Abstract
The use of saliva as a diagnostic sample provides a non-invasive, cost-efficient method of sample collection for disease screening without the need for highly trained professionals. Saliva collection is far more practical and safe compared with invasive methods of sample collection, because of the infection risk from contaminated needles during, for example, blood sampling. Furthermore, the use of saliva could increase the availability of accurate diagnostics for remote and impoverished regions. However, the development of salivary diagnostics has required technical innovation to allow stabilization and detection of analytes in the complex molecular mixture that is saliva. The recent development of cost-effective room temperature analyte stabilization methods, nucleic acid pre-amplification techniques and direct saliva transcriptomic analysis have allowed accurate detection and quantification of transcripts found in saliva. Novel protein stabilization methods have also facilitated improved proteomic analyses. Although candidate biomarkers have been discovered using epigenetic, transcriptomic, proteomic and metabolomic approaches, transcriptomic analyses have so far achieved the most progress in terms of sensitivity and specificity, and progress towards clinical implementation. Here, we review recent developments in salivary diagnostics that have been accomplished using genomic, transcriptomic, proteomic and metabolomic approaches.
Saliva: a biospecimen for non-invasive and accurate disease detection
The discovery that saliva contains molecular profiles that reflect systemic diseases has opened the doors to a new non-invasive diagnostic methodology: 'salivary diagnostics'. Using saliva in diagnosis is not only practical and non-invasive, but at times is also proving to be more accurate than available alternatives. These advantages might facilitate early detection of many diseases and improve clinical management.
Saliva comprises the secretions of the parotid, submandibular and sublingual glands, hundreds of minor salivary glands and gingival crevice fluid. Saliva functions include food digestion, bolus formation, lubrication and taste facilitation and immune-related functions mediated by secreted antimicrobial peptides [1] and immunoglobulins [2]. However, these components constitute only a small part of saliva. Many molecular species have now been found, but their functions in the oral cavity remain largely unknown.
Cell-free saliva (CFS) has been found to contain over a thousand proteins that are thought to be involved in a wide range of biological functions [3], as well as mRNA and microRNA (miRNA) transcripts [4-6], and metabolites [7,8]. Detecting changes in the salivary concentrations of these molecules has allowed the detection of oral and systemic diseases. Recent developments in genomic, proteomic and metabolomic approaches have facilitated sensitive and high-throughput analysis of saliva and are proving increasingly useful for diagnostics. In this article, we review recent advances in the identification of salivary biomarkers using these approaches, and the implications for diagnostic applications.
Saliva biomarker study design and clinical trial design
Early studies of salivary diagnostics for breast cancer assessed the use of proteins, including c-erbB-2 (human epidermal growth factor receptor 2, a receptor tyrosine kinase involved in cell growth), VEGF (vascular endothelial growth factor, a signal protein involved in vasculogenesis and angiogenesis), EGF (epidermal growth factor, which promotes cell growth, division and differentiation), and CEA (carcinoembryonic antigen, a glycoprotein tumor marker involved in cell adhesion) [9-13]. However, these studies investigated the presence of known serum biomarkers in saliva, rather than the potential existence of unknown biomarkers. Subsequently, de novo transcriptomic, proteomic and metabolomic approaches [5-8,14-17] have allowed the discovery and validation of salivary biomarker profiles for breast, oral, pancreatic and lung cancers, and periodontitis.
Stringent testing of biomarker performance, including verification and validation, is necessary before clinical application. Although there have been dramatic advances in biotechnology and biomarker research in the last decade, there have been relatively few successful clinical trials of biomarkers, compared with therapeutic research. Therapeutic research utilizes randomized placebo-controlled blinded clinical trials with internationally agreed standards in the evaluation of definitive clinical endpoints and outcomes. Analogous standards for biomarker research did not exist until 2008 with the proposal of prospective specimen collection and retrospective blinded evaluation (PRoBE) study design [18]. PRoBE incorporates prospective specimen collection from the target population, collected in a blinded fashion with no knowledge about patient outcomes. After outcome has been determined, patients and control subjects are selected randomly and their specimens tested. We expect that the application of PRoBE will greatly facilitate the transfer of salivary diagnostics to the clinic.
In the following sections we review recent advances made in the salivary biomarker field using genomic, epigenomic, transcriptomic, proteomic and metabolomic approaches. Tables 1 and 2 describe biomarkers that have been characterized and might be potentially useful for clinical applications.
Table 1.
Promising oral, and head and neck cancer salivary biomarkers discovered using epigenomics, transcriptomics, proteomics and metabolomics
| Approach | Markers | Sensitivity/specificity (%)/AUC | |
|---|---|---|---|
| Epigenomics | Candidate from previous study, Q-MSP analysis (HNSCC) [29] | DAPK, DCC, MINT-31, TIMP-3, p16, MGMT, CCNA1 | NR |
| Candidate gene selection based on a previous study, Q-MSP discovery and validation (HNSCC) [23] | MINT31, CCNA1, DCC, DAPK, p16 | 34.1/91.8/0.63 | |
| MINT31, CCNA1, p16 | 24.0/97.1/0.61 | ||
| Candidate from previous study, Q-MSP discovery and validation (HNSCC) [25] | KIF1A, EDNRB | 77.4/93.1/NR | |
| Candidate from previous study, Q-MSP assessment (HNSCC) [26] | EDNRB | 65/51/0.61 | |
| Discovery by methylation array (OSCC) [27] | GABRB3_E42_F, IL11_P11_R, INSR_P1063_R, NOTCH3_E403_F, NTRK3_E131_F, PXN_P308_F | 77/83/NR | |
| ADCYAP1_P455_R, CEBPA_P706_F, EPHA5_E158_R, FGF3_E198_R, HLF_E192_F, IL11_P11_R, INSR_P1063_R, NOTCH3_E403_F | 69/96/NR | ||
| ERBB4_P255_F, IL11_P11_R, PTCH2_P37_F, TMEFF1_P234_F, TNFSF10_E53_F, TWIST1_P44_R | 62/100/NR | ||
| Discovery with HumanMethylation27 DNA assay, validation with Q-MSP [30] | HOXA9, NID2 | 50/90/0.77 | |
| Transcriptomics | Microarray discovery and qPCR validation (OSCC) [4] | IL8, IL1B, OAZ1, SAT | 91/91/0.95 |
| qRTPCR and ELISA validation of previously reported candidates [67] | IL-8, SAT, H3F3A, S100P | 71/89/0.81 | |
| IL-8, IL-1B, SAT, OAZ1 | 79/77/0.86 | ||
| IL-8, IL-1B, SAT, DUSP1 | 80/77/0.85 | ||
| IL-8, IL-1B, S100P, OAZ1 | 64/86/0.78 | ||
| IL-8, SAT, OAZ1, S100P | 87/56/0.75 | ||
| Discovery and validation by RT-preamp-qPCR (OSCC) [6] | miR-200a | NR/NR/0.65 | |
| miR-125a | NR/NR/0.62 | ||
| Candidate gene selection based on previous study, qRT-PCR quantification [57] | miR-31 | 80/68/0.82 | |
| Proteomics | Discovery by C4 RP-LC and capillary reversed-phase LC with quadruple time-of-flight MS and validation by ELISA and immunoblotting (OSCC) [15] | M2BP, profilin, CD59, MRP14, catalase | 90/83/0.93 |
| ELISA assessment and qPCR confirmation [65] | IL-8 | 86/97/0.98 | |
| Combination of proteomic/transcriptomics | Reproducibility study of validated biomarkers using ELISA and qRT-PCR (OSCC) [46] | Proteins: IL-1B, IL-8 and M2BP | |
| mRNAs: IL-8, IL-1B, SAT1, S100P | 0.89/0.78/0.86, 0.67/0.96/0.85, 0.82/0.84/0.88 for OSCC total/T1-T2/T3-T4 respectively | ||
| Metabolomics | ULC/Q-TOF-MS (OSCC) [8] | Valine, lactic acid | 86.5/82.4/0.89 |
| Discovery by CE-TOF-MS-based metabolomics [7] | Taurine, piperidine and a peak at 120.0801 m/z | NR/NR/0.87 | |
AUC, area under curve; CE-TOF-MS, capillary electrophoresis time-of-flight mass spectrometry; ELISA, enzyme-linked immunosorbent assay; HNSCC, head and neck squamous cell carcinoma; LC, liquid chromatography; MS, mass spectrometry; NR, not reported; OSCC, oral squamous cell carcinoma; Q-MSP, quantitative methylation-sensitive PCR; qPCR, quantitative PCR; qRT-PCR, quantitative reverse transcription PCR; RT-preamp-qPCR, reverse transcription preamplification quantitative PCR; ULC/Q-TOF-MS, ultraperformance liquid chromatography coupled with quadruple/time-of-flight mass spectrometry; C4 RP-LC, C4 reversed-phase liquid chromatography.
Table 2.
