Abstract
Gene regulatory factors encoded by the nuclear genome are essential for mitochondrial biogenesis and function. Some of these factors act exclusively within the mitochondria to regulate the control of mitochondrial transcription, translation and other functions. Others govern the expression of nuclear genes required for mitochondrial metabolism and organelle biogenesis. The peroxisome proliferator-activated receptor gamma coactivator-1 (PGC-1) family of transcriptional coactivators plays a major role in transducing and integrating physiological signals governing metabolism, differentiation and cell growth to the transcriptional machinery controlling mitochondrial functional capacity. Thus, the PGC-1 coactivators serve as a central component of the transcriptional regulatory circuitry that coordinately controls the energy-generating functions of mitochondria in accordance with the metabolic demands imposed by changing physiological conditions, senescence, and disease.
Keywords: Mitochondria, Peroxisome proliferator-activated receptor gamma coactivator 1 (PGC-1), Peroxisome proliferator-activated receptor (PPAR), transcription, gene regulation, metabolism
Regulation of mitochondrial biogenesis
A set of nuclear-encoded factors coordinately regulate mitochondrial mass and function in response to a host of energy and growth demands, including cellular metabolic stress, such as the constant production of reactive oxygen species (ROS). Dysregulation of mitochondrial function has broad implications for human disease including diabetes, heart failure and neurodegeneration. Thus, there is a high level of interest in developing therapeutic strategies aimed at modulating the regulatory pathways that control mitochondrial function and biogenesis. The nuclear-encoded transcription factors and coactivators that govern mitochondrial function serve as one major focus. Here we review recent advances in our understanding of this regulatory circuitry and its potential role in integrating mitochondrial biogenesis with cellular stress responses.
Transcriptional circuitry controlling mitochondrial biogenesis
Factors acting upon the mitochondrial genome
The transcription and translation of the mitochondrial genome is dependent upon a host of nuclear-encoded gene products (Figure 1; Box 1). Mitochondrial DNA (mtDNA) transcription requires a single RNA polymerase (POLRMT, see glossary) that shares sequence similarity with yeast mitochondrial and T3/T7 bacteriophage polymerases, two stimulatory transcription factors (Tfam, TFB2M) and a termination factor (MTERF1) [1, 2]. Transcription occurs bi-directionally from divergent promoters termed LSP and HSP within the D-loop regulatory region. Because of the semi-autonomous nature of mitochondria, interest has focused on a number of nuclear-encoded gene products that act exclusively within the organelle to regulate the mitochondrial genetic system. The first of these was Tfam, a high mobility group (HMG)-box protein that stimulates bidirectional transcription through specific promoter recognition [3].
Figure 1. Nuclear-encoded factors acting within mitochondria.
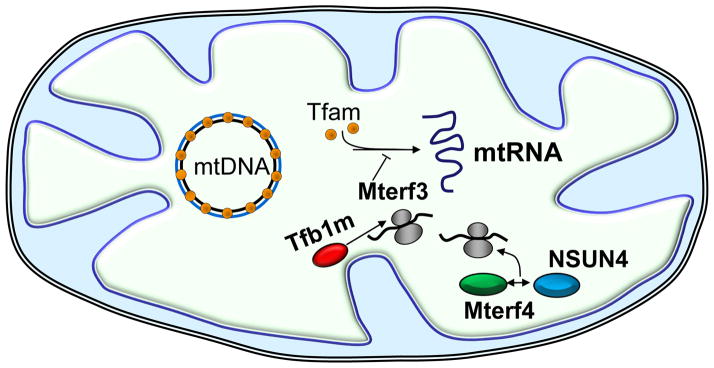
Targeted gene disruptions in mice (knockouts) have helped define the functions of several nuclear genes encoding products that function within mitochondria (as described in the text) including: Tfam (orange spheres), binds the mtDNA at multiple sites and functions in both mtDNA maintenance and transcription initiation; Mterf3, functions as a negative regulator of mtDNA transcription; Tfb1m (red ellipse) and Mterf4 (green ellipse), participate in mitochondrial ribosome assembly. Tfb1m is a dimethyltransferase that catalyzes the adenine dimethylation of the small ribosomal RNA required for ribosome assembly and translation. Similarly, a complex containing Mterf4 and the rRNA methyltransferase, NSUN4 (blue ellipse), participates in the assembly of the large ribosomal subunit.
Box 1. Mitochondrial structure and function.
Mitochondria engage an array of biochemical activities and are the major sites of oxidative ATP production in eukaryotic cells [94]. The organelle consists of a soluble matrix bounded by a double membrane, an inner ion-impermeable membrane, and an outer membrane permeable to molecules of molecular mass up to approximately 5 kd. The electron donors, NADH and FADH2, derived from the oxidation of acetyl-CoA, are utilized by the electron transport chain of the mitochondrial inner membrane to establish an electrochemical proton gradient across the membrane. The resulting proton motive force, comprised of both a voltage potential and a pH gradient, is used by the membrane bound ATP synthase to drive the synthesis of ATP [94] or by uncoupling proteins to generate heat [95]. Although the mitochondrial electron transport chain efficiently delivers electron pairs to molecular oxygen, the terminal acceptor, occasionally molecular oxygen becomes partially reduced by a single electron forming superoxide anion, a highly reactive and toxic species.
According to the endosymbiont hypothesis, mitochondria originated from the engulfment of aerobic eubacteria by a primordial anaerobic eukaryote, an event possibly coincident with the origin of eukaryotic cells [96]. As a result, the organelle has its own genetic system with several bacteria-like features including a compact circular DNA genome (mtDNA), a simple transcription system that produces multigenic RNA transcripts, and a translational apparatus with antibiotic sensitivities similar to prokaryotic cells. Gene loss to the nucleus over evolutionary time left the mammalian organelle with only 37 genes. Only 13 mitochondrial genes encode proteins, all of which are essential subunits of the respiratory chain. The remaining genes specify tRNAs and rRNAs required for protein translation within the mitochondrial matrix [97]. Nearly the same small complement of mitochondrial genes exists over the entire evolutionary spectrum and, thus, nuclear genes control mitochondrial transcription, translation and DNA replication. They also provide the vast majority of gene products required for the biochemical functions and molecular architecture of the organelle.
