Abstract
Dental caries remains the most common chronic childhood disease. Despite strong evidence of genetic components, there have been few studies of candidate genes and caries. In this analysis we tried to assess genetic and environmental factors contributing to childhood caries in the Iowa Fluoride Study. Environmental factors (age, sex, race, tooth-brushing frequencies and water fluoride level) and three dental caries scores (d2fs-total, d2fs-pit/fissure, and d2fs-smooth surface) were assessed in 575 unrelated children (mean age=5.2 years). Regression analyses were applied to assess environmental correlates. The Family Based Association Test was used to test genetic associations for 23 SNP markers in 7 caries candidate genes on 333 Caucasian parent-child trios. We evaluated the associations between caries status and the level of both single SNPs and multiple SNPs (haplotype) respectively. Permutation procedure was performed for correction of inflated type-I errors due to multiple testing. Age, tooth-brushing frequencies and water fluoride level were significantly correlated to at least one carious score. Caries on pit and fissure surfaces was substantially higher than on smooth surfaces (61% vs. 39%). SNPs in three genes (DSPP, KLK4, and AQP5) showed consistent associations with protection against caries. Of note, KLK4 and AQP5 were also highlighted by subsequent haplotype analysis. Our results support the concept that genes can modify the susceptibility of caries in children. Replication analysis in independent cohorts is highly needed in order to verify the validity of our findings.
Keywords: dental caries, genetics, children, candidate gene, association analysis
Introduction
Dental caries, a multifactorial complex disease [Shuler, 2001], remains the most common chronic disease of childhood. Although a decline in dental caries rates in the United States and other industrialized nations in the primary dentition was found until the mid-1980s, later reports have suggested that this decline has slowed or even reversed in the U.S. and elsewhere ([Burt, 1994; Downer, 1994]. This has also been supported by recent NHANES data (NHANES surveillance summaries on oral health, 2005). Further, billions of dollars are spent for the treatment of dental caries every year and the costs keep increasing. The etiology of dental caries involves a complex interplay of environmental and genetic factors. [Hunter, 1988] Epidemiological studies have tried for many years to understand fully the mechanisms of this disease, with the eventual goal of prevention. Thus, identifying the underlying genetic and environmental risk factors is a crucial step toward that goal.
Pioneering twin studies investigating the heritability (i.e., proportion of variation due to genes) of dental caries in children have clearly supported the key role of genetics in tooth decay [Liu et al., 1998; Bretz et al., 2006]. Caries heritability estimates for children based on twins range from 64–85%. In our recent heritability study based on larger families [Wang et al., 2010], heritability of dental caries in the primary dentition was over 50%. Also, we found that genes affecting caries might be different for the primary and permanent teeth. These findings have reinforced the importance of studying genetic components of dental caries in primary teeth separately.
Numerous efforts on gene mapping have been made so far to identify specific genetic loci contributing to caries susceptibility [Werneck et al., 2010]. Results from previous studies have highlighted features involved in processes related to development of caries in the following four categories: 1) Salivary composition and flow: Saliva contains components that can directly kill cariogenic bacteria. It is also rich in calcium and phosphates which are actively involved in the tooth enamel re-mineralization process. The physical flow of saliva helps to dislodge pathogens (viruses, bacteria, and yeast) from teeth and mucosal surfaces. Saliva can also cause microbes to clump together so that they can be swallowed before they become firmly attached. [Stookey, 2008]; 2) Tooth morphology: Tooth morphology refers to the number and shape of cusps, ridges, grooves, and even the tooth size as a whole. Malposition of the teeth, deep anatomy grooves, and areas of retention due to the natural morphology of the tooth structure can cause difficulties in tooth brushing, and fluoride penetration, and thus be considered as caries risk factors, especially for pit and fissure caries.[Guzman-Armstrong, 2005]; 3) Dietary and taste preferences: In general, there are multiple potential effects of diet. Diet can influence the amount and type of plaque formation and debris and the presence of relative numbers of cariogenic microorganisms on tooth surfaces. The interactions of the cariogenic potential of foods (e.g., sucrose), the frequency of eating and the physical state (or type) of the diet all can affect individually or jointly the carious process; [Wendell et al., 2010]. On the other hand, taste preference affects dietary habits. Dietary choices often are guided by how foods taste. For example, children especially love sweets and dislike bitter tastes and they eat more of the foods they like best [Drewnowski, 1997]; 4) Enamel and dentin formation: dental caries itself is a destructive process causing decalcification of the tooth enamel and leading to continued destruction of enamel and dentin, and eventually the cavitation of the tooth. Demineralization of enamel leads to caries and in contrast, enamel remineralization can help prevent caries. Thus, genes regulating enamel/dentin formation make significant contributions to caries experience. There are two plausible theories which explain how enamel genes may associate with caries lesion development: they interact with oral bacteria (such as S mutans) to affect caries susceptibility [Patir et al., 2008] and/or they enhance the enamel thickness and enamel fluoride concentration to protect teeth against caries. [Keene et al., 1980]
Besides genetics, environmental determinants also play a crucial role in caries susceptibility in children [Verrips et al., 1992; Lukacs and Largaespada, 2006]. Demographic parameters, including age, sex and ethnicity, are important predictors for caries. In general, older children have more caries because their teeth are exposed to the environment and at risk longer; girls are typically found to exhibit higher caries prevalence rates than boys, mostly due to earlier eruption of their teeth and food preferences [Ferraro and Vieira, 2010]. Moreover, caries prevalence was observed to vary among different racial groups, with non-Hispanic whites having the lowest caries prevalence and severity (NHANES, 2005). However, this could be partly due to different genetic factors as well. In addition, other environmental factors such as tooth-brushing behaviors, water fluoride levels and family education level are acknowledged to be protective factors for primary teeth [Levy et al., 2003; Ferreira et al., 2007; Menghini et al., 2008].