Promising salivary biomarkers for other diseases discovered using transcriptomics, proteomics and metabolomics
| Approach | Markers | Sensitivity/specificity (%)/AUC | |
|---|---|---|---|
| Transcriptomics | Affymetrix array discovery and qRT-PCR validation (pancreatic cancer) [5] | KRAS, MBD3L2, ACRV1, DPM1 | 90.0/95.0/0.97 |
| Microarray discovery and qRT-PCR verification and pre-validation (lung cancer) [84] | CCNI, EGFR, FGF19, FRS2, GREB1 | 93.75/82.8/0.93 | |
| ELISA assessment (periodontal disease) [81] | MIP-1α | 94/92.7/0.94 | |
| Multiplex protein array discovery. Markers for distinguishing high and low responders to treatment for gingivitis (periodontal disease) [61] | IL-6, IL-8 | NR/NR/0.81 (HR) and 0.72 (LR) | |
| Proteomics | Two-dimensional gel electrophoresis and LC-MS-MS (lung cancer) [59] | Calprotectin, AZGP1, haptoglobin hp2 | 88.5/92.3/0.9 |
| Combination proteomic/transcriptomic approaches | Discovery by 2D-DIGE and RT-PCR/Affymetrix, validation by qRT-PCR (breast cancer) [14] | mRNAs: CSTA, TPT1, IGF2BP1, GRM1, GRIK1, H6PD, MDM4, S100A8 | |
| Protein: CA6 | 83/97/92% accuracy | ||
| Metabolomics | Discovery by CE-TOF-MS-based metabolomics (pancreatic cancer) [7] | Leucine with isoleucine, tryptophan, valine, glutamic acid, phenylalanine, glutamine, aspartic acid | NR/NR/0.99 |
| Discovery by CE-TOF-MS-based metabolomics (breast cancer) [7] | NR | NR/NR/0.97 | |
| Discovery by SERS (lung cancer) [17] | Unidentified peak wavelengths; 822, 884, 909, 925, 1009, 1,077, 1,369, 1,393, 1,721 cm-1 | 94/81/86% accuracy |
AUC, area under curve; CE-TOF-MS, capillary electrophoresis time-of-flight mass spectrometry; ELISA, enzyme-linked immunosorbent assay; HR, high responder; LC-MS-MS, liquid chromatography-tandem mass spectrometry; LR, low responder; NR, not reported; qRT-PCR, quantitative reverse transcription PCR; RT-PCR, reverse transcription PCR; SERS, surface-enhanced Raman spectroscopy; 2D-DIGE, two-dimensional difference gel electrophoresis.
The salivary genome and epigenome
Salivary genetic and epigenetic analysis provides insight into the presence of invading pathogens, and aberrant gene transcription profiles that directly reflect pathological genetic processes pertaining to, for example, cancers. This section discusses the most recent advances in genetic and epigenetic biomarker research. Table 1 gives details of promising genetic and epigenetic salivary biomarkers reported for oral, and head and neck cancers.
The salivary genome consists of DNAs representing the genome of the individual, oral microbiota and infecting DNA viruses. The quality and yield of DNA that can be obtained from saliva is relatively good compared with blood and urine, can be used for genotyping, amplification or sequencing [19], and can be stored long-term without significant degradation [20]. As such, salivary DNA is a robust analyte for diagnostics, but is limited to reflecting presence or absence of specific genes, alteration to sequences (mutation) and methylation status, and cannot provide information on upregulation and downregulation of gene expression.
The potential for salivary diagnosis of Epstein-Barr virus (EBV)-associated nasopharyngeal carcinoma (NPC) using quantitative PCR (qPCR) was reported in 2011 [21]. This method achieved a detection rate of 80%, significantly less sensitive than serological methods (>90%). Also results were somewhat inconsistent with previous reports, as viral load was significantly higher in saliva post-therapy compared with pre-therapy. The authors suggested that this might have been due to decreased salivary flow rates caused by radiotherapy. However, higher levels of EBV DNA in T3/T4 stage NPC compared with T1/T2 stage NPC were reported, which is in agreement with other studies [21].
Aberrant methylation of tumor suppressor genes is common in cancer cells, and in oral squamous cell carcinoma (OSCC) hypermethylation has been linked with several cancer-related alterations of dysplastic oral epithelium [22]. Promoter hypermethylation has been reported in premalignant OSCC lesions and in head and neck squamous cell carcinoma (HNSCC), showing potential as a biomarker for early detection of primary and relapsing OSCC or HNSCC [23-28] (Table 1). Furthermore, methylation array analysis of DNA extracts from preoperative saliva, postoperative saliva, and tissue samples from patients with OSCC and from saliva from normal individuals showed significant differences in methylation patterns between preoperative and postoperative OSCC and between preoperative OSCC and control saliva samples. Gene panels were constructed that had low-to-moderate sensitivity and good-to-excellent specificity for OSCC [27] (Table 1).
Quantitative methylation specific PCR (Q-MSP) has shown promise for the development of biomarkers for HNSCC. Carvalho et al. [29] studied tumor-suppressor gene hypermethylation in salivary rinses from pre-treatment HNSCC patients using Q-MSP. More than half of the study population was found to have methylation of at least one of the selected genes and had significantly reduced overall survival (Table 1). Early epigenetic attempts have been limited to approaches utilizing candidate genes or cell-culture-based discovery, with validation using well-characterized pathology specimens from homogeneous cohorts. Guerrero-Preston et al. [30] sought to overcome these limitations by using a new study design. To identify differentially methylated genes that could distinguish OSCC or HNSCC tumors from normal tissue, they used clinically defined samples from populations with different risk profiles, high-density promoter methylation platforms, publicly available expression arrays, two-stage design ('discovery screen' and 'prevalence screen') and Q-MSP in a phase I biomarker development trial. This approach aims to improve phase I trials, so that only robust biomarkers are assessed in phase II trials. The discovery screen found that promoter methylation in KIF1A, HOXA9, NID2 and EDNRB genes had a moderate-to-substantial correlation with clinical diagnosis. The prevalence screen showed a near perfect correlation between HOXA9 and NID2 promoter hypermethylation and histological diagnosis. The combined analysis of these two genes resulted in 94% sensitivity, 97% specificity and an area under the receiver operating characteristic curve (AUC) of 0.97 in tissue samples. However, the performance was far lower using saliva samples, with very low sensitivity [30] (Table 1).
The Human Oral Microbe Identification Microarray (HOMIM) has recently been developed comprising an oligonucleotide microarray based on 16S rRNA, and has allowed profiling and monitoring of changes in the oral microbiota [31]. Alterations in the oral bacterial profile have been found to correlate with several diseases, including pancreatic, oral and lung cancer [32], colonic neoplasia and extracolonic malignancy [33], cardiovascular and cerebrovascular disease [34-36], and preterm birth [37], and these findings are being explored as potential salivary diagnostic approaches.
The salivary transcriptome
mRNAs and miRNAs are secreted from cells into the extracellular milieu and can be found in biofluids that are distant to the cellular sources [6,38,39]. In a diseased state, transcription of specific mRNAs and miRNAs is altered. Although the validity of salivary RNAs for the development of biomarkers with diagnostic potential initially received some criticism [40], this approach is now widely accepted. However, the precise sources of salivary RNAs and other molecules remain unclear (for further discussion see Box 1).
Standard procedures for salivary mRNA analysis previously required low temperatures and time-consuming and costly mRNA isolation, which impeded clinical application. Now, simple methods of stabilizing mRNA in saliva samples have been developed, allowing room temperature storage without stabilizing agents, and sample analysis without further processing: so-called 'direct-saliva-transcriptomic-analysis' [41]. However, this approach still involves centrifugation. An alternative method has been described [42], but was based on the use of a costly stabilizing agent. As such, neither method is completely practical for all applications. Salivary and plasma mRNAs can be surprisingly stable. The mechanism that confers such stability is currently unknown; however, AU-rich element (ARE) sequence motifs in the 3' UTR regions of transcripts can recruit ARE-binding proteins, thereby controlling stability [43].
Microarray technology has allowed high-throughput saliva analysis and is the current gold standard for identifying saliva transcripts. The salivary transcriptome is profiled using microarrays and validated with qPCR. However, due to the low concentration of some biomarkers and occasional small sample volume, further innovations in technology have been required. Hu et al. [44] has overcome these limitations by two developments: a universal mRNA-amplification method for microarray discovery, and a multiplex pre-amplification method for qPCR validation. Moreover, the multiplex pre-amplification method allows the simultaneous detection of many transcripts, providing cost-effective screening and quantitative measurement using a relatively small amount of pre-amplified product. In the following sections, we discuss advances in transcriptomic analysis of mRNAs and miRNAs using saliva samples.
mRNA
Characterization of mRNA profiles in body fluids provides insights into gene transcription in normal and disease states. There have been several advances in transcriptomic salivary mRNA biomarker characterization in the last 4 years. As discussed in this section, salivary mRNA biomarkers have been pursued for a number of diseases, including OSCC, primary Sjögren's syndrome, pancreatic cancer and ovarian cancer, and they have proved to be sensitive, specific and accurate.
