Abstract
MicroRNAs have important functions in the nervous system through post-transcriptional regulation of neurogenesis genes. Here we show that microRNA let-7d, which has been implicated in cocaine addiction and other neurological disorders, targets the neural stem cell regulator TLX. Overexpression of let-7d in vivo reduced neural stem cell proliferation and promoted premature neuronal differentiation and migration, a phenotype similar to those induced by TLX knockdown or overexpression of its negatively-regulated target, microRNA-9. We found a let-7d binding sequence in the tlx 3′ UTR and demonstrated that let-7d reduced TLX expression levels in neural stem cells, which in turn, up-regulated miR-9 expression. Moreover, co-expression of let-7d and TLX lacking its 3′ UTR in vivo restored neural stem cell proliferation and reversed the premature neuronal differentiation and migration. Therefore, manipulating let-7d and its downstream targets could be a novel strategy to unravel neurogenic signaling pathways and identify potential interventions for relevant neurological disorders.
Neural stem cells are self-renewing, multipotent precursor cells in the nervous system1. Although much effort has been devoted to understanding the mechanisms of neural stem cell proliferation and self-renewal, the mechanisms they use to differentiate and then subsequently mature into neurons are not well characterized. Recently, it has been discovered that microRNAs (miRNAs) can affect multiple processes in the cell including cell proliferation, differentiation, and development2,3. On average, miRNAs are 20–22 bp in length and act by binding to the 3′-terminus of their target mRNAs to inhibit gene expression through mRNA degradation or suppression of translation2,3. We and others have shown that miRNAs play an important role in neurogenesis; they regulate a diverse number of target genes that have been directly implicated in the neurogenic process4,5,6.
One of the founding members of the miRNA family is lethal-7 (let-7), which plays many important roles in normal development and diseases7,8,9,10,11,12,13. Recently, the functions played by let-7 miRNAs in the nervous system have begun to be identified. During brain development, let-7a can regulate the neuronal differentiation of neural precursors14. We and others have shown that in both embryonic and aging brains, the self-renewal of neural stem cells is negatively regulated by let-7b15,16. Another member is let-7d, whose expression is highly enriched in the brain and there is evidence to suggest that it plays a role in reward circuit and addiction17,18. However, the role that let-7d plays in the control of neural stem cell fate and neurogenesis is unknown.
Previously, we showed that TLX plays a critical role in regulating the maintenance and self-renewal of adult neural stem cells19,20. The neural stem cells in the hippocampus that are TLX-positive contribute to learning and memory acquisition in adult mice21, whereas the TLX-positive neural cells in the subventricular zones of adult brains are the relatively quiescent stem cells22,23,24. TLX also regulates neural development by controlling the cell cycle progression of neural precursor cells in the developing brain25,26,27. Mechanistically, TLX regulates genes that control cell cycle and cell proliferation by recruiting histone modifying enzymes to their promoters28,29. In addition, TLX promotes neural stem cell self-renewal by activating canonical Wnt signaling30. Furthermore, under hypoxia, TLX maintains the proliferation of adult hippocampal neural progenitors through its regulation of Oct4 expression, and induces Mash1 to activate neuronal lineage commitment31,32. Recently, we showed that TLX can regulate the cell fate determination of neural stem cells by repressing the expression of miR-9 and miR-137. These miRNAs in turn regulate their own downstream target genes, including TLX. The expression of TLX can be inhibited by miR-9 and let-7b, both of which can bind to the tlx 3′ UTR15,33,34. Whether TLX interacts with other miRNAs, including let-7d, and what the functional outcome of these interactions could be, remains unknown.
In this study we show that let-7d functions as a key regulator of neurogenesis through its regulation of the expression of the TLX and miR-9 cascade in the mouse brain. When overexpressed, let-7d inhibited neural stem cell proliferation, accelerated neural differentiation and enhanced neuronal migration, a phenotype similar to those induced by both TLX-knockdown and overexpression of miR-9 in embryonic mouse brains.
