Abstract
Aldehyde Dehydrogenase (ALDH) genes are increasingly associated with stem / progenitor cell status but their role in the maintenance of pluripotency remains uncertain. In a screen conducted for downstream Ngn3 target genes using ES derived pancreas progenitors we identified Aldh1b1, encoding a mitochondrial enzyme, as one of the genes strongly up regulated in response to Ngn3 expression. We found both by in situ hybridization and immunofluorescence using a specific antibody that ALDH1B1 is exclusively expressed in the emerging pancreatic buds of the early embryo (9.5 dpc) in a Pdx1 dependent manner. Around the time of secondary transition, ALDH1B1 expression was restricted in the tip tripotent progenitors of the branching epithelium and in a subset of the trunk epithelium. Expression in the latter was Ngn3 dependent. Subsequently, ALDH1B1 expression persisted only in the tip cells that become restricted to the exocrine lineage and declined rapidly as these cells mature. In the adult pancreas we identified rare ALDH1B1+ cells that become abundant following pancreas injury in either the caerulein or streptozotocin paradigms. Blocking ALDH catalytic activity in pancreas embryonic explants resulted in reduced size of the explants and accelerated differentiation suggesting for the first time that ALDH activity may be necessary in the developing pancreas for the maintenance and expansion of progenitor pools.
Keywords: aldehyde dehydrogenase, pancreas stem and progenitor cells, NGN3, PDX1, pancreas injury, centroacinar cells
INTRODUCTION
The identification of stem/progenitor cell markers, particularly in adult tissues, is an important goal for regenerative medicine. Identification of specific markers for stem/progenitor cells is essential for their isolation and study as well as for the enrichment of ES cell or iPS cell derived populations for specific stem/progenitor cells. In the pancreas, the question of whether multipotent pancreatic progenitors persist into adulthood, as well as their possible anatomical location have been highly contested issues.
Genetic lineage tracing based on CRE recombinase and combined with functional analyses using a variety of pancreas injury and regeneration models have not yet pinpointed an adult progenitor cell for all pancreatic lineages (Furuyama et al., 2011; Inada et al., 2008;Kopinke et al., 2011; Kopinke and Murtaugh, 2010; Kopp et al., 2011b; Solar et al., 2009). Obviously, this does not exclude the possibility that an as yet unknown gene may specifically mark such pancreas stem/progenitor cells. Additionally, unidentified extracellular signals may be necessary for the expansion, survival and differentiation of stem/progenitor cells following injury and regeneration. The conflicting results regarding the presence of progenitor cells and their differentiation potential may be indicative of the presence of the required signal(s) in sub-optimal concentrations. Furthermore, bona fide stem/progenitor cells may not be identifiable by the expression of a single gene but by the combinatorial expression of several genes, possibly specific transcription factors (Kopp et al., 2011a)). In this case only the development and use of highly sophisticated lineage tracing methods will allow the unambiguous identification of this population. Genes strongly expressed in progenitor pools during development are prime candidates as markers for adult stem/progenitor cells. Ideal markers should allow the isolation of the target cell population with minimal damage and be highly specific to allow for unambiguous lineage tracing of descendant cells.
The pancreas develops at the posterior foregut region of the definitive endoderm and is first morphologically obvious around 9.5 dpc with the emergence of the dorsal and ventral pancreatic buds (Spooner et al., 1970). Induction of the pancreatic fates is mediated from signals emanating from the adjacent mesodermal, notochord and dorsal aorta tissues. Retinoic acid (RA), FGF and BMP signaling and inhibition of SHH signaling (Dessimoz et al., 2006; Hebrok et al., 1998;Kim and Melton, 1998; Kumar et al., 2003; Martin et al., 2005; Molotkov et al., 2005;Stafford and Prince, 2002; Wells and Melton, 2000) play key roles in the emergence of the pancreatic primordia. All pancreatic cell types are derived from this pool of early pancreatic progenitor cells that co-express the transcription factors PDX1, PTF1a and SOX9 (Kawaguchi et al., 2002; Seymour et al., 2007).
The pancreatic buds thicken and then start expanding at around 10.5 dpc by branching morphogenesis (Puri and Hebrok, 2007). Successive rounds of branching and growth result in a tree-like epithelial network surrounded by mesenchyme. As branching morphogenesis proceeds, expression of Ptf1a and Cpa1 become restricted to the tips of the branching epithelium. Lineage tracing suggest that the tips retain their multipotentiality until 13.5 dpc, at which point resident CPA1+ cells become dedicated acinar progenitors (Zhou et al., 2007). The extension of the tips leaves behind trunk cells expressing Nkx6.1, Nkx6.2, Sox9 and Hnf1b. Hnf1b is expressed exclusively in all trunk cells and HNF1b+ cells remain multipotent up until around 13.5 dpc. The reciprocal repression between Ptf1a and Nkx6 transcription factors acts as a switch in multipotent progenitors directing them to either the acinar or the ductal /endocrine fate (Schaffer et al., 2010). This is reflected in the switch of the multipotent trunk HNF1b+ cells to exclusive duct /endocrine progenitors cells (Solar et al., 2009). NGN3+ endocrine progenitor cells arise from the trunk epithelium, delaminate and migrate into the mesenchyme to differentiate into the islet cells of the mature organ leaving behind acinar progenitors (Cole et al., 2009; Gouzi et al., 2011; Rukstalis and Habener, 2007).
Thus the emerging picture of stem/progenitor cells is a highly dynamic one, whereby as cells mature and become restricted in their developmental potential, expression of distinct transcription factors segregates to different progenitor populations and then differentiated cells. For example, based on its expression pattern in embryonic and adult pancreas, but also on its unique expression in various stem/progenitor cell domains (Kopp et al., 2011a), Sox9 was postulated as a possible marker of stem/progenitor cells in the adult pancreas. Careful lineage tracing analysis showed that whereas the Sox9+ cell domain remains multipotent up until birth, this multipotency is rapidly lost during the first three weeks of age (Kopp et al., 2011b).
The existence of a small population of PDX1, PTF1a, SOX9 and NKX6.1 co-positive cells at the interface of the tip and the trunk regions raised the possibility that this population were the precursors of the centro-acinar cells that are thought to represent the facultative stem/progenitor cell of the adult pancreas (Kopp et al., 2011a). ALDH activity is being increasingly associated with stem/progenitor cell status (Balber, 2011). It is hypothesized that this activity constitutes a significant determinant of stem / progenitor cell survival through its ability to detoxify metabolic byproducts and potentially cytotoxic molecules (Alison et al., 2010; Gasparetto et al., 2012), but this hypothesis has not been genetically tested. Thus putative progenitor centroacinar cells were isolated from adult pancreata on the basis of their high aldehyde dehydrogenase (ALDH) activity and shown to generate multiple pancreatic cell types in vitro or upon transplantation into embryonic pancreas explants. Strong expression of the aldehyde dehydrogenase genes Aldh1a1 and Aldh1a7 in this cell population was demonstrated by qPCR, but the capacity of the isolated cells to generate endocrine and exocrine cells was not directly associated with Aldh1a1 and/or Aldh1a7 function. Using a broad specificity (polyclonal) ALDH1 antibody, ALDH immunoreactivity, restricted at the tips of the branching epithelium, was detected at embryonic stages 12.5 dpc and 14.5 dpc (Rovira et al., 2010).
In a screen conducted for downstream NGN3 target genes using ES derived pancreas progenitors we identified a novel aldehyde dehydrogenease gene, Aldh1b1 (encoding a mitochondrial enzyme) as one of the genes strongly up regulated in response to Ngn3 expression (Serafimidis et al., 2008). We then found both by in situ hybridization and immunofluorescence using an ALDH1B1-specific antibody (Stagos et al., 2010) that it is exclusively expressed in the emerging pancreatic buds of the early embryo (9.5 dpc) in a Pdx1 dependent manner. Expression of Aldh1b1 and Aldh1a1 were the strongest among expression of several Aldh genes examined around the time of secondary transition. Expression of both genes dropped in Ngn3 null 14.5 dpc pancreata suggesting that together they may play a key role in the development of the epithelium and the endocrine lineage in particular. Aldh1b1 expression was restricted in the tip tripotent progenitors of the branching epithelium and in a subset of the trunk epithelium. Expression in the latter was Ngn3 dependent. Later, Aldh1b1 expression persisted only in the tip cells that are restricted to the exocrine lineage and declined rapidly as these cells matured. In the adult pancreas we identified rare ALDH1B1+ cells that became abundant following pancreas injury and regeneration. Blocking ALDH activity in pancreas embryonic explants resulted in reduced size of the explants and accelerated differentiation, suggesting for the first time that ALDH activity may be necessary in the developing pancreas for the maintenance and expansion of progenitor pools.
MATERIALS AND METHODS
Animal strains
Animal studies were conducted in accordance with international guidelines and after ethical approval of the competent Veterinary Service of Athens. Null mouse mutants were use for the Ngn3 (Gradwohl et al., 2000), and the Pdx1 (Holland et al., 2002) genes. Mouse genotyping procedures were as described for Ngn3 (Gradwohl et al., 2000), and Pdx1 (Holland et al., 2002).
