Summary
The Septum Initiation Network (SIN) regulates multiple functions during late mitosis to ensure successful completion of cytokinesis in S. pombe. One mechanism by which the SIN promotes cytokinesis is by inhibiting a competing polarity pathway called the MOR [1], which is required for initiation of polarized growth following completion of cytokinesis [2]. Mutual antagonism between the two NDR kinase pathways, SIN and MOR, is required to coordinate cytoskeletal rearrangements during the mitosis-interphase transition. To determine how the SIN regulates the MOR pathway, we developed a proteomics approach that allowed us to identify multiple substrates of the SIN effector kinase, Sid2, including the MOR pathway components Nak1 kinase and an associated protein Sog2. We show that Sid2 phosphorylation of Nak1 causes removal of Nak1 from the SPBs, which may both relieve Nak1 inhibition of the SIN, and block MOR signaling by preventing interaction of Nak1 with the scaffold protein Mor2. Because the SIN and MOR are conserved in mammalian cells (Hippo and Ndr1/2 pathways respectively), this work may provide important insight into how the activities of these essential pathways are coordinated.
Results and Discussion
Identification of Sid2 substrates
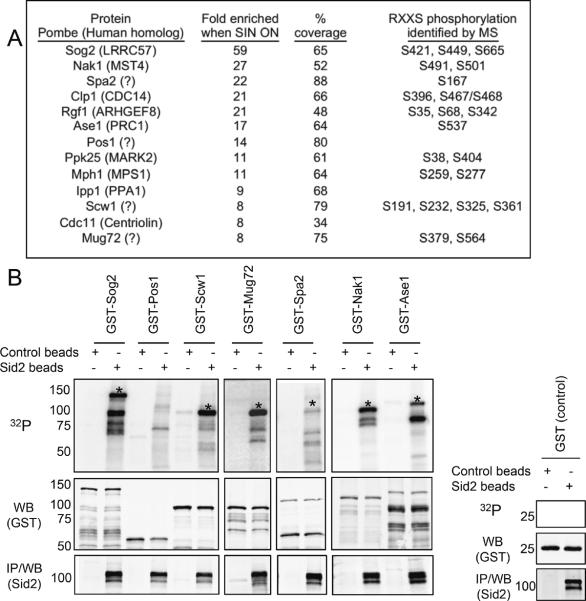
To determine how the SIN performs its various functions in late mitosis including MOR inhibition, we developed an approach to identify new Sid2 substrates. Sid2 family kinases preferentially phosphorylate the consensus sequence RXXS (where `S' becomes phosphorylated) [3, 4], which, in its phosphorylated form, is also the core consensus binding site for 14-3-3 proteins [5, 6]. Because Sid2 phosphorylation of Clp1 at RXXS sites is required for the 14-3-3 protein Rad24 to bind Clp1 [3], we hypothesized that other Sid2 substrates might be identified through SIN-dependent interaction with Rad24. Since many kinases besides Sid2 create 14-3-3 binding sites, Rad24-TAP protein complexes were purified from cells with constitutively active SIN (cdc16-116) and cells with inactive SIN (sid1-239, cdc11-123) for comparison. Protein samples were digested and analyzed by 2D LC-tandem mass spectrometry (LC-MS/MS) to identify Rad24 binding partners. The abundance (spectral counts) of each Rad24 interactor was normalized to Rad24 abundance for each experiment and then averaged over 2 biological replicates. The ratio of individual protein abundance in SIN “ON” (cdc16-116) versus SIN “OFF” (sid1-239, cdc11-123) cells was calculated, revealing many proteins significantly enriched in SIN “ON” conditions, with the top 13 proteins enriched at least 8 fold (Figure 1A, Table S1). Validating our methodology, the two known Sid2 substrates Clp1 and Cdc11 were among our top hits [3, 4]. Most other top hits were plausible Sid2 targets with annotated roles in contractile ring and septum assembly, spindle checkpoint, and/or mitosis. In addition to the identification of peptides, LC-MS/MS analysis also revealed phosphorylation sites on predicted Sid2 motifs (RXXS) in most of the putative SIN targets (Figure 1A, Table S2, Figure S1).
