Abstract
Translesion synthesis (TLS) is a pathway in which specialized, low-fidelity DNA polymerases are used to overcome replication blocks caused by DNA damage. The use of this pathway often results in somatic mutations that can drive carcinogenesis. Rev1 is a TLS polymerase found in all eukaryotes that plays a pivotal role in mediating DNA-damage induced mutagenesis. It possesses a BRCA1 C-terminal (BRCT) domain that is required for its function. The rev1-1 allele encodes a mutant form of Rev1 with a G193R substitution in this domain, which reduces DNA damage-induced mutagenesis. Despite its clear importance in mutagenic TLS, the role of the BRCT domain is unknown. Here, we report the X-ray crystal structure of the yeast Rev1 BRCT domain and show that substitutions in residues constituting its phosphate-binding pocket do not affect mutagenic TLS. This suggests that the role of the Rev1 BRCT domain is not to recognize phosphate groups on protein binding partners or on DNA. We also found that the G193 residue is located in a conserved turn region of the BRCT domain, and our in vivo and in vitro studies suggest that the G193R substitution may disrupt Rev1 function by destabilizing the fold of the BRCT domain.
INTRODUCTION
DNA damage presents a serious problem during DNA replication, because DNA synthesis by the high-fidelity, replicative DNA polymerases is blocked by DNA lesions. One way to overcome these blocks is translesion synthesis (TLS). In TLS, low-fidelity DNA polymerases, which have evolved to accommodate DNA lesions, are used to replicate through the damage [1-8]. Because of their low fidelity and lack of proofreading activity, TLS polymerases generate mutations. This is problematic, because an accumulation of somatic mutations drives carcinogenesis. It has previously been shown that the knockdown of TLS polymerases reduces carcinogen-induced tumor formation [9]. Moreover, TLS polymerases promote acquired-chemoresistance, and inhibition of TLS polymerases enhances the efficacy of some chemotherapeutics [10, 11]. Therefore, TLS polymerases are potential chemopreventative and chemotherapeutic drug targets [9-12].
In eukaryotes, the TLS polymerases Pol ζ and Rev1 often function together and play critical roles in mediating mutagenic TLS [13-15]. For example, in the budding yeast S. cerevisiae, these proteins are responsible for >90% of ultraviolet (UV) radiation-induced mutations, and in human cells, knockdown of either protein reduces UV-induced mutagenesis nearly 75% [16-19]. The role of Pol ζ in generating these mutations is well understood. Although Pol ζ is not able to efficiently incorporate nucleotides directly opposite sites of DNA damage, it is able to extend from nucleotides incorporated either correctly or incorrectly opposite the damaged base by another TLS polymerase [1, 15, 20, 21]. Amino acid substitutions that reduce the efficiency of nucleotide incorporation by Pol ζ in vitro, reduce DNA damage-induced mutagenesis in vivo [22].
The role of the Rev1 protein in generating mutations, however, is less clear. Rev1 is able to efficiently incorporate nucleotides opposite several common types of DNA damage including abasic sites, N2-adducted guanines, and N6-ethinoadenines [23-26]. In most cases the catalytic activity of Rev1 is dispensable for mutagenesis. Thus, Rev1 plays a critical non-catalytic function in mutagenic TLS that likely involves mediating a key protein-protein interaction [27]. In addition to the polymerase domain, Rev1 also contains at least two other functional regions: a C-terminal domain and an N-terminal BRCT domain. The C-terminal domain of Rev1 interacts with other TLS DNA polymerases and is thought to promote mutagenic TLS by acting as a protein scaffold [28-33]. The BRCT domain of Rev1 also plays a critical role in mutagenic TLS. The rev1-1 allele encodes a mutant form of Rev1 with a G193R substitution in the BRCT domain, which nearly eliminates UV-induced mutagenesis in yeast [34]. Targeted deletion of the Rev1 BRCT domain in mice also reduces UV-induced mutagenesis and nearly eliminates mutagenesis at thymine dimers [35]. Despite its clear importance, the function of the Rev1 BRCT domain is unknown.
BRCT domains are typically found in proteins involved in the DNA damage response, and these domains have been associated with several activities including the binding of phosphorylated proteins, the binding of non-phosphorylated proteins, and the binding of DNA [36-38]. A previous study showed that the yeast Rev1 BRCT domain binds phosphorylated peptides with a higher affinity than non-phosphorylated peptides in vitro [39]. Another study demonstrated that the yeast and mouse Rev1 BRCT domains are capable of binding phosphate groups on DNA primer-template junctions in vitro [40]. It is unknown if either of these activities are required for its function in mutagenic TLS in cells. Nor is it known if the G193R substitution disrupts either of these activities.
To gain insight into the role of the Rev1 BRCT domain in TLS, we determined its X-ray crystal structure and carried out structure-function analyses of key residues within the domain. We found that the yeast Rev1 BRCT domain has a canonical BRCT domain phosphate-binding pocket, but substitutions of conserved residues within this binding pocket did not lead to a reduction of UV-induced mutagenesis in vivo. This suggests that recognition of phosphate groups on either proteins or on DNA by the BRCT domain of Rev1 is not required for mutagenic TLS. By contrast, even conservative substitutions of Gly-193 resulted in substantial defects in UV-induced mutagenesis similar to the G193R substitution. Our analysis of the structure suggested that the substitution of residues at this position might destabilize the fold of the BRCT domain. Consistent with our structural analysis, we found that only the substitution of residues designed to disrupt the stability of the BRCT domain led to defects in induced mutagenesis. Based on these and other results, we conclude that that G193R substitution destabilizes the yeast Rev1 BRCT domain and that proper folding of the domain is necessary for its function in the TLS pathway.
EXPERIMENTAL PROCEDURES
Protein expression and purification
The yeast Rev1 G193R mutant protein (residues 1-746), which is lacking 239 amino acid residues from its C-terminus, but contains the N-terminal BRCT domain and the polymerase domain, was expressed and purified as described previously for the wild-type protein [41]. We used this truncated protein because it is over-expressed to a greater extent than is the full length protein and because this truncation does not affect catalytic activity [42]. Briefly, the mutant protein was expressed in the yeast strain BJ5464 harboring the pKW385 plasmid which encoded the Rev1 mutant protein fused in frame with glutathione-s-transferase (GST). The protein was purified by affinity chromatography using a glutathione column (GE Healthcare). The Rev1 mutant protein was eluted from the column by incubation with Prescission Protease (GE healthcare) at 4°C overnight. This resulted in the removal of the GST tag, which remained bound to the column. The concentration of purified protein was determined by UV spectroscopy.
