Abstract
Congenital myasthenic syndromes are a heterogeneous group of inherited disorders that arise from impaired signal transmission at the neuromuscular synapse. They are characterized by fatigable muscle weakness. We performed linkage analysis, whole-exome and whole-genome sequencing to determine the underlying defect in patients with an inherited limb-girdle pattern of myasthenic weakness. We identify ALG14 and ALG2 as novel genes in which mutations cause a congenital myasthenic syndrome. Through analogy with yeast, ALG14 is thought to form a multiglycosyltransferase complex with ALG13 and DPAGT1 that catalyses the first two committed steps of asparagine-linked protein glycosylation. We show that ALG14 is concentrated at the muscle motor endplates and small interfering RNA silencing of ALG14 results in reduced cell-surface expression of muscle acetylcholine receptor expressed in human embryonic kidney 293 cells. ALG2 is an alpha-1,3-mannosyltransferase that also catalyses early steps in the asparagine-linked glycosylation pathway. Mutations were identified in two kinships, with mutation ALG2p.Val68Gly found to severely reduce ALG2 expression both in patient muscle, and in cell cultures. Identification of DPAGT1, ALG14 and ALG2 mutations as a cause of congenital myasthenic syndrome underscores the importance of asparagine-linked protein glycosylation for proper functioning of the neuromuscular junction. These syndromes form part of the wider spectrum of congenital disorders of glycosylation caused by impaired asparagine-linked glycosylation. It is likely that further genes encoding components of this pathway will be associated with congenital myasthenic syndromes or impaired neuromuscular transmission as part of a more severe multisystem disorder. Our findings suggest that treatment with cholinesterase inhibitors may improve muscle function in many of the congenital disorders of glycosylation.
Keywords: congenital myasthenic syndrome, ALG2, ALG14, mutation, N-linked glycosylation
Introduction
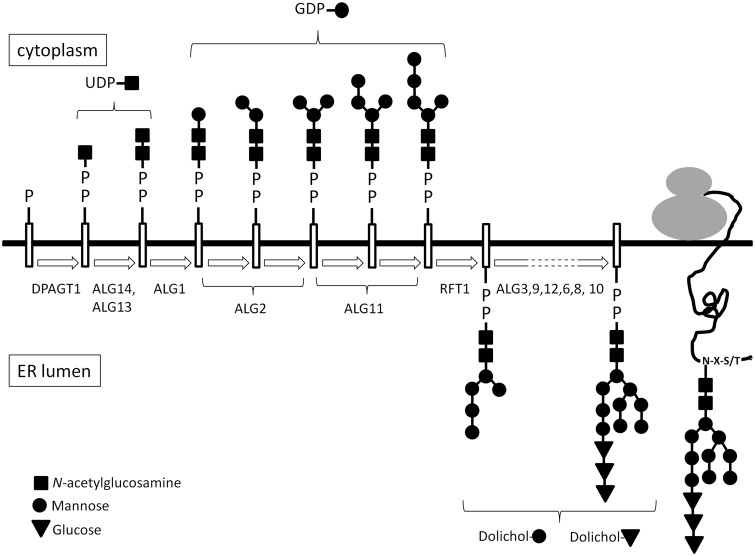
Mutations in genes that encode proteins involved in the asparagine-linked (N-linked) glycosylation pathway underlie a spectrum of disorders known as congenital disorders of glycosylation (CDG) (Jaeken and Matthijs, 2007; Jaeken, 2011). Many membrane-bound or exported proteins are modified by the addition and processing of glycans that can be crucial for protein folding and function, intracellular localization and transport and protein stability (Larkin and Imperiali, 2011). Defects of these critical biological processes may affect many different organs, especially the nervous system, muscles and the intestines (Freeze, 2006).The broad spectrum of both symptoms and severity often makes diagnosis of these disorders challenging. DPAGT1 encodes dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosamine phosphotransferase 1 (Bretthauer, 2009). It is a transmembrane endoplasmic reticulum protein that catalyses the first committed step of the core glycan assembly, the addition of GlcNAc-1-P from cytoplasmic UDP-GlcNAc to dolichol-P. Rare mutations identified in DPAGT1 have been found to underlie a severe multisystems disorder [CDG1J (MIM 608093)] typically with delayed development, microcephaly, intractable seizures and learning difficulties (Marek et al., 1999; Wu et al., 2003; Wurde et al., 2012). However, more recently, mutations in DPAGT1 have also been shown to be a cause of a limb-girdle form of congenital myasthenic syndrome (Belaya et al., 2012). The congenital myasthenic syndromes are inherited disorders of signal transmission at the neuromuscular synapse (Chaouch et al., 2012; Engel, 2012). They are most commonly autosomal recessive, and are characterized by fatigable muscle weakness, with the pattern of the muscle weakness varying according to which gene is mutated and the underlying molecular mechanism. The congenital myasthenic syndrome due to DPAGT1 mutations is a disabling disorder with typically a limb-girdle distribution of muscle weakness, tubular aggregates on muscle biopsy and patients who show a beneficial response to anticholinesterase medication (Belaya et al., 2012). Analysis of patient muscle biopsies and molecular and biochemical studies suggest that the major pathogenic mechanism is reduced numbers of acetylcholine receptors at the motor endplate. To date, at least 16 different genes have been identified in which mutations can cause a congenital myasthenic syndrome (Chaouch et al., 2012; Engel, 2012), but additional cases occur in which the genetic cause is unknown. In particular, a number of patients who show a characteristic predominant weakness of proximal muscle groups lack a genetic diagnosis. Here, we use linkage analysis, whole-exome and whole-genome sequencing to identify mutations in two additional genes, ALG14 and ALG2, encoding early components of the N-linked glycosylation pathway (Fig. 1) that underlie congenital myasthenic syndromes. A single report of heteroallelic mutations in one individual in ALG2 has previously been reported (Thiel et al., 2003), which caused a severe multisystem disorder, termed CDG type Ii (MIM 607906). Our findings expand the phenotypic spectrum of the CDGs, demonstrating that multiple genes encoding components in this biologically crucial and complex pathway have the potential to harbour mutations that cause syndromes with symptoms largely restricted to defective neuromuscular transmission.
Figure 1.
Diagrammatic representation of the N-linked glycosylation pathway. ER = endoplasmic reticulum.
Materials and methods
Patient data
Patient consent for use of data was obtained with ethical approval OXREC B: 04.OXB.017 and Oxfordshire REC C 09/H0606/74. Genomic DNA was extracted from peripheral blood using a Nucleon kit (Gen-Probe Life Sciences Ltd).
Whole-exome sequencing and analysis
For Family 1, whole-exome capture was performed from 3 µg of genomic DNA using Agilent SureSelect Human All Exon Kit v2. The captured libraries were sequenced on Illumina HiSeq platforms using 51 bp paired end reads. Sequence data were mapped to human genome build hg19 using Novoalign software (Novocraft Technologies). To avoid artefacts, we filtered out the duplicate reads generated as a result of PCR amplification. Only reads that mapped uniquely to the genome were used for further analysis. Visualization of sequence data was performed using GBrowse (Stein et al., 2002) and the UCSC genome browser (Kent et al., 2002). Variants were called using the Samtools program (Li et al., 2009). To exclude common variants, we filtered the data against the dbSNP132 database (Sherry et al., 2001) (unless they were annotated as medically-associated single nucleotide polymorphisms). This filtering narrowed the list of variants to 1511 or 1548 variants per exome (Supplementary Table 1). We then used ANNOVAR software (Wang et al., 2010) to functionally annotate the variants and thereby identify non-synonymous substitutions, splicing mutations or mutations in 3′UTR or 5′UTR.
For Family 2, genomic DNA was fragmented by adaptive focused acoustics (Covaris). Libraries were constructed using the TruSeqExome enrichment capture technology (Illumina) as per the manufacturer’s protocol in a technical service provided by Eurofins MWG Operon (www.eurofinsdna.com). Sequencing was performed on the Illumina HiSeq2000 platform as 2 × 100 bp paired end reads. Raw sequencing reads were aligned to the consensus genome (hg19), sorted and variants called using SAMtools and optimized for indel calling using Dindel. The resulting list of variants were visualized and assessed using the UCSC Genome Browser. All sequencing was performed using bi-directional fluorescent sequencing on an ABI 3730 XL 96 capillary sequencer, with BigDye Version 3.1 chemistry.
