Abstract
Protein modification by SUMO (small ubiquitin-related modifier) has emerged as an essential regulatory mechanism in eukaryotes. Even though the molecular mechanisms of SUMO conjugation/deconjugation are conserved, the number of SUMO machinery components and their degree of conservation are specific to each organism. In the present paper, we show data contributing to the notion that the four expressed Arabidopsis SUMO paralogues, AtSUMO1, 2, 3 and 5, have functionally diverged to a higher extent than their human orthologues. We have explored the degree of conservation of these paralogues and found that the surfaces involved in E1-activating enzyme recognition, and E2-conjugating enzyme and SIM (SUMO-interacting motif) non-covalent interactions are well conserved in AtSUMO1/2 isoforms, whereas AtSUMO3 shows a lower degree of conservation, and AtSUMO5 is the most divergent isoform. These differences are functionally relevant, since AtSUMO3 and 5 are deficient in establishing E2 non-covalent interactions, which has not been reported for any naturally occurring SUMO orthologue. In addition, AtSUMO3 is less efficiently conjugated than AtSUMO1/2, and AtSUMO5 shows the lowest conjugation level. A mutagenesis analysis revealed that decreases in conjugation rate and thioester-bond formation are the result of the non-conserved residues involved in E1-activating enzyme recognition that are present in AtSUMO3 and 5. The results of the present study support a role for the E1-activating enzyme in SUMO paralogue discrimination, providing a new mechanism to favour conjugation of the essential AtSUMO1/2 paralogues.
Keywords: conjugation rate, E1-activating enzyme, E2-conjugating enzyme, non-covalent interaction, poly-small ubiquitin-related modifier (SUMO) chain, small ubiquitin-related modifier (SUMO)
INTRODUCTION
In plants, regulation of protein activity by SUMO (small ubiquitin-related modifier) attachment is a post-translational modification that has been shown to be essential during seed development and to have a major role in abiotic and biotic stress responses [1]. A common property between plants and animals is that the SUMOylation system appears to be a target for pathogenic effectors [2-5], as well as the accumulation of SUMO conjugates in response to heat and oxidative stresses [6,7]. But there are also biological processes specific to plants in which SUMO has a relevant role, such as flowering [8], phosphate starvation [9], drought responses [10] and the abscisic acid signalling pathway [11,12], a hormone that mediates plant responses to environmental stresses and a key regulator of plant growth and development.
SUMO is covalently attached to target proteins by the sequential action of E1-activating, E2-conjugating and E3-ligase enzymes [13]. SUMO activation is mediated by a heterodimeric enzyme consisting of a large subunit, SAE (SUMO-activating enzyme) 2, and a small subunit, SAE1. The SAE2 subunit contains the adenylation, catalytic cysteine, UFD (ubiquitin fold) and C-terminal functional domains [14]. The adenylation domain is responsible for SUMO recognition and adenylation of the C-terminus of SUMO. After adenylation, the catalytic cysteine thiol group attacks the SUMO C-terminal adenylate resulting in the formation of a thioester bond between the E1 and SUMO, in a mechanism that involves a rotation of the cysteine domain [15]. At this stage, SUMO can be transferred to the E2-conjugating enzyme. SUMO-charged E2 is competent to transfer SUMO to the target lysine residue in the substrate, although this reaction is facilitated by E3-ligase enzymes in vitro and in vivo [16,17].
During conjugation, SUMO molecules establish non-covalent interactions with the E1-activating and the E2-conjugating enzymes. Structural studies have identified eleven residues on the HsSUMO (human SUMO) surface that establish contacts with the E1-activating enzyme and which, presumably, are responsible for conferring modifier specificity [14]. SUMO can be attached to the target protein as a monomer or polymer, and polySUMO chains have been shown to act as signals to promote ubiquitination of the SUMO-modified substrate targeting it for proteasomal degradation [18]. SUMO chain growth is dependent on the presence of a SUMOylation consensus site at the SUMO N-terminal tail, and polymerization is facilitated by non-covalent interactions between SUMO and the E2-conjugating enzyme [19,20]. In vitro, the chain length is modulated by the relative abundance of HsSUMO2/3, which can build SUMO chains, compared with HsSUMO1, which does not have a SUMOylation consensus site and that could function as a chain terminator [21].
The consequences of covalent SUMO attachment to target proteins are very variable and include regulation of subcellular localization, protein activity and stability, and protein–protein interactions. At the molecular level, the SUMOylation outcome is achieved through the interaction with specific effectors that contain a SIM (SUMO-interaction motif). Most SIMs consist of a hydrophobic core of three to four aliphatic residues flanked by acidic residues [22,23]. Structural and functional studies determined that a hydrophobic groove surrounded by basic residues in SUMO is crucial for SIM interaction. Interestingly, the relative position between the hydrophobic groove and the basic residues differ among HsSUMO isoforms and this could confer SUMO–SIM interaction specificity [24,25]. During conjugation, SUMO paralogue selection can be mediated by SIM-dependent recruitment of targets to SUMO thioester-charged E2 and/or SUMO-modified E2 [26,27]. Moreover, SIMs have been identified in E3 ligases and shown to regulate ligase activity and localization [28–30].
In plants, much less is known about the molecular mechanisms that regulate SUMOylation, and the complexity of the SUMOylation components is apparently higher when compared with other organisms. In Arabidopsis, expression has been detected for the four SUMO paralogues AtSUMO1, AtSUMO2, AtSUMO3 and AtSUMO5. Among them AtSUMO1 and 2 are the most closely related isoforms. Previous studies have shown that Arabidopsis SUMO paralogues do not serve as equivalent substrates of AtSUMO proteases, referred to as ULP (ubiquitin-like protein-specific protease). The four proteases AtULP1a, c, d and AtESD4 (Arabidopsis early is short days 4) displayed similar peptidase and isopeptidase activities towards AtSUMO1 and AtSUMO2 isoforms, although none of them showed a significant activity towards AtSUMO5. Only AtULP1a exhibited a poor peptidase activity towards AtSUMO3 and no isopeptidase activity at all [31–33]. In addition, in non-quantitative assays, only AtSUMO1, 2 and 3 were shown to be conjugated to the yeast substrate PCNA (proliferating-cell nuclear antigen), and the capacity to form polymeric chains was displayed exclusively by AtSUMO1 and AtSUMO2 [32]. At first glance, it seemed that the situation of the AtSUMO1 and 2 isoforms resembles that of the human SUMO2 and 3 isoforms according to their capacity to form polymeric chains, as opposite to the HsSUMO1 and the AtSUMO3 isoforms that are conjugated as monomers. But, in contrast, homology studies failed to cluster human and Arabidopsis isoforms according to their ability or inability to polymerize [11,32]. In addition, AtSUMO3 and 5 are not capable of complementing the lethal double-mutant atsumo1atsumo2 plants [34], whereas SUMO1-knockout mice are viable. These results suggest that mammalian SUMO1 and SUMO2/3 have partially redundant functions [35,36], in contrast with AtSUMO paralogues that seem to have developed more divergent functions [34,37].