Real Time PCR
Six (12.5 dpc) or four (14.5 dpc) embryonic pancreata were pooled. Total RNA isolation and cDNA preparation were according to standard procedures (Serafimidis et al., 2008). Real Time PCR primers were designed using the Beacon Designer software, Real Time PCR reactions were performed with SYBR-GREENER (Invitrogen) using an ABI PRISM 7000 machine and primary results were analyzed using the machine’s software. Absolute expression values were calculated using the ΔCt method (expression = 2 −(Ct sample − Ct β-actin)) and multiplied by 1000. At least three reactions for each gene at each time point were carried out. Primers used were evaluated by inspection of the dissociation curve and were as follows: Aldh1b1 F: AGCCTCTGTTCAAGTTCAAG, R: CCTTAAAGCCTCCGAATGG, Aldh2 F: GTAGACAAGGCAGTGAAG, R: GGTACGAGATGACATAAGG, Aldh1a1 F: TATATGATGTTGTCAGCCCAGTG, R: ATGTTCACCCAGTTCTCTTCC, Aldh1a2 F: TGGAAATGGGAGAGAAATGG, R: GGCAAGAGTGGTGAATGG, Aldh1a3 F: CCAGTGTGAGGTAGAGAATGTG, R: GTGTTGAGGGAGGAAAGAAGG, Aldh1a7 F: TTAGCAGCAGGAGTCTTCAC, R: ACAGGGACAGCCAAATAGC, Aldh8a1 F: ATTCCTCGGTCTGTTCTG, R: TGGCTATCACGGTATTCC, β-actin F: TGGCTCCTAGCACCATGA, R: CCACCGATCCACACAGAG. The relative efficiency of the primers for Aldh1a1, Aldh1a2 and Ald1b1 was calculated by plotting the threshold cycle (Ct) as a function of log10 concentration of the target sequences that were contained in the template plasmids carrying the cDNAs of the three genes (Sup. Fig 1; x-axis is plotted as a function of the −log pMol of target sequence). The slope of the trend line generated is a function of PCR efficiency, with a slope of −3.32 indicating 100% efficiency (Ballester et al., 2004). Linear regression analysis showed very good correlation coefficients and indicated that efficiency (Ballester et al., 2004) of the three primer pairs was high and very similar (Sup. Fig 1).
In situ antisense RNA hybridization
To generate an Aldh1b1 riboprobe plasmid, cDNA was generated from Ngn3 expressing ESTet-On/Ngn3 derived pancreas progenitor cells (Serafimidis et al., 2008) and PCR amplification (annealing temperature 61 0C, 35 cycles) using specific forward (5’GGCGAGTCTAGACTCAAGAGAGTCACC 3’) and reverse (5’CACTCTGGATCCAGCCACCG 3’) primers incorporating XbaI and BamHI sites, respectively was employed to amplify a 910 bp fragment of the Aldh1b1 cDNA. The cDNA was subsequently cloned in the respective sites of pBS SK+ and the antisense RNA probe was generated using standard procedures. RNA in situ hybridization and tyramide signal amplification were carried out as described (Serafimidis et al., 2011).
Mouse embryo pancreas dissections and immunofluorescence
Mouse pancreata were fixed, cryoprotected and cryosectioned at 12 µm using standard procedures and immunofluorescence was carried out as described (Serafimidis et al., 2011). Primary antibodies used were: rabbit anti–ALDH1B1 (1:500) (Stagos et al., 2010) rabbit anti-NGN3 (1:200; gift from H. Edlund), rat anti-E-CADHERIN (1:400; Zymed), rat anti-CYTOKERATIN 19 (anti-CK-19; 1:250; DSHB), mouse anti-INSULIN (1:1,000; Sigma), mouse anti-GLUCAGON (1:500; Sigma), mouse anti-BrdU (1:200 DAKO), mouse anti-PDX1 (1:300, DSHB), rabbit anti-AMYLASE (1:300, Sigma), rabbit anti-SOX9 (1:500; Chemicon), PNA-FITC lectin (1:100, Sigma), DBA-Rhodamine (1:100, Vector Laboratories). Secondary antibodies were anti-mouse, anti-rabbit, and anti-rat Alexa-488-, Alexa-633-, and Alexa-568-conjugated goat antibodies (1:500; Molecular Probes).
Organotypic cultures of embryonic pancreata
Dorsal pancreatic buds were dissected at 11.5 dpc and cultured for 2.5 or 6 days in a drop (40 µl) of DMEM/F12 (Gibco) supplemented with penicillin -streptomycin-glutamine (Gibco) and non-essential amino acids (Gibco). Diethylaminobenzaldehyde (DEAB) (Sigma) and / or RA were used at 100 µM or 25 nM respectively and the medium was renewed daily. BrdU (BD Biosciences) was used at 10 uM 2 hrs prior to fixation. Cultured pancreata were processed for immunofluorescence on cryosections using standard procedures. All sections from at least three pancreata for each experimental condition were used for quantitation. Images for each section were taken and analyzed as described (Serafimidis et al., 2011) to determine differentiation corresponding to each pancreatic lineage as well as BrdU incorporation for the mesenchymal and epithelial cell populations. Epithelial cells were defined as E-CADHERIN+ and mesenchymal cells as E-CADHERIN− cells.
Pancreatic injury and regeneration
Acute pancreatitis (AP) was induced by six hourly intraperitoneal caerulein (Sigma-Aldrich) injections at 50 µg/kg of body weight in 0.9% NaCl, following a fasting period of 12h, and pancreata were harvested 21 hrs after the last injection. For β cell ablation, streptozotocin (Sigma-Aldrich) was administered intraperitoneally at 50mg/kg of body weight following 12 hrs of fasting. Streptozotocin was dissolved in 50 mM NaCitrate pH 4.5 and was prepared fresh just before each injection.
Results
Aldh1b1 is expressed in the early pancreatic buds in a Pdx1 dependent manner
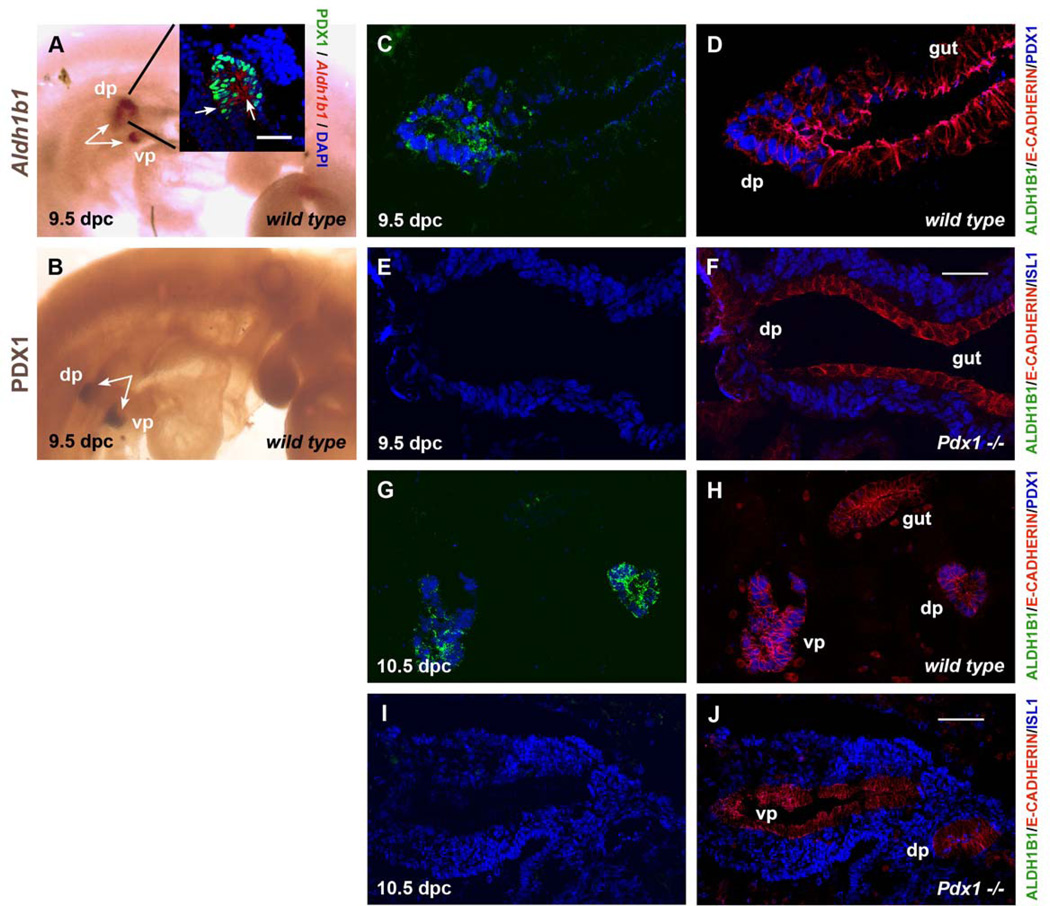
We first examined whether Aldh1b1 is expressed at the early bud stages of pancreas development using whole mount in situ hybridization. At e9.5, Aldh1b1 expression was similar to that of Pdx1, a homeobox gene expressed in the developing pancreas anlagen, and was exclusively and specifically expressed in both dorsal and ventral pancreatic buds (Fig. 1A, B). Combined Aldh1b1 in situ hybridization with PDX1 immunofluorescence analysis of transverse sections at the level of the developing pancreatic buds revealed that Aldh1b1 is expressed in all cells that have no contact with the mesenchyme (inner cell layers) and some cells of the external layer of the developing pancreatic buds (Fig. 1A, inset). Double Aldh1b1 in situ/ALDH1B1 immunofluorescence experiments confirmed antibody specificity as all ALDH1B1+ cells were expressing Aldh1b1 (Sup. Fig. 2A, B). Double immunofluorescence experiments showed that the mitochondrial ALDH1B1 and the transcription factor PDX1 were expressed in the same cells at both 9.5 dpc (Fig. 1C, D) and 10.5 dpc (Fig. 1G, H). Pdx1 expression is dispensable for induction of the pancreatic buds but absolutely essential for their subsequent development (Jonsson et al., 1994;Offield et al., 1996). We found that Aldh1b1 expression in the developing pancreatic buds was Pdx1 dependent as its expression was completely abolished in the Pdx1 null mutants at both 9.5 dpc (Fig. 1E, F) and 10.5 dpc (Fig. 1I, J). Thus Aldh1b1 is expressed in the early pancreatic buds that consist of multipotent pancreas progenitors in a Pdx1-dependent manner.