Taken together, genetic and environmental factors play crucial roles in susceptibility and resistance to this complex disease, and thus both should be considered in any study of dental caries. Considering the higher caries prevalence and evidence for a stronger genetic influence in primary versus permanent dentitions, conducting a genetic epidemiology study concerning caries in a younger age cohort is particularly meaningful. Hence, the purpose of the current study was to assess genetic and environmental determinants of primary tooth caries in the Iowa Fluoride Study cohort.
Subjects and Methods
Study Sample
The recruitment of the Iowa Fluoride Study cohort has been described previously [Levy et al., 2003]. Five hundred and seventy-five unrelated children (300 girls and 275 boys) and their parents were included in our analyses. Most children were aged 4 to 7 years old, with two outliers (age = 2.8 and 7.6, respectively). The mean and standard deviation of age was 5.2±0.4 years. Among them, 95% of these children were from Caucasian families, 2% of them were African American and 3% from other racial/ethnic groups.
Phenotype
Phenotype data on dental caries was assessed on children, but not their parents. A trained and calibrated dentist performed a caries examination on each participant. Procedures and criteria have been described in detail previously [Warren et al., 2006]. Briefly, the assessment included dental caries status for each surface on all erupted primary teeth, i.e., sound, decayed (d) or filled (f), and examinations were conducted with portable equipment (chair, headlight and the Denlite System). Examinations were primarily visual, with explorer use to confirm caries found. Radiographs were not taken. On primary molars, pit-and-fissure decay and fillings were distinguished from smooth surface ones. The missing (m) component was not included in our dental caries scores due to possible misidentification of exfoliated primary and unerupted permanent teeth in children. Also, although scored separately, we did not include non-cavitated (d1, white-spot/incipient) lesions. Finally, we included the following three dental caries scores in our genetic analysis as phenotypes: (1) total number of tooth surfaces with frank cavitated or filled caries experience (d2fs-total), (2) pit and fissure surfaces with caries experience (d2fs-pit/fissure) and (3) caries experience of all other tooth surfaces (d2fs-smooth-surface). These scores were dichotomized in the downstream analyses as cases (children with scores ≥ 1) and controls (scores = 0). The phenotypic correlations among above three caries scores were moderately high (r2 ~ 0.5–0.8).
Genotype
We genotyped 23 SNPs (single nucleotide polymorphisms) from 7 candidate genes for 333 Caucasian parent-child trios. (Table 1). Genes involved in tooth development, enamel mineralization and tooth morphology, and salivary buffering were selected for analysis, with the premise that some of the genes involved in these basic biological processes may also play a role in caries susceptibility/resistance. Genotyping was performed using TaqMan assays, amplified in a 384-well thermocycler and semi-automatically analyzed on an Applied Biosystems (ABI) 7900 DNA Analyzer. TaqMan assays were selected using an ABI SNP browser that displays haplotype block structure characterized in the four HapMap populations (http://www.hapmap.org). In addition to HapMap, we also used the following public databases: (http://www.ncbi.nlm.nih.gov/SNP/) and (http://bioinfo.weizmann.ac.il/cards/index/shtml) to identify SNPs. SNPs were chosen based on a balance between availability characteristics of the SNP and knowledge about LD block structure from the HapMap Project. To optimize genotyping information, SNPs were selected from different haplotype blocks. Priority was given to functional and nonsynonymous SNPs. PCR primer sets were purchased from ABI. PCR primer conditions were obtained from the ABI website (http://snp.ims.u=tokyo.ac.jp). Selected SNPs were genotyped, along with no template controls, two positive control reference individuals and several internal replicates in order to determine genotyping error rates. The inheritance pattern of each marker was checked using PedCheck (O’Connell, 1998). Triads with unresolved Mendelian inconsistencies were removed from the analysis. All 23 SNPs were consistent with Hardy-Weinberg Equilibrium (HWE) and the minor allele frequencies (MAF) ≥0.04. (MAF of 21 SNPs between 0.12 and 0.46; MAF for 2 SNPs at ENAM gene = 0.04). Further, when compared to Hapmap (latest release #28) CEU (samples with Northern and Western European ancestry) data, the MAF of all SNPs were comparable to the reference population except for SNP rs11728679 from SPP1 gene. (Minor allele C with MAF=0.41 in our data; minor allele T with MAF=0.16 in Hapmap CEU).
