Abstract
Chronic lung diseases (CLDs), including chronic obstructive pulmonary disease (COPD), are the second leading cause of death worldwide. The first report of database-driven drug discovery in carefully phenotyped COPD specimens has now been published in Genome Medicine, combining gene expression data in defined emphysematous areas with connectivity-map-based compound discovery. This joint effort may lead the way to novel and potentially more efficient concepts of personalized drug discovery for COPD in particular, and CLD in general.
See research article http://genomemedicine.com/content/4/8/67/abstract
Chronic lung diseases and chronic obstructive pulmonary disease: a need for innovation
Chronic lung diseases (CLDs), including chronic obstructive pulmonary disease (COPD), asthma, lung cancer, neonatal chronic lung disease and pulmonary fibrosis, are the leading diseases worldwide with regard to mortality, prevalence and socioeconomic burden [1,2]. The devastating impact of these diseases is largely due to increased environmental effects on lung health in recent years (for example, air pollution, allergens, and cigarette smoke or related particles), limited understanding of their mechanisms of pathogenesis, and consequently, insufficient therapeutic options. Importantly, COPD is the only leading cause of death with increasing incidence among the top diseases: by 2020, more than 6 million deaths every year are projected to be due to COPD [2].
COPD is defined as 'a decreased airflow that is not fully reversible' and is classically diagnosed by lung function testing (FEV1, forced expiratory volume in one second). Pathologically, COPD is characterized by airway inflammation, loss of functional alveolar tissue, irreversible airflow obstruction and loss of lung function [3,4]. It has two main pathological features: small airway disease (SAD; which includes airway inflammation with increased mucous production, activation of immune cells, airway wall remodeling and peribronchiolar fibrosis) and emphysema (defined as the destruction of the distal alveolar architecture due to distal airspace enlargement). Ultimately, both features can lead to a loss of functional alveolar epithelium and impaired lung function, with an apparent inability of the lung to self-repair.
It has long been known that a decline in lung function does not accurately correlate with the degree of emphysema or SAD. It has therefore been a major challenge to definitively phenotype COPD patients with regard to their underlying dominant pathological process (that is, emphysema or SAD), and this has hampered clinical study design and drug discovery. Until recently, approaches for the accurate correlation of gene expression signatures with histological subtypes of COPD were not available, and this has prevented the molecular characterization of cell populations involved in dominant pathogenic processes at different stages of COPD/emphysema. This is in striking contrast to the extensive profiling of idiopathic pulmonary fibrosis (IPF), which has resulted in a clear gene expression signature in the lungs and blood of individuals with IPF, accelerating biomarker identification and disease stratification for this disease in recent years [5].
Definition of a strong disease signature
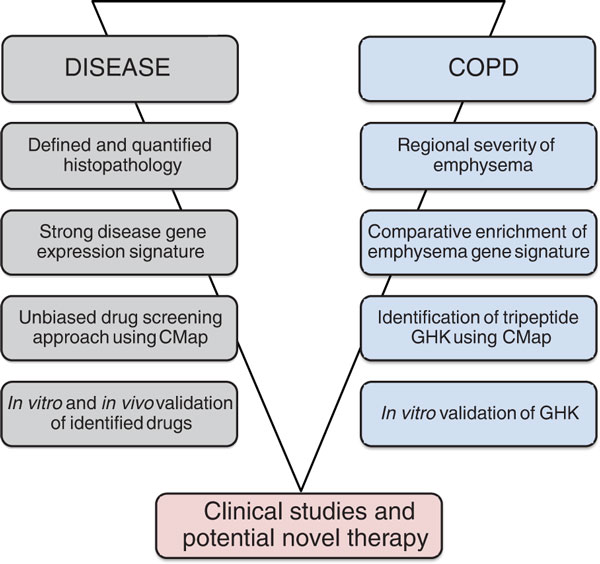
A recent study by Campbell et al. published in Genome Medicine now appears to overcome these challenges in COPD [6]. The study is remarkable for the following reasons: (1) gene signatures in different grades of emphysema were derived from regions of quantitatively assessed pathology (by micro-computed tomography and stereology); (2) these signatures were extensively compared with available COPD datasets published by different groups; (3) these validated signatures were then queried using the Connectivity Map (CMap) [7,8] for compounds that might be capable of reversing the emphysematous phenotype; and (4) one identified compound (the tripeptide GHK) was then tested in vitro for proof-of-principle (Figure 1).
