Abstract
Subtropical oceanic gyres are the most extensive biomes on Earth where SAR11 and Prochlorococcus bacterioplankton numerically dominate the surface waters depleted in inorganic macronutrients as well as in dissolved organic matter. In such nutrient poor conditions bacterioplankton could become photoheterotrophic, that is, potentially enhance uptake of scarce organic molecules using the available solar radiation to energise appropriate transport systems. Here, we assessed the photoheterotrophy of the key microbial taxa in the North Atlantic oligotrophic gyre and adjacent regions using 33P-ATP, 3H-ATP and 35S-methionine tracers. Light-stimulated uptake of these substrates was assessed in two dominant bacterioplankton groups discriminated by flow cytometric sorting of tracer-labelled cells and identified using catalysed reporter deposition fluorescence in situ hybridisation. One group of cells, encompassing 48% of all bacterioplankton, were identified as members of the SAR11 clade, whereas the other group (24% of all bacterioplankton) was Prochlorococcus. When exposed to light, SAR11 cells took 31% more ATP and 32% more methionine, whereas the Prochlorococcus cells took 33% more ATP and 34% more methionine. Other bacterioplankton did not demonstrate light stimulation. Thus, the SAR11 and Prochlorococcus groups, with distinctly different light-harvesting mechanisms, used light equally to enhance, by approximately one-third, the uptake of different types of organic molecules. Our findings indicate the significance of light-driven uptake of essential organic nutrients by the dominant bacterioplankton groups in the surface waters of one of the less productive, vast regions of the world's oceans—the oligotrophic North Atlantic subtropical gyre.
Keywords: SAR11, Prochlorococcus, light stimulation, flow cytometric sorting, radioisotope tracing, ATP and amino-acid uptake
Introduction
Over half of the bacterioplankton inhabiting the oceans occupy the thin surface layer that is regularly exposed to solar rays. However, beyond CO2 fixation rates by phototrophs the effect of light on the metabolism of the dominant microorganisms remains uncertain. Photoheterotrophic microorganisms have been defined as microorganisms that use light as their energy source and organic compounds as their carbon source with anoxygenic phototrophs as a typical example (Madigan, 2012). However, in the last two decades the use of the term was broadened to incorporate other microorganisms that could harvest light energy as well as utilise organic molecules (see Beja and Suzuki, 2008; Zubkov, 2009 for a review).
Three main groups of marine photoheterotrophic bacteria were identified: (i) chlorophyll-containing cyanobacteria, such as Prochlorococcus (Chisholm et al., 1988) and Synechococcus (Waterbury et al., 1979), (ii) aerobic anoxygenic bacteriochlorophyll-containing bacteria, such as members of the Roseobacter clade (Beja et al., 2002) and (iii) proteorhodopsin (PR)-containing bacteria that use a rhodopsin-based system (Beja et al., 2000) like the SAR11 alphaproteobacterial clade (Giovannoni et al., 2005a). The ability of those phylogenetic and metabolically diverse bacteria to harness light energy might impose a benefit in a range of physiological functions. However, the effect of light on PR-containing microorganisms is controversial (Fuhrman et al., 2008) and its role in SAR11, the most abundant microorganism in the surface ocean (Morris et al., 2002) remains unresolved (Steindler et al., 2011). Recently, it was shown that energy starved PR-containing Candidatus Pelagibacter ubique HTCC1062, a member of the SAR11 clade, increased the transport of amino acids and cellular ATP content after being exposed to light, but light had no effect during active growth (Steindler et al., 2011).
The influence of light on the uptake rates of organic molecules by natural photoheterotrophic bacterioplankton populations in the oligotrophic ocean, however, is poorly known (Beja and Suzuki, 2008), and has only been evaluated using amino acids as tracers (Church et al., 2004; Michelou et al., 2007; Mary et al., 2008). We previously showed preliminary evidence that light significantly stimulated the transport of amino acids by Prochlorococcus and heterotrophic non-pigmented low nucleic acid (LNA)-containing bacterioplankton, as discernible by flow cytometry. These two groups closed the budget of total bacterioplankton light-enhanced uptake of amino acids (Mary et al., 2008). On the other hand, Michelou et al. (2007) did not evaluate LNA bacteria and could not budget the total bacterioplankton light-enhanced amino-acids uptake.
The LNA bacterioplankton population in the surface open ocean includes the SAR11 clade. However, on average 40% of the LNA cells have not yet been phylogenetically assigned further than to bacteria (Mary et al., 2008; Hill et al., 2010; Schattenhofer et al., 2011). Considering the small size, (0.01–0.05 μm3, Rappe et al., 2002; Malmstrom et al., 2004) and the streamlined genome (1.3 Mbp, Giovannoni et al., 2005b) of SAR11, they likely have few ribosomes. Therefore, their detection by fluorescence in situ hybridisation (FISH) could be hampered (Amann and Fuchs, 2008) even when horseradish peroxidase-labelled oligonucleotide probes are used in combination with catalysed reported deposition (CARD) of fluorescently labelled tyramides.
The objective of this study is to directly compare the light-stimulated uptake of two essential biomass building blocks, nucleotides and amino acids, by SAR11 and Prochlorococcus in the North Atlantic subtropical gyre. We aim to compare the effect of light on the uptake rates of two simple organic nutrients that have distinctly different transport mechanisms. 5′-Nucleotides, such as ATP, are dephosphorylated extracellularly and then the phosphate and adenosine monomers transported (Bengis-Garber and Kushner, 1982; Bengis-Garber, 1983; Wanner, 1996; Sebastian and Ammerman, 2011) whereas amino acids are directly taken up by the cells via high-affinity ABC transporters. We hypothesise that light enhances transport of essential nutrients by SAR11 bacteria and Prochlorococcus, inhabiting surface waters of the oligotrophic North Atlantic subtropical gyre, in autumn when the water column becomes more stratified and depleted in inorganic macronutrients.
Materials and methods
Sampling and bacterioplankton enumeration
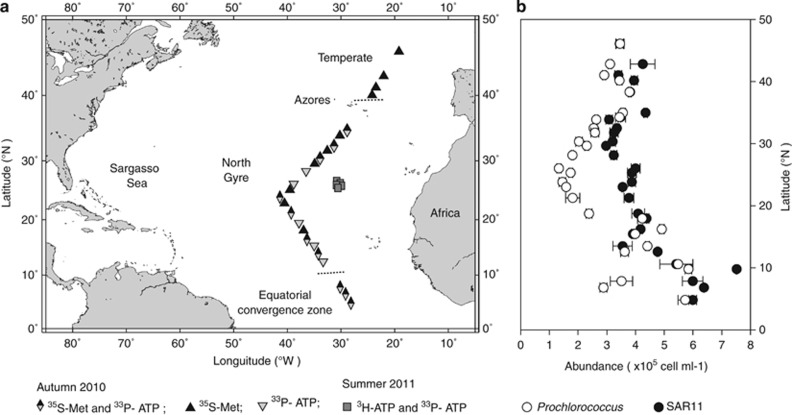
Experimental work was performed on board the Royal Research Ship James Cook (cruise no. JC53, October–November 2010) as part of the Atlantic Meridional Transect programme, and on board the Royal Research Ship Discovery (cruise no. D369, August–September 2011; Figure 1). At each station, samples were collected from 20 m depth with a sampling rosette of 20-l Niskin bottles mounted on a conductivity-temperature-depth profiler. Samples were collected in 1 l thermos flasks (washed with 10% v/v HCl) in the dark and processed immediately. A depth of 20 m was chosen because it represents the mixed layer and it was the shallowest depth unaffected by the ship's movement, including thrusting, that could artificially affect microbial metabolism in nutrient-depleted stratified surface waters. The effect of light on metabolic rates was evaluated using photosynthetically active radiation light spectra (400–700 nm), because at 20 m the penetration of ultraviolet is low.
Figure 1.
