, 14] suggest that aberrations of the histone code are a common mechanism in the initiation, maintenance or progression of cancer. The development of pharmacological inhibitors of PcG-gene components, including the EZH2 inhibitors 3-Deazane-planocin (DZNep) [1] and GSK126 [13], has demonstrated promising preliminary results inhibiting cancer-induced H3K27me3-repressive marks, and sensitizing cells to apoptotic agents [1]. Given the relatively high frequency of MLL2 mutations in MB, we focused our efforts on characterizing the prognostic use and biological effect of somatic mutations affecting modifiers of the histone code, and global changes in the chromatin state mediated by H3K4me3 and H3K27me3 patterns across a combined total of 463 tumors.
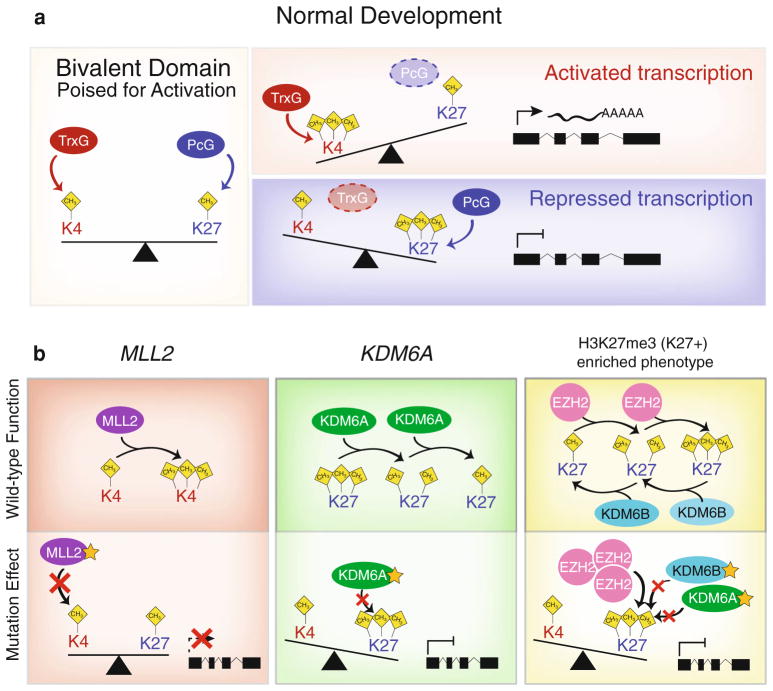
Fig. 5.
Schematic representation of normal developmental Trithorax (trxG) and Polycomb (PcG) group proteins and the effects of MLL2 and KDM6A mutations on chromatin state and transcriptional activation. a Representation of the balancing act of bivalent domains in which Trithorax (trxG) and Polycomb (PcG) group proteins counteract each other to maintain genes poised for activation. Activation of gene expression through a trxG-mediated shift towards H3K4-trimethylation and repressed transcription via PcG-regulated H3K27me3 induce differentiation. b Wild-type and pathogenic roles of MLL2 and KDM6A on chromatin state and transcription. MLL2 functions as H3K4 methyltransferase shunting promoters towards an active state. MLL2 mutations in medulloblastoma largely disrupt the coding sequence through nonsense and frameshift mutations causing premature truncation of the transcript and ablation of the methyltransferase activity preventing the normal activation of gene expression. KDM6A, a H3K27-demethylase, functions as a trxG-protein to inhibit PcG-mediate H3K27me3 marks. KDM6A mutations in medulloblastomas result in the accumulation of H3K27me3, normally removed by the wild-type gene. In Group 3 and 4 medulloblastomas an H3K27me3-enriched phenotype (K27+) is observed, in which overexpression of the H3K27-methyltransferase EZH2 with concomitant down-regulation of both KDM6A and KDM6B demethylases results in a strong shift towards increased H3K27me3
Materials and methods
Patient characteristics
Medulloblastomas derived from 175 (n = 175) patients where sufficient DNA or tissue was available and where robust clinical details were available were included in our sequencing cohort, accurately reflecting the known demographics, molecular subgroups, and histology of this disease. Annotated clinical and histological data for sequenced tumors and TMAs (JHU, n = 50; DKFZ, n = 238) are outlined in Supplementary Table 1.
MLL2 and KDM6A sequencing
MLL2 and KDM6A were sequenced using an exon-capture workflow across 175 tumors which form the discovery sequencing cohort. In brief, capture probes designed against coding sequences were used to pull down sonicated genomic DNA. Barcoding and sequence adaptors were ligated and sequencing was performed using Illumina GAIIX sequencer. Following alignment to the reference genome (Hg18) using BWA alignment software, single nucleotide variants (SNVs) were detected using SNVmix2 (pAB + pBB ≥0.99) and insertions/deletions (INDELs) were detected using SamTools (Consensus quality ≥50). Predicted SNVs were filtered through BC Genome Sciences Centre (GSC) in-house database of normal genomes, which is used to store and process human variation [6]. The database contains variation data from genomes sequenced at BC GSC, the 1,000 Genomes Project, dbSNP132 as well as other publically available genomes. Filtered SNVs were visually inspected using integrative genomic viewer (IGV) and final curated mutations were validated using Sanger sequencing. The validation cohort was composed of four previously published series: Jones et al. [9], Parsons et al. [17], Pugh et al. [19] and Robinson et al. [20] totaling 398 primary medulloblastomas.
