Abstract
Purpose
The issue of potential confounding factors is critical to the development of any approach to radiation biodosimetry, and has not been fully addressed for gene expression-based approaches.
Materials and Methods
As a step in this direction, we have investigated the effect of smoking on the global radiation gene expression response in ex vivo irradiated peripheral blood cells using microarray analysis. We also evaluated the ability of gene expression signatures to predict the radiation exposure level of ex-vivo-exposed samples from smokers and non-smokers of both genders.
Results
We identified 8 genes with a radiation response that was significantly affected by smoking status, and confirmed an effect of smoking on the radiation response of the four and a half LIM domains 2 (FHL2) gene using quantitative real-time polymerase chain reaction. The performance of our previously defined 74-gene signature in predicting the radiation dose to samples in this study was unaffected by differences in gender or smoking status, however, giving 98% correct prediction of dose category. This is the same accuracy as that found in the original study from which the signature was derived, using different donors.
Conclusion
The results support the development of peripheral blood gene expression as a viable strategy for radiation biodosimetry.
Keywords: radiation biodosimetry, peripheral blood, gene expression, microarray, smoking, confounding factors
Introduction
Gene expression profiles that can predict radiation exposure or provide radiation dose estimates are being developed as one potential approach to high-throughput radiation biodosimetry for use in a large-scale radiological event, such as a terrorist attack. Most such approaches use peripheral blood as a radiation-sensitive and relatively accessible tissue, and have measured gene expression in whole blood (Grace et al. 2002, 2003; Amundson et al. 2004; Paul and Amundson 2008), isolated lymphocytes (Amundson et al. 2000; Miller et al. 2002; Dressman et al. 2007; Meadows et al. 2008; Turtoi et al. 2008), or specific blood cell subsets, such as T cells (Mori et al. 2004, 2005). Peripheral blood gene expression profiles indicative of chronic low-dose exposures in radiation workers are also being developed (Fachin et al. 2009; Morandi et al. 2009), but it is not known if such chronic exposures might alter the response observed following an acute exposure.
The utility of the gene expression approach to biodosimetry largely depends on the reproducibility of the responses among individuals and the specificity of those responses for radiation exposure. The influence of other factors on the gene expression signatures being developed for radiation biodosimetry has not been thoroughly investigated, however. Previous studies have indicated that inter-individual variation in peripheral blood gene expression profiles occurs among healthy individuals (Cheung et al. 2003; Whitney et al. 2003; Radich et al. 2004). These variations have been associated with factors including sex, age, and body mass index (Eady et al. 2005). Such variations may also affect the gene expression response of individuals to radiation exposure (Correa and Cheung 2004; Smirnov et al. 2009). For instance, Meadows et al., (Meadows et al. 2008) found very different gene expression profiles in response to radiation in male and female mice. They also reported that gender could affect the accuracy of radiation dose estimation using gene expression profiling.
Gene expression in the peripheral blood can also reflect disease states, such as stroke (Baird 2007), cancer (Martin et al. 2001), various immune or inflammatory conditions (Heller et al. 1997; Patino et al. 2005; Mandel and Achiron 2006; Edwards et al. 2007) and graft rejection (Cadeiras et al. 2008). It can also reflect environmental exposures, including tobacco smoke (Dumeaux et al. 2010). It is not currently clear if such disease- or environmentally-associated gene expression variations might interfere with the prediction of radiation exposure dose using gene expression methods.
Considering smoking as a potential confounder, numerous studies have investigated gene expression in smokers’ alveolar cells and a substantial number of genes have been reported to be differentially expressed in lung cells exposed to smoke (Beane et al. 2007; Chari et al. 2007; Harvey et al. 2007; Lodovici et al. 2007; Landi et al. 2008; Boelens et al. 2009). Some of these genes, such as cyclin-dependent kinase inhibitor 1A (CDKN1A), damage specific DNA binding protein 2 (DDB2) and v-myc myelocytomatosis viral oncogene homolog (MYC), are also known to respond to ionizing radiation. Fewer studies have looked at the effect of smoking on gene expression in the peripheral blood, but gene expression is altered here too (Lampe et al. 2004; van Leeuwen et al. 2007; Dumeaux et al. 2010), although it appears that fewer genes are involved. As most of the smoking-responsive genes have been identified in lung cells exposed directly to smoke, it is not immediately clear if smoking may confound the accuracy of gene expression radiation biodosimetry using peripheral blood. Such potential confounders must be considered, however, as it is possible they could either obscure the baseline expression in unexposed individuals, or even alter the response to radiation in exposed individuals.
In the present study, we have investigated the potential impact of smoking on the gene expression response to radiation exposure, and evaluated the ability of gene expression signatures to predict the radiation exposure level of ex vivo irradiated blood samples in smokers and non-smokers of both genders. Our results suggest that peripheral blood gene expression remains a viable strategy for radiation biodosimetry, as the accuracy of our previously defined 74-gene signature in classifying samples by radiation dose level was unaffected by differences in smoking status or gender.
Materials and Methods
Study participants
Smoker and non-smoker volunteers were recruited with informed consent under a protocol approved by the Institutional Review Board of Columbia University. A total of 12 smokers and 12 non-smokers comprising an equal number of males and females of each participated in this study (Table I). Only healthy individuals with no record of previous radiotherapy or recent radiodiagnostic examination who reported themselves as either never-smokers or one pack or more per day smokers were included in this study.
Table I.
Donor characteristics
| Gender | Age | Smoker | Cigarettes smoked / day |
Years smoked |
|---|---|---|---|---|
| Male | 29 | No | None | None |
| Male | 38 | No | None | None |
| Male | 31 | No | None | None |
| Male | 52 | No | None | None |
| Male | 23 | No | None | None |
| Male | 41 | No | None | None |
| Male | 41 | Yes | 1 pack | 20 |
| Male | 31 | Yes | 1 pack | 12 |
| Male | 52 | Yes | 1 pack | 20 |
| Male | 28 | Yes | 1 pack | 10 |
| Male | 59 | Yes | 1.5 pack | 30 |
| Male | 64 | Yes | 1 pack | 50 |
| Female | 24 | No | None | None |
| Female | 35 | No | None | None |
| Female | 24 | No | None | None |
| Female | 39 | No | None | None |
| Female | 40 | No | None | None |
| Female | 21 | No | None | None |
| Female | 32 | Yes | 1.5 pack | 12 |
| Female | 35 | Yes | 1 pack | 15 |
| Female | 27 | Yes | 1 pack | 3 |
| Female | 55 | Yes | 1 pack | 25 |
| Female | 29 | Yes | 1 pack | 15 |
| Female | 54 | Yes | 1.5 pack | 30 |
Sample collection, processing and ribonucleic acid (RNA) preparation
Peripheral blood from smoker and non-smoker volunteers was drawn into 0.105 mol/l sodium citrate Vacutainer tubes (Becton Dickinson and Company, Franklin Lakes, NJ, United States of America (USA)). The blood was divided into 3 ml aliquots and exposed at a rate of 0.82 Gy/min to 0, 0.1, 0.5, or 2 Gy γ-rays using a Gammacell-40 137Cs irradiator (Atomic Energy of Canada, Limited, Ottawa, Ontario, Canada). After irradiation, blood samples were diluted 1:1 with Roswell Park Memorial Institute 1640 Medium (Mediatech, Herndon, VA, USA) supplemented with 10% heat-inactivated fetal bovine serum (HyClone, Logan, UT, USA), as previously described (Grace et al. 2002), and were incubated for 6 h at 37° C in a humidified incubator with 5% CO2.