Results
Let-7d inhibits neural stem cell proliferation and accelerates differentiation
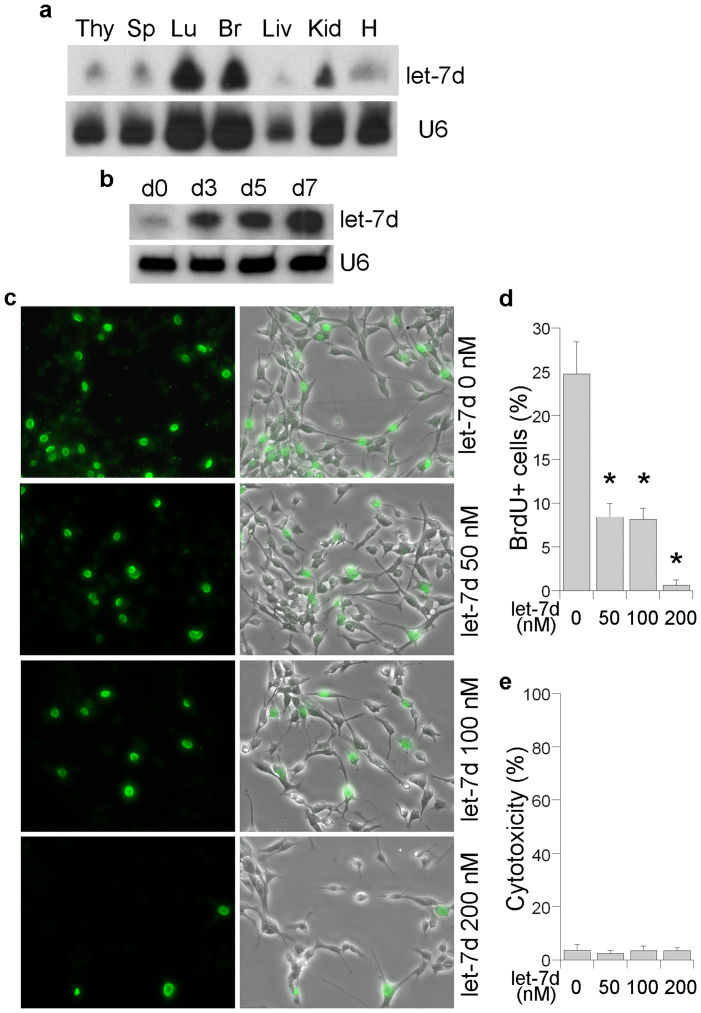
Although the roles of miRNAs have begun to be appreciated in the nervous system, the expression and function of let-7d in mammalian brains has yet to be studied. We show here that let-7d is highly expressed in the mouse brain, lung and kidney (Fig. 1a). As shown by Northern blot analysis, let-7d is expressed in neural stem cells (d0) at a very low level. The expression is increased over a 7-day differentiation time course (Fig. 1b). To investigate the role of let-7d in neural stem cell proliferation, we transfected mouse neural stem cells with synthetic let-7d RNA duplexes using concentrations ranging from 0 to 200 nM, and then treated the transfected cells with 5-bromodeoxyuridine (BrdU). The BrdU-treated cells were then immunostained using a BrdU antibody to determine cell proliferation status. Treatment with let-7d suppressed cell proliferation in a dose-dependent manner, as revealed by the dramatically reduced percentage of BrdU-positive cells in response to increasing concentrations of let-7d (Fig. 1c, d). The decreased BrdU labeling in the let-7d-transfected cells was unlikely to have been caused by cell death because similarly low levels of lactate dehydrogenase (LDH), a marker of cytotoxicity, were detected in both mock and let-7d-treated cells (Fig. 1e). These results indicate that let-7d is expressed in mammalian brains and inhibits neural stem cell proliferation.
Figure 1. Let-7d is expressed in neural stem cells and down-regulates neural stem cell proliferation.
(a). Northern blot analysis of let-7d expression in different mouse tissues. U6 was used as the loading control. (b). Northern blot analysis of let-7d expression in mouse neural stem cells over 7 days of differentiation. Cells at day (d) 0 are the undifferentiated neural stem cells. U6 was used as the loading control. (c). BrdU labeling of neural stem cells transfected with let-7d. The RNA concentrations (from 0 to 200 nM) were indicated and BrdU labeling is shown in green. The merged images of BrdU staining and phase contrast are shown on the right. (d). Quantification of BrdU labeling in neural stem cells transfected with let-7d. The percentage of BrdU+ cells was scored by dividing the BrdU+ cells with total living cells. Error bars are standard deviation of the mean. * p<0.001 by one-way Anova test. (e). Let-7d transfection did not induce cytotoxicity. Neural stem cells were transfected with increasing concentrations of let-7d RNA duplexes, then analyzed for toxicity using LDH assays.