Figure 1. Aldh1b1 is expressed in the pancreatic buds in a Pdx1 dependent manner.
(A, B) In situ whole mount hybridization shows that Aldh1b1 is expressed in both the dorsal and ventral pancreatic buds (dp and vp, respectively) at 9.5 dpc (arrows in A) marked at the same time point by whole mount immunostaining for PDX1 (arrows in B). Combined in situ hybridization for Aldh1b1 expression and PDX1 immunostaining showed co-localisation (insert in A).
(C–F) Triple immunofluorescence at 9.5 dpc for ALDH1B1, E-CADHERIN and PDX1 or ISL1. Both ALDH1B1 and PDX1 proteins are present in the same cells of wt embryos (compare C and D). ALDH1B1 expression was completely lost in 9.5 dpc Pdx1 null embryos (E, F). In E, F mesenchymal cells are marked by ISL1 expression.
(G–J) Triple immunofluorescence at 10.5 dpc for ALDH1B1, E-CADHERIN and PDX1 or ISL1. ALDH1B1 and PDX1 proteins continue to be co-expressed in wt embryos (compare G and H). ALDH1B1 expression was completely lost in 10.5 dpc Pdx1 null embryos (I, J). In I, J mesenchymal cells are marked by Isl1 expression.
Scale bars: A (inlet), C–F 50 um; G–J 100 um.
Aldh1b1 is strongly expressed in the developing pancreatic epithelium and its expression in the trunk epithelium is Ngn3 dependent
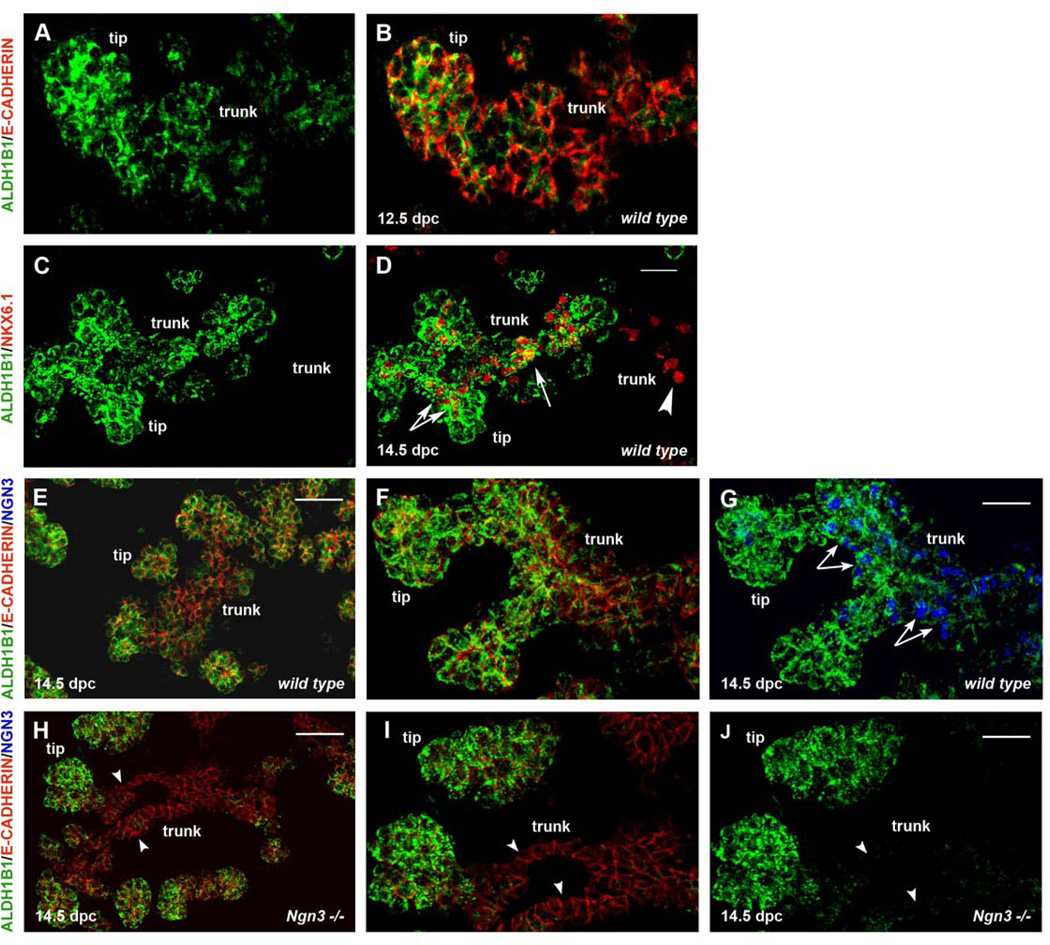
To examine Aldh1b1 expression at subsequent stages of pancreas development we performed in situ hybridization and immunofluorescence experiments at 12.5 and 14.5 dpc. ALDH1B1 expression persisted exclusively at the tips and the trunk of the developing pancreas epithelium at both stages, with expression being stronger at the tips. No expression was detected in the mesenchyme (Fig. 2A, B and 2C–G). Tripotent pancreas progenitors are located at the tips of the pancreatic epithelium up until 13.5 dpc when they become exocrine progenitors (Zhou et al., 2007). At 14.5 dpc NKX6.1+ endocrine / ductal progenitors and Ngn3+ endocrine progenitors are located in the trunk. Additionally, the presence of an NKX6.1+ tripotent progenitor population at the interface between the tip and trunk territories has been postulated (Kopp et al., 2011a). We found that the majority of NKX6.1+ cells were also ALDH1B1+ (86%, n=482). NKX6.1+ cells at the interface between tips and trunk epithelium were all ALDH1B1+, whereas most, but not all NKX6.1+ cells at the trunk were ALDH1B1+ (Fig. 2C, D). Aldh1b1 was initially identified as a gene downstream of Ngn3. Consistently, all trunk ALDH1B1+ cells were NGN3+ (Fig. 2E–G) and Aldh1b1 expression in the trunk was completely and selectively abolished in Ngn3-null pancreata (Fig. 2H–J). Accordingly, overall levels of Aldh1b1 expression were decreased in the Ngn3-null pancreata at 14.5 dpc (Fig. 3; Sup. Table 1). At 16.5 dpc ALDH1B1 expression was restricted at the tips, where exocrine progenitors are located, and its expression was completely abolished by the end of the gestation period (data not shown).
Figure 2. Aldh1b1 is expressed in tip and trunk pancreas progenitors and expression in the latter is Ngn3 dependent.
(A, B) Double immunofluorescence for ALDH1B1 and E-CADHERIN showed that ALDH1B1 is expressed exclusively in the 12.5 dpc pancreatic epithelium in both the tips and the trunks.
(C, D) Double immunofluorescence for ALDH1B1 and NKX6.1 at 14.5 dpc wt embryonic pancreata showed that most (arrows in D) but not all (arrowheads in D) NKX6.1+ trunk cells were also ALDH1B1+.
(E–G) Triple immunofluorescence experiments for ALDH1B1, E-CADHERIN and NGN3 14.5 dpc wt embryonic pancreata showed that all trunk ALDH1B1+ cells were also NGN3+ (arrows in G).
(H–J) Triple immunofluorescence experiments for ALDH1B1, E-CADHERIN and NGN3 at NGN3 null embryonic pancreata showed that expression of ALDH1B1 in the trunk is NGN3 dependent (arrowheads in H–J).
Scale bars: A–B, C–D, F–G, I–J 50 um; E, H 100 um.