Whole-genome sequencing and analysis
Whole-exome sequencing was performed for the index case from Family 3. Detailed methods of the sample preparation, sequencing and analysis are provided in the online Supplementary material. Briefly, 2 µg of DNA was fragmented, libraries constructed and adaptors ligated. Size-selected ligated libraries were amplified by PCR, and sequencing was performed on aHiSeq2000 as 100 paired end. Whole-genome sequencing reads were mapped to the human reference genome (GRCh37d5/hg19) using STAMPY (Lunter and Goodson, 2011) and duplicate reads removed using Picard. Identification of variant sites and alleles was performed with in-house software Platypus (Rimer et al., 2012), which can detect single nucleotide polymorphisms and short (<50 bp) indels. The variants were then processed with a functional annotation pipeline based on the ANNOVAR software package (Wang et al., 2010).
Analysis of ALG14 and ALG2 expression by western blot
IMAGE clones containing complementary DNA for ALG14 (clone number 3689162) and ALG2 (clone number 4698763) were purchased from Source Bioscience Lifesciences and were subcloned into mammalian expression vector pcDNA3.1-hygro (Invitrogen). Mutations were introduced into the respective sequences by site-directed mutagenesis using the Quikchange® kit from Stratagene and the full sequence confirmed by Sanger sequencing. HEK293 cells were transfected with the wild-type or mutant complementary DNAs, and 48 h later, cell lysates were harvested. Cells or biopsy tissue were resuspended in cold lysis buffer (10 mM Tris (pH 7.5), 100 mM NaCl, 1 mM EDTA, 1% Triton™ X-100) containing mammalian protease inhibitor cocktail (Sigma). Lysates were extracted by rotating for 1 h at 4°C followed by centrifugation. Protein concentration was measured using a BCA kit from Pierce, and 20 µg was subject to SDS PAGE and transferred onto polyvinylidene difluoride membrane. The membrane was incubated with the appropriate primary antibody (ALG2 antibody from Aviva Systems Biology and ALG14 antibody from Abgent), washed and incubated with a secondary antibody conjugated to horseradish peroxidase (Dako). ECL (GE Healthcare) was used for visualization.
Silencing of ALG14 expression by small interfering RNA and analysis of acetylcholine receptor expression
Four small interfering RNAs targeted to ALG14 were obtained from Sigma-Aldrich (1, SASI_Hs01_00063857; 4, SASI_Hs02_00364796; 7, SASI_Hs02_00364797; and 10, SASI_Hs02_00364799). Small interfering RNAs were co-transfected with complementary DNA expressing ALG14, using Lipofectamine® RNAiMAX (Invitrogen, 13778) and silencing of expression visualized by western blot. As a control, cells were transfected with scrambled small interfering RNA that did not share significant homology with any sequences in the human genome. Subsequently, small interfering RNAs were transfected with acetylcholine receptor α-, β-, δ- and ε-subunit complementary DNAs into human embryonic kidney (HEK) 293 cells. Surface acetylcholine receptor expression was determined 2 days post-transfection by overlaying the cells in PBS containing 10 nM 125I-α-bungarotoxin and 1 mg/ml of bovine serum albumin for 30 min. Cells were washed four times with PBS and removed from the plate in 60 mM Tris–HCl (pH 7.4), 100 mM NaCl, 1 mM EDTA, 0.5 mM phenylmethylsulphonyl fluoride and 1.25% Triton™ X-100. 125I-α-bungarotoxin binding was determined by gamma counter.
Immunofluorescence labelling of mouse neuromuscular junctions
Leg muscle was frozen in isopentane cooled in liquid N2 for 15 s, sectioned in a transverse plane using a Leica CM1900 cryostat at −18°C and at a thickness of 10–15 µm and mounted on slides coated with 3-aminopropyltriethoxysilane. Sections were left to dry overnight at room temperature. Sections were rinsed in PBS and then permeabilized in 0.1% Triton™ X-100 for 10 min at room temperature. Incubation in 3% bovine serum albumin in PBS for 15 min was used to block non-specific binding before incubation with the primary antibody. A fluorescent anti-rabbit secondary antibody was used for visualization.
Results
Clinical features
Family 1
Cases 1 and 2 (Table 1) are sisters, with no consanguinity in the pedigree. Case 1 had normal motor milestones and early childhood, although she was always poor at sports. Symptoms first became apparent around the age of 7 years, with fatigable proximal weakness and difficulty climbing stairs, and during subsequent years, the weakness gradually progressed, and she would suffer repeated falls. At the age of 18 years, a diagnosis of autoimmune myasthenia gravis was made following a positive tensilon test and clear decrement of compound muscle action potentials with repetitive nerve stimulation on electromyography. However, antibodies to the acetylcholine receptor were not detected, and she did not respond to extensive immunosuppressive therapy, but has shown long-term benefit from anticholinesterase medication. On examination, she had no ptosis, and extraocular movements were full. She had a mild latent divergent squint. Neck and facial muscles were strong, and she had no bulbar involvement. She had generalized limb and truncal weakness and contractures in multiple joints, worse in the upper limbs. Muscle biopsy at age 42 years was largely unremarkable and did not demonstrate the presence of tubular aggregates. She now requires a stick for walking, but her symptoms have remained generally stable during adult life, though she experiences marked exacerbation of weakness during viral illness. Her sister, Case 2, achieved normal motor milestones, though on retrospect, she recalls she was considerably weaker than her peers in childhood. At age 40 years, she developed proximal limb weakness, had difficulty lifting her arms above her head and struggled to climb stairs. A tensilon test was strongly positive, and antibodies to acetylcholine receptor and MuSK were negative. She was treated with immunosuppressive agents for a presumed diagnosis of sero-negative myasthenia gravis, which did not result in improvement. Nevertheless, she showed long-term benefit from treatment with cholinesterase inhibitors. Examination when off treatment revealed mild weakness of neck flexion and moderately severe weakness in proximal and distal muscles of the upper and lower limbs. Neurophysiology showed decrement of compound muscle action potentials (∼19%) on repetitive nerve stimulation, and two-thirds of potential pairs showed increased jitter values on single-fibre electromyography. No muscle biopsy has been performed.
Table 1.
Clinical features of individuals with mutations in ALG14
| Patient | Case 1 | Case 2 |
|---|---|---|
| Current age (years) | 62 | 51 |
| Age when assessed (years) | 61 | 41 |
| Progression | Stable | Stable with deterioration, age 41 years |
| Age at onset (years) | 7 | Childhood |
| Symptoms at onset | Difficulty climbing steps and hills | Difficulty running |
| Bulbar/respiratory symptoms | −/+ but probably not myasthenic | −/− |
| Ptosis | − | Mild right sided |
| Ophthalmoplegia | − | − |
| Strabismus | Minimal latent divergent | Minimal latent divergent |
| Muscle strength, MRC grade | ||
| Face | 5 | 5 |
| Neck flexion/extension | 5, 5 | 5− |
| Shoulder abduction | 4 | 4 |
| Elbow flexion/extension | 4+, 5 | 5/4+ |
| Wrist/finger extension | 4+, 4 | ND/ND |
| Finger/thumb abduction | 4+, 4− | 5/4+ |
| Hip flexion/extension | 5−, 5 | 4+ |
| Knee flexion/extension | 5, ND | 5− |
| Ankle dorsiflexion | 4+ | 4+ |
| Wasting | − | − |
| Contractures | + | − |
| Spine | − | − |
| Decrement on RNS | + | + |
| Abnormal jitter | + | ND |
| Blocking | + | ND |
| Muscle biopsy | No tubular aggregates | ND |
| Treatments used | P, E, D | P |
| Creatine kinase | ND | 38 IU/L (24–170) |
D = 3,4-diaminopyridine; E = ephedrine (bold indicates on-going treatment); ND = not done; P = pyridostigmine; RNS = repetitive nerve stimulation.