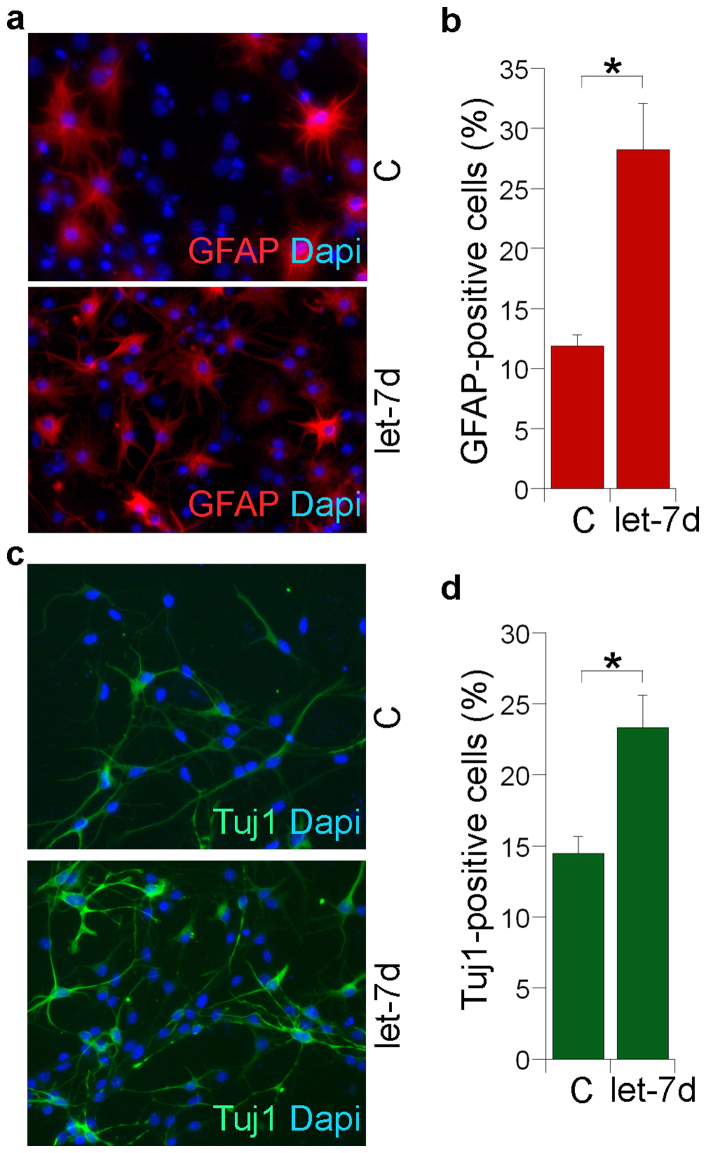
To define the role of let-7d in neural differentiation, neural stem cells transfected with let-7d were induced to differentiate into astroglial cells. When neural stem cells were differentiated for 3 days using forskolin and fetal bovine serum (FBS), the let-7d-treated cells exhibited about 2-fold increase in the GFAP-positive astrocyte populations (Fig. 2a, b). In parallel experiments, let-7d induced a 1.6-fold increase in the Tuj1-positive neuronal populations when neural stem cells were induced with retinoic acid and FBS for 3 days (Fig. 2c, d). These results demonstrate a role for let-7d in promoting neural stem cell differentiation into both neurons and astrocytes.
Figure 2. Let-7d promotes neuronal and astroglial differentiation.
(a). Transfection of let-7d into neural stem cells advanced astroglial differentiation. Let-7d RNA duplexes (100 nM) were transfected into neural stem cells, followed by forskolin and FBS treatment for 3 days. Astrocyte differentiation was evaluated by GFAP staining shown in red. Nuclei dapi staining is shown in blue. (b). Astrocye differentiation was scored as the percentage of GFAP-positive cells over total living cells. (c). Transfection of let-7d into neural stem cells followed by retinoic acid and FBS treatment promoted neuronal differentiation. Neural stem cells were transfected with let-7d and induced to differentiate by treating with retinoic acid and FBS for 3 days. Neuronal differentiation was indicated by Tuj1 staining shown in green. Dapi staining is shown in blue. (d). Neuronal differentiation was quantified as the percentage of Tuj1-positive cells. For both panels (b) and (d), error bars are standard deviation of the mean.* p<0.05 by Student′s t-test.
Let-7d mimics TLX siRNA and miR-9 to promote neuronal differentiation and migration in embryonic mouse brains
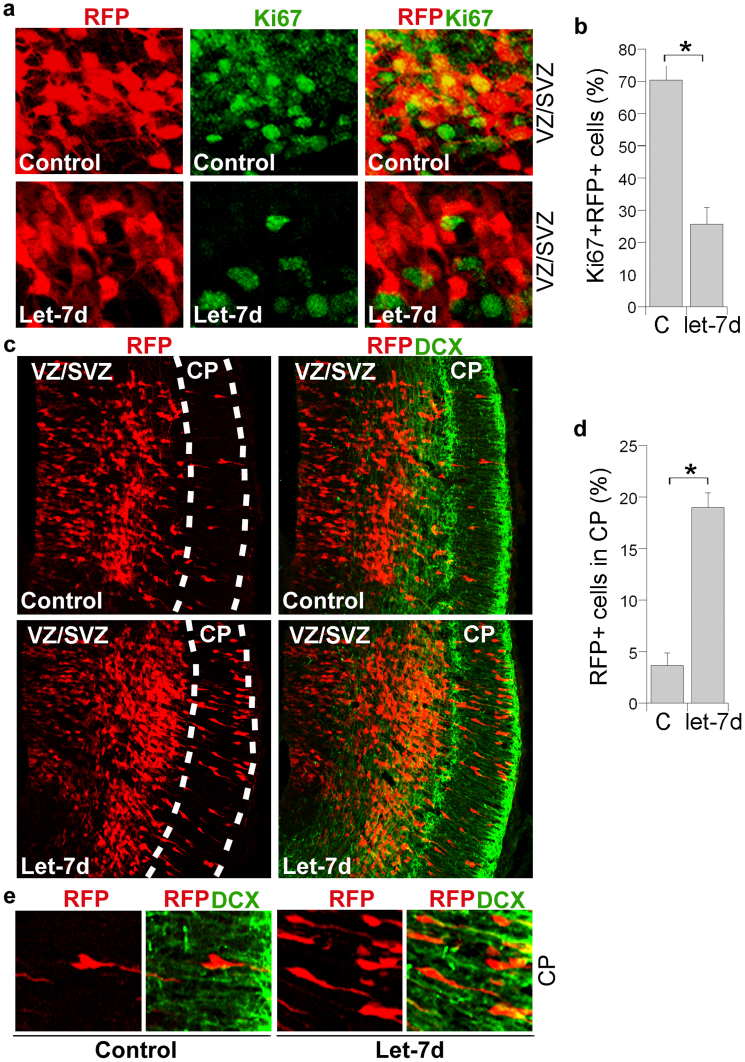
To determine if let-7d regulates neural stem cell proliferation and differentiation in vivo, let-7d RNA duplexes were transfected into neural stem cells in the ventricular zone of E13.5 mouse brains by in utero electroporation. Electroporated brains were analyzed at E15.5. Cells were transfected with let-7d and an RFP reporter. Transfection of let-7d decreased the percent of cells that were labeled by Ki67, a marker of cell proliferation (Fig. 3a, b), but increased the number of transfected cells that migrated to the cortical plate, where differentiated cells reside (Fig. 3c, d). Co-labeling of RFP with the neuronal marker doublecortin (DCX) revealed that the RFP-positive cells that migrated to the cortical plate (CP) are DCX-positive (Fig. 3e), confirming that these cells underwent neuronal differentiation. These results provide in vivo evidence that let-7d inhibits neural stem cell proliferation and