Loss-of-function studies have demonstrated the critical need for the eukaryotic cell to promote mtDNA transcription and mitochondrial function early in embryonic development. Embryonic lethality in germline knockouts with severe respiratory chain defects associated with abundant atypical mitochondria has been observed upon the ablation of Tfam and other nuclear-encoded factors that function within the mitochondria. A germline Tfam knockout mouse exhibited embryonic lethality at E10.5 associated with a severe oxidative phosphorylation defect and a marked reduction in mtDNA content, demonstrating a requirement for Tfam in mtDNA maintenance in vivo [4]. Embryonic lethality also occurs with germline knockout for Mterf3, encoding a negative regulator of mitochondrial transcription initiation [5], Tfb1m, encoding a dimethyltransferase [6], and, most recently, Mterf4, encoding a regulator of ribosome biogenesis and translation through its actions with the rRNA methyltransferase NSUN4 [7]. Loss of these functions is often accompanied by defects in respiratory complexes I, III, IV and V which rely upon mitochondrial-encoded subunits. Interestingly, mice with a homozygous germline knockout of Ant1, encoding an adenine nucleotide translocator, also exhibit ragged red muscle fibers with abundant abnormal mitochondria and their hearts display cardiac hypertrophy with massive mitochondrial proliferation [8]. The mitochondrial proliferation likely represents a compensatory response.
Transcription factors acting upon the nuclear genome
The nuclear respiratory factors, NRF-1 and NRF-2, were the first nuclear transcription factors implicated in the expression of multiple mitochondrial functions in vertebrates. NRF-1, initially identified through its binding to the cytochrome c promoter, functions as a positive regulator of transcription [9]. It acts on genes encoding respiratory subunits [10] as well as Tfam [9]and both TFB isoform genes [11] whose products (as discussed) are major regulators of mitochondrial transcription and ribosome assembly [6, 12, 13]. Human NRF-2 was identified as a multi-subunit transcriptional activator of the cytochrome oxidase subunit IV (COXIV) promoter and its mouse ortholog is GABP, an ETS-domain transcription factor[9]. The human protein has a DNA-binding α subunit and four others (β1,β2, γ1 and γ2), that complex with α to provide an activation domain and to modulate binding affinity. Evidence points to a direct role for both NRF-1 and NRF-2 in the expression of all 10 nucleus-encoded cytochrome oxidase subunits [14, 15]. Both factors also participate in the expression of the mitochondrial import receptor complex and of COX17, a putative cytochrome oxidase assembly factor [9]. Control of the mitochondrial transcription and import machinery by both NRFs may be part of a mechanism coordinating the expression of the respiratory chain with the biogenesis of the organelle itself. Genetic deletion of the NRFs and other nuclear respiratory factors, such as the transcriptional repressor protein YY1, results in peri-implantation lethality [16–18]. NRF-1 null blastocysts failed to progress beyond E6.5 and had depleted mitochondrial DNA levels and diminished mitochondrial membrane potential, consistent with a respiratory chain deficiency [16]. In keeping with a potential link between mitochondrial biogenesis and cell cycle progression, mouse embryo fibroblasts lacking GABPα (NRF-2α) failed to proliferate because of an inability to replicate DNA [19].
Members of the nuclear receptor (NR) superfamily also play a key role in the transcriptional control of nuclear genes encoding mitochondrial enzymes and proteins. The peroxisome proliferator-activated receptor α (PPARα) was the first NR shown to be involved in the transcriptional control of mitochondrial metabolism. Originally discovered as a regulator of genes encoding peroxisomal fatty acid oxidation (FAO) enzymes, PPARα is now known to coordinately regulate nuclear genes encoding mitochondrial FAO enzymes, reviewed in [20]. PPARα is a member of a family of related NRs including the ubiquitously-expressed PPARβ (also known as PPARδ) and PPARγ, an adipose-enriched transcription factor involved in adipocyte differentiation and the target of the insulin-sensitizing thiazolidinediones. The PPARs bind cognate DNA response elements as heterodimers with members of the retinoid X receptor (RXR) family. An interesting feature of the PPARs is that they are activated by a variety of fatty acid-containing lipid ligands, yet, the endogenous ligands for the PPARs have not been fully delineated. The activation of PPARα and PPARβ by lipid ligands provides a mechanism for transducing changes in cellular lipid metabolism to the transcriptional control of mitochondrial FAO, a key source of ATP production in heart and muscle [20].
A second family of NRs, the estrogen-related receptors or ERRs (ERRα, ERRβ, and ERRγ), have also been shown to regulate nuclear genes encoding mitochondrial proteins involved in FAO, TCA cycle, respiratory chain, and oxidative phosphorylation. Functional genomic analysis involving the genome-wide location of transcription factor occupancy has revealed that the ERRs are associated with hundreds of genes controlling all aspects of mitochondrial function [21], including a subgroup involved in FAO that overlap with PPAR targets [22]. In addition, ERRα can regulate the transcription of the PPARα gene itself [23], forming a potential cross-regulatory loop to amplify the subset of ERR targets involved in mitochondrial FAO under certain circumstances. All three ERRs are expressed in mitochondrial-rich tissues such as heart and skeletal muscle, with ERRγ being selectively expressed in oxidative muscles enriched in type I and IIa fibers [24]. ERRα null mice display a number of cardiac deficiencies when stressed by pressure overload but apparently lack any robust defect in organelle biogenesis [25], while ERRγ knockouts exhibit early postnatal lethality associated with a defect in oxidative phosphorylation in the heart [26]. Consistent with these findings, transgenic overexpression of ERRγ in skeletal muscle also promotes an oxidative phenotype with enhanced expression of mitochondrial FAO and respiratory chain genes [24, 27]. These mice also display an increase in the proportion of type I fibers suggesting a coordinate regulation of fiber type determination and oxidative metabolism by ERRγ.