Table 1.
Candidate Gene Marker List*
| Gene | SNP | Base Change | Minor Allele Frequency† | SNP Location/Type |
|---|---|---|---|---|
| Dentin Sialophosphoprotein (DSPP) | rs2615487 | C-T | T (0.28) | intron |
| Tuftelin 1 (TUFT1) | rs3748609 | A-G | C (0.32) | intron |
| Tuftelin 1 (TUFT1) | rs11204846 | A-G | A (0.43) | intron |
| Tuftelin 1 (TUFT1) | rs3748608 | A-G | G (0.30) | intron |
| Tuftelin 1 (TUFT1) | rs7526319 | C-T | T (0.43) | intron |
| Tuftelin 1 (TUFT1) | rs3828054 | A-G | G (0.12) | missense Gln - Arg |
| Tuftelin 1 (TUFT1) | rs6587597 | A-G | G (0.33) | intron |
| Tuftelin 1 (TUFT1) | rs7554707 | G-T | G (0.31) | intron |
| Tuftelin 1 (TUFT1) | rs2337360 | A-G | A (0.43) | intron |
| Secreted Phosphoprotein 1 (SPP1) | rs10516800 | C-G | C (0.25) | nearest gene spp1 |
| Secreted Phosphoprotein 1 (SPP1) | rs6840362 | C-T | T (0.28) | intron |
| Secreted Phosphoprotein 1 (SPP1) | rs10516799 | C-G | G (0.29) | intron |
| Secreted Phosphoprotein 1 (SPP1) | rs11728697 | C-T | C (0.41) | intron |
| Enamelin (ENAM) | rs12640848 | A-G | A (0.33) | intron |
| Enamelin (ENAM) | rs3796704 | A-G | A (0.04) | missense Arg - Gln |
| Enamelin (ENAM) | rs7671281 | C-T | C (0.04) | missense Ile-Thr |
| Aquaporin 5 (AQP5) | rs923911 | A-C | A (0.13) | intron |
| Aquaporin 5 (AQP5) | rs1996315 | A-G | G (0.41) | between AQP6 and AQP5 |
| Matrix Metallopeptidase 20 (MMP20) | rs1784418 | C-T | T (0.47) | intron |
| Matrix Metallopeptidase 20 (MMP20) | rs2245803 | G-T | T (0.36) | missense Lys-Thr |
| Matrix Metallopeptidase 20 (MMP20) | rs7109663 | C-G | C (0.38) | nearest gene MMP20 |
| Kallikrein-related peptidase 4 (KLK4) | rs2235091 | A-G | C (0.38) | intron |
| Kallikrein-related peptidase 4 (KLK4) | rs198969 | C-G | G (0.49) | intron |
: SNP information was retrieved from Hapmap Genome Browser release #28 (http://hapmap.ncbi.nlm.nih.gov/cgi-perl/gbrowse/hapmap28_B36/)
: Allele frequencies were estimated on founders only;
Environmental Covariates
As described previously [Levy et al., 2003], demographic and environmental factors, including age, sex, race, tooth-brushing frequencies and fluoride intake from water were collected and included in our regression models. In brief, demographic information was obtained at the time of recruitment. Through questionnaires, environment factors such as tooth-brushing frequency and the fluoride intake from water (considering intake amounts and the composite fluoride concentration from all major water sources used by children) were reported by parents.
Statistical Analysis
To assess the significance of these environmental factors, we first evaluated them using linear regression (with dependent variable being the counts of tooth surfaces with caries experience) and then with logistic regression (with dependent variable being the binary variable with caries affection status: yes or no) for verification purposes.
To avoid genetic heterogeneity among different ethnic groups, all genetic analyses were limited to Caucasians only. After removing those unrelated and un-genotyped individuals, the final effective sample size in the analysis was reduced to 333 Caucasian parent-offspring trios (two parents plus one child, table 2). The Family Based Association Test (FBAT) was used to test associations between candidate genes and each of the dental caries scores. FBAT is a variation of the standard transmission disequilibrium test (TDT), in which the statistical comparison is between alleles that were transmitted to the offspring from the parents versus those not transmitted. The association test for each single SNP was performed first, followed by haplotype analyses of multiple SNPs within those genes with significant results for one or more individual SNPs. In order to detect allelic associations with both caries susceptibility and caries resistance, we evaluated each caries score twice, first assessing transmission patterns to the children affected with caries (to detect susceptibility alleles) and second assessing transmission to children with no caries (for resistance).
