Abstract
PURPOSE
This study assesses the pharmacokinetics (PK) and pharmacodynamics (PD) of Nrf2-mediated increased expression of Phase II drug metabolizing enzyme (DME) and antioxidant enzymes which represents an important component of cancer chemoprevention in rat lymphocytes following intravenous (i.v.) administration of an anti-cancer phytochemical sulforaphane (SFN)
METHODS
SFN was administered intravenously to four groups of male Sprague-Dawley JVC rats each group comprising four animals. Blood samples were drawn at selected time points. Plasma were obtained from half of the blood samples and analyzed using a validated LC-MS/MS method. Lymphocytes were collected from the remaining blood samples using Ficoll-Paque™ Plus centrifuge medium. Lymphocyte RNAs were extracted, converted to cDNA, and quantitative real-time PCR analyses were performed and fold changes were calculated against those at time zero for the relative expression of Nrf2-target genes of phase II DME/antioxidant enzymes. PK-PD modeling was conducted based on Jusko’s indirect response model (IDR) using GastroPlus™ and Bootstrap Method.
RESULTS
SFN plasma concentration declined biexponentially and the pharmacokinetic parameters were generated. Rat lymphocyte mRNA expression levels showed no change for GSTM1, SOD, NF-κB, UGT1A1, or UGT1A6. Moderate increases (2-5 folds) over the time zero were seen for HO-1, Nrf2, and NQO1, and significant increase (> 5 folds) for GSTT1, GPx1, and Maf. PK-PD analyses using GastroPlus™ and Bootstrap method provided reasonable fitting for the PK and PD profiles and parameter estimates.
CONCLUSION
Our present study shows that SFN could induce Nrf2-mediated phase II DME/antioxidant mRNA expression for NQO1, GSTT1, Nrf2, GPx, Maf, and HO-1 in rat lymphocytes after i.v. administration, suggesting that Nrf2-mediated mRNA expression in lymphocytes may serve as surrogate biomarkers. The PK-PD IDR model simultaneously linking the plasma concentrations of SFN and the PD response of lymphocyte mRNA expression is valuable for quantitating Nrf2 mediated effects of SFN. This study may provide a conceptual framework for future clinical PK-PD studies of dietary cancer chemopreventive agents in human.
Keywords: sulforaphane, pharmacokinetics, pharmacodynamics, lymphocyte, phase II genes, Nrf2
INTRODUCTION
Sulforaphane (SFN, 4-methylsulfinylbutyl isothiocyanate) is a naturally occurring isothiocyanate, which was first identified from broccoli extracts as a principal inducer of the NAD(P)H quinone oxidoreductase 1 (NQO1) or quinone reductase (QR) activity.1 Subsequently, numerous cell and animal studies have demonstrated its strong anti cancer chemopreventive effects.2
SFN is metabolized through the mercapturic acid pathway, starting with GSH conjugation by glutathione S-transferase (GST) and subsequently generating SFN-cysteine followed by SFN-N-acetylcysteine (NAC).3 After identified as a potential cancer chemopreventive agent,2 SFN has been studied as a modulator for phase I metabolism through direct inhibition of cytochrome P450 enzymes (CYP) or regulation of their mRNA transcription,4 as a potent inducer of Phase II drug metabolism enzymes (DME)/antioxidant enzymes, and as an inhibitor of DME-mediated activation of carcinogens via the antioxidant response element (ARE)-mediated gene expression. These ARE-mediated genes include NQO1, GST, and antioxidant enzymes heme oxygenase (HO-1), and other Phase II DME genes.5 Regulation of ARE-target Phase II DME/antioxidant genes for the detoxification of carcinogens/reactive oxygen nitrogen species (RONS) is typically mediated by the transcription factor, nuclear factor E2-related factor 2 (Nrf2).6
Nrf2, a member of the basic leucine-zipper (bZIP) NF-E2 family, is typically sequestrated in the cytosol of the cell by Kelch-like ECH-associated protein 1 (Keap1), a cysteine rich protein interacting with Nrf2 in its dimeric form under basal unstimulated condition.7 SFN appears to react with the thiol groups of Keap1 and to promote Nrf2 dissociation from Keap1. Subsequently, Nrf2 translocates into the nucleus, forming heterodimer with a group of nuclear bZIP proteins, Maf proteins. The Maf proteins, lacking the transactivation domain, enhances the binding of Nrf2/Maf to the ARE cis-acting enhancer located in the promoter region of a battery of cytoprotective Phase II DME/antioxidant genes.8, 9 Nrf2 and Nrf2-target genes are tightly linked to cancer chemoprevention.6, 10 For instance, genetic knock-out (KO) of Nrf2 would render the animals more prone to cancer induced by carcinogens and that chemopreventive agent such as SFN would lose its cancer chemopreventive effectiveness in Nrf2 KO mice.11 The functions of Nrf2 and Nrf2’s downstream target genes including GSTs, HO-1, and NQO1 have been shown to be important for protection against oxidative stress or chemical-induced cellular damage in liver, lung, as well as in prevention of cancer in the GI tract.12 In one of our previous studies,13 SFN was studied in the liver of Nrf2 wild-type (+/+) and Nrf2 knockout (−/−) mice. Genes induced by SFN in the wild-type mice but not in the Nrf2-knockout (KO) mice were classified as Nrf2-dependent SFN-inducible genes. The Nrf2-target genes that are positively regulated at inducible level by Nrf2 have been identified to be potential chemoprevention surrogate markers.6, 10 Therefore, the Nrf2-mediated phase II/antioxidant gene expression can be linked to cancer chemoprevention based on these previous studies and may serve as valuable biomarkers or surrogate markers in clinical trials.
Numerous studies have been conducted with SFN on the cancer preventive blocking mechanisms, and suppression via anti-proliferative mechanisms.14 Metabolism, bioavailability, pharmacokinetics, preclinical and clinical studies on SFN have also been conducted to better understand its performance in vitro and in vivo.14 We have previously reported the in vivo pharmacokinetics and liver gene expression profiles using 4,967 oligonucleotides microarray analysis after oral gavage dosing of 50 μmol SFN in the rats,15 and pharmacokinetics and pharmacodynamics of broccoli sprouts that generates SFN on the suppression of prostate cancer in TRAMP mice.16 However, no study thus far has involved the simultaneous linking of the pharmacokinetics (PK) and pharmacodynamics (PD) of SFN, especially in the lymphocytes, an easily accessible tissue. In addition, there is no report directly linking plasma concentration of SFN and lymphocyte gene expression which potentially could be a valuable cancer chemoprevention surrogate biomarker for clinical studies of SFN or other cancer chemopreventive agents. In this study, we report the PK in rat plasma and the PD of Phase II DME/antioxidant gene expression in rat lymphocytes following intravenous (i.v.) administration of 25 mg/kg of SFN in the rats.