The yeast Rev1 BRCT domain (residues 159-250) was over-expressed and purified using the IMPACT kit (New England Biolabs). The coding sequence of the Rev1 BRCT domain was cloned into the protein expression plasmid pTXB1 to yield pKW384, which expresses this domain as a fusion protein containing a C-terminal tag consisting of an intein and a chitin-binding domain. The fusion protein was produced in E.coli Rosetta-2 (DE3) cells (Invitrogen) and was purified by affinity chromatography using a chitin column (New England Biolabs). The Rev1 BRCT domain was eluted from the chitin column by incubation in a buffer containing 50 mM DTT at 25°C overnight. This resulted in the removal of the intein and chitin-binding domain, which remained bound to the column. The Rev1 BRCT domain was further purified using size exclusion chromatography using a Superdex 75 column (GE Healthcare). The concentration of the purified protein was determined by UV spectroscopy.
DNA and nucleotide substrates
The synthetic oligonucleotides used as substrates in this study were a 24-mer primer strand with the sequence: 5′-GCC TCG CAG CCG TCC AAC CAA CTC and a 45-mer template strand with the sequence: 5′-GGA CGG CAT TGG ATC GAC CTG GAG TTG GTT GGA CGG CTG CGA GGC (Integrated DNA Technologies). Oligonucleotides were gel purified and their concentrations were determined using UV spectroscopy. The primer strand was 32P-labeled on the 5′ end using (γ-32P)ATP (Sigma) and polynucleotide kinase (New England Biolabs). The primer strand and template strand (1 μM each) were annealed in 25 mM Tris·Cl (pH 7.5) and 100 mM NaCl by heating to 90 °C for 2 min. followed by slow cooling to room temperature over several hours. Labeled and annealed DNA substrates were stored at 4°C for up to one week. Solutions of each dNTP (100 mM) were purchased from New England Biolabs and stored in 10 μL aliquots at −80 °C.
Pre-steady state kinetics
Analysis of the kinetics of nucleotide incorporation by the Rev1 G193R mutant protein was carried out as described previously for the wild-type Rev1 protein [23]. Reactions were performed in a buffer containing 25 mM Tris·Cl (pH 7.5), 5 mM MgCl2, 5 mM DTT, and 10% glycerol at 30°C using a rapid chemical quench flow apparatus (KinTek). The Rev1 G193R mutant protein (800 nM) was pre-incubated with various concentrations of 32P-labeled DNA (10–100 nM), and reactions were initiated by the addition of 1 mM dCTP. Reactions were quenched after various time points ranging from 10 ms to 5 s and were resolved on a 15% polyacrylamide sequencing gel containing 8 M urea. The radiolabeled gel band intensities were quantified using a Phosphorimager (GE Healthcare). The concentrations of extended product (P) were graphed as a function of time (t). The amplitude of the burst phase (A), the observed first-order rate constants for nucleotide incorporation during the burst phase (kobs), and the observed rate for the steady state phase (vss) were obtained from the best fit of the data to the burst equation:
The amplitudes of the burst phases (A) were graphed as a function of DNA concentration ([DNA]). The apparent dissociation constant for the DNA (Kd·DNA) and the active site concentration of Rev1 ([Rev1]) were obtained from the best fit of the data to the quadratic form of the binding equation:
Crystallization and structural determination of the Rev1 BRCT domain
Crystallization of the Rev1 BRCT domain was performed using the hanging drop method with 200 nL drops that were setup with a mosquito® crystallization robot (TTP Labtech). Optimally diffracting crystals were obtained by combining an equal volume of protein (8 mg/mL) with a reservoir solution containing 0.1 M HEPES (pH 7) and 20% PEG 2,000 (w/v). Crystals formed within 24 hours at 18°C. Protein crystals were flash cooled in liquid nitrogen and data were collected at 100 K on a Rigaku rotating anode X-ray source. The data were processed and scaled to a resolution of 1.96 Å using XDS and SCALA, respectively [43, 44], and the space group was determined to be P21. Molecular replacement was performed using the structure of the human Rev1 BRCT domain (PDB id 2EBW) and PHASER [45]. Simulated annealing was carried out using PHENIX to remove any structural bias prior to any refinement [46]. Further refinement of the data was done using PHENIX and model building was carried out using Coot [47]. The coordinates have been deposited in the PDB with ID number 4ID3.
Genetic complementation studies
The full length REV1 gene with an additional 1,000 bp of yeast genomic DNA containing the native REV1 promoter was cloned into the low copy number CEN/ARS LEU2 yeast shuttle vector pRS315 to yield pKW386. The QuikChange Lightening Site-Directed Mutagenesis Kit (Stratagene) was used to introduce base substitutions into pKW386 to generate the following constructs that encode the Rev1 mutant proteins: pKW387 (N174A), pKW388 (R181A), pKW389 (K217A), pKW390 (R181A/K217A), pKW391 (L182A), pKW392 (E186A), pKW393 (L190A), pKW394 (K194A), pKW395 (F195A), pKW396 (G193V), pKW397 (G193D), pKW398 (G193R), and pKW399 (W231A).
To measure UV sensitivity, overnight cultures of the rev1Δ strain (Open Biosystems) harboring plasmids pKW386 to pKW399 were washed and diluted in water and then plated on synthetic complete medium lacking leucine. The plated cells were exposed to various doses of UV radiation and incubated at 30°C in the dark for 3 to 4 days. The percent of cell survival was obtained by comparing the number of colonies on the irradiated plates to the number of colonies on non-irradiated control plates. To measure UV-induced mutagenesis, cells were treated the same way except that they were placed on synthetic complete media lacking leucine and arginine, but containing canavanine. The frequency of CAN1S to can1r forward mutations was obtained by comparing the number of colonies on irradiated plates lacking arginine and containing canavanine to those on irradiated plates containing arginine and lacking canavanine. All genetic complementation experiments were duplicated at least four times to ensure reproducibility, and the data points shown are the averages of at least three independent experiments. The standard deviations for each of these data points are included in Table S1 and Table S2 (Supporting Information).
RESULTS
Impact of the yeast Rev1 BRCT domain on the activity of the polymerase domain
The BRCT domain of Rev1 plays an essential, yet enigmatic role in mutagenic TLS. A single amino acid substitution within the Rev1 BRCT domain (G193R), which is encoded by the rev1-1 allele, nearly eliminates UV-induced mutagenesis in yeast [34]. A previous study showed that the nucleotide incorporation activity of the full-length Rev1 G193R mutant protein is reduced by approximately 50% compared to the wild-type Rev1 protein [27]. This suggests that the BRCT domain may support the catalytic activity of the polymerase domain, perhaps through an intramolecular interaction. Thus we have carried out pre-steady state kinetic analyses of the Rev1 G193R mutant protein to understand how disruptions in the BRCT domain impact the mechanism of nucleotide incorporation and the DNA binding affinity.