Table 2.
Summary of Sample Size in FBAT by Dental Caries Scores Status
| Caries Score | All surfaces | Pit & Fissure Surfaces | Smooth Surfaces |
|---|---|---|---|
| dfs=0 | 251 | 263 | 282 |
| dfs≥1 | 82 | 70 | 51 |
Permutation procedures were conducted in PLINK (plink Version 1.07, http://pngu.mgh.harvard.edu/~purcell/plink/) to generate empirical significance levels after correction of type-I error inflations from multiple testing. We only permuted phenotypes while leaving the genotypes unchanged for each person and within each family. Thus, the LD structure as well as the Mendilian consistence across markers remained the same. In order to generate the empirical significance level under the null hypothesis (i.e., no association between disease trait and marker), every phenotype was permuted for 1000 times followed by the association analyses between all permuted phenotypes and each marker (both single SNP and haplotype). Empirical p-values are calculated as (R+1)/(N+1) where R is the number of times the permuted test is greater than the observed test in our discovery analysis; N is the number of permutations (N=1000 in current study). Threshold of P=0.05 is used as the cutoff for statistical significance across all analyses.
Results
Environmental Correlates
We first used multiple linear and logistic regressions to evaluate the effects of assessed environmental factors in 575 children of all races (Table 3). Further, the analysis limited to 333 Caucasian children demonstrated very similar results (data not shown). As expected, age is positively correlated with all three caries scores in both regression models, except for pit and fissure caries with logistic regression. Figure 1 depicts in detail how the caries experiences increase by age across all three surface types. For example, the median score for smooth surface caries is 2 for children age 5 or under; this number jumps to 3 in children aged between 5 to 6 and 4 in age 6 or older. Of note, among all surface caries, pit and fissure surfaces accounted for 61% vs. 39% smooth surface caries. This is in agreement with the common observation that pit and fissure caries are more common in children.
Table 3.
P-values in Regression Analyses for Environmental and Demographic Variables Associated with Caries Scores*
| Caries Score | Age | Sex | Race | Tooth-Brushing Frequency | Fluoride Intake |
|---|---|---|---|---|---|
| D2fs-total | + (0.002/0.07) | + (0.26/0.14) | + (0.70/0.99) | − (0.02/0.07) | − (0.002/0.02) |
| D2fs-pit/fissure | + (0.006/0.13) | + (0.31/0.29) | + (0.67/0.99) | − (0.08/0.13) | − (0.003/0.008) |
| D2fs-smooth-surface | + (0.002/0.002) | + (0.28/0.17) | + (0.76/0.99) | − (0.01/0.17) | − (0.003/0.002) |
: “+” represents positive association (risk) and “−” represents negative association (protective) between covariates and caries phenotype; Entries in the table are in the format as (p-value in linear/logistic regressions); significant P values are in bold; Males and Caucasians samples were set as reference groups when studying the sex and race effects respectively.
Figure 1.
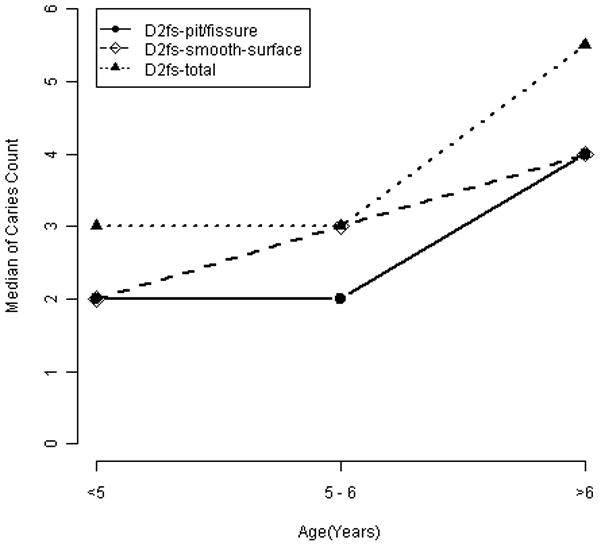
Median caries across age group by surface types *
*: Statistics are based on children with any caries experience (i.e., at least 1 caries).
Unlike previous studies which identified higher caries prevalence rates in girls, there is no significant sex difference for caries in our cohort. Similarly, there is no significant difference observed for different race groups. However, this might be due to insufficient sample size for non-Caucasian children (6%). Frequent tooth brush and higher fluoride intake from water were both found in our study to act as protective factors against caries. (Table 3)
Single SNP association analysis
Next, we performed genetic association analysis testing the possible associations between single SNPs and each caries score. As shown in Table 4, SNPs in three genes (DSPP, AQP5, and KLK4) showed consistent associations with protection against caries, for all three caries scores (nominal P≤0.05). Notably, for KLK4, the minor allele (G) was associated with increased caries risk for phenotype d2fs-smooth-surface, which supports the opposite A allele being protective. After multiple testing corrections by permutation, 6 out of total 10 significant observations above remained significant (empirical P≤0.05). In particular, association signals for SNP rs2615487 at DSPP gene are significant for all 3 caries scores before and after the permutation adjustment.