RNA was prepared using the PerfectPure RNA blood kit (PerfectPure, Gaithersburg, MD, USA) following the manufacturer’s recommendations. This protocol differentially lyses red and white blood cells in whole blood to preferentially deplete globin messenger RNA (mRNA). Remaining globin mRNA was further reduced using GLOBINclear (Ambion Inc., Austin, TX, USA) to remove both α- and β-globin. The RNA was quantified using a NanoDrop-1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA), and quality was monitored with the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). All RNA samples had RNA integrity numbers between 7 and 9.3 (mean, 8.4) and 260/280 absorbance ratios of between 2.0 and 2.4 (mean, 2.09). RNA was stored at −80 °C until use.
Microarray hybridization and data acquisition
Cyanine-3 (Cy3)-labeled complementary RNA (cRNA) was prepared from 0.5 µg input RNA using Agilent’s One-Color Quick Amp labeling kit (Agilent Technologies,) according to the manufacturer’s instructions, followed by purification of cRNA by RNAeasy column (QIAGEN, Valencia, CA, USA). Cy3 dye incorporation and yield of cRNA was checked with a NanoDrop ND-1000 Spectrophotometer (Thermo Scientific). The specific activity of all cRNA samples ranged from 9.5 to 15.8 (mean 12.2). For each sample, 1.65 µg cRNA was fragmented and hybridized to Agilent’s whole genome microarrays (G4112A, Agilent Technologies) at 65 °C for 17 hr with rotation, followed by washing according to Agilent’s recommendations. Microarray slides were immediately scanned with the Agilent Scanner (G2404B, Agilent Technologies) using the recommended settings. The scanned images were analyzed with Feature Extraction 9.1 (Agilent Technologies) using default parameters for background correction and flagging of non-uniform features.
Microarray data analysis
Background corrected hybridization intensities were imported into the National Institutes of Health Biometric Research Branch’s (BRB)-ArrayTools Version 3.8.0 beta (Simon et al. 2007), log2 transformed and normalized using the median over the entire array. Features that were non-uniform outliers or not significantly above background intensity in 25% or more of the samples, or that did not change at least 1.5-fold from the median value in at least 20% of the experiments were filtered out. The above filtering criteria yielded 11,486 features that were used in subsequent analyses. The microarray data is available through the National Center for Biotechnology Information’s Gene Expression Omnibus using accession number GSE23515.
We used class comparison in BRB-ArrayTools to identify genes that were differentially expressed across the radiation doses, using a random-variance F test. Genes were considered statistically significant if their p value was less than 0.001. The false discovery rate (FDR) for each of the differentially expressed genes was also estimated as a measure of the expected frequency of false-positive results, but was not used as the basis for gene selection. The 2-way mixed model Analysis of Variance (ANOVA) from BRB-ArrayTools Plug-ins was used to identify genes with radiation-dose-dependent expression that was affected by smoking or gender. The two-way mixed model ANOVA combines one independent sample factor (Factor A, in this case radiation dose) and one correlated groups factor (Factor B, either smoking or gender), evaluating the main effects of Factor A, Factor B, and their interaction (AB). The F-test is used for statistical significance testing.
Multidimensional scaling (MDS) was performed in BRB-ArrayTools to represent high-dimensional data (e.g., a selected gene expression signature) graphically in low dimensions. The Euclidian distance metric was used to compute a distance matrix and the principal components of the gene expression signature. Each sample was then represented by a single point and the distance between two points indicated the overall similarity of those two samples. The first three principal components of gene expression were used as axes to generate a plot. Data was also visualized using hierarchical clustering in BRB-ArrayTools. The Euclidean distance metric and average linkage were used to cluster genes and generate a heat map.
The BRB-ArrayTools Class Prediction Tool was also used to build classifiers for predicting the radiation dose received by each sample. Diagonal linear discriminant analysis (DLDA), 3-Nearest Neighbors (3NN) and nearest centroid (NC) algorithms were used to predict the dose exposure class of individual samples using gene expression profiles derived from the present study, and most importantly, to test our previously-published 74-gene signature (Paul and Amundson 2008) using leave-one-out cross-validation to compute the misclassification rate. The cross-validated misclassification error rate was estimated as a permutation p-value by randomly permuting the class labels for all samples, and repeating the entire cross-validation process. The p value represents the proportion of 1,000 random permutations that produced a cross-validated misclassification rate at least as low as that obtained with the actual class labels. Sensitivity of prediction for each class (N) was calculated as (number of Class N samples predicted to belong to Class N)/(total number of class N samples), and the specificity was calculated as (number of non-N samples predicted as non-N)/(total number of non-N samples).
Taqman reverse transcription polymerase chain reaction (RT-PCR)
Gene-specific primers and probes were designed with the aid of Applied Biosystems’ Primer Express software (Applied Biosystems, Foster City, CA, USA) and GeneScript Corporation’s online TaqMan Primer Design software (GeneScript Corporation, Piscataway, NJ, USA) as follows: actin, beta (ACTB), 5’-CACTCTTCCAGCCTTCCTTC-3’ (Forward), 5’-GGATGTCCACGTCACACTTC-3’ (Reverse), 5’-TGCCACAGGACTCCATGCCC-3’ (Probe). DDB2, 5’-CAGGACACGGAAGTGAGAGA-3’ (Forward), 5’-CAAATCGCCACCTCTGCTTG-3’ (Reverse), 5’-TCCAAGGCCTTGTCTGGCCC-3’ (Probe), apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H (APOBEC3H), 5’-TGACTTCATCAAGGCTCACG-3’ (Forward), 5’-ACCTGGGATCCACACAGAAG-3’ (Reverse), 5’-CATCTTCGCCTCCCGCCTGT-3’ (Probe), four and a half LIM domains 2 (FHL2), 5’-TCACAGCTCGCGATGACTT-3’ (Forward), 5’-CAGCACACTTCTTGGCATACAAG-3’ (Reverse), 5’-TGCCTACTGCCTGAACTGCTTC-3’ (Probe). Sequences for CDKN1A were the same as those used previously (Amundson et al. 2004). Primers and probes were synthesized by Operon Biotech, Inc. (Huntsville, AL, USA) with the probes containing 6-FAM (carboxyfluorescein) at the 5’-end and BHQ1 (Black Hole Quencher 1) at the 3’-end. 500-ng total RNA was reverse transcribed to complementary deoxyribonucleic acid (cDNA) using the High-Capacity cDNA Archive Kit (Applied Biosystems) according to the manufacturer’s instructions. The real-time PCR reactions were performed with the ABI 7900HT Fast Real-Time PCR System (Applied Biosystems) using Applied Biosystems’ Universal PCR Master Mix (Applied Biosystems) following the manufacturer’s recommendations. Standard curves were generated to optimize the amount of input cDNA for measurement of each gene (5 or 10 ng). All samples were run in duplicate and repeated 3 times on different days for each gene. The relative fold induction of target genes was calculated by the comparative cycle threshold (ΔΔCT) method with ACTB used for normalization. To analyze the statistical significance of smoking or gender on the expression of individual genes, a 2-way mixed model ANOVA was performed using Prism 5 (GraphPad Software, La Jolla, CA, USA).