(a) Map showing the study area in the North Atlantic Ocean during the Atlantic Meridional Transect 20 cruise in autumn 2010 and the LINK cruise in summer 2011. Symbols indicate the stations at which light/dark sorting experiments were carried out. The boundaries of the oceanic provinces were identified with the Synechococcus distribution (Hartmann et al., 2012) indicated with short dash lines. (b) Latitudinal distribution of the abundance of SAR11 and Prochlorococcus during autumn 2010. Error bars show s.e.
Samples (1.6 ml) were fixed with 1% (w/v) paraformaldehyde (PFA) and stained with SYBR Green I DNA dye (Marie et al., 1997; Zubkov et al., 2000). Bacterioplankton, the cyanonobacteria Prochlorococcus and LNA bacteria (Supplementary Figure 1) as well as Synechococcus were enumerated with a FACSort flow cytometer (Becton Dickinson, Oxford, UK). Yellow-green 0.5 and 1.0 μm reference beads (Fluoresbrite Microparticles, Polysciences, Warrington, PA, USA) were used in all analyses as an internal standard for both fluorescence and flow rates. The absolute concentration of beads in the stock solution was determined using syringe pump flow cytometry (Zubkov and Burkill, 2006).
Ambient concentrations and turnover rates of methionine and ATP
Ambient concentrations as well as microbial uptake rates of the amino-acid methionine and of ATP were measured using isotopic dilution time-series incubations (Zubkov et al., 2004, 2007), referred to below as bioassays. ℒ-[35S] methionine (specific activity >1000 Ci mmol−1, Hartmann Analytic GmbH, Braunschweig, Germany) was added at a concentration of 0.05 nℳ and diluted with unlabelled ℒ-methionine (Sigma Aldrich, Dorset, UK) using a dilution series spanning the range 0.05–1.0 nℳ.
We compared the uptake rate of the phosphate and of the adenosine monomers of ATP by using two radiotracers: α-3333P-ATP where the phosphorus in the α position is labelled, and [2,5′,8-3H]-ATP (3H-ATP) where the adenosine is labelled. [α 33P]-ATP (specific activity >3000 Ci mmol−1, Hartmann Analytic GmbH) was added at a concentration of 0.05 or 0.1 nℳ and diluted with non-labelled ATP-disodium salt hydrate (Sigma Aldrich) using a dilution series in the range 0.1–2.0 nℳ. Incubated samples were fixed after 10, 20, 30 and 40 min with 1% (w/v) PFA final concentration. [2,5′,8-3H]–ATP (specific activity 51.5 Ci mmol−1, PerkinElmer Inc., MA, USA) was added at a concentration of 0.1–1.0 nℳ. Samples were fixed after 15, 30, 45 and 60 min. Fixed samples were filtered onto 0.2-μm-pore size polycarbonate filters and washed twice with 4 ml of deionised water. Radioactivity retained on the filters was measured as counts per minute using a liquid scintillation counter (Tri-Carb 3100TR, Perkin-Elmer, Beaconsfield, UK). Calculations of substrate concentration, uptake rate and turnover time were performed as described previously (Zubkov et al., 2007).
Light and dark incubations
The experimental set up for the light and dark uptake measurements was a dark room illuminated only by a very dim light of <1 μmol per photons per m2 per s. Light incubation experiments were placed in a 6-l water tank illuminated by a warm white light-emitting diode array (Photon Systems Instruments, Drasov, Czech Republic). Dark uptake experiments were placed in a water tank covered with two layers of black bags that kept the experiment in fully dark conditions. Undetectable CO2 fixation by eukaryotic cells confirmed the absence of light in the dark experiment (Hartmann, Zubkov personal communication). All experiments were placed in the light and in the dark incubators simultaneously. Temperature in both tanks was maintained within 1 °C of sea surface water temperature by circulating water through the tanks using a refrigerated bath (Grant Instruments, Cambridge, UK).
In autumn 2010, samples were incubated in polypropylene crystal clear microcentrifuge tubes (Starlab, Milton Keynes, UK) according to Mary et al. (2008). The tubes transmitted 72% of the light at 400 nm, increasing approximately linearly to 82% at 700 nm (Mary et al., 2008). The light-emitting diode light array was adjusted to an intensity of 500 μmol per photons per m2 per s. Therefore, the intensity at which samples were incubated was between 350 and 410 μmol per photons per m2 per s. In summer 2011, samples were incubated in borosilicate glass bottles (Pyrex, SciLabware, Staffordshire, UK) at 300 μmol per photons per m2 per s. Borosilicate glass bottles were soaked overnight with 10% (v/v) HCl, rinsed three times with deionised water and three times with the seawater sample before incubations. There was no difference in the bacterioplankton groups observed by flow cytometry between samples incubated (10 h) in microcentrifuge tubes and those in bottles (data not shown). For high-throughput experiments, microcentrifuge tubes presented the advantage that they are sterile and disposable despite their modest light absorbance. The light intensity of the light-emitting diode array was measured using a PAR quantum sensor (Skye, Powys, UK), and intensity was chosen within the range of ambient light at 20 m at noon, measured with the PAR sensor mounted on the conductivity-temperature-depth profiler.
Microbial light and dark uptake
During autumn 2010, light and dark uptake of ATP and methionine were evaluated at 12 and 22 stations, respectively. In each case, three to five independent time points were measured. A volume of 0.05 nℳℒ-[35S] methionine and 0.1–0.2 nℳ of unlabelled methionine, or 0.05–0.1 nℳ of [α 33P]-ATP and 0.45–0.8 nℳ of unlabelled ATP was placed into tubes and 1.6 ml of seawater was added to the experimental vials. Samples were fixed with 1% (w/v) PFA after 30, 60, 90, 120 and occasionally 150 min and processed as described in the previous section. We calculated the difference between the uptake in the light and in the dark in all paired experiments, and applied a one sample t-test, to test whether the difference between light and dark uptake was significantly higher than zero. All time points measured were tested independently. If data were not normally distributed (Shapiro–Wilk test), it was natural log transformed before computing light and dark differences.
In autumn 2010, four 1.6 ml samples were inoculated with 0.5 nℳℒ-[35S] methionine or 0.1–0.4 nℳ [α33P]-ATP, and incubated for 2 h in the light or dark before being fixed with 1% (w/v) PFA as described above. In summer 2011, parallel measurements of [2,5′,8-3H]–ATP and [α 33P]-ATP uptake were performed. The drawback of the latter is that the specific activity of the tritium label is low (51.5 Ci mmol−1) and therefore long (10 h) incubations were required to achieve cell labelling sufficient for detection in sorted cells. As the turnover of the internal ATP is very fast (Chapman and Atkinson, 1977; Winn and Karl, 1984) long incubations might lead to recycling of ATP. Therefore, we compared the uptake rates of both substrates, but chose the 33P-ATP for extensive testing of light-enhanced uptake because of its high-specific activity (3000 Ci mmol−1). Between 4.8 and 8 ml of seawater sample was incubated with 0.8 nℳ [2,5′,8-3H]–ATP or 0.3 nℳ [α 33P]-ATP in borosilicate glass bottles. To monitor that microbial uptake was linear, subsamples were fixed every 1 or 2 h and filtered as described above. In all experiments performed, bacterioplankton uptake in the light and dark was linear (r2>0.98 in eight independent experiments; see Supplementary Figure 2).
Flow cytometric cell sorting of radioactively labelled bacterioplankton cells
Flow cytometric cell sorting of radioactively labelled bacterioplankton cells was performed on board the ship within 12 h of fixation. Cells were flow-sorted from SYBR Green I DNA-stained samples (Supplementary Figure 1) by a FACSort flow cytometer (Becton Dickinson) using single-cell sort mode at a rate of 10–250 particles s−1. For each experiment, four proportional numbers of cells of total bacterioplankton, LNA bacteria and Prochlorococcus cells were sorted. At three stations, where their abundance was sufficiently high to warrant radiotracer flow sorting (Figure 1), Synechococcus were sorted from stained samples by discriminating them from other cells using their characteristic orange phycoerythrin autofluorescence. Sorted cells were directly collected onto 0.2-μm-pore size polycarbonate filters, washed twice with 4-ml deionised water and the radioactivity retained on the filters radioassayed as described above. The 3H-ATP experiments were counted using an ultra-low-level liquid scintillation counter (1220 Quantulus, Wallac, Finland) to improve the sensitivity of tritium detection. The mean cellular tracer uptake of each group was determined as the slope of the linear regression of radioactivity against the number of sorted cells, resulting in the uptake of an average cell (c.p.m. per cell).
