Abstract
DNA rearrangements such as sister chromatid exchanges (SCEs) are sensitive indicators of genomic stress and instability, but they are typically masked by single-cell sequencing techniques. We developed Strand-seq to independently sequence parental DNA template strands from single cells, making it possible to map SCEs at orders-of-magnitude greater resolution than was previously possible. On average, murine embryonic stem (mES) cells exhibit eight SCEs, which are detected at a resolution of up to 23 bp. Strikingly, Strand-seq of 62 single mES cells predicts that the mm9 mouse reference genome assembly contains at least 17 incorrectly oriented segments totaling nearly 1% of the genome. These misoriented contigs and fragments have persisted through several iterations of the mouse reference genome and have been difficult to detect using conventional sequencing techniques. The ability to map SCE events at high resolution and fine-tune reference genomes by Strand-seq dramatically expands the scope of single-cell sequencing.
Genomic instability is a major driving force of tumor evolution and produces copy number variations (CNVs), mutations, loss of heterozygosity and aneuploidy1. The resulting genomic heterogeneity can give proliferative and survival advantages to subsets of cells that then undergo clonal expansion2. Though existing single-cell deep-sequencing techniques can identify clonal expansions by CNV signatures of individual tumor cells3, these signatures are a readout of past genomic events that have been propagated in a significant proportion of cells in the population. Insight into the mechanisms driving tumor evolution will require single-cell methods that more directly assess genome instability and genomic rearrangements.
SCEs are the result of double-strand breaks (DSBs) repaired by homologous recombination pathways, and their accumulation is an early indicator of genomic instability4. SCEs are a diagnostic phenotype for genotoxic stresses5 and cancer-prone genetic instability syndromes such as Bloom’s syndrome6. Despite the perceived importance of SCEs, it has not been possible to identify them in single cells using high-resolution sequencing approaches.
Here we report the development of Strand-seq, a single-cell sequencing technique that identifies the original parental DNA template strands in daughter cells following cell division. The method uses bromodeoxyuridine (BrdU) incorporation in the nascent strand during DNA replication followed by selective degradation of the nascent strand to isolate the template strand for construction of directional sequencing libraries.
Using Strand-seq, we identified and mapped SCEs in mES cells at a resolution orders of magnitude greater than was previously possible7,8. In addition, we identified aneuploidy events and CNVs in single mES cells arising from a single replication round. Notably, Strand-seq identified misoriented contigs and fragments in the current mouse reference genome assembly (mm9) that totaled nearly 25.57 Mb, or roughly 1% of the genome. SCEs and contig misorientations are undetectable using conventional sequencing techniques, thus highlighting the advantage of Strand-seq in identifying and characterizing genomic instability and in fine-tuning reference genome assembly. We also demonstrate that Strand-seq can be used to assay single-cell template-strand inheritance on a genome-wide scale. We anticipate that Strand-seq will be useful for haplotyping and detection of genomic rearrangements such as inversions and translocations that are more difficult to detect in the absence of directional information.
RESULTS
Strand-seq library construction and data visualization
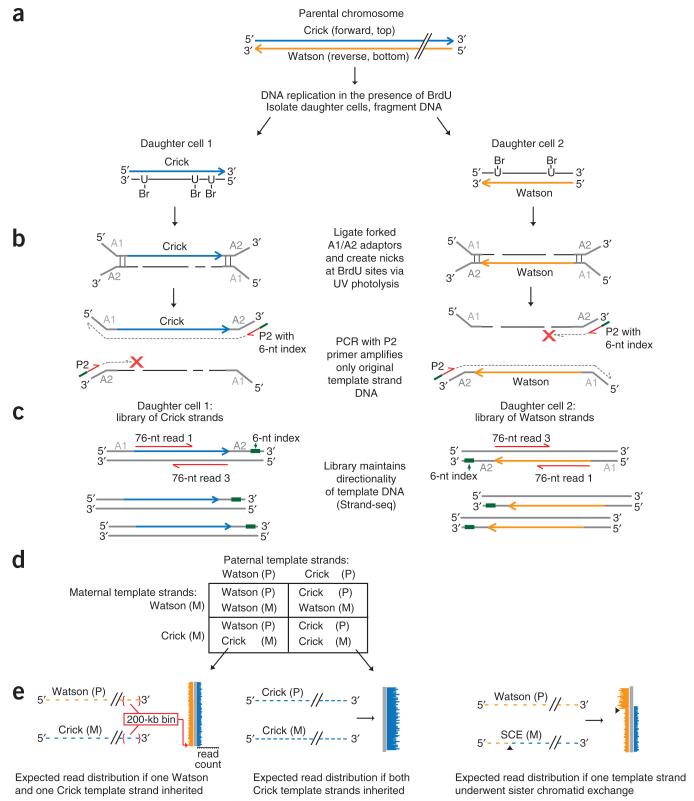
Strand-seq identifies parental DNA template strands in daughter cells following DNA replication and cell division. We previously designated these template strands as Crick or Watson, corresponding to the top (forward, plus) and bottom (reverse, minus) strands, respectively, in the mouse reference genome9 (Fig. 1a). To perform Strand-seq, we cultured C2 mES cells (from an inbred C57BL/6 background) in the presence of BrdU for one round of DNA replication to create hemi-substituted genomic DNA. We then sorted single daughter cells at the subsequent G1 stage of the cell cycle on the basis of the expression of a modified Fucci fluorescent cell-cycle reporter construct10 or by synchronization of the parental cells following G2 arrest11 (Supplementary Fig. 1). We fragmented the DNA by micrococcal nuclease digestion and performed custom-indexed Illumina library construction (Fig. 1a,b). Prior to PCR amplification, we nicked the newly formed BrdU-substituted strands by treatment with Hoechst 33258 and UV light. The subsequent PCR amplified only the original intact DNA template strand, resulting in libraries in which the original genomic directionality was maintained (Fig. 1b,c). This allowed us to identify the original parental template strands from paired short sequencing reads (Fig. 1c).