Family 2
Cases 3, 4, 5 and 6 are siblings now aged 23, 17, 12 and 3 years, respectively, from a consanguineous marriage of second cousins from Saudi Arabia (clinical features are summarized in Table 2). They share a similar phenotype with a clinical diagnosis of Ullrich’s congenital muscular dystrophy. They presented with delayed motor milestones and hypotonia. Cases 3, 4 and 5 showed slowly progressive deterioration and are now wheelchair dependent. Mild learning difficulty is apparent. Neurological examination revealed generalized muscle weakness, more pronounced in proximal over distal muscles, absent reflexes, proximal joint contractures and distal joint laxity noted in wrist, fingers and ankles. There was mild facial weakness, but no ptosis or ocular involvement. Bilateral pes planus deformity of both feet was present in each case. A high arched palate was noted in all. Laboratory investigations showed normal haematological indices, electrolytes, urea and creatine, creatine kinase and liver function tests. Cases 3 and 4 had normal brain MRI results and showed mild respiratory insufficiency due to muscle weakness but have not required assisted ventilation so far. For Cases 3 and 4, on electromyography, repetitive nerve stimulation showed decrement of compound muscle action potentials and significantly increased jitter on single-fibre electromyography. Case 4 underwent a biopsy of the quadriceps muscle at the age of 6 years, which revealed myopathic features on histology and histochemistry. Case 6 presented with hypotonia, diminished reflexes and delayed motor milestones; at the age of 15 months, she could not stand or crawl.
Table 2.
Clinical features of individuals with mutations in ALG2
| Patient | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 |
|---|---|---|---|---|---|
| Current age (years) | 23 | 17 | 12 | 3 | 59 |
| Age when assessed (years) | 20 | 14 | 9 | 15 months | 59 |
| Age at onset (years) | 22 months | Infancy | Infancy | 15 months | 4 |
| Progression | Slow | Slow | Slow | Slow | Slow |
| Symptoms at onset | Delayed motor milestones, hypotonia | Delayed motor milestones | Delayed motor milestones, hypotonia | Delayed motor milestones, hypotonia | Waddling gait, falls |
| Never achieved ambulation | + | + | + | + | − |
| Bulbar/respiratory symptoms | −/+ | −/+ | −/− | −/− | −/− |
| Ptosis | − | − | − | − | − |
| Ophthalmoplegia | − | − | − | − | − |
| Facial weakness (mild) | + | + | + | + | − |
| Pattern of muscle weakness | Proximal > distal | Proximal > distal | Proximal > distal | Proximal > distal | Proximal > distal |
| Contractures | Knees 80o | Knees 50o | Knees 30o | − | − |
| Distal joint laxity | + | + | + | − | − |
| Spine abnormality | − | − | − | − | Lumbar hyperlordosis |
| Decrement on RNS | + | + | ND | ND | + |
| Abnormal jitter | + | + | ND | ND | + |
| Blocking | ND | ND | ND | ND | + |
| Muscle biopsy | ND | Variation in fibre size | ND | ND | Variation in fibre size. Increase in internal nucleation. Myopathic. Tubular aggregates |
| Type 1 predominance | |||||
| Myopathic | |||||
| Treatments used | − | − | − | − | P |
| CK (normal range 38−174 IU/L) | 86 IU/L | 82 IU/L | 128 IU/L | 269 IU/L | Variable |
| age 53: 267 IU/L (24−195) | |||||
| age 59: 123 IU/L (55−170) |
CK = creatine kinase; ND = not done; P = pyridostigmine (bold indicates on-going treatment); RNS = repetitive nerve stimulation.
Family 3
Case 7 is white European of Italian descent whose parents are first cousins (clinical features summarized in Table 2). Symptoms became clearly apparent around the age of 4 years with falls and difficulty rising from the floor or climbing stairs. A diagnosis of muscular dystrophy was made at that time. His symptoms became worse at the age of 10 years when he was unable to walk unaided. He was re-diagnosed with myasthenia in his teens and started taking pyridostigmine, which greatly helped him, though he was never able to run and still required support. Now at age 60 years, he is able to walk 15–20 m independently. Symptoms are exacerbated by stress and infections. On examination, he has a waddling gait with marked lumbar lordosis. There is scapular winging. He can rise from the floor using Gower’s manoeuvre. Facial and neck muscles are strong, and he has full extraocular muscle movement with no ptosis. Limb weakness is most severe proximally in all limbs and is mild in distal upper limb. On electromyography, repetitive nerve stimulation with the patient off treatment showed ∼67% decrement of compound muscle action potentials. In addition, marked abnormality was seen on volitional single-fibre electromyography with two-thirds of potential pairs showing increased jitter or blocking. Tubular aggregates in type II fibres were detected on electron microscopy of a muscle biopsy taken at the age of 50 years. Non-specific myopathic features of moderate variability of fibre size and shape and several internal nuclei were also noted.
Identification of mutations in ALG14
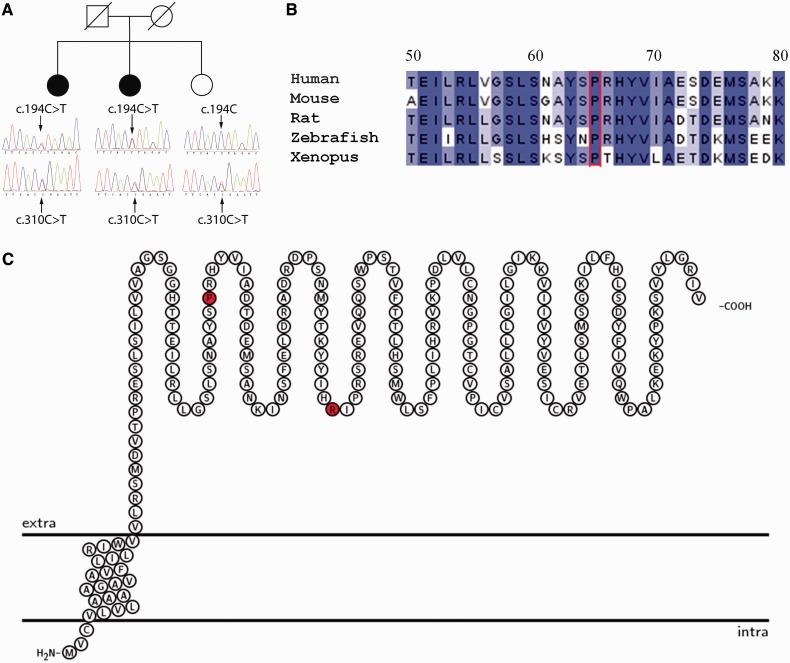
DNA from Cases 1 and 2, with a clinical diagnosis of probable congenital myasthenic syndrome, but who had tested negative for mutations in known congenital myasthenic syndrome associated genes, was subjected to next-generation sequence analysis. Filtering of the sequence data allowed us to limit the number of variants of interest to 333 or 356 per exome, respectively. Nearly all of the congenital myasthenic syndromes are commonly inherited in an autosomal recessive manner, and therefore, we focused on the genes that had either homozygous variants or contained two or more heterozygous variants in the same gene. Among these, 54 genes had potential mutations in both siblings. We excluded all variants that were present in our in-house database of 14 exomes from cases with unrelated disorders. This allowed us to eliminate all but one gene—ALG14 [RefSeq NM_144988.3], which encodes N-linked glycosylation 14 homologue (Saccharomyces cerevisiae). Both siblings had two mutations in the gene: c.194C>T (p.Pro65Leu) and c.310C>T (p.Arg104*). We confirmed the presence of both mutations in the genome of the analysed individuals using Sanger sequencing (Fig. 2A). To analyse whether the mutations in ALG14 segregate with disease, we performed Sanger sequencing of ALG14 in the unaffected sibling (the only available family member). The sibling carried only one of the mutations—c.310C>T, confirming that the two mutations were likely to be inherited on separate chromosomes from two different parents. Thus, the segregation of the mutations in the family is consistent with the mutations being disease causing (Fig. 2A). None of the ALG14 variants described earlier in the text were present in dbSBP 135 (Sherry et al., 2001) or 1000 Genomes (The 1000 Genomes Project Consortium, 2010) databases. Notably, one of the mutations (c.310C>T) was present in the exome variant server (Exome Variant Server, 2012). However, the frequency of this mutation is very low—0.0077% (or 1 in 13 005 alleles) in the general population or 0.0116% (1 in 8599 alleles) in the European American population. The family we describe is of European origin, and thus the frequencies described on the exome variant server are valid for our study. Such low frequency of the variation incidence is consistent with a pathogenic mutation.
Figure 2.
(A) Segregation of mutations ALG14 c.194C > T, p.Pro65Leu and c.310C > T, p.Arg104* within Family 1. (B) Conservation of ALG14 protein sequence within the vicinity of the p.Pro65Leu mutation. Alignment of protein sequence flanking Pro65 (red) from several species was carried using ClustalW. Colours indicate percentage identity and were introduced using JalView. Residue numbering is according to the human protein. (C) Representation of the ALG14 protein transmembrane topology and location of mutations. Protein structure was visualized using TEXtopo (Beitz, 2000).