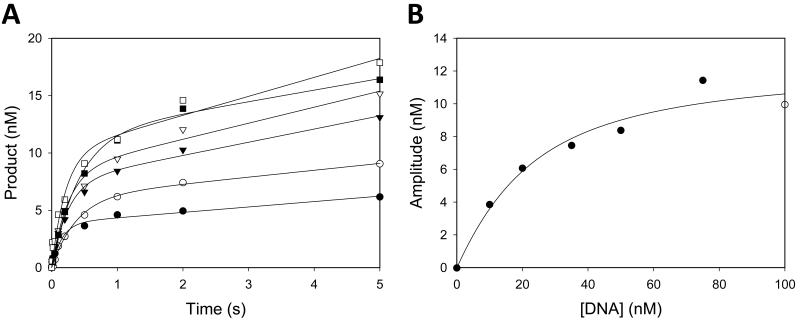
In a previous study, we examined the kinetics of nucleotide incorporation for the wild-type Rev1 protein, and we found that the kinetics were biphasic [23]. This means that the nucleotide is incorporated faster in the first enzyme turnover than in subsequent turnovers. The Rev1 G193R mutant protein also displays biphasic (or burst) kinetics (Fig. 1A). Because the amplitude of the burst phase is proportional to the active Rev1-DNA complex formed during the pre-incubation period, the apparent dissociation constant for the Rev1-DNA complex (Kd·DNA) can be obtained by measuring the burst amplitudes at various DNA concentrations. Thus we graphed the burst amplitude as a function of DNA concentration, and the best fit of the data yielded a Kd·DNA equal to 16 ± 5 nM (Fig. 1B). Thus the affinity of the Rev1 G193R mutant protein for DNA does not differ significantly from the wild-type protein, which had a Kd·DNA equal to 10 ± 2 nM [23].
Figure 1.
Pre-steady state kinetics of the Rev1 G193R mutant protein. (A) The Rev1 mutant protein (800 nM) was incubated with various concentrations of the DNA substrate (10 nM (●), 20 nM (○), 35 nM (▼), 50 nM (▽), 75 nM (■), and 100 nM (□)) and then mixed with dCTP (1 mM). The amount of nucleotide incorporation was plotted as a function of time, and the solid lines represent the best fits of the data to the burst equation. (B) The burst amplitudes were plotted as a function of DNA concentration, and the solid line represent the best fit of the data to the quadratic form of the binding equation. The Kd·DNA was equal to 16 ± 5 nM and the concentration of active Rev1 mutant protein was equal to 13 ± 1 nM.
The observed rate of nucleotide incorporation in the first enzyme turnover (kobs) for the Rev1 G193R mutant protein was at 10 ± 4 s−1. This is very similar to the kobs for the wild-type protein, which was 16 ± 1 s−1 [23]. Thus neither the DNA binding affinity nor the rate of the nucleotide-incorporation step account for the lower catalytic activity observed with the Rev1 G193R mutant protein. Interestingly, the active site concentrations of the two proteins, which correspond to the fraction of Rev1 molecules capable of catalyzing nucleotide incorporation, were quite different. We previously showed that the wild-type Rev1 protein has an active site concentration that is approximately 7% of the total protein concentration, and we concluded that this is due to a high fraction of catalytically inactive Rev1 protein that does not bind the DNA substrate [23]. Here, we find that the Rev1 G193R mutant protein has an active site concentration that is approximately 1.6% of the total protein concentration (Fig. 1B). This difference in active site concentrations does fully account for the previously reported differences in catalytic activity between the wild-type and mutant proteins. These results indicate that the G193R substitution does not affect DNA binding or nucleotide incorporation by the Rev1 polymerase domain, but does appear to have a significant effect on the ability of the protein to maintain a stable, catalytically active conformation.
Structure of the yeast Rev1 BRCT domain
To gain insight into the function of the yeast Rev1 BRCT domain, we determined its X-ray crystal structure to a resolution of 1.96 Å (Table 1). There are two BRCT domain molecules per asymmetric unit. The buried solvent accessible surface area between these two molecules in the asymmetric unit and between these molecules and symmetry related neighbors is less than 390 Å2, which suggests that the Rev1 BRCT domain is a monomer in the crystal. This is consistent with size exclusion chromatography results, which show that this domain is a monomer in solution (data not shown). This domain (comprising residues 164-250) has the typical BRCT domain fold with a core of four parallel beta strands flanked by three alpha helices (Fig. 2). Overall, its structure is similar to the structures of other single-copy BRCT domains. Direct comparisons of the structure of the yeast Rev1 BRCT domain to the structures of other single-copy BRCT domains is shown in Fig. S1 (Supporting Information).
Table 1.
Data collection and refinement statistics.
| (A) Data collection statistics | |
| Resolution (Å) | 17.4– 1.96 |
| Wavelength (Å) | 1.54 |
| Space Group | P21 |
| Cell (Å, °) | a = 32.0, b = 82.5, c = 32.7 α = 90.0°, β = 91.5°, γ = 90.0° |
| Completeness (%) a | 94.1 (85.1) |
| Redundancy a | 2.8 (2.5) |
| <I/σ(I)> a | 19.5 (4.7) |
| Rmerge (%) a | 2.7 (25.5) |
| (B) Refinement statistics | |
| Resolution range (Å) | 17.4-1.96 |
| R (%) | 19.08 |
| Rfree (%) | 23.32 |
| rms bonds (Å) | 0.008 |
| rms angles (°) | 1.096 |
| Number of protein atoms | 1428 (186 AA residues) |
| Number of water molecules | 120 |
| Average B factors | |
| Protein | 35.7 |
| Solvent | 39.4 |
| Wilson B factor | 30.4 |
| Ramachandran analysis (%) | |
| Most favored | 97.65 |
| Allowed | 2.35 |
Values in parentheses are for the highest resolution shell
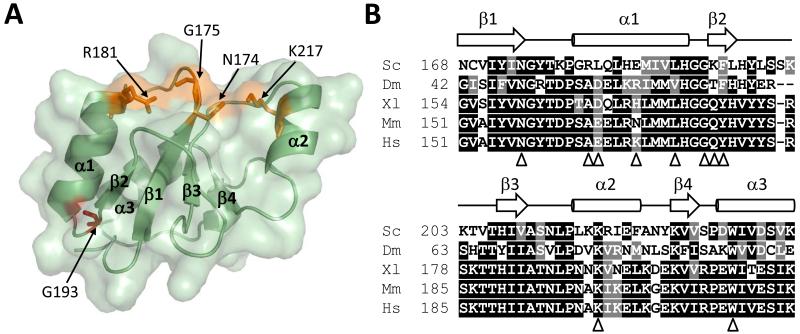
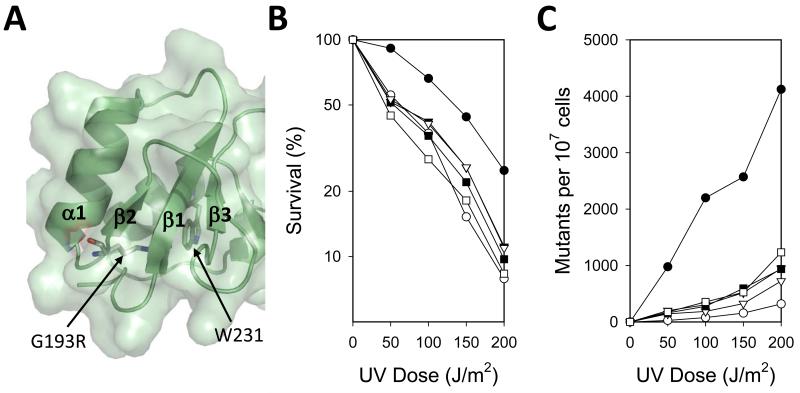
Figure 2.