Results
Differential gene expression in irradiated blood samples
Global gene expression was measured in human peripheral white blood cells (WBC) at 6 h after ex vivo exposure to doses of 0, 0.1, 0.5 and 2 Gy γ-rays. A total of 24 experiments were performed using different donors for each experiment. The donors included equal numbers of smokers and non-smokers from each gender. Agilent whole genome microarrays were hybridized using the one-color protocol to identify genes differentially expressed across radiation doses or to distinguish gene expression responses that varied between smokers and non-smokers and between males and females.
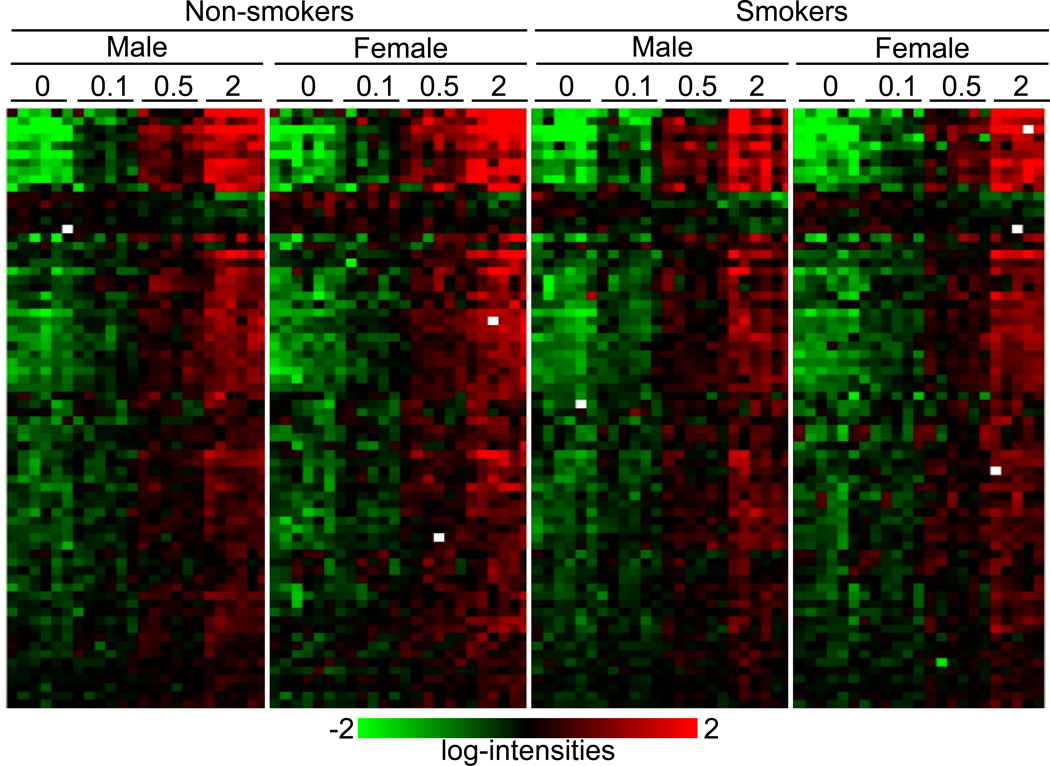
We first used the class comparison feature of BRB-ArrayTools to identify genes that were differentially expressed across all four radiation doses (0, 0.1, 0.5 and 2 Gy), and identified 289 genes that were differentially expressed (p < 0.001). All the genes found at this level of significance were calculated to have a false discovery rate (FDR) < 4% (Supplementary Table 1). Of these 289 genes responding significantly to radiation doses between 0.1 and 2 Gy, 35% (100 genes) were previously found to be dose responsive in the WBC of an independent group of donors between 0.5 and 8 Gy at 6 h after ex vivo irradiation (Paul and Amundson 2008). The relatively modest overlap may be due in part to the difference in the dose ranges used in the two studies. In the earlier work, we had also identified a signature of 74 sequences, which predicted radiation exposure level (0, 0.5, 2, 5, or 8 Gy) at both 6 and 24 hours after exposure. All but 10 of the sequences from that gene set were represented among the 289 genes responding here to the lower dose range. The dose-response of all the genes from the 74-gene consensus signature is illustrated by a hierarchically clustered heatmap of the genes (Figure 1). The gene names and annotations in cluster order are available in Supplementary Table 2. A radiation dose response is clearly evident across the 74-gene radiation signature, without any obvious patterns related to smoking status or gender.
Figure 1.
Average linkage clustering of expression of the genes from the ex vivo-derived 74-gene signature (Paul and Amundson 2008). Radiation dose (in Gy), smoking status and gender of the donors are indicated across the top of the figure. Each row represents an individual gene and each column an individual donor’s sample. Green indicates the lowest expression and red the highest expression. White squares indicate missing data (genes in a specific sample that do not pass the filtering criteria.) Annotation of the genes in cluster order is available in Supplementary Table 2.
Potential effects of smoking
To look for potential confounding of radiation biomarkers by smoking, we first performed a class comparison for differentially expressed genes between smokers and non-smokers using only the sham-irradiated control samples. This analysis identified 43 genes significantly differentially expressed between smokers and non-smokers (p < 0.001) (Supplementary Table 3), but with fairly high FDR (12–25%). Among the 43 potential smoking-modulated genes identified, only one (FHL2) was also a member of the 289-gene radiation dose-responsive set, and none were included in the previous 74-gene signature. To investigate further if smoking could alter the response of genes to radiation, we used BRB-ArrayTools’ 2-way mixed model ANOVA to identify genes that responded to radiation differently in smokers and non-smokers. This analysis identified eight genes with radiation dose responses modified by smoking status (p < 0.001) (Supplementary Table 3). Again, only FHL2 was common with the present 289-gene radiation dose-responsive gene-list, and none of the eight were included in the previous 74-gene set.
Potential effects of gender
We next used the same analyses to look for altered gene expression between genders that might also affect the performance of radiation biomarkers. Class comparison identified 62 genes as differentially expressed (p <0.001 and FDR <18%; 15 genes with FDR <5%) between males and females (Supplementary Table 4). Of these 62 genes, four (lysosomal-associated membrane protein 3 (LAMP3), tumor protein p53 inducible protein 3 (TP53I3), THC2695815, and THC2518594) were also found in the 289-gene set distinguishing dose. We also looked for genes with radiation dose responses that differed between males and females using BRB-ArrayTools’ 2-way mixed model ANOVA. This analysis identified 14 genes differentially modulated across radiation doses that also showed an effect of gender (Supplementary Table 4), with only one gene (APOBEC3H) also identified as a significant radiation dose responsive gene (in both the previous 74-sequence signature and the 289-gene dose-responsive set identified in this study).