Several additional nuclear transcription factors have been linked to the expression of the respiratory apparatus including YY1, MEF2 and c-myc. YY1 was first associated with the expression of cytochrome oxidase subunits Vb and VIIc [28, 29] and subsequent analysis of 723 human core promoters revealed a preponderance of YY1 sites in nuclear genes encoding ribosomal subunits and mitochondrial proteins [30]. Functional studies have demonstrated that YY1 is an important component of nutrient sensing by the mammalian target of rapamycin (mTOR). Inhibition of mTOR by rapamycin decreased the expression of ERRα and the NRFs and blocked the interaction between YY1 and one of its coactivators, PGC-1α. In addition, YY1 silencing diminished both mitochondrial gene expression and respiration [31]. Tissue-specific expression of at least some muscle-specific cytochrome oxidase subunits depend upon MEF2/Ebox recognition sites [32]. NRF-1 regulates MEF2A in muscle possibly accounting for indirect NRF-1 control over the expression of muscle-specific MEF2 target genes [33]. Finally, c-myc acts on the expression of certain NRF-1 target genes through a canonical NRF-1 binding site resulting in the sensitization of cells to apoptosis [34]. Expression of a dominant-negative allele of NRF-1 blocked the induction of apoptosis by c-myc.
It is important to note that many target genes for ERR and the other nuclear factors controlling mitochondrial function have been identified largely by functional genomic analysis. Although functional associations have been elucidated in some cases [35, 36], there is a relative paucity of information regarding the direct and quantitative effects of a given factor on mitochondrial protein expression, complex assembly or metabolic flux through a specific pathway. This may be especially significant for the respiratory complexes which are thought to be in excess and thus must suffer substantial reductions in expression before the threshold for a respiratory defect is crossed [37].
Integration of mitochondrial biogenic regulatory pathways: The PGC-1 coactivators
A major breakthrough in the understanding of how transcription factors controlling mitochondrial biogenesis are co-regulated came with the discovery of PPARγ coactivator-1α(PGC-1α). PGC-1α was first identified as a coactivator of the adipocyte-enriched nuclear receptor, PPARγ, in brown adipocytes [38]. PGC-1α is a member of a family of transcriptional coactivators that includes the closely-related PGC-1β [39, 40] and more distant relative, PGC-1 related coactivator (PRC) [10]. All three PGC-1 members are characterized by an N-terminal activation domain near leucine-rich LXXLL motifs that mediate interaction with nuclear receptors, an RNA recognition domain (RRM) and a host cell factor-1 (HCF) binding domain. In addition, PGC-1α contains an RNA splicing domain (RS), the function of which has not been fully defined.
Forced overexpression studies conducted in cultured adipocyte lines [38], cardiac myocytes [41] and in conditional, tissue-specific, transgenic mice [42] have shown that PGC-1α is capable of driving virtually all aspects of mitochondrial biogenesis, including activation of respiratory chain and FAO genes, increased mitochondrial number, and augmentation of mitochondrial respiratory capacity. As shown in Figure 2, PGC-1α exerts these effects through direct interaction with, and coactivation of PPARs, ERRs, and NRF-1/NRF-2 among other transcription factors [43]. PGC-1β has been shown to interact with and coactivate many of the same transcription factors as PGC-1α[40]. As such, it is not surprising that it appears that the downstream gene targets have significant overlap with PGC-1α [44, 45]. However, it is likely that at least a subset of the upstream regulatory pathways controlling the expression and activity of PGC-1αand PGC-1β, including those involved in cell- and tissue-specific expression, are distinct.
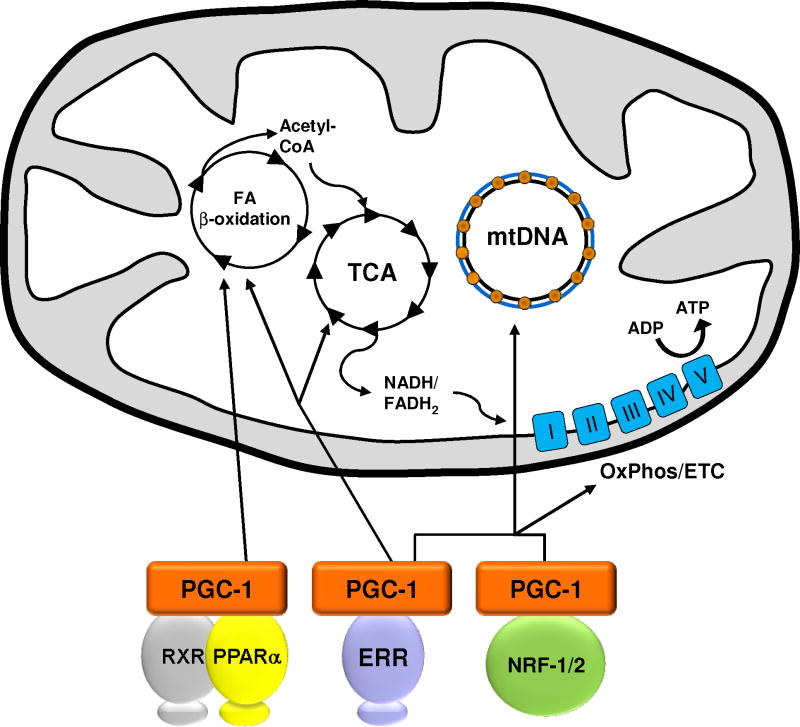
Figure 2. Control of mitochondrial biogenesis by PGC-1.
Interaction between PGC-1 and specific transcription factors orchestrates major functions of mitochondria including fatty acid β-oxidation, the tricarboxylic acid cycle (TCA), mtDNA replication and oxidative phosphorylation and the electron transport chain (OxPhos/ETC) in addition to biogenesis of this organelle. PPARα interacts with its binding partner, the retinoid X receptor (RXR), to control the expression of many fatty acid β-oxidation enzymes. NRF-1 and NRF-2 contribute to the maintenance of mtDNA and the expression of multiple components of the ETC. ERR members regulate the expression of virtually all functions of the mitochondria including those shown here.