MATERIALS AND METHODS
Animal and Drug Treatments
Male Sprague-Dawley rats weighing between 250 and 300 g with jugular vein cannulae (JVC) were purchased from Hilltop Lab Animals Inc. (Scottdale, PA). The animals were housed in the Animal Care Facility of Rutgers University under 12 h light-dark cycles with free access to food and water. Upon arrival, the rats were given AIN-76A diet (Research Diets, New Brunswick, NJ) free of antioxidant and acclimatized to the laboratory conditions for 3 days. Due to the small amount of lymphocyte in the blood and the limited amount of blood that can be drawn at each time point from a rat, four groups of rats each comprising four animals (total n=16 rats) were given SFN in 0.9% saline solution via i.v. bolus dose of 25 mg/kg through the jugular vein cannulae followed by flushing with one volume of saline solution. Blood samples (~300 μL) were collected alternately for each rat at 0, 2, 5, 15, 30, 45 minutes, 1, 1.5, 2, 4, 6, 8, 12, 16, or 24 hours following SFN administration such that the total collected blood was under 10% of the rat’s total blood volume. The blood samples within a group (n = 3 to 4 rats) at each time point were mixed together and an approximate 1.0 mL of blood would be used for the reliable collection of plasma and lymphocytes. Plasma was separated immediately from half of the collected blood sample by centrifugation and stored at −80°C until analysis. Lymphocytes were extracted immediately from the remaining blood samples using Ficoll-Paque™ PLUS density gradient centrifugation medium (GE Healthcare Life Sciences, Piscataway, NJ) and were dissolved in Qiagen RNeasy® Mini Kit buffer RLT (lysis buffer) (Valencia, CA). The buffer RLT samples containing lysed lymphocytes were frozen at −80°C until analyses.
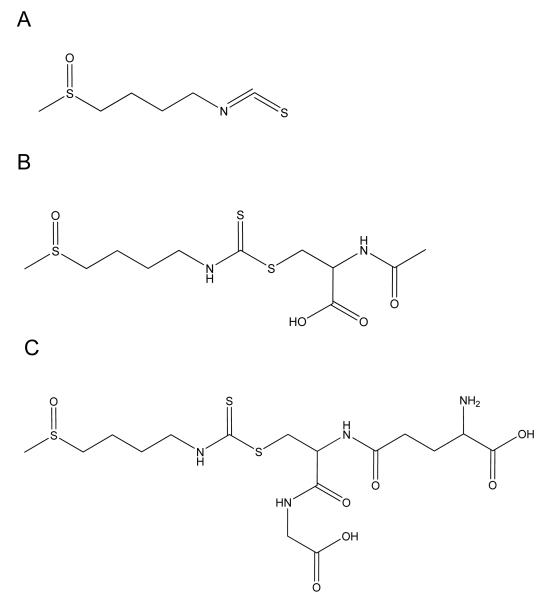
Plasma and Bioanalytical Analysis Plasma-drug concentrations were determined using LC-MS/MS tandem mass spectroscopy (MicroMass Quattro Ultima). The method was validated following the Food and Drug Administration (FDA)’s Guidance for Industry for Bioanalytical Method Validation in a separate study.17 Briefly, 50 μL of plasma samples were precipitated using methanol containing 0.1% TFA. Internal standard, sulforaphene was added and the mixtures were vortexed and centrifuged at 10,000 g at 4°C for 3 minutes. The supernatants were collected and dried on a stream of nitrogen and then reconstituted with acetonitrile:water (50/50,v/v) mixture and passed through nylon filters (Analytical Sales and Services, Pompton Plains, NJ). The LC-MS/MS system was composed of an Agilent 1100 HPLC system equipped with Develosil® 150 × 4.6 mm 5 μm C30 column. SFN, its major metabolites SFN-NAC and SFN GSH (see chemical structures in Figure 1) were detected in MRM mode and quantitated by peak area ratio. MassLynx™ version 3.5 was used for data processing. The quantitation limit of the method was validated at 1 ng/mL for SFN, 10 ng/mL for SFN-GSH and for SFN-NAC. The pharmacokinetic data were analyzed using GastroPlus™ version 6.0.
Figure 1.
Structures of A) Sulforaphane and its major metabolites B) Sulforaphane-NAC, and C) Sulforaphane-GSH.
Lymphocyte RNA and qRT-PCR The lymphocyte RLT buffer solutions were thawed and further processed to extract the total RNA following Qiagen RNeasy® Mini Kit protocol. The total RNA concentrations were measured using Invitrogen (Carlsbad, CA) Quant-It™ reagents and its purity was verified using spectrophotometer A260/A280 method. Same amount of RNA (~ 250 ng) were used for reverse-transcription to obtain cDNA with Applied Biosystems™ Taqman® Reagent and oligo DT. Quantitative real-time PCR (qRT-PCR) analyses were conducted using Applied Biosystems™ SYBR® Green Master Mix (Foster City, CA). The qRT-PCR analyses were performed on an Applied Biosystems™ PRISM® 7900HT following the established laboratory protocol of delta-delta Ct method.18 Thermal cycling was done in triplicate for each cDNA sample according to the following profile: 2 minutes at 50.0°C, 10 minutes at 95°C for the reverse transcriptase reaction, followed by 40 real-time PCR cycling of 10 seconds at 95°C, 30 seconds at 55°C, and 1 minute at 68.0°C, with a subsequent final dissociation stage of 15 seconds at 95.0°C, 15 seconds at 60.0°C, and 15 seconds at 95.0°C. The Relative Quantitation (RQ) results were processed with Applied Biosystems™ Sequence Detection System software (SDS) version 2.0 and Relative Quantitation (RQ) Manager software version 1.2 to obtain the relative mRNA expressions at each time point against their respective expression at time zero. The house keeping gene β-actin was used for the mRNA level normalization. The oligonucleotide primers used for quantitative real-time PCR (qRT-PCR) were designed using Nucleotide from www.ncbi.nlm.nih.gov and PrimerQuest (SM) from Integrated DNA Technologies (www.idtdna.com, Coralville, IA) with 120 to 200 bp in amplicon size, as in previous studies in our lab.15 The gene expression pharmacodynamic data were processed with GastroPlus™ 6.0 Indirect Response (IDR) Model (Jusko)19 as described below.