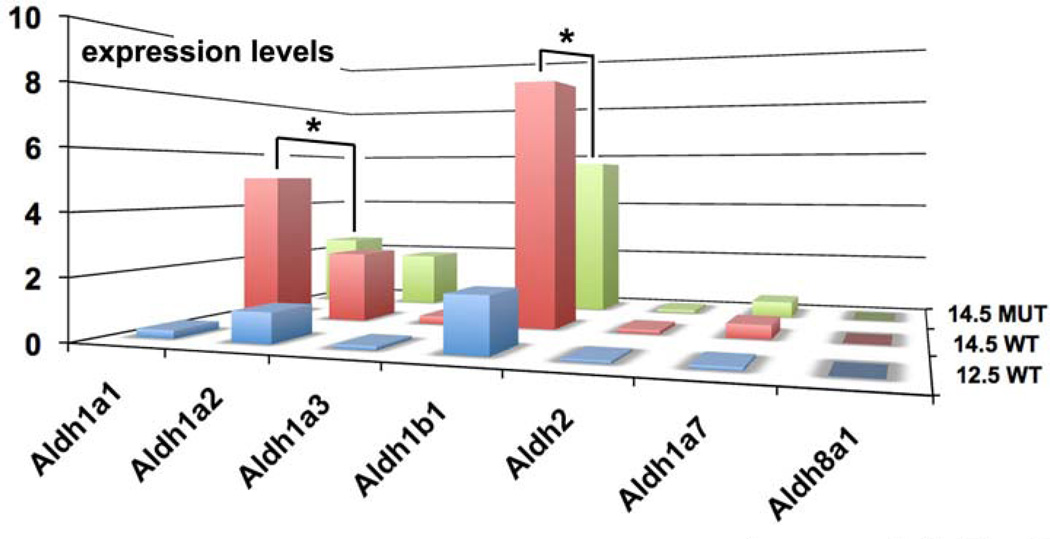
Figure 3. Aldh1b1 is strongly expressed in the developing pancreas compared to other Aldh genes.
Real Time PCR in 12.5 and 14.5 dpc wt and 14.5 Ngn3 null pancreata was performed for all known Raldh genes as well as for Aldh1b1 and the closely related Aldh2. Aldh1b1 appears to be expressed in higher levels compared to Aldh1a1 and Aldh1a2. Aldh1b1 and Aldh1a1 expression levels drop in 14.5 dpc Ngn3 null pancreata in a statistically significant manner (*, p < 0.05).
Whereas the role of Aldh1a2 in pancreas bud induction is through the supply of RA (Martin et al., 2005; Molotkov et al., 2005), the expression and possible role of other Aldh genes during subsequent stages of pancreas development remains obscure. Thus we compared expression levels of Aldh1b1 with expression levels of (a) Aldh2, a gene encoding an Aldh1b1 related mitochondrial ALDH that clears toxic aldehydes (Chen et al., 2008), (b) Raldh genes encoding enzymes necessary for RA generation in vivo such as Aldh1a1, Aldh1a2 and Aldh1a3 (Duester, 2008), (c) Aldh1a7, a gene encoding an enzyme that it may also act to clear toxic aldehydes (Hsu et al., 1999; Montplaisir et al., 2002) and (d) Aldh8a1, a gene that may also be encoding an RA generating enzyme (Lin et al., 2003). At 12.5 dpc only Aldh1b1, and to a lesser extent Aldh1a2, were expressed to appreciable levels (Fig. 3, Sup Table 1). At 14.5 dpc expression of both Aldh1a1 and Aldh1b1 was strongly induced and Aldh1a2 expression was slightly stronger compared to the earlier stage examined. Expression of Aldh1a2 most likely corresponded to mesenchymal cells (Tulachan et al., 2003) whereas, based on published data and the results presented above, Aldh1a1 and Aldh1b1 expression corresponded exclusively to the developing epithelium (Ostrom et al., 2008; Tulachan et al., 2003) (Fig. 3, Sup. Table 1). Aldh1b1 expression dropped substantially in the 14.5 dpc Ngn3 null pancreata reflecting the loss of the Aldh1b1 expressing NGN3+ endocrine progenitor cells and confirming that Aldh1b1 expression in the epithelium is partly Ngn3 dependent. Consistent with the epithelial expression of Aldh1a1 and epithelial alterations in the Ngn3 null embryonic pancreata (Magenheim et al., 2011) its expression also decreased significantly in the Ngn3 null pancreata. In contrast, and consistent with its postulated expression in the mesenchyme (Tulachan et al., 2003), expression of Aldh1a2 was not affected in the mutants (Fig. 3, Sup. Table 1). Primer efficiency was essentially the same for Aldh1b1 and Aldh1a1 primer pairs and efficiency of the Aldh1a2 primers was only slightly higher (Sup. Fig. 1).
Taken together, the data suggested that Aldh1b1 is expressed in different pancreas progenitor pools during the expansion and branching of the pancreatic epithelium. Its expression is at least comparable and most likely higher than that of Aldh1a1 and definitely higher than that of Aldh1a2 around the time of secondary transition, suggesting an important role in the development of the pancreatic epithelium.
ALDH activity is implicated in maintaining the progenitor status and balancing the relative development of the pancreas lineages
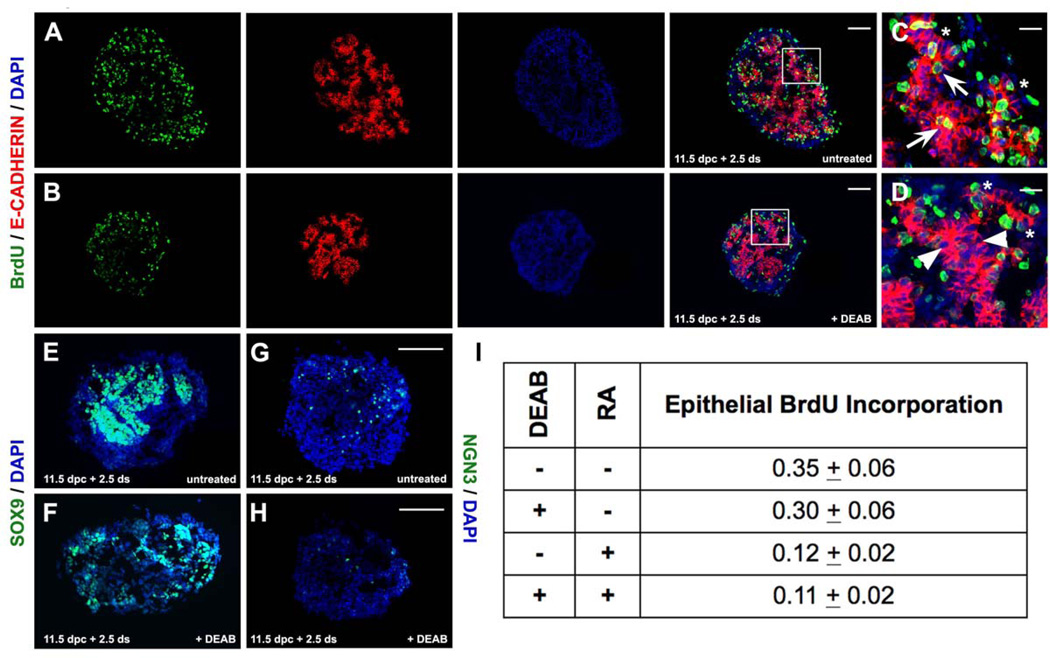
To address the role of ALDH enzymatic activity in progenitor cell expansion and/or cell lineage specification during pancreas development we used drop explant cultures of 11.5 dpc embryonic pancreata. Dorsal pancreata were dissected and cultured in a drop of defined medium for six days. At the end of that period, cells representing all three lineages developed reproducibly, suggesting that this approach could be used to assess the contribution of the ALDH enzymatic activity in early embryonic pancreas growth and differentiation. We blocked ALDH activity by including in the culture medium 100 uM DEAB, a specific competitive reversible inhibitor of ALDH enzymes (Marchitti et al., 2008). At the end of the six-day culture period, quantification of the DAPI signal indicated that DEAB treated pancreas explants (n=7) were reduced in size by half when compared to untreated ones (n=7, p<0.05). There was no evidence of excessive cell death (data not shown). To assess whether the size reduction was due to a reduction in mitotic rate we cultured 11.5 dpc pancreata for a period of 60 hrs and labeled mitotic cells in the presence of 10 uM BrdU 2hrs prior to fixation. We quantitated BrdU incorporation in the epithelium (E-CADHERIN+ domain) compared to the rest of the developing pancreas (E-CADHERIN− domain) by immunofluorescence. BrdU incorporation in the epithelium of pancreata cultured in the presence of 100 uM DEAB was preferentially reduced by approximately 15% whereas BrdU incorporation in the mesenchyme was not affected (Fig. 4A, B, I). Since DEAB is a universal inhibitor of ALDH activity (Marchitti et al., 2008), including RALDHs, we assessed whether the decrease in mitotic cells was due to inhibition of RA generation, by supplementing the explant cultures with 25 nM RA. Strikingly, the proliferation rate dropped even further with the addition of RA, suggesting that the DEAB effect was not through inhibition of RA generation. Inclusion of both DEAB and RA in the same explant cultures did not produce additional synergistic effects, suggesting that they act in independent, parallel pathways (Fig. 4I).