The third PGC-1 family member, PGC-1 related coactivator (PRC), was discovered by a database search for sequence similarities with the carboxy-terminal RS domain and RNA recognition motif of PGC-1α [23]. Delineation of the entire PRC amino acid sequence revealed additional similarities with PGC-1α and PGC-1β including an acidic amino-terminal activation domain, an LXXLL coactivator signature motif and a central proline rich region. Like PGC-1α and β, PRC binds nuclear transcription factors implicated in the expression of mitochondrial function including NRF-1, CREB and ERRα [11, 46, 47]. PRC silencing in cultured cells results in reduced respiratory chain expression and ATP production along with abundant abnormal mitochondria [48, 49]. These phenotypes resemble those observed upon tissue-specific disruption of Tfam, Tfb1m, Mterf3 and Mterf4 whose products are localized to mitochondria and are required by the mitochondrial genetic system [4, 5, 7, 50]. PRC can also direct mitochondrial biogenesis through its interaction with and activation of ERRα in cultured thyroid cells [51].
PRC expression during the initiation of cell growth is characteristic of the immediate early genes [23, 46], a family of genes that orchestrates the early response to growth factors [52, 53]. Notably, PRC expression peaks during the first day of embryoid body formation [54], an event reminiscent of the early induction of PRC in response to serum growth factors in cultured cells [23, 46]. A germline knockout of the PRC gene in mice results in peri-implantation lethality [54], a phenotype similar to that observed in mouse knockouts of NRF-1, and several other essential transcription factors associated with mitochondrial biogenesis [9]. Thus, PRC contrasts with the other PGC-1 family members in that it is essential for early embryonic development. Among other growth-related functions, PRC may support the high rate of mitochondrial transcription occurring during early embryogenesis.
PGC-1 coactivators are inducible responders to cellular energetic and metabolic stress
Integration of mitochondrial biogenesis and metabolic signaling
The PGC-1 coactivators are dynamically regulated at the level of mRNA and protein expression in response to a variety of signaling pathways (e.g. AMPK, CaMK) involved in cellular growth, differentiation, and energy metabolism [2, 55]. These include pathways that often converge on the CREB-dependent induction of PGC-1α gene transcription in response to cold exposure, nutrient deprivation and exercise [56]. The importance of this response has been confirmed in PGC-1knockout mice. Mice with germline targeted disruptions of either PGC-1α [57, 58] or PGC-1β [59, 60] are viable and fertile with essentially normal energy expenditure, mitochondrial abundance and morphology. However, under physiological stress the single PGC-1 knockouts exhibit significant phenotypes. For example, the PGC-1α-deficient mouse is unable to maintain body temperature upon cold exposure and has reduced capacity for maintaining cardiac output in response to physiological and pathophysiologic stress [57, 58, 61]. PGC-1β knockout mice also exhibit mitochondrial dysfunction under stress conditions with diminished expression of nuclear-encoded respiratory proteins [45, 59]. In striking contrast to the singly PGC-1α- and PGC-1β-deficient mice, PGC-1α/β double knockout mice die of cardiac failure within 24 hours of birth [45]. The lethal heart failure of these mice was due to arrest in the known perinatal surge in mitochondrial biogenesis. The results indicate that the two coactivators provide complementary functions in supporting perinatal cardiac mitochondrial biogenesis and function.
Metabolic signaling through PGC-1α occurs, in part, through post-translational modifications (Figure 3). For example, PGC-1α is directly regulated by deacetylation (SIRT1) and phosphorylation (AMPK) in response to changes in nutrient or energy depletion [62, 63]. The SirT and AMPK pathways can also cooperate in promoting calcium dependent mitochondrial biogenesis in myocytes [64, 65]. The control of PGC-1α by AMPK and SIRT1 is suggestive of a compelling link between mitochondrial biogenesis and metabolic signaling pathways. In mice, ablation of SIRT1 renders the animals impervious to the effects of calorie restriction whereas its overexpression promotes the beneficial effects of CR even when the mice are fed normally [66]. Pathways that inhibit PGC-1 signaling have also been unveiled. Acetylation of PGC-1α by GCN5 (Figure 3) represses its activity through target transcription factors [67]. The coactivator SRC-3 induces GCN5 expression in response to caloric excess thereby inhibiting PGC-1α and energy expenditure [68].
Figure 3. Proposed integration of energy-generating and antioxidant pathways by PGC-1α.
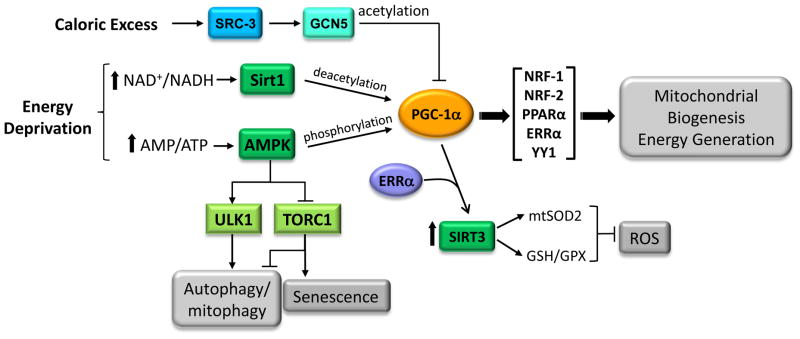
PGC-1α is activated via post-transcriptional phosphorylation by AMPK or by deacetylation via SIRT1 in response to nutrient deprivation. Induction or activation of the coactivator can enhance mitochondrial biogenesis and oxidative function through the coactivation of multiple transcription factors involved in respiratory gene expression (see Figure 2). PGC-1α activation may also promote an antioxidant environment by coactivating ERRα to induce SIRT3, a mitochondrial sirtuin that has been implicated in ROS detoxification. In addition, AMPK promotes autophagy through direct phosphorylation of ULK1 or suppression of the TORC1 kinase complex. Inhibition of the TORC1 pathway also has been shown to have a negative effect on senescence. Increased autophagy, enhanced ROS detoxification and inhibition of TORC1 are all associated with health and longevity. Under conditions of caloric excess, SRC-3 induces GCN5 which inactivates PGC-1α through acetylation.