Immunohistochemistry
TMAs obtained from the German Cancer Research Centre (DKFZ) and Johns Hopkin’s University (JHU) were stained using antibodies directed towards H3K4me3 (1:1,000 CIT, No. 9751; Cell Signaling Technology, Danvers, MA) H3K27me3 (1:10,000 CIT and PEP 1 min, No. 07-449, Millipore, Billerica, MA) and MLL2 (1:1,000, HPA035977, Sigma Aldrich, St. Louis, MO). Stained TMAs were scored in two semi-quantitative manners (negative, positive or super-negative, weak positive, positive) by two independent reviewers (A.K. and A.M.D.) who were blinded to clinical and molecular patient variables.
Copy number and expression analysis
Single nucleotide polymorphisms (SNP) 100 K and 500 K arrays were processed on a series of 201 primary medulloblastomas as previously described [16]. Similarly, Affymetrix Exon 1.0ST arrays were used to analyze the transcriptome of 103 primary tumors. Array analysis and molecular subgroup assignment were performed as previously described [15]. Both expression sets were previously deposited in GEO under GSE14437 and GSE21140.
Results
MLL2 mutations occur in a subgroup-independent manner
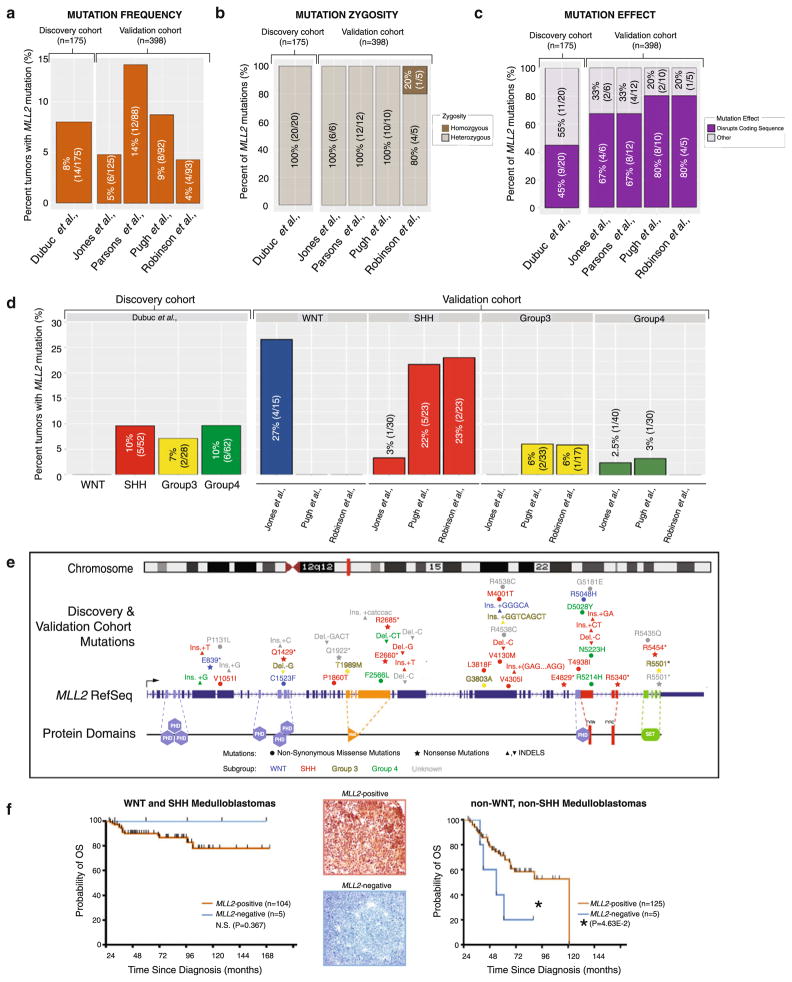
We performed targeted sequencing (Supplementary Fig. S1) on a clinically annotated discovery cohort of 175 MBs, reflecting the spectrum of demographics, molecular subgroups, and histological subtypes of the disease (Supplementary Table S1) [11]. We verified our results using a validation cohort of 398 previously sequenced medulloblastomas from four independent centers [9, 17, 19, 20]. In our discovery series, eight percent (8 %, 14/175) of tumors harbored MLL2 mutations independent of age or histological subtypes (Fig. 1a; Supplementary Table S2). No homozygous mutations were identified (Fig. 1b, Supplementary Fig. S2a) and 45 % (9/20) of those mutations detected disrupted the coding sequence through nonsense and frameshift mutations (Fig. 1c; Supplementary Table S2). While the mutation frequency (Fig. 1a) and zygosity (Fig. 1b) across our validation cohort supported our discovery cohort, there was a 22–35 % increase in the frequency of damaging mutations (Fig. 1c). MLL2 mutations occurred in a similar frequency across non-WNT subgroups in our discovery cohort (Fig. 1d); however, there was considerable variability in the molecular distribution across the validation cohort, with an increased incidence of MLL2 mutations in WNT tumors reported by Jones et al. [9], whereas studies by Pugh et al. [19] and Robinson et al. [20] have a greater incidence of SHH MLL2 mutations. Across the combined discovery and validation series (n = 573), the SHH subgroup has the highest total number of tumors with MLL2 mutations (n = 13) and represents the only subgroup where >1 MLL2 mutation per tumor genome was identified (Supplementary Fig. S2c). No MLL2 mutational hotspots exist and only a single recurrent mutation (1/53, R5501*) was identified across the combined cohort (Fig. 1e). MLL2 mutational status, however, provided no prognostic or diagnostic utility (Supplementary Fig. S3a).
Fig. 1.