PK/PD Model Development
The time-course of the pooled SFN plasma concentration was fitted according to the following two compartment system of differential equations (1), and (2) and the resulting PK profile was input into the PD model (3) below:
| (1) |
| (2) |
where kpc, kcp represent the inter-compartment rate constants between the central (c) and peripheral (p) compartments; CL, total clearance from central compartment; Vc, volume of central compartment; Ac and Ap, amount of drug in central and peripheral compartment, respectively.
The pharmacodynamics (PD) response (R) investigated in this study is the relative expression levels of Phase II/antioxidant gene expression. An indirect response model with stimulation of input (kin) by treatment is employed to describe the response time profile (PD). The equation is shown below:
| (3) |
where the stimulatory function (i.e., the stimulation of kin) was given by the function of
| (4) |
for all Phase II/Antioxidant genes investigated in this study. The initial condition is defined as E(0)=1. The parameter symbols are defined in the Abbreviations. All pharmacokinetic and pharmacodynamic parameters were estimated by nonlinear regression analysis using the maximum likelihood estimator in GastroPlus™.
Evaluation of Pharmacodynamic Parameters and Confidence Intervals by Bootstrap Methods In order to calibrate the values estimated by GastroPlus™ version 6.0 above, we next applied a bootstrap in conjunction with least square methods.20 This method offers a robust estimation of the parameter values as well as associated confidence intervals as we have previously demonstrated in the context of indirect response modeling.21 The advantage of this method is that it can take each replicate at one experimental time point into consideration instead of estimation based only on the average of all the replicates at one time point. For each bootstrap run at each single time point, sampling with replacement is performed on all the replicates at that time point whose number may vary based on the data collection from experiment. Each bootstrap sample was used for estimating a set of parameters for these four parameters Smax, SC50, kin and kout as prescribed by the Dr. Jusko’s indirect response model.19 , the mean of the multiple bootstrap estimates (1,000 runs in this study) is reported as the most likely parameter value,22 in which i denotes bootstrap iteration.
The confidence intervals for each of the estimated parameters were calculated by applying the percentile method. The estimated confidence interval for each parameter was denoted as , where subscript l and u respectively denote the lower and the upper limits of the vector of estimated parameter value β which was approximated by α central confidence interval. The 100 (α/2) and 100 (1-α/2) percentile values of the 1000 estimation values are used as the upper and lower confidence limits for a parameter. The probability α (0<α<1) indicates a 100α% confidence that . In this study, α was chosen as 0.05, then 95% confidence limits for β based on 1,000 bootstrap replications were given by and largest estimates of β.22 The bootstrap simulation and prediction in this study were conducted using MATLAB.
RESULTS
Pharmacokinetics of SFN
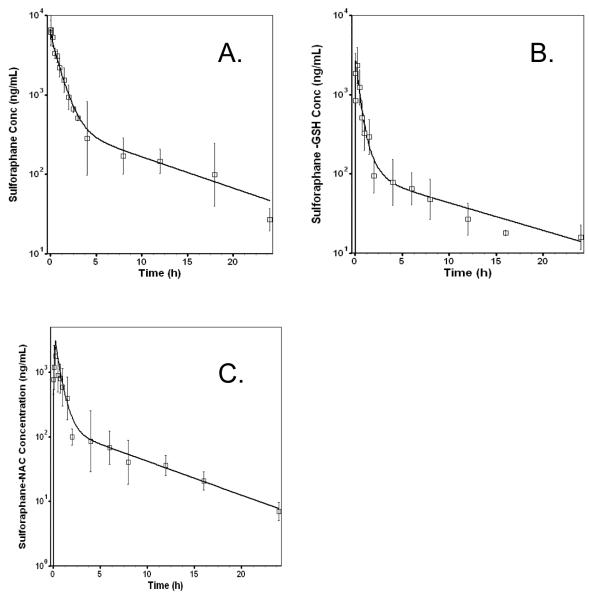
SFN and its major metabolites SFN-GSH and SFN-NAC concentration–time profiles are displayed in Figure 2. The two-compartment PK model estimated parameters are listed in Table 1 for SFN, and in Supplemental Data for the metabolites of SFN-NAC and SFN-GSH. The pharmacokinetics parameters reveal that the two compartmental PK model fitted well for the plasma concentration versus time profiles of SFN, SFN-NAC, and SFN-GSH, while the %CV of the estimated pharmacokinetic parameters were relatively low. As in earlier studies, SFN forms SFN-GSH conjugate first, followed by SFN-NAC.23 Both metabolites may exert their biological activities.1, 24 However, in our previous study, SFN has been found to be the major form where only 12.5% and 9.1% of the SFN dose are in the forms of SFN-GSH and SFN-NAC based on AUC molar ratioes.17 Therefore, in this study, we limit our modeling effort to SFN. The software generated two-compartment PK parameter estimates of SFN were then used as input of PK to fit the SFN-PK PD data as described below.
Figure 2.
Concentration-time profile of sulforaphane (A) after 25 mg/kg i.v. administration of sulforaphane saline solution in rats. Major metabolites sulforaphane-GSH (B) and sulforaphane-NAC (C) formed after sulforaphane administration. The pharmacokinetics parameters of the metabolites can be found in Supporting Information.
Table 1.