Figure 1.
A template for new drug discovery. The general strategy used by Campbell et al. [6] is outlined on the left. The detailed findings of each step from the paper are depicted on the right.
In a joint venture by several leading COPD groups, a comprehensive approach was then used to assess gene expression patterns in lung regions of COPD specimens with defined severities of emphysema [6]. The venture included groups with outstanding expertise in micro- and macro-imaging of the lung, gene expression analysis, clinical phenotyping, and most importantly, bioinformatics. The authors made use of a recently published novel method to assess regional severity of emphysema by determining the mean linear intercept between alveolar walls using micro-computed tomography [4]. They identified a set of 127 differentially regulated genes that were significantly associated with the degree of emphysema in the lung. Importantly, this signature was further validated using gene expression data from other cross-sectional studies of COPD. This signature of 127 genes was significantly enriched among COPD-regulated genes in four out of five previously published data sets, validating this set as a strong signature for lung emphysema.
Importantly, enrichment analysis of gene functions revealed an over-representation of genes involved in B-cell receptor signaling and reduced expression of genes involved in transforming growth factor (TGF)-β signaling. Indeed, an increasing number of CD79A-positive B cells correlated with the severity of emphysema, as shown by immunohistochemical analysis. Using an elegant bioinformatics approach, the authors further validated these gene sets by comparing them with a defined TGF-β activation signature, using seven publicly available gene expression studies on TGF-β-induced gene expression. Interestingly, both the induction of B-cell receptor signaling and downregulation of TGF-β activation were not fully appreciated in previous COPD profiling studies. Thus, strong and validated disease signatures depend on the stringency of data analysis and comparison with apparently unrelated, publicly available data sets from other studies.
Unbiased identification of new drugs
Another strength of the study by Campbell and colleagues [6] lies in the use of the enriched emphysema gene expression signature for an unbiased drug screening approach by in silico connectivity mapping. CMap exploits the transcriptome as a 'universal language' describing cellular responses [7,8] to distinct drugs to connect drug discovery, biology and disease phenotypes. Classically, to exploit CMap, a disease phenotype, represented by a strong gene expression signature (in this case emphysema), is compared with gene expression profiles of cells that have been treated with distinct drugs (here GHK) for various exposure times. While the initial CMap reference catalogue comprised gene expression profiles of 164 drugs, the current catalogue contains 1,309 Food and Drug Administration (FDA)-approved small molecules that have been evaluated in five cancer cell lines [8]. CMap can thus be used as an unbiased in silico screening approach to identify drugs that have either positive or negative connectivity scores, implicating these drugs either as potential inducers of disease phenotypes or therapeutics thereof.
Campbell and co-workers used their enriched emphysema signature and screened for compounds that demonstrated connectivity with regard to gene expression changes observed in emphysema. Connectivity analysis was performed with high stringency by double fitting of the two gene expression signatures, that is, induced with severity of emphysema and deactivated TGF-β signaling. They identified the tripeptide GHK, a natural tripeptide involved in wound healing, which showed inverse connectivity. Importantly, in experimental studies using GHK, it partially reversed emphysema-associated phenotypes in fibroblasts from COPD patients.
Future directions
It will now be of utmost interest and importance to see whether further animal experiments will confirm that GHK is indeed able to revert disease in vivo, using relevant animal models of emphysema or COPD. If this is shown to be the case, clinical studies will then surely investigate whether GHK has clinical potential for individuals with COPD with a dominant emphysematous phenotype. It will be interesting to see whether such a comparative bioinformatics approach will generate new and/or overlapping gene expression signatures in lung, blood or bronchoalveolar lavage samples not only in COPD, but also in other heterogeneous CLD, such as asthma or chronic neonatal lung disease.