Table 4.
Single SNP Association Analyses with Dental Caries Scores*
| Caries Scores | Association Model | # of informative families | DSPP rs2615487 | AQP5 rs1996315 | KLK4 rs2235091 |
|---|---|---|---|---|---|
| D2fs-total | Risk | 49 | C(0.17) | G(0.41) | G(0.18) |
| Protective | 147 | T(0.0002/0.0009) | A(0.02/0.08) | A(0.008/0.02) | |
| D2fs-pit/fissure | Risk | 41 | C(0.24) | G(0.21) | G(0.38) |
| Protective | 154 | T(0.0004/0.002) | A(0.05/0.07) | A(0.004/0.03) | |
| D2fs-smooth surface | Risk | 28 | C(0.33) | G(0.89) | G(0.02/0.13) |
| Protective | 166 | T(0.002/0.004) | A(0.008/0.02) | A(0.02/0.08) |
: Entries in the table are the associated SNP alleles and their corresponding nominal P values and empirical P values by permutation test (in format of nominal P/empirical P); Empirical P values are reported only for those significant observations (nominal P≤0.05, highlighted in bold).
Haplotype Analysis
We conducted association tests between caries scores and haplotypes of 2 or more adjacent SNPs within the same gene. Interestingly, the haplotype analyses not only verified the importance of markers from previous single SNP analyses, but also revealed additional positive association signals: 1) For 2 SNPs (rs923911 and rs1996315) in AQP5, the first SNP demonstrated no association signals alone. However, when combined with the second SNP, both haplotypes (CA and CG) showed a consistent protective effect against caries for all 3 caries scores before multiple comparison correction. Both haplotypes remained significant for smooth surface caries after permutation adjustments (Table 5–A); 2) For 2 SNPs (rs2235091 and rs198969) in KLK4, we observed very similar patterns as were found in single SNP analysis. We identified two “protective haplotypes” (AC or GC) against all 3 caries scores and one “risk haplotype” (GG) for smooth surface caries (Table 5–B). After multiple comparison corrections, only “protective haplotype” GC and “risk haplotype” GG remained significant.
Table 5-A.
Haplotype Association Analyses of AQP5 Genes with Dental Caries Scores*
| Caries score | Effect | AQP5 Haplotype
|
p-value | |
|---|---|---|---|---|
| rs923911 allele | rs1996315 allele | |||
| D2fs-total | Protective | C | A | 0.01/0.09 |
| Protective | C | G | 0.02/0.17 | |
| D2fs-pit/fissure | Protective | C | A | 0.04/0.25 |
| Protective | C | G | 0.04/0.48 | |
| D2fs-smooth-surface | Protective | C | A | 0.003/0.01 |
| Protective | C | G | 0.004/0.03 | |
In 5-A, 5-B and 5-C, we report both nominal P values and the corresponding empirical P values by permutation test (in format of nominal P/empirical P); Significant observations (P≤0.05) are highlighted in bold.
Table 5-B.
Haplotype Association Analyses of KLK4 Genes with Dental Caries Scores
| Caries score | Effect | KLK4 Haplotype
|
p-value | |
|---|---|---|---|---|
| rs2235091 allele | rs198969 allele | |||
| D2fs-total | Protective | A | C | 0.04/0.40 |
| D2fs-pit/fissure | Protective | A | C | 0.03/0.63 |
| Protective | G | C | 0.0007/0.01 | |
| D2fs-smooth-surface | Protective | G | C | 0.002/0.01 |
| Risk | G | G | 0.03/0.01 | |
Discussion
This project is one of the first comprehensive genetic studies of dental caries aimed at better understanding the underlying environmental and genetic components by screening the significant environment correlates and investigating individual SNPs and SNP haplotypes within candidate genes. All highlighted genes reported in this paper have strong implications for their biological plausibility in pathways potentially affecting dental caries.