MLL2 mutations in medulloblastoma are independent of molecular subgroup and lack prognostic utility. a Incidence of MLL2 mutations across a discovery (n = 175) (left) and validation cohort (n = 398) (right). b Percent distribution of MLL2 homozygous versus heterozygous mutations based on allele frequency, identifies a single homozygous mutation (1/53) across the combined cohorts. c Analysis of the functional consequences of MLL2 mutations highlights a significant fraction of discovery (45 %) and validation (67–80 %) single nucleotide variants that disrupt the MLL2 coding sequence through frameshift and nonsense mutations ablating enzymatic methyltransferase activity. d Subgroup-specific examination of MLL2 mutations describes the extensive variability across both sequencing cohorts suggesting MLL2 mutations occur independent of molecular subgroup. e Schematic representation of the MLL2 mutational landscape in medulloblastoma indicates single nucleotide variants scattered throughout the coding sequence and the absence of a mutational hotspot. A single recurrent mutation (R5501*) was identified across 53 MLL2 mutations. f MLL2 immunohistochemical (IHC) analysis provides no prognostic utility based on presence/absence of MLL2 protein expression for WNT and SHH subgroups (left), whereas loss of MLL2 protein expression was significantly (P < 4.63E–2) associated with survival for Group 3 and Group 4 (i.e., non-WNT, non-SHH) tumors (right)
An expression analysis reveals that MLL2 has a relatively lower expression in WNT and SHH tumors as compared with Group 3 and Group 4 medulloblastomas (Supplementary Fig. S2d) and MLL2-immunonegativity was a predictor of unfavorable outcome across non-WNT/non-SHH MBs (P < 4.63E–2; Fig. 1f). Inactivation of MLL2 appears to be restricted to the level of the nucleotide (i.e., mutational) as no copy number alterations (i.e., deletions, 1.5 < MLL2 CN >2.75) were observed in a large cohort of primary tumors analyzed (n = 201) [16] (Supplementary Fig. S2e).
Genomic and mutational inactivation of KDM6A in Group 4 medulloblastomas
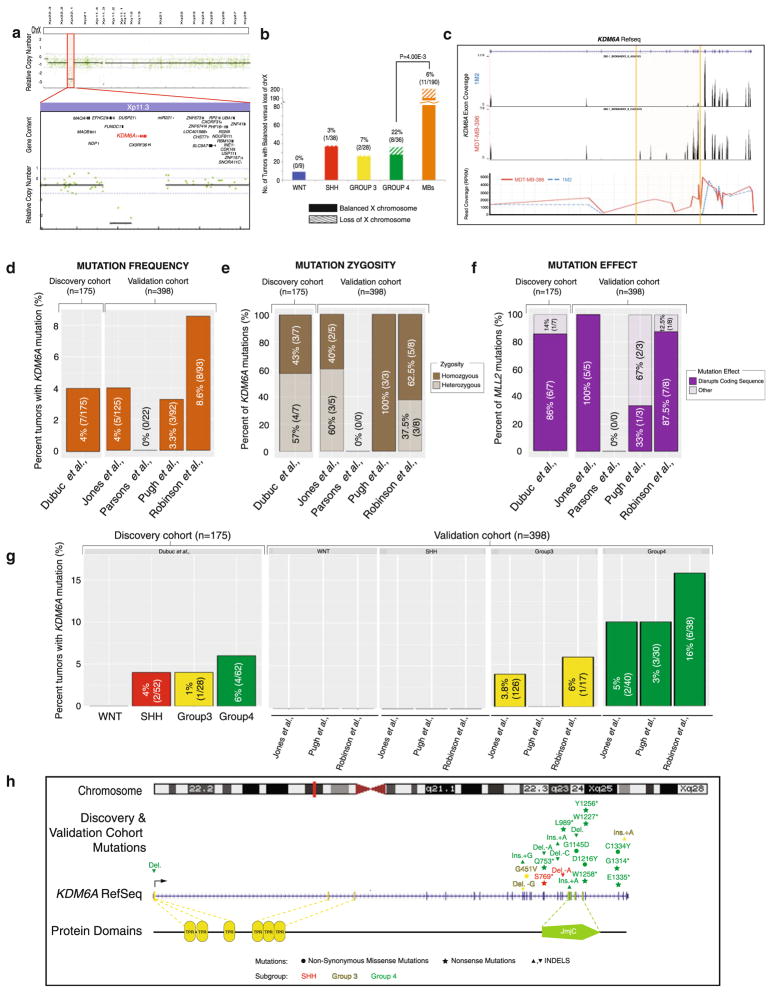
A previous characterization of copy number alterations in the medulloblastoma genome identified a focal deletion targeting KDM6A (0.5 %, 1/201) located on Xp11.3 (Fig. 2a). In addition, broad genetic loss of the X-chromosome frequently occurs in MB, targeting the inactive X-chromosome [10]. We examined the subgroup-specificity of this cytogenetic event, demonstrating a significant (P < 4.00E–3) enrichment in Group 4 MBs, affecting 22 % (8/36) of tumors (Fig. 2b). Given previous reports suggesting that both deletions and mutations lead to KDM6A inactivation [8], we pursued a targeting sequencing approach to identify the incidence and nature of KDM6A SNVs in medulloblastoma.
Fig. 2.