Structure of the yeast Rev1 BRCT domain. (A) The BRCT domain structure (green) is shown with the phosphate-binding pocket (orange) and G193 residue (red) highlighted. (B) Amino acid sequences of the Rev1 BRCT domain of S. cerevisiae (Sc), D. melanogaster (Dm), X. laevis (Xl), M. musculus (Mm), and H. sapiens (Hs) are shown. Black boxes represent identical amino acid residues, and gray boxes represent similar amino acid residues. The secondary structural elements are indicated. Triangles indicate the positions of amino acid substitutions used in the genetic complementation studies.
Consistent with previous work showing that the Rev1 BRCT domain preferentially interacts with phosphorylated peptides and the phosphate groups on DNA, we found that this domain has a conserved, canonical BRCT domain phosphate-binding pocket comprised of residues Asn-174, Gly-175, Arg-181, and Lys-217 (Fig. 2B, Fig. 3A). The configuration of the phosphate-binding pocket is similar to that of the yeast RFC1 BRCT domain, which interacts with the phosphate groups on DNA [48, 49].
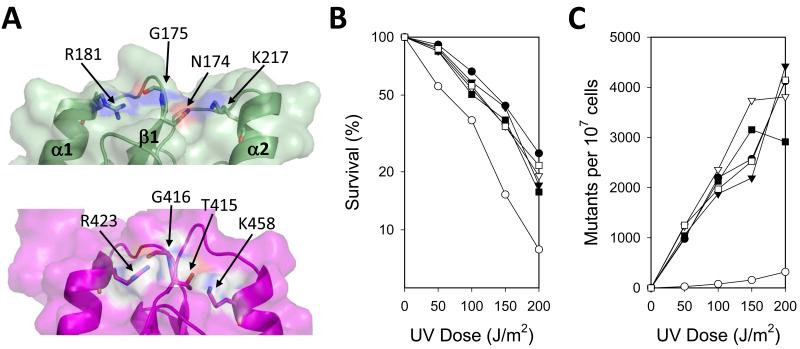
Figure 3.
Genetic complementation studies of Rev1 mutant proteins with substitutions of residues in the phosphate-binding pocket. (A) The phosphate-binding pocket of the Rev1 BRCT domain (green) is shown in comparison to the RFC1 BRCT domain phosphate-binding pocket (2EBU.pdb; maroon). The locations of residues Asn-174, Gly-175, Arg-181, and Lys-217 in the Rev1 BRCT domain and Thr-415, Gly-416, Arg-423, and Lys-458 in the RFC1 BRCT domain are indicated. (B) The UV sensitivity of rev1Δ yeast strains harboring plasmids producing either wild-type Rev1 (●), no Rev1 (○), the N174A mutant protein (▼), the R181A mutant protein (▽), the K217A mutant protein (■), or the R181A/K217A double mutant protein (□) is shown. The percentage of cells surviving each UV dose is graphed. (C) The UV-induced mutagenesis of these same yeast strains is shown. The number of CAN1S to can1r forward mutations per 107 surviving cells for each UV dose is graphed.
Functional significance of the yeast Rev1 BRCT domain phosphate-binding pocket
To assess the functional significance of the phosphate-binding pocket of the yeast Rev1 BRCT domain, we carried out genetic complementation studies. We examined whether full-length Rev1 mutant proteins with substitutions in the phosphate-binding pocket could complement the UV sensitivity of the rev1Δ strain. We tested four Rev1 mutant proteins: N174A, R181A, K217A, and the R181A/K217A double mutant protein (Fig. 2B). Residues corresponding to these are directly involved in phosphate binding in the BRCT domains of other proteins [38]. We found that all of the strains producing these Rev1 mutant proteins had nearly the same level of resistance to UV radiation as did the strain producing the wild-type protein (Fig. 3B). All of these strains were more resistant than the negative control strain containing the empty vector, which did not produce Rev1.
We next examined whether these Rev1 mutant proteins could complement the defect in UV-induced mutagenesis of the rev1Δ strain. All of the strains producing these Rev1 mutant proteins had nearly the same frequencies of CAN1S to can1r forward mutations as did the strain producing the wild-type protein (Fig. 3C). By contrast, the frequency of UV-induced mutagenesis was reduced by nearly 95% in the empty vector control strain. Taken together, these data show that the phosphate-binding pocket of the Rev1 BRCT domain is dispensable for its function in TLS in vivo.
The G193R mutation destabilizes the yeast Rev1 BRCT domain
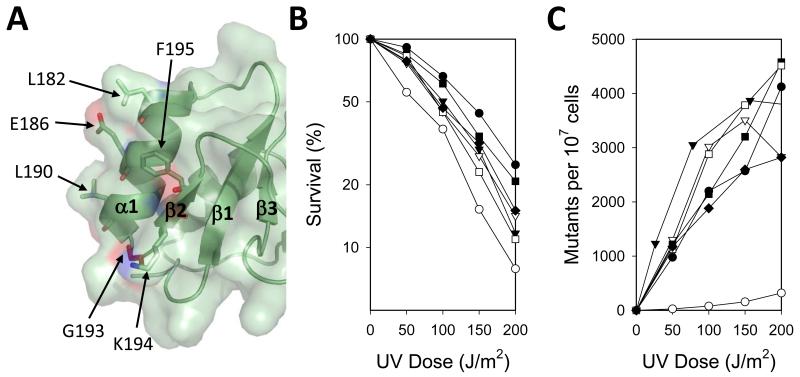
To better understand how the G193R substitution disrupts the function of the Rev1 BRCT domain, we made additional amino acid substitutions in and around Gly-193 and carried out genetic complementation studies. We reasoned that if Gly-193 is part of a protein-protein interaction surface, then amino acid substitutions near Gly-193, particularly along the solvent exposed face of helix α1 and strand β2, should disrupt the function of the BRCT domain in a manner similar to that of the G193R substitution. The solvent exposed residues on helix α1 have been shown to be involved in protein-protein interactions in BRCT domains from several other proteins [50-52]. Thus we examined five substitutions in this region of the protein: L182A, E186A, L190A, K194A, and F195A (Fig. 2B, Fig. 4A). We found that all of the strains producing these Rev1 mutant proteins had sensitivity to UV radiation in between that of the wild-type protein and the vector control (Fig. 4B). All of these strains, however, had nearly the same frequencies of CAN1S to can1r forward mutations as did the strain producing the wild-type protein (Fig. 4C). This suggests that these residues of the Rev1 BRCT domain are not necessary for its function in mutagenic TLS in vivo.