Figure 1. Identification and validation of Sid2 kinase candidate substrates.
(A) Proteins most enriched in Rad24-TAP purifications when the SIN is activated and any RXXS phosphorylation sites identified by LC-MS/MS are shown (see also Supplemental Tables 1 and 2). (B) Sid2 phosphorylates candidate substrates in vitro. Sid2 kinase assays were performed using the indicated substrates purified from bacteria as GST-fusions. Substrate-GST fusions or GST alone were incubated with (Sid2 beads) or without (control beads) Sid2. Half of the kinase reaction was used to detect phosphorylation by autoradiography (32P) and half was used in Western blots to determine the levels of substrate (GST) and kinase (Sid2). Asterisks mark phosphorylation of the indicated full-length substrate.
We next tested whether the SIN “ON” enriched proteins could be directly phosphorylated by the Sid2 kinase in vitro. Seven of the top candidate Sid2 substrates could be purified as soluble recombinant proteins and all but one of these was phosphorylated by Sid2 kinase purified from yeast (Figure 1B). The exception was Pos1, which was likely co-purified with its binding partner and Sid2 substrate Spa2 (Kathy Gould, unpublished observation).
Nak1 and Sog2 are part of a complex whose SPB localization is regulated by Sid2
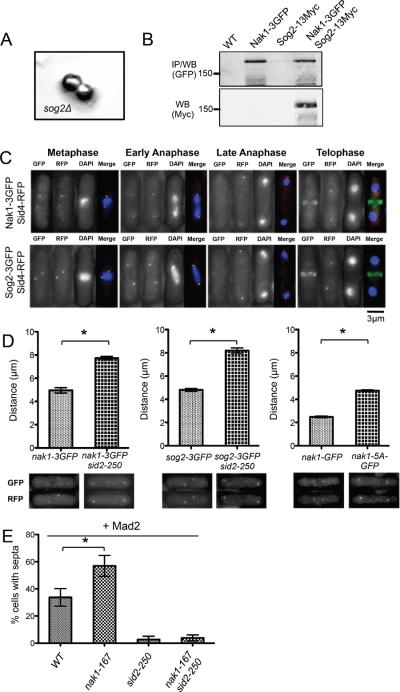
We next sought to use the information acquired about Sid2 substrates to understand how the SIN inhibits the MOR pathway. Our previous studies indicated that the SIN inhibits the MOR by blocking the ability of Nak1 to phosphorylate and activate the Orb6 kinase [2]. Thus it was striking that we identified Nak1, and a predicted Nak1 binding partner (Sog2) (based on the known function of its homolog in budding yeast [7]) as top hits in our screen for Sid2 substrates. Further analysis showed that sog2, like nak1, is an essential gene and germinating sog2Δ spores had a round cell morphology like other MOR pathway mutants (Figure 2A, [1, 8]. Nak1 and Sog2 are in a complex (Figure 2B), and show very similar localization patterns; they are localized to SPBs in early mitosis, then leave the SPBs in anaphase when the SIN gets activated, and localize to the division site late in cytokinesis [9] and Figure 2C). To test if the SIN regulates Nak1 and Sog2 localizations, we observed their GFP fusions in a sid2-250 mutant after incubation at the semi-permissive temperature of 33°C (incubation at full restrictive temperature (36°C) impaired GFP fluorescence). Interestingly, both proteins persisted at the SPBs longer in anaphase in sid2-250 mutants compared to wild-type cells and the distance between the SPBs when either SPB contained either Nak1 or Sog2 was significantly longer in sid2-250 mutants (Figure 2D). These results raise two questions: 1) what does Nak1 do at SPBs in early mitosis? and 2) why is it important for the SIN to remove Nak1 from the SPBs in anaphase?
Figure 2.