accelerates neuronal differentiation. Of interest to us were that the phenotypes induced by transfection of let-7d into embryonic mouse brains is similar to that induced by in utero knockdown of TLX expression27 or overexpression of miR-9 in embryonic mouse brains33. Introducing either a vector expressing TLX small hairpin interference RNAs or miR-9 RNA duplexes into the ventricular zone of E13.5 mouse brains led to precocious neuronal differentiation and migration27,33, resembling the phenotypes we observed after in utero electroporation of let-7d (Fig. 3c-e). These observations led us to look for a potential link between let-7d and TLX/miR-9.
Figure 3. In utero electroporation of let-7d promotes neuronal differentiation and migration in embryonic mouse brains.
(a). In utero electroporation of let-7d into E13.5 mouse brains reduced the proliferating cell population in the ventricular zone and subventricular zone (VZ/SVZ). The Proliferating cells were labeled by Ki67. (b). Quantification of Ki67+RFP+ cells. The percentage of Ki67+RFP+ cells over total RFP+ cells in the VZ/SVZ of let-7d-electroporated brains were calculated and plotted. Error bars are standard deviation of the mean.* p<0.01 by Student's t-test. (c). Transfection of let-7d into E13.5 mouse brains induced precocious cell migration to the cortical plate. The transfected cells were shown in red due to the expression of the RFP reporter. The let-7d mutant RNA was used as the control RNA. CP stands for cortical plate; DCX stands for doublecortin. (d). Quantification of transfected cells (RFP+ cells) that migrated to the CP. C stands for the let-7d mutant control RNA. Error bars are standard deviation of the mean.* p<0.05 by Student's t-test. (e). Enlarged images of the RFP+DCX+ cells in the CP of control RNA or let-7d-transfected brains. The RFP+DCX+ cells showed yellow processes due to the merge of red and green colors.
Let-7d modulates the expression of TLX and miR-9
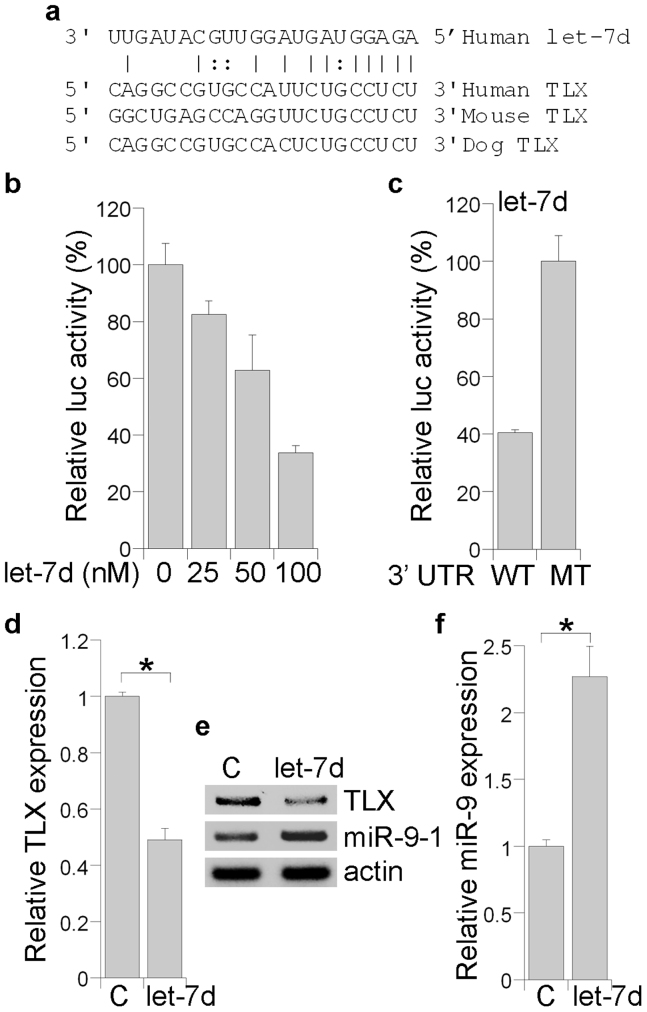
Using the Miranda (http://www.microrna.org) algorithm35,36, we identified a let-7d binding site in the 3′ UTR of the tlx gene, which is conserved in human, mouse and dog tlx sequences (Fig. 4a). To determine if let-7d inhibits TLX expression by targeting the tlx 3′ UTR, HEK 293 cells were transfected with the let-7d RNA duplexes together with a luciferase reporter upstream of the mouse tlx 3′ UTR, which contains the predicted let-7d recognition site and its surrounding nucleotide sequences. Let-7d repressed the luciferase reporter in a dose-dependent manner (Fig. 4b). Furthermore, disruption of the sequence complementarity between let-7d and the tlx 3′ UTR abolished the ability of let-7d to repress the luciferase reporter (Fig. 4c). These results strong imply that let-7d inhibits TLX expression via base-pairing to its 3′ UTR.