To gain new insights into the complex Arabidopsis SUMOylation system, we have assessed whether AtSUMO1, 2, 3 and 5 have distinct molecular properties that might influence their in vivo conjugation and biological function. We have found that Arabidopsis SUMO isoforms have heterogeneous properties at different molecular levels. AtSUMO1 and 2 were competent to interact non-covalently with their cognate E2-conjugating enzyme AtSCE1 (Arabidopsis SUMO-conjugating enzyme 1), whereas AtSUMO3 and AtSUMO5 did not conserve this property. Mutagenesis analysis revealed that the single residue Asp63, conserved in the AtSUMO1/2 surface but not in AtSUMO3 and AtSUMO5, is essential for non-covalent interactions with E2, and that it is necessary for polySUMO chain formation. It is even more significant the fact that SUMO isoforms differed in their conjugation rate, AtSUMO1/2 being the most efficiently conjugated paralogues, AtSUMO3 was less efficiently conjugated and AtSUMO5 showed the lowest conjugation level. A mutagenesis analysis showed that the lower conjugation rates of AtSUMO3 and AtSUMO5 were related to changes in the SUMO residues involved in the E1 interaction, which also affected thioester-bond formation. These results suggest that the first step in the SUMO-conjugation cascade would have a regulatory role in SUMO paralogue discrimination. Overall, the results of the present study suggest that AtSUMO1/2 might be the most efficiently conjugated SUMO isoforms in vivo, and we postulate that this could constitute a molecular mechanism to assure conjugation of the essential AtSUMO1/2 paralogues compared with the non-essential AtSUMO3 and 5.
EXPERIMENTAL
Cloning, expression and protein purification
AtSUMO isoforms, AtSAE2, AtSAE1a/b and AtCAT3 (Arabidopsis catalase isoform 3) were amplified by PCR from cDNA obtained from 2-week-old plants grown on MS (Murashige and Skoog) plates under LD (light/dark) at 22°C (Superscript® III reverse transcriptase from Invitrogen and Pfu DNA polymerase from Stratagene). AtSAE1a/b were cloned into pET15b (Novagen) to encode a native polypeptide, and AtSAE2, AtSCE1 [11], AtSUMO1-(1–93), AtSUMO2-(1–92), AtSUMO3-(1–93) and AtSUMO5-(1–103) were cloned into pET28a (Novagen) to encode an N-terminal His6-fusion protein. AtCAT3Ct-(419–472) (Ct is C-terminal tail) was cloned into pGEX-6P1 (Amersham) to encode an N-terminal GST (glutathione transferase)-fusion protein. Plasmids were transformed individually, or co-transformed in the case of AtSAE2- and AtSAE1a/b-containing plasmids, into Escherichia coli strain BL21 Codon Plus RIL (Stratagene). Cultures (1–4 litres) were incubated at 37°C until they reached an A600 of 0.6–0.8, and protein expression was induced by adding 0.1 mM IPTG (isopropyl β-D-thiogalactopyranoside) for 4 h at 30°C. Cells were harvested and resuspended in 50 mM Tris/HCl (pH 8.0), 20% (w/v) sucrose, 350 mM NaCl, 20 mM imidazole, 0.1% Nonidet P40, 1 mM PMSF, 1 mM 2-mercaptoethanol, 1 μg/ml leupeptin, 1 μg/ml pepstatin and 50 μg/ml DNAse. Protein extracts were prepared by sonication [five pulses of 1 min and 2–3 power setting (Sonifier 250, Branson)] and clarified by centrifugation (18000 g for 30 min at 4°C). Purification via IMAC–Sepharose resin (GE Healthcare) or glutathione–Sepharose (GE Healthcare) was performed according to the manufacturer’s instructions. SDS/PAGE analysis of the purified proteins is shown in Supplementary Figure S1 (at http://www.BiochemJ.org/bj/436/bj4360581add.htm).
Yeast two-hybrid experiments
Yeast expression constructs pGBKT7:AtSCE1/AtUBC10 and pGADT7:AtSUMO1-(1–93)/AtUBI have been generated previously [11]. AtSUMO3-(1–93) and AtSUMO5-(1–103) were cloned into pGADT7 AD (Clontech) to encode an N-terminal GAL4-activation domain fusion protein. AtSUMO1 and AtSUMO3 mutant alleles were generated by QuikChange® site-directed mutagenesis (Stratagene). Plasmids, as indicated, were co-transformed into the yeast strain HF7c using the lithium acetate method as described in the Clontech Yeast Protocols Handbook. Transformed yeast culture was plated on to permissive SD (synthetic dextrose) medium complemented with histidine. A single clone per transformation was selected, disaggregated by vortex agitation in SD medium without amino acids, and serial dilutions were performed (1, 1:8, 1:32 and 1:64). Aliquots (5 μl) of each dilution were sowed on non-selective (SD medium complemented with histidine) or selective (SD medium not complemented with histidine) plates. After incubation for 2 days at 30°C, protein interactions were analysed using histidine auxotrophy as a selective marker.
In vitro pull-down assay
His–AtSCE1 (100 μM) and AtSUMO1/D63N (25 μM) were incubated in 40 μl of binding buffer [20 mM Tris/HCl (pH 8.0), 50 mM NaCl and 20 mM imidazole] for 5 h at 4°C. Next, 10 μl of Ni2+-IMAC–Sepharose resin was added to the binding mixture, and the mixture was incubated for 30 min at 4°C. The binding mixture was transferred to micro bio-spin chromatography columns (Bio-Rad) and the resin was washed four times with 20 μl of binding buffer. The proteins bound to the resin were eluted with 20 μl of binding buffer containing 300 mM imidazole. The input (0.8 μl) and eluate (3.5 μl) fractions were separated by SDS/PAGE and either stained with Coomassie Fluor Orange (Molecular Probes, C-33250) or subjected to immunoblot analysis with anti-SUMO1 antibodies, as indicated.
Polyclonal anti-AtSUMO1 antibody production
Polyclonal antibodies were raised against the purified N-terminal His6-fusion protein of AtSUMO1-(1–93). Purified protein (1 mg) was resolved by SDS/PAGE, the gel was stained with Coomasie Blue and the gel slice containing His–AtSUMO1 was used to immunize rabbits (Cocalico Biological). A 1:1000 dilution of the serum produced was used in the immunoblot analyses.
In vitro SUMO conjugation, polymeric chain formation and E1-thioester assays
In conjugation assays, we used the C-terminal tail of the AtCAT3 (residues 419–472) fused to GST, GST–AtCAT3Ct. Reactions were carried out at the indicated temperatures in 25 μl reaction mixture volumes containing 1 mM ATP, 50 mM NaCl, 20 mM Hepes (pH 7.5), 0.1% Tween 20, 5 mM MgCl2, 0.1 mM DTT (dithiothreitol), 2 μM SUMO, 0.5 μM AtSAE2/AtSAE1a, 0.5 μM AtSCE1 and 5 μM GST–AtCAT3Ct. After the specified incubation time, reactions were stopped by the addition of protein-loading buffer, boiled for 5 min and 10 μl aliquots were resolved by SDS/PAGE. Polymeric chain formation reactions were performed at 37°C in the same reaction buffer as SUMO-conjugation assays and in the presence of 100 μM AtSUMO1, 1 μM AtSAE2/AtSAE1a and 10 μM AtSCE1. Reaction products were detected by immunoblot analysis with anti-GST polyclonal anti-bodies (Sigma, G7781) or with anti-AtSUMO1 polyclonal antibodies, as indicated. E1-thioester assays were performed at 30°C in 50 μl reaction mixture volumes containing 1 mM ATP, 50 mM NaCl, 20 mM Hepes (pH 7.5), 0.1% Tween 20, 5 mM MgCl2, 0.1 mM DTT, 10 μM SUMO and 5 μM AtSAE2/AtSAE1a. At the indicated time points, 15 μl aliquots were removed and analysed by SDS/PAGE followed by Coomasie Fluor Orange staining according to manufacturer’s protocol (Molecular Probes, C-33250). As a thioester-bond formation control, an aliquot of each reaction was treated with 100 mM DTT before loading on to polyacrylamide gels.