Since the release of CMap, several studies have shown the feasibility of this drug-screening approach for the identification of new therapeutics, drug repurposing, and the prediction of off-target or side effects, as reviewed by Qu and Rajpal [8]. Recently, Wang and colleagues [9] applied a similar in silico approach to screen for candidate therapeutic compounds for lung adenocarcinoma. The CMap approach also offers the possibility of predicting synergies of drug combinations. In the future, the use of CMap might also add to our understanding of disease pathogenesis by connecting gene expression profiles of drugs that specifically inhibit single pathways with transcriptional profiles of disease phenotypes.
The use of gene expression profiling in histologically quantified tissue specimens with CMap querying reveals an exciting new approach to perform knowledge-based drug screening. Biomedical research has classically involved the tedious mechanistic understanding of single molecular targets and their interactions as a starting point for lead compound identification and subsequent optimization, but future research might move towards a more empirical analysis of gene expression signatures in combination with drug signatures.
In summary, the approach described herein (Figure 1) might facilitate lead compound identification and repurposing of drugs. Drug discovery is currently estimated to take approximately 15 years, with 90% of drugs failing to move beyond early clinical testing stages [10]. Therefore, such an approach is expected to save precious time and money, and ultimately might have the potential to decrease disease burden more effectively.
Abbreviations
CLD: chronic lung disease; CMap: connectivity map; COPD: chronic obstructive pulmonary disease; IPF: idiopathic pulmonary fibrosis; SAD: small airway disease; TGF: transforming growth factor.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Silke Meiners, Email: silke.meiners@helmholtz-muenchen.de.
Oliver Eickelberg, Email: oliver.eickelberg@helmholtz-muenchen.de.
Acknowledgements
The authors are supported by the Helmholtz Association.
References
- Cosio MG, Saetta M, Agusti A. Immunologic aspects of chronic obstructive pulmonary disease. N Engl J Med. 2009;360:2445–2454. doi: 10.1056/NEJMra0804752. [DOI] [PubMed] [Google Scholar]
- Stockley RA, Mannino D, Barnes PJ. Burden and pathogenesis of chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2009;6:524–526. doi: 10.1513/pats.200904-016DS. [DOI] [PubMed] [Google Scholar]
- McDonough JE, Yuan R, Suzuki M, Seyednejad N, Elliott WM, Sanchez PG, Wright AC, Gefter WB, Litzky L, Coxson HO, Paré PD, Sin DD, Pierce RA, Woods JC, McWilliams AM, Mayo JR, Lam SC, Cooper JD, Hogg JC. Small-airway obstruction and emphysema in chronic obstructive pulmonary disease. N Engl J Med. 2011;365:1567–1575. doi: 10.1056/NEJMoa1106955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuder RM, Petrache I. Pathogenesis of chronic obstructive pulmonary disease. J Clin Invest. 2012;122:2749–2755. doi: 10.1172/JCI60324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards TJ, Kaminski N, Baribaud F, Flavin S, Brodmerkel C, Horowitz D, Li K, Choi J, Vuga LJ, Lindell KO, Klesen M, Zhang Y, Gibson KF. Peripheral blood proteins predict mortality in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2012;185:67–76. doi: 10.1164/rccm.201101-0058OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JD, McDonough JE, Zeskind JE, Hackett TL, Pechkovsky DV, Brandsma CA, Suzuki M, Gosselink JV, Liu G, Alekseyev YO, Xiao J, Zhang X, Hayashi S, Cooper JD, Timens W, Postma DS, Knight DA, Lenburg ME, Hogg JC, Spira A. A gene expression signature of emphysematous lung destruction and its reversal by the tripeptide GHK. Genome Med. 2012;4:67. doi: 10.1186/gm368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet J-P, Subramanian A, Ross KN, Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA, Lander ES, Golub TR. The Connectivity Map: using gene expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–3510. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Qu XA, Rajpal DK. Applications of Connectivity Map in drug discovery and development. Drug Discovery Today. 2012. http://dx.doi.org/10.1016/j.drudis.2012.07.017 [DOI] [PubMed]
- Wang G, Ye Y, Yang X, Liao H, Zhao C, Liang S. Expression-based in silico screening of candidate therapeutic compounds for lung adenocarcinoma. PLoS One. 2011;6:E14573. doi: 10.1371/journal.pone.0014573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirota M, Dudley JT, Kim J, Chiang AP, Morgan A, Sweet-Cordero A, Sage J, Butte AJ. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med. 2011;3:96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]