Abstract
Regulation of both systemic and cellular iron homeostasis requires the capacity to sense iron levels and appropriately modify the expression of iron metabolism genes. These responses are coordinated through the efforts of several key regulatory factors including F-box and Leucine-rich Repeat Protein 5 (FBXL5), Iron Regulatory Proteins (IRPs), Hypoxia Inducible Factor (HIF), and ferroportin. Notably, the stability of each of these proteins is regulated in response to iron. Recent discoveries have greatly advanced our understanding of the molecular mechanisms governing iron-sensing and protein degradation within these pathways. It has become clear that iron’s privileged roles in both enzyme catalysis and protein structure contribute to its regulation of protein stability. Moreover, these multiple pathways intersect with one another in larger regulatory networks to maintain iron homeostasis.
Keywords: F-box and Leucine-rich Repeat Protein 5, hemerythrin domain, iron homeostasis, Iron Regulatory Proteins, ferroportin, Hypoxia Inducible Factor, prolyl hydroxylase
1. Introduction
Due to the abundance and chemical properties of iron, life on Earth evolved to depend on this transition metal [1]. Nearly every prokaryote and all eukaryotes require iron for their viability [2]. On account of iron’s ease in donating and accepting electrons, iron is employed by a variety proteins to carry out oxidation-reduction reactions [3]. Iron-utilizing proteins are necessary for critical biological processes such as the transport and storage of oxygen, deoxyribonucleotide synthesis, chromatin regulation, and oxidative phosphorylation [4]. Though required for sustaining life, iron can have deleterious consequences when present in excess. Copious free iron generates radical species through Fenton chemistry, causing oxidative damage to proteins, nucleic acids, and lipids [5]. Consequently, the acquisition, export, usage, and storage of iron are regulated in most organisms in order to maintain iron homeostasis [6]. Aberrant iron homeostasis is the principal pathogenic factor in multiple human diseases. Due to the high iron demand of red blood cells, iron deficiency causes anemia in approximately a billion individuals worldwide [3]. At the other end of the iron-related disorders spectrum is hemochromatosis, a disease of iron overload, that can lead to heart failure, diabetes, and cirrhosis [6]. Thus, the challenge for humans is to maintain sufficient iron levels to sustain life while avoiding the potentially detrimental effects of excess iron [3]. Due to their multicellular nature and complex tissue organization, mammals regulate iron homeostasis at both the systemic and cellular levels. In contrast to bacteria and yeast that largely regulate iron metabolism genes at the level of transcription [7–9], multiple posttranscriptional mechanisms are critical for maintaining mammalian iron homeostasis [3–6]. In particular, several key iron regulatory proteins differentially accumulate in response to changes in iron availability as a consequence of dynamic proteolysis. Our understanding of the mechanisms underlying iron-sensing and regulated protein degradation has greatly advanced during the last several years.
2. Iron-Responsive Regulation of Ferroportin Proteolysis
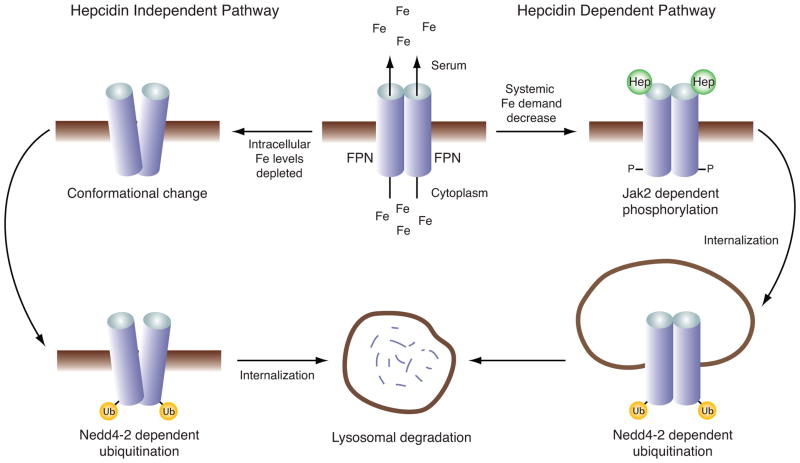
The iron export protein ferroportin is an essential regulator of systemic iron homeostasis (reviewed in this issue of BBA-Molecular Cell Research []). Following uptake dietary iron can be stored in the cytosol or effluxed into the plasma via the actions of ferroportin, a membrane bound dimer localized to the basolateral side of intestinal epithelial cells. Ferroportin is also expressed on the cell surface of hepatocytes and macrophages where it plays a key role in releasing stored iron and iron recycled from phagocytosed erythrocytes, respectively [4]. Ferroportin stability is posttranslationally regulated by the hormone hepcidin, a circulating member of the defensin family of peptides that is secreted primarily by the liver [10]. When serum iron levels are sufficient to satisfy systemic demand, the gene encoding hepcidin (HAMP) is upregulated and hepcidin is secreted into the blood where it binds ferroportin [11]. This interaction induces a structural change within ferroportin, promoting the recruitment of the Jak2 kinase and its subsequent phosphorylation [12]. Phosphorylated ferroportin is internalized and ubiquitinated by the E3 ligase Nedd4-2 [13]. Ubiquitinated ferroportin is processed through the endosome and subsequently delivered to the lysosome where it is degraded (Fig. 1), diminishing iron export to effectively reduce serum iron levels [14, 15]. Transcription of the hepcidin gene is attenuated when systemic demand for iron is high, ultimately stabilizing ferroportin and increasing iron efflux into the serum [15–17].
Fig. 1.
Model of hepcidin (Hep) dependent and independent regulation of ferroportin (FPN).
Ferroportin has also been recently shown to be directly regulated by iron via a hepcidin-independent mechanism in cultured cells [13]. When cytosolic iron levels are depleted, ferroportin is believed to undergo a conformational change due to the lack of iron binding and transport. This alternate conformation renders ferroportin susceptible to ubiquitination, which is dependent on Nedd4-2 and its interacting partner Nfdip-1. Following Nedd4-2/Nfdip-1 dependent ubiquitination, ferroportin is internalized and ostensibly degraded in the lysosome (Fig. 1). Interestingly, a C. elegans ferroportin homolog undergoes iron limiting internalization and degradation, suggesting that this mechanism of controlling iron homeostasis evolutionarily precedes hepcidin, which is only expressed in vertebrates [13]. In this context, ferroportin behaves as an iron sensor whose ensuing conformational change contributes to the regulation of iron homeostasis.
3. Hypoxia Inducible Factor and Iron Homeostasis
Many cellular processes such as oxidative phosphorylation and chromatin remodeling are dependent on both iron and O2 [18, 19]. Therefore, it is not surprising that cells have evolved to coordinate the regulation of iron and oxygen metabolism. Similar to iron, O2 levels must be maintained in an optimal range that is sufficient for critical cellular processes, while minimizing oxidative damage generated in the presence of excess oxygen [20]. In mammalian cells, oxygen homeostasis is maintained in part by the dimeric transcription factor Hypoxia Inducible Factor (HIF). HIF contains a constitutively expressed β-subunit and a regulatedα-subunit. HIF-α is preferentially degraded under normoxic conditions, precluding formation of an active HIF heterodimer. Under hypoxic conditions the α-subunit is stabilized, allowing its interaction with the β-subunit and subsequent induction of an assortment of genes to elicit adaptive responses to low O2 availability [21, 22].