Identification of focal deletions of KDM6A, and mutations that are enriched in Group 4 tumors. a Analysis of medulloblastoma copy number aberrations reveals one (0.5 %, 1/203) tumor with a homozygous deletion spanning KDM6A. b Subgroup-specific distribution of X-chromosome loss across 190 primary medulloblastomas studied by Affymetrix 500 K SNP arrays demonstrates a significant (P = 4.00E−3) enrichment of this cytogenetic event in Group 4 medulloblastomas (22 %, 8/36). c Targeted exon-capture and sequencing identifies a medulloblastoma (0.6 %, 1/175, 1M2) with a KDM6A intragenic deletion spanning exons 5 through 13. d Frequency and percent of KDM6A mutations across a discovery cohort of 175 primary tumors (left) and a validation cohort of 398 tumors (right) reveals mutations in 4–8 % of tumors. e Allele frequency inference of KDM6A mutation zygosity highlights the abundance of homozygous mutations across both cohorts. f Percent distribution of KDM6A mutations which disrupt the coding sequence (frameshift or nonsense mutations) versus non-disruptive nucleotide changes (missense and in-frame INDELs) accents the frequent disruptive nature of KDM6A mutations across the discovery and validation cohorts. g Subgroup-association of KDM6A mutations demonstrates predominant targeting of Group 4 medulloblastomas. h Mutational landscape of KDM6A across 573 tumors (combined discovery and validation cohort) calls attention to the 3′ localized, damaging and Group 4-enriched nature of KDM6A single nucleotide variants
Our approach identified one additional KDM6A deletion (0.6 %, 1/175), spanning exons 5 through 13 (Fig. 2c), while somatic mutations were identified in 4 % (7/175) of MBs (Fig. 2d). KDM6A mutations were often homozygous (57 %, 4/7) (Fig. 2e) causing premature termination of the transcript (nonsense), or frameshift mutations in the majority of cases (86 %, 6/7) (Fig. 2f). No correlation was observed between KDM6A mutations and age, gender, histology, or M-stage (Supplementary Table S3) in the discovery cohort, results which were confirmed across the validation cohort. Interestingly, KDM6A mutations occur predominantly in Group 4 medulloblastomas (Fig. 1g), localized to the 3′ coding exons (Fig. 2h).
Deregulation of the histone code across multiple subgroups of medulloblastoma
MLL2 and KDM6A mutations were mutually exclusive (P = 8.60E–6) suggesting that the trxG-mediated histone code is affected in at least 12 % (21/175) of MBs within the discovery and 11 % (45/398) of the validation cohort based on our mutational analysis alone (Fig. 3a). A single tumor (1/66) across the combined cohorts was detected with mutations affecting both genes (Fig. 3a, right); however, the MLL2 mutation present within this tumor was a missense (D5028Y) mutation that did not disrupt the coding sequence.
Fig. 3.
H3K4me3 and H3K27me3 staining reveals deregulation of the histone code in a significant fraction of medulloblastomas. a Analysis of MLL2 and KDM6A mutations demonstrates the mutually exclusivity (P = 8.60E–6) and mutational inactivation of chromatin-modifier genes in 12 % (21/175) of the discover cohort. The validation cohort recapitulated these findings (P = 4.85E–10); however, a single tumor was identified with both MLL2 and KDM6A mutations. Importantly, the MLL2 mutation in this case was a missense variant with no known deleterious effect to protein functionality. b H3K4me3 (K4) and H3K27me3 (K27) immunostaining results identifies populations of medulloblastomas with modifications from the expected (K4+/K27+) staining patterns in 23.5 % (53/222) of tumors analyzed, suggesting deregulation of the histone code is a common occurrence in medulloblastoma (left). The molecular distribution of medulloblastomas with specific histone marks (right). c Survival analysis of medulloblastomas with normal (K4+/K27+) or aberrant (K4+/K27−; K4−/K27+; K4−/K27−) histone code demonstrates distinct clinical differences with improved outcome associated with K4+/K27− and dismal outcome associated with dual negativity (K4−/K27). d Subgroup-specific analysis of chromatin marks demonstrates the clinical significance associated with H3K4me3 and H3K27me3 staining patterns are driven by Group 3 and Group 4 medulloblastomas
Modifications of chromatin state represent a recurrent and important theme in the pathogenesis of MB. Therefore, we evaluated immunostaining of H3K4me3 and H3K27me3, the chromatin effectors of MLL2 and KDM6A, in two independent tumor cohorts (JHU, n = 50; DKFZ, n = 238). In both patient series, loss of H3K4me3 demonstrated a trend towards decreased survival (Supplementary Fig. S5a). By contrast, no clear trends were observed regarding H3K27me3 staining patterns across the two cohorts (Supplementary Fig. S5b). Given that trxG proteins function as a complex to increase transcriptionally active H3K4me3 marks while simultaneously removing H3K27me3 repressive marks, we overlaid these two histone modifications, where this information was available (n = 220).
While the majority of MBs stain positive for both H3K4me3 and H3K27me3 (K4+/K27+; 76 %, 167/220), a significant fraction of samples (24 %, 53/220) demonstrated isolated or combined loss of these chromatin marks (Fig. 3b). This deregulation of the normal histone code occurs across all molecular subgroups of MB. Importantly, there is a significant difference in overall survival of H3K4me3-positive/H3K27me3-negative (K4+/K27−) staining sections versus dual negative (K4−/K27−; P < 7.60E−3) or dual positive (K4+/K27+; P < 3.37E−2) samples (Fig. 3d). The improved outcome associated with K4+/K27− chromatin state appears to be driven by Group 3 and Group 4 MBs (P < 7.20E−3 versus dual positive; P < 4.40E−3 versus dual negative), as no survival differences associated with the chromatin state were observed in WNT or SHH MBs.