Mitotic localization of Nak1 and Sog2 to the SPB is inhibited by the SIN
(A) Phenotype of sog2Ä cells. Diploid cells heterozygous for the sog2Δ∷ura4+ mutation were sporulated, the tetrads dissected and allowed to germinate. Haploid cells carrying sog2Δ∷ura4+ divided once or twice then stopped dividing and died with a round phenotype. An example of the sog2Δ∷ura4+ phenotype is shown. (B) Sog2-13Myc co-immunoprecipitates with Nak1-3GFP. Extracts were prepared from cells of the indicated genotypes following overnight growth in YE at 25°C. Nak1-3GFP was immunoprecipitated using an anti-GFP antibody and the immunoprecipitates were analyzed by immunoblotting for GFP and Myc. (C) Localization of Nak1-3GFP and Sog2-3GFP during mitosis. Sid4-RFP was used as a marker for SPBs. Cells were grown at 25°C in Y E, fixed in methanol and then imaged. Representative images are shown. GFP (Green), RFP (Red) and DAPI (Blue) are shown in merged images. (D) Sid2 promotes loss of Nak1 and Sog2 from anaphase SPBs. Upper panels depict the average distance between SPBs showing localization of the indicated proteins. Lower panels show representative images of the cells examined above. Sid4-RFP marked SPBs. Cells were grown at 25°C in YE followed by a shift to 33°C for 2h, fixed in methanol and then imaged (left and middle panels). In the right panel, nak1-GFP and nak1-5A-GFP cells were grown overnight in YE at 25°C and then fixed and imaged. Error bars denote SD values obtained from the average distance measured from three separate experiments. Statistical analysis using paired t tests (brackets) indicates that the difference in SPB separation between the control and the experimental values is statistically significant (all p-values <0.007, a minimum of 25 cells were scored for every experiment). (E) Nak1 inhibits SIN dependent ectopic metaphase septation. Cells of the indicated genotypes were grown at 25°C in the absence of thiamine for 16 hou rs to induce expression of the mad2 gene under control of the full strength thiamine inducible promoter (nmt1) and then shifted to 36°C for 3 hours. Cells were then fixed in methanol, imaged and a minimum of 100 cells were scored for the presence of ectopic septa. Error bars denote SD values obtained from percentages measured from three separate experiments. Statistical analysis using paired t tests (brackets) indicates that the difference in ectopic septation between the WT control and the nak1-167 cells is statistically significant (p-value=0.0182).
The SIN is normally kept inactive in early mitosis to keep cells from initiating cytokinesis before chromosome segregation has initiated [4, 10]. We therefore reasoned that SPB-localized Nak1 may inhibit the SIN (which also localizes to SPBs) in early mitosis, and SIN dependent removal of Nak1 from the SPBs in anaphase may enhance SIN activity. To test whether Nak1 inhibits SIN activity in early mitosis, Nak1 was inactivated using the nak1-167 mutation in cells that were first arrested in metaphase by overexpression of the spindle checkpoint protein Mad2 [11]. Although some wild-type cells eventually begin to septate despite the metaphase arrest, premature septation was significantly increased in nak1-167 mutant cells (Figure 2E). The ectopic septation in nak1-167 cells was blocked by the sid2-250 mutation (Figure 2E) indicating that increased septation in nak1-167 mutants can be attributed to premature SIN activation. Together these experiments are consistent with a model where Nak1 helps prevent premature SIN activation in metaphase, and SIN dependent removal of Nak1 from the SPBs in anaphase promotes full SIN activation.