Figure 4.
Genetic complementation studies of Rev1 mutant proteins with substitutions of residues near Gly-193. (A) Close up view of the location of Gly-193 is shown. The locations of residues Leu-182, Glu-186, Leu-190, Gly-193, Lys-194, and Phe-195 are indicated. (B) The UV sensitivity of rev1Δ yeast strains harboring plasmids producing either wild-type Rev1 (●), no Rev1 (○), the L182A mutant protein (▼), the E186A mutant protein (▽), the L190A mutant protein (■), the K194A mutant protein (□), and the F195A mutant protein (◆) is shown. The percentage of cells surviving each UV dose is graphed. (C) The UV-induced mutagenesis of these same yeast strains is shown. The number of CAN1S to can1r forward mutations per 107 surviving cells for each UV dose is graphed.
Analysis of the structure showed that, assuming no alterations to the position of the backbone, the substituted arginine side chain at position 193 would project directly into the hydrophobic core of the domain leading to serious steric clashes (Fig. 5A). This raised the possibility that the G193R substitution destabilized the fold of the BRCT domain. To test this idea, we examined two additional substitutions of Gly-193: G193V and G193D. We also examined a substitution in a nearby conserved tryptophan residue (W231A), which when substituted destabilizes the fold of BRCT domains in other systems [53, 54]. Trp-231 is located on helix α3 and points into the hydrophobic core of the domain (Fig. 5A). All of the strains producing these Rev1 mutant proteins had sensitivities to UV radiation similar to that of the strain producing the G193R mutant protein and the vector control strain (Fig. 5B). Similarly, all of these strains had nearly the same defect in UV-induced mutagenesis as the strain producing the G193R mutant protein and the vector control strain (Fig. 5C). Thus, these substitutions prevent Rev1 from functioning in mutagenic TLS in vivo.
Figure 5.
Genetic complementation studies of Rev1 mutant proteins with substitutions of Gly-193 or W231A. (A) Close up view of the location of Gly-193 is shown. The location of Trp-231 is indicated. (B) The UV sensitivity of rev1Δ yeast strains harboring plasmids producing either wild-type Rev1 (●), no Rev1 (○), the G193V mutant protein (▼), the G193D mutant protein (▽), the G193R mutant protein (■), and the W231A mutant protein (□) is shown. The percentage of cells surviving each UV dose is graphed. (C) The UV-induced mutagenesis of these same yeast strains is shown. The number of CAN1S to can1r forward mutations per 107 surviving cells for each UV dose is graphed.
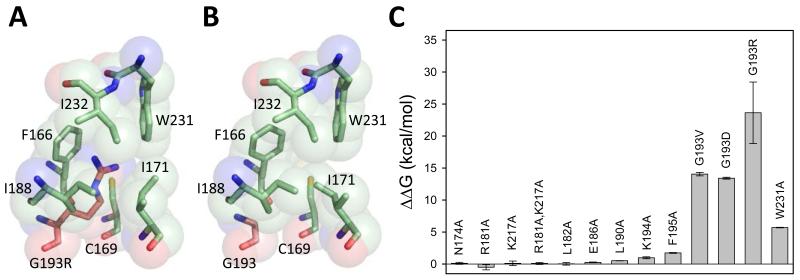
To test whether these amino acid substitutions destabilized the fold of the yeast Rev1 BRCT domain, we attempted to express and purify the G193R, the G193V, the G193D, and the W231A mutant BRCT domains for in vitro studies. However, we were unable to do so. These mutant BRCT domains were over-expressed at greatly reduced levels as chitin-binding-domain fusion proteins. These fusion proteins bound the chitin column, but we were unable to elute any significant amount of soluble mutant BRCT domains upon cleavage of the fusion protein bound to the column. The small amount of soluble BRCT domain that was released from the column was aggregated and eluted in the void volume during size exclusion chromatography. Taken together, these results strongly suggested that all of these substitutions destabilize the fold of the BRCT domain.
Because we were unable to express and purify these mutant BRCT domains, we carried out molecular modeling. To predict the changes in the stability of the BRCT domain resulting from each of these amino acid substitutions, we used the computer algorithm FoldX [55]. We calculated the ΔΔG of unfolding of the mutant BRCT domain versus the wild-type BRCT domain for all of the mutant proteins examined in this study (Fig. 6). As is commonly known, the absolute energies calculated by FoldX are not precise, but the relative energies are quite reliable. We found that the G193V, G193D, G193R, and W231A mutant Rev1 BRCT domains were all substantially less stable than the wild-type Rev1 BRCT domain. All of the other mutant Rev1 BRCT domains that we examined were nearly as stable as the wild-type domain. In fact, there is a direct correlation between the calculated stability of the Rev1 BRCT domain and the in vivo function of the mutant Rev1 proteins. Thus, these molecular modeling studies combined with the experimental work described above clearly show that the lack of function of these mutant Rev1 proteins results from the destabilization of the Rev1 BRCT domain.
Figure 6.
Molecular modeling of the mutant Rev1 BRCT domains. (A) Model of hydrophobic core of the Rev1 G193R mutant BRCT domain generated by FoldX. The side chain of Arg-193 (red) projects into the core of the domain and contacts various hydrophobic residues (green). (B) Model of the hydrophobic core of the wild-type Rev1 BRCT domain generated by FoldX. (C) Plot of the ΔΔG of unfolding of the mutant Rev1 BRCT domains versus the wild-type Rev1 BRCT domain. These values represent the average and standard deviation of five independent calculations using FoldX.
DISCUSSION
The yeast Rev1 protein has three structural elements flanked by regions that are likely intrinsically disordered. There is an N-terminal region that contains a BRCT domain, the catalytic core that is comprised of a polymerase domain and a polymerase associated domain (PAD), and the C-terminal region that contains two ubiquitin-binding motifs (UBMs) plus a C-terminal domain (CTD). The X-ray crystal structure of the catalytic core has been previously determined, and this structure shows that this domain catalyzes nucleotide incorporation utilizing a remarkable protein template-directed mechanism of incoming nucleotide selection [42, 56, 57]. The NMR structure of a close homolog of the two UBM motifs has been determined, and its binding site on ubiquitin has been mapped [58]. These motifs are thought to mediate interactions of Rev1 and ubiquitin-modified PCNA. Recently, the X-ray crystal structure of the Rev1 CTD bound to the Rev7 subunit of Pol ξ has been determined [32, 33]. This domain is thought to allow Rev1 to function as a scaffold to bind other TLS polymerases [28, 30, 59]. The X-ray crystal structure of the Rev1 BRCT domain reported here now completes the set of structures for all the folded domains of the Rev1 protein. Although the Rev1 BRCT domain plays a central role in mutagenic TLS, its function is unknown.