Identification of an H3K27me3-enriched (K27+) phenotype in Group 4 medulloblastomas
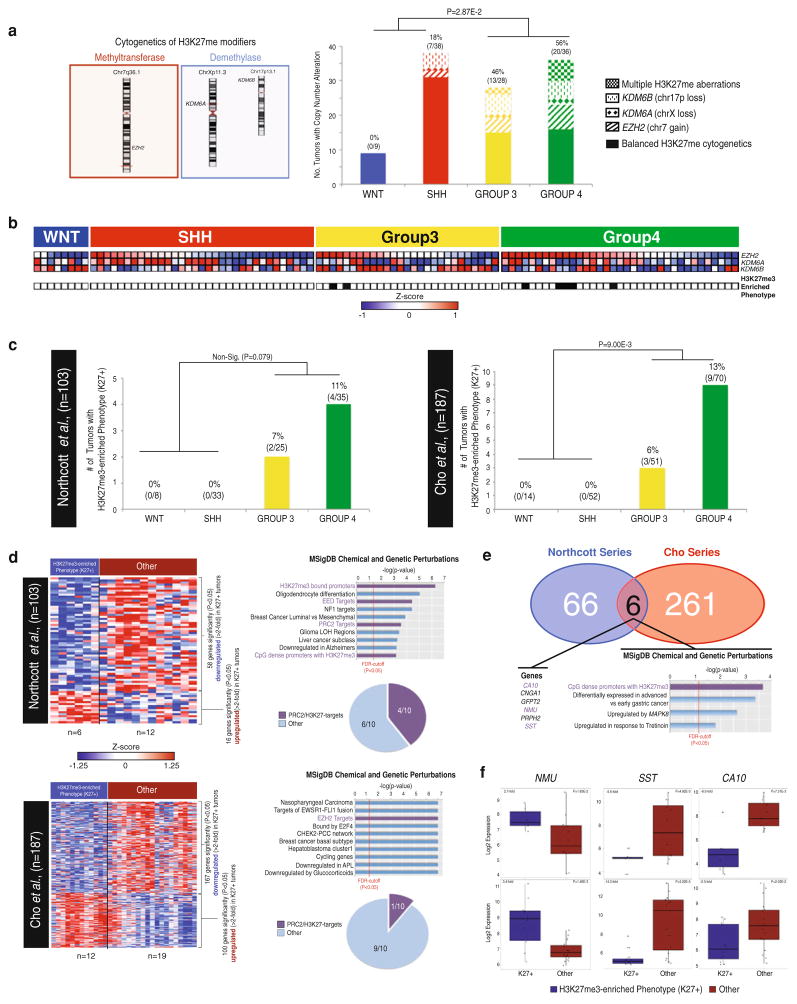
Given the mutational and genomic enrichment for KDM6A inactivation in Group 4 MBs, we examined the transcriptional and copy number status of other known modifiers of H3K27. EZH2, an H3K27-methyltransferase, has previously been reported as significantly over-expressed and amplified in Group 4 MBs [1]. In addition, KDM6B, an H3K27-demethylase, was previously reported as mutated (0.3 %, 1/398) in MBs [9, 17, 19, 20]. At the level of the genome, EZH2 (chr7q36.1) and KDM6B (17p13.1) undergo frequent copy number alterations with chromosome 7 gains and single copy loss of chr17p occurring in 14 % (29/201) and 15 % (30/210) of tumors, respectively. These aberrations, in conjunction with the aforementioned loss of KDM6A (chrXp11.3), affect 31 % (62/201) of all MBs. A subgroup-specific analysis demonstrates a significant enrichment (P < 2.87E–2) in Group 3/Group 4 MBs, with 46 % (13/28) of Group 3 and 56 % (20/36) of Group 4 MBs tumors displaying one or multiple cytogenetic events converging on H3K27 modifiers (Fig. 4a). These cytogenetic alterations were concomitant with gene expression changes for both EZH2 and KDM6A (Supplementary Fig. 6).
Fig. 4.
Identification of a genomic and transcriptional H3K27me3-enrichment phenotype (K27+) in Group 4 medulloblastomas. a Cytogenetic distribution of H3K27me methyltransferases (EZH2, chr7q36.1) and demethylases (KDM6A, chrXp11.3; KDM6B chr17p13.1) (left) and a subgroup-specific analysis of cytogenetic aberrations resulting in the accumulation of H3K27me3 identifies a significant (P = 2.87E−2) enrichment in Group 3 and Group 4 medulloblastomas versus WNT and SHH tumors (right). b Over-expression of H3K27-methyltransferases and down-regulation of H3K27-demethylases reveals a H3K27-methylator phenotype (K27+) occurring exclusively in Group 3 and Group 4 medulloblastomas across 103 primary medulloblastomas. c Discovery and validation transcriptome cohorts comprising 290 tumors demonstrate a statistically significant enrichment of an H3K27-methylator phenotype (K27+) in Group 3 and Group 4 MB. d Transcriptome-wide analysis of significantly (P < 0.05) and differentially (>2-fold) expressed genes in H3K27-methylator phenotype (K27+) versus non-K27+ medulloblastomas. Molecular Signature (MSigDB) analysis reveals extensive deregulation of PRC2-target genes in the discovery (40 %) and validation (10 %) cohorts. e Comparison of significantly (P < 0.05) and differentially (>2-fold) expressed genes across the discovery and validation transcriptome cohort reveals a small number of overlapping genes. Many of the overlapping genes (50 %, 3/5) have previously been implicated as H3K27me3-targets. f Dot plot analysis of genes putatively regulated by H3K27me3 methylation across both the discovery and validation transcriptome cohorts highlights the significantly differential fold changes and possible utility for future PRC2-therapy-related diagnostics
Correlation of EZH2, KDM6A and KDM6B expression demonstrates a strong H3K27me3-enrichment (K27+) phenotype (EZH2-high; KDM6A/KDM6B-low) exclusively in Group 3 (7 %, 2/27) and Group 4 tumors (11 %, 5/35) as compared with WNT (0/8) and SHH (0/33) subgroups (Fig. 4b, c). These findings were reproducible in an independent, non-overlapping MB dataset (n = 187), identifying a similar correlation between K27+ and Group 3 and Group 4 tumors (Fig. 4c, right). We examined the transcriptional consequences of the K27+ phenotype, by comparing K27+ versus non-enriched tumors. Molecular Signature Database (MsigDB) analysis revealed a significant over-representation of PRC2/H3K27me3 targets in genes demonstrating significant (P < 0.05) and differentially (>2-fold) expression across both transcriptome datasets (Fig. 4d). While the two transcriptome series have limited overlap in the genes commonly associated with the K27+ phenotype (Fig. 4e), the strong differential expression of common candidate markers suggest that diagnostic tools could be rapidly developed to identify populations of Group 3 and Group 4 MBs with a H3K27m3-enriched phenotype (Fig. 4f).