Figure 1.
Principle of single-cell DNA template strand sequencing. (a) Top: a single parental chromosome before DNA synthesis is shown with the Crick (blue) and Watson (orange) strands. Bottom: following DNA replication in the presence of BrdU, each sister chromatid with one original template strand and one complementary strand containing BrdU will segregate into one of the daughter cells. (b) DNA is fragmented and ligated to universal forked adaptors; UV photolysis creates nicks at BrdU sites, preventing PCR amplification of newly formed strands but allowing amplification of the original intact template strand. (c) The resulting libraries are directional, containing the template strand in its original genomic orientation in all amplified fragments. Multiple single-cell libraries containing unique 6-nucleotide (nt) index sequences (green lines in b) are pooled and sequenced on an Illumina platform. Two 76-nt reads from both directions (red lines) will read both the original template strand (always read 1) and the complementary strand (always read 3). (d) Possible combinations of maternal (M) and paternal (P) template strands inherited by daughter cells. (e) Expected read distribution from diploid Strand-seq libraries from inbred mouse cells. Watson and Crick reads (orange and blue lines) are binned and mapped to either side of a chromosome ideogram (gray). SCEs are expected to show a switch from both Watson and Crick reads to either Watson or Crick alone (right; arrowheads indicate SCE interval).
The nicking of BrdU-substituted DNA before PCR amplification is essential to identify parental template strands and renders Strand-seq incompatible with whole-genome amplification methods3. Strand-seq identifies parental template strands, which can be useful for haplotyping studies. However, the use of an inbred mouse strain precluded the identification of a parent of origin for any autosomal homolog in this study.
We constructed 66 indexed single-cell libraries from sorted cells (62 Strand-seq libraries and 4 standard whole-genome shotgun (WGS) libraries) that were checked for size distribution (Supplementary Fig. 2) and then pooled and sequenced on an Illumina platform (Fig. 1c and Online Methods). The number of sequence reads per library after quality filters were applied (see Online Methods) ranged from 60 to 1,457 reads per Mb, which translated to genomic coverage of 0.64%–6.46% for single-cell Strand-seq libraries (3.16% mean) and 4.8%–8.2% for WGS libraries (6.22% mean). The compiled genomic coverage of all 62 Strand-seq libraries was 65.56%, with ~30% of the genome covered by two or more reads. Pileups from these compiled libraries showed a periodicity consistent with nucleosomal fragments as input material (data not shown).
Each read aligned to either the forward or reverse direction of the reference genome, which corresponds to the original Crick and Watson strands, respectively. With the exception of the sex chromosomes, C2 mES cells from inbred mice have two identical parental homologs of each chromosome (Fig. 1d), and reads from the template strands of both homologs from a single cell mapped to the same reference chromosome. We binned aligned reads into nonoverlapping 200-kb segments and plotted these bins as colored horizontal lines along an ideogram of each chromosome (Fig. 1e). The length of these lines depends on the number of reads within the bin (Supplementary Fig. 3). If a daughter cell inherited both Crick template strands from both parental homologues, then only blue lines are shown. If both Watson and Crick template strands were inherited, then both blue and orange lines are shown (Fig. 1e). We identified SCEs resulting from mixing of template and newly formed strands during homologous recombination–based resolution of DSBs12 as points along the chromosome ideograms where reads mapping to both Watson and Crick strands switch to reads mapping to either the Watson or the Crick strand (Fig. 1e and Supplementary Fig. 4) while maintaining a consistent average read count (Supplementary Fig. 3).