Phosphorylation of Nak1 by the Sid2 kinase is required for SIN mediated inhibition of polarized growth
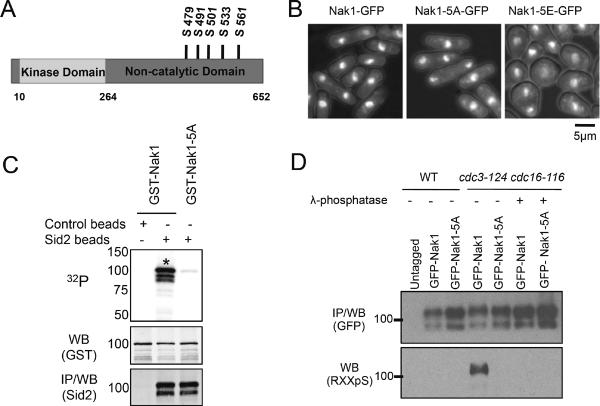
Because the SIN interferes with the ability of Nak1 to activate its downstream kinase Orb6 [2] we sought to understand how Sid2 phosphorylation affects Nak1. Nak1 contains 7 Sid2 consensus phosphorylation motifs (RXXS), two of which (S491, S501) we identified as phosphorylated in vivo by LC-MS/MS (Figure S1). Three other Sid2 consensus sites (S479, S533, S561) were identified in phosphoproteomic mass spectrometry studies [12–14]. These 5 sites, which cluster in an 82 amino acid region of the Nak1 C-terminal non-catalytic domain (Figure 3A), were mutated to either alanine (nak1-5A) or glutamic acid (nak1-5E) to create non-phosphorylatable or phospho-mimetic mutants respectively. Since we expected Sid2 phosphorylation to inhibit Nak1, it was not surprising that the Nak1-5A protein was functional, as judged by its ability to rescue the viability and shape defects of the nak1Δ deletion mutant, whereas the nak1Δ cells expressing Nak1-5E were viable but had defects in cell shape (Figure 3B). Unlike Nak1, recombinant Nak1-5A could not be phosphorylated by Sid2 in vitro (Figure 3C). We also examined the in vivo phosphorylation status of Nak1-5A compared to Nak1 using phospho-specific antibodies that recognize the RXXS motif. Western blot analysis of Nak1 immunoprecipitates from asynchronously growing wild-type cells and cells in which the SIN pathway was activated using the cdc16-116 mutation showed that wild-type Nak1, but not Nak1-5A, was phosphorylated on RXXS sites specifically in cells with activated SIN signaling (Figure 3D).
Figure 3. The Nak1-5A mutant cannot be phosphorylated by Sid2.
(A) Diagram of Nak1. The five Sid2 phosphorylation sites (RXXS) mutated to either alanine (A) or glutamic acid (E) are indicated (see Supplemental Table S2 and Supplemental Figure S1). (B) Rescue of the nak1Δ deletion mutant by the Nak1 phosphorylation site mutants. Plasmids for the Nak1 phosphorylation site mutants were integrated into nak1Δ heterozygous diploids, sporulated, and clones carrying the Nak1 phosphorylation site mutants and the nak1Δ deletion were isolated and grown at 36°C for 4h. (C) Sid2 ph osphorylates GST-Nak1 in vitro, but not GST-Nak1-5A. Kinase assays were performed on GST-Nak1 and GST-Nak1-5A as described in Figure 1B. (D) GFP-Nak1, but not GFP-Nak1-5A is phosphoryated in vivo. GFP-Nak1 and GFP-Nak1-5A were immunoprecipitated from extracts prepared from cells of the indicated genotypes expressing GFPNak1 or GFP-Nak1-5A from a thiamine repressible promoter. Cells were grown in the absence of thiamine for 16 hours, then the SIN pathway was activated by shifting cdc3-124 cdc16-116 cells to 36°C for 3h. In addition, extracts were prepared from cells with activated SIN for treatment with ë-phosphatase following Immunoprecipitation. Levels of Nak1 RXXpS phosphorylation and GFP-Nak1 were detected by western blotting with anti-GFP antibody and a phospho-specific antibody against to the RXXS motif respectively.
