Abstract
Multilevel interactions of the plant hormones ethylene and auxin coordinately and synergistically regulate many aspects of plant growth and development. This study isolated the AUXIN RESISTANT1 (AUX1) allele aux1rcr1 (RCR1 for REVERSING CTR1-10 ROOT1) that suppressed the root growth inhibition conferred by the constitutive ethylene-response constitutive triple response1-10 (ctr1-10) allele. The aux1rcr1 mutation resulted from an L126F substitution at loop 2 of the plasma membrane-associated auxin influx carrier protein AUX1. aux1rcr1 and the T-DNA insertion mutant aux1-T were both defective in auxin transport and many aspects of the auxin response. Unexpectedly, expression of the auxin-response reporter DR5:GUS in the root apex was substantially prevented by the aux1rcr1 but not the aux1-T mutation, even in the presence of the wild-type AUX1 allele. Following treatment with the synthetic auxin 1-naphthaleneacetic acid (NAA), DR5:GUS expression in aux1rcr1 and aux1-T occurred mainly in the root apex and mature zone. NAA-induced DR5:GUS expression in the root apex was markedly prevented by ethylene in genotypes with aux1rcr1 but not in aux1-T genotypes and the wild type. The effect of aux1rcr1 on DR5:GUS expression seemed to be associated with AUX1-expressing domains. Green fluorescence protein-fused aux1rcr1 was localized in the cytoplasm and probably not to the plasma membrane, indicating important roles of the Lys126 residue at loop 2 in AUX1 targeting. The possible effects of aux1rcr1 on DR5:GUS expression are discussed.
Key words: AUX1, Arabidopsis, DR5:GUS, auxin, ethylene, root gravitropism
Introduction
Ethylene and auxin are plant hormones coordinately regulating various aspects of plant growth and development. Ethylene is perceived by a small family of ethylene-receptor members. The biochemical nature of the receptor signalling mechanism is unknown. Current studies suggest that ethylene-receptor signalling is mediated by the physical interaction of the receptor histidine kinase (HK) domain and the mitogen-activated protein kinase kinase kinase CONSTITUTIVE TRIPLE-RESPONSE1 (CTR1) to suppress ethylene responses (Clark et al., 1998; Huang et al., 2003). Recent studies suggest that members of the ethylene receptor can cooperatively mediate the ethylene signal to an alternative pathway independent of CTR1 (Gao et al., 2008; Chen et al., 2010; Liu and Wen, 2012; Qiu et al., 2012; Xie et al., 2006, 2012). Auxin is perceived by a small family of TRANSPORTER INHIBITOR INSENSITIVE1 (TIR1)-related F-box proteins and functions as a ‘molecular glue’ to facilitate the association of the receptor and auxin/indole acetic acid (AUX/ IAA) proteins that negatively modulates the expression of AUXIN RESPONSE FACTORs (ARFs). The association of TIR1 and AUX/IAAs facilitates AUX/IAA polyubiquination, which subjects AUX/IAAs to 26S proteosome-mediated degradation, and the repression of ARF expression is alleviated (Dharmasiri et al., 2005; Dos Santos Maraschin et al., 2009; Parry et al., 2009; Tan et al., 2007). Activation of ARFs directs the expression of genes responsive to auxin.
Arabidopsis etiolated seedlings produce a long hypocotyl and root when grown without exogenous ethylene. With ethylene treatment, the curvature in the apical region is exaggerated, and the hypocotyl and root elongation is inhibited. Ethylene-induced seedling growth alterations are collectively called the seedling triple-response phenotype (Guzman and Ecker, 1990). Many ethylene-induced growth alterations depend on auxin, and seedlings of some mutants defective in auxin biosynthesis or transport show altered triple-response phenotype. ETHYLENE INSENSITIVE ROOT1 (EIR1) encodes the auxin efflux carrier protein PIN-FORMED2 (PIN2), and the eir1/pin2 loss-of-function mutation impacts on root gravitropism and prevents ethylene-induced root growth shortening (Roman et al., 1995; Luschnig et al., 1998; Muller et al., 1998). HOOKLESS1 (HLS1) encodes an N-acetyltransferase, and the hls1 loss-of-function mutation prevents the ethylene-induced apical hook formation. A suppressor screen for hls1 led to the identification of hookless1 suppressor1 (hss1), which is defective in the auxin-response transcription factor AUXIN RESPONSE FACTOR2 (Lehman et al., 1996; Li et al., 2004). AUXIN RESISTANT1 (AUX1) encodes an auxin influx carrier protein that associates with the plasma membrane (PM) depending on the endoplasmic reticulum protein AUXIN RESISTANT4 (AXR4) (Dharmasiri et al., 2006). Loss-of-function mutations of aux1 prevent the ethylene-induced root shortening and apical hook formation (Stepanova et al., 2007; Vandenbussche et al., 2010). Previously, weak ethylene insensitive (wei) mutants were isolated by a genetic screen for components that involve ethylene signalling (Alonso et al., 2003). With ethylene treatment, the seedling root was longer for wei1, wei2, wei7 and wei8 mutants than for the wild type. WEI1 encodes the auxin receptor protein TIR1, and WEI2, WEI7 and WEI8 are involved in auxin biosynthesis (Alonso et al., 2003; Stepanova et al., 2005, 2007, 2008). The ethylene-insensitive mutants ethylene insensitive2 (ein2) and ein3 are resistant to the inhibition of lateral root initiation by auxin, which indicates that both ethylene signalling and auxin responses are essential for this inhibitory effect (Ivanchenko et al., 2008). Measurement of auxin transport suggests that ethylene promotes long-distance polar auxin transport through AUX1 and results in the negative effect of ethylene on lateral root formation (Negi et al., 2008). The involvement of auxin in growth alterations induced by ethylene suggests that the interplay of the two hormones coordinately controls many aspects of plant growth and development.