To compare light-enhanced uptake between stations, the absolute uptake of an average LNA bacteria and Prochlorococus cell (nmol per cell per hour) was computed using the uptake of an average sorted cell, the total uptake in a given light or dark experiment and the microbial uptake rates at ambient concentrations as follows:
(uptake of sorted cells, c.p.m. per cell/total uptake, c.p.m. l−1) × microbial uptake rate, nmol l−1 h−1
The total amount of substrate incorporated by microorganisms (total uptake, c.p.m. l−1) was measured by filtering three subsamples from the sorting experiment onto 0.2-μm-pore size polycarbonate filters and measuring the radioactivity retained on the filters. To compute the absolute uptake in the light the microbial uptake rate at ambient concentration, which was measured in the dark, was multiplied by a light/dark factor determined in parallel for each experiment (Figure 2). The uptake rate of methionine and ATP per Prochlorococcus and LNA cell was multiplied by the number of molecules in 1 mol (Avogadro's constant).
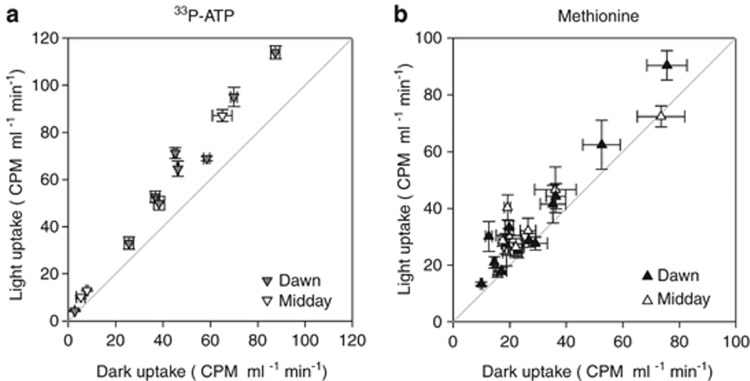
Figure 2.
Scatter plot comparison of the total bacterioplankton uptake of (a) ATP and (b) methionine in the light and dark. Error bars show s.e. of independent time points measurements. The light grey line indicates the unity line.
Flow cytometric cell sorting for CARD-FISH
CARD-FISH was performed on sorted cells to identify the bacterioplankton groups in which uptake rates were measured. Triplicate 1.6 ml samples were fixed with PFA (1% w/v final concentration), for 1 h at room temperature, subsequently flash frozen in liquid nitrogen and stored at −80 °C. Aliquots were thawed on ice and stained with SYBR Green as described above. LNA bacteria and Prochloroccocus cells were sorted on a sterilised FACScalibur flow cytometer (Becton Dickinson) with sheath fluid filtered through a 0.1-μm cartridge filter (Pall corporation, Port Washington, NY, USA). Approximately 1 × 105 cells were sorted and directly filtered onto polycarbonate filters (type GTTP, 13 mm diameter, 0.2 μm pore size, Millipore, Eschborn, Germany) placed on top of a cellulose acetate support filter (0.45 μm pore size, Sartorius, Goettingen, Germany). CARD-FISH was performed as described previously (Pernthaler et al., 2004) with the following modifications: cells were permeabilised with lysozyme (10 mg ml−1) in 0.05 ℳ EDTA, pH 8.0; 0.1 ℳ Tris-HCl, pH 8.0 buffer for 1 h and subsequently for 30 min with 60 U achromopeptidase (Sigma Aldrich) per ml of buffer (10 mℳ NaCl, 10 mℳ Tris-HCl, pH 8.0) at 37 °C. Filters were hybridised overnight at 46 °C with horseradish peroxidase-labelled oligonucleotide probes (Biomers, Ulm, Germany) at varying formamide concentrations depending on the probe used (Supplementary Table 1). Probes (50 ng μl−1) and buffer were mixed in a 1:300 ratio. The probe-delivered horseradish peroxidase was detected with fluorescently labelled tyramide Alexa488 (Life Technologies, Carlsbad, CA, USA) at a ratio of 1:1000 in amplification buffer. All cells were stained with 4,6-diamidino-2-phenylindole (1 mg ml–1). Hybridised and 4,6-diamidino-2-phenylindole-stained cells were enumerated using an Axioskope II epifluorescence microscope (Zeiss, Jena, Germany).
Results
Microbial uptake of the organic molecules ATP and methionine, in surface waters of the North Atlantic Ocean, was significantly higher in light incubations compared with dark incubations (Figures 2a and b). Total microbial 33P-ATP uptake in the light was 27±2% higher than in the dark (n=12, all experiments; Figure 2a). For methionine, it was 19±2% higher in the light than in the dark (in 20 out of 22 experiments). For both substrates, a significant increase in uptake was detected within 30 min of light exposure (t-test, P<0.001). All consecutive time points (up to 150 min) were also significantly higher in the light than in the dark (t-test, P<0.001). Flow sorting of labelled cells showed that bacterioplankton were primarily responsible for the light-enhanced uptake. There was also a good agreement in the proportion that light increased uptake of an average bacterioplankton cell (27±4%, ATP and 21±3%, methionine) and of the total microorganisms retained on filters (values shown above). No statistical difference was detected between measurements (ATP: t-test P=0.55, methionine t-test P<0.17, F-statistic higher than F-critical for both substrates; Supplementary Figures 3a and b).