High-resolution sister-chromatid-exchange mapping
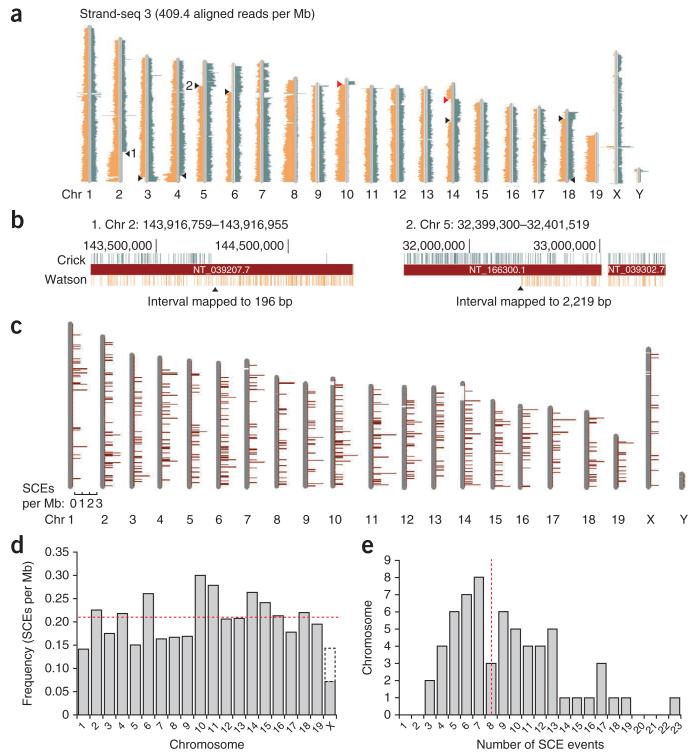
We mapped paired-end sequence reads from all Strand-seq and WGS libraries (Fig. 2a; Supplementary Data contains the ideograms of all 66 individual libraries). No-cell controls that underwent all steps of library construction averaged 17.1 reads per Mb, indicating few contaminating reads in our single-cell libraries (Supplementary Fig. 5). Within the 62 Strand-seq libraries, we identified SCE events and mapped each exchange interval (Fig. 2b and Online Methods). Because we could not distinguish between parental homologs in this inbred mouse strain, the resolution of the exchange region was an approximation. However, we expect it to be within an order of magnitude of our calculations because reads were distributed uniformly across the genome. Strand-seq of non-inbred strains or human cells will further improve the power of SCE analysis because single-nucleotide polymorphisms and haplotype mapping13,14 can help identify the parent of origin of the exchanged chromatid.
Figure 2.
DNA template strand libraries mapped to mouse chromosomes (chr) reveal SCEs. (a) Strand-seq library 3 shows Watson and Crick read distributions according to the template strands inherited from each parental homolog. SCEs (black arrowheads) are in the interval between reads that map to both Watson and Crick strands and reads which map to either strand alone. Complete switches from Watson-only to Crick-only template strand reads (red arrowheads) are potentially misoriented contigs in the reference genome (see Fig. 3). (b) The interval between Watson and Crick reads flanking the SCE can be estimated at the base-pair level in higher-resolution screenshots from the UCSC genome browser. SCE intervals 1 and 2 from a are 196 bp and 2,219 bp, respectively (see also supplementary Fig. 7). (c) All 529 SCE events in 62 Strand-seq libraries were placed into 1-Mb bins and mapped to ideograms of each chromosome. (d) Frequency of SCEs per megabase is normalized for each chromosome. The average frequency across the genome for these wild-type mES cells is 0.21 SCEs per Mb (dashed red line). Note that the actual frequency of SCEs in gray shading represents a diploid content for all autosomes and a haploid X. The SCE frequency for a diploid X (female cell) is extrapolated (white region). (e) Distribution of SCEs in Strand-seq libraries. All libraries contained at least three SCEs, with an average of eight SCEs per cell (dashed red line).
We binned SCEs into nonoverlapping 1-Mb regions and mapped them to chromosome ideograms (Fig. 2c). SCEs were distributed along the length of each chromosome, occasionally with multiple SCE events per chromosome (Supplementary Fig. 6). A total of 517 autosomal SCE events in the 62 Strand-seq libraries were mapped to all chromosomes at a frequency of 0.21 SCE events per Mb of sequence (Fig. 2d). Twelve chromosome X SCEs were also observed, which appeared as a complete switch from Watson to Crick reads as there is only one copy of X in these male cells (Supplementary Fig. 6). The 517 autosomal SCEs were evenly distributed across the genome (Fig. 2c) with no significant clustering or deserts at a variety of bin sizes as compared to a Poisson distribution background model (P = 0.2297 for 1-Mb bin size, data not shown). On average, eight SCEs per cell were identified (Fig. 2e), which corresponds with counts of spontaneous SCEs in wild-type mES cells in previously published cytogenetic studies15,16. Whereas SCE mapping resolution using cytogenetic banding is on the order of several megabases7,8, Strand-seq showed a median resolution of 5.97 kb, and one SCE event mapped to within 23 bp of the actual breakpoint (Supplementary Fig. 7). The high resolution of SCE interval mapping allows more detailed analysis of the sequences and genes surrounding the exchange interval (Supplementary Fig. 8).
Identifying misoriented regions in mm9 genome assembly
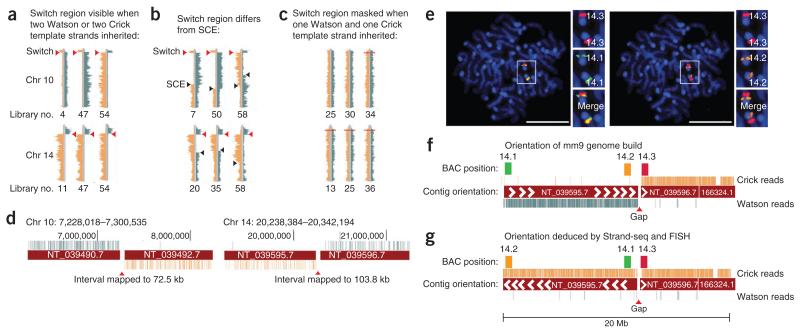
We observed a striking and complete switch in template strands at exactly the same interval in chromosomes 10 and 14 (Fig. 3a and Supplementary Data) in every library in which that region inherited both Watson or both Crick template strands (a total of 24 libraries for chromosome 10 and 27 libraries for chromosome 14). The switch from two Crick to two Watson template strands cannot be explained by SCEs or translocations, as the same event would have had to occur on both parental homologs at the same location, in multiple cells. A monosomy combined with an SCE such as that observed for chromosome X (Supplementary Fig. 6) could also be ruled out because we observed typical-looking SCE events on the same chromosomes exhibiting the switches (Fig. 3b). In addition, the average read depth for chromosomes 10 and 14 in all of these libraries did not support aneuploidy (Supplementary Data). Note that these switch regions are not evident if one Watson and one Crick template strand each were inherited by the daughter cell (Fig. 3c).