HIF-α is degraded when O2 is plentiful via the ubiquitin-proteasome system (UPS) [23, 24]. In this regulatory system a trio of factors, designated E1, E2, and E3, catalyze the transfer of polyubiquitin chains to proteins. Polyubiquitinated proteins are degraded in the macromolecular proteasome complex in order to maintain protein homeostasis [25, 26]. Recognition of HIF-α under normoxic conditions by the product of the von Hippel Lindau tumor suppressor gene (pVHL), a component of the E3 ubiquitin ligase responsible for its polyubiquitination, requires the action of a family of HIF prolyl hydroxylases. As their name suggests, these enzymes hydroxylate proline residues on HIF-α in an O2- and iron-dependent manner [27–30]. These metabolic requirements are conferred in part by the role of iron in facilitating the binding and activation of the O2 substrate consumed during hydroxylation (Fig. 2). As a result, depletion of cellular iron stores upon addition of iron chelators abolishes hydroxylase activity and promotes HIF-α stabilization [20]. Increased stabilization of HIF-α is also observed in mice maintained on a low iron diet [31]. Due to the requirement of iron for their activity, it has been hypothesized that these hydroxylases may function as physiological iron sensors [18, 20, 32]. Moreover, it is becoming increasingly evident that HIF itself plays a direct role in iron homeostasis as a number of HIF target genes mediate iron metabolism [31, 33–41].
Fig. 2.
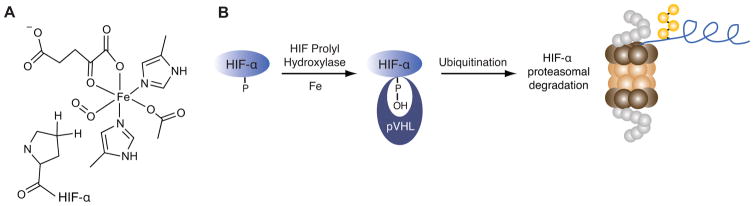
Proteolytic regulation of HIF requires iron. (A) The HIF prolyl hydroxylase active site utilizes iron to bind dioxygen in order to hydroxylate proline residues on HIF-α. (B) Proteasomal degradation of HIF-α is mediated by the iron binding HIF prolyl hydroxylases that facilitate the interaction between pVHL and HIF-α.
4. Iron Responsive Protein Degradation Mediates Cellular Iron Homeostasis
Iron Regulatory Proteins 1 and 2 (IRP1 and 2) bind RNA stem loop structures, known as iron responsive elements (IREs), found in a cohort of genes involved in iron uptake, storage, and utilization (reviewed in this issue of BBA-Molecular Cell Research []). This IRE/IRP system has emerged as the central regulatory pathway for the maintenance of mammalian cellular iron homeostasis [2]. The quintessential IRE-containing genes are those encoding ferritin and transferrin receptor 1 (TfR1). TfR1 is largely responsible for iron uptake from the serum while ferritin stores and sequesters excess cellular iron [42]. IRP binding to the IRE on the 5′ untranslated region (UTR) of ferritin inhibits ribosomal binding and decreases ferritin translation. In the case of TfR1, IRP binding to multiple IREs within its 3′ UTR prevents endonucleolytic mRNA degradation thereby increasing expression. However, IRPs fail to interact with IREs under iron replete conditions, promoting ferritin translation and reduced TfR1 mRNA stability. Through effects on these and other genes, the IRE/IRP system coordinates the posttranscriptional regulation of factors mediating iron metabolism in order to maintain cellular iron homeostasis.
The IRE binding activities of the two IRPs are regulated by distinct mechanisms. When bioavailable iron is abundant, IRP1 assembles an iron-sulfur (Fe-S) cluster (holo-IRP1) and functions as a cytosolic aconitase enzyme, interconverting two intermediates of the TCA cycle, citrate and isocitrate [43, 44]. Apo-IRP1, enriched in cells where iron levels are low and iron-sulfur cluster synthesis is presumably compromised, is competent to bind IREs in the UTRs of genes such as ferritin and TfR1. In contrast, IRP2 is regulated at the posttranslational level via the UPS. IRP2 protein levels decrease substantially in cells treated with iron due to polyubiquitination of the protein with concomitant proteasomal degradation. Under iron limiting conditions however, the IRP2 protein is stabilized and preferentially accumulates, enabling its interaction with IRE-containing mRNAs [45–47]. In addition to being regulated by bioavailable iron levels, IRP2 is regulated in an O2-dependent manner. Under normoxic conditions, IRP2 has a half-life of ~6 hours which increases to greater than 12 hours in hypoxic cells [48].
The physiological importance of both IRPs is reflected by the embryonic-lethal phenotype observed for global Irp1−/−Irp2−/− mice [49]. IRP2 null mice display inappropriate regulation of IRE-containing genes in many tissues, mild neurodegeneration, and microcytic anemia [50–52]. In comparison to IRP2 knockout mice, misregulation of iron metabolism only occurs in brown fat and kidneys of Irp1−/− mice while no other overt phenotypes were reported [51]. These data suggest that IRP2 may play a larger role in regulating iron homeostasis in vivo.
While it has been known for decades that IRP1 can be regulated as a function of its Fe-S cluster binding activity, delineation of the mechanisms controlling IRP2 regulation has proven more elusive [3, 53, 54]. Specifically, the E3 ubiquitin ligase that targets IRP2 for degradation and the mechanisms cells use to sense iron levels and correlate changes of this metabolite to differences in IRP2 stability have only recently been revealed.
5. FBXL5 is an Iron-sensing E3 Ubiquitin Ligase that Regulates Iron Homeostasis
An increased understanding of how the UPS controls the maintenance of iron homeostasis via regulation of IRP2 came with the discovery of an IRP2 E3 ubiquitin ligase. This E3 ligase, F-box and leucine-rich repeat protein 5 (FBXL5), was independently identified by two laboratories [55, 56]. Using a proteomics approach, IRP2 was identified as an interacting partner of a substrate-trapping FBXL5 variant [56]. In an alternative approach, FBXL5 was identified in an RNAi screen for regulators of iron-dependent IRP2 degradation [55]. FBXL5 is a member of the protein family defined by a conserved ~40–50 amino acid F-box domain [57]. F-box proteins assemble into Skp1/Cul1/F-box (SCF) E3 ubiquitin ligases complexes composed of the scaffolding proteins CUL1, RBX1, and SKP1. SKP1 recruits the F-box containing subunit of the E3 ligase complex. The function of the F-box protein is critical as its employs additional domains to tether substrates to the multimeric SCF complex for polyubiquitination. RBX1 is a ring-box protein that binds an activated E2 conjugating enzyme, which transfers ubiquitin to the SCF complex target protein [25, 58].