AMPK serves as a link between PGC-1α-regulated mitochondrial biogenesis and the target of rapamycin complex 1 (TORC1). Downregulation of TORC1 is thought to promote autophagy, including mitophagy, to provide nutrients under adverse metabolic conditions. AMPK can inhibit cell growth and promote autophagy through its inhibition of TORC1 by direct phosphorylation of the TORC1 subunit raptor (regulatory associated protein of mTOR) [69] and by phosphorylation of the upstream regulator, TSC2 [70]. In addition, AMPK can stimulate the destruction of defective mitochondria through mitophagy by activating the ULK complex through direct phosphorylation [71, 72]. Thus, AMPK can potentially enhance the synthesis of new mitochondria through its activation of PGC-1α and at the same time promote the clearance of defective mitochondria while suppressing cell growth. The results are suggestive of an intricate inter-relationship between the transcriptional control of mitochondrial biogenesis through PGC-1α and its target transcription factors and key metabolic signaling pathways.
Integration of mitochondrial function and oxidative stress
Significant evidence points toward the central role of mitochondria in human aging and disease. The free radical theory of aging, originally advanced by Harman [73], attributes oxidant production by the mitochondrial respiratory chain as the essential feature of a vicious cycle of free radical damage to cellular constituents, leading to tissue aging. In addition, according to the Warburg hypothesis, a defining feature of cancer is diminished mitochondrial function accompanied by aerobic glycolysis, the propensity to high glycolytic activity in the presence of oxygen [74]. Indeed, mitochondrial deficiencies are now linked to the major classes of age-related disease including diabetes, neurodegenerative disease, cardiac disease and cancer [75–78]. Emerging evidence suggests that the PGC-1 coactivators link mitochondrial biogenesis and the response to oxidant stress. SIRT3 is a member of the sirtuin family of deacetylases that is localized to mitochondria where it modifies key enzymatic activities in optimizing metabolic function [79]. PGC-1α coactivates the expression of SIRT3 through ERRα which binds the SIRT3 proximal promoter [80, 81]. Importantly, SIRT3 is required for the induction of several components of the ROS detoxifying machinery by PGC-1α. Since PGC-1α is activated by SIRT1-mediated deacetylation in response to nutrient deprivation [82], PGC-1α can potentially coordinate nutrient sensing pathways and the oxidant scavenging mechanisms facilitated by SIRT3. This relationship may contribute to the positive effects on longevity associated with the restriction of caloric intake [83].
PRC may also function as a sensor of metabolic stress (Figure 4). The robust upregulation of PRC protein levels in response to multiple metabolic insults mediates the induction of a battery of genes involved in inflammation, cell stress, and proliferation [84]. The PRC-dependent inflammatory/stress genes include pro-inflammatory molecules (e.g. IL-1α, IL-8, cyclooxygenase 2) as well as members of the proline-rich protein family (SPRR2D and SPRR2F). The latter are associated with the response to DNA damage and exit from the cell cycle and also provide a protective anti-oxidant barrier to cellular damage, thereby promoting tissue remodeling [85, 86]. Several of the genes are also associated with the inflammatory microenvironment in a number of human cancers [77, 87] and with cellular senescence [88, 89]. This is consistent with the observations that c-myc is co-induced along with PRC by chemical uncoupling [84] and that PRC is markedly elevated in several human tumors [9,90]. Interestingly, the induction of PRC by respiratory chain uncoupler and the associated inflammatory/stress response is sensitive to antioxidant [84]. By contrast, PGC-1α promotes an antioxidant environment through SIRT3 in the context of the increased mitochondrial fatty acid oxidation precipitated by nutrient limitation [62]. This may exert positive effects on energy balance and longevity through increased mitochondrial biogenesis and ROS detoxification. However, under conditions of severe stress, possibly resulting from genetic mutations, environmental toxins, or oncogene activation, dysregulation of PRC may contribute to the induction of an inflammatory/stress response. It is tempting to speculate that the balance between the PGC-1α/SIRT3 and PRC pathways is reflected by the reduced levels of SIRT3 in human cancer cells [92, 93].
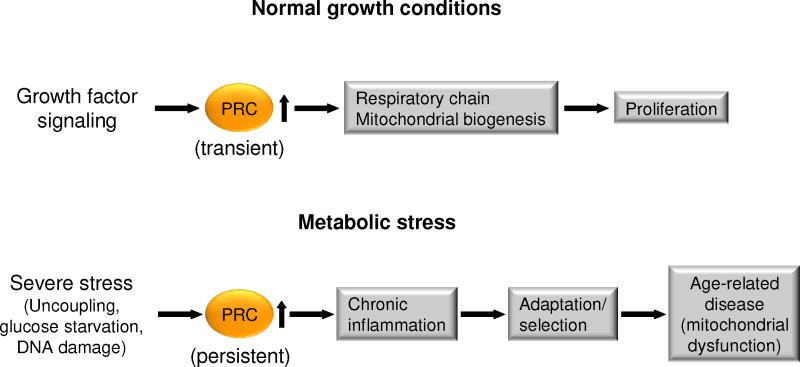
Figure 4. PRC response to growth and metabolic stress.
Under normal growth conditions, PRC acts as an immediate early gene product that is transiently elevated upon the initiation of cell growth. The transient induction of PRC and other immediate early genes is an early event in the program leading to cell proliferation. Under conditions of severe stress, resulting from chemical uncoupling or energy starvation, PRC is induced constitutively in cultured cells as a dysregulated response. This latter response is a departure from its normal expression pattern and leads to the induction of a battery of genes associated with the chronic inflammatory response. The induction of this inflammatory response is thought to be adaptive at the cellular level but may be maladaptive at the organismal level and thus associated with a number of age-related diseases.
Concluding Remarks
Regulatory factors encoded by the nuclear genome are essential to the control of mitochondrial biogenesis and function. The PGC-1 family of coactivators plays an integrative role in linking developmental and physiological signals to a key subset of transcription factors directing the coordinate expression of nuclear and mitochondrial genes. This regulatory circuitry is critical for adjusting mitochondrial function to meet the changing energetic demands for cellular growth, differentiation and development (including longevity). Recent evidence also suggests that PGC-1 family members serve as sensors of metabolic stress. PGC-1α, as a substrate for both AMPK and SIRT1, can promote an antioxidant response to nutrient deprivation, possibly contributing to the longevity-enhancing effects of caloric restriction. PRC functions as an immediate-early gene during cell cycle entry but may also orchestrate an oxidant-sensitive inflammatory response to severe metabolic stress. Collectively, these results indicate that the PGC-1 family and their target transcription factors participate in the fine tuning of both the energy-generating machinery in response to nutrient availability and defenses against metabolic stress. It will be of interest to explore the pathophysiologic implications of these molecular regulatory connections to the cellular redox environment associated with longevity, senescence and disease. A future challenge for the field is to assess and develop effective strategies to modulate this circuitry as a therapeutic intervention for the wide array of diseases that involve mitochondrial dysfunction such as heart failure, diabetes and neurologic diseases.