Figure 3.
Strand-seq identifies contig orientation errors in the mouse reference genome. (a) A complete switch from Watson to Crick reads is observed in chromosome (chr) 10 and 14 in all Strand-seq libraries in which two Watson or Crick template strands are inherited (red arrowheads). See supplementary data for full genomic ideograms of indicated libraries. (b) Switches are not due to a monosomy of these chromosomes because typical-looking SCE switches are also observed (black arrowheads). (c) Switches are not apparent if both Watson and Crick templates are inherited. (d) The interval between the switched reads always maps to the same unbridged gaps on chromosome 10 and chromosome 14 in the reference genome. (e) Metaphase mES cells hybridized simultaneously with three fluorescently labeled BAC probes 14.1 (green), 14.3 (red) and 14.2 (orange). Scale bar, 5 μm. (f) Signals from probes 14.1 and 14.3 should be distinct according to the orientation of contigs flanking the gap in the reference genome; however, the fluorescence signals overlap (e, left), whereas signals from probes 14.2 and 14.3 are distinct (e, right). (g) Corrected orientation of mm9 as inferred from Strand-seq and confirmed by FISH.
One possible explanation for these observations is that the orientation of the contigs nearest to the centromeres of chromosomes 10 and 14 was incorrectly assigned in the reference assembly. We found that in all cases, the template strand switches mapped to the same unbridged gaps between contigs in the mm9 reference genome for both chromosome 10 and 14 (Fig. 3d). Unbridged gaps are variable-sized regions of unknown sequence that are difficult to map because they contain complex segmental duplications and repetitive regions. Consequently, the relative orientations of contigs directly flanking these gaps have not been confirmed and are classified as unknown.
The mm9 genome build contains 186 unbridged gaps. To test whether Strand-seq can correctly predict misoriented contigs, we performed FISH17 using two BAC probes specific for genomic regions on either end of the chromosome 14 contig and a third BAC probe on the neighboring contig, which served as a reference point (Fig. 3e–g and Supplementary Fig. 9). Probes 14.3 and 14.1 are predicted to be 11.40 Mb apart in mm9, but the probe signals overlapped in our FISH analysis, suggesting adjacency (Fig. 3e,f). Probes 14.3 and 14.2 are predicted to be 0.64 Mb apart but showed distinct fluorescence signals, indicating that they are separated by at least several megabases and do not directly flank the gap as in the reference genome (Fig. 3e,f). The results of the FISH analysis of chromosome 10 are similar, thus supporting our hypothesis of contig orientation errors (Supplementary Fig. 9).
To confirm that these findings are not genomic rearrangements unique to the C2 background, we repeated FISH analysis in 3T3 murine fibroblasts with a Swiss albino genetic background and obtained identical results (Supplementary Fig. 9). These findings suggest that the orientation of the contigs NT_039490.7 on chromosome 10 and NT_039595.7 on chromosome 14 in mm9 should be reversed (Fig. 3g). We also observed smaller regions of complete template strand switches (Supplementary Fig. 10). In total, 17 contig fragments totaling nearly 1% of the genome are predicted to be incorrectly oriented according to Strand-seq (Table 1), ranging in size from 166.8 kb to 13.1 Mb (Supplementary Table 1). Most of these fragments are much smaller than the 2-Mb resolution limit of FISH.
Table 1.
Misoriented genomic regions of mm9 genome assembly
| Classification | No. of fragments |
Size (bp) | Proportion of genome (%) |
|---|---|---|---|
| Misoriented | 17 | 25,482,119 | 0.97 |
| Unknown orientation | 18 | 6,192,462 | 0.22 |
| Correctly oriented | 148 | 2,654,895,218 | 98.81 |
Orientations of genomic regions of mm9 genome assembly as classified by Strand-seq.
Comparison to previous releases of the mouse reference genome showed that some predicted fragment misorientations were corrected in subsequent assemblies, whereas others remain unresolved (Supplementary Fig. 11). We observed these misoriented fragments in every library with a Watson-only or Crick-only template-strand inheritance pattern in these regions, with no discrepancies (Supplementary Table 1a). We were unable to determine the orientation of 18 unbridged fragments (totaling 0.22% of the genome) because of poor coverage or complex segmental duplications that prevented strand-specific alignment of short sequencing reads in those regions (Supplementary Table 1b). This analysis confirms that the remaining 148 genomic fragments that flank unbridged gaps are correctly oriented in the reference genome, effectively ‘bridging’ these gaps. Of note, Strand-seq libraries reveal SCEs and misoriented fragments, whereas WGS libraries mask such features (Supplementary Fig. 12); Strand-seq is therefore a valuable tool for fine-tuning reference genome assemblies.