Discussion
Our investigation sought to characterize the clinical and biological characteristics of medulloblastomas with mutations affecting MLL2 and/or its binding partner KDM6A. Our results demonstrate that MLL2 mutations occur in 5–14 % of tumors independent of molecular subgroup or histological subtype. These mutations frequently ablate the enzymatic methyltransferase activity in one copy of MLL2 through heterozygous frameshift and nonsense mutations. The combined discovery and validation cohort confirms a lack of mutational hot spots and the heterozygous nature of MLL2 mutations. Our results highlight that while MLL2 mutations occur in a subgroup-independent manner, geographical variations in the mutation penetrance and subgroup-specificity exist. Specifically, MLL2 mutations identified in European patients (profiled in the Jones et al. [9] study) were identified in a high percentage of WNT tumors and a much lower frequency of SHH tumors. These trends are reversed in North American patients where no MLL2 mutations were identified in WNT tumors and a much higher incidence of SHH tumors. MLL2 mutations provide no prognostic or diagnostic utility, however, MLL2-immunonegativity significantly associated with decreased overall survival in non-WNT, non-SHH medulloblastomas. The near absence (1/53) of homozygous MLL2 mutations highlights a possible oncogenic dependence on the maintenance of H3K4-methylation in tumor cells. Given the established role of MLL2 as a critical requirement for oocyte survival and H3K4me3 maintenance [2, 7], our results suggest that further silencing of MLL2 may represent a synthetic lethal therapeutic avenue in subsets of MBs with MLL2 mutations. While this therapeutic approach would offer a tumor-specific effect, based on the remaining active copy of MLL2 in normal cells, the lack of mutational hot spots and size of the MLL2 coding sequence makes this approach particularly challenging.
Mutational analysis of the MLL2-binding partner KDM6A revealed mutations in 4–8 % of sequenced tumors, predominantly in Group 4 medulloblastomas. The vast majority (>80 %) of mutations disrupted the coding sequence and many homozygous mutations were identified. KDM6A is also affected at a copy number level via intragenic deletions or loss of the entire X-chromosome, which is most prominently observed in Group 4 tumors. Our results suggest that KDM6A inactivation can occur through multiple, independent cancer-specific changes including mutations and copy number alterations, and are particularly prevalent in Group 4 medulloblastomas.
MLL2 and KDM6A mutations principally occurred in a mutually exclusive pattern suggesting a minimum of 11–12 % of medulloblastomas have defects in the trxG-mediated histone code. Immunohistochemical analyses of H3K4me3 (K4+) and H3K27me3 (K27+) identifies a significant fraction of tumors with deregulation of Trithorax (trxG) and Polycomb (PcG) suggesting that additional molecular aberrations, beyond MLL2 and KDM6A mutations, likely induce deregulation of the histone code in medulloblastoma. Overlaying these histone marks identified populations of tumors with improved (K4+/K27−) and dismal (K4−/K27−) outcomes. In future studies, the use of chromatin-immunoprecipitation (ChIP) will permit the identification of cancer-induced changes to the histone code at specific promoters, and may provide a greater understanding of the transcriptional consequences of epigenetic modifications. Our findings suggest that chromatin deregulation occurs across all medulloblastoma subgroups, and therefore might represent an attractive target for therapy in a substantial percentage of medulloblastoma patients.
In addition, Group 3 and Group 4 medulloblastomas display distinct changes in their chromatin state, through the acquisition of mutational, copy number and transcriptional aberrations resulting in a net H3K27me3-enriched (K27+) phenotype with consequent down-regulation of PRC2-target genes. These tumors may be identified using surrogate markers of PRC2 activity based on gene expression of NMU, SST and CA10, respectively. Conservatively, K27+ tumors affect a minimum 10 % of Group 4 medulloblastomas, in which EZH2 is up-regulated with concomitant down-regulation of both KDM6A and KDM6B. However, 57 % of Group 4 tumors demonstrate EZH2 up-regulation with decreased expression of either demethylase (KDM6A or KDM6B). Following the identification of the four distinct medulloblastoma molecular subgroups, much effort has been placed on elucidating subgroup-specific and less toxic therapeutics. While this approach has been promising for SHH tumors [21], and less crucial for WNT tumors, where long-term survival currently exceeds 90 % [25], the outcome for Group 3 and Group 4 tumors remains bleak. Thus, the need for targetable pathways is pressing. Epigenetic modifiers have considerable promise in the treatment of medulloblastoma, suppressing the initiation of tumorigenesis in mouse models of this disease [4], and resulting in enhanced apoptosis via pharmacological targeting of EZH2 activity in MB cell lines [1]. Given the reported success of histone deacetylase inhibitor (HDACi) in preclinical setting [3], our results suggest that histone modifying therapies may represent a promising therapeutic avenue for the treatment of aggressive medulloblastoma subgroups.