Acknowledgments
This work was supported by NIH grants (R01 DK045416, R01 HL058493, and R01 HL101189 [D.P.K.]), and by a National Institute of General Medical Sciences grant (R01 GM32525-28 [R.C.S]). We wish to thank Teresa Leone for critical reading and Lorenzo Thomas for assistance in preparation of the manuscript.
Glossary
- AMP-activated protein kinase (AMPK)
an energy-sensing kinase that directly phosphorylates and enhances the activity of PGC-1
- Dimethyladenosine transferase 1, mitochondrial (Tfb1m)
a mitochondrial methyltransferase that dimethylates 12S rRNA and controls the stability or assembly of the mitochondrial ribosome
- Estrogen related receptors (ERRs)
a family of orphan nuclear receptors that are involved in the regulation of virtually all aspects of mitochondrial function and biogenesis
- General control of amino-acid synthesis (GCN5)
an acetyltransferase that acetylates and inhibits activity of PGC-1α and β
- Nuclear respiratory factor 2 (NRF2) or GA-binding protein (GABP)
a multi-subunit transcription factor that is involved in cytochrome oxidase expression and the nuclear control of mitochondrial function
- Nuclear respiratory factor 1 (NRF-1)
a transcription factor that functions as a homodimer and activates the expression of key metabolic genes regulating cellular growth and nuclear genes required for respiration, heme biosynthesis, and mitochondrial DNA transcription and replication
- Peroxisome proliferator-activated receptors (PPARs)
a family of ligand-activated nuclear receptors that regulate various aspects of lipid and fatty acid metabolism as well as cellular differentiation
- PGC-1 related coactivator (PRC)
a third member of the PGC-1 family that also binds transcription factors important for mitochondrial biogenesis and function
- PPARγ coactivators (PGCs)
transcriptional coactivators that bind to a number of target transcription factors involved in cellular energy metabolism and mitochondrial function including PPARs, ERRs and NRF-1, -2
- RNA polymerase (POLRMT)
a mitochondrial DNA-directed RNA polymerase
- Transcription factor A, mitochondrial (Tfam)
also called mtTFA, is a mitochondrial transcription factor that is a key activator of mitochondrial transcription and participates in mitochondrial genome replication
- Yin-Yang 1 (YY1)
a ubiquitously-expressed transcription factor that directs histone deacetylases and histone acetyltransferases to a promoter in order to activate or repress gene expression
Footnotes
DISCLOSURES
D.P.K. serves on Scientific Advisory Boards for Johnson & Johnson, Pfizer, and Eli Lilly & Company.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bonawitz ND, et al. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 2.Scarpulla RC. Transcriptional paradigms in Mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 3.Fisher RP, Clayton DA. Purification and characterization of human mitochondrial transcription factor 1. Molecular and Cellular Biology. 1988;8:3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larsson NG, et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nature genetics. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 5.Park CB, et al. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell. 2007;130:273–285. doi: 10.1016/j.cell.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 6.Metodiev MD, et al. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell metabolism. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Camara Y, et al. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell metabolism. 2011;13:527–539. doi: 10.1016/j.cmet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Graham BH, et al. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nature genetics. 1997;16:226–234. doi: 10.1038/ng0797-226. [DOI] [PubMed] [Google Scholar]
- 9.Scarpulla RC. Nucleus-encoded regulators of mitochondrial function: Integration of respiratory chain expression, nutrient sensing and metabolic stress. Biochimica et biophysica acta. 2011 doi: 10.1016/j.bbagrm.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes and Development. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 11.Gleyzer N, et al. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Molecular and Cellular Biology. 2005;25:1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seidel-Rogol BL, et al. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nature genetics. 2003;33:23–24. doi: 10.1038/ng1064. [DOI] [PubMed] [Google Scholar]
- 13.Litonin D, et al. Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. The Journal of biological chemistry. 2010;285:18129–18133. doi: 10.1074/jbc.C110.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ongwijitwat S, et al. Nuclear respiratory factor 2 senses changing cellular energy demands and its silencing down-regulates cytochrome oxidase and other target gene mRNAs. Gene. 2006;374:39–49. doi: 10.1016/j.gene.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 15.Dhar SS, et al. Nuclear respiratory factor 1 regulates all ten nuclear-encoded subunits of cytochrome c oxidase in neurons. Journal of Biological Chemistry. 2008;283:3120–3129. doi: 10.1074/jbc.M707587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huo L, Scarpulla RC. Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice. Mol Cell Biol. 2001;21:644–654. doi: 10.1128/MCB.21.2.644-654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ristevski S, et al. The ETS transcription factor GABPalpha is essential for early embryogenesis. Molecular and Cellular Biology. 2004;24:5844–5849. doi: 10.1128/MCB.24.13.5844-5849.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donohoe ME, et al. Targeted disruption of mouse yin yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang ZF, et al. The Ets transcription factor GABP is required for cell-cycle progression. Nature Cell Biology. 2007;9:339–346. doi: 10.1038/ncb1548. [DOI] [PubMed] [Google Scholar]
- 20.Madrazo JA, Kelly DP. The PPAR trio: regulators of myocardial energy metabolism in health and disease. J Mol Cell Cardiol. 2008;44:968–975. doi: 10.1016/j.yjmcc.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 21.Eichner LJ, Giguere V. Estrogen related receptors (ERRs): a new dawn in transcriptional control of mitochondrial gene networks. Mitochondrion. 2011;11:544–552. doi: 10.1016/j.mito.2011.03.121. [DOI] [PubMed] [Google Scholar]