Dose-response of individual genes
The ANOVA analysis of our microarray data indicated that the radiation dose response of FHL2 might be altered by smoking, and that of APOBEC3H might be altered by gender. We used quantitative real-time RT-PCR (qRT-PCR) to validate the radiation dose response of these potential confounding genes. In addition, the expression of CDKN1A (Lam et al. 2006) and DDB2 (Lodovici et al. 2007), well-characterized radiation response genes also included in our 74-gene signature, has been reported to be altered in the lung cells of smokers. Although our analysis did not suggest an effect of smoking on the radiation response of these genes in WBC, we also validated their responses in the present study using qRT-PCR.
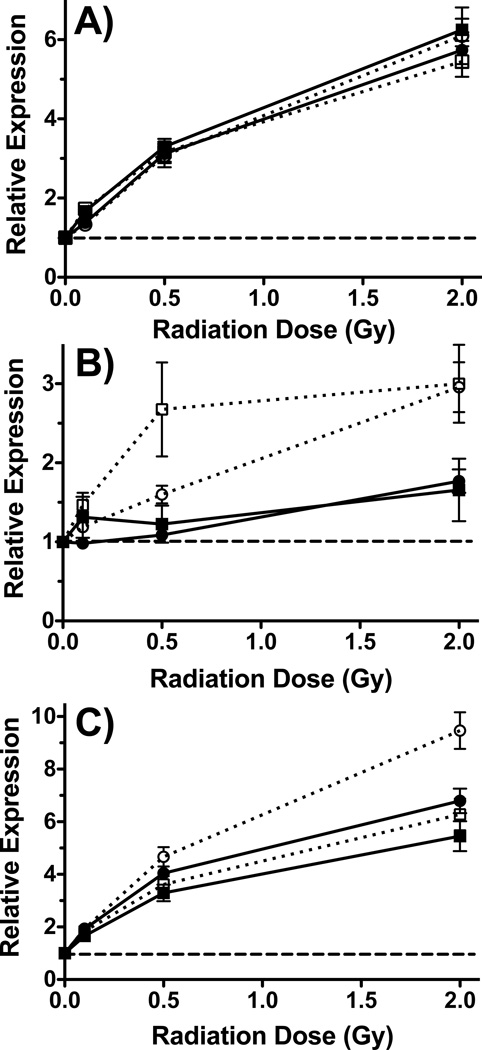
As predicted from the microarray analyses, there was no difference between smokers and non-smokers in the radiation dose response of CDKN1A and DDB2 as measured by qRT-PCR (Figure 2A). In contrast, the dose-response of FHL2 was significantly affected by smoking as measured by both microarray (2-way ANOVA, p<0.0001) and qRT-PCR (2-way ANOVA, p=0.0025) (Figure 2B). Similarly, APOBEC3H responded significantly more to radiation in females than in males as measured by microarray (2-way ANOVA, p=0.0005), and showed a similar trend of gender effect in the qRT-PCR data (Figure 2C). The influence of gender on the radiation response of APOBEC3H was not statistically significant (2-way ANOVA, p=0.16) by qRT-PCR, however.
Figure 2.
Dose response of individual genes measured by microarray and qRT-PCR. A) Relative expression of DDB2 (squares) and CDKN1A (circles) in smokers (open symbols) and non-smokers (closed symbols) measured by qRT-PCR. B) Relative expression of FHL2 in smokers (open symbols) and non-smokers (closed symbols) measured by microarray (circles) and qRT-PCR (squares). C) Mean relative expression of APOBEC3H in females (open symbols) and males (closed symbols) measured by microarray (circles) and qRT-PCR (squares). All points are the mean induced expression (n=12), and error bars are standard error. Induced expression is relative to the levels in untreated controls (dashed lines).
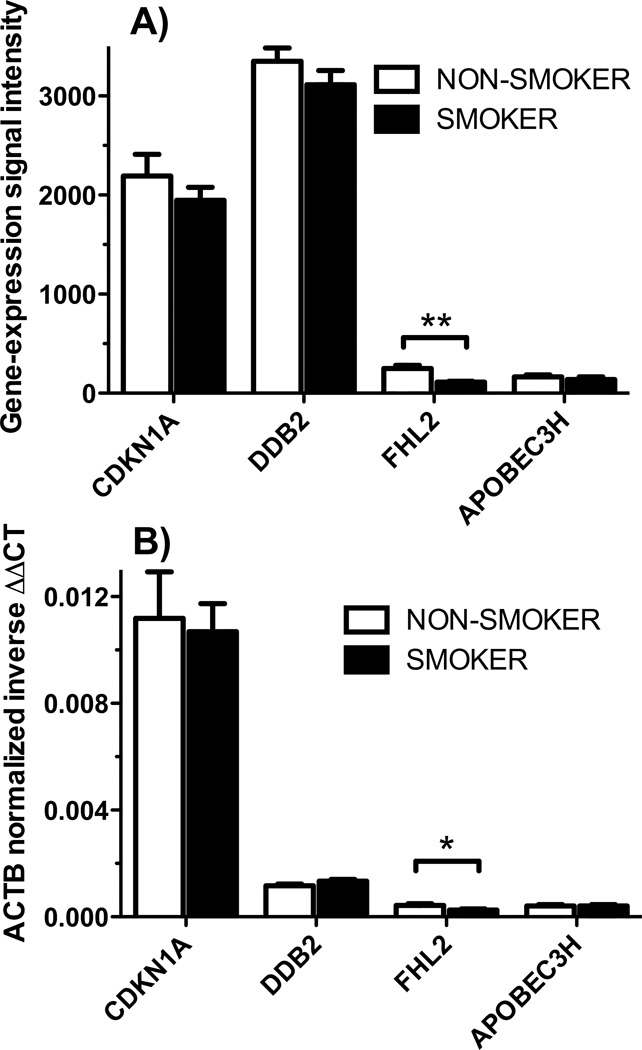
We also compared the baseline expression of these four genes between smokers and non-smokers (Figure 3). No significant differences were observed in baseline expression of CDKN1A, DDB2, or APOBEC3H between smokers and non-smokers. The expression of FHL2 in the absence of radiation exposure, however, was significantly different between smokers and non-smokers as measured by both microarray (Figure 3A) (t-test, p = 0.0004) and qRT-PCR (Figure 3B) (t-test, p = 0.037). There were no gender-associated differences in baseline expression of any of these genes.
Figure 3.
Baseline gene expression of individual genes in smokers and non-smokers was compared by A) microarray gene expression signal intensities normalized to the median array or B) ACTB-normalized inverse ΔΔCT from qRT-PCR. The white bars represent the mean of gene expression measurements for all non-smokers (n=12), and black bars represent the mean of gene expression for all smokers (n=12). Error bars represent standard error. Only FLH2 showed significantly different expression between smokers and non-smokers by either microarray (** p=0.0004) or qRT-PCR (* p=0.037).
Prediction of radiation dose to individual samples
We next used the gene expression data from this study to build classifiers to predict the dose exposure class (0, 0.1, 0.5 or 2 Gy) of individual samples. We used diagonal linear discriminant analysis (DLDA), 3-nearest neighbors (3NN), and nearest centroid (NC) algorithms with leave-one-out cross validation. Using an optimized α (10−6), we achieved 99% correct classification by DLDA, 97% by 3NN and 99% by NC. The permutation p value for the cross-validated misclassification error rate was p < 0.001. Most importantly, we also used our previously identified 74-gene set to build classifiers using the same algorithms, and tested the ability of this previously-defined signature to predict the dose received by samples from smokers and non-smokers of both genders. All three of the algorithms classified 98% of samples correctly using the previously-defined signature, and the p-value of the misclassification error rate was again p<0.001. The sensitivity and specificity of the DLDA classifiers are summarized in Table II.