Supplementary Material
Acknowledgments
MDT is supported by a CIHR Clinician Scientist Phase II award. MDT and WW are supported by a grant from the National Institutes of Health (R01CA148699) and from The Pediatric Brain Tumor Foundation. Marc Remke is funded by the Mildred-Scheel Foundation/German Cancer Aid. We thank Susan Archer for assistance with technical writing.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00401-012-1070-9) contains supplementary material, which is available to authorized users.
Contributor Information
Adrian M. Dubuc, Division of Neurosurgery, Arthur & Sonia Labatt Brain, Tumour Research Centre, The Hospital for Sick Children, Toronto, ON, Canada. Program in Developmental & Stem Cell Biology, The Hospital for Sick Children, 555 University Avenue, Toronto, ON, Canada. Department of Laboratory Medicine & Pathobiology, University of Toronto, Toronto, ON, Canada
Marc Remke, Division of Neurosurgery, Arthur & Sonia Labatt Brain, Tumour Research Centre, The Hospital for Sick Children, Toronto, ON, Canada. Program in Developmental & Stem Cell Biology, The Hospital for Sick Children, 555 University Avenue, Toronto, ON, Canada. Department of Laboratory Medicine & Pathobiology, University of Toronto, Toronto, ON, Canada.
Andrey Korshunov, CCU Neuropathology, German Cancer Research Centre (DKFZ), Heidelberg, Germany. Department of Neuropathology, University of Heidelberg, Heidelberg, Germany. Division of Pediatric Neurooncology, German Cancer Research Centre (DKFZ), Heidelberg, Germany.
Paul A. Northcott, Division of Pediatric Neurooncology, German Cancer Research Centre (DKFZ), Heidelberg, Germany
Shing H. Zhan, Genome Sciences Centre, British Columbia Cancer Agency, Vancouver, BC, Canada
Maria Mendez-Lago, Genome Sciences Centre, British Columbia Cancer Agency, Vancouver, BC, Canada.
Marcel Kool, Division of Pediatric Neurooncology, German Cancer Research Centre (DKFZ), Heidelberg, Germany.
David T. W. Jones, Division of Pediatric Neurooncology, German Cancer Research Centre (DKFZ), Heidelberg, Germany
Alexander Unterberger, Division of Neurosurgery, Arthur & Sonia Labatt Brain, Tumour Research Centre, The Hospital for Sick Children, Toronto, ON, Canada. Program in Developmental & Stem Cell Biology, The Hospital for Sick Children, 555 University Avenue, Toronto, ON, Canada.
A. Sorana Morrissy, Division of Neurosurgery, Arthur & Sonia Labatt Brain, Tumour Research Centre, The Hospital for Sick Children, Toronto, ON, Canada. Program in Developmental & Stem Cell Biology, The Hospital for Sick Children, 555 University Avenue, Toronto, ON, Canada.
David Shih, Division of Neurosurgery, Arthur & Sonia Labatt Brain, Tumour Research Centre, The Hospital for Sick Children, Toronto, ON, Canada. Program in Developmental & Stem Cell Biology, The Hospital for Sick Children, 555 University Avenue, Toronto, ON, Canada. Department of Laboratory Medicine & Pathobiology, University of Toronto, Toronto, ON, Canada.
John Peacock, Division of Neurosurgery, Arthur & Sonia Labatt Brain, Tumour Research Centre, The Hospital for Sick Children, Toronto, ON, Canada. Program in Developmental & Stem Cell Biology, The Hospital for Sick Children, 555 University Avenue, Toronto, ON, Canada. Department of Laboratory Medicine & Pathobiology, University of Toronto, Toronto, ON, Canada.
Vijay Ramaswamy, Division of Neurosurgery, Arthur & Sonia Labatt Brain, Tumour Research Centre, The Hospital for Sick Children, Toronto, ON, Canada. Program in Developmental & Stem Cell Biology, The Hospital for Sick Children, 555 University Avenue, Toronto, ON, Canada. Department of Laboratory Medicine & Pathobiology, University of Toronto, Toronto, ON, Canada.
Adi Rolider, Division of Neurosurgery, Arthur & Sonia Labatt Brain, Tumour Research Centre, The Hospital for Sick Children, Toronto, ON, Canada. Program in Developmental & Stem Cell Biology, The Hospital for Sick Children, 555 University Avenue, Toronto, ON, Canada.
Xin Wang, Division of Neurosurgery, Arthur & Sonia Labatt Brain, Tumour Research Centre, The Hospital for Sick Children, Toronto, ON, Canada. Program in Developmental & Stem Cell Biology, The Hospital for Sick Children, 555 University Avenue, Toronto, ON, Canada. Department of Laboratory Medicine & Pathobiology, University of Toronto, Toronto, ON, Canada.
Hendrik Witt, Division of Pediatric Neurooncology, German Cancer Research Centre (DKFZ), Heidelberg, Germany.
Thomas Hielscher, Division of Pediatric Neurooncology, German Cancer Research Centre (DKFZ), Heidelberg, Germany.