The structure of the yeast Rev1 BRCT domain has guided our structure-function studies of this domain. This has allowed us to test several recent models regarding the role of the BRCT domain. First, it was previously shown that the yeast Rev1 BRCT domain binds phosphorylated peptides in vitro with higher affinity than it binds non-phosphorylated peptides [39]. This led to the model that the Rev1 BRCT domain interacts with a phosphorylated binding partner. Our genetic complementation studies with yeast Rev1 mutant proteins with substitutions in the conserved phosphate-binding pocket show that any such interactions are not necessary for the in vivo function of Rev1. Thus, interactions with a phosphorylated binding partner are unlikely to be the principal function of this domain in mutagenic TLS. Second, previous studies showed that the yeast and mouse Rev1 BRCT domains bind DNA in vitro by interacting with recessed 5′ phosphates at primer-template junctions [40]. This led to the model that Rev1 is recruited to stalled replication forks via the BRCT domain interacting with downstream 5′ primer-template junctions. Again, our genetic complementation results with yeast Rev1 mutant proteins with substitutions in the phosphate-binding pocket argue that this interaction is unlikely to be the principal function of this domain in TLS. Third, we considered additional models in which the BRCT domain supports the function of other domains in the Rev1 protein, such as the catalytic domain or the CTD, via intra-molecular interactions. With respect to the catalytic domain, our pre-steady state kinetics studies of the Rev1 G193R mutant protein shows that disrupting the BRCT domain does not affect DNA binding or nucleotide incorporation by Rev1. This suggests that the BRCT domain does not support the function of the catalytic domain, although it does have some effect on the ability of Rev1 to maintain a stable, catalytically active conformation. In any event, mutagenic TLS does not require the catalytic activity of Rev1. Thus supporting the function of the catalytic domain is not the principal function of the BRCT domain. With respect to the CTD, a previous study showed that the Rev1 CTD is sufficient for interacting with the Rev7 subunit of Pol ξ, another TLS polymerase [28]. This implies that supporting the function of the CTD is not the principal function of this domain.
The rev1-1 encoded G193R protein is defective in mutagenic TLS. Our structural and functional analyses of the BRCT domain strongly suggest that this substitution leads to a destabilizing of the domain rendering the Rev1 protein non-functional in vivo. If one were to fix the position of the backbone atoms around Gly-193, it would not be possible to substitute any other amino acid residue for this glycine, because the side-chain would be protruding into the hydrophobic core of this domain making severe steric clashes (Fig. 5A, Fig. 6A). Thus, in the case of the G193R, G193V, and G193D substitutions, the protein backbone in this region would have to re-orient significantly, and we believe that this destabilizes the BRCT domain. Support for this notion comes from three observations. First, while it is possible to obtain full-length Rev1 with the G193R substitution, we were unable to obtain isolated Rev1 BRCT domains with the G193R, G193V, and G193D substitutions due to lower levels of expression and problems with protein solubility and aggregation. These problems were not observed in the case of the wild-type Rev1 BRCT domain. Second, we made a Rev1 mutant protein with a W231A substitution, which we chose because it was expected to destabilize the BRCT domain. Substitutions of this conserved tryptophan destabilize the BRCT domains in other proteins [53, 54]. This substitution significantly impaired the ability of Rev1 to support mutagenic TLS. Moreover, we were unable to isolate the Rev1 BRCT domain with the W231A substitution due to problems with protein solubility and aggregation. Third, molecular modeling showed that G193V, G193D, G193R, and W231A mutant Rev1 BRCT domains were substantially less stabile than the wild-type Rev1 BRCT domain. Taken together, our results show that all of these amino acid substitutions destabilize the Rev1 BRCT domain and only these substitutions impaired the ability of Rev1 to support mutagenic TLS. Thus we conclude that destabilization of the BRCT domain by these substitutions is what disrupts Rev1 function in vivo. It should be noted that in the case of the G193R substitution, the expression levels of Rev1 mutant protein in cells is not affected [60].
So why is the BRCT domain of Rev1 required for mutagenic TLS? It is likely that this domain interacts with another protein in a manner that is not dependent on this binding partner’s phosphorylation state. The identity of this binding partner is unknown, but there are some interesting candidates suggested by the literature. For example, a previous study showed that when the BRCT domain of Dbf4 (Dumbbell forming 4), the regulatory subunit of the Cdc7 checkpoint kinase, was replaced by other BRCT domains, only the Rev1 BRCT domain was able to fully substitute with respect to function in vivo [61]. These data suggest that Dbf4 BRCT domain and the Rev1 BRCT domain share a common binding partner. The binding of Dbf4 to Rad53 is well characterized, suggesting the Rad53 may be the critical interacting partner of the Rev1 BRCT domain [62]. Incidentally, this may explain why RAD53 influences the nuclear foci formation of Rev1 [63]. Another possible binding partner is PCNA, a key replication accessory factor. Using a pull-down assay one group showed that the BRCT domain of both yeast and mouse Rev1 interacts with PCNA, and the G193R mutation in the yeast Rev1 BRCT domain blocks this interaction [64]. They also demonstrated that the localization of Rev1 to DNA replication foci was reduced in cells expressing Rev1 mutant proteins with a BRCT domain deletion or the G193R mutation. However, these results remain controversial as other groups have been unable to detect direct interactions between the Rev1 BRCT domain and PCNA [40, 65]. Moreover, PCNA stimulates the catalytic activity of the Rev1 G193R mutant protein showing that this functional interaction remains intact even when the BRCT domain is disrupted [66]. In any event, more work will be required to identify the Rev1 BRCT domain binding partner.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Christine Kondratick, Lynne Dieckman, Elizabeth Boehm, and Bret Freudenthal for valuable discussions.
ABBREVIATIONS
- BRCT
BRCA1 C-terminal
- CTD
C-terminal domain
- dNTP
deoxynucleoside triphosphates
- DTT
dithiothreitol
- PAD
polymerase associated domain
- PCNA
proliferating cell nuclear antigen
- Pol
polymerase
- TLS
translesion synthesis
- UBM
ubiquitin-binding motif
- UV
ultraviolet
Footnotes
The project described was supported by Award Number GM081433 from the National Institute of General Medical Sciences to M.T.W. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health
SUPPORTING INFORMATION
The supporting information includes one figure showing the comparison of the yeast Rev1 BRCT domain to other BRCT domains (Figure S1), a table providing the mean and standard deviations of the UV survival experiments (Table S1), and a table providing the mean and standard deviations of the UV mutagenesis experiments (Table S2). This material is available free of charge via the Internet at http://pubs.acs.ord.