Uptake rates of adenosine and phosphorus moieties
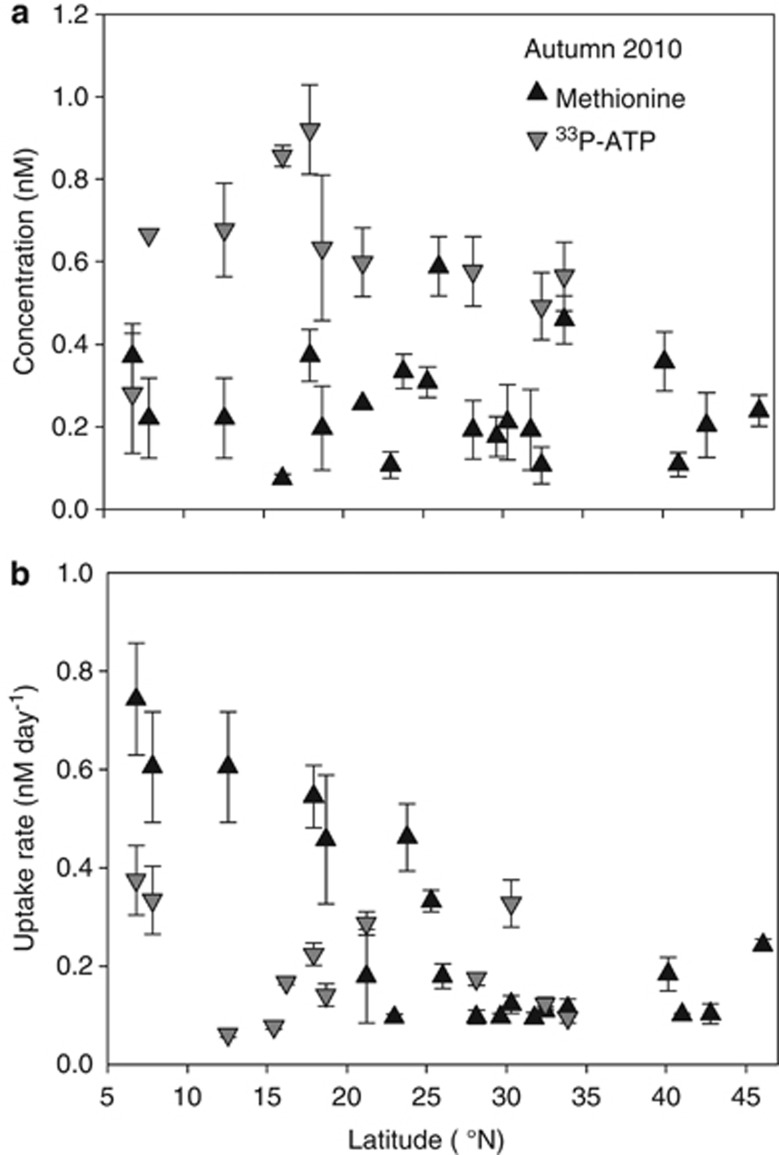
To investigate whether microorganisms consume ATP as an organic phosphorus source or as a nucleoside precursor (adenosine), we compared the uptake rates of ATP using 33P-ATP and 3H-ATP tracers. Parallel measurements with both tracers showed that for the same added concentration of tracer the turnover time of the pool of 3H-ATP was significantly faster than for 33P-ATP (Supplementary Figure 4). The total microbial uptake rate was significantly higher for 3H-ATP (0.243±0.04 nℳATP per day) than for 33P-ATP (0.098±0.019 nℳ ATP per day, P-value <0.05). However, the concentration of nucleotides in seawater measured with both substrates was statistically similar (0.262±0.06 nℳ for 33P-ATP and 0.259±0.05 nℳ for 3H-ATP, respectively, P-value >0.05), showing applicability of both tracers for bioassay of ambient ATP concentration. ATP concentration and uptake rates measured with 33P-ATP in the same region in 2010 but later in the autumn were higher (0.69±0.29 nℳ and 0.18±0.10 nℳ per day, respectively, Figures 3a and b). The methionine concentrations and uptake rates were in the same range as ATP, 0.24±0.13 nℳ and 0.27±0.19 nℳ per day, respectively (Figures 3a and b).
Figure 3.
Latitudinal distribution of bacterioplankton uptake rates and bioavailable methionine and ATP along the Atlantic Meridional Transect 20 transect during autumn 2010. (a) Bioavailable concentration and (b) total bacterioplankton uptake rates of methionine and 33P-ATP. Error bars show s.e.
Molecular identification of flow-sorted cells
CARD-FISH was performed on flow-sorted cells to identify the groups for which uptake rates were measured. High nucleic acid-containing bacteria, based on SYBR Green DNA staining, that had virtually undetectable chlorophyll autofluorescence, were phylogenetically affiliated with Prochlorococcus, in agreement with our previously reported results (Zubkov et al., 2007). An average of 88±4% (n=10) of total 4,6-diamidino-2-phenylindole-stained cells conferred signals with the Prochlorococcus-specific probe PRO405 (West et al., 2001). Interestingly, almost all the LNA bacteria comprised cells affiliated to the SAR11 clade. Thus, 93±6% (n=9) of total 4,6-diamidino-2-phenylindole-stained cells (Table 1) were positively identified using a set of probes that target different regions of the SAR11 rRNA ((Morris et al., 2002) and this study, Supplementary Table 1). Consequently, from this point onwards we refer to LNA bacteria as SAR11. Prochlorococcus and SAR11 numerically dominated bacterioplankton in surface waters of the North Atlantic subtropical gyre: the abundance of the former was, on average, 2.2±0.9 × 105 cells ml−1 whereas the abundance of the latter was 4.2±1.1 × 105 cells ml−1 (Figure 1b), representing 24±5% and 48±6% of total bacterioplankton, respectively.
Table 1. Composition of bacterioplankton in flow cytometrically sorted groups as relative abundance (% DAPI-stained cells) with oligonucleotide probes detailed in Supplementary Table 1.
|
Sample |
LNA |
Prochlorococcus |
|||
|---|---|---|---|---|---|
| Latitude (°N) | Longitude (°W) | EUB I–III | SAR11-mix | EUB I–III | PRO405 |
| 34.21 | 29.72 | ND | ND | 93 | 88 |
| 32.43 | 31.8 | ND | 84 | 94 | 83 |
| 31.73 | 32.56 | 92 | 92 | ND | 82 |
| 28.11 | 36.51 | 93 | 98 | 95 | 90 |
| 23.77 | 41.11 | 88 | 97 | 93 | 86 |
| 21.21 | 39.29 | 90 | 96 | 94 | 86 |
| 16.19 | 35.8 | 88 | 98 | 94 | 85 |
| 13.47 | 33.95 | 90 | 96 | 97 | 95 |
| 7.82 | 30.16 | 80 | 89 | 94 | 93 |
| 4.8 | 28.16 | 84 | 84 | 93 | 93 |
| Mean±s.d. | 88±4 | 93±6 | 94±1 | 88±4 | |
Abbreviations: DAPI, 4,6-diamidino-2-phenylindole; EUB I-III, Bacteria; LNA, low nucleic acid; ND, not determined.
SAR11 and Prochlorococcus uptake rates
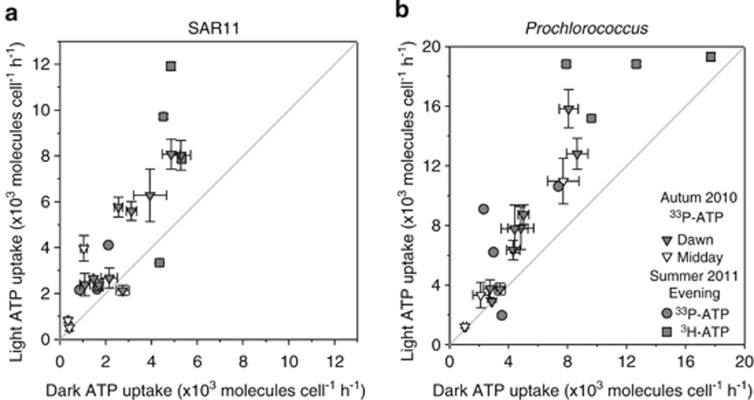
To assess whether light increased the transport of simple organic molecules into SAR11 cells, tracer-labelled cells were flow-sorted and their cellular tracer uptake was compared with tracer uptake by flow-sorted Prochlorococcus cells. On average, SAR11 cells had lower ATP uptake rates than Prochlorococcus cells, whereas methionine uptake rates for the two cell types were comparable (Table 2). SAR11 33P-ATP uptake was, on average, significantly lower than Prochlorococcus both in the light and the dark. Similarly, 3H-ATP uptake by SAR11 cells was significantly lower than 3H-ATP uptake by Prochlorococcus cells in both the light and dark. For both SAR11 and Prochlorococcus, the uptake rate of the adenosine moiety was two to three times faster than the uptake rate of the phosphorus groups of ATP (Figure 4). However, the SAR11 methionine uptake rates were, on average, comparable to those of Prochlorococcus, both in the light and dark (Figure 5). On average, SAR11 and Prochlorococcus accounted for 43% and 68% of total bacterioplankton ATP and methionine uptake, respectively (Supplementary Figures 3c and d).