Figure 4. Let-7d regulates TLX and miR-9 expression.
(a). The sequence complementarity between let-7d and the tlx 3′ UTR. The vertical lines indicate perfect pairing, whereas the “:” indicates a G-U wobble. (b). Let-7d represses the tlx 3′ UTR luciferase reporter in a dose-dependent manner. (c). Mutation in the tlx 3′ UTR that disrupts the sequence complementarity with let-7d relieved let-7d-mediated repression of the luciferase reporter. WT and MT represent the wild type and mutant tlx 3′ UTR reporter, respectively. (d). Let-7d inhibited TLX mRNA expression, analyzed by real-time RT-PCR analysis. C stands for the control RNA for panels d–f. Error bars are standard error of the mean.* p<0.005 by Student's t-test. (e). Let-7d suppressed TLX expression and induced miR-9-1 expression in neural stem cells, analyzed by RT-PCR analysis. (f). Increased expression of mature miR-9 in let-7d-transfected neural stem cells, measured by real-time RT-PCR. Error bars are standard error of the mean.* p<0.05 by Student's t-test.
To determine if let-7d regulates endogenous TLX expression in neural stem cells, cells were transfected with the let-7d RNA duplexes, then analyzed by RT-PCR to determine the expression levels of TLX (Fig. 4d, e). The levels of TLX mRNA were considerably decreased in let-7d transfected cells when compared to that of control RNA-transfected cells (Fig. 4d, e). Previously, we showed that TLX reduces mature miR-9 expression by repressing the transcription of its primary precursor miR-9-133. Because let-7d inhibited TLX expression (Fig. 4d, e), we next tested if let-7d regulated the levels of the primary and mature miRNA forms of miR-9. In contrast to decreased TLX expression, the level of the miR-9 primary precursor miR-9-1 was elevated in let-7d-transfected cells (Fig. 4e). Accordingly, there was a significant increase in mature miR-9 level in let-7d-treated cells detected by real-time RT-PCR analysis (Fig. 4f). These results demonstrate that let-7d inhibits TLX expression but promotes miR-9 expression, presumably due to reduced expression of TLX, which normally suppresses miR-9-1 transcription.
In utero electroporation of TLX rescues let-7d-induced neural phenotypes
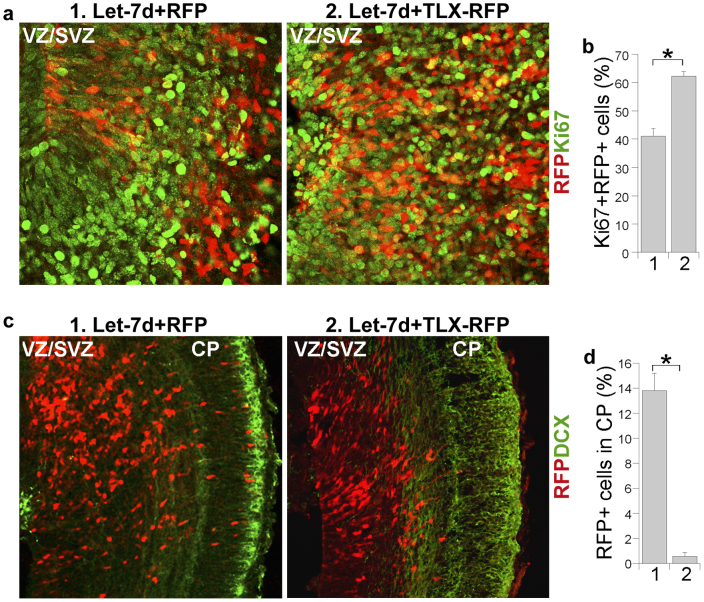
To determine if let-7d promotes neurogenesis by inhibiting TLX expression in vivo, we co-transfected let-7d RNA duplexes with a TLX expression vector that lacks its 3′ UTR by in utero electroporation into the ventricular zone of E13.5 mouse brains. Because the TLX expression vector carries a RFP reporter, the transfected cells were labeled in red (Fig. 5). Co-transfection of let-7d with an empty RFP vector was included as the control. Co-delivery of the TLX expression vector with let-7d (let-7d + TLX-RFP) into E13.5 mouse brains rescued the reduced levels of neural stem cell proliferation, as revealed by increased Ki67 staining (Fig. 5a, b). In contrast, the number of RFP+ cells that migrated to the cortical plate was reduced considerably in let-7d + TLX-RFP-transfected brains, when compared to that in the let-7d + RFP-transfected control brains (Fig. 5c, d). These results suggest that TLX is an important target for let-7d to inhibit neural stem cell proliferation and promote neuronal differentiation and migration in the brain.