Table II.
Performance of classifiers predicting dose level.
| Current study (α=10−6) | |||
|---|---|---|---|
| Dose (Gy) | Sensitivity | Specificity | |
| 0 | 1 | 1 | |
| 0.1 | 1 | 1 | |
| 0.5 | 1 | 0.986 | |
| 2 | 0.957 | 1 | |
| 74-gene signature | |||
| Dose (Gy) | Sensitivity | Specificity | |
| 0 | 1 | 1 | |
| 0.1 | 1 | 0.986 | |
| 0.5 | 0.957 | 0.986 | |
| 2 | 0.957 | 1 | |
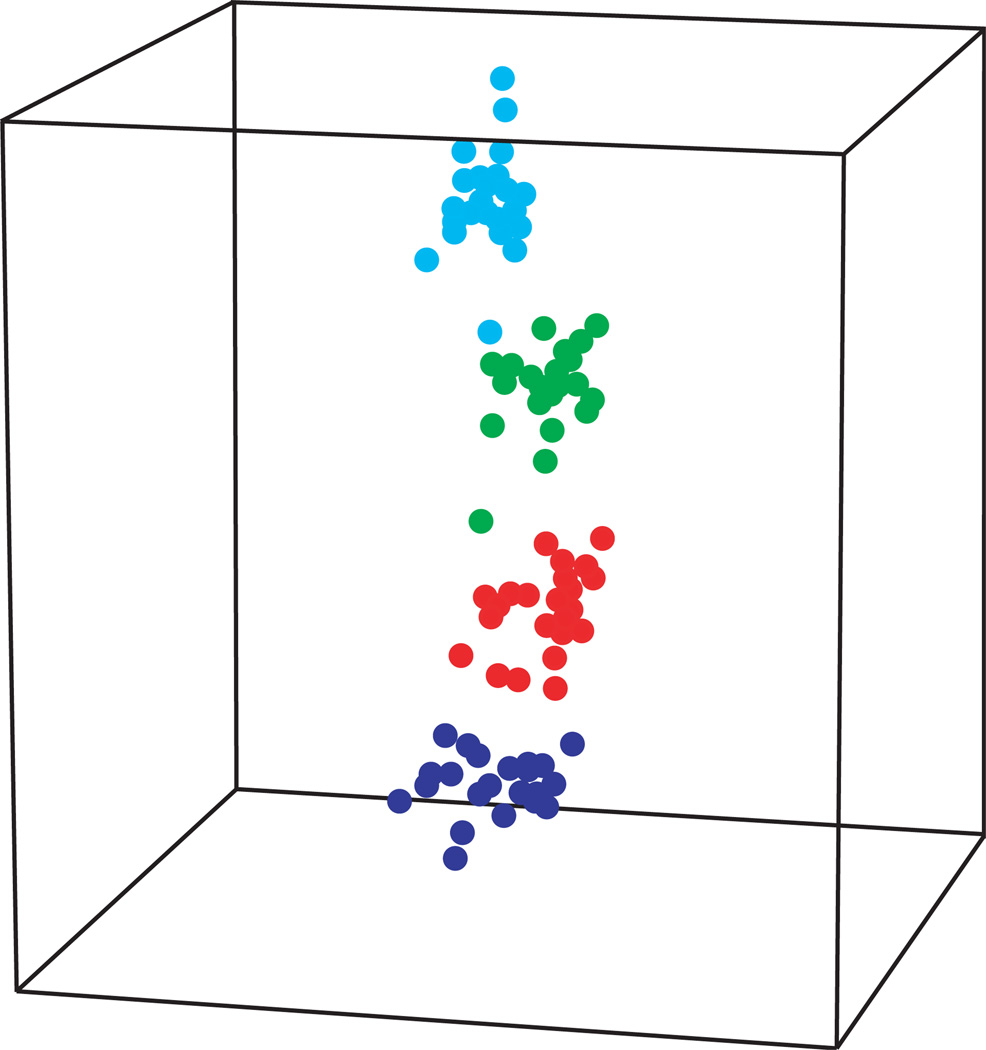
The separation of the samples by dose using the 74-gene signature is illustrated graphically using multi-dimensional scaling (MDS) (Figure 4). MDS is a visualization tool that allows reduced-dimension graphic representation of high dimensional data while preserving pair-wise relationships between samples. In Figure 4, each spot represents a sample and the distance between two spots reflects the overall similarity of all 74 genes in the signature. The MDS plot illustrates a clear dose-response trend of the 74-gene signature across all samples irrespective of gender or smoking habit, and despite the lower dose range of the exposures used in this study.
Figure 4.
Multidimensional scaling plot showing separation of samples by dose using the ex vivo-derived 74-gene signature (Paul and Amundson 2008). The axes represent the first three principle components of gene expression, and each point represents the relative expression of all 74 genes in an individual sample, with the distance between two points reflecting the overall similarity or difference between those two samples. Points are colored according to dose: 0 Gy (blue), 0.1 Gy (red), 0.5 Gy (green), and 2 Gy (cyan).
Discussion
In the current study, we investigated the potential impact of the confounders smoking and gender on the ability of WBC gene expression profiles to predict radiation exposure dose. Various classifiers built from the gene expression data correctly predicted samples as being exposed to 0, 0.1, 0.5 or 2 Gy with 97–99% accuracy, regardless of smoking status or gender.
We had previously identified a 74-gene signature (Paul and Amundson 2008) that robustly distinguished radiation doses across a window of time (6 to 24 h) and dose (0, 0.5, 2, 5, and 8 Gy). In the present study, we focused on lower doses as we reasoned that if smoking constitutively altered transcription levels, confounding might be more likely among the controls and radiation exposures at the low end of the dose range. We found that classifiers built using the previously identified 74-gene signature performed similarly to those built using the gene expression data from the present study. Classifiers built from the 74-gene set could predict radiation doses of 0, 0.1, 0.5 or 2 Gy with 98% accuracy, the same performance found in the original study, despite the lower dose range used here, the use of independent donors, and the addition of smoking as a potential confounding variable. A trend has been emerging in predictive biomarker studies suggesting that application of a predictive signature to a new set of independent samples often results in a drastic loss of predictive power (Michiels et al. 2005; Buyse et al. 2006; Koscielny 2010). The ability of the 74-gene set to accurately classify samples from independent donors by radiation dose, especially considering the lower dose range and addition of the smoking variable, is thus quite remarkable. We have also recently reported on the conservation of performance of the same ex vivo derived 74-gene signature in patients undergoing total body irradiation. In that study, the ex vivo signature predicted the in vivo exposure dose correctly in 98% of samples (Paul et al. 2011), supporting the usefulness of the ex vivo model for developing clinically relevant gene expression signatures. Further work is obviously needed in this area, but the observed conservation of signature performance may be due, at least in part, to the robust and dose-dependent nature of the radiation response, as compared to the potentially more varied and subtle gene expression differences that may be involved in other complex physiological processes, such as tumor development or metastasis.