Cynthia Hawkins, Division of Neurosurgery, Arthur & Sonia Labatt Brain, Tumour Research Centre, The Hospital for Sick Children, Toronto, ON, Canada. Program in Developmental & Stem Cell Biology, The Hospital for Sick Children, 555 University Avenue, Toronto, ON, Canada. Department of Laboratory Medicine & Pathobiology, University of Toronto, Toronto, ON, Canada.
Rajeev Vibhakar, Department of Pediatrics, The Children’s Hospital and University of Colorado, Anschutz Medical Campus, Colorado, USA.
Sidney Croul, Department of Pathology, University Health Network, University of Toronto, Toronto, ON, Canada.
James T. Rutka, Division of Neurosurgery, Arthur & Sonia Labatt Brain, Tumour Research Centre, The Hospital for Sick Children, Toronto, ON, Canada. Department of Laboratory Medicine & Pathobiology, University of Toronto, Toronto, ON, Canada
William A. Weiss, Helen Diller Family Comprehensive Cancer Centre, University of California, San Francisco, CA, USA
Steven J. M. Jones, Genome Sciences Centre, British Columbia Cancer Agency, Vancouver, BC, Canada
Charles G. Eberhart, Department of Pathology, Johns Hopkins University, Baltimore, MD, USA
Marco A. Marra, Genome Sciences Centre, British Columbia Cancer Agency, Vancouver, BC, Canada
Stefan M. Pfister, Division of Pediatric Neurooncology, German Cancer Research Centre (DKFZ), Heidelberg, Germany
Michael D. Taylor, Email: mdtaylor@sickkids.ca, Division of Neurosurgery, Arthur & Sonia Labatt Brain, Tumour Research Centre, The Hospital for Sick Children, Toronto, ON, Canada. Program in Developmental & Stem Cell Biology, The Hospital for Sick Children, 555 University Avenue, Toronto, ON, Canada. Department of Laboratory Medicine & Pathobiology, University of Toronto, Toronto, ON, Canada
References
- 1.Alimova I, Venkataraman S, Harris P, Marquez VE, Northcott PA. Targeting the enhancer of zeste homologue 2 in medulloblastoma. Int J Cancer. 2012 doi: 10.1002/ijc.27455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreu-Vieyra CV, Chen R, Agno JE, Glaser S, Anastassiadis K, et al. MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing. PLoS Biol. 2010;8(8):e1000453. doi: 10.1371/journal.pbio.1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bots M, Johnstone RW. Rational combinations using HDAC inhibitors. Clin Cancer Res. 2009;15(12):3970–3977. doi: 10.1158/1078-0432.CCR-08-2786. [DOI] [PubMed] [Google Scholar]
- 4.Ecke I, Petry F, Rosenberger A, Tauber S, Monkemeyer S, et al. Antitumor effects of a combined 5-aza-2′ deoxycytidine and valproic acid treatment on rhabdomyosarcoma and medulloblastoma in Ptch mutant mice. Cancer Res. 2009;69(3):887–895. doi: 10.1158/0008-5472.CAN-08-0946. [DOI] [PubMed] [Google Scholar]
- 5.Ellison DW, Onilude OE, Lindsey JC, Lusher ME, Weston CL, et al. beta-Catenin status predicts a favorable outcome in childhood medulloblastoma: the United Kingdom Children’s Cancer Study Group Brain Tumour Committee. J Clin Oncol. 2005;23(31):7951–7957. doi: 10.1200/JCO.2005.01.5479. [DOI] [PubMed] [Google Scholar]
- 6.Fejes AP, Khodabakhshi AH, Birol I, Jones SJ. Human variation database: an open-source database template for genomic discovery. Bioinformatics. 2011;27(8):1155–1156. doi: 10.1093/bioinformatics/btr100. [DOI] [PubMed] [Google Scholar]
- 7.Glaser S, Schaft J, Lubitz S, Vintersten K, van der Hoeven F, et al. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development. 2006;133(8):1423–1432. doi: 10.1242/dev.02302. [DOI] [PubMed] [Google Scholar]
- 8.van Haaften G, Dalgliesh GH, Davies H, Chen L, Bignell G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41(5):521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones DT, Jager N, Kool M, Zichner T, Hutter B, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488(7409):100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinico-pathological features. PloS One. 2008;3(8):e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kool M, Korshunov A, Remke M, Jones DT, Schlanstein M, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123(4):473–484. doi: 10.1007/s00401-012-0958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lafay-Cousin L, Bouffet E, Hawkins C, Amid A, Huang A, et al. Impact of radiation avoidance on survival and neurocognitive outcome in infant medulloblastoma. Curr Oncol. 2009;16(6):21–28. doi: 10.3747/co.v16i6.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012 doi: 10.1038/nature 11606. [DOI] [PubMed] [Google Scholar]
- 14.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29(11):1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Northcott PA, Nakahara Y, Wu X, Feuk L, Ellison DW, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41(4):465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331(6016):435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfaff E, Remke M, Sturm D, Benner A, Witt H, et al. TP53 mutation is frequently associated with CTNNB1 mutation or MYCN amplification and is compatible with long-term survival in medulloblastoma. J Clin Oncol. 2010;28(35):5188–5196. doi: 10.1200/JCO.2010.31.1670. [DOI] [PubMed] [Google Scholar]
- 19.Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488(7409):106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488(7409):43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361(12):1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128(4):735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 23.Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol. 2011;12(12):799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- 24.Tamayo P, Cho YJ, Tsherniak A, Greulich H, Ambrogio L, et al. Predicting relapse in patients with medulloblastoma by integrating evidence from clinical and genomic features. J Clin Oncol. 2011;29(11):1415–1423. doi: 10.1200/JCO.2010.28.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123(4):465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.