Table 2. Average ATP and methionine uptake rate of bacterioplankton (Bpl), SAR11, Prochlorococcus (Pro) and Synechococcus (Syn) in the light, dark and light-enhanced uptake.
|
Average uptake rate |
Light enhanced(%) | ||||
|---|---|---|---|---|---|
| Light | Dark | Light enhanced | |||
| × 103 molecules per cell per hour | |||||
| Group | |||||
| 33P-ATP | |||||
| Bpl | 7.7±0.8 | >* | 5.6±0.6 | 2.1±0.6 | 36±4 |
| SAR11 | 3.5±0.6 | >** | 2.1±0.3 | 1.4±0.3 | 35±6 |
| PRO | 7.4±1.0 | >** | 4.6±0.6 | 2.8±0.5 | 33±5 |
| Syn | 96.7±38 | < | 166±68 | No light enhancement | |
| Group | |||||
| 3H-ATP | |||||
| SAR11 | 8.2±1.8 | > | 4.8±0.2 | 3.4±1.7 | 29±20 |
| PRO | 18.0±0.9 | > | 11.9±2.1 | 6.0±1.9 | 34±10 |
| Group | |||||
| Methionine | |||||
| Bpl | 11.9±1.5 | >** | 8.7±1.1 | 3.1±0.5 | 21±3 |
| SAR11 | 8.8±1.1 | >** | 5.8±0.8 | 2.9±0.4 | 32±5 |
| PRO | 8.8±1.4 | >** | 5.7±1.1 | 3.1±0.7 | 34±4 |
*P-value <0.05; **<0.01.
Figure 4.
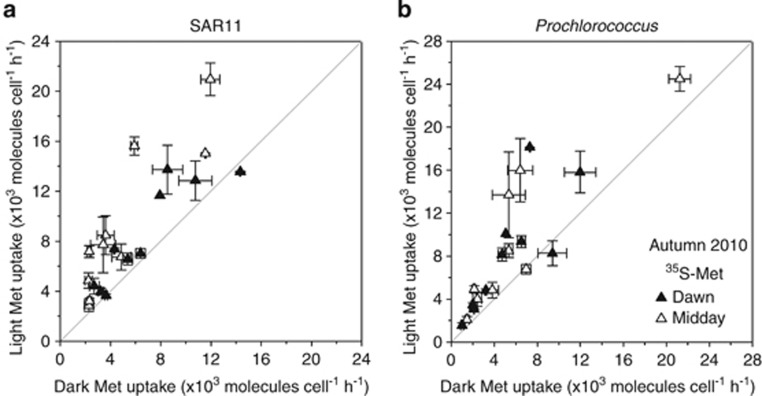
Scatter plot comparison of (a) SAR11 and (b) Prochlorococcus 3H-ATP and 33P-ATP uptake rates in the light and dark. Uptake in the light was significantly higher (P<0.05) than in the dark in (a) 17 of 21 and (b) 16 of 20 experiments. The light grey line indicates the unity line.
Figure 5.
Scatter plot comparison of (a) SAR11 and (b) Prochlorococcus methionine uptake rates in the light and dark. Uptake in the light was significantly higher (P<0.05) than in the dark in (a) 16 of 20 and (b) 15 of 19 experiments. The light grey line indicates the unity line.
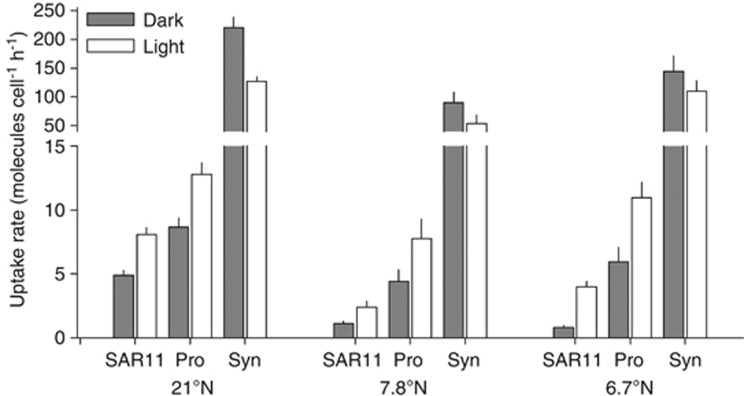
The cyanobacteria Synechococcus depicted a significantly higher uptake of ATP than SAR11 and Prochlorococcus (Figure 6, Table 2), contributing to up to 35% of the bacterioplankton ATP uptake. However, the Synechococcus uptake rate was not enhanced by light (Figure 6). The uptake of other non-sorted groups, mainly high nucleic acid-containing bacteria with low scatter (high nucleic acid-ls, HNA-ls, Supplementary Figure 1), was not significantly different in the light or dark (paired t-test P>0.05, Supplementary Figure 5). The uptake of the HNA-ls group was calculated by subtracting the sum of Prochlorococcus and SAR11 groups from the total bacterioplankton uptake.
Figure 6.
33P-ATP uptake rate of SAR11, Prochlorococcus (Pro) and Synechococcus (Syn) in three selected stations, showing uptake rate in the light and in the dark.
Effect of light on SAR11 and Prochlorococcus bacterioplankton
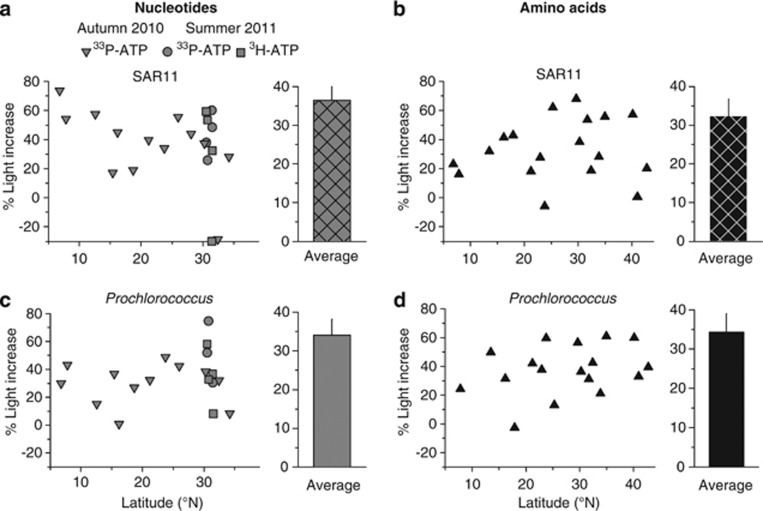
SAR11, as well as Prochlorococcus, showed significant light-enhanced 3H-ATP, 33P-ATP (Figure 4) and methionine uptake (Figure 5; paired t-test, P-value <0.001 for both populations and substrates). On average, SAR11 increased uptake of ATP in the light by 1.4±0.3 and 3.4±1.7 × 103 molecules per cell per hour for 33P-ATP and 3H-ATP, respectively, and by 2.9±0.4 × 103 molecules per cell per hour for methionine. Prochlorococcus showed a similar increase in ATP (2.8±0.5 and 6.0±1.9 × 103 molecules per cell per hour for 33P-ATP and 3H-ATP, respectively), and methionine (3.1±0.7 × 103 molecules per cell per hour) uptake in the light. The lower absolute values of light-stimulated substrate uptake in SAR11 compared with Prochlorococcus reflect the lower absolute uptake rates of the former. However, light-stimulated uptake of both substrates amounted to a similar proportion in both SAR11 and Prochlorococcus. Thus, exposure to light stimulated SAR11 uptake of 3H-ATP, 33P-ATP and methionine by 29±20%, 35±6% and 32±5%, respectively (Figures 7a and b). For Prochlorococcus, the corresponding values were 34±10%, 33±5% and 34±4%, respectively (Figures 7c and d).
Figure 7.
Comparison of relative light-enhanced uptake rates of nucleotides and amino acids by (a and b—correspondingly) SAR11 and (c and d) Prochlorococcus: latitudinal variability (left column) and mean values (right column). Hatch pattern corresponds to SAR11.
The total light-enhanced ATP and methionine uptake was budgeted to assess whether the groups that were not sorted contributed to the light-enhanced uptake. The average light-enhanced uptake of a cell was multiplied by its abundance relative to total bacterioplankton. The contribution of the SAR11 population to the light-enhanced ATP uptake was 17% whereas for Prochlorococcus it was 9%, which equals the bacterioplankton light-enhanced uptake. Similarly, for methionine the contribution of the SAR11 population to the light-enhanced uptake was 16% and 9% for Prochlorococcus, which also accounts for the total bacterioplankton light-enhanced uptake. Therefore, SAR11 and Prochlorococcus were the two bacterioplankton groups for which uptake was enhanced when exposed to light in the oligotrophic North Atlantic gyre.