Pharmacokinetic parameters of SFN in rat plasma using GastroPlus™. Noncompartmental analysis parameters: AUC, area under the curve; MRT, mean residence time; Vss, volume of distribution at steady state; t1/2, terminal half life; Two-compartment model based parameters: kcp, kpc, central and peripheral intercompartmental rate constants; Vc, volume of distribution of central compartment; and CL, clearance.
| Parameter | Unit | Value | %CV |
|---|---|---|---|
| Non-compartmental Parameters | |||
| AUC 0-24 | ng*h/mL | 9272 | |
| AUC 0-inf | ng*h/mL | 9787 | |
| MRT | h | 4.2 | |
| Vss | L | 3.7 | |
| t1/2 | h | 7.6 | |
| 2-Compartment Model Based Parameters | |||
| kcp | 1/h | 0.425 | 54.65% |
| kpc | 1/h | 0.156 | 63.33% |
| Vc | L | 1.235 | 30.21% |
| CL | L/h | 0.848 | 14.67% |
qRT-PCR of mRNA from Lymphocytes and PK/PD Relationships
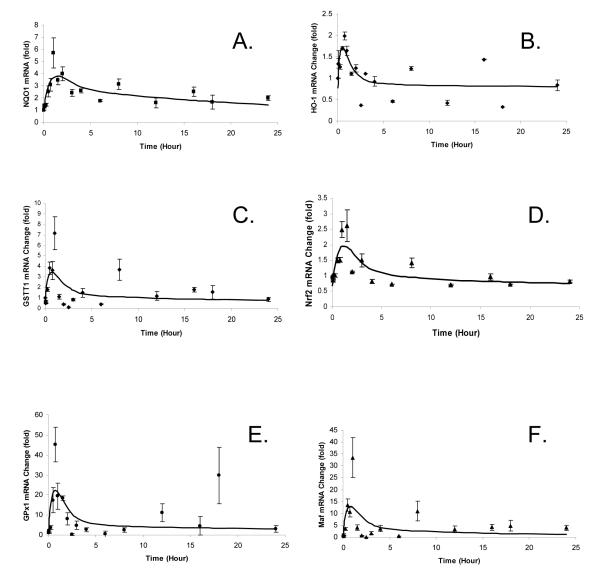
The mRNA expression of the selected Phase II DME/antioxidant genes was quantitated by qRT-PCR. Using Dr. Jusko’s indirect response model Type III – stimulation of kin as described in Equation 3, the PK/PD relationships between SFN concentration and various genes’ expression levels were fitted using GastroPlus™ and the results are shown in Figure 4. The observed profiles (fold changes of the mRNA expression versus time) for NQO1, GPx-1, GSTT1, Nrf2, HO-1, and the small protein Maf (a protein heterodimerizes with Nrf2 in the nucleus) show the expression of these Phase II and related genes in the lymphocytes are induced by SFN treatment. The estimated pharmacodynamic parameters are shown in Table 2. The SC50 values range from 0.26 μg/mL (1.47 μM) for NQO1 (most sensitive) to 8 μg/mL (45.2 μM) for HO-1 (least sensitive) and Smax ranges from 1.55-fold for HO-1 (least responsive) to 39.7-fold for GPx (most responsive). kin and kout are similar for all the phase II DME/antioxidant genes, suggesting their kinetics of production and degradation appears to be similar.
Figure 4.
Pharmacokinetic/Pharmacodynamic profiles of mRNA expression change with time in folds for NQO1, GPx-1, GSTT1, Nrf2, HO-1, and Maf. Lines represent model predicted values. Observed data are presented in mean ± SE.
Table 2.
Pharmacodynamic analysis of Phase II mRNA expression driven by SFN using Type III Indirect Model for NQO1, GSTT1, Maf, GPx, Nrf2, and HO-1. Data are presented as mean ± S.D. (N = 10~12)
| mRNA | Smax | SC50 (μg/mL) | kin (1/hr) |
kout (1/hr) |
Max. effect time (h) |
|---|---|---|---|---|---|
| NQO1 | 3.87 ± 0.67 | 0.26 ± 0.05 | 1.96 ± 0.20 | 2.10 ± 0.23 | 1.60 |
| GSTT1 | 7.78 ± 1.76 | 1.50 ± 0.86 | 1.82 ± 0.35 | 2.41 ± 0.64 | 1.04 |
| Maf | 25.0 ± 3.85 | 1.37 ± 0.12 | 1.76 ± 0.25 | 2.32 ± 0.09 | 1.04 |
| GPx | 39.7 ± 1.58 | 4.82 ± 0.34 | 4.49 ± 0.21 | 3.14 ± 0.07 | 0.72 |
| Nrf2 | 2.92 ± 2.10 | 1.54 ± 3.18 | 1.37 ± 1.04 | 1.78 ± 1.68 | 1.20 |
| HO-1 | 1.55 ± 6.56 | 8.00 ± 49.2 | 4.79 ± 8.14 | 4.26 ± 7.59 | 0.56 |
The mRNAs for the other Nrf2-mediated Phase II DME/antioxidant genes, GSTM1, UGT1A1, SOD, NF-κB, and UGT1A6 show no measurable changes after SFN treatment at the dose administered in the study.
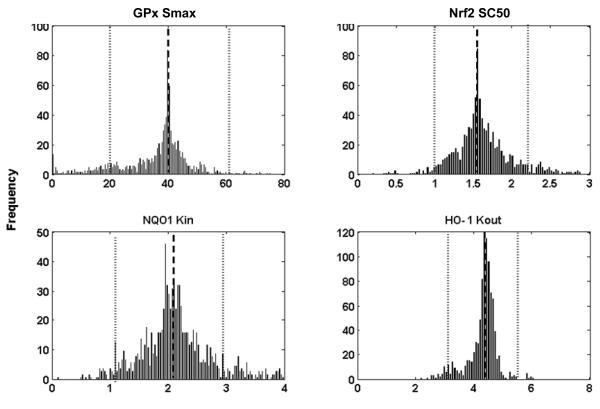
Bootstrap Confirmation
The estimation of the values of the four parameters for each gene and the confidence intervals for the parameters are shown in Table 3. Histograms of 1,000 bootstrap estimates of the four parameters for four selected genes are presented in Figure 5. The estimated values obtained from GastroPlus™ version 6.0 (Table 2) are similar to those estimated by using the bootstrap method (Table 3), which indicates that the parameters are well estimated.
Table 3.