We were also able to detect genomic duplications and aneuploidy in both our Strand-seq and WGS libraries without PCR amplification of input material (Supplementary Fig. 13). The accumulation of aneuploidy is a well-known phenomenon in continually cultured mES cells18, and 17 of our 66 total libraries displayed at least one aneuploidy event (Supplementary Data). For example, one cell (library 4) showed a duplicated region in chromosome 4 as well as trisomy of chromosome 5 and monosomy for chromosome 10. These duplication and aneuploidy events were evident in both the Strand-seq and WGS library constructed from the same single cell (Supplementary Fig. 13), indicating that our libraries can assess genomic CNVs in single cells19 without the bias that could be introduced by PCR amplification of genomic DNA20.
DISCUSSION
Single-cell DNA template strand sequencing (Strand-seq) provides high-resolution maps of SCEs, identifies other indicators of genomic instability such as aneuploidy and CNVs, and identifies misoriented fragments in the mouse reference genome assembly. The contribution of SCEs to tumor heterogeneity is considered secondary to that of other chromosomal abnormalities such as translocations and CNVs, likely because SCEs are thought to be error-free recombination events ensuing from replication-fork collapse. However, unequal crossing over in SCEs can lead to CNVs, loss of heterozygosity and aneuploidy1. Importantly, a high number of SCEs is an indicator of accumulation of DSBs during replication, a symptom of replication stress due to collapsed replication forks, or the inability of the DNA repair pathways to suppress homologous recombination to repair DSBs (as in Bloom’s syndrome)5. Therefore, SCE mapping at high resolution will be a valuable contribution to the analysis of tumor evolution and the progression of genomic instability in replicating cells.
Although we cannot exclude the contribution of BrdU to the formation of DSBs or to the resolution of SCEs in our approach (nor in traditional cytogenetic assays of SCEs requiring two rounds of BrdU incorporation)5, Strand-seq can be used to finely map spontaneous SCEs in cells that undergo replication stress from genotoxic or chemotherapeutic agents, radiation, mutations in DNA repair and recombination pathways, or other genomic instability events. Unlike cytogenetic techniques, Strand-seq can provide in-depth analysis of fragile sites or other characteristics of genomic sequences surrounding breakpoint regions. In addition, the method requires only one mitotic cycle in the presence of BrdU, which is ideal for studies of SCE in vivo.
We have demonstrated that Strand-seq can be used to orient unbridged contigs that can occur in regions that are difficult to assemble, such as complex segmental duplications and repetitive regions. This study provides contig orientation information for 99.78% of the genome assembly from a relatively modest data set (Supplementary Fig. 10c). The importance of correctly oriented contigs is highlighted by disease association studies that rely on the correct location of markers to identify candidate genes—the results of which could be complicated by regions that are misoriented. In our study, the misoriented contig on chromosome 14 is large enough to show a discrepancy between physical and genetic map distance, which has been erroneously attributed to a break-down in linkage disequilibrium due to meiotic recombination21. It will be important to confirm the orientation of fragments in other genomes, including those flanking the 271 unbridged gaps present in the human genome.
Strand-seq is the ideal technique to study template strand inheritance in order to test nonrandom segregation of sister chromatids, as was proposed for chromosome 7 in mES cells22. However, the prevalence of SCEs as well as aneuploidy events in all the single cells that we sequenced prevented the assignment of Watson or Crick template strands for many chromosomes (Supplementary Fig. 14). Nevertheless, if we exclude these chromosomes from analysis, we find no deviation from a random segregation pattern for chromosome 7 in mES cells as judged by χ2 analysis (Supplementary Table 2 and data not shown). The occurrence of SCEs also suggests that it is not valid to use small probes to represent the template strands of entire chromosomes (as in recent template-strand segregation studies9,23) because the mixing of template and nontemplate strands in SCEs is ignored (Supplementary Fig. 14c). Furthermore, unless stem cells are demonstrated to completely suppress SCEs, it is not possible to claim completely asymmetric template-strand segregation to support, for example, the immortal strand hypothesis24,25.
Other expected applications of Strand-seq are the phasing of alleles to establish parental haplotypes13,14 and the mapping of inversions, translocations and other chromosomal abnormalities26,27 in single cells without using the large amounts of input material or the depth of sequencing currently required in existing sequencing approaches28,29. When one Watson and one Crick template strand is inherited from each parent, those strands are already phased because they originate from different parental chromosomes. We expect that Strand-seq will serve as a powerful tool to study genetic rearrangements in single cells during development, cancer and aging.
ONLINE METHODS
Cell culture
Undifferentiated wild-type murine embryonic stem cells (C2, C57BL/6 background) were cultured as described9. Murine embryonic fibroblasts were grown in DMEM-FCS. For preparation of metaphase cells, colcemid (Sigma-Aldrich, 0.1 μg/ml) was added 1 h before harvest. Trypsinized cells were treated with 0.075 M KCl for 10 min before fixation with 3:1 methanol/acetic acid using standard cytogenetic procedures. Fixed cells were stored at −20 °C.