Discussion
Here, we present experimental evidence collected in two consecutive years that in the oligotrophic North Atlantic Ocean subtropical gyre Prochlorococcus cyanobacteria and LNA bacterioplankton, solely comprising SAR11 alphaproteobacteria cells, use light to enhance their uptake of simple organic substrates and do so in equal measure.
Taxonomic identification of LNA bacteria, as discriminated by flow cytometry, has so far remained incomplete (Mary et al., 2006; Hill et al., 2010; Schattenhofer et al., 2011). We show here that virtually all cells in the LNA bacterioplankton are phylogenetically affiliated with the SAR11 clade (Table 1). We improved the detection of SAR11 in the LNA fraction by the application of a combination of six different horseradish peroxidase-labelled probes and a helper oligonucleotide probe (Fuchs et al., 2000) specifically targeting different positions in the SAR11 rRNA (Supplementary Table 1). This approach significantly increased the number and intensity of the probe-conferred signals, which allowed better quantification of the clade. The simultaneous comparison of the in situ light-driven uptake of two organic molecules, and the combination with their phylogenetic identification revealed that, despite significant differences in absolute uptake rates (Figures 4 and 5), the relative response to light by natural populations of SAR11 and Prochlorococcus was remarkably similar (Figure 7).
Light-enhanced uptake of nucleotides and amino acids by Prochlorococcus and SAR11
The aerobic oxygenic cyanobacterium Prochlorococcus has a chlorophyll-based light-harvesting complex and fixes CO2 (Chisholm et al., 1988, 1992). However, in the oligotrophic surface ocean, Prochlorococcus might invest in cyclic photophosphorylation and channel the energy generated to heterotrophically import essential building blocks such as nucleotides (Figure 4b; Michelou et al., 2011) and different essential amino acids (Figure 5b; (Zubkov et al., 2003; Church et al., 2004; Michelou et al., 2007; Mary et al., 2008). The other common marine cyanobacterium Synechococcus is also able to take up organic molecules but its photoheterotrophy is less consistent. For example, a higher proportion of Synechococcus cells than of SAR11 cells took up ATP in the Eastern Mediterranean Sea, depleted in phosphate (Sebastian et al., 2012). In this study, it was shown that Synechococcus cells took up significantly more ATP than either SAR11 or Prochlorococcus cells (Table 2), however, in the present study Synechococcus uptake rate of ATP was light insensitive (Figure 6) whereas the uptake of amino acids by Synechoccocus cells did increase when exposed to light (Mary et al., 2008). The differences in light stimulation are probably a result of differential uptake of organic molecules by Synechococcus (Cuhel and Waterbury, 1984; Willey and Waterbury, 1989; Beja and Suzuki, 2008) that warrant further field investigation.
In the non-pigmented heterotrophic bacterium SAR11, it is likely that light stimulation of methionine and nucleotides uptake rates is linked to the functioning of a PR proton pump. PR is broadly distributed within the SAR11 clade (Giovannoni et al., 2005a; Campbell et al., 2008). However, expression levels are not consistently linked to dark or light conditions (Giovannoni et al., 2005a; Cottrell and Kirchman, 2009; Steindler et al., 2011). The expression of PR even after long periods in the dark might enable cells to respond quickly when light becomes available. SAR11, together with Prochlorococcus, significantly increased their uptake of ATP and methionine within the first 30 min of incubation, both in samples taken at dawn after long periods in the dark and at midday under bright sunlight (Figures 2, 4 and 5). The short time response suggests that light has a direct photophysiological effect on these bacterial cells rather than an indirect one through uptake of organic molecules exuded by photo-stimulated phytoplankton (Karl et al., 1998; Church et al., 2004).
The relative influence of light on the transport of methionine, and adenosine and phosphorus monomers from nucleotides was also remarkably similar (Figure 7), despite significantly different uptake rates between molecules and bacterial groups (Figures 4 and 5), and different uptake mechanisms.
The uptake of 33P-ATP involves the extracellular dephosphorylation by alkaline phosphatase (E.C. 3.1.3.1; Wanner, 1996) or 5′-nucleotidase (E.C. 3.1.3.5; Bengis-Garber, 1985) and the uptake of the phosphate groups. The uptake of the 3H-ATP could either reflect the transport of the intact molecule or of the ribose backbone. Likely, 3H-ATP is taken up by the cells after the phosphate groups have been cleaved from the ribose (Bengis-Garber, 1983, 1985). Bacterial membranes, with the exception of the ones from obligate intracellular bacteria, seem not to be permeable to the intact ATP molecule (Daugherty et al., 2004; Schmitz-Esser et al., 2004). Moreover, if 3H-ATP was taken up with the phosphate groups, as AMP or ADP, then the uptake rates would be comparable between 33P-ATP and 3H-ATP, as we used α-33P-ATP. However, our results show that Prochlorococcus and SAR11 inhabiting the oligotrophic North Atlantic gyre take up the adenosine monomers two to three times faster than the phosphate groups (Figure 4, Supplementary Figure 4). This indicates that they might be scavenging nucleotides as a nucleoside source rather than for phosphorus (Casey et al., 2009; Michelou et al., 2011), which is rather taken from the inorganic phosphate pool (Zubkov et al., 2007). The coupling between the hydrolysis of nucleotides and uptake could be loose, and not all hydrolysed phosphate might be taken up by the cells (Ammerman and Azam, 1985). The energy harnessed from light may be directed to enhance the enzymatic activity of the 5′-nucleotidase or alkaline phosphatases that results in a higher uptake of both phosphate and adenosine by Prochlorococcus and SAR11. Genes encoding phosphatases with potential 5′-nucleotidase activity are known from Prochlorococcus and Synechococcus (Moore et al., 2005; Scanlan et al., 2009; Kathuria and Martiny, 2011), and in SAR11 (Gilbert et al., 2008; Kathuria and Martiny, 2011).
Clearly, Prochlorococcus and SAR11 have a different evolutionary history and lifestyle. However, in surface waters of the oligotrophic ocean they have converged to benefit from abundant light to import sparse organic molecules at a comparable magnitude. Under such nutrient-depleted conditions both phototrophic or heterotrophic metabolism converge on photoheterotrophy, allowing microorganisms to use a resource as abundant as light and direct it to the necessary energy-dependent functions like nutrient acquisition. The energy harnessed from light, by either PR or chlorophyll, can be directed to several cellular processes to promote an increase in enzymatic activity for dephosphorylating nucleotides (Figures 4 and 6) and a general uptake of organic matter (Figures 5 and 6; (Michelou et al., 2007; Mary et al., 2008; Steindler et al., 2011)).
It seems plausible that photoheterotrophy benefits microorganisms inhabiting the least productive ecosystems, such as the North (Figure 1) or South Atlantic subtropical gyres (Michelou et al., 2007; Mary et al., 2008) or the North Pacific Ocean (Church et al., 2004), but might not be significant in more productive seas (Alonso-Saez et al., 2006; Ruiz-Gonzalez et al., 2012). When nutrient availability is high, as in coastal waters, SAR11 uptake of amino acids could even be inhibited by light (Alonso-Saez et al., 2006; Ruiz-Gonzalez et al., 2012), or have no effect, as in Candidatus Pelagibacter ubique cells during active growth (Steindler et al., 2011). A PR-based light harvesting could provide SAR11 cells with extra energy to import organic compounds in oceanic areas where dissolved organic matter as well as essential inorganic macronutrients, like nitrogen and phosphorus, are present at exceedingly low concentrations.