Figure 4. Blocking of ALDH activity in mouse embryonic pancreas explants reduces mitosis in the epithelium.
(A–D) Double immunofluorescence in untreated (A, C) and DEAB treated (B, D) pancreata for E-Cadherin and BrdU showed that BrdU incorporation apparent in the trunk epithelium of control pancreata (arrows in C) was greatly reduced in the trunk epithelium of DEAB treated pancreata. Areas marked by squares in A, B are represented in high magnification in C, D respectively.
(E, F) Immunofluorescence in untreated (E) and DEAB treated (F) pancreata showed a drop in SOX9 expression following DEAB exposure.
(G, H) Immunofluorescence in untreated (G) and DEAB treated (H) pancreata showed a drop in NGN3 expression following DEAB exposure.
(I) Epithelial BrdU incoroporation was reduced in DEAB treated pancreata. Inclusion of RA further reduced incorporation to levels similar by the inclusion of RA alone (p<0.02).
Scale bars: A–D, E–F, G–J 100 um; C–D 25 um.
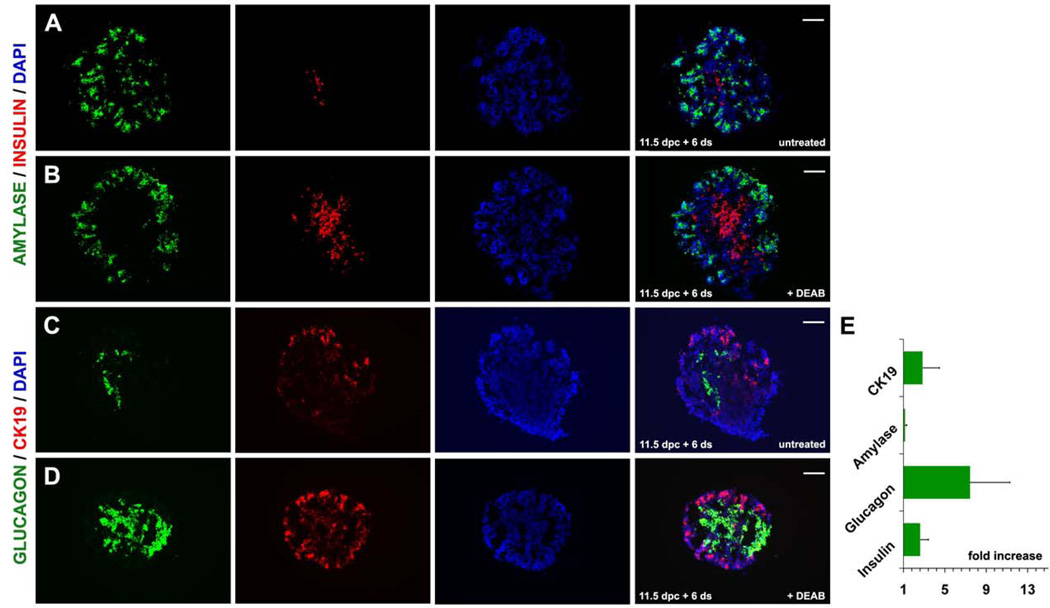
Interestingly, BrdU incorporation in the DEAB treated explants was concentrated at the developing tips of the epithelium (Fig. 4A–D) suggesting that loss of ALDH activity could be affecting preferentially the endocrine and ductal lineages emanating from the trunk epithelium. Accordingly, we then examined whether the reduction of the mitotic rate in the epithelium and primarily in the trunk resulted in precocious differentiation and whether it perturbed the relative balance among pancreas lineages. We indeed found that after 60 hrs in culture, expression of SOX9 was lower and expression of NGN3 was dramatically lower in treated compared to control pancreata (Fig. 4E–H). Furthermore, the relative number of the main endocrine cell subpopulations, insulin+ and glucagon+ cells, expanded by two and seven fold respectively, whereas the smaller SOMATOSTATIN+ population was not affected (Fig. 5A, B, E and data not shown). Surprisingly, expression levels of the endocrine hormone genes did not change appreciably in the DEAB treated pancreata suggesting complex effects of ALDHs on endocrine cell maturation. The number of ductal CK19+ cells also expanded by three fold (Fig. 5C–E). In contrast, the relative number of amylase+ cells did not change (Fig 5I, J, K). Thus, blocking ALDH enzymatic activity resulted in a reduction of the mitotic rate, and consequently explant size, and alteration of the relative balance of pancreatic lineages.
Figure 5. Blocking of Aldh activity in mouse embryonic pancreas explants accelerates differentiation and changes the relative linage distribution.
(A, B) Double immunofluorescence for AMYLASE and INSULIN in untreated (A) and DEAB treated (B) pancreata showed an increase in the number of INSULIN+ cells (depicted graphically in E).
(C, D) Double immunofluorescence for CK19 and glucagon in untreated (C) and DEAB treated (D) pancreata showed a strong increase in the number of GLUCAGON+ cells (depicted graphically in E).
Scale bars: A–D 100 um.
Rare ALDH1B1+ cells persist in the adult pancreas and expand vigorously in two different models of pancreas injury
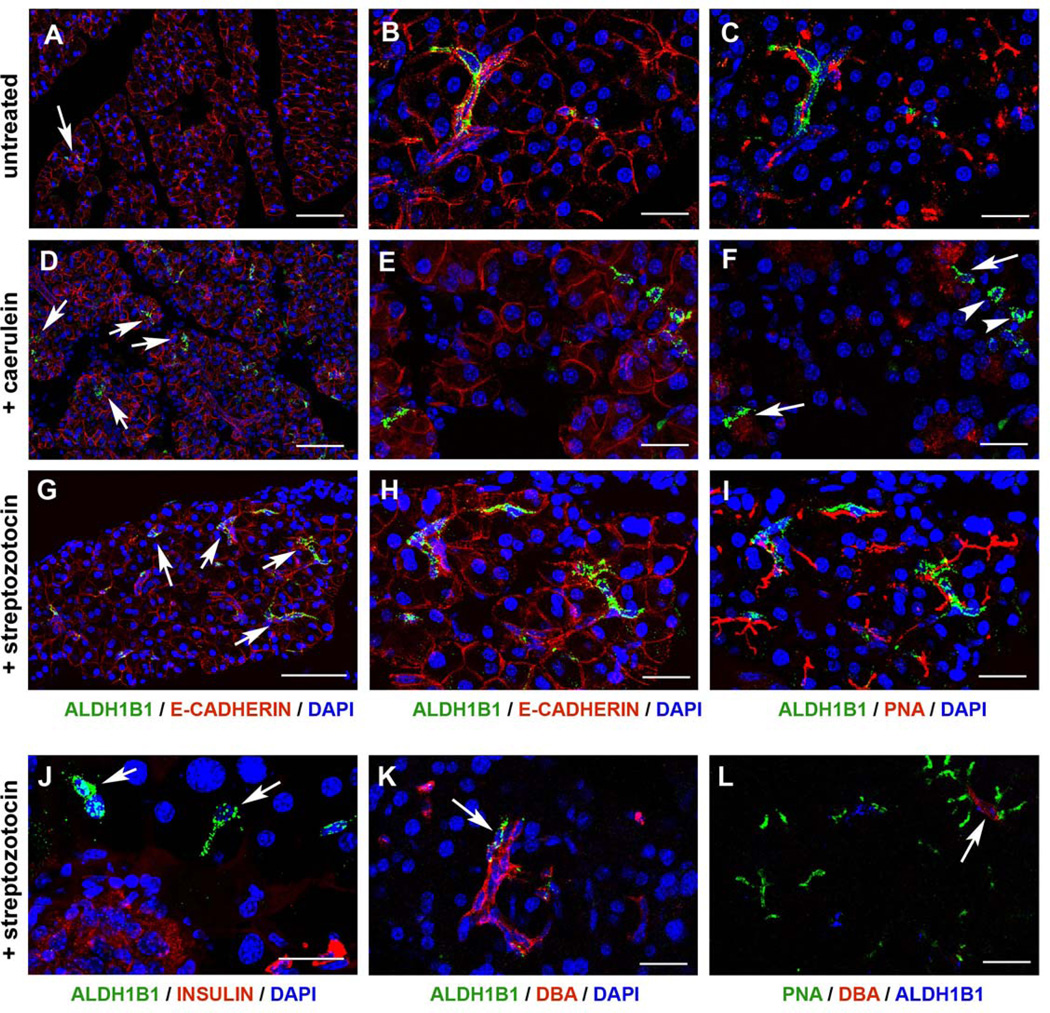
Strong Aldh1b1 expression ceases by birth (data not shown). However, expression persisted in the adult stages in very rare elongated cells that were PNA+/DBA− (93%, n=27) suggesting that they were centroacinar-like cells. We did not detect any ALDH1B1+/DBA+ cells (Fig. 6A–C) whereas very few ALDH1B1+ cells were PNA−/DBA− (7%, n=27).
Figure 6. Very rare ALDH1B1+ cells persist in the adult pancreas but their number expands vastly following either caerulein or streptozotocin insults.
(A–C) Triple immunofluorescence of wt, untreated pancreata showed the presence of rare elongated cells expressing ALDH1B1 (A–C, arrows in A), E-CADHERIN (A, B) and PNA (C).