Values of the parameters calculated by bootstrap sampling in conjunction with least square method
| mRNA | Parameter | Mean | Minimum | Maximum | 5% Percentile | 95% Percentile |
|---|---|---|---|---|---|---|
| NQO1 | Smax | 3.54 | 0.00 | 18.57 | 1.19 | 4.99 |
| SC50 (μg/mL) | 0.74 | 0.00 | 9.07 | 0.06 | 2.59 | |
| kin (1/hr) | 2.33 | 0.09 | 29.80 | 1.12 | 3.93 | |
| kout (1/hr) | 2.08 | 0.12 | 22.19 | 1.08 | 3.02 | |
| GPx | Smax | 39.09 | 0.00 | 98.12 | 19.43 | 62.58 |
| SC50 (μg/mL) | 9.57 | 0.00 | 100.00 | 1.36 | 40.96 | |
| kin (1/hr) | 8.01 | 0.30 | 100.00 | 1.94 | 23.80 | |
| kout (1/hr) | 3.74 | 0.04 | 99.45 | 1.11 | 7.27 | |
| Nrf2 | Smax | 2.81 | 0.74 | 9.25 | 1.38 | 4.21 |
| SC50 (μg/mL) | 1.63 | 0.19 | 5.78 | 1.05 | 2.42 | |
| kin (1/hr) | 1.22 | 0.54 | 3.19 | 0.80 | 1.87 | |
| kout (1/hr) | 1.65 | 0.65 | 3.82 | 1.14 | 2.23 | |
| HO-1 | Smax | 1.97 | 0.84 | 4.53 | 1.32 | 2.67 |
| SC50 (μg/mL) | 8.00 | 2.09 | 30.19 | 7.25 | 8.59 | |
| kin (1/hr) | 5.07 | 2.25 | 96.18 | 3.10 | 5.51 | |
| kout (1/hr) | 4.94 | 2.28 | 100.00 | 3.03 | 5.45 |
Figure 5.
Histograms of 1,000 bootstrap estimates of 4 parameters of four representative genes. The bars represent frequency. The average bootstrap estimator values of parameters are indicated by a dashed line and its lower and upper confidence limits β1(0.05),βu(0.05) are represented by dotted lines respectively.
DISCUSSION
SFN, an isothiocyanate (ITC), is a well known indirect antioxidant that induces Nrf2 dependent phase 2 DME/antioxidant enzymes. It has been listed as one of the thirty four anti-carcinogenesis agents by the National Library of Medicine (NLM), as well as one of the most potent inducers of Phase II DME among the many naturally occurring dietary phytochemical compounds.25 Our previous unpublished study results showed that i.v. doses of 10 and 25 mg/kg demonstrated linear pharmacokinetic behaviors in rats, and i.v. doses at 50 mg/kg or higher exerted short term toxicity or even death. Therefore, the high yet tolerable dose level at 25 mg/kg was selected in this study to establish the PK-PD relationship in rat lymphocytes for more reliable parameter estimation. The dose level of 25 mg/kg of sulforaphane in rats via i.v., which is not achievable from food supply, could be used as an equivalent dose level in clinical treatment. The results show that, induction of Nrf2 was modest and was not as significant as some of the other Phase II genes (NQO1, GSTT1, or GPx1). Danilov and colleagues previously found that pretreatment with SFN for 48 hours decreased cultured rat cortical astrocyte cell death and increases Nrf2 mRNA expression by about 3.44 folds measured by RT-PCR.26 The Nrf2 induction level in our study appears to be similar to this and other studies.27, 28
NQO1 detoxifies quinone and its derivatives to protect cells against redox cycling and oxidative stress. Though characterized as Phase I protein with cytochrome p450s as well as Phase II enzyme, NQO1 has been reported to be a highly inducible enzyme and is regulated by the Keap1/Nrf2/ARE pathway.29 Many chemicals induce NQO1 which are subsequently shown to protect against the toxic and carcinogenic effects caused by a wide array of carcinogens in vivo. SFN has been shown to strongly induce NQO1.2, 30 Regulation of NQO1 via Keap1/Nrf2/ARE pathway was recently reviewed by Dinkova et al.31 In our present study, SFN induces NQO1 mRNA expression by about four folds in rat lymphocytes with the maximum effect achieved at time of ~1.6 hour after i.v. administration. The fast response could be potentially achieved via fast-acting transcription factors such as Nrf2 which would not require protein synthesis but nuclear translocation for the transcription activation of its target genes.
Musculo-aponeurotic factor (Maf) is a small protein which partners with bZIP transcription factors including Nrf2 and binds to the ARE and initiates transcription of Phase II DME/antioxidant genes.32 After i.v. SFN administration, Maf mRNA increases significantly in rat lymphocytes with a maximum effect time of ~1.0 hour, similar to that of Nrf2 (~1.2 hour). This increase may facilitate its protein synthesis and subsequent heterodimerization with Nrf2 to further enhance the transactivation of Nrf2 downstream target genes such as Phase II DME.
HO-1 controls heme degradation and accumulation of iron, bilirubin, and carbon monoxide (CO) which would dampen oxidative damage in the gastrointestinal tissues/cells.33 HO-1 has been shown to be directly regulated by Nrf2,34 although studies also found that other mechanisms of transcriptional regulation are known to exist for HO-1, e.g., Bach1.35-37 Since HO-1 is regulated by multiple mechanisms in addition to Nrf2, other Nrf2 target genes would be needed to quantify as biomarkers for Nrf2 activation.38 As shown in our present study, although increased mRNA expression of HO-1 is observed in rat lymphocytes after SFN treatment, the HO-1 mRNA expression appears to display larger variation from the predicted values and could possibly be explained by its multi-mechanism transcription regulation, and further study would be needed to thoroughly understand the HO-1 expression mechanisms and their contributions.
Glutathione (GSH) synthesis is regulated by Nrf2/Nrf1 via the ARE, activator protein 1 (AP-1), and nuclear factor kappa B (NF-κB).39 Glutathione S-transferases GSTM1 and GSTT1 are the two most studied subtypes of GST. In this study, we found that GSTM1 did not show any obvious change after SFN administration, however, GSTT1 was moderately induced in rat lymphocytes 1 hour after SFN administration. This observation indicates that different subtypes of GSTs are having different responses to SFN in rat lymphocytes.
In this study, the Nrf2-mediated genes were selected based on the current understanding of this signaling pathway. Nrf2 is sequestered by Keap1; upon entering the nucleus, Nrf2 forms heterodimer with Maf. NQO1, GST, UGT, HO-1, SOD, and GPx1 have been previously reported in other tissues and they are well established downstream genes of Nrf2;10, 12, 40 NF-κB and Nrf2 may cross-talk with each other and may be of interest as well.41 Typically, changes in mRNA expression occur in the timeframe of hours.42 Therefore, a 24-hour data collection is considered sufficiently long enough to observe the changes of primary mRNA expression whether induced or suppressed by a drug, especially in this in vivo study with i.v. administration.