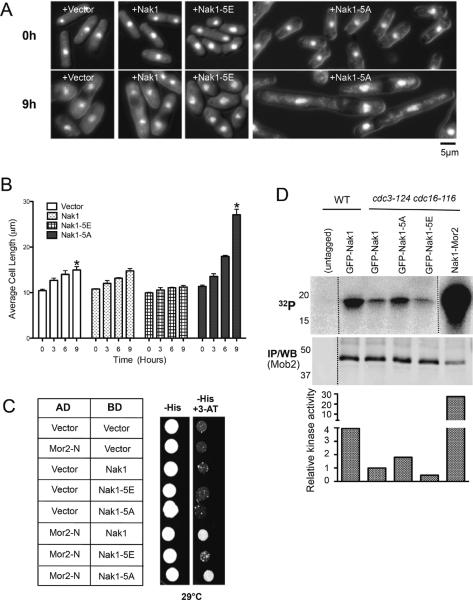
SIN activation causes cessation of polarized cell growth by inhibiting the MOR pathway [2]. To determine whether Sid2 phosphorylation of Nak1 mediates its inhibition of the MOR, we tested if Nak1-5A could bypass SIN-mediated inhibition of polarized cell growth. Wild-type Nak1, Nak1-5E, or Nak1-5A were expressed in cdc16-116 mutant cells where the SIN is constitutively active (Note that for all experiments where the SIN is activated, the cdc3-124 mutation, which blocks septum formation [15], was also present to block non-specific effects on cell growth triggered by ectopic septation upon SIN activation). Nak1-5A but not wild-type Nak1 or Nak1-5E bypassed SIN-mediated inhibition of cell elongation (Figure 4A–B). Cells expressing the non-phosphorylatable form of Nak1 were approximately twice as long as control cells (Figure 4A). Furthermore, unlike Nak1 or Nak1-5E, Nak1-5A expression in cells with active SIN was able to partially restore Orb6 kinase activity (Figure 4D), which is normally inhibited by the SIN [2]. These results indicate that the SIN inhibits Orb6 kinase activity and MOR mediated polarized growth at least in part through phosphorylation of the Nak1 kinase.
Figure 4. Elimination of Sid2 phosphorylation of Nak1 bypasses SIN mediated inhibition of polarized growth.
(A–B) Expression of Nak1-5A bypasses SIN inhibition of cell elongation. cdc16-116 cdc3-124 cells with the indicated plasmids were grown at 25°C in the absence of thiamine for 16 hours (to induce expression of the indicated proteins) and then shifted to 36°C to activate the SIN. Sampl es were collected every 3 hours. (A) Images representing the indicated cells from part at 0 hour and 9 hour time points are shown. Cells were stained with DAPI and visualized by a combination of fluorescence and DIC microscopy. (B) Cell lengths of 50 cells were measured were measured for each time point. Error bars denote SD values obtained from the average cell lengths measured from three separate experiments. Statistical analysis using paired t tests (as indicated by asterisks) shows that the difference in cell lengths between cells expressing the vector control and the Nak1-5A construct was found to be statistically significant (p-value <0.001). (C) Nak1, but not the Nak1-5E mutant shows a physical interaction with the Mor2-N terminus by yeast two-hybrid analysis. S. cerevisiae Y190 cells expressing the Mor2-N terminus (1-1095 amino acid residues) fused to the Gal4 activation domain (AD) along with either Nak1, Nak1-5E or Nak1-5A fused to the Gal4 DNA-binding domain (BD) were spotted on SD agar plates lacking histidine +/− 3-aminotriazole (3AT). Cell growth was observed after they were cultured at 29°C for 3 days. (D) Nak1-5A and t he Nak1-Mor2 fusion cause Orb6 kinase activation even when the SIN is active. All strains except the control (untagged) express the indicated Nak1 proteins and Mob2-13Myc, which was used to pull down its associated kinase Orb6. The temperature sensitive cdc3-124 cdc16-116 background was used to activate the SIN. Cells were grown at 25°C then shifted to 36°C for 3 hours. Orb6 immune complex kinase assays were performed by first pulling down the Orb6 regulatory subunit Mob2. Myelin basic protein (MBP) was used as substrate. Half of the kinase reaction was used to detect phosphorylation by autoradiography (32P) and half was used in Western blots to determine the levels of Mob2. (lower panel) Orb6 kinase activity for each strain was normalized to Mob2 level and relative to the activity of GFP-Nak1 expressing cdc3-124 cdc16-116 cells.
We also examined the localization of Nak1-5A to determine if SIN dependent removal of Nak1 from the SPBs in anaphase depended on direct phosphorylation of Nak1. This analysis showed that Nak1-5A persisted on the SPB longer in anaphase than the wild-type protein, consistent with Sid2 phosphorylation displacing Nak1 from the SPB (Figure 2D).