ALG14 mutation Pro65Leu causes reduced expression in HEK 293 cells
In yeast, ALG14 interacts with ALG7 (in humans the orthologue is DPAGT1), and together ALG7/13/14 form a multiglycosyltransferase that catalyses the first two steps in the biosynthesis of LLO precursor for N-glycan assembly (Gao et al., 2005, 2008; Lu et al., 2012). Human variant c.310C>T introduces a premature stop codon after amino acid residue Arg104, truncating the protein by 112 amino acid residues. The second variant, c.194C>T, encodes a non-synonymous substitution Pro65Leu. Pro65 is a conserved residue (Fig. 2B), and substitution to a leucine is predicted by Polyphen2 (HumVar) to be probably damaging with a score of 0.966. To assess whether this substitution affects protein expression, the c.194C>T variant was introduced into complementary DNA encoding human ALG14 (Fig. 2C), and expression of wild-type or mutant protein visualized by western blot probed with an ALG14-specific antibody following transfection into HEK293 cells. The Pro65Leu mutation severely reduced the expression of ALG14, suggesting that it is likely to be pathogenic (Fig. 3).
Figure 3.
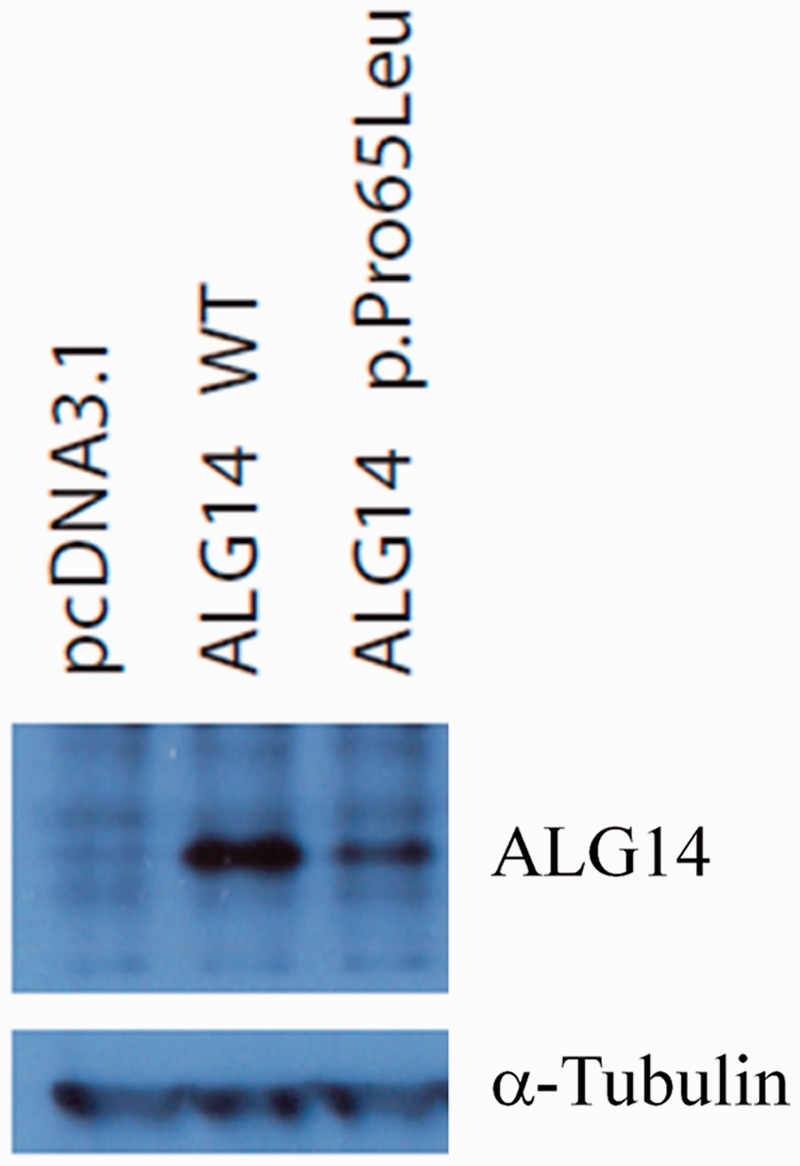
Expression of ALG14 wild-type (WT) and p.Pro65Leu in HEK 293 cells. Complementary DNA expression constructs were transfected into HEK293 cells, and 48 h later, cells were lysed and subjected to western blot analysis using an antibody to ALG14. α-tubulin was used as a loading control.
Small interfering RNA targeted at ALG14 messenger RNA reduced cell-surface expression of acetylcholine receptor
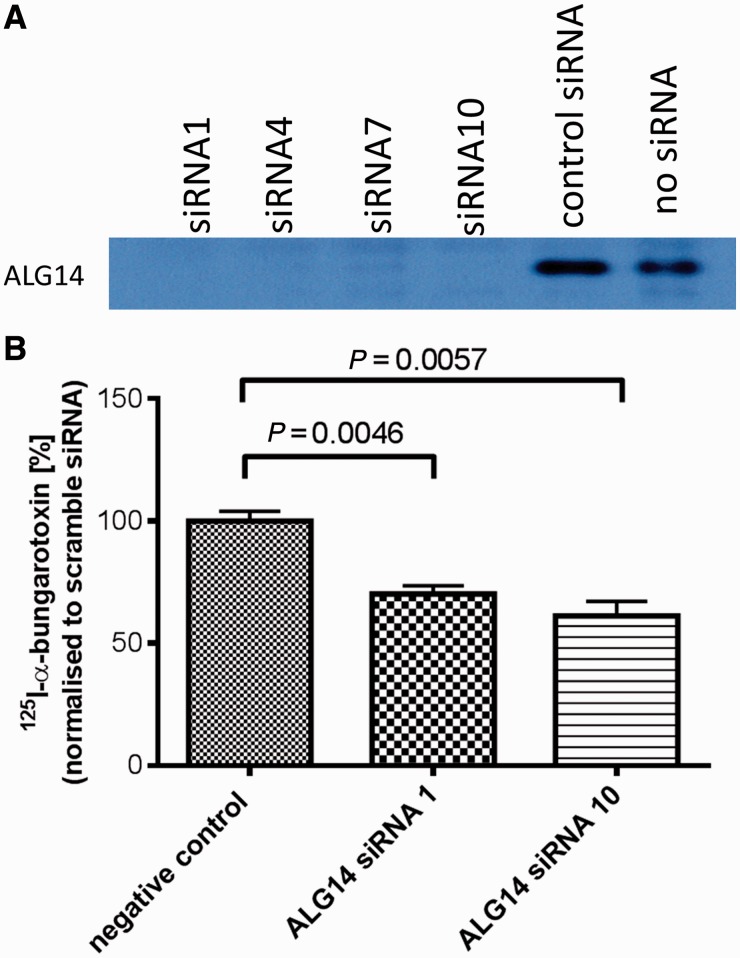
Adult acetylcholine receptor is a pentamer consisting of 2α, 1β, 1δ and 1ε subunits that are assembled in the endoplasmic reticulum before being exported to the plasma membrane. All four subunits of acetylcholine receptor are N-glycosylated (Shoji et al., 1992). N-linked glycosylation is required for the insertion of acetylcholine receptors into the plasma membrane through the regulation of subunit stability, folding, assembly and intracellular transport (Gehle and Sumikawa, 1991; Gehle et al., 1997). To establish whether ALG14 is important for cell-surface expression of acetylcholine receptors, we used an α-bungaratoxin binding assay to measure the levels of acetylcholine receptor that are expressed on the cell surface. First, we confirmed available small interfering RNAs (Sigma-Aldrich) could efficiently silence ALG14 messenger RNA that was overexpressed in transfected HEK293 cells (Fig. 4A). Subsequently two separate small interfering RNAs were used to silence endogenous ALG14 in HEK293 cells that were co-transfected with complementary DNA encoding the human acetylcholine receptor α, β, δ and ε subunits. Cell-surface expression of acetylcholine receptors, detected by 125I-α-bungaratoxin, was reduced to >50% of control (Fig. 4B).
Figure 4.