Some genes, such as UGT1A1, UGT1A6, SOD, GSTM1, and NF-κB did not show any obvious changes in the mRNA expression levels in this study. UGT is responsible for glucuronidation and is involved in an important pathway to eliminate the xenobiotics. In a study investigating resveratrol’s modulation of DME in the lymphocyte of healthy human, UGT1A1 and GST activities were minimally affected.43 This appears to be consistent with the observation in our study. SOD plays an anti-oxidant role in the cell. In a recent study, blueberry caused mean SOD activity increase from 1.51 U/mg (control saline group) to 1.63 U/mg (blueberry juice treatment group) in mouse liver cells.44 Although the increase is statistically significant in that study, similar relative increase may not be captured in the lymphocytes in our study. NF-κB has been a regulator of genes that control immune response, inflammation, cell proliferation, and cell survival. Its possible cross-talk with Nrf2 appears to be inconclusive.41 In this study, NF-κB showed no obvious mRNA change and further studies are needed to investigate its possible role in Nrf2 pathway.
Bootstrap method is a self-sustaining process that proceeds without any external instructional entries. In applying this method in our study, it provided secondary evaluation of the pharmacodynamics parameters estimate generated by GastroPlus™ and the results obtained from both methods were similar.
In the present study, a single-dose of SFN and its effects on mRNA of phase II/antioxidant enzymes in lymphocytes have been evaluated. However, the single dose level may have restricted the pharmacological and the PK-PD modeling assessment. While the time zero (pre-treatment) gene expression level in blood samples collected at different hours of a day were estimated, circadian effects of mRNA expression of the phase II and anti-oxidant enzymes may need to be evaluated in future pre-clinical studies. In addition, after the lymphocytes were extracted by the Ficoll-Paque™ PLUS medium, a measurement of lymphocyte cell amount and using the same number of the lymphocytes may help to reduce the variability of mRNA measurement. Since the blood samples obtained from four rats were mixed together in this study, the variability in the pharmacokinetics and pharmacodynamics measurement may be different from those obtained from individual rats.
In conclusion, in this study, we conducted pharmacokinetic and pharmacodynamic assessment after i.v. administration of SFN in rats. Pharmacodynamically, SFN induced Nrf2-mediated phase II DME/antioxidant mRNA expression of NQO1, GSTT1, Nrf2, GPx, Maf, and HO-1 in rat lymphocytes after i.v. administration, suggesting that Nrf2-mediated mRNA expression in lymphocytes may serve as potential surrogate biomarkers. We linked the plasma concentrations of SFN and the levels of Nrf2-mediated mRNA expression levels by applying the IDR PD models. As the plasma concentration and mRNA levels were measured from the same pooled blood samples, the resulting pharmacokinetic and pharmacodynamic relations could be described simultaneously over the time course of the study. The easily accessible tissue of the blood, a very unique tissue in animals, coupled with the modern LC-MS/MS and quantitative real-time PCR, would make such a direct PK-PD link possible. The preclinical study approach presented in this report may provide a framework for future clinical studies in evaluating a drug candidate for its possible effect on lymphocyte gene expression, e.g., as in those recent clinical studies by using SFN containing broccoli sprouts.45-47 Extracting lymphocytes from human blood can be easily executed using commercial kits. Analyzing the lymphocyte gene expression changes, paired with pharmacokinetics studies, our study approach may lead to a better understanding of a drug’s initial pharmacological effects in the body, and a possible PK-PD modeling and simulation in clinical studies.
Supplementary Material
Figure 3.
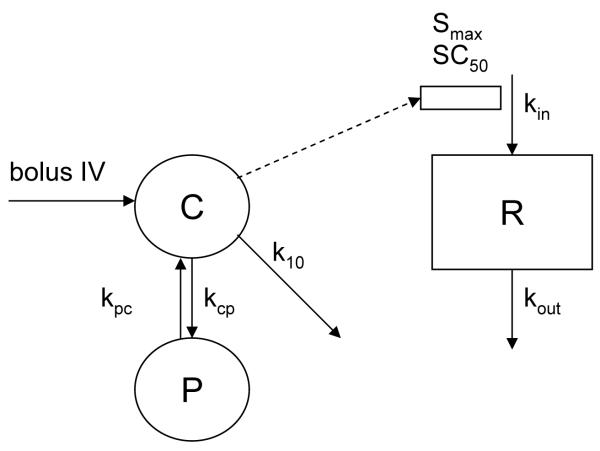
Pharmacokinetic/Pharmacodynamic modeling of Jusko indirect stimulation of kin. The model was first described as Indirect Response Model III by Dayneka.et al 1993.19 C represents central compartment; P, peripheral compartment; R, response of mRNA expression level change over that at initial (t=0).
Acknowledgments
The authors thank the past and present members of Kong’s lab for meaningful discussions, especially Dr. Guoxiang Shen. Thanks also go to Dr. Amin Nomeir of Merck Research Laboratories (retired) for his insightful comments. The authors appreciate Simulations Plus, Inc. for providing extended evaluation of the PDPlus® module to allow the completion of this work.
Grant Support This work is supported in part by NIH R01 CA073674, R01 CA118947, and R01 CA094828 (A.N. Kong).