Transcriptomic biomarkers for primary Sjögren's syndrome have undergone preclinical validation [45], and mRNA biomarkers for OSCC previously discovered in a US cohort [4] have been found to detect OSCC in a cohort of different ethnicity [46]. Discovery and validation of transcriptomic salivary biomarkers has been performed for breast cancer using Affymetrix HG-U133-Plus-2.0 array discovery and preclinical validation in an independent cohort using RT-qPCR [14]. Applying PRoBE design [18], Zhang et al. [5] identified salivary mRNA biomarkers for pancreatic cancer with excellent sensitivity and specificity. Importantly, these biomarkers could also distinguish pancreatic cancer from chronic pancreatitis with excellent sensitivity and specificity (both 96.7%). Also, the ovarian cancer salivary transcriptome profile was discovered in a clinical case controlled study using Affymetrix HG-U133-Plus-2.0 array and then validated with qPCR by Lee et al. [47] (Table 2).
Recently, Spielmann et al. [48] used massively parallel sequencing to determine nucleotide sequences for each salivary RNA. The salivary transcriptome of unstimulated CFS and whole saliva (WS) from healthy human subjects was profiled: 25% of the sequences from CFS corresponded with the human genome and approximately 30% corresponded with the human oral microbiome. In CFS and WS, more than 4,000 genes and non-coding functional sequences of human and microbial origin combined were detected.
miRNA
miRNAs are short (19 to 25 nucleotides) RNA transcripts associated with post-transcriptional regulation by the RNA-induced silencing complex [49,50]. These RNAs have been well characterized and found to play roles in cell growth, differentiation, apoptosis, pathogen-host interactions and stress responses and immune function, and are found in saliva [51-53]. Just as with salivary mRNA, salivary miRNA is surprisingly stable, and this was recently reported to be due to it being harbored in exosomes [54] (also see Box 1).
miRNAs are differentially expressed in several cancer cell types compared with normal cells, with observed differences that range from ten to over a hundredfold. Changes in mRNA levels in cancer cells are comparatively small, and miRNA has been found to be more useful for characterizing solid tumor types than mRNA. miRNA has been shown to accurately distinguish poorly differentiated tumors, whereas mRNA profiling on the same samples produced highly inaccurate results [52,55,56]. Therefore, miRNA cancer biomarkers are potentially very powerful, and if miRNA-based approaches also reveal similarly differential profiles using saliva samples they may become very useful in salivary diagnostics.
Park et al. [6] found significantly reduced levels of miRNAs miR-125a and miR-200a (known tumor suppressors) in the saliva of oral cancer patients compared with controls. Recent work has also shown that salivary miR-31 (implicated in tumorigenesis) was significantly elevated in all stages of oral cancer, and that salivary miR-31 was more abundant than blood miR-31, indicating the oral tumor origin of this biomarker [57].
The salivary proteome
The use of surface-enhanced laser desorption/ionization time-of-flight (SELDI-TOF) mass spectrometry (MS), which allows rapid, high-throughput detection of proteins in minute sample volumes without pre-processing, has been reported for several diseases [58]. Recently, analysis of saliva for protein biomarker discovery has mainly been performed using two-dimensional difference gel electrophoresis (2D-DIGE) coupled with MS (which can identify around 300 proteins in a sample [59]), and liquid chromatograpy-MS (LC-MS) based techniques (which can identify more than 1,050 proteins in a sample; reviewed in [60]). Thus, liquid chromatographic separation appears to resolve protein species more precisely than gel electrophoresis methods. Recently, a multiplex protein array was also employed, providing high-throughput analysis [61]; however, this method requires some prior knowledge of likely analytes.
Despite these advances, the discovery and validation of salivary protein biomarkers provide challenges. Proteins generally have short half-lives, although there can be large differences in observed stability. Both the nature of peptides and the milieu of the oral cavity make salivary proteins vulnerable to degradation. Thus, protein-based salivary diagnostics require immediate processing and/or analysis of saliva samples, or the use of freezers and costly protease inhibitors. In a research setting, these requirements can be easily met. However, in clinical settings protein stabilization without freezers and specialized machinery may be required. Our laboratory has developed protein stabilization methods that allow storage of saliva samples at room temperature, without the need for expensive reagents and equipment [62,63].
Several different classes of salivary protein biomarkers have been reported. Endothelin-1, a vasoconstrictor, was reported as a potential biomarker for OSCC development in patients with oral lichen planus [64]. Interleukins IL-8 and IL-1β, glycoprotein M2BP (thought to be involved in natural killer cell activity enhancement, tumor suppression, interleukin production and cell aggregation), profilin (actin-binding protein, important for temporal and spatial control of actin microfilament growth), CD59 (complement regulatory protein), MRP14 (a protein involved in regulating myeloid cell function) and catalase (catalyses the conversion of hydrogen peroxide to water and oxygen) have been reported as salivary biomarkers for oral cancer [15,65-69], and immunoglobulins have long been described as salivary biomarkers for HIV infection [70,71].
The salivary metabolome
The metabolome is the complement of small-molecule metabolites. Similar to the transcriptome and proteome, the metabolome changes continually and any single profile is a snapshot reflecting gene and protein expression. Metabolomic investigations can generate quantitative data for metabolites in order to elucidate metabolic dynamics related to disease state and drug exposure [72].
Using capillary electrophoresis time-of-flight MS, Sugimoto et al. [7] identified metabolites for detecting oral, breast and pancreatic cancer, and periodontal disease. Multiple logistic regression models yielded AUCs of 0.865 for oral cancer, using a profile consisting of three oral-cancer-specific metabolites: taurine, piperidine and a peak with a mass-to-charge ratio (m/z) of 120.0801. A breast cancer profile consisting of taurine and lysine achieved an AUC of 0.973. A model consisting of a pancreatic-cancer-specific metabolite signature comprising leucine with isoleucine, tryptophan, valine, glutamic acid, phenylalanine, glutamine and aspartic acid achieved an AUC of 0.993. Periodontal diseases achieved an AUC of 0.969 using a panel of only two markers, but the specific markers were not reported [7] (Tables 1 and 2). Using ultraperformance LC with quadrupole/time-of-flight MS and multivariate statistical analysis, Wei et al. [8] demonstrated that a salivary metabolic profile comprising valine, lactic acid and phenylalanine could discriminate OSCC from controls and oral leukoplakia with good-to-excellent sensitivity, specificity and accuracy (Table 1). However, a limitation of metabolomics compared with genomics, transcriptomics and proteomics is the inability at times to specifically identify the differentially expressed metabolites, as exemplified by these findings [7,8,17] (Tables 1 and 2).
Salivary diagnostics
Early-stage disease detection is imperative for successful therapy for most cancers, and the ability to quickly identify suspicious lesions will also greatly reduce hospital burdens. In the future, salivary diagnostics is expected to facilitate rapid, easily accessible and non-invasive clinical diagnosis, thus allowing more cases of disease to be detected at early stages and decreasing mortality caused by oral and systemic cancers, and infectious diseases.
Oral disease detection
Since the 1990s salivary diagnostics have been developed for oral diseases, allowing monitoring of periodontal disease and assessment of caries risk [73-75]. In recent years there have been many reports of potential salivary biomarkers for periodontal diseases found by candidate approaches, and conventional proteomics has been found to be highly accurate for detecting periodontitis as well as oral, head and neck cancers.
Several candidate approaches have elucidated biomarkers for periodontitis and responses to therapy. Gursoy et al. [76] compared the concentration of five specific salivary proteins, and the presence of five oral pathogens in patients with advanced periodontal disease and healthy controls. They found an association of IL-1β and oral pathogens with periodontitis. Using enzyme-linked immunosorbent assay (ELISA), patients with chronic periodontitis were found to exhibit significantly lower IL-17 and significantly higher IL-18 saliva concentrations compared with healthy individuals [77].
In a response to therapy study, salivary biomarkers for periodontitis were assessed in adults with chronic periodontitis, in which 33 participants received oral hygiene instructions alone and 35 participants had conventional periodontal treatment comprising scaling and root planning combined with oral hygiene instructions. Saliva samples collected at intervals were analyzed for several proteins, and it was found that the levels of IL-1β, macrophage inflammatory protein 1α (MIP-1α, a chemokine produced by macrophages after bacterial endotoxin stimulation, which activates granulocytes), matrix metalloproteinase-8 (MMP-8, a collagen cleaving enzyme found in connective tissue and involved in degradation of extracellular matrix in both normal and disease processes) and osteoprotegerin (OPG, a decoy receptor that reduces the production of osteoclasts) detected in saliva reflected disease severity and response to therapy and may have potential use in monitoring periodontal disease [78].
Nomura et al. [16] found that a combination of alanine aminotransferase (a transaminase that catalyses two steps of the alanine cycle) level and Porphyromonas gingivalis ratio could indicate periodontal disease progression with 40% sensitivity and 96% specificity. Lee et al. [61] analyzed saliva samples using a multiplex protein array for selected biomarkers implicated in host defense, inflammation, tissue destruction and angiogenesis, and measured subgingival bacterial proportions by checkerboard DNA-DNA hybridization. The authors devised a system to discriminate 'high' and 'low' responders to treatment for gingivitis. Salivary IL-6 and IL-8 levels provided the best distinction between high and low responders (Table 2).