(D–F) The number of ALDH1B1+ cells expanded by 20 – fold in response to acute pancreatitis as shown by triple immunofluorescence for ALDH1B1 (D–F, arrows in D), E-CADHERIN (D, E) and PNA (F). Most ALDH1B1+ cells were PNA+ (F, arrows) but some were PNA− (F, arrowheads).
(G–I) The number of ALDH1B1+ cells expanded by 10 – fold in response to β cell ablation as shown by triple immunofuorescence for ALDH1B1 (G–I, arrows in G), E-CADHERIN (G, H) and PNA (I).
(J–L) ALDH1B1+ cells occasionally surrounded β cell depleted islets (J, arrows). Occasionally, ALDH1B1+/DBA+ (K, arrow) or ALDH1B1+/DBA+/PNA+ (L, arrow) cells were observed.
Scale bars: A, D, G 100 um; B–C, E–F, H–I, J–L 25 um.
Genetic, surgical or pharmacological insults can induce pancreas regeneration characterized by upregulation of the expression of progenitor markers and variable degrees of tissue repair. Administration of caerulein results in excessive secretion and auto-activation of digestive enzymes as well as inflammation leading to extensive death of the acinar cells. Remarkably, the pancreas is able to recover from such an insult through regeneration and return to normal histology and function (Jensen et al., 2005; Strobel et al., 2007). We evaluated the behavior of the rare centroacinar-like ALDH1B1+ cells in the context of acute pancreatitis induced by successive caerulein injections. Pancreata were analyzed for ALDH1B1 expression 21 hrs after the last injection. Strikingly, and in contrast to the very low abundance of ALDH1B1+ cells in untreated adult pancreata (Fig. 6A), the number of ALDH1B1+ cells expanded approximately 20 fold following caerulein-induced pancreatitis (Fig. 6D). Additionally, the percentage of ALDH1B1+ cells that were either PNA+/DBA+ or PNA−/DBA− increased as compared to untreated controls (to 6% and 14% respectively, n=47). (Fig 6D–F and data not shown). ALDH1B1+ cells did not express either PDX1 or NKX6.1 (data not shown). However, consistent with the notion that these were progenitor cells, ALDH1B1+ cells did incorporate BrdU (Sup. Fig. 3A).
We also investigated whether an insult directed at the endocrine compartment had a comparable result. To ablate β cells we delivered streptozotocin intraperitoneally at 50mg/kg of body weight, once per day, for 3 consecutive days and pancreata were analyzed fourteen days after the last injection. Similarly to the caerulein treated pancreata, the number of ALDH1B1+ cells increased 10-fold following streptozotocin treatment (Fig. 6G). In contrast to caerulein treatment, streptozotocin treatment did not appaear to alter the percentages of ALDH1B1+ cells that were PNA+/DBA− or PNA−/DBA− (Fig. 6H, I, K, L). Some ALDH1B1+ cells did incorporate BrdU suggesting, that at least some ALDH1B1+ cells retained progenitor status (Sup. Fig. 3B), but ALDH1B1+ cells were not positive for either PDX1 or NKX6.1 (data not shown). Remarkably ALDH1B1+ cells were occasionally seen surrounding β cell depleted islets (Fig. 6J).
Taken together these data showed that very rare ALDH1B1+ cells persisted in the adult pancreas. The ALDH1B1+ cell population expanded greatly following two different insults ablating the acinar and β cell population suggesting that ALDH1B1 expression marks stem/progenitor cells in the adult pancreas.
Discussion
Deregulation of pancreatic homeostasis can lead to diabetes or pancreatic cancer, both diseases with very high prevalence and high associated human cost. Generating insulinogenic β cells from pluripotent stem cells or inducing pancreas regeneration from endogenous stem cells to replenish the pool of β cells could provide new effective treatments for diabetes. The existence of pancreatic stem cells and their contribution to cancer remains controversial. In the mouse, putative adult pancreas stem/progenitor cells were isolated as cells with high ALDH activity from the centroacinar compartment and had the capacity to generate endocrine and exocrine cells in culture (Rovira et al., 2010). Strikingly, experiments in a genetically engineered mouse model of pancreatic cancer suggested that cells in the same location might be the cells of origin of pancreatic cancer (Esposito et al., 2007; Guerra et al., 2007). The identification of specific marker(s) for these cells may help to devise strategies for β cell replenishment in diabetes or stopping pancreatic cancer progression.
Here we report the identification of a novel Aldh gene, Aldh1b1, expressed in the early embryo pancreatic buds and during subsequent development in different progenitor pools. Rare ALDH1B1+ cells persisted in the adult but their number expanded greatly after pancreas injury targeting either the β cell population or the acinar cell population. We propose that Aldh1b1 is a pancreas progenitor marker. The elucidation of its function may aid in devising strategies to stimulate in vivo expansion and differentiation of these cells.
The ALDH superfamily consists of proteins with complex expression patterns and diverse, often unknown, substrates (Jackson et al., 2011). ALDH catalytic activity, determined by Aldefluor assay, is being increasingly associated with stem / progenitor cell status (Balber, 2011) but also cancer stem cells (Alison et al., 2010). Lack of specific antibodies for distinct ALDH proteins has not allowed the unequivocal assignment of specific ALDHs to distinct stem / progenitor populations. Here we demonstrated that ALDH1B1 persists in pancreas progenitor pools during development and in rare cells in the adult pancreas. A possible role of ALDHs in stem/progenitor cells may relate to the regulation of reactive oxygen species (ROS) levels. Stem and progenitor cells need to precisely regulate levels of ROS, which may damage DNA but also regulate cell proliferation and differentiation in some types of tissue stem cells (Kobayashi and Suda, 2012). Oxidative stress resulting from the accumulation of ROS can be detrimental to stem cells as it may result to accumulation of DNA damage thus permanently impairing the function of terminally differentiated descendant cells. Thus, systems that neutralize ROS should be fine tuned according to the requirements of different specific stem cell type and ALDHs are part of the cellular response that protects against oxidative stress (Black et al., 2012; Brocker et al., 2010). Under physiological conditions, the principal source of ROS are the mitochondria (Balaban et al., 2005) and therefore, in pancreas progenitors, the mitochondrial ALDH1B1 could be participating in reducing the load of toxic aldehydes, possibly in conjunction with the cytosolic ALDH1A1, to levels compatible with stem / progenitor status.
So far, three Aldh genes have been found to be expressed during pancreas development. Aldh1a2 (aka Raldh2) encodes the main retinoic acid generating enzyme and plays a key role in the induction of the dorsal pancreatic bud in the mouse. At early stages it is expressed in the dorsal pancreatic mesenchyme, and signals to adjacent mesenchymal cells to induce Isl1 and endodermal cells to induce Prox1 and Pdx1 and acquire dorsal pancreatic bud fates. Lack of functional Aldh1a2 results in loss of the dorsal pancreatic bud and this can be rescued by maternal supplementation of RA (Martin et al., 2005; Molotkov et al., 2005). This function is evolutionarily conserved as loss of function mutations of the zebrafish Aldh1a2 resulted in a similar phenotype (Stafford and Prince, 2002). During subsequent developmental stages, Aldh1a2 is expressed at low levels in the mesenchyme but not in the developing epithelium (Tulachan et al., 2003). Aldh1a1 (aka Raldh1) is detected in the branching epithelium (Ostrom et al., 2008;Tulachan et al., 2003). Aldh1a1 null mice were viable and fertile and showed no overt pancreas related phenotype (Fan et al., 2003; Matt et al., 2005). Thus whereas RA is indispensable for the induction of the pancreas anlagen, its involvement in subsequent stages of pancreas development remains to be demonstrated. Our findings demonstrate that another Aldh gene, Aldh1b1, may be implicated in pancreas development. At 9.5 dpc it is exclusively and strongly expressed in both pancreatic buds. At subsequent stages its expression remains strong and, based on qPCR analysis, stronger than either Aldh1a1 or Ald1a2. Aldh1b1 nulls are viable and fertile with no obvious pancreas associated defects (Vasiliou V, Gavalas A unpublished).
Two models may account for the role of Aldh1b1 and Aldh1a1 in the development of the pancreatic epithelium. One postulates a sequential role for the corresponding proteins; the first promoting the progenitor state via as yet unknown mechanisms, and the second promoting differentiation, possibly through the local generation of retinoic acid. Supplementation of the explant cultures with physiological concentrations of RA promoted differentiation but this finding does not necessarily imply a role of RA in vivo. The other model would postulate that the two proteins overlap functionally, possibly participating in the ROS detoxification process. The inactivation of all ALDHs in pancreatic explant cultures by DEAB suggested that the net effect of ALDH activity is to promote the progenitor state, but it cannot exclude the possibility that ALDH1A1 and ALDH1B1 may function in opposing capacities to balance progenitor pool maintenance and differentiation. Elucidating the developmental mechanisms controlling this balance will provide valuable cues for the efficient differentiation of PS derived pancreas progenitors into mature endocrine cells. Given the absence of any overt pancreas associated phenotype in either the Aldh1a1 or the Aldh1b1 null mutants (Vasiliou V, Gavalas A unpublished), we favor the hypothesis that these two ALDHs overlap functionally.