A number of genes that were radiation responsive in this or our previous peripheral blood study (Ras-related associated with diabetes (RRAD), calcium binding tyrosine-(γ)-phosphorylation regulated (CABYR), caveolin 1 (CAV1), apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (APOBEC3G), harakiri (HRK), like-glycosyltransferase (LARGE), methyltransferase like 7A (METTL7A), nuclear receptor subfamily 4, group A, member 3 (NR4A3), triggering receptor expressed on myeloid cells 2 (TREM2), tetratricopeptide repeat domain 21A (TTC21A), CDKN1A, DDB2 and MYC) have previously been reported to be differentially expressed in alveolar cells from smokers’ lungs compared to those from non-smokers (Chari et al. 2007; Harvey et al. 2007; Lodovici et al. 2007; Landi et al. 2008; Boelens et al. 2009). Three of these genes, CDKN1A, DDB2 and MYC, were included in our previous ex vivo 74-gene signature (Paul and Amundson 2008), and might have been of concern as potential confounders. However, we did not detect any significant differences in basal expression or radiation response in WBC of any of these genes by microarray, or of CDKN1A or DDB2 by qRT-PCR. Fewer studies have considered gene expression differences in peripheral WBC of smokers and non-smokers, and none of the genes from the 74-gene set have been reported to be differentially expressed in WBC. Among genes reported to be up regulated in the WBC of smokers, only superoxide dismutase 2, mitochondrial (SOD2) (van Leeuwen et al. 2007), killer cell immunoglobulin-like receptor, two domains, short cytoplasmic tail, 4 (KIR2DS4) and thromboxane A synthase 1 (TBXAS1) (Dumeaux et al. 2010) were also responsive to radiation in prior WBC studies. It seems likely that smoking-related altered expression of most genes may require direct exposure to cigarette smoke, such as would be experienced by alveolar cells.
Although we observed some differences in peripheral WBC baseline gene expression between smokers and non-smokers, and between males and females, very few of these genes were also significantly responsive to radiation exposure. The radiation response of only one gene (FHL2) was found to be altered by smoking. Another gene (APOBEC3H) seemed to respond to radiation differently in males and females based on microarray measurements, but the difference was not significant by pRT-PCR. Since very few of the radiation response genes were affected by these variables, such potential confounding genes could easily be excluded from biodosimetric signatures. This also suggests that confounding by smoking or gender would not be expected to negatively affect classification of radiation dose using gene expression, as indeed it did not in this study. Because our study looked at only a small number of individuals (24), however, and considered only 1–1.5 pack-a-day current smokers, it is still possible that other aspects of the smoking variable, such as previous smokers, lighter or heavier smokers than those included here, or total number of pack-years smoked could affect the gene expression response to radiation. It is also possible that smoking could affect the radiation signature in only a subset of individuals with a specific genetic make-up or other interacting lifestyle or health factor. Large-scale population studies will ultimately be necessary to determine if such effects might impact radiation dosimetry signatures.
Gender-related variation in WBC gene expression in healthy human populations has also been previously reported (Whitney et al. 2003) and could potentially limit the accuracy of WBC-based gene expression radiation biodosimetry. In a murine radiation biodosimetry study, Meadows et al., (Meadows et al. 2008) found a strong gender specificity for radiation responses, with only 2 genes in common between the male and female WBC signatures of a 50 cGy exposure. Although their different male- and female-specific signatures did perform relatively well in classifying samples from the opposite gender, they concluded that gender-related differences could be a concern for distinguishing low dose exposures from unirradiated controls. Studies of other endpoints, including DNA strand breaks, DNA methylation (Pogribny et al. 2004), and expression of micro-RNA (Ilnytskyy et al. 2008) have also reported very different responses to radiation in male and female mice. In contrast, in human peripheral WBC, we have found that gene expression can accurately predict exposure to 0, 0.1, 0.5 or 2 Gy doses irrespective of gender. Of 289 radiation dose responsive genes identified in the present study, only five genes (LAMP3, TP53I3, THC2695815, THC2518594 and APOBEC3H) were also differentially expressed between males and females, but with high FDR. Since the Meadows study indicated little similarity between the radiation responsive genes in male and female mice, but did show a fair ability of the very distinct signatures to classify samples from the opposite gender, we also re-analyzed our data to identify radiation dose classifiers using data only from males or only from females in order to see if our human classifiers could show a similar gender specificity. We found that classifiers derived using data from a single gender performed equally well when applied to unknown samples from the same or opposite gender (Table III and Supplementary Tables 5 and 6). We also found an 88% overlap in the genes contained in the two signatures. Thus it appears that gender may have less influence on the radiation responsiveness of gene expression in humans than in mice. It is also possible that the different modes of irradiation (ex vivo versus in vivo) and different analyses applied in the two studies may have contributed to the differing results.
Table III.
Accuracy of classifiers derived from only one gender.
| Percent Correct Classificationa | ||
|---|---|---|
| Male Classifiersb | Male Samples | Female Samples |
| DLDA | 98% | 100% |
| 3NN | 94% | 98% |
| NC | 94% | 100% |
| Female Classifiersc | Male Samples | Female Samples |
| DLDA | 98% | 100% |
| 3NN | 98% | 95% |
| NC | 96% | 100% |
Percentage of samples of unknown radiation dose assigned to the correct dose category using leave-one-out cross validation.
Classification by different algorithms using signature based only on data from male samples.
Classification by different algorithms using signature based only on data from female samples.
Previous studies have indicated that inter-individual variation in gene expression in peripheral blood generally occurs within populations of healthy individuals (Cheung et al. 2003; Whitney et al. 2003; Radich et al. 2004; Dumeaux et al. 2010) and may therefore limit the practical use of gene expression for radiation biodosimetry. Further biodosimetry studies are still needed to understand potential variations in larger populations, and should include people with diverse clinical conditions, different lifestyle factors, ethnic backgrounds, and so on. Conditions like extreme exercise, infection or septic shock, have also been shown to alter gene expression in peripheral blood (Zieker et al. 2005; Ramilo et al. 2007; Wong et al. 2009; Nakamura et al. 2010), and should also be considered for their potential to confound dosimetric estimates based on gene expression. In addition, the potential effect of combined injuries (involving radiation exposure and physical trauma, such as lacerations, broken bones or burns) should also be accounted for in the development of gene expression biodosimetry, especially as such injuries might be expected in a large-scale radiological disaster.
Although there is clearly further work to be done, our results demonstrate that a large number of genes responded to radiation exposure in a dose-dependent manner in a group of donors comprising two potential confounding variables. Most importantly, our previously defined 74-gene signature performed dose classification of the samples in the present study equally as well as did the gene set selected here. The 74-gene signature was based on the responses of non-smokers, who were different individuals from those included in the present study. Those genes were also selected from responses to a higher range of doses, including 5 and 8 Gy exposures not used in the present study, and not including the 0.1 Gy dose, which was nonetheless effectively classified here. These findings support the continued development of a gene expression approach for radiation biodosimetry in large heterogeneous populations, and suggest that the effects of gender or smoking status are not likely to meaningfully confound dose estimates.
Supplementary Material
Acknowledgements
We thank the nursing staff of the Department of Radiation Oncology at the Columbia University Medical Center for drawing blood from our volunteers. Analyses were performed using BRB-ArrayTools developed by Dr. Richard Simon and Amy Peng Lam.