The DSPP (a.k.a. dentin sialophosphoprotein) gene encodes two principal proteins of the dentin extracellular matrix of the tooth: the preproprotein is secreted by odontoblasts and cleaved into dentin sialoprotein and dentin phosphoprotein. Dentin phosphoprotein is thought to be involved in the biomineralization process of dentin.[Kim et al., 2005a] Defects in this gene are the cause of many diseases such as dentinogenesis imperfecta type 1 (DGI1) and dentin dysplasia type 2 (DTDP2), both of which are autosomal dominant disorders. [Bhandari and Pannu, 2008; Lee et al., 2009] Researchers also found that mutations in the the DSPP gene lead to the production of abnormal DSPP-derived proteins or reduce the amount of these proteins in developing teeth. As a result, teeth have abnormally soft dentin. Teeth with defective dentin are discolored, weak, and prone to breakage and decay. Lee and colleagues (2006) showed that when caries reaches the dentin, the dentin-pulp complex is stimulated to form sclerotic dentin as a defensive mechanism and the DSPP gene is possibly involved in this response.[Lee et al., 2006]. In addition, another evidence also revealed that, in families with DSPP gene mutation, the softer malformed dentin is always associated with elevated risk of oral cavity.[Kim et al., 2005a]
Aquaporins encode a series of homologous membrane proteins that function as highly selective water channels. Aquaporin-5 (AQP5) is uniquely present in lacrimal and salivary glands, where it accounts for generation of saliva, tears and pulmonary secretions. [Smith et al., 1999] Also, this gene together with Aquaporin-4 has been identified as playing a role in extracellular matrix hydration during tooth development. The most striking findings of about AQP5 related to tooth decay were from Culp and colleagues[Culp et al., 2005], who observed that in AQP5 deficient (AQP5 −/−) mice, there was a significant caries susceptibility increase accompanied by reduced salivary flow. Evidence indicated the low salivary flow was caused by decreased water content of saliva.
Another gene, KLK4 was also highlighted in our study. It has been well characterized in previous study as playing pivotal roles in enamel mineralization.[Lu et al., 2008] The Kallikreins 4 (KLK4) gene is one of the fifteen kallikrein subfamily members located in a cluster on chromosome 19. The expression pattern of a similar mouse protein supports its role in the degradation of enamel proteins. During the enamel development process, MMP20 (the gene which has border-line significant P value (0.05<P<0.1) in our study, data not shown) is expressed in earlier stages of enamel development, such as the secretory and transition stages. Conversely, KLK4 is expressed later during the transition through maturation stages as the enamel hardens. The principal functions of MMP-20 and KLK4 in dental enamel formation are to facilitate the orderly replacement of organic matrix with mineral, generating an enamel layer.[Bartlett et al., 2004] Harder, less porous enamels are in turn more resistant to tooth decay. Interestingly, MMP20 and, KLK4 are implicated in hereditary enamel defects known broadly as amelogenesis imperfecta (AI) [Ozdemir et al., 2005]. Individuals with AI typically exhibit structurally abnormal enamel, which is undermineralized, and often susceptible to fracture and caries formation.
Several limitations of the study warrant discussion. First, the analytic method in this paper is a candidate gene approach where it is inherently limited by our a priori understanding of the biology of caries. A more comprehensive genome-wide approach is warranted to identify additional regions of the genome likely to contain caries genes. This hypothesis-free approach will allow for the identification of genes that may not have been obvious choices for involvement in the caries process. In this regard, our research collaboration has received funding through the NIH Genes and Environment Initiative to perform a caries genome-wide-association analysis of 550,000 SNPs within the Iowa, West Virginia, and Pennsylvania study cohorts. Second, the SNP selection is quite arbitrary and does not well-represent the full genetic variation within the gene. While SNPs were selected to provide the most genetic information, that is high heterozygosity and/or being within a large haplotype block, they likely do not represent all genetic variation for a given candidate gene. Hence lack of positive findings should be interpreted as inconclusive results. Third, applying permutation approach to correct false positive results from multiple comparisons will in no double help filter out spurious results caused by type-I error inflation. However, risk of generating possible false negative results is probably the down side of this approach (or all approaches for multiple comparison correction). This is more likely when the sample size is smaller, which is the case of our data. In our preliminary association analysis based on just nominal P values (before multiple testing correction procedures), there are a few SNPs from two more genes being highlighted, the MMP20 and TUFT1 (data not shown). MMP20 involves in the breakdown of extracellular matrix of both dentin and enamel and hence could affect the carious resistance of both tissues. The latter one, TUFT1 is also a possible carious gene due to its important role during the development and mineralization of enamel through the tooth development, but its precise function is still unclear [Deutsch et al., 2002]. Unfortunately, neither of these genes became significant after the penalty of multiple comparisons. Therefore, replication of our analysis using independent data with larger sample size is a definitely imperative task. We are currently performing the similar analysis in a cohort with over 2,300 participants from more than 700 families from the Center for Oral Health Research in Appalachia (COHRA) project. Fourth, due to the low caries frequency in our children cohort, we may not have enough power to detect the genetic loci associated with increased caries risk. This perhaps can explain why we observed much more caries protective alleles. Finally, as is well-known, dental caries experience is a chronic, cumulative disorder for which time is a very critical component that should be considered in any related research. Like most of the other studies, the cross-sectional design of our analysis work has a variety of limitations. Our previous finding that genetic determinants affecting caries might vary by dentition makes us believe different sets of genes can influence the susceptibility to caries at different time periods, even within the same dentition. Thus, studying the trajectory patterns of caries, especially how they related to the underlying genetic background, is quite valuable and exciting. Fortunately, the intrinsic longitudinal design of the Iowa Fluoride Study has offered this potential in our future work.