Figure 5. Suppression of TLX expression is critical for let-7d function in inhibition of neural stem cell proliferation and promotion of neuronal differentiation and migration.
(a). Co-expression of TLX lacking its 3′ UTR with let-7d (let-7d + TLX-RFP) restored let-7d-reduced levels of cell proliferation in the germinal zone of embryonic mouse brains. VZ/SVZ stands for ventricular zone and subventricular zone. The transfected cells were RFP+, shown in red. The proliferating cells were Ki67+, shown in green. (b). Percentage of the Ki67+RFP+ cells over total RFP+ cells in let-7d + RFP-transfected brains (1) and let-7d + TLX-RFP-transfected brains (2). Error bars are standard deviation of the mean. * p<0.05 by Student′s t-test. (c). Co-expression of TLX lacking its 3′ UTR reversed let-7d-induced premature cell migration to the cortical plate. The transfected cells were RFP+, shown in red. DCX staining (green) was included to show the brain structure. CP stands for cortical plate. (d). Quantification of transfected cells (RFP+) that migrated to the CP in let-7d + RFP (1) and let-7d + TLX-RFP (2)- electroporated brains. *p<0.001 by Student's t-test. Error bars are standard deviation of the mean for both panels (b) and (d).
Discussion
Although we have begun to define the roles played by miRNAs during neurogenesis4, our understanding of how miRNAs regulate the biology of neural stem cells is still very limited. Specifically, there have been no studies yet that define the role played by let-7d in neural stem cell maintenance and neurogensis. In this paper, we demonstrated for the first time that let-7d can regulate multiple aspects of neurogenesis, including neural stem cell proliferation, differentiation, and neuronal migration in the mammalian brain. Overexpression of let-7d inhibited the proliferation of neural stem cells and, instead, promoted neuronal differentiation and outward migration. Moreover, we showed that let-7d regulated these effects on neurogenesis by modulating the TLX and miR-9 expression cascade.
We and others have shown that the brain-specific miR-9 and miR-124 miRNAs, the brain-enriched miR-137 miRNA, and the ubiquitously-expressed let7a and let-7b miRNAs, all play roles in neurogenesis during various stages of development14,15,16,33,34,37,38,39,40,41,42,43,44,45,46. Their expression is coordinated in the brain to regulate neurogenic events that range from the proliferation and self-renewal of neural stem cells to neuronal differentiation, migration, maturation, and functional integration. Furthermore, since miRNAs usually fine-tune the expression of their target genes, inhibition of the same target genes, such as TLX, by multiple miRNAs, including miR-9 and let-7, may lead to more dramatic effect on target gene expression. Regulation of target gene expression then dictates the roles of these miRNAs in modulating specific processes of cell fate control and function.
In this study, we showed that overexpression of let-7d inhibited neural stem cell proliferation and promoted neuronal differentiation and migration in embryonic mouse brains, a phenotype that is similar to that induced by knockdown of TLX expression. The phenotypical resemblance between let-7d overexpression and TLX knockdown suggested to us that let-7d might affect TLX expression. Indeed, our results indicate that let-7d can bind to the 3′ UTR of tlx mRNA to reduce its expression levels. Moreover, we demonstrated that co-electroporation of let-7d and TLX without its 3′ UTR restored the levels of cell proliferation and reversed the increased neuronal differentiation and migration in vivo induced by overexpression of let-7d alone. These results indicate that TLX serves as a key target for let-7d and that let-7d modulates TLX expression to regulate neurogenesis.
The brain-specific miRNA miR-9 has been extensively studied in neurogenesis (see review by Lang and Shi5). Electroporation of miR-9 into mouse brains in utero decreased neural stem cell proliferation and promoted neuronal differentiation and migration33. In a different cellular context, expression of miR-9 enhanced proliferation and inhibited migration of early stage human neural progenitor cells during neural differentiation of human embryonic stem cells in vitro38. Even more recently, a study of ectopic expression of FoxP2, a miR-9 and miR-132 target gene, in cortical neurons suggested that miR-9 can promote neuronal migration47. In this study, we showed that when let-7d was overexpressed, TLX expression was inhibited, which in turn, induced the expression of miR-9, a result consistent with our previous observation that TLX represses miR-9 expression33. Indeed, overexpressing let-7d in embryonic mouse brains enhanced neuronal differentiation and migration, which is phenotypically similar to what we see when miR-9 is electroporated in utero and overexpressed in embryonic mouse brains33. Our observations for these miRNAs are supported by the recent study in which a loss of target gene repression by miR-9 inhibited neuronal migration47.