SUMO-conjugation rate quantification
Reaction products were detected using ECL (enhanced chemiluminescence) and Western blot detection reagents (GE Healthcare), and the signal was acquired with the LAS-3000 imaging system and quantified with Multi Gauge V3.0 (Fujifilm). Signals were normalized against known amounts of GST included in each blot. When data are represented by relative units, SUMO-conjugation or E1-thioester rates are referred to the average value calculated using all the rates obtained in each independent experiment.
Bioinformatics
Sequence alignments were performed using the protein multiple alignment software MUSCLE [38] and alignments were edited with GeneDoc software (www.psc.edu/biomed/genedoc). Protein structure models were generated using the SWISS-MODEL workspace [40] on automated mode. AtSUMO1/3/5 and AtSCE1 models were generated using PDB code 2PE6 [2.40 Å (1 Å = 0.1 nm)] or PDB code 2IY1 (2.46 Å) as templates. Models were assembled and images were generated using PyMOL (http://www.pymol.org).
Accession numbers
The assigned accession numbers for the genes studied are as follows: At4g26840 (AtSUMO1), At5g55160 (AtSUMO2), At5g55170 (AtSUMO3), At2g32765 (AtSUMO5), At2g21470 (AtSAE2), At4g24940 (AtSAE1a), At5g50580 (AtSAE1b), At3g57870 (AtSCE1), At4g02890 (AtUBI), At5g53300 (AtUBC10) and At1g20620 (AtCAT3).
RESULTS
Conservation of E1-, E2- and SIM-interacting residues among AtSUMO paralogues
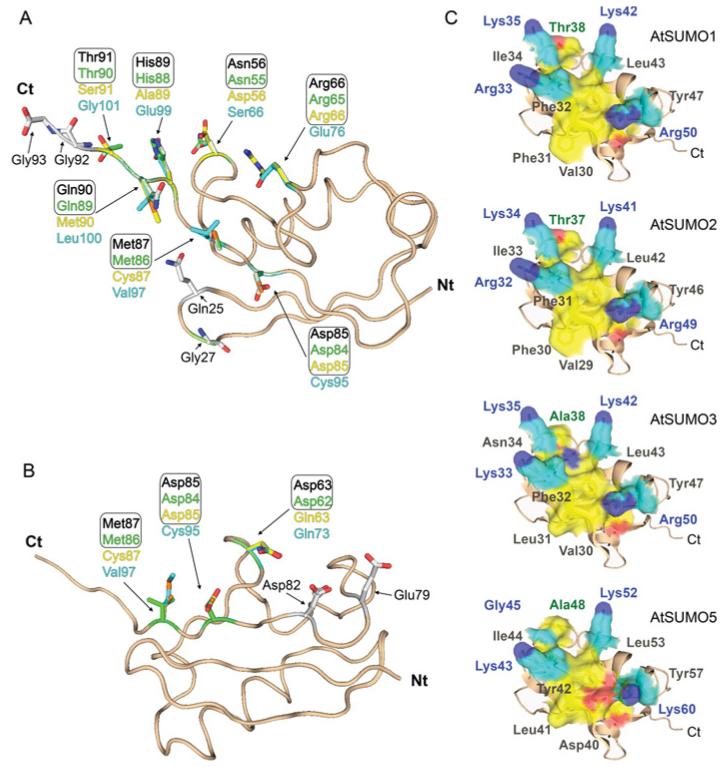
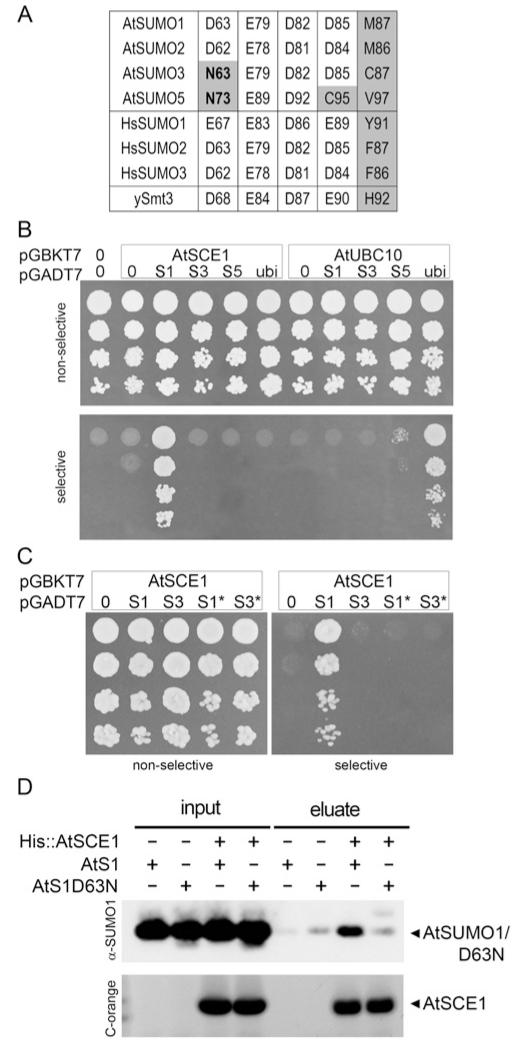
The overall degree of homology among SUMO paralogues ranges from 83% sequence identity between AtSUMO1 and AtSUMO2, to 42% and 30% sequence identity between AtSUMO1 and AtSUMO3 or AtSUMO5 respectively. Interestingly, these differences are also present in the degree of conservation found between residues involved in E1 and E2 non-covalent interactions. AtSUMO1 and AtSUMO2 have identical amino acid residues at the positions involved in non-covalent interactions with the E1-activating and E2-conjugating enzymes. In contrast, AtSUMO3 and AtSUMO5 have more divergent interacting surfaces. AtSUMO3 has 55% and 75% degree of conservation of E1- and E2-interacting residues respectively, whereas AtSUMO5 is the most divergent isoform, showing a degree of conservation of 36% and 56% for E1- and E2-interacting residues (Figures 1A and 1B, and Supplementary Figure S2 at http://www.BiochemJ.org/bj/436/bj4360581add.htm). Previous structural studies identified 11 residues in the HsSUMO1 surface involved in E1 interactions [14]. Taken as reference positions in AtSUMO1, the four residues, Gln25, Gly27, Gly92 and Gly93, are identical in all Arabidopsis paralogues. Among the others, two are divergent only in the AtSUMO5 isoform, Arg66 and Asp85, and five are not conserved in either AtSUMO3 or AtSUMO5, Asn56, Met87, His89, Gln90 and Thr91 (Figures 1A and 5A). All of them are identical between AtSUMO1 and 2. With regard to the residues involved in E2 non-covalent interactions, those residues establishing lateral chain contacts with the E2-conjugating enzyme are identical across AtSUMO1/2, HsSUMO2/3 and yeast ySmt3, except for the residue Met87 in AtSUMO1, which is highly variable (Figures 1B and 2A). The deduced consensus motif considering the most frequent residues would be Asp63/Glu63, Glu79, Asp82 and Asp85/Glu85 (the residue numbering is for AtSUMO1). When we analysed AtSUMO3, it is remarkable that the acidic Asp63/Glu63 residue in the consensus sequence is substituted with a polar asparagine residue. This change is also present in the most divergent SUMO isoform AtSUMO5 that, in addition, has substitutions in Asp77/Glu77 and Asp85/Glu85 by histidine and cysteine respectively (Figures 1B and 2A).
Figure 1. Conservation analysis of residues on AtSUMO paralogue surfaces involved in non-covalent interactions.
AtSUMO paralogue structures, aspredicted by the SWISS-MODEL comparative protein modelling server, are shown as a ribbon representations. Residues contained in the N-terminus tail and those absent in the mature forms were not included in the modelling. Residues involved in E1 (A) and E2 (B) non-covalent interactions are shown in stick representation. Residues corresponding to AtSUMO1, AtSUMO2, AtSUMO3 and AtSUMO5 are shown in black, green, yellow and blue respectively. (C) Same structure model as in (A) but showing residues involved in SIM interactions in surface representation. Basic residues and the groove-forming residues are coloured cyan and yellow respectively. Side-chain nitrogen and oxygen atoms are coloured blue and red.