Proteomic and microbial studies have been used to successfully identify biomarkers for periodontal disease. Choi et al. [79] searched for potential protein biomarkers for periodontitis in gingival crevicular fluid using LC-tandem MS (LC-MS-MS). Azurocidin (an antibiotic protein of azurophil granules with chemotactic activity) was identified as upregulated in gingival crevicular fluid, but not in saliva. ELISA was then used to verify upregulation of azurocidin, identifying the latter as a candidate biomarker for inflammatory periodontal disease [79]. Using SELDI-TOF MS, Hart et al. [80] characterized the oral flora and salivary proteome in caries-free and caries-active children, and found that a combination model of microbial-proteomic data was far more accurate than microbial or proteomic models alone. Their best model had a 6% test error, with >92% sensitivity, and >95% specificity.
Al-Sabbagh et al. [81] investigated salivary levels of bone-remodeling-related molecules in chronic periodontitis patients using ELISA. MIP-1α levels were found to be 18-fold higher in periodontitis patients than normal and clinical measures correlated significantly with MIP-1α levels, and achieved excellent sensitivity, specificity and accuracy (Table 2). In a study of dysplastic oral leukoplakia in relation to tobacco habits and periodontitis, Sharma et al. [82] found that increasing IL-6 levels correlated with increasing severity of dysplasia.
Discovery and validation of salivary biomarkers for OSCC followed, and has also been confirmed across ethnic backgrounds [46]. Many salivary biomarkers, identified using candidate as well as 'omic approaches (including metabolomic, transcriptomic (miRNA and mRNA), proteomic, epigenomic and microbial) have been described for oral, and head and neck cancers [4,6-8,22,23,25-29,46,57,64,65,67,83].
A comparison of salivary biomarkers for OSCCs and HNSCCs can be seen in Table 1. Several single marker candidates for OSCC have been reported, including EDNRB gene (which encodes endothelin receptor type B, a G-protein-coupled receptor that activates a phosphatidylinositol-calcium second messenger system) hypermethylation [25,26], miRNAs miR-200a, miR-125a and miR-31 [6,57], and IL-8 protein [65], but only IL-8 has been shown to be a sensitive, specific and accurate single marker for OSCC and could prove to be a cost-efficient analyte for OSCC detection.
Several biomarker panels have also been investigated for OSCC detection. An mRNA panel has been validated with excellent sensitivity, specificity and accuracy and could be considered 'second place' to IL-8 [67]. Interestingly, proteomic-transcriptomic combination panels have achieved good sensitivity, specificity and accuracy, but are lagging behind the transcriptomic panel in these performance measures [46]. A five-protein panel achieved higher sensitivity, lower specificity and similar accuracy to IL-8 alone [15]. Metabolomic markers for OSCC have also achieved good accuracy, but have not been validated [7,8]. Many epigenetic salivary biomarker panels have been reported and some have achieved 100% specificity, but accompanied by low sensitivity (62% [27]; Table 1), and have not been as stringently tested as proteomic and transcriptomic biomarkers.
Systemic disease detection
More recently, advances in biotechnology, genomics and proteomics have extended the range of salivary diagnostics to systemic disease monitoring. Possibly the most attractive attribute of salivary diagnostics is its implication in systemic disease diagnosis, due to the invasive nature and in some cases poor accuracy of current standard diagnostic methods [5].
Available tests for systemic cancers include invasive and expensive procedures (such as magnetic resonance imaging, biopsy, X-ray, computed tomography scan, exfoliative cytology, positron emission tomography scan and endoscopy). This, combined with significantly worse prognosis associated with late diagnosis, makes the prospect of salivary diagnostics particularly valuable in oncology. Available diagnostic methodologies for other systemic diseases, such as infections, have similar problems. In this section, we will discuss progress in the use of salivary biomarker-based approaches for diagnosis of pancreatic cancer, lung cancer and breast cancer and finally other systemic diseases.
Pancreatic cancer
A significant milestone in salivary diagnostics was reached by Zhang et al. [5], with a prospective sample collection and retrospective double-blinded validation study that showed that a salivary transcriptome profile could detect early stage resectable pancreatic cancer. The use of the salivary transcriptome for detecting pancreatic cancer was also found to outperform currently used blood-based tests in terms of sensitivity and specificity, but did not allow sensitive detection of pancreatitis, unlike some blood-based tests. Farrell et al. [32] showed that variations in oral microbiota could possibly be used to detect pancreatic cancer and pancreatitis. Also, a metabolomic approach [7] has identified pancreatic-cancer-specific salivary metabolomic biomarkers that could distinguish pancreatic cancer from oral and breast cancer and normal controls. Although both the transcriptomic and the metabolomic biomarkers achieved excellent accuracy for this disease, only the transcriptomic panel has been validated (Table 2).
Lung cancer
Currently, lung cancer may be detected with chest X-ray and computed tomography, and diagnosis is confirmed via biopsy. Thus, lung cancer detection is limited by stage of disease, and more than 75% of cases are diagnosed in the late stages, significantly reducing survival rate. Lung cancer is often asymptomatic or presents with non-specific symptoms, so there is considerable need for non-invasive approaches for earlier detection of disease [59,84].
Using 2D-DIGE and MS, Xiao et al. [59] performed proteomic analysis of saliva samples from lung cancer patients. Sixteen candidate biomarkers were discovered and further verified. Three candidate markers (calprotectin, a calcium and zinc binding protein, zinc-α-2-glycoprotein, a lipolysis stimulating protein, and haptoglobin hp2, a protein that binds free plasma hemoglobin) achieved good sensitivity and excellent specificity and accuracy (Table 2).
Zhang et al. [84] profiled the salivary lung cancer transcriptome and discovered and pre-validated a panel of seven highly discriminatory mRNAs: BRAF (which encodes the protein B-Raf, and is involved in directing cell growth), CCNI (which encodes the protein cyclin-I, and binds activated cyclin-dependant kinase 5), EGFR (epidermal growth factor receptor), FGF19 (fibroblast growth factor 19, involved in many biological processes, including morphogenesis and cell growth, and tumor growth and invasion), FRS2 (fibroblast growth factor receptor substrate 2, an adapter protein that links fibroblast growth factor and nerve growth factor receptors to downstream signaling pathways), GREB1 (growth regulation by estrogen in breast cancer 1, an early response gene in the estrogen receptor pathway), and LZTS1 (leucine zipper putative tumor suppressor 1, a putative tumor suppressor gene). A panel consisting of five of these markers differentiated lung cancer from normal subjects with 93.75% sensitivity and 82.81% specificity (Table 2).
Surface-enhanced Raman spectroscopy (SERS) was recently employed by Li et al. [17] to identify biomarkers for lung cancer. SERS revealed nine peaks (assigned to amino acids and nucleic acid bases) that could distinguish samples from patients with lung cancer and controls with 86% accuracy, 94% sensitivity and 81% specificity. Thus, for lung cancer, both transcriptomic and proteomic approaches are proving highly useful for biomarker development, and metabolomic approaches are also showing promise (Table 2).
Breast cancer
Today, breast cancer detection employs physical examination and imaging techniques. Emerging technologies such as molecular analysis of nipple fluid aspirate and ductal lavage samples [85] provide improved accuracy and potentially earlier diagnosis, but are invasive and limited to high-risk patients. Several earlier studies indicated the potential for salivary proteomic detection of breast cancer and its potential use in monitoring response to treatment, and the use of single markers, such as salivary protein c-erbB-2, has also been investigated [9-12].
Cao et al. [86] investigated the salivary proteome of breast cancer patients compared with healthy individuals. Saliva samples were analyzed with isobaric tags for relative and absolute quantitation (iTRAQ) technology coupled with LC-MS-MS. Of 464 identified proteins, 9 were associated with breast cancer and had a 1.5-fold upregulation or downregulation. Recently, preclinical validation of transcriptomic and proteomic salivary biomarkers with diagnostic power for breast cancer or ovarian cancer has also been reported [14,47]. However, the metabolomic approach to breast cancer detection has shown better results than the combination proteomic-transcriptomic approach in terms of accuracy (Table 2).
Other systemic diseases
Advances in the use of salivary biomarkers for detecting autoimmune diseases, systemic microbial infections and diabetes have been described recently [2,45,70,87]. Menezes et al. [88] investigated hepatitis C virus (HCV) loads in saliva and serum samples from HCV-infected individuals using qPCR, and found that HCV RNA could be consistently detected in saliva of HCV-infected individuals, although at significantly lower levels than in serum.
Border et al. [89] performed label-free differential protein expression analysis using multidimensional LC-MS-MS to characterize the proteome of saliva collected from edentulous type 2 diabetic patients compared with non-diabetic controls. Several proteins, PLUNC (palate, lung and nasal epithelium carcinoma-associated protein, a secretory protein expressed in the upper airways and nasopharyngeal regions), GAPDH (glyceraldehyde-3-phosphate dehydrogenase, an enzyme that catalyses the sixth step of glycolysis), SAA (serum amyloid A, an adipolipoprotein associated with high density lipoproteins in plasma), serotransferrin (an iron-binding transport protein), PIP (prolactin-induced protein, an aspartyl protease that inhibits T-lymphocyte programmed cell death), A2MG (α2-macroglobulin, a broad protease inhibitor), and cystatin S and cystatin SN (protein inhibitors of cysteine proteinases), were found to have diagnostic potential for type 2 diabetes but require further testing to determine clinical value.