- 22.Dufour CR, et al. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell metabolism. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Andersson U, Scarpulla RC. PGC-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Molecular and Cellular Biology. 2001;21:3738–3749. doi: 10.1128/MCB.21.11.3738-3749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narkar VA, et al. Exercise and PGC-1alpha-independent synchronization of type I muscle metabolism and vasculature by ERRgamma. Cell metabolism. 2011;13:283–293. doi: 10.1016/j.cmet.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huss JM, et al. The nuclear receptor ERRα is required for the bioenergetic and functional adaptation to cardiac pressure overload. Cell metabolism. 2007;6:25–37. doi: 10.1016/j.cmet.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 26.Alaynick WA, et al. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell metabolism. 2007;6:13–24. doi: 10.1016/j.cmet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 27.Rangwala SM, et al. Estrogen-related receptor gamma is a key regulator of muscle mitochondrial activity and oxidative capacity. The Journal of biological chemistry. 2010;285:22619–22629. doi: 10.1074/jbc.M110.125401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basu A, et al. Regulation of murine cytochrome oxidase Vb gene expression in different tissues and during myogenesis - Role of a YY-1 factor- binding negative enhancer. Journal of Biological Chemistry. 1997;272:5899–5908. doi: 10.1074/jbc.272.9.5899. [DOI] [PubMed] [Google Scholar]
- 29.Seelan RS, Grossman LI. Structural organization and promoter analysis of the bovine cytochrome c oxidase subunit VIIc gene - A functional role for YY1. Journal of Biological Chemistry. 1997;272:10175–10181. doi: 10.1074/jbc.272.15.10175. [DOI] [PubMed] [Google Scholar]
- 30.Xi H, et al. Analysis of overrepresented motifs in human core promoters reveals dual regulatory roles of YY1. Genome Res. 2007;17:798–806. doi: 10.1101/gr.5754707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cunningham JT, et al. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 32.Wan B, Moreadith RW. Structural characterization and regulatory element analysis of the heart isoform of cytochrome c oxidase VIa. Journal of Biological Chemistry. 1995;270:26433–26440. doi: 10.1074/jbc.270.44.26433. [DOI] [PubMed] [Google Scholar]
- 33.Ramachandran B, et al. Nuclear respiratory factor 1 controls myocyte enhancer factor 2A transcription to provide a mechanism for coordinate expression of respiratory chain subunits. Journal of Biological Chemistry. 2008;283:11935–11946. doi: 10.1074/jbc.M707389200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrish F, et al. c-MYC apoptotic function is mediated by NRF-1 target genes. Genes Dev. 2003;17:240–255. doi: 10.1101/gad.1032503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mootha VK, et al. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci U S A. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schreiber SN, et al. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci U S A. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossignol R, et al. Threshold effect and tissue specificity. Implication for mitochondrial cytopathies. Journal of Biological Chemistry. 1999;274:33426–33432. doi: 10.1074/jbc.274.47.33426. [DOI] [PubMed] [Google Scholar]
- 38.Puigserver P, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 39.Kressler D, et al. The PGC-1-related protein PERC is a selective coactivator of estrogen receptor alpha. Journal of Biological Chemistry. 2002;277:13918–13925. doi: 10.1074/jbc.M201134200. [DOI] [PubMed] [Google Scholar]
- 40.Lin J, et al. PGC-1α: A novel PGC-1-related transcription coactivator associated with host cell factor. Journal of Biological Chemistry. 2002;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- 41.Lehman JJ, et al. Peroxisome proliferator-activated receptor g coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russell LK, et al. Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circ Res. 2004;94:525–533. doi: 10.1161/01.RES.0000117088.36577.EB. [DOI] [PubMed] [Google Scholar]
- 43.Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Espinoza DO, et al. Dual modulation of both lipid oxidation and synthesis by peroxisome proliferator-activated receptor-gamma coactivator-1alpha and -1beta in cultured myotubes. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2010;24:1003–1014. doi: 10.1096/fj.09-133728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai L, et al. Transcriptional coactivators PGC-1alpha and PGC-lbeta control overlapping programs required for perinatal maturation of the heart. Genes and Development. 2010;22:1948–1961. doi: 10.1101/gad.1661708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vercauteren K, et al. PGC-1-related coactivator (PRC): immediate early expression and characterization of a CREB/NRF-1 binding domain associated with cytochrome c promoter occupancy and respiratory growth. Mol Cell Biol. 2006;26:7409–7419. doi: 10.1128/MCB.00585-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vercauteren K, et al. PGC-1-related Coactivator Complexes with HCF-1 and NRF-2{beta} in Mediating NRF-2(GABP)-dependent Respiratory Gene Expression. Journal of Biological Chemistry. 2008;283:12102–12111. doi: 10.1074/jbc.M710150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vercauteren K, et al. Short hairpin RNA-mediated silencing of PRC (PGC-1-related coactivator) results in a severe respiratory chain deficiency associated with the proliferation of aberrant mitochondria. Journal of Biological Chemistry. 2009;284:2307–2319. doi: 10.1074/jbc.M806434200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raharijaona M, et al. PGC-1-related coactivator modulates mitochondrial-nuclear crosstalk through endogenous nitric oxide in a cellular model of oncocytic thyroid tumours. PLoS One. 2009;4:e7964. doi: 10.1371/journal.pone.0007964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Metodiev MD, et al. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell metabolism. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 51.Mirebeau-Prunier D, et al. Estrogen-related receptor alpha and PGC-1-related coactivator constitute a novel complex mediating the biogenesis of functional mitochondria. FEBS J. 2010;277:713–725. doi: 10.1111/j.1742-4658.2009.07516.x. [DOI] [PubMed] [Google Scholar]
- 52.Herschman HR. Primary response genes induced by growth factors and tumor promoters. Annual Review of Biochemistry. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- 53.Winkles JA. Serum- and polypeptide growth factor-inducible gene expression in mouse fibroblasts. Prog Nucleic Acid Res Mol Biol. 1998;58:41–78. doi: 10.1016/s0079-6603(08)60033-1. [DOI] [PubMed] [Google Scholar]
- 54.He X, et al. Peri-implantation lethality in mice lacking the PGC-1-related coactivator protein. Dev Dyn. 2012 doi: 10.1002/dvdy.23769. [DOI] [PubMed] [Google Scholar]