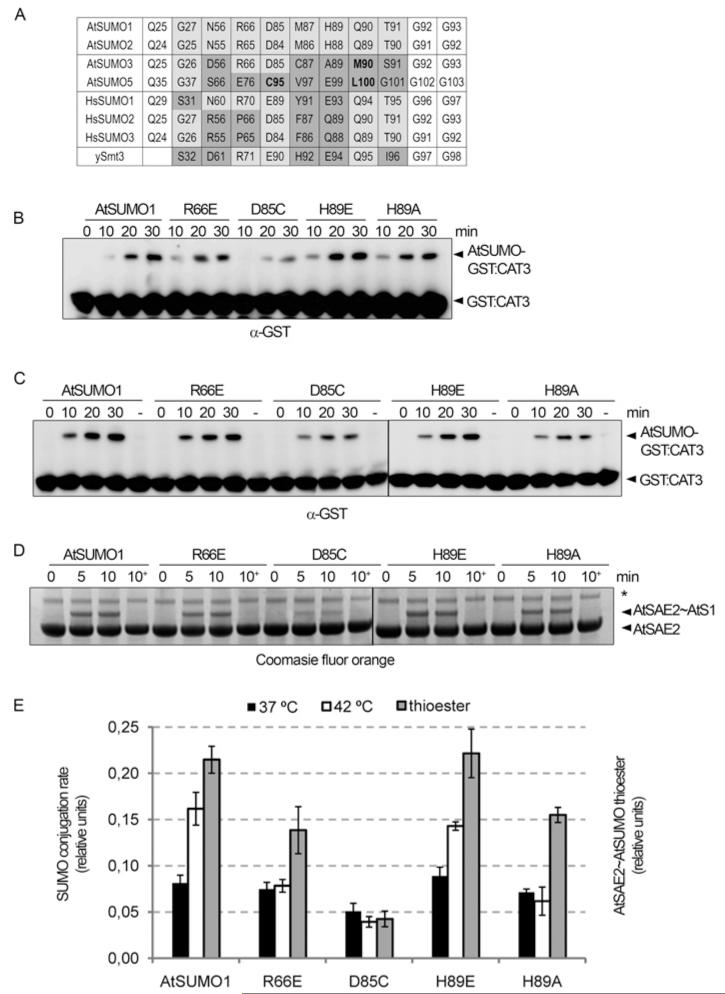
Figure 5. Mutations in the AtSUMO1 residues involved in E1 recognition resulted in SUMO-conjugation defects.
(A) Sequence alignment of Arabidopsis (At), human (Hs) and yeast (y) SUMO residues involved in the SUMO–E1 non-covalent interaction. Non-conserved residues are highlighted in grey and residues exclusively non-conserved in AtSUMO3/5 are in bold. (B) In vitro SUMOylation assays were performed in the presence of AtE1a, AtSUMO1 or the indicated mutant, AtSCE1a and GST–AtCAT3Ct as a substrate. Reaction mixtures were incubated at 37°C and stopped at the indicated time points. Reaction products were resolved by SDS/PAGE and examined by immunoblot analysis with anti-GST antibodies. (C) The same as in (B), but reactions were incubated at 42°C. (D) Time course for AtSAE2–thioester formation using Arabidopsis E1 (AtSAE2–AtSAE1a) and AtSUMO1 and AtSUMO1 mutant variants as substrates. Reaction mixtures were incubated at 30°C and aliquots were removed at the specified time points. In each case, a 10 min aliquot was treated with DTT as a control for thioester-bond formation and this is indicated as 10+. Reaction products were separated by SDS/PAGE and stained with Coomassie Fluor Orange. The asterisk indicates a contaminating protein. (E) Reactions were performed in triplicate and GST–AtCAT3Ct SUMOylation and AtSAE2–thioester formation rates were quantified. Average values of the relative activity among AtSUMO1 variants and the corresponding S.E.M. are plotted.
Figure 2. Non-covalent E2-interaction properties of AtSUMO isoforms.
(A) Sequence alignment of Arabidopsis (At), human (Hs) and yeast (y) SUMO residues involved in SUMO–E2 non-covalent interaction through their lateral chain contacts. Non-conserved residues are highlighted in grey, and residues exclusively non-conserved in AtSUMO3/5 are in bold. (B) Yeast two-hybrid assay to study interactions between AtSUMO1, 2, 3, 5 or ubiquitin and AtSCE1 or AtUBC10. (C) Interaction analysis between the mutant AtSUMO1/D63N (S1*) or AtSUMO3/N63D (S3*) and AtSCE1 as in (B). Native SUMO isoforms were also included as a control. (D) Poly-histidine pull-down assay of AtSUMO1 or the AtSUMO1/D63N mutant variant using His–AtSCE1 as a bait. Incubations in the absence of the bait or the prey were used as negative controls. Aliquots of input and eluate fractions were resolved by SDS/PAGE. AtSUMO1/D63N and AtSCE1 were analysed by immunoblotting or Coomassie Fluor Orange staining (C-orange) respectively.
Another important interacting surface on SUMO involves the second β-sheet and the downstream α-helix, which form a hydrophobic groove flanked by basic residues that accommodates SIMs [42]. Structural studies have determined that aliphatic and aromatic residues constitute this hydrophobic groove, HsSUMO1 Ile34, His35, Phe36, Val38, Leu47 and Tyr51 [23]. Whereas functional studies identified residues required for the role of HsSUMO2 in transcriptional inhibition, which include the hydrophobic Val30 and Ile34 (equivalent to Ile34 and Val38 in HsSUMO1), the polar Thr38 (Thr42 in HsSUMO1), and the four basic residues Lys33, Lys35, Lys42 and Arg50 (Lys37, Lys39, Lys46 and Arg54 in HsSUMO1) [24]. Among the basic residues, HsSUMO1 Lys39 has been proposed to interact with phosphorylated residues located next to the hydrophobic core, which is the essential component of SIMs [22]. The previous functional amino acids identified in HsSUMO1 and HsSUMO2 are conserved in AtSUMO1/2, suggesting that these paralogues will share the molecular basis for the SIM interaction as their human orthologues. In contrast, major changes are present in AtSUMO3 and AtSUMO5. The hydrophobic Val30 and Ile34 residues shown to be necessary for transcriptional repression in HsSUMO2 are substituted by an acidic residue in AtSUMO5 (Asp40) and a polar residue (Asn34) in AtSUMO3 respectively. In addition, the polar Thr38 in HsSUMO2, which is also necessary for transcriptional repression, is substituted by the hydrophobic alanine residue in both AtSUMO3 and AtSUMO5 (positions 38 and 48 respectively). Finally, the basic Lys39 in HsSUMO1 proposed to interact with phosphorylated residues in the target is substituted by an uncharged glycine residue in AtSUMO5 (Figure 1C). Considering that no SIM-containing targets have been identified in Arabidopsis, we have focused on the functional analysis of E1 and E2 non-covalent interacting residues in Arabidopsis SUMO paralogues, according to their role in conjugation.