A modified Fucci reporter construct was cloned by linking the cell-cycle reporters from the pFucci-G1 Orange and pFucci-S/G2/M expression vectors (MBL International) with a self-cleaving T2A peptide30. The Fucci construct was transfected into C2 cells using Effectene Reagent (Qiagen), and cells were selected using puromycin and repeated FACS sorting. Cycling between cell-cycle colors was confirmed by acquisition of time-lapse movies on a Coolsnap HQ digital camera attached to an inverted microscope (IX70 Olympus) fitted to a DeltaVision RT imaging system (Applied Precision) equipped with appropriate filter sets. Movies confirm ES-cell accumulation of mAG during the S, G2 and M stages of the cell cycle, punctuated by cytokinesis and followed by mKO fluorescence in the G1 daughter cells (data not shown). BrdU (Invitrogen) was added to semiconfluent cultures at a final concentration of 40 μM for 8–12 h before harvest.
G2 synchronization of mES cells
C2 ES cells alone or with the Fucci reporter construct were synchronized at the G2 phase by treatment with 10 μM (final) RO-33066 (ref. 11) for 4 h, which was followed by release into 40 μM (final) BrdU for 16 h.
FACS sorting and genomic DNA fragmentation
To analyze DNA content, 10 μg/ml Hoechst 33342 (Sigma-Aldrich) was added to the cell culture 30 min before harvest. The dye was also present in the FACS buffer. Cells were trypsinized, resuspended in phosphate-buffered saline with 2% FCS and sorted on a BD Influx cell sorter (BD Cytopeia) equipped with two tunable Coherent I305C argon lasers and a Cobolt Jive 50 561-nm diode laser.
Single cells were sorted directly into 100 μl lysis buffer (nuclei isolation buffer, NucleiEZ kit, Sigma) in flexible unskirted PCR plates (Bio-Rad) fitted into a rigid plate holder for sorting and spinning. Plates were immediately spun in a 4 °C prechilled centrifuge at 500g for 5 min to pellet nuclei. Plates were carefully removed from adaptors, and 90 μl cell-lysate supernatant was removed slowly and carefully using a long flexible gel-loading tip in order to avoid aspirating the nucleus. Next, 40 μl of 1.25× micrococcal nuclease (MNase) master mix (62.5 mM Tris-HCl pH 7.9, 6.25 mM CaCl, 0.03125 U/μl MNase enzyme, New England Biolabs) was added to each well containing a nucleus (as well as to no-cell negative-control wells containing only lysis buffer). Reactions were mixed 20–30 times using a pipettor and incubated at room temperature for 5 min. Reactions were stopped by adding 5.5 μl 100 mM EDTA (10 mM final) and mixing 20–30 times with a pipettor. The digested chromatin was transferred from the PCR plate into clean microcentrifuge tubes. Each well was rinsed with 100 μl buffer EB (Qiagen) and added to each tube. DNA was extracted by adding an equivalent amount (155 μl) of 25:24:1 ultrapure phenol:chloroform:isoamyl alcohol (Invitrogen) to each tube, mixing well and spinning at 13,000 r.p.m. for 5 min at room temperature in a benchtop microcentrifuge. Then 150 μl of the top aqueous layer containing extracted DNA fragments was removed to a clean microcentrifuge tube and precipitated with 0.1 vol. 3 M sodium acetate solution (Sigma-Aldrich) and 2.5 vol. 100% ethanol (EMD) with 1.5 μl linear polyacrylamide (GeneElute LPA, Sigma-Aldrich) added as a coprecipitant. Tubes were incubated at −20 °C for 20 min and centrifuged at 14,000 r.p.m. for 30 min. at 4 °C. Supernatant was carefully removed, and the pellet was washed once with 70% ethanol and then dried at room temperature. DNA was reconstituted in 20 μl EB for library construction.
DNA template strand library construction
Library construction for the Illumina sequencing platform was performed using a modified paired-end protocol (Illumina). This involved end-repair and A-tailing of fragmented DNA followed by ligation to Illumina PE adaptors and PCR amplification. At each step in the process, reactions were purified using either phenol:chloroform:isoamyl alcohol extraction followed by ethanol precipitation or solid-phase reversible immobilization paramagnetic beads (Agencourt AMPure, Beckman Coulter). 1 μM of Illumina PE adaptors were ligated to A-tailed DNA fragments at a final concentration of 33.5 nM for 15 min at room temperature using 5,000 units of Quick T4 ligase (New England Biolabs). Ligation products were purified using 0.8 vol. Agencourt AmpureXP magnetic beads (Beckman-Coulter) and eluted in 11 μl or 22 μl EB buffer (Qiagen). To create nicks in the BrdU substituted DNA strands, eluted DNA was incubated with 10 ng/μl Hoechst 33258 (Sigma-Aldrich) for 15 min at room temperature in clear 0.25-ml PCR tubes (Rose Scientific) protected from light. PCR tubes were then uncapped, and DNA was treated with UV for 15 min (the calculated dose was 2.7 × 103 J/m2). Nicked DNA was then used as a template for PCR using Phusion HF master mix (NEB) and primers PE 1.0 (Illumina) and a custom multiplexing PCR primer 5′-CAAGCAGAAGACGGCATACGAGATNNNNNNNCGGT CTCGGCATTCCTGCTGAACCGCTCTTCCGATCT-3′, where ‘NNNNNN’ was replaced with unique fault-tolerant hexamer barcodes. The PCR program was as follows: initial denaturation of 98 °C, 30 s; 15 cycles of (98 °C, 10 s; 65 °C, 30 s; 72 °C, 30 s); and final extension of 72 °C, 5 min. PCR products were purified using 0.8 vol. AmpureXP beads and eluted in 11 μl EB. 1 μl library was run on an Agilent High Sensitivity chip (Agilent) to check size distribution before pooling for sequencing.