Recent studies have suggested a role for let-7d in cocaine addiction and attention deficit hyperactivity disorder (ADHD)17,18,48. Defining the role of let-7d in the functions of normal neural stem cells and neurogenesis, and identifying its downstream target genes may help uncover the pathological mechanisms of these neurological disorders. Furthermore, manipulating the expression of let-7d and its downstream targets could identify potential interventions for these disorders in the future.
Methods
Neural stem cell culture, differentiation, immunostaining, cytotoxicity analysis
Mouse neural stem cell isolation, culture and differentiation were performed as described19. Immunostaining was performed using antibodies for BrdU (Accurate; diluted 1:10,000), Tuj1 (Covance, 1:2,500), and GFAP (Advance Immuno; 1:1,000), as described19. Cytotoxicity of let-7d transfection was assessed using LDH assay as described15.
DNA construct preparation
The luciferase reporter containing the 1.4 kb mouse TLX 3′ UTR was constructed as described15. The pEF-TLX-pUb-RFP (TLX-RFP) expression vector was constructed by placing the TLX coding region without its 3′UTR under the elongation factor EF1α promoter and expressing the RFP reporter under the ubiquitin promoter.
Transfection and reporter assays
Transfection of RNA duplexes alone or together with plasmid DNAs were performed as described15. The sequence of the wild type let-7d RNA duplex sense strand is: 5′-AGA GGU AGU AGG UUG CAU AGU-3′. The sequence of the mutant let-7d control RNA duplex sense strand is: 5′-UCU CCU UCU UGG UUG UGU GGU U-3′.
RT-PCR and Northern blotting of let-7d
Mature miR-9 expression was determined by real-time RT-PCR using TaqMan MicroRNA Assay kit (ID 583, Applied Biosystem) and U6 was used as the internal control (ID 001973, Applied Biosystems). Real-time RT-PCR for TLX was performed using SYBR Green PCR master mix (Bio-Rad) and StepOnePlus system (Applied Biosystem) and actin was used as the normalization control. Primers for TLX real-time RT-PCR are TLX forward: 5′-GGT TCA GAC AGC TCC GAT TAG A-3′ and TLX reverse: 5′-TGG AGA GCG GCA ATG GCG GCA GC-3′. Semi-quantitative RT-PCR of TLX and miR-9 primary precursor miR-9-1 was performed as described33. Northern blotting of let-7d was performed as described33 using let-7d antisense probe: 5′-ACT ATG CAA CCT ACT ACC TCT-3′.
In utero electroporation
All animal experiments were performed under the IACUC protocol approved by the City of Hope Institutional Animal Care and Use Committee. In utero electroporation was carried out as described27,33,15. Brain sections were immunostained with antibodies for Ki67 (1:500, Lab Vision) and DCX (1:400, Santa Cruz).
Author Contributions
Y.S. designed research; C.Z., G.S., P.Y., S.L. and Y.S. performed research; C.Z. and Y.S. analyzed data; and Y.S. wrote the paper.
Acknowledgments
We thank Dr. M. Morgan for expert editing, G. Asuelime, Q. Cui, and M. Smith for critical comments. C.Z. was supported by a California Institute for Regenerative Medicine Visiting Scholar Award. G.S. is a Herbert Horvitz Postdoctoral Fellow. This work was supported by NIH NINDS R01 NS059546, RC1 NS068370, California Institute for Regenerative Medicine TR2-01832 and RB4-06277.