Measuring energy harvesting and storage in bacterioplankton cells is challenging (Zubkov, 2009) therefore the costs and benefits of utilising light energy by aerobic anoxygenic bacteria and PR-containing bacteria have been estimated using bioenergetics models (Kirchman and Hanson, 2012). The net energy gain seems to be sufficient to meet maintenance costs by the former and insufficient to meet the maintenance costs by the latter except situations when high light intensities are combined with a large number of PR molecules imbedded in cellular membrane. Such situation is plausible in the subtropical North Atlantic gyre, where light intensities are regularly high (up to 800 μmol per photons per m2 per s measured when samples for this study were taken) and the extra energy harvested from light could advantage PR-containing microorganisms like SAR11.
It is likely then, that PR- or Chl a-based photoheterotrophy provides a large fraction of cellular energy requirements (equally large—one third for amino-acid and nucleotide uptake) for the SAR11 clade and Prochlorococcus to outperform other bacterioplankton groups in essential nutrient acquisition in the least productive and most extensive ecosystems on Earth like the North Atlantic subtropical gyre.
Acknowledgments
We thank the captains, officers and crew aboard the Royal Research Ship James Cook and the Royal Research Ship Discovery for their help during both cruises. We are grateful to J Wulf for advice on FISH analyses. This study was supported by the UK Natural Environment Research Council through Research grants NE/H005196/1 and NE/H007083/1, as well as NE/E016138/1, NE/G005125/1, the Oceans 2025 Core Programme of the National Oceanography Centre and Plymouth Marine Laboratory and the European Commission Seventh Framework Programme through the GreenSeas Collaborative Project (FP7-ENV-2010 contract 265294). This is Atlantic Meridional Transect publication no. 224.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alonso-Saez L, Gasol JM, Lefort T, Hofer J, Sommaruga R. Effect of natural sunlight on bacterial activity and differential sensitivity of natural bacterioplankton groups in northwestern Mediterranean coastal waters. Appl Environ Microbiol. 2006;72:5806–5813. doi: 10.1128/AEM.00597-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R, Fuchs BM. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat Rev Microbiol. 2008;6:339–348. doi: 10.1038/nrmicro1888. [DOI] [PubMed] [Google Scholar]
- Ammerman JW, Azam F. Bacterial 5′-Nucleotidase in aquatic ecosystems—a novel mechanism of phosphorus regeneration. Science. 1985;227:1338–1340. doi: 10.1126/science.227.4692.1338. [DOI] [PubMed] [Google Scholar]
- Beja O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- Beja O, Suzuki MT.2008Photohetrotrophic marine prokaryotesIn: Kirchman DL (ed)Microbial Ecology of the Oceans2nd edn.John Wiley & Sons, Inc.132–157. [Google Scholar]
- Beja O, Suzuki MT, Heidelberg JF, Nelson WC, Preston CM, Hamada T, et al. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature. 2002;415:630–633. doi: 10.1038/415630a. [DOI] [PubMed] [Google Scholar]
- Bengis-Garber C. Uptake of adenosine in a marine bacterium is not an active-transport process. FEBS Lett. 1983;160:31–36. doi: 10.1016/0014-5793(83)80930-2. [DOI] [PubMed] [Google Scholar]
- Bengis-Garber C. Membrane-bound 5′-Nucleotidase in marine luminous bacteria—biochemical and immunological properties. Can J Microbiol. 1985;31:543–548. doi: 10.1139/m85-101. [DOI] [PubMed] [Google Scholar]
- Bengis-Garber C, Kushner DJ. Role of membrane-bound 5′-nucleotidase in nucleotide uptake by the moderate halophile Vibrio costicola. J Bacteriol. 1982;149:808–815. doi: 10.1128/jb.149.3.808-815.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BJ, Waidner LA, Cottrell MT, Kirchman DL. Abundant proteorhodopsin genes in the North Atlantic Ocean. Environ Microbiol. 2008;10:99–109. doi: 10.1111/j.1462-2920.2007.01436.x. [DOI] [PubMed] [Google Scholar]
- Casey JR, Lomas MW, Michelou VK, Dyhrman ST, Orchard ED, Ammerman JW, et al. Phytoplankton taxon-specific orthophosphate (Pi) and ATP utilization in the western subtropical North Atlantic. Aquat Microb Ecol. 2009;58:31–44. [Google Scholar]
- Chapman AG, Atkinson DE. Adenine nucleotide concentrations and turnover rates. Their correlation with biological activity in bacteria and yeast. Adv Microb Physiol. 1977;15:253–306. doi: 10.1016/s0065-2911(08)60318-5. [DOI] [PubMed] [Google Scholar]
- Chisholm SW, Frankel SL, Goericke R, Olson RJ, Palenik B, Waterbury JB, et al. Prochlorococcus marinus nov. gen. nov. sp.: an oxyphototrophic marine prokaryote containing divinyl chlorophyll a and chlorophyll b. Arch Microbiol. 1992;157:297–300. [Google Scholar]
- Chisholm SW, Olson RJ, Zettler ER, Goericke R, Waterbury JB, Welschmeyer NA. A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature. 1988;334:340–343. [Google Scholar]
- Church MJ, Ducklow HW, Karl DA. Light dependence of [3H]leucine incorporation in the oligotrophic North Pacific Ocean. Appl Environ Microbiol. 2004;70:4079–4087. doi: 10.1128/AEM.70.7.4079-4087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell MT, Kirchman DL. Photoheterotrophic microbes in the Arctic Ocean in summer and winter. Appl Environ Microbiol. 2009;75:4958–4966. doi: 10.1128/AEM.00117-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuhel RL, Waterbury JB. Biochemical composition and short term nutrient incorporation patterns in a unicellular marine cyanobacterium, Synechococcus (WH7803) Limnol Oceanogr. 1984;29:370–374. [Google Scholar]
- Daugherty RM, Linka N, Audia JP, Urbany C, Neuhaus HE, Winkler HH. The nucleotide transporter of Caedibacter caryophilus exhibits an extended substrate spectrum compared to the analogous ATP/ADP translocase of Rickettsia prowazekii. J Bacteriol. 2004;186:3262–3265. doi: 10.1128/JB.186.10.3262-3265.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs BM, Glockner FO, Wulf J, Amann R. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 2000;66:3603–3607. doi: 10.1128/aem.66.8.3603-3607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, Schwalbach MS, Stingl U. Opinion—Proteorhodopsins: an array of physiological roles. Nat Rev Microbiol. 2008;6:488–494. doi: 10.1038/nrmicro1893. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Muhling M, Joint I. A rare SAR11 fosmid clone confirming genetic variability in the ‘Candidatus Pelagibacter ubique' genome. ISME J. 2008;2:790–793. doi: 10.1038/ismej.2008.49. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Bibbs L, Cho JC, Stapels MD, Desiderio R, Vergin KL, et al. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature. 2005a;438:82–85. doi: 10.1038/nature04032. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005b;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- Hartmann M, Grob C, Tarran GA, Martin AP, Burkill PH, Scanlan DJ, et al. Mixotrophic basis of Atlantic oligotrophic ecosystems. Proc Natl Acad Sci USA. 2012;109:5756–5760. doi: 10.1073/pnas.1118179109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill PG, Zubkov MV, Purdie DA. Differential responses of Prochlorococcus and SAR11 dominated bacterioplankton groups to atmospheric dust inputs in the tropical Northeast Atlantic Ocean. FEMS Microbiol Lett. 2010;306:82–89. doi: 10.1111/j.1574-6968.2010.01940.x. [DOI] [PubMed] [Google Scholar]