E2 non-covalent interactions with SUMO isoforms
In order to assess the effect of changes in residues involved in SUMO–E2 non-covalent interactions, we performed yeast two-hybrid assays. In these experiments, the capacity of the yeast strain HF7c to grow in the absence of histidine was used as a marker for the interaction between proteins. Previous studies have shown that AtSUMO1 and 2 were capable of interacting with AtSCE1 in similar assays [11]. We found that histidine auxotrophy was restored only when AtSCE1a was co-transformed with AtSUMO1, but not with AtSUMO3 or AtSUMO5. In these experiments, AtSUMO1 was used as a positive control and Arabidopsis ubiquitin was used as a negative control. When AtSCE1 was replaced by the ubiquitin-conjugating enzyme AtUBC10, we observed that histidine auxotrophy was conferred only when ubiquitin was co-expressed, consistent with the specificity of the system (Figure 2B). The results of the present study demonstrate that AtSUMO3/5 are not competent to interact with AtSCE1, suggesting the existence of a change in the SUMO-interacting surface that might be common to both paralogues. As described above, the central aspartate residue in AtSUMO1 and 2 (Asp63 and Asp62) is replaced by an asparagine residue at the equivalent position in AtSUMO3 and 5 (Asn63 and Asn73) (Figures 1B and 2A). To test the role of this divergent residue, we generated the mutants AtSUMO1/D63N and AtSUMO3/D63N and assessed their capacity to interact with AtSCE1. None of these mutant isoforms interacted with AtSCE1 in yeast two-hybrid assays (Figure 2C), indicating that the presence of an aspartate residue at position 63 is essential, but not sufficient, for SUMO–E2 non-covalent interactions. To further evaluate the essential role of Asp63 in non-covalent interactions with E2, we performed in vitro pull-down assays. In these assays, His–AtSCE1 was incubated in the presence of AtSUMO1 or AtSUMO1 D63N. After binding to a Ni2+ -charged resin, His–AtSCE1 was eluted with imidazole and we observed that AtSUMO1 was co-eluted with His–AtSCE1. In contrast, a small amount of AtSUMO1/D63N was present in the elution fraction to the same extent as in the negative control, where His–AtSCE1 was omitted.
Identification and validation of AtCAT3 as a bona fide SUMO target in vitro
In order to reconstitute a complete Arabidopsis SUMOylation system that allowed us the biochemical characterization of the SUMO paralogues, we aimed to identify an endogenous SUMO target. Since SUMOylation is involved in oxidative stress responses, we analysed whether oxidative stress scavengers could be SUMO targets. We found that AtCAT3 contained a SUMOylation consensus site at its C-terminal domain (Supplementary Figure S3A at http://www.BiochemJ.org/bj/436/bj4360581add.htm). Previous studies have determined that SUMO-conjugation sites are located in an extended structure on the surface of the target protein in order to be accessible to the SUMOylation machinery [43]. To determine the position of the putative AtCAT3 SUMOylation site on the quaternary structure, we performed the AtCAT3 structure prediction using the Exiguobacterium oxidotolerans catalase structure as a template (PDB code 2J2M). The model generated model indicated that the putative SUMO acceptor lysine residue Lys423 in AtCAT3 is fully exposed at the protein surface (Supplementary Figures S3B and S3C). Validation of AtCAT3 as a SUMO substrate was performed by in vitro SUMOylation reactions containing the AtCAT3 C-terminal domain, which includes the predicted SUMOylation site (GST–AtCAT3Ct; Supplementary Figure S3D), in the presence of the reconstituted Arabidopsis SUMOylation system, AtSAE2, AtSAE1a, AtSCE1 and AtSUMO1. As a result, we detected SUMO conjugation to AtCAT3Ct in an ATP-dependent manner. In addition, the mutant AtCAT3Ct/K423R was unable to accept SUMO (Supplementary Figure S3D). These results validate AtCAT3 as a SUMO target and identify Lys423 as the acceptor site for SUMO modification. The advantage of using AtCAT3Ct as a substrate for in vitro reactions as opposed to other targets described in the literature is that it does not require the presence of an E3 ligase in order to be modified, which simplifies the biochemical analysis of SUMO conjugation.
AtSUMO1 Asp63 is necessary for efficient polySUMO chain formation
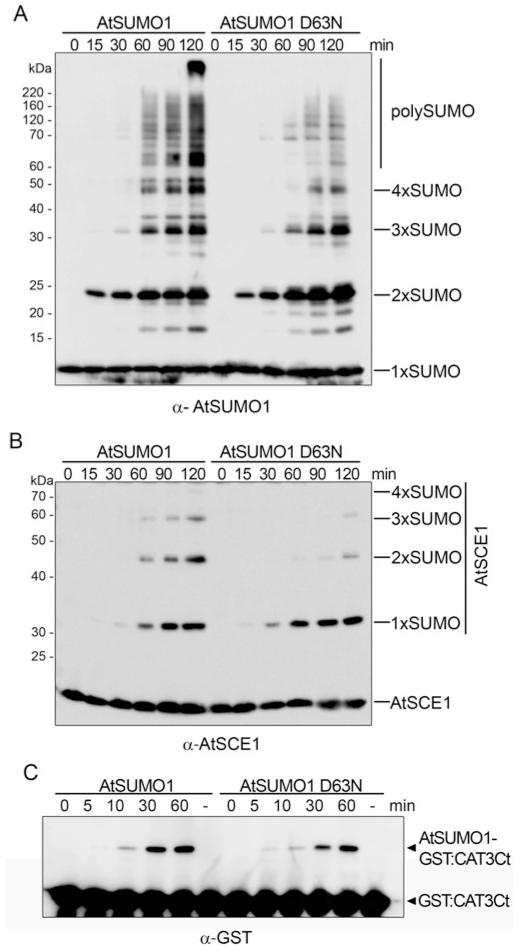
Since AtSUMO1/D63N prevented AtSCE1 non-covalent interactions, we tested whether this mutation affected polySUMO chain formation. In vitro polySUMO chain-formation assays were performed in the presence of AtSAE2/AtSAE1a, AtSCE1, and the native or mutated SUMO form. Under these conditions, the native AtSUMO1 isoform efficiently built polymeric chains and it was also conjugated to AtSCE1, on which polySUMO chains were also formed. When Asp63 was replaced by an asparagine residue, a reduction in polySUMO chain formation was observed. This defect was more evident from the second conjugation cycle, independently of whether polySUMO chains in the strict sense or polySUMO chains built on AtSCE1 were analysed (Figures 3A and 3B). These results indicate that the mechanism to build polySUMO chains is conserved in Arabidopsis and that, presumably, the naturally occurring E2-non-interacting SUMO isoforms AtSUMO3 and AtSUMO5 will not interfere with polyAtSUMO1/2 chain formation in vivo. Consistent with a main role for SUMO–E2 non-covalent interactions in polySUMO chain formation, the mutant AtSUMO1/D63N was conjugated to the target AtCAT3Ct with the same efficiency as the native AtSUMO1 (Figure 3C).
Figure 3. PolySUMO chain formation is dependent on Asp63 of AtSUMO1.
In vitro SUMOylation assays were performed in the presence of AtSAE2/AtSAE1a, AtSUMO1 or AtSUMO1/D63N and AtSCE1. Reaction mixtures were incubated at 37°C and stopped at the specified time points. Reaction products were resolved by SDS/PAGE and examined by immunoblot analysis with anti-AtSUMO1 (A) or anti-AtSCE1 (B) antibodies. (C) In vitro SUMOylation assays were performed in the presence of AtCAT3Ct as a substrate. Reaction mixtures were incubated at 37°C, aliquots were removed at the specified time points and reaction products were analysed by immunoblot analysis with anti-GST antibodies. The molecular mass in kDa is indicated on the left-hand side.