Illumina sequencing
Libraries were pooled for sequencing, and the 200- to 400-bp size range was purified away from adaptor ligation artifacts on an 8% Novex TBE PAGE gel (Invitrogen). DNA quality was assessed and quantified using an Agilent DNA 1000 series II assay (Agilent) and Nanodrop 7500 spectrophotometer (Nanodrop) and subsequently diluted to 10 nM. The final concentration was confirmed using a Quant-iT dsDNA HS assay kit and Qubit fluorometer (Invitrogen). For sequencing, clusters were generated on the Illumina cluster station (GAIIx) or cBOT (Hiseq2000), and paired-end 76-nt reads were generated using v4 sequencing reagents on the Illumina GAIIx (v4) or Hiseq2000 (SBSxx) platform following the manufacturer’s instructions. Between the paired 76-nt reads, a third 7-bp read was performed using the custom sequencing primer 5′-GATCGGAAGAGCGG TTCAGCAGGAATGCCGAGACCG-3′ to sequence the hexamer barcode. Image analysis, base-calling and error calibration were performed using Illumina’s genome-analysis pipeline.
Bioinformatic analysis
Indexed paired-end .qseq files were aligned to the mouse reference genome (mm9) using bwa31, and custom scripts were used to split the resulting .bam files by index and to add the chastity flag. The resulting .bam files were sorted and filtered for duplicates (which removes both single-end and dual-end duplicates) and low-quality alignments (q < 20) using Samtools Version 0.1.10 (ref. 32). We developed a pipeline, BAIT (bioinformatic analysis of inherited templates), that parsed the bam files on the basis of the strand directionality assigned to each read. Reads that mapped to the ‘+’ strand from the first PET (paired-end tag) and the ‘−’ strand reads from the second PET were classified as Watson reads, and reads that mapped to the – strand from the first PET and the + strand from the second PET were classified as Crick reads. These data were plotted as separate histograms against ideograms of mouse chromosomes, with reads counted in 200-kb bins across each chromosome. Additional files in .bed format were plotted over the ideograms to represent sequence gaps and contig orientations. The number of reads mapping to Watson or Crick for each chromosome were summed, and the number of reads per megabase for each chromosome was calculated and printed below the ideograms. Normalized counts per megabase were determined by calculating the sum of both Watson and Crick reads for all autosomes and dividing by the length of the autosomes (in megabases). Any chromosomes in which read counts were 0.66× lower or 1.33× higher than the normalized count were classified as monosomies or trisomies, respectively. SCE events were defined as the interval in which there was a switch from reads mapping to both Watson and Crick strands to reads mapping to just one of the strands, without a corresponding change in the total number of reads such that the sum of Watson and Crick reads remained constant. Our criteria further stipulated that there must be ten consecutive Watson-only or Crick-only reads after the interval switch to count the switch as an SCE or to confirm fragment or contig orientation. To verify SCE and misorientation events, the SCE and misoriented contig interval coordinates were also converted to .bed files using BEDtools33 and uploaded to the UCSC genome browser to identify genomic features and genome build features, such as contigs, and to determine suitable BACs for FISH probes.
Fluorescence in situ hybridization analysis
Metaphase chromosomes from C2 ES cells and prematurely condensed chromosomes34 from murine 3T6 fibroblast cells were prepared and used for three-color FISH. BAC probes from chr 10 or chr 14 were labeled using a nick translation kit (Abbott Molecular) with Spectrum-Green dUTP (probe 10.1: RP23-38N9 and probe14.1: RP23-452I3), Spectrum-Orange dUTP (probe 10.2: RP23-128M21 and probe 14.2: RP23-154F13) and Red dUTP (probe 10.3: RP24-258P4 and probe14.3: RP23-255D5) according to manufacturer instructions. Hybridization and image analysis were performed as described previously17.
Fluorescence microscopy, image acquisition and selection
Fluorescence signals were captured on an Axioplan microscope (Zeiss) equipped with filters for DAPI, FITC, Cy3, Cy5 and Texas Red (Chroma Technology and Semrock) using an Axiocam MRm digital camera controlled by Metasystems ISIS software (Altlussheim). Alternatively, images were acquired on a Coolsnap HQ digital camera attached to an inverted microscope (IX70 Olympus) fitted to an imaging system (DeltaVision RT, Applied Precision) equipped with similar filter sets. Grayscale (12-bit) images at the wavelengths of interest were acquired through a high–numerical aperture 63×/1.4-N.A. or 60×/1.4-N.A. oil-immersion lens.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Brind’Amour and S. Rentas for discussions and J. Schein and C. Carter (Genome Sciences Centre) for BACs. We also thank K. Gan for help with preliminary MNase experiments. U.N. was supported by a Fellowship for Prospective Researchers from the Swiss National Science Foundation (project no. PBBEP3_131554). Work in the Hirst laboratory is supported by Canadian Institutes of Health Research grant RMF-92093. Work in the Lansdorp laboratory is supported by grants from the Canadian Institutes of Health Research (RMF-92093 and 105265), the US National Institutes of Health (R01GM094146) and the Terry Fox Foundation (018006). P.M.L. is a recipient of an Advanced Grant from the European Research Council.