Sid2 phosphorylation of Nak1 inhibits its interaction with the scaffold protein Mor2
To further test whether Sid2 phosphorylation of Nak1 could explain our previous observation that Sid2 inhibits the ability of Nak1 to activate Orb6 [2]. Both Nak1 and Orb6 kinase interact with a drosophila furry-like protein called Mor2 [16, 17]. Mor2 functions as a scaffold that allows Nak1 to activate Orb6 [18]. We hypothesized that Sid2 phosphorylation of Nak1 might block the Nak1-Mor2 interaction. If this were the case, then fusing Nak1 to Mor2 would bypass SIN inhibition of cell elongation. Therefore we constructed a Nak1-Mor2 fusion (Figure S2A) that was expressed on a plasmid using a thiamine repressible promoter. This fusion rescued both nak1-167 and mor2-286 mutants (under repressed conditions) indicating that both proteins in the fusion were functional (data not shown). Next, its expression was induced in cells with activated SIN (cdc3-124 cdc16-116) (Figure 4D and Figure S2B–C). Expression of the fusion caused a huge increase in Orb6 kinase activity even when the SIN is active (Figure 4D). Although SIN activation blocked cell elongation in cells with vector control, Nak1 or Mor2 alone, or co-expression of unfused Nak1 and Mor2, expression of the Nak1-Mor2 fusion caused a remarkable increase in cell length under both induced and repressed conditions (Figure S2B–C, and data not shown). Cells expressing the Nak1-Mor2 fusion grew more than three times the length of control cells after 9h of SIN activation, supporting the model that the SIN inhibits cell elongation by interfering with Nak1-Mor2 interaction.
To test more directly whether Sid2 mediated Nak1 phosphorylation disrupted the Nak1-Mor2 interaction, we utilized the phospho-mimetic mutant Nak1-5E. Although Nak1-5E could not support polarized growth or bypass SIN-mediated inhibition of cell elongation (Figure 3B and Figure 4A–B), fusion of Nak1-5E to Mor2 (Nak1-5E-Mor2) did bypass SIN inhibition of polarized cell growth similar to the Nak1-Mor2 fusion (Figure S2B–C). These results are consistent with the idea that phosphorylation of Nak1 by Sid2 inhibits its ability to interact with Mor2. To directly test this hypothesis in another manner, we examined whether phosphomimetic mutations in Nak1 could disrupt the interaction between Nak1 and Mor2. Although we could not observe interaction between endogenous Nak1 and Mor2 by co-immunoprecipitation, as with a previous study we could observe an interaction between the two proteins by 2-hybrid analysis [17]. While we observed an interaction between Nak1 (or Nak1-5A) and the Mor2 amino-terminus as previously reported, this interaction was greatly reduced in the Nak1-5E mutant (Figure 4C). Together these results support the model that Sid2-mediated phosphorylation of Nak1 blocks its interaction with the Mor2 scaffold.
The SIN has multiple essential functions in late mitosis mediated by its effector kinase Sid2, but our understanding of these events has been hampered by our lack of knowledge of Sid2 substrates. Our proteomics screen for Sid2 targets yielded many promising candidate mediators of SIN signaling. Although some of the candidate substrates may be indirectly regulated by Sid2, we expect many are direct targets. Two previously identified Sid2 substrates (Clp1 and Cdc11) [3, 4] were hits in our screen, validating our methodology and many of the other candidates, including the MOR component Nak1, are phosphorylated on Sid2 consensus sites in vivo, and can be phosphorylated by Sid2 in vitro. We expect that characterizing other hits from our screen will illuminate how the SIN regulates additional mitotic events. Particularly compelling candidates for follow-up studies include Rgf1, Scw1, and Mph1. Rgf1 is a guanine exchange factor for Rho1, a GTPase essential for cytokinesis that regulates actomyosin ring assembly, and septum deposition [19, 20], and a multi-copy suppressor of SIN mutants [21]. Mutants of the RNA binding protein Scw1 suppress SIN mutants, suggesting that it could be inhibited by the SIN [22, 23]. And finally, Mph1 (Mps1 in budding yeast and humans), is a spindle checkpoint kinase and the SIN inhibits the spindle checkpoint through an unknown mechanism [24, 25].