Downregulation of ALG14 reduces surface expression of acetylcholine receptor in transfected HEK293 cells. (A) Silencing of ALG14 expressed in HEK 293 cells by small interfering RNAs (siRNA) targeted at ALG14 messenger RNA. Small interfering RNAs and the ALG14 expression construct were transfected into HEK 293 cells, and 48 h later, ALG14 expression analysed by western blot. (B) Two small interfering RNAs (1 and 10) targeted at ALG14 messenger RNA or ‘scrambled’ small interfering RNA were transfected into HEK 293 cells. Twenty-four hours later, the same cells were transfected with complementary DNAs encoding the human acetylcholine receptor α, β, δ and ε subunits, and 48 h later, cell-surface 125I-α-bungarotoxin binding was determined (n = 3).
Identification of mutations in ALG2
Family 2
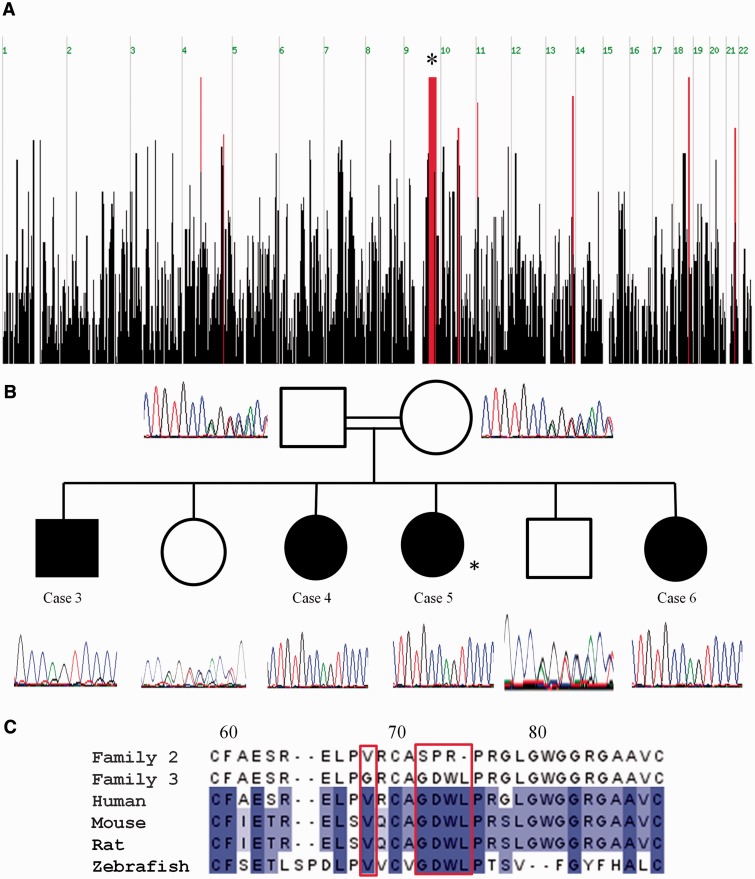
Consanguinity in the family of Cases 3, 4, 5 and 6 allowed us to apply a genome-wide linkage approach using AffymetrixGeneChip Human Mapping 250 K Nsp microarray, with a technical service provided by DNAVision SA (www.dnavision.be). Regions of homozygosity were analysed using AutoSNPa. A 27 Mb homozygous candidate interval was identified on chromosome 9 (9q31.1; genomic location 100114051-105435311) that contained 283 genes (Fig. 5A). Exome sequencing was used to identify variants in the candidate interval (for detailed methods of the sample preparation, sequencing and analysis, see Supplementary material). A filtering algorithm was applied to exclude all but the most likely pathogenic variants (Supplementary material), which were verified by PCR amplification and Sanger sequencing. A frequency >1% in the Exome Variant Server and the 1000 Genomes Project ruled out all of these variants as the causal mutation. Forty-two exons in the candidate interval had <10-fold sequencing read depth. PCR amplification and Sanger sequencing of these 42 exons revealed an indel in exon 1 of the ALG2 gene (NM.033087.3); c.214_226delGGGGACTGGCTGCinsAGTCCCCGGC p.72_75delGDWLinsSPR, which can be expressed more simply as c.214_226delinsAGTCCCCGGC, p.72_75delinsSPR (Fig. 5B).
Figure 5.
(A) Graphical representation of homozygous regions shared by Cases 3, 4 and 5 (autozygosity analysis performed before the birth of Case 6) generated using the AutoSNPa program. Shared blocks of homozygous single nucleotide polymorphisms are shown as red bars. The most significant homozygous region is on 9q31.1 (genomic location 100114051–105435311) is 27 Mb long and contains 283 genes (denoted by asterisk). (B) Sequence analysis in the c.214_226delGGGGACTGGCTGCinsAGTCCCCGGC p.72_75delGDWLinsSPR region for members of Family 2. Cases 3–6 are homozygous for this indel, whereas their parents (second cousins) and unaffected siblings are heterozygous. Asterisk denotes the individual in which exome sequencing was performed (Case 5). (C) Location of mutations for Families 2 and 3 in ALG2. ALG2 amino acid residues p.Val68 and p.72_75GDWL are conserved across species. Alignment of protein sequence flanking p.72_75GDWL from several species was carried out using ClustalW. Colours indicate per cent identity and were introduced using JalView. Residue numbering is according to the human protein.
Family 3
For Case 7, the DNA sample was subject to whole genome sequencing (Supplementary material). Heterozygous variants were excluded because the patient was from a consanguineous marriage. Variants present in dbSNP132 database were also excluded, giving 226 interesting homozygous variants (Supplementary Table 2). None of these variants were within genes already known to cause congenital myasthenic syndromes (CHRNA1, CHRNB, CHRND, CHNRE, CHRNG, DOK7, RAPSN, MUSK, CHAT, COLQ, AGRN, LAMB2, GFPT1 and DPAGT1).
Aligned sequence data were visualized with GBrowse (Stein et al., 2002). Further filtering involved comparison with the union file from WGS500 Data Freeze 2, which includes 107 individual samples and 21 594 778 unique entries; scrutinizing for misalignment (which was usually owing to segment duplication) and for reads that were not unambiguously homozygous; and exclusion of variants on the X chromosome (as there was no evidence that the disease was X-linked). Three non-synonymous homozygous variants remained in ALG2, DEFA4 and PLEKHA5. We had previously identified patients with a similar phenotype with pathogenic variants in DPAGT1 (Belaya et al., 2012), a gene that encodes a protein involved in N-linked glycosylation. ALG2 encodes a closely associated protein in this pathway, asparagine-linked glycosylation 2 (ALG2).
Analysis of ALG2 mutations
Variant c.214_226delinsAGTCCCCGGC, p.72_75delinsSPR segregates with disease in the family (Fig. 5B), with all affected individuals homozygous for this mutation, whereas the parents and unaffected siblings are heterozygous. Interestingly, the inserted snippet is the reverse complement of the sequence spanning the mutant locus, but displaced by two nucleotides upstream. It is absent in the Exome Sequencing Project, 1000 Genomes Project, dbSNP132 and in 96 in-house control exomes. To provide further evidence that this indel in ALG2 was the causal mutation, exome data were interrogated in their entirety, rather than with restriction to the candidate interval on chromosome 9 (Supplementary material). Exonic variants (13 879) were filtered according to zygosity and absence from dbSNP. Of the 12 variants that remained, PCR and Sanger sequencing showed all were excluded as the causal variant either owing to non-segregation (eight variants) or false positives (four variants). Variant c.214_226delinsAGTCCCCGGC, p.72_75delinsSPR removes a highly conserved GDWL motif (Fig. 5C) of the glycosyltransferase 4-like domain, and inserts three different amino acids, SPR.