Footnotes
Accession codes. Sequencing data have been deposited in the Sequence Read Archive: SRA055924.
Note: Supplementary information is available in the online version of the paper.
Methods and any associated references are available in the online version of the paper
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Bishop AJ, Schiestl RH. Homologous recombination and its role in carcinogenesis. J. Biomed. Biotechnol. 2002;2:75–85. doi: 10.1155/S1110724302204052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 5.Wilson DM, III, Thompson LH. Molecular mechanisms of sister-chromatid exchange. Mutat. Res. 2007;616:11–23. doi: 10.1016/j.mrfmmm.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 6.Wu L. Role of the BLM helicase in replication fork management. DNA Repair (Amst.) 2007;6:936–944. doi: 10.1016/j.dnarep.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 7.Kato H. Spontaneous sister chromatid exchanges detected by a BUdR-labelling method. Nature. 1974;251:70–72. doi: 10.1038/251070a0. [DOI] [PubMed] [Google Scholar]
- 8.Allen JW, Latt SA. Analysis of sister chromatid exchange formation in vivo in mouse spermatogonia as a new test system for environmental mutagens. Nature. 1976;260:449–451. doi: 10.1038/260449a0. [DOI] [PubMed] [Google Scholar]
- 9.Falconer E, et al. Identification of sister chromatids by DNA template strand sequences. Nature. 2010;463:93–97. doi: 10.1038/nature08644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakaue-Sawano A, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 11.Vassilev LT, et al. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. USA. 2006;103:10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helleday T. Pathways for mitotic homologous recombination in mammalian cells. Mutat. Res. 2003;532:103–115. doi: 10.1016/j.mrfmmm.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 13.Yang H, Chen X, Wong WH. Completely phased genome sequencing through chromosome sorting. Proc. Natl. Acad. Sci. USA. 2011;108:12–17. doi: 10.1073/pnas.1016725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan HC, Wang J, Potanina A, Quake SR. Whole-genome molecular haplotyping of single cells. Nat. Biotechnol. 2011;29:51–57. doi: 10.1038/nbt.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tateishi S, et al. Enhanced genomic instability and defective postreplication repair in RAD18 knockout mouse embryonic stem cells. Mol. Cell. Biol. 2003;23:474–481. doi: 10.1128/MCB.23.2.474-481.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaco I, Canela A, Vera E, Blasco MA. Centromere mitotic recombination in mammalian cells. J. Cell Biol. 2008;181:885–892. doi: 10.1083/jcb.200803042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trask BJ, et al. Studies of metaphase and interphase chromosomes using fluorescence in situ hybridization. Cold Spring Harb. Symp. Quant. Biol. 1993;58:767–775. doi: 10.1101/sqb.1993.058.01.084. [DOI] [PubMed] [Google Scholar]
- 18.Rebuzzini P, et al. Karyotype analysis of the euploid cell population of a mouse embryonic stem cell line revealed a high incidence of chromosome abnormalities that varied during culture. Cytogenet. Genome Res. 2008;121:18–24. doi: 10.1159/000124377. [DOI] [PubMed] [Google Scholar]
- 19.Liang Q, Conte N, Skarnes WC, Bradley A. Extensive genomic copy number variation in embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2008;105:17453–17456. doi: 10.1073/pnas.0805638105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalisky T, Quake SR. Single-cell genomics. Nat. Methods. 2011;8:311–314. doi: 10.1038/nmeth0411-311. [DOI] [PubMed] [Google Scholar]
- 21.Cox A, et al. A new standard genetic map for the laboratory mouse. Genetics. 2009;182:1335–1344. doi: 10.1534/genetics.109.105486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armakolas A, Klar AJ. Cell type regulates selective segregation of mouse chromosome 7 DNA strands in mitosis. Science. 2006;311:1146–1149. doi: 10.1126/science.1120519. [DOI] [PubMed] [Google Scholar]
- 23.Rocheteau P, Gayraud-Morel B, Siegl-Cachedenier I, Blasco MA, Tajbakhsh S. A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell. 2012;148:112–125. doi: 10.1016/j.cell.2011.11.049. [DOI] [PubMed] [Google Scholar]
- 24.Cairns J. Somatic stem cells and the kinetics of mutagenesis and carcinogenesis. Proc. Natl. Acad. Sci. USA. 2002;99:10567–10570. doi: 10.1073/pnas.162369899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lansdorp PM. Immortal strands? Give me a break. Cell. 2007;129:1244–1247. doi: 10.1016/j.cell.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 26.Kloosterman WP, et al. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum. Mol. Genet. 2011;20:1916–1924. doi: 10.1093/hmg/ddr073. [DOI] [PubMed] [Google Scholar]
- 27.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szymczak AL, et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat. Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bezrookove V, et al. Premature chromosome condensation revisited: a novel chemical approach permits efficient cytogenetic analysis of cancers. Genes Chromosom. Cancer. 2003;38:177–186. doi: 10.1002/gcc.10268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.