Loss of FBXL5 expression in human cells causes inappropriate accumulation of IRP2 with concomitant misregulation of IRE containing genes under iron replete conditions [55, 56]. FBXL5 interacts with IRP2 in human cells and recombinant SCFFBXL5 polyubiquitinates IRP2 in vitro [55, 56]. Taken together, these observations suggest that FBXL5 is a bona fide IRP2 E3 ligase, required for proper regulation and maintenance of cellular iron homeostasis. IRP1 can similarly be regulated via FBXL5 in an iron-dependent fashion when iron-sulfur cluster assembly is otherwise compromised, although the physiological relevance of this proteolytic regulation is unknown [55, 56, 59, 60].
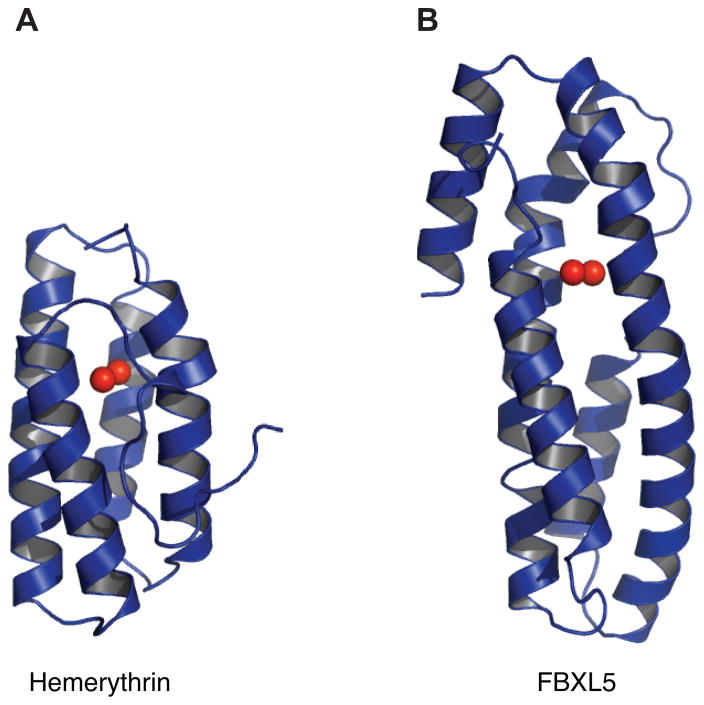
Intriguingly, FBXL5 is regulated in a reciprocal fashion to IRP2. FBXL5 accumulates under conditions of excess iron, while it is destabilized when either iron or O2 becomes limiting [55, 56]. Both iron- and oxygen-dependent control of FBXL5’s stability are mediated through its N terminus, predicted to encode an iron-binding hemerythrin-like domain [55, 56]. Hemerythrin (Hr) domains from marine invertebrates and bacteria are well characterized though no vertebrate Hr domains had previously been identified [61–67]. In invertebrates, Hr containing proteins frequently function as O2 transport and storage proteins similar to the mammalian proteins hemoglobin and myoglobin [68]. In addition to these roles, it has been postulated that Hr proteins function as iron and/or O2 sensors [63, 69, 70]. Canonical Hr domains are characterized by a helical bundle fold containing four α helices and an O2-binding diiron center. The binuclear iron center is typically ligated by seven amino acids comprised of five histidines, one aspartic acid, and one glutamic acid. The first iron atom (Fe1) of the diiron center is coordinated by three histidines and the second iron (Fe2) is coordinated by two histidines with the acidic residues bridging both atoms. The diiron center is also bridged by a solvent-derived μ-hydroxo group in the deoxy state (Fig. 3A). Hr domains can reversibly bind dioxygen at the Fe2 site. Bound O2 is reduced and the ensuing peroxide species forms a hydrogen bond with the resultant μ-oxo group and is further stabilized by hydrophobic residues lining the binding pocket [68, 71, 72].
Fig. 3.
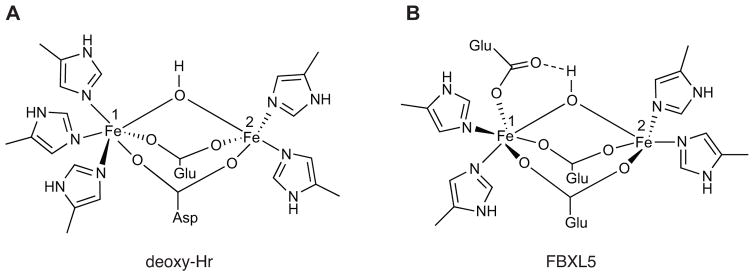
Schematics of the diiron center from the canonical deoxy Hr (A) and the unorthodox FBXL5 Hr (B).
Recently, X-ray crystallography has confirmed that the N terminus of human FBXL5 contains a hemerythrin-like domain [73]. Analogous to bacterial and invertebrate Hr domains, the FBXL5 Hr domain features a diiron center assembled within an up-down α-helical bundle supported by an atypical fifth helix. Moreover, the helical bundle is substantially longer (~58 Å) in FBXL5 than in previously characterized domains (~40–50 Å) [61, 63] (Fig. 4). FBXL5’s diiron center also differs from canonical Hrs by employing only four coordinating histidines. In place of the fifth histidine ligating Fe1, the FBXL5 Hr employs a glutamic acid that appears to form an unprecedented hydrogen bond with both Fe1 and the μ-oxygen species (Fig. 3B). Another distinctive aspect of the FBXL5 Hr domain is the absence of observable O2 binding at the diiron center despite crystallization in the presence of ambient oxygen [73]. Stopped-flow spectroscopy also suggests the domain may not bind O2 in solution. Although the FBXL5 Hr has a hydrophobic pocket juxtaposed to the diiron center that resembles canonical Hr domains, the dimensions of the FBXL5 pocket are much smaller and therefore dioxygen-binding may not be physically possible in this uncommon vertebrate Hr.
Fig. 4.
Structure of the FBXL5 Hr domain compared to the classical domain. Ribbon representations of the Hr domain from T. dyscritum (A; PDB code 1HMD) and the hemerythrin-like domain from H. sapien FBXL5 (B; PDB code 3V5Y).