- 55.Fernandez-Marcos PJ, Auwerx J. Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis. Am J Clin Nutr. 2011;93:884S–8890. doi: 10.3945/ajcn.110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 57.Lin J, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 58.Leone TC, et al. PGC-1α deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:0672–0687. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lelliott CJ, et al. Ablation of PGC-1beta results in defective mitochondrial activity, thermogenesis, hepatic function, and cardiac performance. PLoS Biol. 2006;4:e369. doi: 10.1371/journal.pbio.0040369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sonoda J, et al. PGC-1beta controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proc Natl Acad Sci US A. 2007;104:5223–5228. doi: 10.1073/pnas.0611623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arany Z, et al. Transcriptional coactivator PGC-1α controls the energy state and contractile function of cardiac muscle. Cell metabolism. 2005;1:259–271. doi: 10.1016/j.cmet.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 62.Gerhart-Hines Z, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO Journal. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jager S, et al. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci US A. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Canto C, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iwabu M, et al. Adiponectin and AdipoR1 regulate PGC-1alpha and mitochondria by Ca(2+) and AMPK/SIRT1. Nature. 2010;464:1313–1319. doi: 10.1038/nature08991. [DOI] [PubMed] [Google Scholar]
- 66.Donmez G, Guarente L. Aging and disease: connections to sirtuins. Aging Cell. 2010;9:285–290. doi: 10.1111/j.1474-9726.2010.00548.x. [DOI] [PubMed] [Google Scholar]
- 67.Kelly TJ, et al. GCN5-mediated transcriptional control of the metabolic coactivator PGC-1beta through lysine acetylation. Journal of Biological Chemistry. 2009;284:19945–19952. doi: 10.1074/jbc.M109.015164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coste A, et al. The genetic ablation of SRC-3 protects against obesity and improves insulin sensitivity by reducing the acetylation of PGC-1{alpha} Proc Natl Acad Sci U S A. 2008;105:17187–17192. doi: 10.1073/pnas.0808207105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Inoki K, et al. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 71.Egan DF, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim J, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature Cell Biology. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harman D. Free radical theory of aging: dietary implications. Am J Clin Nutr. 1972;25:839–843. doi: 10.1093/ajcn/25.8.839. [DOI] [PubMed] [Google Scholar]
- 74.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 75.Lee YJ, et al. Inflammation and Alzheimer’s disease. Arch Pharm Res. 2010;33:1539–1556. doi: 10.1007/s12272-010-1006-7. [DOI] [PubMed] [Google Scholar]
- 76.Baker RG, et al. NF-kappaB, inflammation, and metabolic disease. Cell metabolism. 2011;13:11–22. doi: 10.1016/j.cmet.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mantovani A, et al. Molecular pathways and targets in cancer-related inflammation. Annals of Medicine. 2010;42:161–170. doi: 10.3109/07853890903405753. [DOI] [PubMed] [Google Scholar]
- 78.Grivennikov SI, Karin M. Inflammation and oncogenesis: a vicious connection. Curr Opin Genet Dev. 2010;20:65–71. doi: 10.1016/j.gde.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Verdin E, et al. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends in Biochemical Sciences. 2010;35:669–675. doi: 10.1016/j.tibs.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Giralt A, et al. Peroxisome proliferator-activated receptor-gamma coactivator- 1alpha controls transcription of the Sirt3 gene, an essential component of the thermogenic brown adipocyte phenotype. Journal of Biological Chemistry. 2011;286:16958–16966. doi: 10.1074/jbc.M110.202390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kong X, et al. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS One. 2010;5:e11707. doi: 10.1371/journal.pone.0011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dominy JE, Jr, et al. Nutrient-dependent regulation of PGC-1alpha’s acetylation state and metabolic function through the enzymatic activities of Sirt1/GCN5. Biochim Biophys Acta. 2009;1804:1676–1683. doi: 10.1016/j.bbapap.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bell EL, Guarente L. The SirT3 Divining Rod Points to Oxidative Stress. Mol Cell. 2011;42:561–568. doi: 10.1016/j.molcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gleyzer N, Scarpulla RC. PGC-1-related coactivator (PRC), a sensor of metabolic stress, orchestrates a redox-sensitive program of inflammatory gene expression. Journal of Biological Chemistry. 2011 doi: 10.1074/jbc.M111.291575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vermeij WP, Backendorf C. Skin cornification proteins provide global link between ROS detoxification and cell migration during wound healing. PLoS One. 2010;5:e11957. doi: 10.1371/journal.pone.0011957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vermeij WP, et al. ROS quenching potential of the epidermal cornified cell envelope. J Invest Dermatol. 2011;131:1435–1441. doi: 10.1038/jid.2010.433. [DOI] [PubMed] [Google Scholar]
- 87.Grivennikov SI, et al. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Orjalo AV, et al. Cell surface-bound IL-1alpha is an upstream regulator of the senescence-associated IL-6/IL-8 cytokine network. Proc Natl Acad Sci US A. 2009;106:17031– 17036. doi: 10.1073/pnas.0905299106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Davalos AR, et al. Senescent cells as a source of inflammatory factors for tumor progression. Cancer Metastasis Rev. 2010;29:273–283. doi: 10.1007/s10555-010-9220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Savagner F, et al. PGC-1-related coactivator and targets are upregulated in thyroid oncocytoma. Biochemical and Biophysical Research Communications. 2003;310:779–784. doi: 10.1016/j.bbrc.2003.09.076. [DOI] [PubMed] [Google Scholar]
- 91.Scarpulla RC. Nucleus-encoded regulators of mitochondrial function: Integration of respiratory chain expression, nutrient sensing and metabolic stress. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagrm.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim HS, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41– 52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Finley LW, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Balaban RS. Regulation of oxidative phosphorylation in the mammalian cell. American Journal of Physiology: Cell Physiology. 1990;258:C377–C389. doi: 10.1152/ajpcell.1990.258.3.C377. [DOI] [PubMed] [Google Scholar]
- 95.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 96.Gray MW, et al. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 97.Wallace DC. A Mitochondrial Paradigm of Metabolic and Degenerative Diseases, Aging, and Cancer: A Dawn for Evolutionary Medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]