It was surprising to find that qPCR reactions did not detect appreciable changes in the endocrine hormone transcripts in control and DEAB treated pancreata. There could be several explanations for this result. It is possible that although the numbers of endocrine cells are increased, the amount of transcript produced per cell may be reduced in response to DEAB treatment. Alternatively, blocking ALDH activity may alter endocrine cell maturation either by increasing the rates of hormone translation from pre-existing mRNA transcripts, or by stabilizing the already synthesized hormones and preventing their degradation. The exact mechanisms by which ALDHs exert their effects on endocrine cell maturation remain to be elucidated.
The existence of facultative stem / progenitor cells in the adult pancreas remains controversial. Extensive studies using lineage tracing have not pinpointed such a cell population. A possible caveat of these studies is that mostly markers of mature cells have been used for the lineage tracing, possibly excluding specialized progenitor cells. On the other hand combined expression of several transcription factors may define this subpopulation, making it very difficult to detect unequivocally (Kopp et al., 2011a). Recently, putative adult pancreas stem / progenitor cells were isolated from the interface of ducts with the exocrine compartment using FACS and the Aldefluor fluorescence assay that labels cells with ALDH enzymatic activity. These cells had the capacity to generate endocrine and exocrine cells in vitro and were shown to express certain stem / progenitor markers and three Aldh genes, namely Aldh1a1/a2/a7. The number of these cells greatly increased after caerulein treatment that induces pancreatitis and sets in motion a regenerative response (Rovira et al., 2010). Here we show the population of Aldh1b1+ cells expands greatly following injuries directed against either the endocrine or acinar compartments. In both cases cells are proliferative and mostly E-cadherin+ / PNA+ suggesting an origin from the acinar compartment. The relatively fast appearance of Aldh1b1+ cells following injury, particularly after caerulein treatment, suggests that they originate from cells that have dedifferentiated, rather than the preexisting small population of Aldh1b1+ cells. Furthermore, the expansion of this population in two different models of pancreatic injury suggests that Aldh1b1 is part of a general injury response in the adult pancreas and it will be important to identify the signals driving this expansion. Intriguingly, following caerulein treatment, and in contrast to streptozotocin treatment, ALDH1B+ that were either DBA+/PNA+ or DBA−/PNA− appeared suggesting a differential response of ALDH1B1+ cells to distinct insults. To evaluate the relative importance of ALDHs in pancreas progenitors and regeneration it will be important to assess which, if any, ALDH activity is required for the multipotency of the ALDH active cells and for the pancreas regenerative response.
ALDH1B1 appears unique among other ALDHs because it may continuously and specifically mark stem / progenitor cells in the developing and adult pancreas. Interestingly, ALDH1B1 has been identified as a potential colon cancer biomarker (Chen et al., 2011). Often, the same signaling pathways used in development are utilized during regeneration but also for cancer stem cell propagation. Having a single gene specifically expressed in stem cells during development, the mature organ and development of cancer will allow for the isolation and comparative analysis of the corresponding populations. This, combined with functional studies of Aldh1b1, may accelerate the discovery of approaches to enhance pancreas regeneration and halt the progression of pancreatic cancer.
Supplementary Material
HIGHLIGHTS.
Aldh1b1 was expressed in the early pancreatic buds in a Pdx1-dependent manner
Aldh1b1 persisting expression in the tips and trunks was partly Ngn3-dependent.
ALDH activity in pancreatic explants was necessary for growth and lineage allocation.
Rare centoacinar-like Aldh1b1+ cells persisted in the adult pancreas.
Aldh1b1+ cells increased dramatically after regeneration-inducing insults.
Acknowledgements
We would like to thank our colleagues for critically reviewing this manuscript. This work was supported by an EFSD / Eli Lilly grant to AG and in part by the National Institutes of Health grants R01 EY17963 and R21 AA017754 to VV and R01 DK082590 to LS. The authors have no conflicts of interest related to the work detailed within this manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alison MR, Guppy NJ, Lim SM, Nicholson LJ. Finding cancer stem cells: are aldehyde dehydrogenases fit for purpose? J Pathol. 2010;222:335–344. doi: 10.1002/path.2772. [DOI] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Balber AE. Concise review: aldehyde dehydrogenase bright stem and progenitor cell populations from normal tissues: characteristics, activities, and emerging uses in regenerative medicine. Stem Cells. 2011;29:570–575. doi: 10.1002/stem.613. [DOI] [PubMed] [Google Scholar]
- Ballester M, Castello A, Ibanez E, Sanchez A, Folch JM. Real-time quantitative PCR-based system for determining transgene copy number in transgenic animals. Biotechniques. 2004;37:610–613. doi: 10.2144/04374ST06. [DOI] [PubMed] [Google Scholar]
- Black W, Chen Y, Matsumoto A, Thompson DC, Lassen N, Pappa A, Vasiliou V. Molecular mechanisms of ALDH3A1-mediated cellular protection against 4-hydroxy-2-nonenal. Free Radic Biol Med. 2012;52:1937–1944. doi: 10.1016/j.freeradbiomed.2012.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocker C, Lassen N, Estey T, Pappa A, Cantore M, Orlova VV, Chavakis T, Kavanagh KL, Oppermann U, Vasiliou V. Aldehyde dehydrogenase 7A1 (ALDH7A1) is a novel enzyme involved in cellular defense against hyperosmotic stress. J Biol Chem. 2010;285:18452–18463. doi: 10.1074/jbc.M109.077925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Budas GR, Churchill EN, Disatnik MH, Hurley TD, Mochly-Rosen D. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science. 2008;321:1493–1495. doi: 10.1126/science.1158554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Orlicky DJ, Matsumoto A, Singh S, Thompson DC, Vasiliou V. Aldehyde dehydrogenase 1B1 (ALDH1B1) is a potential biomarker for human colon cancer. Biochem Biophys Res Commun. 2011;405:173–179. doi: 10.1016/j.bbrc.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole L, Anderson M, Antin PB, Limesand SW. One process for pancreatic beta-cell coalescence into islets involves an epithelial-mesenchymal transition. J Endocrinol. 2009;203:19–31. doi: 10.1677/JOE-09-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessimoz J, Opoka R, Kordich JJ, Grapin-Botton A, Wells JM. FGF signaling is necessary for establishing gut tube domains along the anterior-posterior axis in vivo. Mech Dev. 2006;123:42–55. doi: 10.1016/j.mod.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito I, Seiler C, Bergmann F, Kleeff J, Friess H, Schirmacher P. Hypothetical progression model of pancreatic cancer with origin in the centroacinar-acinar compartment. Pancreas. 2007;35:212–217. doi: 10.1097/mpa.0b013e31805d0190. [DOI] [PubMed] [Google Scholar]
- Fan X, Molotkov A, Manabe S, Donmoyer CM, Deltour L, Foglio MH, Cuenca AE, Blaner WS, Lipton SA, Duester G. Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Mol Cell Biol. 2003;23:4637–4648. doi: 10.1128/MCB.23.13.4637-4648.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama K, Kawaguchi Y, Akiyama H, Horiguchi M, Kodama S, Kuhara T, Hosokawa S, Elbahrawy A, Soeda T, Koizumi M, Masui T, Kawaguchi M, Takaori K, Doi R, Nishi E, Kakinoki R, Deng JM, Behringer RR, Nakamura T, Uemoto S. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet. 2011;43:34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- Gasparetto M, Sekulovic S, Zakaryan A, Imren S, Kent DG, Humphries RK, Vasiliou V, Smith C. Varying levels of aldehyde dehydrogenase activity in adult murine marrow hematopoietic stem cells are associated with engraftment and cell cycle status. Exp Hematol. 2012;0:0–0. doi: 10.1016/j.exphem.2012.05.014. [DOI] [PubMed] [Google Scholar]
- Gouzi M, Kim YH, Katsumoto K, Johansson K, Grapin-Botton A. Neurogenin3 initiates stepwise delamination of differentiating endocrine cells during pancreas development. Dev Dyn. 2011;240:589–604. doi: 10.1002/dvdy.22544. [DOI] [PubMed] [Google Scholar]
- Gradwohl G, Dierich A, LeMeur M, Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C, Schuhmacher AJ, Canamero M, Grippo PJ, Verdaguer L, Perez-Gallego L, Dubus P, Sandgren EP, Barbacid M. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Hebrok M, Kim SK, Melton DA. Notochord repression of endodermal Sonic hedgehog permits pancreas development. Genes Dev. 1998;12:1705–1713. doi: 10.1101/gad.12.11.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AM, Hale MA, Kagami H, Hammer RE, MacDonald RJ. Experimental control of pancreatic development and maintenance. Proc Natl Acad Sci U S A. 2002;99:12236–12241. doi: 10.1073/pnas.192255099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu LC, Chang WC, Hoffmann I, Duester G. Molecular analysis of two closely related mouse aldehyde dehydrogenase genes: identification of a role for Aldh1, but not Aldh-pb, in the biosynthesis of retinoic acid. Biochem J. 1999;339(Pt 2):387–395. [PMC free article] [PubMed] [Google Scholar]