Recent molecular approaches have provided insights into the mechanisms controlling FBXL5 regulation [73]. Mutation of the iron-ligating residues in the Hr domain results in constitutive destabilization of FBXL5 demonstrating that Fe binding is necessary for proper regulation of FBXL5. Under iron replete conditions when the protein accumulates at high levels, FBXL5 exhibits low levels of polyubiquitination. Conversely, when iron is limiting, FBXL5 displays pronounced polyubiquitination and subsequent degradation that is dependent on a functional E1 ubiquitin-activating enzyme and the proteasome. Moreover, residues 77–81 within the Hr domain appear to comprise part of a regulatory element, or degron, mediating FBXL5’s differential stability. Taken together, these multiple lines of evidence suggest a model in which FBXL5’s N-terminal Hr domain senses iron and regulates FBXL5 stability (Fig. 5). According to this model, when intracellular iron levels are abundant the Hr domain binds iron and adopts a conformation in which its degron is less accessible. Degron sequestration results in FBXL5 stabilization, formation of an SCFFBXL5 E3 ligase complex, and polyubiquitination of IRP2 with subsequent proteasomal degradation. Conversely, when bioavailable iron levels are low the FBXL5 Hr domain is unable to form a diiron center, resulting in a structural change that exposes the regulatory element targeting FBXL5 for polyubiquitination and degradation. Iron-dependent regulation of FBXL5 is predicted to be critical as it controls IRP2 expression levels, which in turn regulates key iron homeostasis genes. Thus FBXL5, via its Hr domain, enables cells to gauge bioavailable iron levels and control IRP2 accumulation accordingly, forming a tightly regulated circuit in the maintenance of iron homeostasis.
Fig. 5.
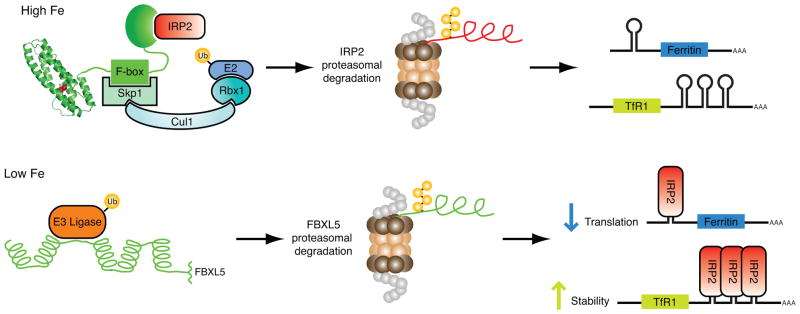
Model of how the N-terminal Hr domain controls iron homeostasis through regulation of FBXL5 stability with concomitant control of IRP2 degradation.
The finding that FBXL5’s stability is controlled via an iron-binding regulatory switch in its Hr domain is particularly significant. This type of ligand-dependent regulation has been observed with multiple F-box proteins in plants and algae. For example, binding of jasmonate to the plant F-box protein COIl, enhances the interaction between COI1 and JAZ proteins [74, 75]. In red algae, Fbx3 binds Mg-protoporphyrin IX which negatively regulates binding to its substrate cyclin 1 [76]. However, FBXL5 is the first example of a metazoan F-box protein to function in an analogous ligand-regulated manner.
As with ferroportin, iron binding to the Hr domain is postulated to induce a conformational change that increases ubiquitination and degradation of FBXL5. Such regulation is also reminiscent of the yeast ER protein Pca1, which functions as an ATPase and exporter of the toxic metal cadmium. In the absence of cadmium, a degron is exposed and Pca1 is targeted for proteasomal degradation. However when cadmium levels are high, Pca1 binds cadmium and elicits a structural change purported to mask a degron and spare the protein from degradation. Upon cadmium binding, the Pca1-Cd complex may be excreted from the cell in order to reduce cadmium levels [77]. Both FBXL5’s and Pca1’s stabilities are dependent on binding of their cognate metals to control degron accessibility. Moreover, both proteins functions as sensors to maintain homeostasis of their respective metals.
While these results indicate that FBXL5 is necessary for maintaining proper iron homeostasis in cultured cells [55, 56], they do not address the role of SCFFBXL5 in regulating iron homeostasis in vivo. A forward genetics screen in mice generated a FBXL5 mutant strain where methionine 127 had been changed to lysine [78]. As shown in the FBXL5 Hr structure, M127 is positioned very closely (~4 Å) to both the bridging μ-oxo atom and Fe2 [73]. Mutation of the basic lysine residue likely interferes with assembly of the diiron center, constitutively destabilizing FBXL5. Mice homozygous for this mutation display embryonic lethality, suggesting that a functional FBXL5 is required for murine development [78].
Using a targeted approach, it was recently reported that global disruption of the FBXL5 gene in mice results in death at E8.5 [79]. Importantly, these Fbxl5−/− embryos feature aberrant iron accumulation and increased oxidative stress that are likely responsible for lethality. Consistent with FBXL5’s importance for IRE-containing gene regulation, Fbxl5−/− embryos have elevated levels of TfR1 mRNA. To demonstrate that misregulation of IRE containing genes due to uncontrolled IRP2 accumulation causes lethality in Fbxl5−/− embryos, Fbxl5+/−Irp2+/− mice were intercrossed to generate Fbxl5−/−Irp2−/− mice. Interestingly, simultaneous loss of IRP2 rescues the FBXL5 null phenotype as these mice develop normally and are fertile, indicating that control of IRP2 expression via FBXL5 is required for the maintenance of iron homeostasis and viability during murine development. To investigate the function of FBXL5 in adult mice, liver specific knockout mice were also generated [79]. These mice displayed increased liver iron accumulation and elevated hepatic IRP2 and TfR1 protein levels. Additionally, mice lacking FBXL5 expression in the liver manifested decreased hepcidin expression and increased serum iron levels compared to wild-type littermates. When fed a high iron diet, the majority of these mice accumulate even more iron within the liver in conjunction with increased oxidative stress and steatosis. Though no other organs were observed with iron overload, most of these mice die within one day due to the ensuing liver failure [79]. Together, these data indicate that FBXL5 expression is required for maintaining both cellular and systemic iron homeostasis in vivo.
The discovery that FBXL5 plays an integral role in maintaining iron homeostasis represents an important step in the metals biology field. However, FBXL5 research is still in its infancy and much remains to be resolved. Current data indicates that IRP2 and apo-IRP1 are SCFFBXL5 substrates [55, 56, 79] though p150Glued, a protein involved in vesicular transport, has also been reported to be a FBXL5 substrate [80]. A key step in understanding FBXL5’s functions will be to delineate its complete repertoire of substrates. Due to the iron-responsiveness of FBXL5, future studies identifying additional FBXL5 targets could reveal that this E3 plays a role in regulating iron and/or oxygen homeostasis independent of the IRP/IRE regulatory system.
While it has been clearly demonstrated that FBXL5 directly binds iron, the mechanism by which the metal gets loaded into the Hr domain remains unknown. Some metalloenzymes rely on chaperones for incorporation of the correct metal into their active site [81, 82]. Recently, PCBP1, has been identified as a putative chaperone that delivers iron to both ferritin and the HIF hydroxylases [83, 84]. It is plausible that FBXL5 may also require an ancillary protein for insertion of iron into its Hr domain. Another critical question is how FBXL5 is regulated in an iron dependent manner via the UPS. Specifically, is there a dedicated E3 ligase that tags the protein for proteasomal degradation when iron is limiting and the FBXL5 degron is exposed?