This work was supported by the Center for High-Throughput Minimally-Invasive Radiation Biodosimetry, National Institute of Allergy and Infectious Diseases grant No. U19 AI067773.
Footnotes
Declaration of Interest
The authors report no conflicts of interest.
References
- Amundson SA, Grace MB, McLeland CB, Epperly MW, Yeager A, Zhan Q, Greenberger JS, Fornace AJ., Jr Human in vivo radiation-induced biomarkers: gene expression changes in radiotherapy patients. Cancer Research. 2004;64:6368–6371. doi: 10.1158/0008-5472.CAN-04-1883. [DOI] [PubMed] [Google Scholar]
- Amundson SA, Shahab S, Bittner M, Meltzer P, Trent J, Fornace AJ., Jr Identification of potential mRNA markers in peripheral blood lymphocytes for human exposure to ionizing radiation. Radiation Research. 2000;154:342–346. doi: 10.1667/0033-7587(2000)154[0342:iopmbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Baird AE. Blood genomics in human stroke. Stroke. 2007;38:694–698. doi: 10.1161/01.STR.0000250431.99687.7b. [DOI] [PubMed] [Google Scholar]
- Beane J, Sebastiani P, Liu G, Brody JS, Lenburg ME, Spira A. Reversible and permanent effects of tobacco smoke exposure on airway epithelial gene expression. Genome Biology. 2007;8:R201. doi: 10.1186/gb-2007-8-9-r201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelens MC, van den Berg A, Fehrmann RS, Geerlings M, de Jong WK, te Meerman GJ, Sietsma H, Timens W, Postma DS, Groen HJ. Current smoking-specific gene expression signature in normal bronchial epithelium is enhanced in squamous cell lung cancer. Journal of Pathology. 2009;218:182–191. doi: 10.1002/path.2520. [DOI] [PubMed] [Google Scholar]
- Buyse M, Loi S, van't Veer L, Viale G, Delorenzi M, Glas AM, d'Assignies MS, Bergh J, Lidereau R, Ellis P, Harris A, Bogaerts J, Therasse P, Floore A, Amakrane M, Piette F, Rutgers E, Sotiriou C, Cardoso F, Piccart MJ. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. Journal of the National Cancer Institute. 2006;98:1183–1192. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- Cadeiras M, Bayern M, Burke E, Dedrick R, Gangadin A, Latif F, Shazad K, Sinha A, Tabak EG, Marboe CC, Califano AC, Deng MC. Gene expression profiles of patients with antibody-mediated rejection after cardiac transplantation. Journal of Heart and Lung Transplantation. 2008;27(8):932–934. doi: 10.1016/j.healun.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Chari R, Lonergan KM, Ng RT, MacAulay C, Lam WL, Lam S. Effect of active smoking on the human bronchial epithelium transcriptome. BMC Genomics. 2007;8:297. doi: 10.1186/1471-2164-8-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung VG, Conlin LK, Weber TM, Arcaro M, Jen KY, Morley M, Spielman RS. Natural variation in human gene expression assessed in lymphoblastoid cells. Nature Genetics. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- Correa CR, Cheung VG. Genetic variation in radiation-induced expression phenotypes. American Journal of Human Genetics. 2004;75:885–890. doi: 10.1086/425221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressman HK, Muramoto GG, Chao NJ, Meadows S, Marshall D, Ginsburg GS, Nevins JR, Chute JP. Gene expression signatures that predict radiation exposure in mice and humans. PLoS Medicine. 2007;4:e106. doi: 10.1371/journal.pmed.0040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumeaux V, Olsen KS, Nuel G, Paulssen RH, Borresen-Dale AL, Lund E. Deciphering normal blood gene expression variation--The NOWAC postgenome study. PLoS Genetics. 2010;6:e1000873. doi: 10.1371/journal.pgen.1000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eady JJ, Wortley GM, Wormstone YM, Hughes JC, Astley SB, Foxall RJ, Doleman JF, Elliott RM. Variation in gene expression profiles of peripheral blood mononuclear cells from healthy volunteers. Physiological Genomics. 2005;22:402–411. doi: 10.1152/physiolgenomics.00080.2005. [DOI] [PubMed] [Google Scholar]
- Edwards CJ, Feldman JL, Beech J, Shields KM, Stover JA, Trepicchio WL, Larsen G, Foxwell BM, Brennan FM, Feldmann M, Pittman DD. Molecular profile of peripheral blood mononuclear cells from patients with rheumatoid arthritis. Molecular Medicine. 2007;13:40–58. doi: 10.2119/2006-000056.Edwards. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachin AL, Mello SS, Sandrin-Garcia P, Junta CM, Ghilardi-Netto T, Donadi EA, Passos GA, Sakamoto-Hojo ET. Gene expression profiles in radiation workers occupationally exposed to ionizing radiation. Journal of Radiation Research (Tokyo) 2009;50:61–71. doi: 10.1269/jrr.08034. [DOI] [PubMed] [Google Scholar]
- Grace MB, McLeland CB, Blakely WF. Real-time quantitative RT-PCR assay of GADD45 gene expression changes as a biomarker for radiation biodosimetry. International Journal of Radiation Biology. 2002;78:1011–1021. doi: 10.1080/09553000210158056. [DOI] [PubMed] [Google Scholar]
- Grace MB, McLeland CB, Gagliardi SJ, Smith JM, Jackson WEr, Blakely WF. Development and assessment of a quantitative reverse transcription-PCR assay for simultaneous measurement of four amplicons. Clinical Chemistry. 2003;49:1467–1475. doi: 10.1373/49.9.1467. [DOI] [PubMed] [Google Scholar]
- Harvey BG, Heguy A, Leopold PL, Carolan BJ, Ferris B, Crystal RG. Modification of gene expression of the small airway epithelium in response to cigarette smoking. Journal of Molecular Medicine. 2007;85:39–53. doi: 10.1007/s00109-006-0103-z. [DOI] [PubMed] [Google Scholar]
- Heller RA, Schena M, Chai A, Shalon D, Bedilion T, Gilmore J, Woolley DE, Davis RW. Discovery and analysis of inflammatory disease-related genes using cDNA microarrays. Proceedings of the National Academies of Science (USA) 1997;94:2150–2155. doi: 10.1073/pnas.94.6.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilnytskyy Y, Zemp FJ, Koturbash I, Kovalchuk O. Altered microRNA expression patterns in irradiated hematopoietic tissues suggest a sex-specific protective mechanism. Biochemical and Biophysical Research Communications. 2008;377:41–45. doi: 10.1016/j.bbrc.2008.09.080. [DOI] [PubMed] [Google Scholar]
- Koscielny S. Why most gene expression signatures of tumors have not been useful in the clinic. Science Translational Medicine. 2010;2:14ps2. doi: 10.1126/scitranslmed.3000313. [DOI] [PubMed] [Google Scholar]
- Lam DC, Girard L, Suen WS, Chung LP, Tin VP, Lam WK, Minna JD, Wong MP. Establishment and expression profiling of new lung cancer cell lines from Chinese smokers and lifetime never-smokers. Journal of Thoracic Oncology. 2006;1:932–942. [PubMed] [Google Scholar]
- Lampe JW, Stepaniants SB, Mao M, Radich JP, Dai H, Linsley PS, Friend SH, Potter JD. Signatures of environmental exposures using peripheral leukocyte gene expression: tobacco smoke. Cancer Epidemiology Biomarkers and Prevention. 2004;13:445–453. [PubMed] [Google Scholar]
- Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, Murphy SE, Yang P, Pesatori AC, Consonni D, Bertazzi PA, Wacholder S, Shih JH, Caporaso NE, Jen J. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodovici M, Luceri C, De Filippo C, Romualdi C, Bambi F, Dolara P. Smokers and passive smokers gene expression profiles: correlation with the DNA oxidation damage. Free Radical Biology and Medicine. 2007;43:415–422. doi: 10.1016/j.freeradbiomed.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Mandel M, Achiron A. Gene expression studies in systemic lupus erythematosus. Lupus. 2006;15:451–456. doi: 10.1191/0961203306lu2332oa. [DOI] [PubMed] [Google Scholar]
- Martin KJ, Graner E, Li Y, Price LM, Kritzman BM, Fournier MV, Rhei E, Pardee AB. High-sensitivity array analysis of gene expression for the early detection of disseminated breast tumor cells in peripheral blood. Proceedings of the National Academies of Science (USA) 2001;98:2646–2651. doi: 10.1073/pnas.041622398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows SK, Dressman HK, Muramoto GG, Himburg H, Salter A, Wei Z, Ginsburg GS, Chao NJ, Nevins JR, Chute JP. Gene expression signatures of radiation response are specific, durable and accurate in mice and humans. PLoS One. 2008;3:e1912. doi: 10.1371/journal.pone.0001912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels S, Koscielny S, Hill C. Prediction of cancer outcome with microarrays: a multiple random validation strategy. Lancet. 2005;365:488–492. doi: 10.1016/S0140-6736(05)17866-0. [DOI] [PubMed] [Google Scholar]
- Miller AC, Luo L, Chin WK, Director-Myska AE, Prasanna PG, Blakely WF. Proto-oncogene expression: a predictive assay for radiation biodosimetry applications. Radiation Protection and Dosimetry. 2002;99:295–302. doi: 10.1093/oxfordjournals.rpd.a006789. [DOI] [PubMed] [Google Scholar]
- Morandi E, Severini C, Quercioli D, Perdichizzi S, Mascolo MG, Horn W, Vaccari M, Nucci MC, Lodi V, Violante FS, Bolognesi C, Grilli S, Silingardi P, Colacci A. Gene expression changes in medical workers exposed to radiation. Radiation Research. 2009;172:500–508. doi: 10.1667/RR1545.1. [DOI] [PubMed] [Google Scholar]
- Mori M, Benotmane MA, Tirone I, Hooghe-Peters EL, Desaintes C. Transcriptional response to ionizing radiation in lymphocyte subsets. Cellular and Molecular Life Sciences. 2005;62:1489–1501. doi: 10.1007/s00018-005-5086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Benotmane MA, Vanhove D, van Hummelen P, Hooghe-Peters EL, Desaintes C. Effect of ionizing radiation on gene expression in CD4+ T lymphocytes and in Jurkat cells: unraveling novel pathways in radiation response. Cellular and Molecular Life Sciences. 2004;61:1955–1964. doi: 10.1007/s00018-004-4147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Kobayashi M, Sugino T, Kajimoto O, Matoba R, Matsubara K. Effect of exercise on gene expression profile in unfractionated peripheral blood leukocytes. Biochemical and Biophysical Research Communications. 2010;391:846–851. doi: 10.1016/j.bbrc.2009.11.150. [DOI] [PubMed] [Google Scholar]
- Patino WD, Mian OY, Kang JG, Matoba S, Bartlett LD, Holbrook B, Trout HHr, Kozloff L, Hwang PM. Circulating transcriptome reveals markers of atherosclerosis. Proceedings of the National Academies of Science (USA) 2005;102:3423–3428. doi: 10.1073/pnas.0408032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Amundson SA. Development of gene expression signatures for practical radiation biodosimetry. International Journal of Radiation Oncology Biology and Physics. 2008;71:1236–1244. doi: 10.1016/j.ijrobp.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Barker CA, Turner HC, McLane A, Wolden SL, Amundson SA. Prediction of in vivo radiation dose status in radiotherapy patients using ex vivo and in vivo gene expression signatures. Radiation Research in press. 2011 doi: 10.1667/RR2420.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogribny I, Raiche J, Slovack M, Kovalchuk O. Dose-dependence, sex- and tissue-specificity, and persistence of radiation-induced genomic DNA methylation changes. Biochemical and Biophysical Research Communications. 2004;320:1253–1261. doi: 10.1016/j.bbrc.2004.06.081. [DOI] [PubMed] [Google Scholar]
- Radich JP, Mao M, Stepaniants S, Biery M, Castle J, Ward T, Schimmack G, Kobayashi S, Carleton M, Lampe J, Linsley PS. Individual-specific variation of gene expression in peripheral blood leukocytes. Genomics. 2004;83:980–988. doi: 10.1016/j.ygeno.2003.12.013. [DOI] [PubMed] [Google Scholar]
- Ramilo O, Allman W, Chung W, Mejias A, Ardura M, Glaser C, Wittkowski KM, Piqueras B, Banchereau J, Palucka AK, Chaussabel D. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–2077. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Lam A, Li M-C, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-Array Tools. Cancer Informatics. 2007;2:11–17. [PMC free article] [PubMed] [Google Scholar]
- Smirnov DA, Morley M, Shin E, Spielman RS, Cheung VG. Genetic analysis of radiation-induced changes in human gene expression. Nature. 2009;459:587–591. doi: 10.1038/nature07940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turtoi A, Brown I, Oskamp D, Schneeweiss FH. Early gene expression in human lymphocytes after gamma-irradiation-a genetic pattern with potential for biodosimetry. International Journal of Radiation Biology. 2008;84:375–387. doi: 10.1080/09553000802029886. [DOI] [PubMed] [Google Scholar]
- van Leeuwen DM, van Agen E, Gottschalk RW, Vlietinck R, Gielen M, van Herwijnen MH, Maas LM, Kleinjans JC, van Delft JH. Cigarette smoke-induced differential gene expression in blood cells from monozygotic twin pairs. Carcinogenesis. 2007;28:691–697. doi: 10.1093/carcin/bgl199. [DOI] [PubMed] [Google Scholar]
- Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA, Brown PO. Individuality and variation in gene expression patterns in human blood. Proceedings of the National Academies of Science (USA) 2003;100:1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HR, Cvijanovich N, Allen GL, Lin R, Anas N, Meyer K, Freishtat RJ, Monaco M, Odoms K, Sakthivel B, Shanley TP. Genomic expression profiling across the pediatric systemic inflammatory response syndrome, sepsis, and septic shock spectrum. Critical Care Medicine. 2009;37:1558–1566. doi: 10.1097/CCM.0b013e31819fcc08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieker D, Zieker J, Dietzsch J, Burnet M, Northoff H, Fehrenbach E. CDNA-microarray analysis as a research tool for expression profiling in human peripheral blood following exercise. Exercise Immunology Review. 2005;11:86–96. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.