SUMO isoforms display a distinct conjugation rate in vitro
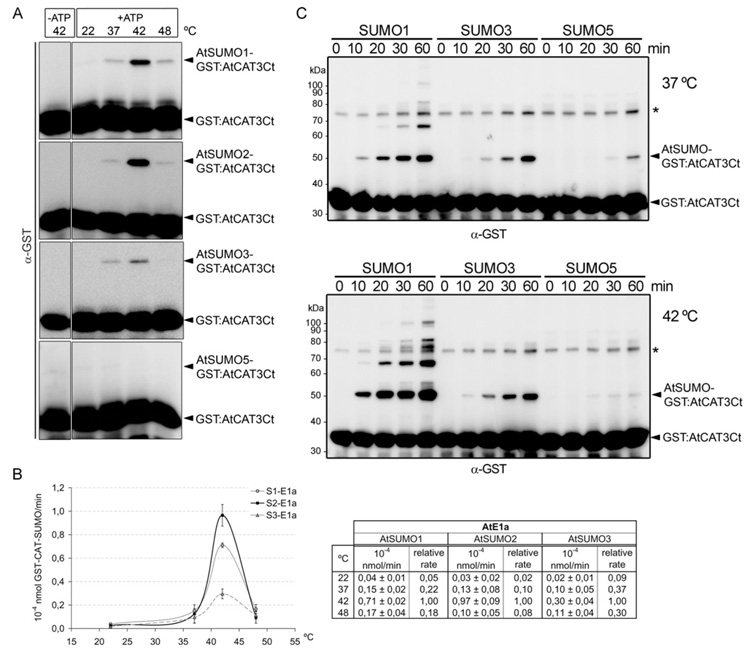
Since AtSUMO isoforms show differences in their capacity to establish non-covalent interactions with their cognate E2-conjugating enzyme, we explored the possibility that they could also differ in their conjugation rates. First, we chose a short incubation time, 10 min, in order to compare the first conjugation cycle. In this way, we avoided conjugation rate underestimation of the isoforms competent for polySUMO chain formation. We were also interested in analysing the effect of the incubation temperature on conjugation rate, which could be biologically relevant since SUMO conjugates accumulate massively upon heat stress. Reaction mixtures were incubated at 22, 37, 42 and 48°C, and we observed that the highest reaction rate was achieved at 42°C (Figure 4A). Under these experimental conditions, we did not observe conjugation of AtSUMO5. All other isoforms were conjugated to AtCAT3 at different rates. In general, AtSUMO1 and 2 were better conjugated than AtSUMO3, the highest difference being observed at 42°C. At this temperature, the AtSUMO1 and AtSUMO2 conjugation rate is 2.4- and 3.2-fold higher than AtSUMO3 respectively (Figure 4B). Next, we performed a time-course analysis that allowed us to detect AtSUMO5 conjugation after 60 min incubation at 37°C. At 42°C, AtSUMO5 conjugation was very weak, and differences in the conjugation level between AtSUMO1 and AtSUMO5, as well as between AtSUMO1 and AtSUMO3, were more pronounced than at 37°C (Figure 4C).
Figure 4. In vitro conjugation properties of AtSUMO isoforms.
(A) In vitro SUMOylation assays were performed in the presence of AtE1a, AtSUMO1, AtSUMO2, AtSUMO3 or AtSUMO5, AtSCE1a and GST–AtCAT3Ct as a substrate. Reaction mixtures were incubated at the indicated temperature and stopped after incubation for 10 min. Reaction products were resolved by SDS/PAGE and examined by immunoblot analysis with anti-GST antibodies. (B) Reactions were performed in triplicate and the GST–AtCAT3Ct SUMOylation rate was quantified. Values are means ± S.D. The Table containing the data plotted into the graph is shown to the right-hand side. (C) A time course of SUMO conjugation in vitro reactions were performed at 37°C (top panel) and 42°C (bottom panel). Reaction mixtures were set up as described in (A). The asterisk indicates a contaminating protein. The molecular mass in kDa is indicated on the left-hand side.
Non-conserved residues involved in the E1 interaction are responsible for differences in the SUMO paralogue conjugation rate
As has been described above, SUMO residues involved in E1 recognition are not evenly conserved between AtSUMO isoforms. In order to evaluate the effect of these changes in conjugation rate, we focused on those positions that were not conserved only in the less conjugated isoform AtSUMO5, which corresponds to AtSUMO1 Arg66 and Asp85. We also focused on AtSUMO1 His89 which is not conserved either in AtSUMO3 or AtSUMO5, and whose equivalent position in Nedd8 has previously been reported to confer modifier specificity [44]. We mutated residues Arg66, Asp85 and His89 of AtSUMO1 to those present in AtSUMO3 and AtSUMO5 at the equivalent positions, and analysed their conjugation rates (Figures 5B and 5C). At 42°C, the most dramatic effect was observed in D85C followed by H89A and R66E mutants, which showed 24%, 40% and 50% of the native AtSUMO1 rate respectively. The H89E substitution had a smaller effect and showed 90% of the native AtSUMO1 activity. Similar to what we observed during the conjugation analysis of SUMO isoforms, at 37°C differences in conjugation levels were smaller, although their behaviour was similar, and the D85C mutation was the most affected. These mutants, with the exception of the H89E substitution, also showed SUMO–E1 thioester-bond formation defects, and the D85C mutation was the most affected. These results suggest that the conjugation level reduction of these mutants was the result of E1-interaction defects.
DISCUSSION
The molecular consequences of protein modification by SUMO will be dependent on the specific SUMO paralogue that is attached to the target, and major effort has been put into elucidating the mechanisms involved in SUMO paralogue specificity. Among these mechanisms, ULP-mediated deconjugation and SIM-mediated conjugation have been proposed to facilitate SUMO paralogue selection [45]. In the present study we have described results supporting a role of the E1-activating enzyme in SUMO paralogue discrimination in Arabidopsis. The analysis of the molecular properties of the four AtSUMO isoforms expressed indicates that they have diverged to a higher degree than their human orthologues, and that the essential AtSUMO1 and AtSUMO2 are the most functionally conserved isoforms.
Non-covalent interactions between SUMO and its cognate E2-conjugating enzyme is an intrinsic property of the system. Initial studies showed that this property is conserved in AtSUMO1 and 2 [11] but, surprisingly, our results showed that AtSUMO3 and 5 do not retain the capacity to interact with their cognate E2, AtSCE1. This situation is unique to the Arabidopsis system, since all Hs-SUMO isoforms and yeast Smt3 interact efficiently with their cognate E2-conjugating enzyme. We have also identified AtSUMO1 Asp63 as an essential residue for establishing E2 non-covalent interactions. But the fact that AtSUMO3 Asn63 substitution with aspartate did not confer competence for this interaction suggests that the inability of AtSUMO3/5 to interact with AtSCE1 could be the result of two types of amino acid changes: the loss of essential residues and the appearance of residues that would be detrimental for this interaction. SUMO–E2 non-covalent interactions have been proposed to be involved in SUMO chain formation of human SUMO2 and 3 isoforms, which also contain a SUMO attachment site in their N-terminal tail [19,20]. In this model, HsSUMO1 would function as a chain terminator, since it does not comprise an acceptor lysine residue for another SUMO molecule, whereas it retains the capacity to interact non-covalently with E2 enzyme. In Arabidopsis, a SUMOylation consensus site is only present in those paralogues capable of interacting non-covalently with AtSCE1, which are AtSUMO1/2, correlating with their ability to form polymeric chains. On the other hand, considering their inability to interact with AtSCE1, it is not clear whether AtSUMO3/5 would have a role as polyAtSUMO1/2 chain terminators, as has been proposed for the HsSUMO1, suggesting that other molecular mechanisms might regulate polySUMO chain length in Arabidopsis. Supporting this, recent proteomic studies have failed to identify AtSUMO3/5 peptides in purified AtSUMO1 conjugates [46].