The molecular details defining the FBXL5-IRP substrate interactions are also unresolved. Initial indications that IRP1, unlike IRP2, was not subject to iron-dependent degradation, focused attention on a 73 amino acid insertion within IRP2 as a likely determinant for E3 ligase recognition [85, 86]. However, subsequent studies demonstrated that this region was dispensable for IRP2 regulation. [48, 53, 55]. More likely, IRP1 and IRP2 share a common degron that can be selectively masked in a conformation adopted by IRP1 following assembly of its Fe-S cluster. FBXL5 contains six predicted leucine rich repeats [55], which are known to function as substrate recognition motifs in other F-box proteins [87]. However, which, if any of these repeats are required for FBXL5 substrate binding is an open question. Intriguingly, iron bioavailability also appears to regulate the physical interaction between FBXL5 and IRP2 [55, 56], suggesting there may be additional iron-sensing mechanisms that control this E3 ligase subunit independent of its Hr domain.
Diverging from canonical Hr domains, the FBXL5 Hr does not appear to directly bind O2. Nevertheless, accumulation of FBXL5 and its Hr domain in cells is regulated by oxygen availability via the UPS. Delineation of the molecular mechanisms underlying FBXL5 oxygen-responsiveness, and its physiological relevance, may reveal novel insights into the crosstalk between oxygen and iron homeostasis. In addition to liver-specific FBXL5 ablation, mice lacking FBXL5 in the intestine and brain will be pivotal in determining whether FBXL5 plays a role in iron absorption and neurodegeneration, respectively.
6. Conclusion
Multiple studies indicate that iron homeostasis is governed in part through regulated proteolysis of ferroportin, HIF, IRPs, and FBXL5. Iron binding to the FBXL5 Hr domain regulates its stability, which has reciprocal affects on IRPs degradation. When bioavailable iron levels are sufficient for enzymatic activity, HIF Prolyl Hydroxylases modify the transcription factor HIF, mediating its degradation. When systemic demand for iron is attenuated, hepcidin expression increases and it is released into the circulation where it binds ferroportin to triggers its degradation. In an alternative cell autonomous pathway, low cytosolic iron levels are directly sensed by ferroportin and result in a conformational change promoting hepcidin-independent degradation (Fig. 6).
Fig. 6.
Interconnections in the proteolytic pathways responsible for maintaining iron homeostasis. FBXL5 and HIF Prolyl Hydroxylases function as ostensible iron-sensors and promote the degradation of IRPs and HIF, respectively. FBXL5 is degraded when its Hr domain is prevented from binding iron. Ferroportin degradation is initiated upon hepcidin binding or following low cytosolic iron availability. These diverse regulatory factors are intertwined at multiple levels including IRPs negatively regulating HIF-α and ferroportin translation, and HIF-α inhibiting transcription of hepcidin.
While ferroportin, HIF, IRPs, and FBXL5 play critical roles in regulating cellular and systemic iron homeostasis, it is becoming apparent that there are multiple interconnections among the disparate pathways (Fig. 6). For example, HIF-2α contains an IRE in its 5′ UTR [88]. Similar to the regulation of ferritin, IRP binding to its IRE under conditions of iron deficiency inhibits HIF-2α translation, while under iron replete conditions its expression is derepressed. HIF-2α displays high expression levels in the kidney where it controls expression of erythropoietin, a hormone that stimulates the production of erythrocytes [89]. The IRP/IRE regulatory system may temper red blood cell production under conditions where iron is limiting to downregulate the production of heme- and/or hemoglobin-deficient erythrocytes [88]. Ferroportin expression is also regulated posttranscriptionally via an IRE its 5′ UTR. This phenotype is highlighted in intestine specific Irp1−/−Irp2−/− mice. These mice, despite increased hepcidin expression, display a paucity of bioavailable intestinal iron levels in part due to the overexpression of ferroportin [90]. Ferroportin expressing cells such as intestinal epithelia, which function to maintain systemic iron homeostasis, may rely on proper IRP/IRE regulation to protect themselves against excessive iron efflux that could lead to iron deficiency [91]. As noted above, several HIF target genes, including hepcidin, are involved in maintaining iron homeostasis. Interestingly, HIF is believed to transcriptionally repress HAMP as loss of pVHL expression in the mouse liver stabilizes HIF-α, attenuating hepcidin expression and stabilizing ferroportin [31]. This mechanism may be important for increasing iron levels in the serum to facilitate erythropoiesis in response to physiological conditions such as hypoxia. Additional studies are needed to advance our understanding of the mechanisms regulating dynamic proteolysis in iron metabolism. For example, degradation of the iron transport protein divalent metal transporter 1 has recently been suggested to be regulated in an iron dependent manner [90, 92, 93]. Lastly, altered expression or activities of many of the regulatory factors discussed here are frequently observed in human disease. Additional factors, such as FBXL5, need to be analyzed in greater detail to determine whether they contribute to iron related disorders or other diseases. Future work has great potential to provide insights into disease etiology and uncover new therapeutic targets for various pathologies.
Acknowledgments
R.K.B. is the Michael L. Rosenberg Scholar in Medical Research and is supported by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund. J.W.T. was supported by the Graduate Programs Initiative from the UT Board of Regents.