Concluding remarks
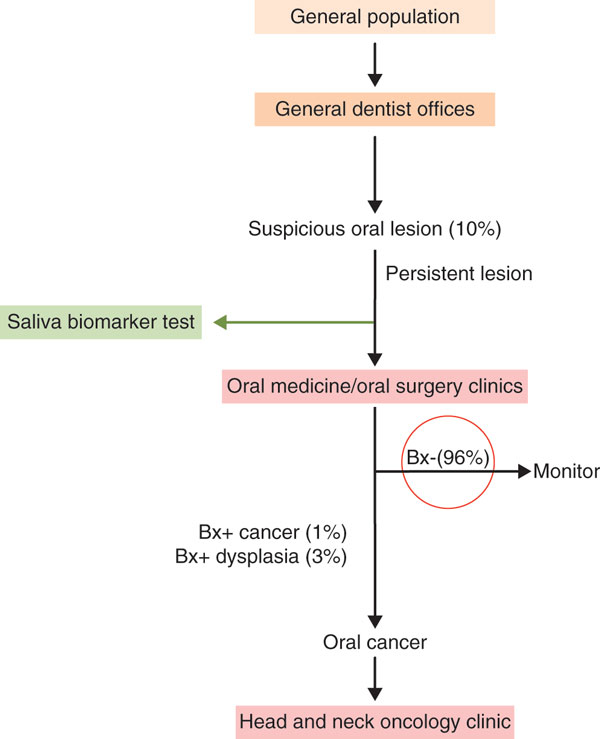
The use of saliva as a diagnostic specimen avoids the pain, anxiety and infection risk associated with traditional methods of specimen collection, such as blood sampling or tissue biopsy. Saliva sampling also facilitates the collection of multiple subsequent samples for disease monitoring. Processing and analysis of saliva is also much easier than blood and other samples. Unlike blood, saliva does not clot and salivary analytes are very stable. Analytes must be cost efficiently and easily stabilized for storage until analysis or transport to the laboratory. Technologies for saliva sampling and stabilization, and point-of-care technology that can allow accurate detection of biomarkers without any processing of saliva samples are currently available or being developed. Using such approaches, salivary diagnostics could be performed without the need for specially trained professionals or be incorporated as part of primary healthcare, greatly reducing burdens on hospitals relating to costly, complex and invasive diagnostic procedures (Figure 1). However, most reports on individual biomarkers and multiple biomarker panels have been only preliminary and require further validation before testing at the clinical level.
Figure 1.
Clinical context of salivary diagnostics. Salivary diagnostics could reduce the number of unnecessary referrals to hospital for invasive, time-consuming and expensive diagnostics. An example situation is shown (with examples of percentages of patients that might be involved in parentheses) for the use of a saliva biomarker (Bx) test, in which a health professional, such as a dentist, observes a suspicious oral lesion. This is followed by screening of saliva, and if appropriate, referral to a specialist. Salivary diagnostics using clinically validated biomarkers can be implemented in the early stages of the clinical management hierarchy (green arrow and box), thus significantly reducing unnecessary referrals to specialists and unnecessary biopsies seen in current clinical approaches, where management follows the black arrows exclusively and results in high (such as 96%) negative test rates at biopsy (Bx-; red circle).
Transcriptomic and proteomic approaches to biomarker development have so far had the most progress towards clinical testing. Salivary biomarkers for a range of cancers, both local and distant to the oral cavity, have been characterized and validated at the pre-clinical level. Certainly, salivary biomarkers for oral diseases such as OSCC are likely to become clinically implemented before those for systemic cancers, as they have been well described and validated and often originate locally to the oral cavity where the biospecimen is collected, lending legitimacy to such biomarkers.
The transfer of scientific knowledge of salivary biomarkers to clinical applications is a challenging process that has rarely resulted in clinical implementation. Understanding the sources of salivary biomarkers (for example, exosomes; Box 1) will be a significant objective in the development of salivary diagnostics, as this will provide the rationale for the use of salivary biomarkers for systemic diseases. Our group and others continue to investigate the mechanisms by which salivary biomarkers appear in saliva, and to develop technologies for detecting biomarkers with high sensitivity and specificity in pursuit of compliance at the FDA level.
Abbreviations
AUC: area under the receiver operating characteristic curve; ARE: AU-rich element; CFS: cell-free saliva; EBV: Epstein-Barr virus; ELISA: enzyme-linked immunosorbent assay; HCV: hepatitis C virus; HNSCC: head and neck squamous cell carcinoma; HOMIM: Human Oral Microbe Identification Microarray; IL: interleukin; LC-MS: liquid chromatography-mass spectrometry; LC-MS-MS: liquid chromatography-tandem mass spectrometry; miRNA: microRNA; MS: mass spectrometry; NPC: nasopharyngeal carcinoma; OSCC: oral squamous cell carcinoma; PCR: polymerase chain reaction; PRoBE: prospective specimen collection and retrospective blinded evaluation; qPCR: quantitative PCR; RT-PCR: reverse transcription PCR; RT-qPCR: reverse transcription quantitative PCR; SELDI-TOF: surface-enhanced laser desorption/ionization time-of-flight mass spectrometry; SERS: surface-enhanced Raman spectroscopy; WS: whole saliva; 2D-DIGE: two-dimensional difference gel electrophoresis.
Competing interests
The authors declare that they have no proprietary, financial or other personal interest of any nature or kind in any product, service and/or company that is presented in this article except for the following: Dr Wong reports proprietary interests in RNAmeTRIX Inc., a molecular diagnostic company, and personal interests in intellectual properties in salivary diagnostics technologies.
Box 1. Salivary biomarkers: where do they come from?
Accumulating evidence suggests that some salivary biomarkers could derive from systemic sources. Gao et al. [90] showed that altered salivary gland tissue production of transcription factors might account for a large proportion of upregulated and downregulated gene expression in the saliva of tumor-bearing mice, and that tumor-derived ectopic nerve growth factor could induce production of the transcription factor Egr-1 in salivary glands. Therefore, the salivary transcriptomic profile might be composed of transcripts originating in distant diseased tissues and transcripts originating in salivary glands, and altered expression levels of these transcripts might be caused by transcription factors originating in distant tissues.
Exosomes
Exosomes might be involved in transport of systemic biomarkers to saliva or transport of effectors of salivary gland transcriptional machinery from blood to the salivary glands. These membrane bound microvesicles of 30 to 100 nm in diameter and endocytic origin [91] are secreted by many cell types in diseased and non-diseased states. Exosomes contain protein, RNA and DNA, and have been found in most body fluids, including saliva [92-95]. Using in vitro methods, studies have shown that exosomes secreted from one cell can be taken up by other cells and can influence the receiving cell's transcriptional signature [92,96,97]. Intercellular communication via exosomes can also occur between normal cells and cancer cells and between cancer cells and distant sites. This has been suggested as a mechanism by which cancer cells prepare distant sites for receiving metastatic cells [98,99]. Exosomes protect their contents from enzymatic degradation, suggesting a role in intercellular transport of molecules [6,97,100]. Similarities between serum and saliva biomarker profiles and the co-existence of exosomes in saliva and serum [101] also suggest the involvement of exosomes in transport of salivary biomarkers. Furthermore, a recent in vitro investigation showed that tumor-derived exosomes can interact with salivary gland cells and activate their transcriptional machinery [102].
Contributor Information
Nicolai J Bonne, Email: nicolai@bonne.no.
David TW Wong, Email: dtww@ucla.edu.