Another remaining question to be addressed was whether AtSUMO paralogues also differed in their conjugation levels. In addition, since SUMO conjugates accumulate dramatically upon heat stress, we were interested in studying the effect of the temperature in conjugation reactions. In previously reported assays, experiments were designed in such a manner that non-quantitative results were obtained (in most cases the incubation time ranged from a few hours to overnight) [32,47]. The results of the present study, using a quantitative SUMOylation assay, showed that AtSUMO1 and 2 isoforms were more efficiently conjugated in comparison with AtSUMO3, whereas AtSUMO5 showed the lowest conjugation level. For the SUMO isoforms tested, with the exception of AtSUMO5, the conjugation rate increased with temperature, and it was striking to observe that the highest activity occurred at 42°C. Even though this temperature is higher than standard environmental conditions, it highlights the robustness of the SUMO-conjugation system. At the same time it suggests that the massive and rapid SUMO conjugation observed in plants upon heat-shock treatments could be mediated, at least in part, by the increasing activity of the conjugation system with temperature. In addition, this effect is more pronounced in the case of AtSUMO1 and AtSUMO2 conjugation, which are the isoforms that are greatly conjugated under heat stress.
Differences in conjugation efficiency among SUMO paralogues are also specific to the Arabidopsis SUMOylation system, since all three human SUMO isoforms have been shown to form E1- and E2-thioester bonds and be conjugated to the substrate RanGAP with the same efficiency [14], suggesting that, in this case, SUMO paralogue selection will be dependent on availability or SIM-mediated interactions. The conservation analysis of the 11 SUMO residues involved in the E1 adenylation domain interaction [14] showed that five and seven residues are not conserved in AtSUMO3 and AtSUMO5 respectively, suggesting that adenylation could be deficient for these paralogues. A mutagenesis analysis revealed that some of these substitutions were responsible for a lower conjugation rate, although the effect was dependent on the nature of the substitution. The most deleterious was the D85C substitution present in AtSUMO5, supporting the previous result indicating that AtSUMO5 was the less-conjugated isoform. Interestingly, this position in HsSUMO1, Glu89, was previously shown to be crucial for E1-thioester formation [19], supporting our results pointing to a critical role of this residue in SUMO–E1 recognition. As to AtSUMO3, the only substitution tested occurs at a position previously proposed to contribute to modifier discrimination [44] and, when introduced in AtSUMO1, it reduced the conjugation rate to a value equivalent to that of AtSUMO3, suggesting that this substitution has a major contribution in AtSUMO3 conjugation-rate reduction. Conjugation defects in these mutants were more pronounced when reaction mixtures were incubated at higher temperatures, similar to what we observed when comparing SUMO paralogues. Moreover, the reduction in conjugation correlated with a reduction in E1-thioester formation, supporting a function of the residues tested in SUMO recognition by the E1. To our knowledge, this natural occurring SUMO paralogue discrimination by the E1-activating enzyme has not been reported previously in any other system.
Among the different studies that have addressed characterization of the Arabidopsis SUMOylation system, in vitro AtSUMO5 conjugation has only been detected to a mammalian substrate, RanGAP, in the presence of the mammalian conjugation system [31]. It is well established that SUMO conjugation is highly regulated by specific protein–protein interactions, and using heterologous systems to test SUMO conjugation might give results that could not be relevant in vivo. This could explain why AtSUMO5 conjugation was facilitated by the SUMO mammalian system, but when we have used the Arabidopsis system, which presumably is more selective, AtSUMO5 was very poorly conjugated. Similarly, a previous report showed that HsSUMO1 was able to interact with the Arabidopsis ubiquitin-conjugating enzyme AtUBC10 in yeast two-hybrid assays. In contrast, in the same assays, neither AtSUMO1 nor AtSUMO2 interacted with AtUBC10, suggesting that selective interactions within the SUMO pathway are more permissive when heterologous SUMOylation components are used [11], and highlighting the importance of using homologous systems in biochemical studies.
In vivo, different evidence points to a predominant role for AtSUMO1/2 paralogues. Early studies have shown that endogenous AtSUMO1 and 2 [7,11] and AtSUMO3 [7] were conjugated in planta, although AtSUMO3 conjugate levels were lower than for AtSUMO1/2. Instead, endogenous AtSUMO5 conjugation has not been observed. In addition, according to the Genevestigator database, atsumo3 and atsumo5 mRNA levels are on average 10-fold lower than atsumo1/2. Furthermore, none of the SUMO-specific proteases identified are competent to process AtSUMO5, and only AtULP1a displays an inefficient peptidase activity towards AtSUMO3, suggesting that, even if expressed at lower levels, it would have to be determined which fraction is present in its mature/conjugable form [31,32]. Finally, the fact that double-mutant atsumo1atsumo2 plants are not viable [34] suggests a biological specialization and, regardless of AtSUMO3 and 5 in vivo function, it seems clear that they cannot compensate for the AtSUMO1/2 loss. A recent study suggests that AtSUMO3 function might differ from AtSUMO1 and AtSUMO2 in flowering and salicylic-acid-dependent responses, although homozygous atsumo3 mutant plants showed normal plant development [37]. Interestingly, a more recent report described the identification of proteins that are specifically conjugated by AtSUMO3 and not AtSUMO1, although the molecular mechanism of this specificity remains elusive [48]. Surprisingly, almost half of the substrates analysed were exclusively modified by AtSUMO3, in contrast with the low levels of AtSUMO3 conjugates previously detected in crude plant extracts.
Overall, different molecular mechanisms seem to converge in order to assure a proper conjugation of the essential Arabidopsis SUMO1/2 isoforms compared with the non-essential AtSUMO3/5. These mechanisms comprise regulation of expression levels, maturation and release from targets, and AtSUMO1/2 are the highest expressed isoforms and the most efficient substrates of the characterized endogenous proteases [31,32], suggesting that most of the endogenous pool of mature SUMO will comprise these two isoforms. In the present study, we provide evidence for the existence of a preferential conjugation of AtSUMO1/2 compared with AtSUMO3/5, which is determined by a role of the E1-activating enzyme in SUMO paralogue discrimination.
Supplementary Material
ACKNOWLEDGEMENTS
We thank C.D. Lima for technical advice and M. Rodríguez-Concepción for critical reading of the manuscript before submission.
FUNDING
The work was supported by the European Research Council [grant number ERC-2007-StG-205927], and the Spanish Ministry of Education and Science [grant numbers BIO2008-01495, CONSOLIDER CSD 2007-00036]. L.C.M. is supported by a predoctoral contract through the CRAG. J.S. is supported by a predoctoral fellowship from the Ministry of Education and Science [grant number BES-2005-6843]. We also thank the ‘Departament d’Innovació, Universitats i Empresa’ from the Generalitat de Catalunya (Xarxa de Referència en Biotecnologia and 2009SGR 09626) for support.
Abbreviations used
- AtCAT3
Arabidopsis catalase isoform 3
- DTT
dithiothreitol
- GST
glutathione transferase
- SD
synthetic dextrose
- SUMO
small ubiquitin-related modifier
- AtSUMO
Arabidopsis SUMO
- HsSUMO
human SUMO
- SAE
SUMO-activating enzyme
- AtSAE
Arabidopsis SAE
- AtSCE1
Arabidopsis SUMO-conjugating enzyme 1
- SIM
SUMO-interacting motif
- ULP
ubiquitin-like protein-specific protease
Footnotes
AUTHOR CONTRIBUTION
Laura Castaño-Miquel performed mutagenesis experiments, characterization of mutant variants, and time-course experiments; Josep Seguí performed the initial characterization of SUMO isoforms; and Maria Lois designed the experiments and wrote the manuscript.