Acknowledgments
Grant Support
Supported by NIH grants R01-DE014889, R01-DE09551, R01-DE12101, R03DE021425, M01-RR00059; Biosciences Advantage program 5R25GM058939-08; and Dr. Levy’s Wright-Bush-Shreves Endowed Professorship
Footnotes
Declaration of Interests
All co-authors have no conflicts of interest.
References
- Bartlett JD, Beniash E, Lee DH, Smith CE. Decreased mineral content in mmp-20 null mouse enamel is prominent during the maturation stage. J Dent Res. 2004;83:909–913. doi: 10.1177/154405910408301204. [DOI] [PubMed] [Google Scholar]
- Bhandari S, Pannu K. Dentinogenesis imperfecta: A review and case report of a family over four generations. Indian J Dent Res. 2008;19:357–361. doi: 10.4103/0970-9290.44543. [DOI] [PubMed] [Google Scholar]
- Bretz WA, Corby PM, Melo MR, Coelho MQ, Costa SM, Robinson M, Schork NJ, Drewnowski A, Hart TC. Heritability estimates for dental caries and sucrose sweetness preference [see comment] Arch Oral Biol. 2006;51:1156–1160. doi: 10.1016/j.archoralbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Burt BA. Trends in caries prevalence in north american children. Int Dent J. 1994;44:403–413. [PubMed] [Google Scholar]
- Culp DJ, Quivey RQ, Bowen WH, Fallon MA, Pearson SK, Faustoferri R. A mouse caries model and evaluation of aqp5−/− knockout mice. Caries Res. 2005;39:448–454. doi: 10.1159/000088179. [DOI] [PubMed] [Google Scholar]
- Deutsch D, Dafni L, Palmon A, Hekmati M, Young MF, Fisher LW. Tuftelin: Enamel mineralization and amelogenesis imperfecta. Ciba Foundation Symposium. 1997;205:135–147. doi: 10.1002/9780470515303.ch10. discussion 147–155. [DOI] [PubMed] [Google Scholar]
- Deutsch D, Leiser Y, Shay B, Fermon E, Taylor A, Rosenfeld E, Dafni L, Charuvi K, Cohen Y, Haze A, Fuks A, Mao Z. The human tuftelin gene and the expression of tuftelin in mineralizing and nonmineralizing tissues. Connect Tissue Res. 2002;43:425–434. doi: 10.1080/03008200290001186. [DOI] [PubMed] [Google Scholar]
- Downer MC. Caries prevalence in the united kingdom. Int Dent J. 1994;44:365–370. [PubMed] [Google Scholar]
- Drewnowski A. Taste preferences and food intake. Annu Rev Nutr. 1997;17:237–253. doi: 10.1146/annurev.nutr.17.1.237. [DOI] [PubMed] [Google Scholar]
- Ferraro M, Vieira AR. Explaining gender differences in caries: A multifactorial approach to a multifactorial disease. International Journal of Dentistry. 2010 doi: 10.1155/2010/649643. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira SH, Beria JU, Kramer PF, Feldens EG, Feldens CA. Dental caries in 0- to 5-year-old brazilian children: Prevalence, severity, and associated factors. Int J Paediatr Dent. 2007;17:289–296. doi: 10.1111/j.1365-263X.2007.00831.x. [DOI] [PubMed] [Google Scholar]
- Guzman-Armstrong S. Rampant caries. J Sch Nurs. 2005;21:272–278. doi: 10.1177/10598405050210050501. [DOI] [PubMed] [Google Scholar]
- Hunter PB. Risk factors in dental caries. Int Dent J. 1988;38:211–217. [PubMed] [Google Scholar]
- Keene HJ, Mellberg JR, Pederson ED. Relationship between dental caries experience and surface enamel fluoride concentration in young men from three optimally fluoridated cities. J Dent Res. 1980;59:1941–1945. doi: 10.1177/00220345800590110301. [DOI] [PubMed] [Google Scholar]
- Kim JW, Hu JC, Lee JI, Moon SK, Kim YJ, Jang KT, Lee SH, Kim CC, Hahn SH, Simmer JP. Mutational hot spot in the dspp gene causing dentinogenesis imperfecta type ii. Hum Genet. 2005;116:186–191. doi: 10.1007/s00439-004-1223-6. [DOI] [PubMed] [Google Scholar]
- Lee SK, Lee KE, Jeon D, Lee G, Lee H, Shin CU, Jung YJ, Lee SH, Hahn SH, Kim JW. A novel mutation in the dspp gene associated with dentinogenesis imperfecta type ii. J Dent Res. 2009;88:51–55. doi: 10.1177/0022034508328168. [DOI] [PubMed] [Google Scholar]
- Lee YL, Liu J, Clarkson BH, Lin CP, Godovikova V, Ritchie HH. Dentin-pulp complex responses to carious lesions. Caries Res. 2006;40:256–264. doi: 10.1159/000092235. [DOI] [PubMed] [Google Scholar]