References
- 1.Crichton RR, Pierre JL. Old iron, young copper: from Mars to Venus. Biometals. 2001;14:99–112. doi: 10.1023/a:1016710810701. [DOI] [PubMed] [Google Scholar]
- 2.Klausner RD, Rouault TA, Harford JB. Regulating the fate of mRNA: the control of cellular iron metabolism. Cell. 1993;72:19–28. doi: 10.1016/0092-8674(93)90046-s. [DOI] [PubMed] [Google Scholar]
- 3.Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 4.Andrews NC, Schmidt PJ. Iron homeostasis. Annu Rev Physiol. 2007;69:69–85. doi: 10.1146/annurev.physiol.69.031905.164337. [DOI] [PubMed] [Google Scholar]
- 5.Muckenthaler MU, Galy B, Hentze MW. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu Rev Nutr. 2008;28:197–213. doi: 10.1146/annurev.nutr.28.061807.155521. [DOI] [PubMed] [Google Scholar]
- 6.De Domenico I, McVey Ward D, Kaplan J. Regulation of iron acquisition and storage: consequences for iron-linked disorders. Nat Rev Mol Cell Biol. 2008;9:72–81. doi: 10.1038/nrm2295. [DOI] [PubMed] [Google Scholar]
- 7.Castells-Roca L, Muhlenhoff U, Lill R, Herrero E, Belli G. The oxidative stress response in yeast cells involves changes in the stability of Aft1 regulon mRNAs. Mol Microbiol. 2011;81:232–248. doi: 10.1111/j.1365-2958.2011.07689.x. [DOI] [PubMed] [Google Scholar]
- 8.Lee JW, Helmann JD. Functional specialization within the Fur family of metalloregulators. Biometals. 2007;20:485–499. doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- 9.Waldron KJ, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Microbiol. 2009;7:25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- 10.Andrews NC. Forging a field: the golden age of iron biology. Blood. 2008;112:219–230. doi: 10.1182/blood-2007-12-077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicolas G, Chauvet C, Viatte L, Danan JL, Bigard X, Devaux I, Beaumont C, Kahn A, Vaulont S. The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J Clin Invest. 2002;110:1037–1044. doi: 10.1172/JCI15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Domenico I, Lo E, Ward DM, Kaplan J. Hepcidin-induced internalization of ferroportin requires binding and cooperative interaction with Jak2. Proc Natl Acad Sci USA. 2009;106:3800–3805. doi: 10.1073/pnas.0900453106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Domenico I, Lo E, Yang B, Korolnek T, Hamza I, Ward DM, Kaplan J. The role of ubiquitination in hepcidin-independent and hepcidin-dependent degradation of ferroportin. Cell Metab. 2011;14:635–646. doi: 10.1016/j.cmet.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 15.De Domenico I, Ward DM, Langelier C, Vaughn MB, Nemeth E, Sundquist WI, Ganz T, Musci G, Kaplan J. The molecular mechanism of hepcidin-mediated ferroportin down-regulation. Mol Biol Cell. 2007;18:2569–2578. doi: 10.1091/mbc.E07-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drakesmith H, Schimanski LM, Ormerod E, Merryweather-Clarke AT, Viprakasit V, Edwards JP, Sweetland E, Bastin JM, Cowley D, Chinthammitr Y, Robson KJ, Townsend AR. Resistance to hepcidin is conferred by hemochromatosis-associated mutations of ferroportin. Blood. 2005;106:1092–1097. doi: 10.1182/blood-2005-02-0561. [DOI] [PubMed] [Google Scholar]
- 17.Pietrangelo A. The ferroportin disease. Blood Cells Mol Dis. 2004;32:131–138. doi: 10.1016/j.bcmd.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Ozer A, Bruick RK. Non-heme dioxygenases: cellular sensors and regulators jelly rolled into one? Nat Chem Biol. 2007;3:144–153. doi: 10.1038/nchembio863. [DOI] [PubMed] [Google Scholar]
- 19.Salahudeen AA, Bruick RK. Maintaining Mammalian Iron and Oxygen Homeostasis: Sensors, Regulation, and Cross-Talk. Ann NY Acad Sci. 2009;1177:30–38. doi: 10.1111/j.1749-6632.2009.05038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peyssonnaux C, Nizet V, Johnson RS. Role of the hypoxia inducible factors HIF in iron metabolism. Cell Cycle. 2008;7:28–32. doi: 10.4161/cc.7.1.5145. [DOI] [PubMed] [Google Scholar]
- 21.Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salceda S, Caro J. Hypoxia-inducible factor 1alpha (HIF-1alpha) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J Biol Chem. 1997;272:22642–22647. doi: 10.1074/jbc.272.36.22642. [DOI] [PubMed] [Google Scholar]
- 23.Hon WC, Wilson MI, Harlos K, Claridge TD, Schofield CJ, Pugh CW, Maxwell PH, Ratcliffe PJ, Stuart DI, Jones EY. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature. 2002;417:975–978. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- 24.Min JH, Yang H, Ivan M, Gertler F, Kaelin WG, Jr, Pavletich NP. Structure of an HIF-1alpha -pVHL complex: hydroxyproline recognition in signaling. Science. 2002;296:1886–1889. doi: 10.1126/science.1073440. [DOI] [PubMed] [Google Scholar]
- 25.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 26.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 28.Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 29.Ivan M, Haberberger T, Gervasi DC, Michelson KS, Gunzler V, Kondo K, Yang H, Sorokina I, Conaway RC, Conaway JW, Kaelin WG., Jr Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor. Proc Natl Acad Sci USA. 2002;99:13459–13464. doi: 10.1073/pnas.192342099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masson N, Willam C, Maxwell PH, Pugh CW, Ratcliffe PJ. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 2001;20:5197–5206. doi: 10.1093/emboj/20.18.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peyssonnaux C, Zinkernagel AS, Schuepbach RA, Rankin E, Vaulont S, Haase VH, Nizet V, Johnson RS. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs) J Clin Invest. 2007;117:1926–1932. doi: 10.1172/JCI31370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith TG, Balanos GM, Croft QP, Talbot NP, Dorrington KL, Ratcliffe PJ, Robbins PA. The increase in pulmonary arterial pressure caused by hypoxia depends on iron status. J Physiol. 2008;586:5999–6005. doi: 10.1113/jphysiol.2008.160960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lok CN, Ponka P. Identification of a hypoxia response element in the transferrin receptor gene. J Biol Chem. 1999;274:24147–24152. doi: 10.1074/jbc.274.34.24147. [DOI] [PubMed] [Google Scholar]
- 34.Mastrogiannaki M, Matak P, Keith B, Simon MC, Vaulont S, Peyssonnaux C. HIF-2alpha, but not HIF-1alpha, promotes iron absorption in mice. J Clin Invest. 2009;119:1159–1166. doi: 10.1172/JCI38499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shah YM, Matsubara T, Ito S, Yim SH, Gonzalez FJ. Intestinal hypoxia-inducible transcription factors are essential for iron absorption following iron deficiency. Cell Metab. 2009;9:152–164. doi: 10.1016/j.cmet.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lakhal S, Schodel J, Townsend AR, Pugh CW, Ratcliffe PJ, Mole DR. Regulation of type II transmembrane serine proteinase TMPRSS6 by hypoxia-inducible factors: new link between hypoxia signaling and iron homeostasis. J Biol Chem. 2011;286:4090–4097. doi: 10.1074/jbc.M110.173096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tacchini L, Bianchi L, Bernelli-Zazzera A, Cairo G. Transferrin receptor induction by hypoxia. HIF-1-mediated transcriptional activation and cell-specific post-transcriptional regulation. J Biol Chem. 1999;274:24142–24146. doi: 10.1074/jbc.274.34.24142. [DOI] [PubMed] [Google Scholar]