References
- Gorr SU. Antimicrobial peptides in periodontal innate defense. Front Oral Biol. 2012;15:84–98. doi: 10.1159/000329673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaka-ur-Rab Z, Abqari S, Shahab T, Islam N, Shukla I. Evaluation of salivary anti-Salmonella typhi lipopolysaccharide IgA ELISA for serodiagnosis of typhoid fever in children. Arch Dis Child. 2012;97:236–238. doi: 10.1136/adc.2011.300622. [DOI] [PubMed] [Google Scholar]
- Denny P, Hagen FK, Hardt M, Liao L, Yan W, Arellanno M, Bassilian S, Bedi GS, Boontheung P, Cociorva D, Delahunty CM, Denny T, Dunsmore J, Faull KF, Gilligan J, Gonzalez-Begne M, Halgand F, Hall SC, Han X, Henson B, Hewel J, Hu S, Jeffrey S, Jiang J, Loo JA, Ogorzalek Loo RR, Malamud D, Melvin JE, Miroshnychenko O, Navazesh M. et al. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J Proteome Res. 2008;7:1994–2006. doi: 10.1021/pr700764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, St John MAR, Zhou X, Kim Y, Sinha U, Jordan RCK, Eisele D, Abemayor E, Elashoff D, Park N, Wong DT. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res. 2004;10:8442–8450. doi: 10.1158/1078-0432.CCR-04-1167. [DOI] [PubMed] [Google Scholar]
- Zhang L, Farrell J, Zhou H, Elashoff D, Akin D, Park N, Chia D, Wong DT. Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology. 2010;138:949–957.e1-7. doi: 10.1053/j.gastro.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, Abemayor E, Wong DT. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 2009;15:5473–5477. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010;6:78–95. doi: 10.1007/s11306-009-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Xie G, Zhou Z, Shi P, Qiu Y, Zheng X, Chen T, Su M, Zhao A, Jia W. Salivary metabolite signatures of oral cancer and leukoplakia. Int J Cancer. 2011;129:2207–2217. doi: 10.1002/ijc.25881. [DOI] [PubMed] [Google Scholar]
- Streckfus CF, Bigler LR, Zwick M. The use of surface-enhanced laser desorption/ionization time-of-flight mass spectrometry to detect putative breast cancer markers in saliva: a feasibility study. J Oral Pathol Med. 2006;35:292–300. doi: 10.1111/j.1600-0714.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- Bigler LR, Streckfus CF, Copeland L, Burns R, Dai XL, Kuhn M, Martin P, Bigler SA. The potential use of saliva to detect recurrence of disease in women with breast carcinoma. J Oral Pathol Med. 2002;31:421–431. doi: 10.1034/j.1600-0714.2002.00123.x. [DOI] [PubMed] [Google Scholar]
- Streckfus C, Bigler L. The use of soluble, salivary c-erbB-2 for the detection and post-operative follow-up of breast cancer in women: the results of a five-year translational research study. Adv Dent Res. 2005;18:17–24. doi: 10.1177/154407370501800105. [DOI] [PubMed] [Google Scholar]
- Streckfus C, Bigler L, Dellinger T, Dai X, Kingman A, Thigpen JT. The presence of soluble c-erbB-2 in saliva and serum among women with breast carcinoma: a preliminary study. Clin Cancer Res. 2000;6:2363–2370. [PubMed] [Google Scholar]
- Navarro MA, Mesia R, Diez-Gibert O, Rueda A, Ojeda B, Alonso MC. Epidermal growth factor in plasma and saliva of patients with active breast cancer and breast cancer patients in follow-up compared with healthy women. Breast Cancer Res Treat. 1997;42:83–86. doi: 10.1023/A:1005755928831. [DOI] [PubMed] [Google Scholar]
- Zhang L, Xiao H, Karlan S, Zhou H, Gross J, Elashoff D, Akin D, Yan X, Chia D, Karlan B, Wong DT. Discovery and preclinical validation of salivary transcriptomic and proteomic biomarkers for the non-invasive detection of breast cancer. PLoS One. 2010;5:e15573. doi: 10.1371/journal.pone.0015573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Arellano M, Boontheung P, Wang J, Zhou H, Jiang J, Elashoff D, Wei R, Loo JA, Wong DT. Salivary proteomics for oral cancer biomarker discovery. Clin Cancer Res. 2008;14:6246–6252. doi: 10.1158/1078-0432.CCR-07-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura Y, Shimada Y, Hanada N, Numabe Y, Kamoi K, Sato T, Gomi K, Arai T, Inagaki K, Fukuda M, Noguchi T, Yoshie H. Salivary biomarkers for predicting the progression of chronic periodontitis. Arch Oral Biol. 2012;57:413–420. doi: 10.1016/j.archoralbio.2011.09.011. [DOI] [PubMed] [Google Scholar]
- Li X, Yang T, Lin J. Spectral analysis of human saliva for detection of lung cancer using surface-enhanced Raman spectroscopy. J Biomed Opt. 2012;17:037003. doi: 10.1117/1.JBO.17.3.037003. [DOI] [PubMed] [Google Scholar]
- Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst. 2008;100:1432–1438. doi: 10.1093/jnci/djn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian TK, Fejerdy P, Csermely P. Salivary genomics, transcriptomics and proteomics: the emerging concept of the oral ecosystem and their use in the early diagnosis of cancer and other diseases. Curr Genomics. 2008;9:11–21. doi: 10.2174/138920208783884900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes AP, Oliveira IO, Santos BR, Millech C, Silva LP, Gonzalez DA, Hallal PC, Menezes AM, Araujo CL, Barros FC. Quality of DNA extracted from saliva samples collected with the Oragene DNA self-collection kit. BMC Med Res Methodol. 2012;12:65. doi: 10.1186/1471-2288-12-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pow EH, Law MY, Tsang PC, Perera RA, Kwong DL. Salivary Epstein-Barr virus DNA level in patients with nasopharyngeal carcinoma following radiotherapy. Oral Oncol. 2011;47:879–882. doi: 10.1016/j.oraloncology.2011.06.507. [DOI] [PubMed] [Google Scholar]
- Shaw R. The epigenetics of oral cancer. Int J Oral Maxillofac Surg. 2006;35:101–108. doi: 10.1016/j.ijom.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Carvalho AL, Jeronimo C, Kim MM, Henrique R, Zhang Z, Hoque MO, Chang S, Brait M, Nayak CS, Jiang WW, Claybourne Q, Tokumaru Y, Lee J, Goldenberg D, Garrett-Mayer E, Goodman S, Moon CS, Koch W, Westra WH, Sidransky D, Califano JA. Evaluation of promoter hypermethylation detection in body fluids as a screening/diagnosis tool for head and neck squamous cell carcinoma. Clin Cancer Res. 2008;14:97–107. doi: 10.1158/1078-0432.CCR-07-0722. [DOI] [PubMed] [Google Scholar]
- Chang X, Monitto CL, Demokan S, Kim MS, Chang SS, Zhong X, Califano JA, Sidransky D. Identification of hypermethylated genes associated with cisplatin resistance in human cancers. Cancer Res. 2010;70:2870–2879. doi: 10.1158/0008-5472.CAN-09-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demokan S, Chang X, Chuang A, Mydlarz WK, Kaur J, Huang P, Khan Z, Khan T, Ostrow KL, Brait M, Hoque MO, Liegeois NJ, Sidransky D, Koch W, Califano JA. KIF1A and EDNRB are differentially methylated in primary HNSCC and salivary rinses. Int J Cancer. 2010;127:2351–2359. doi: 10.1002/ijc.25248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattani KM, Zhang Z, Demokan S, Glazer C, Loyo M, Goodman S, Sidransky D, Bermudez F, Jean-Charles G, McCaffrey T, Padhya T, Phelan J, Spivakovsky S, Bowne HY, Goldberg JD, Rolnitzky L, Robbins M, Kerr AR, Sirois D, Califano JA. Endothelin receptor type B gene promoter hypermethylation in salivary rinses is independently associated with risk of oral cavity cancer and premalignancy. Cancer Prev Res (Phila) 2010;3:1093–1103. doi: 10.1158/1940-6207.CAPR-10-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viet CT, Schmidt BL. Methylation array analysis of preoperative and postoperative saliva DNA in oral cancer patients. Cancer Epidemiol Biomarkers Prev. 2008;17:3603–3611. doi: 10.1158/1055-9965.EPI-08-0507. [DOI] [PubMed] [Google Scholar]
- Righini CA, de Fraipont F, Timsit JF, Faure C, Brambilla E, Reyt E, Favrot MC. Tumor-specific methylation in saliva: a promising biomarker for early detection of head and neck cancer recurrence. Clin Cancer Res. 2007;13:1179–1185. doi: 10.1158/1078-0432.CCR-06-2027. [DOI] [PubMed] [Google Scholar]