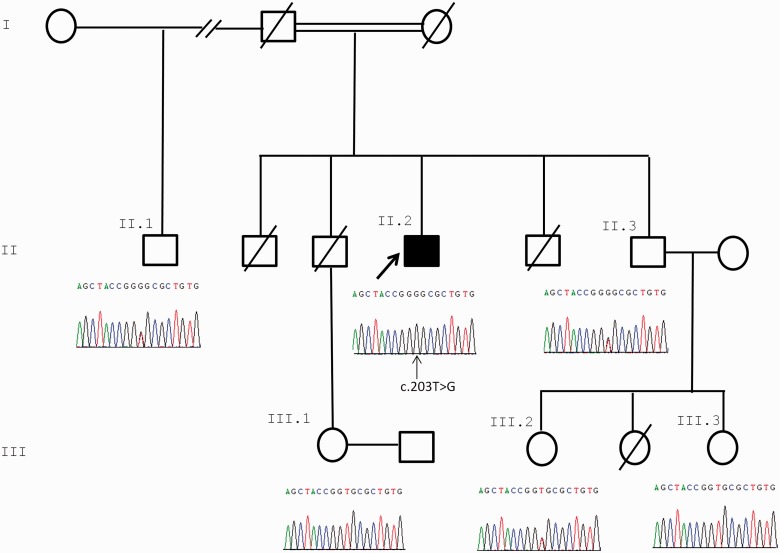
For Case 7, the variant in ALG2, c.203 T>G (p.Val68Gly), was confirmed in the index case by Sanger sequencing of exon 1. Sequencing data from five other non-affected family members were consistent with this variant segregating with disease (Fig. 6A). Restriction digest analysis of amplified exon 1 also confirmed the sequencing data (Supplementary Fig. 1). This variant does not appear in dbSNP build 137, the 1000 Genomes database or on the Exome Variant Server, and therefore is present at a very low frequency in the population. This is consistent with it being a pathogenic mutation causing a rare congenital disorder such as CMS. The predicted amino acid substitution Val68Gly is a non-conservative change, and Val68 is highly conserved across species (Fig. 5C). In addition, Polyphen2 (HumVar) predicts that this substitution is likely to be damaging with a score of 0.996.
Figure 6.
Segregation of ALG2 c.203 T > G with disease within the pedigree of Family 3. Only the index case is homozygous for the variant. Sequence analysis in the c.203C > T region of five unaffected family members is shown. The index case (Case 7) and homozygous mutated nucleotide are indicated with arrows.
Expression of ALG2 harbouring the p.Val68Gly variant
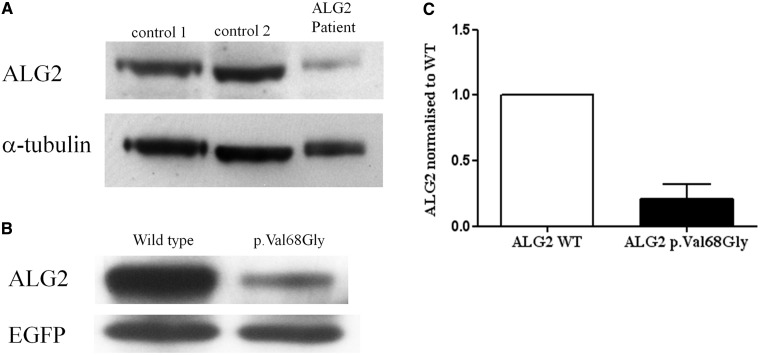
To investigate pathogenicity for p.Val68Gly, we first analysed the protein expression level of ALG2 in a biopsy from the rectus femoris muscle of the patient. Western blots of tissue lysates with an anti-ALG2 antibody (Aviva Systems Biology) revealed that the protein is barely detectable compared with two control muscle biopsies (Fig. 7A). We confirmed that the variant reduces expression levels by transfecting complementary DNA expressing wild-type or mutant ALG2 in HEK 293 cells. Western blot analysis of cell lysates showed again that p.Val68Gly variant severely reduced the expression level of ALG2, in this case to ∼20% of controls (Fig. 7B and C).
Figure 7.
Reduced expression of the ALG2p.Val68Gly variant. (A) Proteins in lysates from two controls muscles biopsies with no N-glycosylation defect or from a muscle biopsy from Case 7 were subject to western blot analysis probed with an antibody against ALG2 (Aviva Systems Biology). (B) HEK 293 cells transfected with wild-type or mutant ALG2 were subject to similar western blot analysis. Transfection efficiency was normalized by co-transfection with EGFP. (C) Quantification of the data from transfected HEK 293 cells (P = 0.0197, t-test, n = 3).
ALG2 and ALG14 are enriched at motor endplates
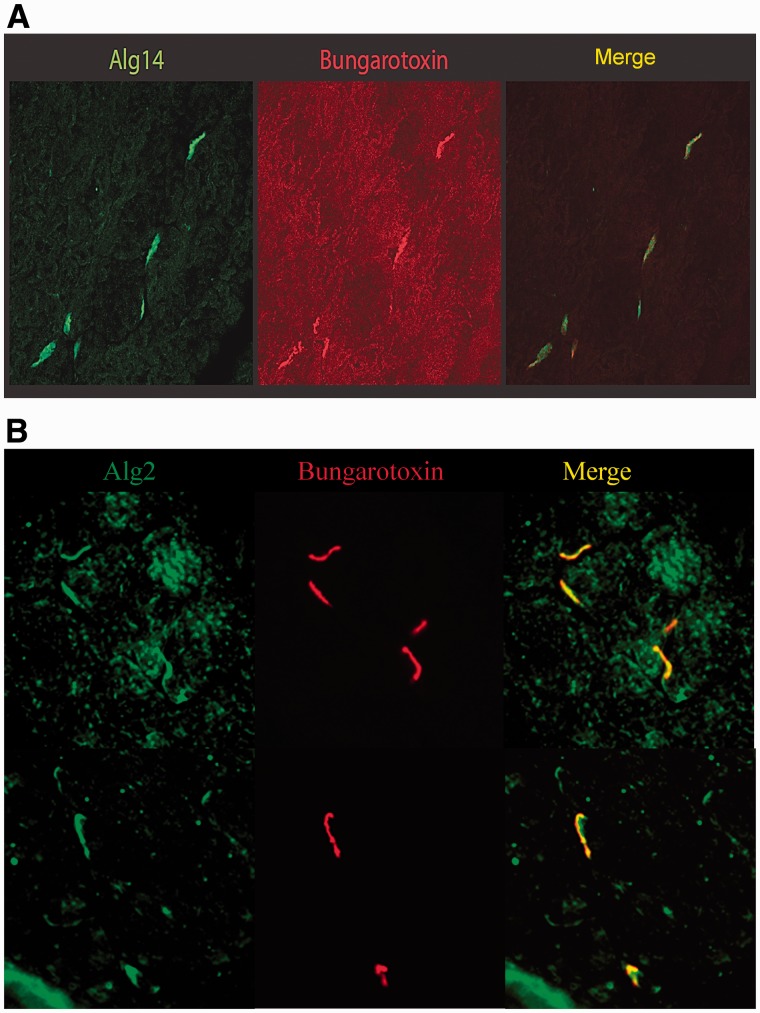
The distribution of ALG14 and ALG2 in mouse muscle sections was probed by staining with the antibodies to ALG14 or ALG2. Strong staining by an anti-ALG14 antibody localizes with α-bungarotoxin, indicating that it is concentrated at endplate regions of muscle (Fig. 8A). Similarly, antibodies to ALG2 show the protein to be enriched in the region of the α-bungarotoxin binding (Fig. 8B).
Figure 8.
Alg14 and Alg2 are enriched at endplate regions in mouse muscle sections. (A) Mouse muscle sections were stained with an anti-Alg14 antibody (Abgent), or (B) with an anti-Alg2 antibody (Santa Cruz Biotechnology). Secondary antibodies were Alexa Fluor®488 conjugated anti-rabbit from Invitrogen (green). AChRs at the neuromuscular junctions were visualized by staining with an Alexa Fluor®594 conjugated α-Bungarotoxin (red).
Discussion
We report two new genes, ALG14 and ALG2, in which mutations underlie congenital myasthenic syndromes. These genes encode components involved in the early steps of N-linked glycosylation (Fig. 1). Our findings, in combination with our recent study of congenital myasthenic syndrome due to DPAGT1 mutations, underscore the importance of N-linked protein glycosylation for proper functioning of the neuromuscular junction, indicate a mechanism that may underlie hypotonia in many of the CDGs and suggest appropriate treatment for the muscle weakness.