- 38.Mukhopadhyay CK, Mazumder B, Fox PL. Role of hypoxia-inducible factor-1 in transcriptional activation of ceruloplasmin by iron deficiency. J Biol Chem. 2000;275:21048–21054. doi: 10.1074/jbc.M000636200. [DOI] [PubMed] [Google Scholar]
- 39.Lee PJ, Jiang BH, Chin BY, Iyer NV, Alam J, Semenza GL, Choi AM. Hypoxia-inducible factor-1 mediates transcriptional activation of the heme oxygenase-1 gene in response to hypoxia. J Biol Chem. 1997;272:5375–5381. [PubMed] [Google Scholar]
- 40.Manalo DJ, Rowan A, Lavoie T, Natarajan L, Kelly BD, Ye SQ, Garcia JG, Semenza GL. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood. 2005;105:659–669. doi: 10.1182/blood-2004-07-2958. [DOI] [PubMed] [Google Scholar]
- 41.Xia X, Lemieux ME, Li W, Carroll JS, Brown M, Liu XS, Kung AL. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc Natl Acad Sci USA. 2009;106:4260–4265. doi: 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rouault TA. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat Chem Biol. 2006;2:406–414. doi: 10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- 43.Kaptain S, Downey WE, Tang C, Philpott C, Haile D, Orloff DG, Harford JB, Rouault TA, Klausner RD. A regulated RNA binding protein also possesses aconitase activity. Proc Natl Acad Sci U S A. 1991;88:10109–10113. doi: 10.1073/pnas.88.22.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Philpott CC, Klausner RD, Rouault TA. The bifunctional iron-responsive element binding protein/cytosolic aconitase: the role of active-site residues in ligand binding and regulation. Proc Natl Acad Sci USA. 1994;91:7321–7325. doi: 10.1073/pnas.91.15.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo B, Phillips JD, Yu Y, Leibold EA. Iron regulates the intracellular degradation of iron regulatory protein 2 by the proteasome. J Biol Chem. 1995;270:21645–21651. doi: 10.1074/jbc.270.37.21645. [DOI] [PubMed] [Google Scholar]
- 46.Guo B, Yu Y, Leibold EA. Iron regulates cytoplasmic levels of a novel iron-responsive element-binding protein without aconitase activity. J Biol Chem. 1994;269:24252–24260. [PubMed] [Google Scholar]
- 47.Samaniego F, Chin J, Iwai K, Rouault TA, Klausner RD. Molecular characterization of a second iron-responsive element binding protein, iron regulatory protein 2. Structure, function, and post-translational regulation. J Biol Chem. 1994;269:30904–30910. [PubMed] [Google Scholar]
- 48.Hanson ES, Rawlins ML, Leibold EA. Oxygen and iron regulation of iron regulatory protein 2. J Biol Chem. 2003;278:40337–40342. doi: 10.1074/jbc.M302798200. [DOI] [PubMed] [Google Scholar]
- 49.Smith SR, Ghosh MC, Ollivierre-Wilson H, Hang Tong W, Rouault TA. Complete loss of iron regulatory proteins 1 and 2 prevents viability of murine zygotes beyond the blastocyst stage of embryonic development. Blood Cells Mol Dis. 2006;36:283–287. doi: 10.1016/j.bcmd.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 50.LaVaute T, Smith S, Cooperman S, Iwai K, Land W, Meyron-Holtz E, Drake SK, Miller G, Abu-Asab M, Tsokos M, Switzer R, 3rd, Grinberg A, Love P, Tresser N, Rouault TA. Targeted deletion of the gene encoding iron regulatory protein-2 causes misregulation of iron metabolism and neurodegenerative disease in mice. Nat Genet. 2001;27:209–214. doi: 10.1038/84859. [DOI] [PubMed] [Google Scholar]
- 51.Meyron-Holtz EG, Ghosh MC, Iwai K, LaVaute T, Brazzolotto X, Berger UV, Land W, Ollivierre-Wilson H, Grinberg A, Love P, Rouault TA. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J. 2004;23:386–395. doi: 10.1038/sj.emboj.7600041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galy B, Ferring D, Minana B, Bell O, Janser HG, Muckenthaler M, Schumann K, Hentze MW. Altered body iron distribution and microcytosis in mice deficient in iron regulatory protein 2 (IRP2) Blood. 2005;106:2580–2589. doi: 10.1182/blood-2005-04-1365. [DOI] [PubMed] [Google Scholar]
- 53.Wang J, Chen G, Muckenthaler M, Galy B, Hentze MW, Pantopoulos K. Iron-mediated degradation of IRP2, an unexpected pathway involving a 2-oxoglutarate-dependent oxygenase activity. Mol Cell Biol. 2004;24:954–965. doi: 10.1128/MCB.24.3.954-965.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zumbrennen KB, Hanson ES, Leibold EA. HOIL-1 is not required for iron-mediated IRP2 degradation in HEK293 cells. Biochim Biophys Acta. 2008;1783:246–252. doi: 10.1016/j.bbamcr.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salahudeen AA, Thompson JW, Ruiz JC, Ma HW, Kinch LN, Li Q, Grishin NV, Bruick RK. An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science. 2009;326:722–726. doi: 10.1126/science.1176326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vashisht AA, Zumbrennen KB, Huang X, Powers DN, Durazo A, Sun D, Bhaskaran N, Persson A, Uhlen M, Sangfelt O, Spruck C, Leibold EA, Wohlschlegel JA. Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science. 2009;326:718–721. doi: 10.1126/science.1176333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 59.Clarke SL, Vasanthakumar A, Anderson SA, Pondarre C, Koh CM, Deck KM, Pitula JS, Epstein CJ, Fleming MD, Eisenstein RS. Iron-responsive degradation of iron-regulatory protein 1 does not require the Fe-S cluster. EMBO J. 2006;25:544–553. doi: 10.1038/sj.emboj.7600954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fillebeen C, Chahine D, Caltagirone A, Segal P, Pantopoulos K. A phosphomimetic mutation at Ser-138 renders iron regulatory protein 1 sensitive to iron-dependent degradation. Mol Cell Biol. 2003;23:6973–6981. doi: 10.1128/MCB.23.19.6973-6981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holmes MA, Le Trong I, Turley S, Sieker LC, Stenkamp RE. Structures of deoxy and oxy hemerythrin at 2.0 Å resolution. J Mol Biol. 1991;218:583–593. doi: 10.1016/0022-2836(91)90703-9. [DOI] [PubMed] [Google Scholar]
- 62.Holmes MA, Stenkamp RE. Structures of met and azidomet hemerythrin at 1.66 Å resolution. J Mol Biol. 1991;220:723–737. doi: 10.1016/0022-2836(91)90113-k. [DOI] [PubMed] [Google Scholar]
- 63.Isaza CE, Silaghi-Dumitrescu R, Iyer RB, Kurtz DM, Jr, Chan MK. Structural basis for O2 sensing by the hemerythrin-like domain of a bacterial chemotaxis protein: substrate tunnel and fluxional N terminus. Biochemistry. 2006;45:9023–9031. doi: 10.1021/bi0607812. [DOI] [PubMed] [Google Scholar]