Abbreviations
- ARE
antioxidant response element
- DME
drug metabolism enzyme;
- GSTM1
glutathione S-transferases mu 1
- GSTT1
glutathione S-transferases theta 1
- HO-1
hemeoxygenase-1
- JVC
jugular vein cannulae
- kin
input turn-over rate
- kout
fractional turn-over rate for loss
- NF-κB
Nuclear factor-kappa-B
- NQO1
NAD(P)H quinone oxidoreductase 1
- Nrf2
nuclear factor-erythroid 2-related factor 2
- qRT-PCR
quantitative real time polymerase chain reaction
- ROS
reactive oxygen species
- Smax
maximum effect attributed to the drug stimulation
- SC50
drug concentration producing 50% of the maximum stimulation achieved at the effect site
- SOD
superoxide dismutase
- UGT1A1
UDP glucuronosyltransferase 1 family, polypeptide A1
- UGT1A6
UDP glucuronosyltransferase 1 family, polypeptide A6
Footnotes
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
Supporting Information Supporting Information (SI) include tables of 2-compartment analysis of SFN-GSH and SFN-NAC in rat plasma. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Conaway CC, Yang YM, Chung FL. Isothiocyanates as cancer chemopreventive agents: their biological activities and metabolism in rodents and humans. Curr Drug Metab. 2002;3:233–255. doi: 10.2174/1389200023337496. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Talalay P, Cho CG, Posner GH. A major inducer of anticarcinogenic protective enzymes from broccoli: isolation and elucidation of structure. Proc Natl Acad Sci U S A. 1992;89:2399–2403. doi: 10.1073/pnas.89.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fahey JW, Zalcmann AT, Talalay P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry. 2001;56:5–51. doi: 10.1016/s0031-9422(00)00316-2. [DOI] [PubMed] [Google Scholar]
- 4.Gross-Steinmeyer K, Stapleton PL, Tracy JH, Bammler TK, Strom SC, Eaton DL. Sulforaphane- and phenethyl isothiocyanate-induced inhibition of aflatoxin B1-mediated genotoxicity in human hepatocytes: role of GSTM1 genotype and CYP3A4 gene expression. Toxicol Sci. 2010;116:422–432. doi: 10.1093/toxsci/kfq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu C, Li CY, Kong AN. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch Pharm Res. 2005;28:249–268. doi: 10.1007/BF02977789. [DOI] [PubMed] [Google Scholar]
- 6.Hu R, Saw CL, Yu R, Kong AN. Regulation of NF-E2-related factor 2 signaling for cancer chemoprevention: antioxidant coupled with antiinflammatory. Antioxid Redox Signal. 2010;13:1679–1698. doi: 10.1089/ars.2010.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keum YS. Regulation of the Keap1/Nrf2 system by chemopreventive sulforaphane: implications of posttranslational modifications. Ann N Y Acad Sci. 2011;1229:184–189. doi: 10.1111/j.1749-6632.2011.06092.x. [DOI] [PubMed] [Google Scholar]
- 8.Motohashi H, O’Connor T, Katsuoka F, Engel JD, Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294:1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 10.Kwak MK, Kensler TW. Targeting NRF2 signaling for cancer chemoprevention. Toxicol Appl Pharmacol. 2010;244:66–76. doi: 10.1016/j.taap.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu C, Huang MT, Shen G, Yuan X, Lin W, Khor TO, Conney AH, Kong AN. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res. 2006;66:8293–8296. doi: 10.1158/0008-5472.CAN-06-0300. [DOI] [PubMed] [Google Scholar]
- 12.Saw CL, Kong AN. Nuclear factor-erythroid 2-related factor 2 as a chemopreventive target in colorectal cancer. Expert Opin Ther Targets. 2011;15:281–295. doi: 10.1517/14728222.2011.553602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu R, Xu C, Shen G, Jain MR, Khor TO, Gopalkrishnan A, Lin W, Reddy B, Chan JY, Kong AN. Gene expression profiles induced by cancer chemopreventive isothiocyanate sulforaphane in the liver of C57BL/6J mice and C57BL/6J/Nrf2 (−/−) mice. Cancer Lett. 2006;243:170–192. doi: 10.1016/j.canlet.2005.11.050. [DOI] [PubMed] [Google Scholar]
- 14.Cheung KL, Kong AN. Molecular targets of dietary phenethyl isothiocyanate and sulforaphane for cancer chemoprevention. Aaps J. 2010;12:87–97. doi: 10.1208/s12248-009-9162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu R, Hebbar V, Kim BR, Chen C, Winnik B, Buckley B, Soteropoulos P, Tolias P, Hart RP, Kong AN. In vivo pharmacokinetics and regulation of gene expression profiles by isothiocyanate sulforaphane in the rat. J Pharmacol Exp Ther. 2004;310:263–271. doi: 10.1124/jpet.103.064261. [DOI] [PubMed] [Google Scholar]
- 16.Keum YS, Khor TO, Lin W, Shen G, Kwon KH, Barve A, Li W, Kong AN. Pharmacokinetics and pharmacodynamics of broccoli sprouts on the suppression of prostate cancer in transgenic adenocarcinoma of mouse prostate (TRAMP) mice: implication of induction of Nrf2, HO-1 and apoptosis and the suppression of Akt-dependent kinase pathway. Pharm Res. 2009;26:2324–2331. doi: 10.1007/s11095-009-9948-5. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Lin W, Shen G, Khor T, Nomeir A, Kong A-N. Development and validation of a liquid chromatography-tandem mass spectrometric method for the simultaneous determination of sulforaphane and its metabolites in rat plasma and its application in pharmacokinetic studies. J Chromatogr Sci. 2011;49:801–806. doi: 10.1093/chrsci/49.10.801. [DOI] [PubMed] [Google Scholar]
- 18.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dayneka NL, Garg V, Jusko WJ. Comparison of four basic models of indirect pharmacodynamic responses. J Pharmacokinet Biopharm. 1993;21:457–478. doi: 10.1007/BF01061691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chrysikopoulos CV, Hsuan P-Y, Fyrillas MM. Bootstrap estimation of the mass transfer coefficient of a dissolving nanaqueous phase liquid pool in porous media. Water Resources Research. 2002;38:8–1-6. [Google Scholar]
- 21.Yang Q, Berthiaume F, Androulakis IP. A quantitative model of thermal injury-induced acute inflammation. Math Biosci. 2010;229:135–148. doi: 10.1016/j.mbs.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Efron B, Tibshirani RJ. An Introduction to the Bootstrap. Chapman and Hall; New York: 1993. [Google Scholar]