Mutations that underlie a genetic disorder have not previously been seen in ALG14. In yeast, ALG14 is a membrane protein and is essential for the recruitment of ALG13 to the endoplasmic reticulum where the two proteins form a heterodimer (Gao et al., 2005, 2008). ALG13 catalyses the second step of LLO synthesis, and this activity requires ALG14 (Gao et al., 2008). ALG14 also interacts with ALG7 (the human orthologue is DPAGT1) (Lu et al., 2012), and together ALG7/13/14 form a multiglycosyltransferase that catalyses the first two steps in the biosynthesis of LLO precursor for N-glycan assembly (Gao et al., 2005, 2008; Lu et al., 2012). ALG13 and ALG14 interact via their C-termini, and truncation of ALG14 by only three amino acids at the C terminus prevents its interaction with ALG13 in yeast (Gao et al., 2008). By analogy, for human orthologues, as well as the possibility of nonsense-mediated decay, it is highly likely the c.310C>T variant is pathogenic, as it leads to a severe truncation of ALG14, which would be unable to recruit ALG13 to the endoplasmic reticulum. The second ALG14 mutation, p. Pro65Leu, results in markedly reduced expression in HEK293 cells. A muscle biopsy was not available to analyse endplate acetylcholine receptor numbers, but we were able to demonstrate in cell culture models that knock-down of ALG14 expression resulted in reduced cell-surface acetylcholine receptor expression, thus providing the mechanistic link for the myasthenic syndrome. Mutations in DPAGT1 can cause reduced endplate acetylcholine receptor (Belaya et al., 2012), and as ALG14 is predicted to be part of a single multiglycosyltransferase complex that includes DPAGT1 (Lu et al., 2012), our cell culture experiments that indicate that reduced ALG14 expression leads to reduced cell-surface acetylcholine receptor expression are consistent both with previous findings for DPAGT1 and with the positive response to cholinesterase inhibitors seen in the patients.
ALG2 is an alpha-1,3-mannosyltransferase that catalyses the second and third mannosylation steps in the N-linked glycosylation pathway (Huffaker and Robbins, 1983; Jackson et al., 1993) (Fig. 1). Human ALG2 is predicted to be a type I transmembrane protein (Haeuptle and Hennet, 2009) with the active site located in the cytosol. Only one individual with a CDG type Ii (MIM 607906) owing to heteroallelic mutations in ALG2 has previously been reported (Thiel et al., 2003). The maternal mutation induced a frameshift (c.1040delG) and produced an enzyme with reduced activity. The paternal mutation (c.393 G>T) gave no detectable transcript. In skin fibroblasts from this patient, accumulation of Man1GlcNAc2-PP-dolichol and Man2GlcNAc2-PP-dolichol was observed, confirming that ALG2 function was impaired. This patient had symptoms often seen in patients with CDG, such as mental and developmental delay, seizures, poor vision, prolonged clotting and delayed myelination. Notably, the patient also had severely delayed motor milestones. The clear segregation of ALG2 variant c.214_226delinsAGTCCCCGGC, p.72_75delinsSPR, with disease in Family 2, provides evidence for pathogenicity. Moreover, the absence of any potential disease-causing variants in the exome and the discovery of this very probably significant deleterious indel in a highly conserved protein encoded by a gene in a candidate interval determined by autozygosity analysis points to ALG2 being the disease gene in this pedigree. Similarly, whole-genome sequencing, from which other variants can be excluded, including those present in introns, enabled the identification of ALG2 mutation p.Val68Gly. Analysis of a muscle biopsy from the patient (in which endplates were not present) demonstrated that the level of ALG2, harbouring the p.Val68Gly mutation, is severely reduced compared with two control muscle biopsies (in which endplates were also absent). This result was also seen in cell culture experiments. Amino acid residue p.Val68 is predicted to be located in the endoplasmic reticulum lumen and would be unlikely to directly affect the catalytic activity of the enzyme. The phenotype of the individual with the p.Val68Gly mutation closely mirrors patients with congenital myasthenic syndrome owing to DPAGT1 mutations, and our results are consistent with impairment, but not complete disruption of N-linked glycosylation.
In addition to the mutations we have described for ALG2, ALG14 and DPAGT1, it is possible that the pathogenic mechanism that underlies congenital myasthenic syndromes due to GFPT1 mutations (Senderek et al., 2011) is owing to reduced availability of one key product of the hexosamine synthetic pathway, uridinediphopho-N-acetylglucosamine (UDP-GlcNAc), as a substrate for DPAGT1. The congenital myasthenic syndromes that result from mutations in these four genes have a similar phenotype of a limb-girdle pattern of muscle weakness with eye, facial and bulbar muscles largely spared, which contrasts with many myasthenic syndromes. Some of our patients presented with permanent and early-onset weakness reminiscent of a congenital myopathy or congenital muscular dystrophy. A decremental response of compound muscle action potentials to repetitive nerve stimulation on electromyography provides evidence for impaired neuromuscular transmission. Tubular aggregates on muscle biopsy is a common feature, although they were not seen in the one ALG14 case that had a muscle biopsy, are not always present in individuals with GFPT1 mutations and were not seen in an early biopsy from one of the ALG2 mutation cases. The importance of a genetic diagnosis is emphasized by the case histories of the sisters with ALG14 mutations who were previously thought to have autoimmune seronegative myasthenia gravis and as a result underwent extensive inappropriate treatments that included steroids, intravenous immunoglobulin, plasmapheresis and thymectomy. Mutations in DPAGT1 (Wu et al. 2003; Wurde et al., 2012) or ALG2 (Thiel et al., 2003) can also underlie severe multisystem disorders. N-glycosylation of proteins is a key cellular function throughout the body, and it has yet to be determined why, in one patient, a severe multisystem disorder such as CDG-Ii (MIM 607906) manifests (Thiel et al., 2003), whereas in the case of congenital myasthenic syndrome reported here, symptoms resulting from the ALG2 mutations were largely restricted to muscle weakness and impaired neuromuscular transmission. ALG14 shows marked localization at endplate regions, and ALG2 is also enriched at the neuromuscular junctions of mouse muscle (Fig. 8A and B) providing additional support for the importance of N-linked glycosylation proteins at the neuromuscular junction. The standard transferrin glycoform analysis (Jeppsson et al., 2007), which is often used to detect defective glycosylation is not abnormal for any of our cases with congenital myasthenic syndrome tested to date, suggesting only modest impairment of N-linked glycosylation. It is possible that generation of endplate acetylcholine receptor is particularly sensitive to deficits in the N-linked glycosylation pathway, whereas more severe forms of CDG result from more damaging mutations (Marklova and Albahri, 2007). Complete knock-down of ALG2 and ALG14 in zebrafish embryos results in more widespread pathology including the brain and the nervous system (unpublished observation). Many other proteins localized to the neuromuscular junction are glycosylated, including proteins involved in receptor clustering and the maintenance of the structure at the neuromuscular synapse (Sanes and Lichtman, 2001), but analysis of individuals with DPAGT1 mutations suggests that reduced numbers of endplate acetylcholine receptor is the key pathogenic mechanism (Belaya et al., 2012). Consistent with this hypothesis, these disorders show a beneficial response to anticholinesterase medication, such as pyridostigmine. In more severe forms of the CDG characterized by multisystem disorders affecting many organs, hypotonia is a frequent feature. Thus, as well as for the subgroup of congenital myasthenic syndromes we describe, it is possible that treatment with anticholinesterase medication will benefit muscle function in a wider spectrum of the N-linked CDG.
Funding
Generation of the Sequencing data was from the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics (see Supplementary data) funded by Wellcome Trust (grant reference 090532/Z/09/Z) and MRC Hub (grant G0900747). K.B. is a fellow of the Wellcome Trust-funded OXION: Ion channels and disease initiative. The authors are grateful for funding from The Medical Research Council, UK (MRC programme grant G0701521 and MRC research grant G1002274), the Muscular Dystrophy Campaign and the Myasthenia Gravis Association.
Supplementary material
Supplementary material is available at Brain online.
Acknowledgements
The authors thank Dr. S. Twigg for help with preparation of samples for exome sequencing. The authors are grateful to Dr. Mohammad M Kabiraj, Armed Forces Hospital, Riyadh, for conducting electrophysiological studies; to Dr. Ahmad Amer Al-Boukai and Dr. Handy H. Hassan, Department of Medical Imaging and Radiology, King Saud University for MRI data; and to Dr. Molham al Rayess for muscle biopsy data in the Saudi Arabian family (Cases 3–6). The authors thank Prof. Volker Straub and Prof Kate Bushby for their interest and advice on the project. They thank the patients and their families for participating in this study and for their consent, which were obtained with ethical approval OXREC B: 04.OXB.017 and Oxfordshire REC C 09/H0606/74.