REFERENCES
- 1.Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: A one- or two-polymerase affair. Genes & Development. 2002;16(15):1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 2.Lehmann AR. Replication of damaged DNA. Cell Cycle. 2003;2(4):300–302. [PubMed] [Google Scholar]
- 3.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annual Review of Biochemistry. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 4.Lehmann AR. Replication of damaged DNA by translesion synthesis in human cells. Febs Letters. 2005;579(4):873–876. doi: 10.1016/j.febslet.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 5.Washington MT, et al. Variations on a theme: Eukaryotic Y-family DNA polymerases. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbapap.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waters LS, et al. Eukaryotic Translesion Polymerases and Their Roles and Regulation in DNA Damage Tolerance. Microbiology and Molecular Biology Reviews. 2009;73(1):134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo C, et al. Y-family DNA polymerases in mammalian cells. Cellular and Molecular Life Sciences. 2009;66(14):2363–2381. doi: 10.1007/s00018-009-0024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nature Reviews Molecular Cell Biology. 2012;13(3):141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dumstorf CA, et al. REV1 Is Implicated in the Development of Carcinogen-Induced Lung Cancer. Molecular Cancer Research. 2009;7(2):247–254. doi: 10.1158/1541-7786.MCR-08-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie K, et al. Error-prone translesion synthesis mediates acquired chemoresistance. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(48):20792–20797. doi: 10.1073/pnas.1011412107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma S, et al. DNA Polymerase zeta Is a Major Determinant of Resistance to Platinum-Based Chemotherapeutic Agents. Molecular Pharmacology. 2012;81(6):778–787. doi: 10.1124/mol.111.076828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watson NB, Mukhopadhyay S, McGregor WG. Translesion DNA replication proteins as molecular targets for cancer prevention. Cancer Letters. 2006;241(1):13–22. doi: 10.1016/j.canlet.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 13.Lawrence CW. Cellular roles of DNA polymerase zeta and Rev1 protein. DNA Repair. 2002;1(6):PII S1568–7864(02)00038-1. doi: 10.1016/s1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- 14.Lawrence CW. Cellular functions of DNA polymerase zeta and Rev1 protein. DNA Repair and Replication. 2004;69:167–203. doi: 10.1016/S0065-3233(04)69006-1. [DOI] [PubMed] [Google Scholar]
- 15.Lawrence CW, Maher VM. Eukaryotic mutagenesis and translesion replication dependent on DNA polymerase zeta and Rev1 protein. Biochem Soc Trans. 2001;29(Pt 2):187–191. doi: 10.1042/0300-5127:0290187. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence CW, Christensen RB. Ultraviolet-induced reversion of cyc1 alleles in radiation-sensitive strains of yeast. III. rev3 mutant strains. Genetics. 1979;92(2):397–408. doi: 10.1093/genetics/92.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawrence CW, Christensen RB. Ultraviolet-induced reversion of cyc1 alleles in radiation-sensitive strains of yeast. I. rev1 Mutant strains. J Mol Biol. 1978;122(1):1–21. doi: 10.1016/0022-2836(78)90104-3. [DOI] [PubMed] [Google Scholar]
- 18.Gibbs PE, et al. The function of the human homolog of Saccharomyces cerevisiae REV1 is required for mutagenesis induced by UV light. Proc Natl Acad Sci U S A. 2000;97(8):4186–4191. doi: 10.1073/pnas.97.8.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibbs PEM, et al. A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase zeta. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(12):6876–6880. doi: 10.1073/pnas.95.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson RE, et al. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406(6799):1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 21.Haracska L, et al. Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites. Genes & Development. 2001;15(8):945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howell CA, Kondratick CM, Washington MT. Substitution of a residue contacting the triphosphate moiety of the incoming nucleotide increases the fidelity of yeast DNA polymerase zeta. Nucleic Acids Research. 2008;36(5):1731–1740. doi: 10.1093/nar/gkn023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pryor JM, Washington MT. Pre-steady state kinetic studies show that an abasic site is a cognate lesion for the yeast Rev1 protein. DNA Repair. 2011;10(11):1138–1144. doi: 10.1016/j.dnarep.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Washington MT, et al. Efficient and error-free replication past a minor-groove N-2-guanine adduct by the sequential action of yeast Rev1 and DNA polymerase zeta. Molecular and Cellular Biology. 2004;24(16):6900–6906. doi: 10.1128/MCB.24.16.6900-6906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi JY, Guengerich FP. Kinetic analysis of translesion synthesis opposite bulky N-2- and O-6-alkylguanine DNA adducts by human DNA polymerase REV1. Journal of Biological Chemistry. 2008;283(35):23645–23655. doi: 10.1074/jbc.M801686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang YB, et al. Response of human REV1 to different DNA damage: preferential dCMP insertion opposite the lesion. Nucleic Acids Research. 2002;30(7):1630–1638. doi: 10.1093/nar/30.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson JR, et al. Evidence for a second function for Saccharomyces cerevisiae Rev1p. Molecular Microbiology. 2000;37(3):549–554. doi: 10.1046/j.1365-2958.2000.01997.x. [DOI] [PubMed] [Google Scholar]
- 28.D’Souza S, Waters LS, Walker GC. Novel conserved motifs in Rev1 C-terminus are required for mutagenic DNA damage tolerance. DNA Repair. 2008;7(9):1455–1470. doi: 10.1016/j.dnarep.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pozhidaeva A, et al. NMR Structure and Dynamics of the C-Terminal Domain from Human Revl and Its Complex with Rev1 Interacting Region of DNA Polymerase eta. Biochemistry. 2012;51(27):5506–5520. doi: 10.1021/bi300566z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wojtaszek J, et al. Multifaceted Recognition of Vertebrate Rev1 by Translesion Polymerases zeta and kappa. Journal of Biological Chemistry. 2012;287(31):26400–26408. doi: 10.1074/jbc.M112.380998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pustovalova Y, Bezsonova I, Korzhnev DM. The C-terminal domain of human Rev1 contains independent binding sites for DNA polymerase eta and Rev7 subunit of polymerase zeta. FEBS Lett. 2012 doi: 10.1016/j.febslet.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kikuchi S, et al. Structural basis of recruitment of DNA polymerase zeta by interaction between REV1 and REV7. J Biol Chem. 2012 doi: 10.1074/jbc.M112.396838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wojtaszek J, et al. Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric Pol zeta and Pol kappa. J Biol Chem. 2012 doi: 10.1074/jbc.M112.394841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larimer FW, Perry JR, Hardigree AA. THE REV1 GENE OF SACCHAROMYCES-CEREVISIAE - ISOLATION, SEQUENCE, AND FUNCTIONAL-ANALYSIS. Journal of Bacteriology. 1989;171(1):230–237. doi: 10.1128/jb.171.1.230-237.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jansen JG, et al. The BRCT domain of mammalian Rev1 is involved in regulating DNA translesion synthesis. Nucleic Acids Research. 2005;33(1):356–365. doi: 10.1093/nar/gki189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez MC, Songyang Z. BRCT Domains: phosphopeptide binding and signaling modules. Frontiers in Bioscience. 2008;13:5905–5915. doi: 10.2741/3125. [DOI] [PubMed] [Google Scholar]