- 64.Kao WC, Wang VC, Huang YC, Yu SS, Chang TC, Chan SI. Isolation, purification and characterization of hemerythrin from Methylococcus capsulatus (Bath) J Inorg Biochem. 2008;102:1607–1614. doi: 10.1016/j.jinorgbio.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 65.Sheriff S, Hendrickson WA, Smith JL. Structure of myohemerythrin in the azidomet state at 1.7/1.3 A resolution. J Mol Biol. 1987;197:273–296. doi: 10.1016/0022-2836(87)90124-0. [DOI] [PubMed] [Google Scholar]
- 66.Stenkamp RE, Sieker LC, Jensen LH, McCallum JD, Sanders-Loehr J. Active site structures of deoxyhemerythrin and oxyhemerythrin. Proc Natl Acad Sci USA. 1985;82:713–716. doi: 10.1073/pnas.82.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Onoda A, Okamoto Y, Sugimoto H, Shiro Y, Hayashi T. Crystal structure and spectroscopic studies of a stable mixed-valent state of the hemerythrin-like domain of a bacterial chemotaxis protein. Inorg Chem. 2011;50:4892–4899. doi: 10.1021/ic2001267. [DOI] [PubMed] [Google Scholar]
- 68.French CE, Bell JM, Ward FB. Diversity and distribution of hemerythrin-like proteins in prokaryotes. FEMS Microbiol Lett. 2008;279:131–145. doi: 10.1111/j.1574-6968.2007.01011.x. [DOI] [PubMed] [Google Scholar]
- 69.Bailly X, Vanin S, Chabasse C, Mizuguchi K, Vinogradov SN. A phylogenomic profile of hemerythrins, the nonheme diiron binding respiratory proteins. BMC Evol Biol. 2008;8:244. doi: 10.1186/1471-2148-8-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiong J, Kurtz DM, Jr, Ai J, Sanders-Loehr J. A hemerythrin-like domain in a bacterial chemotaxis protein. Biochemistry. 2000;39:5117–5125. doi: 10.1021/bi992796o. [DOI] [PubMed] [Google Scholar]
- 71.Nordlund P, Eklund H. Di-iron-carboxylate proteins. Curr Opin Struct Biol. 1995;5:758–766. doi: 10.1016/0959-440x(95)80008-5. [DOI] [PubMed] [Google Scholar]
- 72.Stenkamp RE. Dioxygen and Hemerythrin. Chemical Reviews. 1994;94:715–726. [Google Scholar]
- 73.Thompson JW, Salahudeen AA, Chollangi S, Ruiz JC, Brautigam CA, Makris TM, Lipscomb JD, Tomchick DR, Bruick RK. Structural and Molecular Characterization of the Iron-sensing Hemerythrin-like Domain within F-box and Leucine-rich Repeat Protein 5 (FBXL5) J Biol Chem. 2012 doi: 10.1074/jbc.M111.308684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, Garcia-Casado G, Lopez-Vidriero I, Lozano FM, Ponce MR, Micol JL, Solano R. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 75.Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 76.Kobayashi Y, Imamura S, Hanaoka M, Tanaka K. A tetrapyrrole-regulated ubiquitin ligase controls algal nuclear DNA replication. Nat Cell Biol. 2011;13:483–487. doi: 10.1038/ncb2203. [DOI] [PubMed] [Google Scholar]
- 77.Adle DJ, Wei W, Smith N, Bies JJ, Lee J. Cadmium-mediated rescue from ER-associated degradation induces expression of its exporter. Proc Natl Acad Sci USA. 2009;106:10189–10194. doi: 10.1073/pnas.0812114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ching YH, Munroe RJ, Moran JL, Barker AK, Mauceli E, Fennell T, Dipalma F, Lindblad-Toh K, Abcunas LM, Gilmour JF, Harris TP, Kloet SL, Luo Y, McElwee JL, Mu W, Park HK, Rogal DL, Schimenti KJ, Shen L, Shindo M, Shou JY, Stenson EK, Stover PJ, Schimenti JC. High resolution mapping and positional cloning of ENU-induced mutations in the Rw region of mouse chromosome 5. BMC Genet. 2010;11:106. doi: 10.1186/1471-2156-11-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moroishi T, Nishiyama M, Takeda Y, Iwai K, Nakayama KI. The FBXL5-IRP2 axis is integral to control of iron metabolism in vivo. Cell Metab. 2011;14:339–351. doi: 10.1016/j.cmet.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 80.Zhang N, Liu J, Ding X, Aikhionbare F, Jin C, Yao X. FBXL5 interacts with p150Glued and regulates its ubiquitination. Biochem Biophys Res Commun. 2007;359:34–39. doi: 10.1016/j.bbrc.2007.05.068. [DOI] [PubMed] [Google Scholar]
- 81.Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460:823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- 82.Rosenzweig AC. Metallochaperones: bind and deliver. Chem Biol. 2002;9:673–677. doi: 10.1016/s1074-5521(02)00156-4. [DOI] [PubMed] [Google Scholar]
- 83.Shi H, Bencze KZ, Stemmler TL, Philpott CC. A cytosolic iron chaperone that delivers iron to ferritin. Science. 2008;320:1207–1210. doi: 10.1126/science.1157643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nandal A, Ruiz JC, Subramanian P, Ghimire-Rijal S, Sinnamon RA, Stemmler TL, Bruick RK, Philpott CC. Activation of the HIF Prolyl Hydroxylase by the Iron Chaperones PCBP1 and PCBP2. Cell Metab. 2011;14:647–657. doi: 10.1016/j.cmet.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Iwai K, Drake SK, Wehr NB, Weissman AM, LaVaute T, Minato N, Klausner RD, Levine RL, Rouault TA. Iron-dependent oxidation, ubiquitination, and degradation of iron regulatory protein 2: implications for degradation of oxidized proteins. Proc Natl Acad Sci USA. 1998;95:4924–4928. doi: 10.1073/pnas.95.9.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Iwai K, Klausner RD, Rouault TA. Requirements for iron-regulated degradation of the RNA binding protein, iron regulatory protein 2. EMBO J. 1995;14:5350–5357. doi: 10.1002/j.1460-2075.1995.tb00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 88.Sanchez M, Galy B, Muckenthaler MU, Hentze MW. Iron-regulatory proteins limit hypoxia-inducible factor-2alpha expression in iron deficiency. Nat Struct Mol Biol. 2007;14:420–426. doi: 10.1038/nsmb1222. [DOI] [PubMed] [Google Scholar]
- 89.Warnecke C, Zaborowska Z, Kurreck J, Erdmann VA, Frei U, Wiesener M, Eckardt KU. Differentiating the functional role of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha (EPAS-1) by the use of RNA interference: erythropoietin is a HIF-2alpha target gene in Hep3B and Kelly cells. FASEB J. 2004;18:1462–1464. doi: 10.1096/fj.04-1640fje. [DOI] [PubMed] [Google Scholar]
- 90.Galy B, Ferring-Appel D, Kaden S, Grone HJ, Hentze MW. Iron regulatory proteins are essential for intestinal function and control key iron absorption molecules in the duodenum. Cell Metab. 2008;7:79–85. doi: 10.1016/j.cmet.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 91.Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: regulation of Mammalian iron metabolism. Cell. 2010;142:24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 92.Foot NJ, Leong YA, Dorstyn LE, Dalton HE, Ho K, Zhao L, Garrick MD, Yang B, Hiwase D, Kumar S. Ndfip1-deficient mice have impaired DMT1 regulation and iron homeostasis. Blood. 2011;117:638–646. doi: 10.1182/blood-2010-07-295287. [DOI] [PubMed] [Google Scholar]
- 93.Howitt J, Putz U, Lackovic J, Doan A, Dorstyn L, Cheng H, Yang B, Chan-Ling T, Silke J, Kumar S, Tan SS. Divalent metal transporter 1 (DMT1) regulation by Ndfip1 prevents metal toxicity in human neurons. Proc Natl Acad Sci USA. 2009;106:15489–15494. doi: 10.1073/pnas.0904880106. [DOI] [PMC free article] [PubMed] [Google Scholar]