Glossary
Abbreviations
- CDG
congenital disorder of glycosylation
- HEK
human embryonic kidney
- N-linked
asparagine linked
Web resources
The URLs for data presented herein are as follows:
1000 Genomes Project, http://browser.1000genomes.org
Exome variant server, http://evs.gs.washington.edu/EVS/
dbSNP database, http://www.ncbi.nlm.nih.gov/projects/SNP/
BEDTools, http://code.google.com/p/bedtools/
Platypus, http://www.well.ox.ac.uk/platypus/
Picard, http://www.picard.sourceforge.net/
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org/
ClustalW2, http://www.ebi.ac.uk/Tools/msa/clustalw2/
SAMtools, http://samtools.sourceforge.net/
Dindel, http://www.sanger.ac.uk/resources/software/dindel/
UCSC genome browser, genome.ucsc.edu
AutoSNPa, http://autozygosity.org
References
- Beitz E. T(E)Xtopo: shaded membrane protein topology plots in LAT(E)X2epsilon. Bioinformatics. 2000;16:1050–1. doi: 10.1093/bioinformatics/16.11.1050. [DOI] [PubMed] [Google Scholar]
- Belaya K, Finlayson S, Slater C, Cossins J, Liu W-W, Maxwell S, et al. Mutations in DPAGT1 cause a limb-girdle congenital myasthenic syndrome with tubular aggregates. Am J Hum Genet. 2012;91:193–201. doi: 10.1016/j.ajhg.2012.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretthauer RK. Structure, expression, and regulation of UDP-GlcNAc: dolichol phosphate GlcNAc-1-phosphate transferase (DPAGT1) Curr Drug Targets. 2009;10:477–82. doi: 10.2174/138945009788488369. [DOI] [PubMed] [Google Scholar]
- Chaouch A, Beeson D, Hantai D, Lochmuller H. 186th ENMC international workshop: congenital myasthenic syndromes 24–26 June 2011, Naarden, The Netherlands. Neuromuscul Disord. 2012;22:566–76. doi: 10.1016/j.nmd.2011.12.004. [DOI] [PubMed] [Google Scholar]
- Engel AG. Current status of the congenital myasthenic syndromes. Neuromuscul Disord. 2012;22:99–111. doi: 10.1016/j.nmd.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exome Variant Server. NHLBI Exome Sequencing Project (ESP) Seattle, WA: 2012. Available from: http://evs.gs.washington.edu/EVS/ [Google Scholar]
- Freeze HH. Genetic defects in the human glycome. Nat Rev Genet. 2006;7:537–51. doi: 10.1038/nrg1894. [DOI] [PubMed] [Google Scholar]
- Gao XD, Tachikawa H, Sato T, Jigami Y, Dean N. Alg14 recruits Alg13 to the cytoplasmic face of the endoplasmic reticulum to form a novel bipartite UDP-N-acetylglucosaminetransferase required for the second step of N-linked glycosylation. J Biol Chem. 2005;280:36254–62. doi: 10.1074/jbc.M507569200. [DOI] [PubMed] [Google Scholar]
- Gao XD, Moriyama S, Miura N, Dean N, Nishimura S. Interaction between the C termini of Alg13 and Alg14 mediates formation of the active UDP-N-acetylglucosaminetransferase complex. J Biol Chem. 2008;283:32534–41. doi: 10.1074/jbc.M804060200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehle VM, Sumikawa K. Site-directed mutagenesis of the conserved N-glycosylation site on the nicotinic acetylcholine receptor subunits. Brain Res Mol Brain Res. 1991;11:17–25. doi: 10.1016/0169-328x(91)90016-q. [DOI] [PubMed] [Google Scholar]
- Gehle VM, Walcott EC, Nishizaki T, Sumikawa K. N-glycosylation at the conserved sites ensures the expression of properly folded functional ACh receptors. Brain Res Mol Brain Res. 1997;45:219–29. doi: 10.1016/s0169-328x(96)00256-2. [DOI] [PubMed] [Google Scholar]
- Haeuptle M, Hennet T. Congenital disorders of glycosylation: an update on defects affecting the biosynthesis of dolichol-linked oligosaccharides. Hum Mutat. 2009;30:1628–41. doi: 10.1002/humu.21126. [DOI] [PubMed] [Google Scholar]
- Huffaker T, Robbins P. Yeast mutants deficient in protein glycosylation. Proc Natl Acad Sci USA. 1983;80:7466–70. doi: 10.1073/pnas.80.24.7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson B, Kukuruzinska M, Robbins P. Biosynthesis of asparagine-linked oligosaccharides in Saccharomyces cerevisiae: the alg2 mutation. Glycobiology. 1993;3:357–64. doi: 10.1093/glycob/3.4.357. [DOI] [PubMed] [Google Scholar]
- Jaeken J. Congenital disorders of glycosylation (CDG): It’s (nearly) all in it! J Inherit Metab Dis. 2011;34:853–8. doi: 10.1007/s10545-011-9299-3. [DOI] [PubMed] [Google Scholar]
- Jaeken J, Matthijs G. Congenital disorders of glycosylation: a rapidly expanding disease family. Annu Rev Genomic Hum Genet. 2007;8:261–78. doi: 10.1146/annurev.genom.8.080706.092327. [DOI] [PubMed] [Google Scholar]
- Jeppsson J, Arndt T, Schellenberg F, Wielders JP, Anton RF, Whitfield JB, et al. International federation of clinical chemistry and laboratory medicine working group on standardization of carbohydrate-deficient Transferin (IFCC-WG-CDT). Toward standardization of carbohydrate-deficient transferrin (CDT) measurements: I. Analyte definition and proposal of a candidate reference method. Clin Chem Lab Med. 2007;45:558–62. doi: 10.1515/CCLM.2007.107. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin K, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin A, Imperiali B. The expanding horizons of asparagine-linked glycosylation. Biochemistry. 2011;50:4411–26. doi: 10.1021/bi200346n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Takahashi T, Ohoka A, Nakajima K, Hashimoto R, Miura N, et al. Alg14 organizes the formation of a multiglycosyltransferase complex involved in initiation of lipid-linked oligosaccharide biosynthesis. Glycobiology. 2012;22:504–16. doi: 10.1093/glycob/cwr162. [DOI] [PubMed] [Google Scholar]
- Lunter G, Goodson M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21:936–9. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek KW, Vijay IK, Marth JD. A recessive deletion in the GlcNAc-1-phosphotransferase gene results in peri-implantation embryonic lethality. Glycobiology. 1999;9:1263–71. doi: 10.1093/glycob/9.11.1263. [DOI] [PubMed] [Google Scholar]
- Marklova E, Albahri Z. Screening and diagnosis of congenital disorders of glycosylation. Clin Chim Acta. 2007;385:6–20. doi: 10.1016/j.cca.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Rimer A, Mathieson I, Lunter G, McVean G. 2012. Platypus: An Integrated Variant Caller. Available at: www.well.ox.ac.uk/platypus. [Google Scholar]
- Sanes JR, Lichtman JW. Induction, assembly, maturation and maintenance of a postsynaptic apparatus. Nat Rev Neurosci. 2001;2:791–805. doi: 10.1038/35097557. [DOI] [PubMed] [Google Scholar]
- Senderek J, Muller JS, Dusl M, Strom TM, Guergueltcheva V, Diepolder I, et al. Hexosamine biosynthetic pathway mutations cause neuromuscular transmission defect. Am J Hum Genet. 2011;88:162–72. doi: 10.1016/j.ajhg.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji H, Takahashi N, Nomoto H, Ishikawa M, Shimada I, Arata Y, et al. Detailed structural analysis of asparagine-linked oligosaccharides of the nicotinic acetylcholine receptor from Torpedo California. Eur J Biochem. 1992;207:631–41. doi: 10.1111/j.1432-1033.1992.tb17090.x. [DOI] [PubMed] [Google Scholar]
- Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel C, Schwarz M, Peng J, Grzmil M, Hasilik M, Braulke T, et al. A new type of congenital disorders of glycosylation (CDG-Ii) provides new insights into the early steps of dolichol-linked oligosaccharide biosynthesis. J Biol Chem. 2003;278:22498–505. doi: 10.1074/jbc.M302850200. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Rush JS, Karaoglu D, Krasnewich D, Lubinsky MS, et al. Deficiency of UDP-GlcNAc:Dolichol Phosphate N-Acetylglucosamine-1 Phosphate Transferase (DPAGT1) causes a novel congenital disorder of Glycosylation Type Ij. Hum Mutat. 2003;22:144–50. doi: 10.1002/humu.10239. [DOI] [PubMed] [Google Scholar]
- Wurde AE, Reunert J, Rust S, Hertzberg C, Haverkamper S, Nurnberg G, et al. Congenital disorder of glycosylation type Ij (CDG-Ij, DPAGT1-CDG): extending the clinical and molecular spectrum of a rare disease. Mol Genet Metab. 2012;105:634–41. doi: 10.1016/j.ymgme.2012.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.