- Levy SM, Warren JJ, Broffitt B, Hillis SL, Kanellis MJ. Fluoride, beverages and dental caries in the primary dentition. Caries Res. 2003;37:157–165. doi: 10.1159/000070438. [DOI] [PubMed] [Google Scholar]
- Liu H, Deng H, Cao CF, Ono H. Genetic analysis of dental traits in 82 pairs of female-female twins. Chin J Dent Res. 1998;1:12–16. [PubMed] [Google Scholar]
- Lu Y, Papagerakis P, Yamakoshi Y, Hu JC, Bartlett JD, Simmer JP. Functions of klk4 and mmp-20 in dental enamel formation. Biol Chem. 2008;389:695–700. doi: 10.1515/BC.2008.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs JR, Largaespada LL. Explaining sex differences in dental caries prevalence: Saliva, hormones, and “life-history” etiologies. Am J Human Biol. 2006;18:540–555. doi: 10.1002/ajhb.20530. [DOI] [PubMed] [Google Scholar]
- Luo W, Wen X, Wang HJ, MacDougall M, Snead ML, Paine ML. In vivo overexpression of tuftelin in the enamel organic matrix. Cells Tissues Organs. 2004;177:212–220. doi: 10.1159/000080134. [DOI] [PubMed] [Google Scholar]
- Menghini G, Steiner M, Imfeld T. Early childhood caries--facts and prevention. Ther Umsch. 2008;65:75–82. doi: 10.1024/0040-5930.65.2.75. [DOI] [PubMed] [Google Scholar]
- Ozdemir D, Hart PS, Ryu OH, Choi SJ, Ozdemir-Karatas M, Firatli E, Piesco N, Hart TC. Mmp20 active-site mutation in hypomaturation amelogenesis imperfecta [see comment] J Dent Res. 2005;84:1031–1035. doi: 10.1177/154405910508401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patir A, Seymen F, Yildirim M, Deeley K, Cooper ME, Marazita ML, Vieira AR. Enamel formation genes are associated with high caries experience in turkish children. Caries Res. 2008;42:394–400. doi: 10.1159/000154785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuler CF. Inherited risks for susceptibility to dental caries. J Dent Educ. 2001;65:1038–1045. [PubMed] [Google Scholar]
- Slayton RL, Cooper ME, Marazita ML. Tuftelin, mutans streptococci, and dental caries susceptibility. J Dent Res. 2005;84:711–714. doi: 10.1177/154405910508400805. [DOI] [PubMed] [Google Scholar]
- Smith JK, Siddiqui AA, Modica LA, Dykes R, Simmons C, Schmidt J, Krishnaswamy GA, Berk SL. Interferon-alpha upregulates gene expression of aquaporin-5 in human parotid glands. J Interferon Cytokine Res. 1999;19:929–935. doi: 10.1089/107999099313479. [DOI] [PubMed] [Google Scholar]
- Stookey GK. The effect of saliva on dental caries. J Am Dent Assoc. 2008;139 (Suppl):11S–17S. doi: 10.14219/jada.archive.2008.0347. [DOI] [PubMed] [Google Scholar]
- Verrips GH, Frencken JE, Kalsbeek H, ter Horst G, Filedt Kok-Weimar TL. Risk indicators and potential risk factors for caries in 5-year-olds of different ethnic groups in amsterdam. Community Dent Oral Epidemiol. 1992;20:256–260. doi: 10.1111/j.1600-0528.1992.tb01694.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Shaffer JR, Weyant RJ, Cuenco KT, Desensi RS, Crout R, McNeil DW, Marazita ML. Genes and their effects on dental caries may differ between primary and permanent dentitions. Caries Res. 2010;44:277–284. doi: 10.1159/000314676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren JJ, Levy SM, Broffitt B, Kanellis MJ. Longitudinal study of non-cavitated carious lesion progression in the primary dentition. J Public Health Dent. 2006;66:83–87. doi: 10.1111/j.1752-7325.2006.tb02560.x. [DOI] [PubMed] [Google Scholar]
- Wendell S, Wang X, Brown M, Cooper ME, Desensi RS, Weyant RJ, Crout R, McNeil DW, Marazita ML. Taste genes associated with dental caries. J Dent Res. 2010 doi: 10.1177/0022034510381502. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werneck R, Mira M, Trevilatto P. A critical review: An overview of genetic influence on dental caries. Oral Diseases. 2010;16:613–623. doi: 10.1111/j.1601-0825.2010.01675.x. [DOI] [PubMed] [Google Scholar]