- 23.Kassahun K, Davis M, Hu P, Martin B, Baillie T. Biotransformation of the naturally occurring isothiocyanate sulforaphane in the rat: identification of phase I metabolites and glutathione conjugates. Chem Res Toxicol. 1997;10:1228–1233. doi: 10.1021/tx970080t. [DOI] [PubMed] [Google Scholar]
- 24.Kim BR, Hu R, Keum YS, Hebbar V, Shen G, Nair SS, Kong AN. Effects of glutathione on antioxidant response element-mediated gene expression and apoptosis elicited by sulforaphane. Cancer Res. 2003;63:7520–7525. [PubMed] [Google Scholar]
- 25.Ye L, Zhang Y. Total intracellular accumulation levels of dietary isothiocyanates determine their activity in elevation of cellular glutathione and induction of Phase 2 detoxification enzymes. Carcinogenesis. 2001;22:1987–1992. doi: 10.1093/carcin/22.12.1987. [DOI] [PubMed] [Google Scholar]
- 26.Danilov CA, Chandrasekaran K, Racz J, Soane L, Zielke C, Fiskum G. Sulforaphane protects astrocytes against oxidative stress and delayed death caused by oxygen and glucose deprivation. Glia. 2009;57:645–656. doi: 10.1002/glia.20793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keum YS, Yu S, Chang PP, Yuan X, Kim JH, Xu C, Han J, Agarwal A, Kong AN. Mechanism of action of sulforaphane: inhibition of p38 mitogen-activated protein kinase isoforms contributing to the induction of antioxidant response element-mediated heme oxygenase-1 in human hepatoma HepG2 cells. Cancer Res. 2006;66:8804–8813. doi: 10.1158/0008-5472.CAN-05-3513. [DOI] [PubMed] [Google Scholar]
- 28.Ebert B, Kisiela M, Malatkova P, El-Hawari Y, Maser E. Regulation of human carbonyl reductase 3 (CBR3; SDR21C2) expression by Nrf2 in cultured cancer cells. Biochemistry. 2010;49:8499–8511. doi: 10.1021/bi100814d. [DOI] [PubMed] [Google Scholar]
- 29.Talalay P. Mechanisms of induction of enzymes that protect against chemical carcinogenesis. Adv Enzyme Regul. 1989;28:237–250. doi: 10.1016/0065-2571(89)90074-5. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Tang L. Discovery and development of sulforaphane as a cancer chemopreventive phytochemical. Acta Pharmacol Sin. 2007;28:1343–1354. doi: 10.1111/j.1745-7254.2007.00679.x. [DOI] [PubMed] [Google Scholar]
- 31.Dinkova-Kostova AT, Talalay P. NAD(P)H:quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch Biochem Biophys. 2010;501:116–123. doi: 10.1016/j.abb.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 33.Gibbons SJ, Farrugia G. The role of carbon monoxide in the gastrointestinal tract. J Physiol. 2004;556:325–336. doi: 10.1113/jphysiol.2003.056556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL. Nrf2, a Cap’n’Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem. 1999;274:26071–26078. doi: 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- 35.Igarashi K, Hoshino H, Muto A, Suwabe N, Nishikawa S, Nakauchi H, Yamamoto M. Multivalent DNA binding complex generated by small Maf and Bach1 as a possible biochemical basis for beta-globin locus control region complex. J Biol Chem. 1998;273:11783–11790. doi: 10.1074/jbc.273.19.11783. [DOI] [PubMed] [Google Scholar]
- 36.Reichard JF, Motz GT, Puga A. Heme oxygenase-1 induction by NRF2 requires inactivation of the transcriptional repressor BACH1. Nucleic Acids Res. 2007;35:7074–7086. doi: 10.1093/nar/gkm638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida C, Tokumasu F, Hohmura KI, Bungert J, Hayashi N, Nagasawa T, Engel JD, Yamamoto M, Takeyasu K, Igarashi K. Long range interaction of cis-DNA elements mediated by architectural transcription factor Bach1. Genes Cells. 1999;4:643–655. doi: 10.1046/j.1365-2443.1999.00291.x. [DOI] [PubMed] [Google Scholar]
- 38.Klaassen CD, Reisman SA. Nrf2 the rescue: effects of the antioxidative/electrophilic response on the liver. Toxicol Appl Pharmacol. 2010;244:57–65. doi: 10.1016/j.taap.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu SC. Regulation of glutathione synthesis. Mol Aspects Med. 2009;30:42–59. doi: 10.1016/j.mam.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheung KL, Khor TO, Huang MT, Kong AN. Differential in vivo mechanism of chemoprevention of tumor formation in azoxymethane/dextran sodium sulfate mice by PEITC and DBM. Carcinogenesis. 2010;31:880–885. doi: 10.1093/carcin/bgp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W, Khor TO, Xu C, Shen G, Jeong WS, Yu S, Kong AN. Activation of Nrf2-antioxidant signaling attenuates NFkappaB-inflammatory response and elicits apoptosis. Biochem Pharmacol. 2008;76:1485–1489. doi: 10.1016/j.bcp.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mager DE, Jusko WJ. Development of translational pharmacokinetic-pharmacodynamic models. Clin Pharmacol Ther. 2008;83:909–912. doi: 10.1038/clpt.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chow HH, Garland LL, Hsu CH, Vining DR, Chew WM, Miller JA, Perloff M, Crowell JA, Alberts DS. Resveratrol modulates drug- and carcinogen metabolizing enzymes in a healthy volunteer study. Cancer Prev Res (Phila) 2010;3:1168–1175. doi: 10.1158/1940-6207.CAPR-09-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang YP, Cheng ML, Zhang BF, Mu M, Zhou MY, Wu J, Li CX. Effect of blueberry on hepatic and immunological functions in mice. Hepatobiliary Pancreat Dis Int. 2010;9:164–168. [PubMed] [Google Scholar]
- 45.Kensler TW, Ng D, Carmella SG, Chen M, Jacobson LP, Munoz A, Egner PA, Chen JG, Qian GS, Chen TY, Fahey JW, Talalay P, Groopman JD, Yuan JM, Hecht SS. Modulation of the metabolism of airborne pollutants by glucoraphanin-rich and sulforaphane-rich broccoli sprout beverages in Qidong, China. Carcinogenesis. 2012;33:101–107. doi: 10.1093/carcin/bgr229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riedl MA, Saxon A, Diaz-Sanchez D. Oral sulforaphane increases Phase II antioxidant enzymes in the human upper airway. Clin Immunol. 2009;130:244–251. doi: 10.1016/j.clim.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yanaka A. Sulforaphane enhances protection and repair of gastric mucosa against oxidative stress in vitro, and demonstrates anti-inflammatory effects on Helicobacter pylori-infected gastric mucosae in mice and human subjects. Curr Pharm Des. 2011;17:1532–1540. doi: 10.2174/138161211796196945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.