References
- Gage F. H. Mammalian neural stem cells. Science 287, 1433–1438 (2000). [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature 431, 350–355 (2004). [DOI] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- Shi Y. et al. MicroRNA regulation of neural stem cells and neurogenesis. J Neurosci 30, 14931–14936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang M. F. & Shi Y. Dynamic Roles of microRNAs in Neurogenesis. Front Neurosci 6, 71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asuelime G. E. & Shi Y. The little molecules that could: a story about microRNAs in neural stem cells and neurogenesis. Front Neurosci 6, 176 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli A. E. et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408, 86–89 (2000). [DOI] [PubMed] [Google Scholar]
- Lee C. T., Risom T. & Strauss W. M. Evolutionary conservation of microRNA regulatory circuits: an examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny. DNA Cell Biol 26, 209–218 (2007). [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Meyer J., Borkhardt A. & Tuschl T. New microRNAs from mouse and human. RNA 9, 175–179 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S. M. et al. RAS is regulated by the let-7 microRNA family. Cell 120, 635–647 (2005). [DOI] [PubMed] [Google Scholar]
- Yu F. et al. Let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131, 1109–1123 (2007). [DOI] [PubMed] [Google Scholar]
- Wulczyn F. G. et al. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. Faseb J 21, 415–426 (2007). [DOI] [PubMed] [Google Scholar]
- Rybak A. et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol 10, 987–993 (2008). [DOI] [PubMed] [Google Scholar]
- Schwamborn J. C., Berezikov E. & Knoblich J. A. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136, 913–925 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C. et al. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci USA 107, 1876–1881 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino J., Kim I., Chada K. & Morrison S. J. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell 135, 227–239 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekar V. & Dreyer J. L. microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Mol Cell Neurosci 42, 350–362 (2009). [DOI] [PubMed] [Google Scholar]
- Chandrasekar V. & Dreyer J. L. Regulation of MiR-124, Let-7d, and MiR-181a in the Accumbens Affects the Expression, Extinction, and Reinstatement of Cocaine-Induced Conditioned Place Preference. Neuropsychopharmacology 36, 1149–1164 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. et al. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature 427, 78–83 (2004). [DOI] [PubMed] [Google Scholar]
- Shi Y., Sun G., Zhao C. & Stewart R. Neural stem cell self-renewal. Crit Rev Oncol Hematol 65, 43–53 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. L., Zou Y., He W., Gage F. H. & Evans R. M. A role for adult TLX-positive neural stem cells in learning and behaviour. Nature 451, 1004–1007 (2008). [DOI] [PubMed] [Google Scholar]
- Liu H. K. et al. The nuclear receptor tailless is required for neurogenesis in the adult subventricular zone. Genes Dev 22, 2473–2478 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. K. et al. The nuclear receptor tailless induces long-term neural stem cell expansion and brain tumor initiation. Genes Dev 24, 683–695 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Sun G., Murai K., Ye P. & Shi Y. Characterization of TLX Expression in Neural Stem Cells and Progenitor Cells in Adult Brains. PLoS One 7, e43324 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy K. et al. The Tlx gene regulates the timing of neurogenesis in the cortex. J Neurosci 24, 8333–8345 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenman J. M., Wang B. & Campbell K. Tlx controls proliferation and patterning of lateral telencephalic progenitor domains. J Neurosci 23, 10568–10576 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. et al. Nuclear Receptor TLX Regulates Cell Cycle Progression in Neural Stem Cells of the Developing Brain. Mol Endocrinol 22, 56–64 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G., Yu R. T., Evans R. M. & Shi Y. Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proc Natl Acad Sci USA 104, 15282–15287 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G. et al. Histone demethylase LSD1 regulates neural stem cell proliferation. Mol Cell Biol 30, 1997–2005 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Q. et al. Orphan nuclear receptor TLX activates Wnt/beta-catenin signalling to stimulate neural stem cell proliferation and self-renewal. Nat Cell Biol 12, 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmi M. et al. TLX activates MASH1 for induction of neuronal lineage commitment of adult hippocampal neuroprogenitors. Mol Cell Neurosci 45, 121–131 (2010). [DOI] [PubMed] [Google Scholar]
- Chavali P. L., Saini R. K., Matsumoto Y., Agren H. & Funa K. Nuclear orphan receptor TLX induces Oct-3/4 for the survival and maintenance of adult hippocampal progenitors upon hypoxia. J Biol Chem 286, 9393–9404 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Sun G., Li S. & Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nature Struct Mol Biol 16, 365–371 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G. et al. MiR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat Commun 2, 529 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B. et al. Human MicroRNA targets. PLoS Biol 2, e363 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P. & Burge C. B. Prediction of mammalian microRNA targets. Cell 115, 787–798 (2003). [DOI] [PubMed] [Google Scholar]
- Krichevsky A. M., Sonntag K. C., Isacson O. & Kosik K. S. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells 24, 857–864 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaloy C. et al. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell 6, 323–335 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber J. et al. MiR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med 6, 14 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrt R. D. et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells 28, 1060–1070 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan J., Lee S., Lee B., Lee J. W. & Lee S. K. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev 21, 744–749 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Pfaff S. L. & Gage F. H. A functional study of miR-124 in the developing neural tube. Genes Dev 21, 531–536 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L. C., Pastrana E., Tavazoie M. & Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci 12, 399–408 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorano N. A. & Mallamaci A. Promotion of embryonic cortico-cerebral neuronogenesis by miR-124. Neural Dev 4, 40 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo A. S., Staahl B. T., Chen L. & Crabtree G. R. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460, 642–646 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev E. V., Zhang J., Carrasco M. A. & Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell 27, 435–448 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clovis Y. M., Enard W., Marinaro F., Huttner W. B. & De Pietri Tonelli D. Convergent repression of Foxp2 3'UTR by miR-9 and miR-132 in embryonic mouse neocortex: implications for radial migration of neurons. Development 139, 3332–3342 (2012). [DOI] [PubMed] [Google Scholar]
- Wu L. et al. A novel function of microRNA let-7d in regulation of galectin-3 expression in attention deficit hyperactivity disorder rat brain. Brain Pathol 20, 1042–1054 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]