- Carvalho AL, Henrique R, Jeronimo C, Nayak CS, Reddy AN, Hoque MO, Chang S, Brait M, Jiang WW, Kim MM, Claybourne Q, Goldenberg D, Khan Z, Khan T, Westra WH, Sidransky D, Koch W, Califano JA. Detection of promoter hypermethylation in salivary rinses as a biomarker for head and neck squamous cell carcinoma surveillance. Clin Cancer Res. 2011;17:4782–4789. doi: 10.1158/1078-0432.CCR-11-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Preston R, Soudry E, Acero J, Orera M, Moreno-Lopez L, Macia-Colon G, Jaffe A, Berdasco M, Ili-Gangas C, Brebi-Mieville P, Fu Y, Engstrom C, Irizarry RA, Esteller M, Westra W, Koch W, Califano J, Sidransky D. NID2 and HOXA9 promoter hypermethylation as biomarkers for prevention and early detection in oral cavity squamous cell carcinoma tissues and saliva. Cancer Prev Res (Phila) 2011;4:1061–1072. doi: 10.1158/1940-6207.CAPR-11-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyghe A, Francois P, Charbonnier Y, Tangomo-Bento M, Bonetti EJ, Paster BJ, Bolivar I, Baratti-Mayer D, Pittet D, Schrenzel J. Novel microarray design strategy to study complex bacterial communities. Appl Environ Microbiol. 2008;74:1876–1885. doi: 10.1128/AEM.01722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell JJ, Zhang L, Zhou H, Chia D, Elashoff D, Akin D, Paster BJ, Joshipura K, Wong DT. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. 2012;61:582–588. doi: 10.1136/gutjnl-2011-300784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold JS, Bayar S, Salem RR. Association of Streptococcus bovis bacteremia with colonic neoplasia and extracolonic malignancy. Arch Surg. 2004;139:760–765. doi: 10.1001/archsurg.139.7.760. [DOI] [PubMed] [Google Scholar]
- Joshipura KJ, Douglass CW, Willett WC. Possible explanations for the tooth loss and cardiovascular disease relationship. Ann Periodontol. 1998;3:175–183. doi: 10.1902/annals.1998.3.1.175. [DOI] [PubMed] [Google Scholar]
- Meurman JH. Oral microbiota and cancer. J Oral Microbiol. 2010;2 doi: 10.3402/jom.v2i0.5195. doi:10.3402/jom.v2i0.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison HI, Ellison LF, Taylor GW. Periodontal disease and risk of fatal coronary heart and cerebrovascular diseases. J Cardiovasc Risk. 1999;6:7–11. doi: 10.1177/204748739900600102. [DOI] [PubMed] [Google Scholar]
- Goldenberg RL, Culhane JF. Preterm birth and periodontal disease. N Engl J Med. 2006;355:1925–1927. doi: 10.1056/NEJMe068210. [DOI] [PubMed] [Google Scholar]
- Garcia JM, Garcia V, Pena C, Dominguez G, Silva J, Diaz R, Espinosa P, Citores MJ, Collado M, Bonilla F. Extracellular plasma RNA from colon cancer patients is confined in a vesicle-like structure and is mRNA-enriched. RNA. 2008;14:1424–1432. doi: 10.1261/rna.755908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinzl M, Marberger M, Horvath S, Chypre C. DD3 RNA analysis in urine? A new perspective for detecting prostate cancer. Eur Urol. 2004;46:182–187. doi: 10.1016/j.eururo.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Kumar SV, Hurteau GJ, Spivack SD. Validity of messenger RNA expression analyses of human saliva. Clin Cancer Res. 2006;12:5033–5039. doi: 10.1158/1078-0432.CCR-06-0501. [DOI] [PubMed] [Google Scholar]
- Lee YH, Zhou H, Reiss JK, Yan X, Zhang L, Chia D, Wong DT. Direct saliva transcriptome analysis. Clin Chem. 2011;57:1295–1302. doi: 10.1373/clinchem.2010.159210. [DOI] [PubMed] [Google Scholar]
- Park NJ, Yu T, Nabili V, Brinkman BM, Henry S, Wang J, Wong DT. RNAprotect saliva: An optimal room-temperature stabilization reagent for the salivary transcriptome. Clin Chem. 2006;52:2303–2304. doi: 10.1373/clinchem.2006.075598. [DOI] [PubMed] [Google Scholar]
- Zimmermann BG, Wong DT. Salivary mRNA targets for cancer diagnostics. Oral Oncol. 2008;44:425–429. doi: 10.1016/j.oraloncology.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Zimmermann BG, Zhou H, Wang J, Henson BS, Yu W, Elashoff D, Krupp G, Wong DT. Exon-level expression profiling: a comprehensive transcriptome analysis of oral fluids. Clin Chem. 2008;54:824–832. doi: 10.1373/clinchem.2007.096164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Gao K, Pollard R, Arellano-Garcia M, Zhou H, Zhang L, Elashoff D, Kallenberg CG, Vissink A, Wong DT. Preclinical validation of salivary biomarkers for primary Sjogren's syndrome. Arthritis Care Res (Hoboken) 2010;62:1633–1638. doi: 10.1002/acr.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann O, Kastratovic DA, Dimitrijevic MV, Konstantinovic VS, Jelovac DB, Antic J, Nesic VS, Markovic SZ, Martinovic ZR, Akin D, Spielmann N, Zhou H, Wong DT. Oral squamous cell carcinoma detection by salivary biomarkers in a Serbian population. Oral Oncol. 2011;47:51–55. doi: 10.1016/j.oraloncology.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim JH, Zhou H, Kim BW, Wong DT. Salivary transcriptomic biomarkers for detection of ovarian cancer: for serous papillary adenocarcinoma. J Mol Med (Berl) 2011;90:427–434. doi: 10.1007/s00109-011-0829-0. [DOI] [PubMed] [Google Scholar]
- Spielmann N, Ilsley D, Gu J, Lea K, Brockman J, Heater S, Setterquist R, Wong D. The human salivary RNA transcriptome revealed by massively parallel sequencing. Clin Chem. 2012;58:1314–1321. doi: 10.1373/clinchem.2011.176941. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Zeng Y. Principles of micro-RNA production and maturation. Oncogene. 2006;25:6156–6162. doi: 10.1038/sj.onc.1209908. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Stadler BM, Ruohola-Baker H. Small RNAs: keeping stem cells in line. Cell. 2008;132:563–566. doi: 10.1016/j.cell.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7:e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Lu J, Sun M, Li Z, Zhang H, Neilly MB, Wang Y, Qian Z, Jin J, Zhang Y, Bohlander SK, Le Beau MM, Larson RA, Golub TR, Rowley JD. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc Natl Acad Sci USA. 2007;104:19971–19976. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33:5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Lin SC, Yang C, Cheng HW, Chang KW. Exploiting salivary miR-31 as a clinical biomarker of oral squamous cell carcinoma. Head Neck. 2012;34:219–224. doi: 10.1002/hed.21713. [DOI] [PubMed] [Google Scholar]
- Al-Tarawneh SK, Bencharit S. Applications of surface-enhanced laser desorption/ionization time-of-flight (SELDI-TOF) mass spectrometry in defining salivary proteomic profiles. Open Dent J. 2009;3:74–79. doi: 10.2174/1874210600903010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Zhang L, Zhou H, Lee JM, Garon EB, Wong DT. Proteomic analysis of human saliva from lung cancer patients using two-dimensional difference gel electrophoresis and mass spectrometry. Mol Cell Proteomics. 2012;11:M111.012112. doi: 10.1074/mcp.M111.012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Duan Y. Saliva: A potential media for disease diagnostics and monitoring. Oral Oncol. 2012;48:569–577. doi: 10.1016/j.oraloncology.2012.01.021. [DOI] [PubMed] [Google Scholar]
- Lee A, Ghaname CB, Braun TM, Sugai JV, Teles RP, Loesche WJ, Kornman KS, Giannobile WV, Kinney JS. Bacterial and salivary biomarkers predict the gingival inflammatory profile. J Periodontol. 2012;83:79–89. doi: 10.1902/jop.2011.110060. [DOI] [PubMed] [Google Scholar]
- Xiao H, Wong DT. Method development for proteome stabilization in human saliva. Anal Chim Acta. 2012;722:63–69. doi: 10.1016/j.aca.2012.02.017. [DOI] [PubMed] [Google Scholar]
- Jiang J, Park NJ, Hu S, Wong DT. A universal pre-analytic solution for concurrent stabilization of salivary proteins, RNA and DNA at ambient temperature. Arch Oral Biol. 2009;54:268–273. doi: 10.1016/j.archoralbio.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YS, Rees T, Jordan L, Oxford L, O'Brien J, Chen HS, Wong D. Salivary endothelin-1 potential for detecting oral cancer in patients with oral lichen planus or oral cancer in remission. Oral Oncol. 2011;47:1122–1126. doi: 10.1016/j.oraloncology.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John MA, Li Y, Zhou X, Denny P, Ho CM, Montemagno C, Shi W, Qi F, Wu B, Sinha U, Jordan R, Wolinsky L, Park NH, Liu H, Abemayor E, Wong DT. Interleukin 6 and interleukin 8 as potential biomarkers for oral cavity and oropharyngeal squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2004;130:929–935. doi: 10.1001/archotol.130.8.929. [DOI] [PubMed] [Google Scholar]
- Wei F, Patel P, Liao W, Chaudhry K, Zhang L, Arellano-Garcia M, Hu S, Elashoff D, Zhou H, Shukla S, Shah F, Ho CM, Wong DT. Electrochemical sensor for multiplex biomarkers detection. Clin Cancer Res. 2009;15:4446–4452. doi: 10.1158/1078-0432.CCR-09-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elashoff D, Zhou H, Reiss J, Wang J, Xiao H, Henson B, Hu S, Arellano M, Sinha U, Le A, Messadi D, Wang M, Nabili V, Lingen M, Morris D, Randolph T, Feng Z, Akin D, Kastratovic DA, Chia D, Abemayor E, Wong DT. Prevalidation of salivary biomarkers for oral cancer detection. Cancer Epidemiol Biomarkers Prev. 2012;21:664–672. doi: 10.1158/1055-9965.EPI-11-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai B, Kaur J, Jacobs R, Anand SC. Adenosine deaminase in saliva as a diagnostic marker of squamous cell carcinoma of tongue. Clin Oral Invest. 2011;15:347–349. doi: 10.1007/s00784-010-0404-z. [DOI] [PubMed] [Google Scholar]