This work also clarified another mechanism of crosstalk between the SIN and MOR pathways. We found first that Nak1, perhaps through its SPB localization in early mitosis, helps prevent premature activation of the SIN. Further investigation will be required to determine how Nak1 inhibits the SIN. Second at anaphase onset, Sid2 phosphorylation of Nak1 removes Nak1 from the SPBs, promotes SIN activation and inhibits MOR signaling by blocking interaction of Nak1 with the Mor2 scaffold protein. The SIN may inhibit the MOR through additional mechanisms. An obvious possibility is via phosphorylation of the Nak1 binding partner Sog2 as we identified it as another probable Sid2 substrate. Because SIN and MOR pathway components are conserved in mammalian cells, we expect that these studies will be informative for understanding coordination of the activities of the counterpart Hippo and Ndr1/2 pathways.
Supplementary Material
Highlights
Comparative proteomics identifies novel SIN targets.
Mechanisms of crosstalk between SIN and MOR NDR-kinase pathways.
Nak1 inhibits premature SIN activation in early mitosis.
SIN blocks MOR signaling by inhibiting interaction of Nak1 with the Mor2 scaffold.
Acknowledgements
J.R.M. was supported by NCI T32CA119925. This work was supported by the Howard Hughes Medical Institute of which K.L.G. is an investigator, and National Institutes of Health grant GM058406-14 to D. McCollum. We thank Dai Hirata for strains and plasmids and Liping Ren and Jianqiu Wang for performing LCMS/MS experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gupta S, McCollum D. Crosstalk between NDR kinase pathways coordinates cell cycle dependent actin rearrangements. Cell Div. 2011;6:19. doi: 10.1186/1747-1028-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ray S, Kume K, Gupta S, Ge W, Balasubramanian M, Hirata D, McCollum D. The mitosis-to-interphase transition is coordinated by cross talk between the SIN and MOR pathways in Schizosaccharomyces pombe. J. Cell Biol. 2010;190:793–805. doi: 10.1083/jcb.201002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen C-T, Feoktistova A, Chen J-S, Shim Y-S, Clifford DM, Gould KL, McCollum D. The SIN kinase Sid2 regulates cytoplasmic retention of the S. pombe Cdc14-like phosphatase Clp1. Curr. Biol. 2008;18:1594–1599. doi: 10.1016/j.cub.2008.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feoktistova A, Morrell-Falvey J, Chen J-S, Singh NS, Balasubramanian MK, Gould KL. The fission yeast septation initiation network (SIN) kinase, Sid2, is required for SIN asymmetry and regulates the SIN scaffold, Cdc11. Mol. Biol. Cell. 2012;23:1636–1645. doi: 10.1091/mbc.E11-09-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 6.Mah AS, Elia AEH, Devgan G, Ptacek J, Schutkowski M, Snyder M, Yaffe MB, Deshaies RJ. Substrate specificity analysis of protein kinase complex Dbf2-Mob1 by peptide library and proteome array screening. BMC Biochem. 2005;6:22. doi: 10.1186/1471-2091-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson B, Kurischko C, Horecka J, Mody M, Nair P, Pratt L, Zougman A, McBroom LD, Hughes TR, Boone C, et al. RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol Biol Cell. 2003;14:3782–803. doi: 10.1091/mbc.E03-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maerz S, Seiler S. Tales of RAM and MOR: NDR kinase signaling in fungal morphogenesis. Curr. Opin. Microbiol. 2010;13:663–671. doi: 10.1016/j.mib.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 9.Leonhard K, Nurse P. Ste20/GCK kinase Nak1/Orb3 polarizes the actin cytoskeleton in fission yeast during the cell cycle. J Cell Sci. 2005;118:1033–44. doi: 10.1242/jcs.01690. [DOI] [PubMed] [Google Scholar]
- 10.Guertin DA, Chang L, Irshad F, Gould KL, McCollum D. The role of the sid1p kinase and cdc14p in regulating the onset of cytokinesis in fission yeast. EMBO J. 2000;19:1803–15. doi: 10.1093/emboj/19.8.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He X, Jones MH, Winey M, Sazer S. Mph1, a member of the Mps1-like family of dual specificity protein kinases, is required for the spindle checkpoint in S. pombe. J. Cell. Sci. 1998;111(Pt 12):1635–1647. doi: 10.1242/jcs.111.12.1635. [DOI] [PubMed] [Google Scholar]