- 37.Leung CC, Glover JN. BRCT domains: easy as one, two, three. Cell Cycle. 2011;10(15):2461–2470. doi: 10.4161/cc.10.15.16312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheng ZZ, Zhao YQ, Huang JF. Functional Evolution of BRCT Domains from Binding DNA to Protein. Evol Bioinform Online. 2011;7:87–97. doi: 10.4137/EBO.S7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu XC, et al. The BRCT domain is a phospho-protein binding domain. Science. 2003;302(5645):639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 40.de Groote FH, et al. The Rev1 translesion synthesis polymerase has multiple distinct DNA binding modes. DNA Repair. 2011;10(9):915–925. doi: 10.1016/j.dnarep.2011.04.033. [DOI] [PubMed] [Google Scholar]
- 41.Howell CA, Prakash S, Washington MT. Pre-steady-state kinetic studies of protein-template-directed nucleotide incorporation by the yeast rev1 protein. Biochemistry. 2007;46:13451–13459. doi: 10.1021/bi701429v. [DOI] [PubMed] [Google Scholar]
- 42.Nair DT, et al. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309(5744):2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 43.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallographica Section D-Biological Crystallography. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallographica Section D-Biological Crystallography. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallographica Section D-Biological Crystallography. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 46.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallographica Section D-Biological Crystallography. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 48.Kobayashi M, et al. Characterization of the DNA binding and structural properties of the BRCT region of human replication factor C p140 subunit. Journal of Biological Chemistry. 2006;281(7):4308–4317. doi: 10.1074/jbc.M511090200. [DOI] [PubMed] [Google Scholar]
- 49.Kobayashi M, et al. Structure of the DNA-bound BRCA1 C-terminal Region from Human Replication Factor C p140 and Model of the Protein-DNA Complex. Journal of Biological Chemistry. 2010;285(13):10087–10097. doi: 10.1074/jbc.M109.054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cuneo MJ, et al. The structural basis for partitioning of the XRCC1/DNA ligase III-alpha BRCT-mediated dimer complexes. Nucleic Acids Research. 2011;39(17):7816–7827. doi: 10.1093/nar/gkr419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mueller GA, et al. A comparison of BRCT domains involved in nonhomologous end-joining: Introducing the solution structure of the BRCT domain of polymerase lambda. DNA Repair. 2008;7(8):1340–1351. doi: 10.1016/j.dnarep.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams RS, Green R, Glover JNM. Crystal structure of the BRCT repeat region from the breast cancer-associated protein BRCA1. Nature Structural Biology. 2001;8(10):838–842. doi: 10.1038/nsb1001-838. [DOI] [PubMed] [Google Scholar]
- 53.Zhang XD, et al. Structure of an XRCC1 BRCT domain: a new protein-protein interaction module. Embo Journal. 1998;17(21):6404–6411. doi: 10.1093/emboj/17.21.6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoelzel M, et al. The BRCT domain of mammalian Pes1 is crucial for nucleolar localization and rRNA processing. Nucleic Acids Research. 2007;35(3):789–800. doi: 10.1093/nar/gkl1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schymkowitz J, et al. The FoldX web server: an online force field. Nucleic Acids Research. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nair DT, et al. Protein-template-directed synthesis across an acrolein-derived DNA adduct by yeast rev1 DNA polymerase. Structure. 2008;16(2):239–245. doi: 10.1016/j.str.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 57.Nair DT, et al. DNA Synthesis across an Abasic Lesion by Yeast Rev1 DNA Polymerase. Journal of Molecular Biology. 2011;406(1):18–28. doi: 10.1016/j.jmb.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bomar MG, et al. Unconventional Ubiquitin Recognition by the Ubiquitin-Binding Motif within the Y Family DNA Polymerases iota and Rev1. Molecular Cell. 2010;37(3):408–417. doi: 10.1016/j.molcel.2009.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D’Souza S, Walker GC. Novel role for the C terminus of Saccharomyces cerevisiae Rev1 in mediating protein-protein interactions. Molecular and Cellular Biology. 2006;26(21):8173–8182. doi: 10.1128/MCB.00202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiltrout ME, Walker GC. The DNA Polymerase Activity of Saccharomyces cerevisiae Rev1 is Biologically Significant. Genetics. 2011;187(1):21–35. doi: 10.1534/genetics.110.124172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harkins V, et al. Budding Yeast Dbf4 Sequences Required for Cdc7 Kinase Activation and Identification of a Functional Relationship Between the Dbf4 and Rev1 BRCT Domains. Genetics. 2009;183(4):1269–1282. doi: 10.1534/genetics.109.110155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matthews LA, et al. Saccharomyces cerevisiae Dbf4 Has Unique Fold Necessary for Interaction with Rad53 Kinase. Journal of Biological Chemistry. 2012;287(4):2378–2387. doi: 10.1074/jbc.M111.233973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Conde F, et al. Regulation of tolerance to DNA alkylating damage by Dot1 and Rad53 in Saccharomyces cerevisiae. DNA Repair. 2010;9(10):1038–1049. doi: 10.1016/j.dnarep.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 64.Guo CX, et al. REV1 protein interacts with PCNA: Significance of the REV1 BRCT domain in vitro and in vivo. Molecular Cell. 2006;23(2):265–271. doi: 10.1016/j.molcel.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 65.Ohmori H, et al. SEPARATE ROLES OF STRUCTURED AND UNSTRUCTURED REGIONS OF Y-FAMILY DNA POLYMERASES. In: McPherson A, editor. Advances in Protein Chemistry and Structural Biology. Vol. 78. 2009. pp. 99–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wood A, Garg P, Burgers PMJ. A ubiquitin-binding motif in the translesion DNA polymerase Rev1 mediates its essential functional interaction with ubiquitinated proliferating cell nuclear antigen in response to DNA damage. Journal of Biological Chemistry. 2007;282(28):20256–20263. doi: 10.1074/jbc.M702366200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.