- Korostoff A, Reder L, Masood R, Sinha UK. The role of salivary cytokine biomarkers in tongue cancer invasion and mortality. Oral Oncol. 2011;47:282–287. doi: 10.1016/j.oraloncology.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Emmons W. Accuracy of oral specimen testing for human immunodeficiency virus. Am J Med. 1997;102:15–20. doi: 10.1016/s0002-9343(97)00033-8. [DOI] [PubMed] [Google Scholar]
- Malamud D. Oral diagnostic testing for detecting human immunodeficiency virus-1 antibodies: a technology whose time has come. Am J Med. 1997;102:9–14. doi: 10.1016/s0002-9343(97)00032-6. [DOI] [PubMed] [Google Scholar]
- Spielmann N, Wong DT. Saliva: diagnostics and therapeutic perspectives. Oral Dis. 2011;17:345–354. doi: 10.1111/j.1601-0825.2010.01773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornman KS, Crane A, Wang HY, di Giovine FS, Newman MG, Pirk FW, Wilson TG Jr, Higginbottom FL, Duff GW. The interleukin-1 genotype as a severity factor in adult periodontal disease. J Clin Periodontol. 1997;24:72–77. doi: 10.1111/j.1600-051X.1997.tb01187.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Smith C, Duff GW. Microbiological parameters associated with IL-1 gene polymorphisms in periodontitis patients. J Clin Periodontol. 2000;27:810–818. doi: 10.1034/j.1600-051x.2000.027011810.x. [DOI] [PubMed] [Google Scholar]
- Baughan LW, Robertello FJ, Sarrett DC, Denny PA, Denny PC. Salivary mucin as related to oral Streptococcus mutans in elderly people. Oral Microbiol Immunol. 2000;15:10–14. doi: 10.1034/j.1399-302x.2000.150102.x. [DOI] [PubMed] [Google Scholar]
- Gursoy UK, Kononen E, Uitto VJ, Pussinen PJ, Hyvarinen K, Suominen-Taipale L, Knuuttila M. Salivary interleukin-1beta concentration and the presence of multiple pathogens in periodontitis. J Clin Periodontol. 2009;36:922–927. doi: 10.1111/j.1600-051X.2009.01480.x. [DOI] [PubMed] [Google Scholar]
- Ozcaka O, Nalbantsoy A, Buduneli N. Interleukin-17 and interleukin-18 levels in saliva and plasma of patients with chronic periodontitis. J Periodontal Res. 2011;46:592–598. doi: 10.1111/j.1600-0765.2011.01377.x. [DOI] [PubMed] [Google Scholar]
- Sexton WM, Lin Y, Kryscio RJ, Dawson DR, Ebersole JL, Miller CS. Salivary biomarkers of periodontal disease in response to treatment. J Clin Periodontol. 2011;38:434–441. doi: 10.1111/j.1600-051X.2011.01706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Heo SH, Lee JM, Cho JY. Identification of azurocidin as a potential periodontitis biomarker by a proteomic analysis of gingival crevicular fluid. Proteome Sci. 2011;9:42. doi: 10.1186/1477-5956-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart TC, Corby PM, Hauskrecht M, Hee Ryu O, Pelikan R, Valko M, Oliveira MB, Hoehn GT, Bretz WA. Identification of microbial and proteomic biomarkers in early childhood caries. Int J Dent. 2011;2011:196721. doi: 10.1155/2011/196721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sabbagh M, Alladah A, Lin Y, Kryscio RJ, Thomas MV, Ebersole JL, Miller CS. Bone remodeling-associated salivary biomarker MIP-1alpha distinguishes periodontal disease from health. J Periodontal Res. 2012;47:389–395. doi: 10.1111/j.1600-0765.2011.01445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Bairy I, Pai K, Satyamoorthy K, Prasad S, Berkovitz B, Radhakrishnan R. Salivary IL-6 levels in oral leukoplakia with dysplasia and its clinical relevance to tobacco habits and periodontitis. Clin Oral Invest. 2011;15:705–714. doi: 10.1007/s00784-010-0435-5. [DOI] [PubMed] [Google Scholar]
- Mager DL, Haffajee AD, Devlin PM, Norris CM, Posner MR, Goodson JM. The salivary microbiota as a diagnostic indicator of oral cancer: a descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. J Transl Med. 2005;3:27. doi: 10.1186/1479-5876-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Xiao H, Zhou H, Santiago S, Lee JM, Garon EB, Yang J, Brinkmann O, Yan X, Akin D, Chia D, Elashoff D, Park NH, Wong DT. Development of transcriptomic biomarker signature in human saliva to detect lung cancer. Cell Mol Life Sci. 2012;69:3341–3350. doi: 10.1007/s00018-012-1027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter ER. Analysis of nipple aspirate fluid for diagnosis of breast cancer: an alternative to invasive biopsy. Expert Rev Mol Diagn. 2005;5:873–881. doi: 10.1586/14737159.5.6.873. [DOI] [PubMed] [Google Scholar]
- Cao MQ, Wu ZZ, Wu WK. Identification of salivary biomarkers in breast cancer patients with thick white or thick yellow tongue fur using isobaric tags for relative and absolute quantitative proteomics. Zhong Xi Yi Jie He Xue Bao. 2011;9:275–280. doi: 10.3736/jcim20110307. [DOI] [PubMed] [Google Scholar]
- Rao PV, Reddy AP, Lu X, Dasari S, Krishnaprasad A, Biggs E, Roberts CT, Nagalla SR. Proteomic identification of salivary biomarkers of type-2 diabetes. J Proteome Res. 2009;8:239–245. doi: 10.1021/pr8003776. [DOI] [PubMed] [Google Scholar]
- Menezes GB, Pereira FA, Duarte CA, Carmo TM, Silva Filho HP, Zarife MA, Krieger MA, Reis EA, Reis MG. Hepatitis C virus quantification in serum and saliva of HCV-infected patients. Mem Inst Oswaldo Cruz. 2012;107:680–683. doi: 10.1590/S0074-02762012000500016. [DOI] [PubMed] [Google Scholar]
- Border MB, Schwartz S, Carlson J, Dibble CF, Kohltfarber H, Offenbacher S, Buse JB, Bencharit S. Exploring salivary proteomes in edentulous patients with type 2 diabetes. Mol Biosyst. 2012;8:1304–1310. doi: 10.1039/c2mb05079j. [DOI] [PubMed] [Google Scholar]
- Gao K, Zhou H, Zhang L, Lee JW, Zhou Q, Hu S, Wolinsky LE, Farrell J, Eibl G, Wong DT. Systemic disease-induced salivary biomarker profiles in mouse models of melanoma and non-small cell lung cancer. PLoS One. 2009;4:e5875. doi: 10.1371/journal.pone.0005875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael A, Bajracharya SD, Yuen PS, Zhou H, Star RA, Illei GG, Alevizos I. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16:34–38. doi: 10.1111/j.1601-0825.2009.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser J, Gehrmann U, D'Alexandri FL, Hidalgo-Estevez AM, Wheelock CE, Scheynius A, Gabrielsson S, Radmark O. Exosomes from human macrophages and dendritic cells contain enzymes for leukotriene biosynthesis and promote granulocyte migration. J Allergy Clin Immunol. 2010;126:1032–1040. doi: 10.1016/j.jaci.2010.06.039. 1040.e1-4. [DOI] [PubMed] [Google Scholar]
- Keller S, Ridinger J, Rupp AK, Janssen JW, Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med. 2011;9:86. doi: 10.1186/1479-5876-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasser C, Alikhani VS, Ekstrom K, Eldh M, Paredes PT, Bossios A, Sjostrand M, Gabrielsson S, Lotvall J, Valadi H. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J Transl Med. 2011;9:9. doi: 10.1186/1479-5876-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanisamy V, Wong DT. Transcriptomic analyses of saliva. Methods Mol Biol. 2010;666:43–51. doi: 10.1007/978-1-60761-820-1_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood JL, Pan H, Lanza GM, Wickline SA. Paracrine induction of endothelium by tumor exosomes. Lab Invest. 2009;89:1317–1328. doi: 10.1038/labinvest.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR. Exosomal transfer of proteins and RNAs at synapses in the nervous system. Biol Direct. 2007;2:35. doi: 10.1186/1745-6150-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park NJ, Li Y, Yu T, Brinkman BM, Wong DT. Characterization of RNA in saliva. Clin Chem. 2006;52:988–994. doi: 10.1373/clinchem.2005.063206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Begne M, Lu B, Han X, Hagen FK, Hand AR, Melvin JE, Yates JR. Proteomic analysis of human parotid gland exosomes by multidimensional protein identification technology (MudPIT) J Proteome Res. 2009;8:1304–1314. doi: 10.1021/pr800658c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CS, Wong DT. Breast cancer exosome-like microvesicles and salivary gland cells interplay alters salivary gland cell-derived exosome-like microvesicles in vitro. PloS One. 2012;7:e33037. doi: 10.1371/journal.pone.0033037. [DOI] [PMC free article] [PubMed] [Google Scholar]