- 12.Beltrao P, Trinidad JC, Fiedler D, Roguev A, Lim WA, Shokat KM, Burlingame AL, Krogan NJ. Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol. 2009;7:e1000134. doi: 10.1371/journal.pbio.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson-Grady JT, Villén J, Gygi SP. Phosphoproteome analysis of fission yeast. J. Proteome Res. 2008;7:1088–1097. doi: 10.1021/pr7006335. [DOI] [PubMed] [Google Scholar]
- 14.Koch A, Krug K, Pengelley S, Macek B, Hauf S. Mitotic substrates of the kinase aurora with roles in chromatin regulation identified through quantitative phosphoproteomics of fission yeast. Sci Signal. 2011;4:rs6. doi: 10.1126/scisignal.2001588. [DOI] [PubMed] [Google Scholar]
- 15.Balasubramanian MK, Hirani BR, Burke JD, Gould KL. The Schizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis. J Cell Biol. 1994;125:1289–301. doi: 10.1083/jcb.125.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirata D, Kishimoto N, Suda M, Sogabe Y, Nakagawa S, Yoshida Y, Sakai K, Mizunuma M, Miyakawa T, Ishiguro J, et al. Fission yeast Mor2/Cps12, a protein similar to Drosophila Furry, is essential for cell morphogenesis and its mutation induces Wee1-dependent G(2) delay. EMBO J. 2002;21:4863–74. doi: 10.1093/emboj/cdf495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanai M, Kume K, Miyahara K, Sakai K, Nakamura K, Leonhard K, Wiley DJ, Verde F, Toda T, Hirata D. Fission yeast MO25 protein is localized at SPB and septum and is essential for cell morphogenesis. EMBO J. 2005;24:3012–25. doi: 10.1038/sj.emboj.7600782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hergovich A, Stegert MR, Schmitz D, Hemmings BA. NDR kinases regulate essential cell processes from yeast to humans. Nat Rev Mol Cell Biol. 2006;7:253–64. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- 19.Mutoh T, Nakano K, Mabuchi I. Rho1-GEFs Rgf1 and Rgf2 are involved in formation of cell wall and septum, while Rgf3 is involved in cytokinesis in fission yeast. Genes Cells. 2005;10:1189–1202. doi: 10.1111/j.1365-2443.2005.00908.x. [DOI] [PubMed] [Google Scholar]
- 20.García P, Tajadura V, García I, Sánchez Y. Rgf1p is a specific Rho1-GEF that coordinates cell polarization with cell wall biogenesis in fission yeast. Mol. Biol. Cell. 2006;17:1620–1631. doi: 10.1091/mbc.E05-10-0933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin Q-W, Zhou M, Bimbo A, Balasubramanian MK, McCollum D. A role for the septation initiation network in septum assembly revealed by genetic analysis of sid2-250 suppressors. Genetics. 2006;172:2101–2112. doi: 10.1534/genetics.105.050955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin Q-W, McCollum D. Scw1p antagonizes the septation initiation network to regulate septum formation and cell separation in the fission yeast Schizosaccharomyces pombe. Eukaryotic Cell. 2003;2:510–520. doi: 10.1128/EC.2.3.510-520.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karagiannis J, Oulton R, Young PG. The Scw1 RNA-binding domain protein regulates septation and cell-wall structure in fission yeast. Genetics. 2002;162:45–58. doi: 10.1093/genetics/162.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guertin DA, Venkatram S, Gould KL, McCollum D. Dma1 prevents mitotic exit and cytokinesis by inhibiting the septation initiation network (SIN) Dev. Cell. 2002;3:779–790. doi: 10.1016/s1534-5807(02)00367-2. [DOI] [PubMed] [Google Scholar]
- 25.Fankhauser C, Marks J, Reymond A, Simanis V. The S. pombe cdc16 gene is required both for maintenance of p34cdc2 kinase activity and regulation of septum formation: a link between mitosis and cytokinesis? EMBO J. 1993;12:2697–2704. doi: 10.1002/j.1460-2075.1993.tb05931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.