- Karl DM, Hebel DV, Bjorkman K, Letelier RM. The role of dissolved organic matter release in the productivity of the oligotrophic North Pacific Ocean. Limnol Oceanogr. 1998;43:1270–1286. [Google Scholar]
- Kathuria S, Martiny AC. Prevalence of a calcium-based alkaline phosphatase associated with the marine cyanobacterium Prochlorococcus and other ocean bacteria. Environ Microbiol. 2011;13:74–83. doi: 10.1111/j.1462-2920.2010.02310.x. [DOI] [PubMed] [Google Scholar]
- Kirchman DL, Hanson TE. Bioenergetics of photoheterotrophic bacteria in the oceans. Environ Microbiol Rep. 2012. [DOI] [PubMed]
- Madigan MT.2012Brock Biology of Microorganisms13th edn.Benjamin Cummings: San Francisco [Google Scholar]
- Malmstrom RR, Kiene RP, Cottrell MT, Kirchman DL. Contribution of SAR11 bacteria to dissolved dimethylsulfoniopropionate and amino acid uptake in the North Atlantic ocean. Appl Environ Microbiol. 2004;70:4129–4135. doi: 10.1128/AEM.70.7.4129-4135.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie D, Partensky F, Jacquet S, Vaulot D. Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl Environ Microbiol. 1997;63:186–193. doi: 10.1128/aem.63.1.186-193.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mary I, Heywood JL, Fuchs BM, Amann R, Tarran GA, Burkill PH, et al. SAR11 dominance among metabolically active low nucleic acid bacterioplankton in surface waters along an Atlantic Meridional Transect. Aquat Microb Ecol. 2006;45:107–113. [Google Scholar]
- Mary I, Tarran GA, Warwick PE, Terry MJ, Scanlan DJ, Burkill PH, et al. Light enhanced amino acid uptake by dominant bacterioplankton groups in surface waters of the Atlantic Ocean. FEMS Microbiol Ecol. 2008;63:36–45. doi: 10.1111/j.1574-6941.2007.00414.x. [DOI] [PubMed] [Google Scholar]
- Michelou VK, Cottrell MT, Kirchman DL. Light-stimulated bacterial production and amino acid assimilation by cyanobacteria and other microbes in the North Atlantic Ocean. Appl Environ Microbiol. 2007;73:5539–5546. doi: 10.1128/AEM.00212-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelou VK, Lomas MW, Kirchman DL. Phosphate and adenosine-5′-triphosphate uptake by cyanobacteria and heterotrophic bacteria in the Sargasso Sea. Limnol Oceanogr. 2011;56:323–332. [Google Scholar]
- Moore LR, Ostrowski M, Scanlan DJ, Feren K, Sweetsir T. Ecotypic variation in phosphorus acquisition mechanisms within marine picocyanobacteria. Aquat Microb Ecol. 2005;39:257–269. [Google Scholar]
- Morris RM, Rappe MS, Connon SA, Vergin KL, Siebold WA, Carlson CA, et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- Pernthaler A, Pernthaler J, Amann R.2004Sensitive multicolor fluorescence in situ hybridization for the identification of environmental microorganismsIn: Kowalchuk G, de Bruijn FJ, Head IM, Akkermans ADL, van Elsas JD (eds)Molecular Microbial Ecology Manual2nd edn.Kluwer Academic Publishers: Dordrecht, The Netherlands; 711–726. [Google Scholar]
- Rappe MS, Connon SA, Vergin KL, Giovannoni SJ. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature. 2002;418:630–633. doi: 10.1038/nature00917. [DOI] [PubMed] [Google Scholar]
- Ruiz-Gonzalez C, Simo R, Vila-Costa M, Sommaruga R, Gasol JM. Sunlight modulates the relative importance of heterotrophic bacteria and picophytoplankton in DMSP-sulphur uptake. ISME J. 2012;6:650–659. doi: 10.1038/ismej.2011.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, et al. Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev. 2009;73:249–299. doi: 10.1128/MMBR.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattenhofer M, Wulf J, Kostadinov I, Glockner FO, Zubkov MV, Fuchs BM. Phylogenetic characterisation of picoplanktonic populations with high and low nucleic acid content in the North Atlantic Ocean. Syst Appl Microbiol. 2011;34:470–475. doi: 10.1016/j.syapm.2011.01.008. [DOI] [PubMed] [Google Scholar]
- Schmitz-Esser S, Linka N, Collingro A, Beier CL, Neuhaus HE, Wagner M, et al. ATP/ADP translocases: a common feature of obligate intracellular amoebal symbionts related to Chlamydiae and Rickettsiae. J Bacteriol. 2004;186:683–691. doi: 10.1128/JB.186.3.683-691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian M, Ammerman JW. Role of the phosphatase PhoX in the phosphorus metabolism of the marine bacterium Ruegeria pomeroyi DSS-3. Environ Microbiol Rep. 2011;3:535–542. doi: 10.1111/j.1758-2229.2011.00253.x. [DOI] [PubMed] [Google Scholar]
- Sebastian M, Pitta P, Gonzalez JM, Thingstad TF, Gasol JM. Bacterioplankton groups involved in the uptake of phosphate and dissolved organic phosphorus in a mesocosm experiment with P-starved Mediterranean waters. Environ Microbiol. 2012. [DOI] [PubMed]
- Steindler L, Schwalbach MS, Smith DP, Chan F, Giovannoni SJ. Energy starved Candidatus Pelagibacter ubique substitutes light-mediated ATP production for endogenous carbon respiration. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner BL.1996Phosphorus assimilation and control of the phosphate regulonIn: Neidhardt FC, Curtiss R III, Ingra-Ham JL, Lin ECC, Low KB, Magasanik B et al. (eds)Escherichia coli and Salmonella: Cellular and Molecular Biology ASM Press: Washington, DC; 1357–1381. [Google Scholar]
- Waterbury JB, Watson SW, Guillard RRL, Brand LE. Widespread occurrence of a unicellular, marine, planktonic, cyanobacterium. Nature. 1979;277:293–294. [Google Scholar]
- West NJ, Schonhuber WA, Fuller NJ, Amann RI, Rippka R, Post AF, et al. Closely related Prochlorococcus genotypes show remarkably different depth distributions in two oceanic regions as revealed by in situ hybridization using 16S rRNA-targeted oligonucleotides. Microbiology. 2001;147:1731–1744. doi: 10.1099/00221287-147-7-1731. [DOI] [PubMed] [Google Scholar]
- Willey JM, Waterbury JB. Chemotaxis toward nitrogenous compounds by swimming strains of marine Synechococcus spp. Appl Environ Microbiol. 1989;55:1888–1894. doi: 10.1128/aem.55.8.1888-1894.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn CD, Karl DM. Laboratory calibrations of the [3H]adenine technique for measuring rates of RNA and DNA synthesis in marine microorganisms. Appl Environ Microbiol. 1984;47:835–842. doi: 10.1128/aem.47.4.835-842.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubkov MV. Photoheterotrophy in marine prokaryotes. J Plankton Res. 2009;31:933–938. [Google Scholar]
- Zubkov MV, Burkill PH. Syringe pumped high speed flow cytometry of oceanic phytoplankton. Cytometry A. 2006;69:1010–1019. doi: 10.1002/cyto.a.20332. [DOI] [PubMed] [Google Scholar]
- Zubkov MV, Fuchs BM, Tarran GA, Burkill PH, Amann R. High rate of uptake of organic nitrogen compounds by Prochlorococcus cyanobacteria as a key to their dominance in oligotrophic oceanic waters. Appl Environ Microbiol. 2003;69:1299–1304. doi: 10.1128/AEM.69.2.1299-1304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubkov MV, Mary I, Woodward EMS, Warwick PE, Fuchs BM, Scanlan DJ, et al. Microbial control of phosphate in the nutrient-depleted North Atlantic subtropical gyre. Environ Microbiol. 2007;9:2079–2089. doi: 10.1111/j.1462-2920.2007.01324.x. [DOI] [PubMed] [Google Scholar]
- Zubkov MV, Sleigh MA, Burkill PH, Leakey RJG. Picoplankton community structure on the Atlantic Meridional Transect: a comparison between seasons. Prog Oceanogr. 2000;45:369–386. [Google Scholar]
- Zubkov MV, Tarran GA, Fuchs BM. Depth related amino acid uptake by Prochlorococcus cyanobacteria in the Southern Atlantic tropical gyre. FEMS Microbiol Ecol. 2004;50:153–161. doi: 10.1016/j.femsec.2004.06.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.