- Inada A, Nienaber C, Katsuta H, Fujitani Y, Levine J, Morita R, Sharma A, Bonner-Weir S. Carbonic anhydrase II-positive pancreatic cells are progenitors for both endocrine and exocrine pancreas after birth. Proc Natl Acad Sci U S A. 2008;105:19915–19919. doi: 10.1073/pnas.0805803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson B, Brocker C, Thompson DC, Black W, Vasiliou K, Nebert DW, Vasiliou V. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum Genomics. 2011;5:283–303. doi: 10.1186/1479-7364-5-4-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JN, Cameron E, Garay MV, Starkey TW, Gianani R, Jensen J. Recapitulation of elements of embryonic development in adult mouse pancreatic regeneration. Gastroenterology. 2005;128:728–741. doi: 10.1053/j.gastro.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Kim SK, Melton DA. Pancreas development is promoted by cyclopamine, a hedgehog signaling inhibitor. Proc Natl Acad Sci U S A. 1998;95:13036–13041. doi: 10.1073/pnas.95.22.13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi CI, Suda T. Regulation of reactive oxygen species in stem cells and cancer stem cells. J Cell Physiol. 2012;227:421–430. doi: 10.1002/jcp.22764. [DOI] [PubMed] [Google Scholar]
- Kopinke D, Brailsford M, Shea JE, Leavitt R, Scaife CL, Murtaugh LC. Lineage tracing reveals the dynamic contribution of Hes1+ cells to the developing and adult pancreas. Development. 2011;138:431–441. doi: 10.1242/dev.053843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopinke D, Murtaugh LC. Exocrine-to-endocrine differentiation is detectable only prior to birth in the uninjured mouse pancreas. BMC Dev Biol. 2010;10:38. doi: 10.1186/1471-213X-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp JL, Dubois CL, Hao E, Thorel F, Herrera PL, Sander M. Progenitor cell domains in the developing and adult pancreas. Cell Cycle. 2011a;10:1921–1927. doi: 10.4161/cc.10.12.16010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp JL, Dubois CL, Schaffer AE, Hao E, Shih HP, Seymour PA, Ma J, Sander M. Sox9+ ductal cells are multipotent progenitors throughout development but do not produce new endocrine cells in the normal or injured adult pancreas. Development. 2011b;138:653–665. doi: 10.1242/dev.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Jordan N, Melton D, Grapin-Botton A. Signals from lateral plate mesoderm instruct endoderm toward a pancreatic fate. Dev Biol. 2003;259:109–122. doi: 10.1016/s0012-1606(03)00183-0. [DOI] [PubMed] [Google Scholar]
- Lin M, Zhang M, Abraham M, Smith SM, Napoli JL. Mouse retinal dehydrogenase 4 (RALDH4), molecular cloning, cellular expression, and activity in 9-cis-retinoic acid biosynthesis in intact cells. J Biol Chem. 2003;278:9856–9861. doi: 10.1074/jbc.M211417200. [DOI] [PubMed] [Google Scholar]
- Magenheim J, Klein AM, Stanger BZ, Ashery-Padan R, Sosa-Pineda B, Gu G, Dor Y. Ngn3(+) endocrine progenitor cells control the fate and morphogenesis of pancreatic ductal epithelium. Dev Biol. 2011;359:26–36. doi: 10.1016/j.ydbio.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4:697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Gallego-Llamas J, Ribes V, Kedinger M, Niederreither K, Chambon P, Dolle P, Gradwohl G. Dorsal pancreas agenesis in retinoic acid-deficient Raldh2 mutant mice. Dev Biol. 2005;284:399–411. doi: 10.1016/j.ydbio.2005.05.035. [DOI] [PubMed] [Google Scholar]
- Matt N, Dupe V, Garnier JM, Dennefeld C, Chambon P, Mark M, Ghyselinck NB. Retinoic acid-dependent eye morphogenesis is orchestrated by neural crest cells. Development. 2005;132:4789–4800. doi: 10.1242/dev.02031. [DOI] [PubMed] [Google Scholar]
- Molotkov A, Molotkova N, Duester G. Retinoic acid generated by Raldh2 in mesoderm is required for mouse dorsal endodermal pancreas development. Dev Dyn. 2005;232:950–957. doi: 10.1002/dvdy.20256. [DOI] [PubMed] [Google Scholar]
- Montplaisir V, Lan NC, Guimond J, Savineau C, Bhat PV, Mader S. Recombinant class I aldehyde dehydrogenases specific for all-trans- or 9-cis-retinal. J Biol Chem. 2002;277:17486–17492. doi: 10.1074/jbc.M112445200. [DOI] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Ostrom M, Loffler KA, Edfalk S, Selander L, Dahl U, Ricordi C, Jeon J, Correa-Medina M, Diez J, Edlund H. Retinoic acid promotes the generation of pancreatic endocrine progenitor cells and their further differentiation into beta-cells. PLoS One. 2008;3:e2841. doi: 10.1371/journal.pone.0002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri S, Hebrok M. Dynamics of embryonic pancreas development using real-time imaging. Dev Biol. 2007;306:82–93. doi: 10.1016/j.ydbio.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovira M, Scott SG, Liss AS, Jensen J, Thayer SP, Leach SD. Isolation and characterization of centroacinar/terminal ductal progenitor cells in adult mouse pancreas. Proc Natl Acad Sci U S A. 2010;107:75–80. doi: 10.1073/pnas.0912589107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rukstalis JM, Habener JF. Snail2, a mediator of epithelial-mesenchymal transitions, expressed in progenitor cells of the developing endocrine pancreas. Gene Expr Patterns. 2007;7:471–49. doi: 10.1016/j.modgep.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer AE, Freude KK, Nelson SB, Sander M. Nkx6 transcription factors and Ptf1a function as antagonistic lineage determinants in multipotent pancreatic progenitors. Dev Cell. 2010;18:1022–1029. doi: 10.1016/j.devcel.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafimidis I, Heximer S, Beis D, Gavalas A. GPCR signaling and S1P play a phylogenetically conserved role in endocrine pancreas morphogenesis. Mol Cell Biol. 2011;31:5702–5711. doi: 10.1128/MCB.05702-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafimidis I, Rakatzi I, Episkopou V, Gouti M, Gavalas A. Novel effectors of directed and Ngn3-mediated differentiation of mouse embryonic stem cells into endocrine pancreas progenitors. Stem Cells. 2008;26:3–16. doi: 10.1634/stemcells.2007-0194. [DOI] [PubMed] [Google Scholar]
- Seymour PA, Freude KK, Tran MN, Mayes EE, Jensen J, Kist R, Scherer G, Sander M. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc Natl Acad Sci U S A. 2007;104:1865–1870. doi: 10.1073/pnas.0609217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solar M, Cardalda C, Houbracken I, Martin M, Maestro MA, De Medts N, Xu X, Grau V, Heimberg H, Bouwens L, Ferrer J. Pancreatic exocrine duct cells give rise to insulin-producing beta cells during embryogenesis but not after birth. Dev Cell. 2009;17:849–860. doi: 10.1016/j.devcel.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Spooner BS, Walther BT, Rutter WJ. The development of the dorsal and ventral mammalian pancreas in vivo and in vitro. J Cell Biol. 1970;47:235–246. doi: 10.1083/jcb.47.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford D, Prince VE. Retinoic acid signaling is required for a critical early step in zebrafish pancreatic development. Curr Biol. 2002;12:1215–1220. doi: 10.1016/s0960-9822(02)00929-6. [DOI] [PubMed] [Google Scholar]
- Stagos D, Chen Y, Brocker C, Donald E, Jackson BC, Orlicky DJ, Thompson DC, Vasiliou V. Aldehyde dehydrogenase 1B1: molecular cloning and characterization of a novel mitochondrial acetaldehyde-metabolizing enzyme. Drug Metab Dispos. 2010;38:1679–1687. doi: 10.1124/dmd.110.034678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel O, Dor Y, Stirman A, Trainor A, Fernandez-del Castillo C, Warshaw AL, Thayer SP. Beta cell transdifferentiation does not contribute to preneoplastic/metaplastic ductal lesions of the pancreas by genetic lineage tracing in vivo. Proc Natl Acad Sci U S A. 2007;104:4419–4424. doi: 10.1073/pnas.0605248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulachan SS, Doi R, Kawaguchi Y, Tsuji S, Nakajima S, Masui T, Koizumi M, Toyoda E, Mori T, Ito D, Kami K, Fujimoto K, Imamura M. All-trans retinoic acid induces differentiation of ducts and endocrine cells by mesenchymal/epithelial interactions in embryonic pancreas. Diabetes. 2003;52:76–84. doi: 10.2337/diabetes.52.1.76. [DOI] [PubMed] [Google Scholar]
- Wells JM, Melton DA. Early mouse endoderm is patterned by soluble factors from adjacent germ layers. Development. 2000;127:1563–1572. doi: 10.1242/dev.127.8.1563. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Law AC, Rajagopal J, Anderson WJ, Gray PA, Melton DA. A multipotent progenitor domain guides pancreatic organogenesis. Dev Cell. 2007;13:103–114. doi: 10.1016/j.devcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.