REFERENCES
- 1.Lois LM. Diversity of the SUMOylation machinery in plants. Biochem. Soc. Trans. 2010;38:60–64. doi: 10.1042/BST0380060. [DOI] [PubMed] [Google Scholar]
- 2.Hanania U, Furman-Matarasso N, Ron M, Avni A. Isolation of a novel SUMO protein from tomato that suppresses EIX-induced cell death. Plant J. 1999;19:533–541. doi: 10.1046/j.1365-313x.1999.00547.x. [DOI] [PubMed] [Google Scholar]
- 3.Hotson A, Chosed R, Shu H, Orth K, Mudgett MB. Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol. Microbiol. 2003;50:377–389. doi: 10.1046/j.1365-2958.2003.03730.x. [DOI] [PubMed] [Google Scholar]
- 4.Roden J, Eardley L, Hotson A, Cao Y, Mudgett MB. Characterization of the Xanthomonas AvrXv4 effector, a SUMO protease translocated into plant cells. Mol. Plant Microbe Interact. 2004;17:633–643. doi: 10.1094/MPMI.2004.17.6.633. [DOI] [PubMed] [Google Scholar]
- 5.Castillo AG, Kong LJ, Hanley-Bowdoin L, Bejarano ER. Interaction between a geminivirus replication protein and the plant sumoylation system. J. Virol. 2004;78:2758–2769. doi: 10.1128/JVI.78.6.2758-2769.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 7.Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung DY, Vierstra RD. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J. Biol. Chem. 2003;278:6862–6872. doi: 10.1074/jbc.M209694200. [DOI] [PubMed] [Google Scholar]
- 8.Murtas G, Reeves PH, Fu YF, Bancroft I, Dean C, Coupland G. A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. Plant Cell. 2003;15:2308–2319. doi: 10.1105/tpc.015487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catala R, Ouyang J, Abreu IA, Hu Y, Seo H, Zhang X, Chua NH. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell. 2007;19:2952–2966. doi: 10.1105/tpc.106.049981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lois LM, Lima CD, Chua NH. Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell. 2003;15:1347–1359. doi: 10.1105/tpc.009902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5418–5423. doi: 10.1073/pnas.0811088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ulrich HD. The SUMO system: an overview. Methods Mol. Biol. 2009;497:3–16. doi: 10.1007/978-1-59745-566-4_1. [DOI] [PubMed] [Google Scholar]
- 14.Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 2005;24:439–451. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olsen SK, Capili AD, Lu X, Tan DS, Lima CD. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature. 2010;463:906–912. doi: 10.1038/nature08765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson ES. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 17.Melchior F, Schergaut M, Pichler A. SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem. Sci. 2003;28:612–618. doi: 10.1016/j.tibs.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Geoffroy MC, Hay RT. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell Biol. 2009;10:564–568. doi: 10.1038/nrm2707. [DOI] [PubMed] [Google Scholar]
- 19.Knipscheer P, van Dijk WJ, Olsen JV, Mann M, Sixma TK. Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J. 2007;26:2797–2807. doi: 10.1038/sj.emboj.7601711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capili AD, Lima CD. Structure and analysis of a complex between SUMO and Ubc9 illustrates features of a conserved E2-Ubl interaction. J. Mol. Biol. 2007;369:608–618. doi: 10.1016/j.jmb.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH, Hay RT, Lamond AI, Mann M, Vertegaal AC. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol. Cell Proteomics. 2008;7:132–144. doi: 10.1074/mcp.M700173-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. 2006;281:16117–16127. doi: 10.1074/jbc.M512757200. [DOI] [PubMed] [Google Scholar]
- 23.Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chupreta S, Holmstrom S, Subramanian L, Iniguez-Lluhi JA. A small conserved surface in SUMO is the critical structural determinant of its transcriptional inhibitory properties. Mol. Cell. Biol. 2005;25:4272–4282. doi: 10.1128/MCB.25.10.4272-4282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song J, Zhang Z, Hu W, Chen Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J. Biol. Chem. 2005;280:40122–40129. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- 26.Meulmeester E, Kunze M, Hsiao HH, Urlaub H, Melchior F. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol. Cell. 2008;30:610–619. doi: 10.1016/j.molcel.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 27.Knipscheer P, Flotho A, Klug H, Olsen JV, van Dijk WJ, Fish A, Johnson ES, Mann M, Sixma TK, Pichler A. Ubc9 sumoylation regulates SUMO target discrimination. Mol. Cell. 2008;31:371–382. doi: 10.1016/j.molcel.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 28.Merrill JC, Melhuish TA, Kagey MH, Yang SH, Sharrocks AD, Wotton D. A role for non-covalent SUMO interaction motifs in Pc2/CBX4 E3 activity. PLoS ONE. 2010;5:e8794. doi: 10.1371/journal.pone.0008794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho G, Lim Y, Golden JA. SUMO interaction motifs in Sizn1 are required for promyelocytic leukemia protein nuclear body localization and for transcriptional activation. J. Biol. Chem. 2009;284:19592–19600. doi: 10.1074/jbc.M109.010181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang SH, Sharrocks AD. The SUMO E3 ligase activity of Pc2 is coordinated through a SUMO interaction motif. Mol. Cell. Biol. 2010;30:2193–2205. doi: 10.1128/MCB.01510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chosed R, Mukherjee S, Lois LM, Orth K. Evolution of a signalling system that incorporates both redundancy and diversity: Arabidopsis SUMOylation. Biochem. J. 2006;398:521–529. doi: 10.1042/BJ20060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colby T, Matthai A, Boeckelmann A, Stuible HP. SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol. 2006;142:318–332. doi: 10.1104/pp.106.085415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chosed R, Tomchick DR, Brautigam CA, Mukherjee S, Negi VS, Machius M, Orth K. Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases. J. Biol. Chem. 2007;282:6773–6782. doi: 10.1074/jbc.M608730200. [DOI] [PubMed] [Google Scholar]
- 34.Saracco SA, Miller MJ, Kurepa J, Vierstra RD. Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 2007;145:119–134. doi: 10.1104/pp.107.102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evdokimov E, Sharma P, Lockett SJ, Lualdi M, Kuehn MR. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J. Cell Sci. 2008;121:4106–4113. doi: 10.1242/jcs.038570. [DOI] [PubMed] [Google Scholar]
- 36.Zhang FP, Mikkonen L, Toppari J, Palvimo JJ, Thesleff I, Janne OA. SUMO-1 function is dispensable in normal mouse development. Mol. Cell. Biol. 2008;28:5381–5390. doi: 10.1128/MCB.00651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Den Burg HA, Kini RK, Schuurink RC, Takken FL. Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell. 2010;22:1998–2016. doi: 10.1105/tpc.109.070961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reference deleted
- 40.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 41.Reference deleted
- 42.Kerscher O. SUMO junction-what’s your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8:550–555. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–356. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- 44.Walden H, Podgorski MS, Huang DT, Miller DW, Howard RJ, Minor DL, Jr, Holton JM, Schulman BA. The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol. Cell. 2003;12:1427–1437. doi: 10.1016/s1097-2765(03)00452-0. [DOI] [PubMed] [Google Scholar]
- 45.Wilkinson KA, Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 2010;428:133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2010;107:16512–16517. doi: 10.1073/pnas.1004181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Budhiraja R, Hermkes R, Muller S, Schmidt J, Colby T, Panigrahi K, Coupland G, Bachmair A. Substrates related to chromatin and to RNA-dependent processes are modified by Arabidopsis SUMO isoforms that differ in a conserved residue with influence on desumoylation. Plant Physiol. 2009;149:1529–1540. doi: 10.1104/pp.108.135053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elrouby N, Coupland G. Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc. Natl. Acad. Sci. U.S.A. 2010;107:17415–17420. doi: 10.1073/pnas.1005452107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.