The biologically active auxin IAA is transported across cells distantly. IAA may exist in two forms: the charged IAA– and the protonated IAAH. With the acidic condition in the apoplast (pH ~5.5) outside the cell, a portion of IAA is protonated (IAAH) and can pass passively through the PM into the cell, whereas the charged IAA– cannot. The PM-associated AUX1 and its homologs Like AUX1s (LAXs) are the auxin influx carriers that transport the charged IAA– to the cytoplasm (Carrier et al., 2008; Péret et al., 2012; Robert and Friml, 2009; Zažímalová et al., 2010). About 17% of IAA freely enters the cells and 83% is de-protonated (Blakeslee et al., 2005; Zažímalová et al., 2010). The pH is nearly neutral in the cytoplasm, and IAA is de-protonated and exists as IAA–. The de-protonated IAA– is trapped in the cytoplasm and cannot permeate the membrane; it requires auxin transporters such as PIN proteins to exit the cell. With the polarized localization of different PIN proteins, IAA is transported in a polar fashion, the so-called polar auxin transport (Kerr and Bennett, 2007; Robert and Friml, 2009; Zažímalová et al., 2010) for acropetal and basipetal auxin transport and thus modulation of various growth and development events (Blakeslee et al., 2005).
To isolate components that involve synergistic functions by ethylene and auxin, we isolated the reversing ctr1-10 root1 (rcr1) mutation that suppresses root growth inhibition of the hypomorphic ctr1-10. rcr1 is allelic to AUX1, and is designated aux1rcr1. The aux1rcr1 mutation but not the T-DNA insertion allele aux1-T, prevented expression of the DR5:GUS construct in the root apex. The aux1rcr1 isoform predominantly localized in the cytoplasm, whereas AUX1 localized to the PM. AUX1 loops 1 and 3 but not loop 2 have been predicted to be involved in AUX1 functioning (Swarup et al., 2004). The residue Lys126 in AUX1 loop 2 could involve correct AUX1 targeting to the PM. The aux1rcr1 mutation may prevent DR5:GUS expression, possibly by affecting nuclear auxin signalling.
Materials and methods
Plant materials and seedling germination and growth conditions
The ctr1-10 (SALK_122868.46.30.n) and aux1-T (CS859699) mutants were from the Arabidopsis Biological Resource Center. For seed germination and seedling growth, Arabidopsis seeds were stratified on Murashige and Skoog (MS) salt-containing agar (0.8% agar, pH 5.8) for 72h at 4 °C and then germinated at 22 °C. For ethylene treatment, ethylene gas (20 µl l–1) was applied. For auxin treatment, the auxin concentrations were as indicated. Seedling hypocotyl and root measurements were carried out with VideoTesT (Moscow) as described previously (Zhou et al., 2007); more than 30 individual seedlings were scored for each treatment, and the measurement was presented as mean ±standard deviation (SD). The cloning of transgenes in this study is described in Supplementary Data S1 at JXB online.
Quantitative RT-PCR (qRT-PCR) and PCR-based genotyping
qRT-PCR of the expression of CTR1, AUX1, and aux1rcr1 involved use of StepOne Plus™ (ABI). The primer and sequence information for qRT-PCR is given in Supplementary Data S1.
Root gravity response assay
Seeds were surface sterilized, stratified in the dark at 4°C for 72h, and germinated vertically on 0.5× strength MS (0.5× MS) salt-containing agar with constant illumination at 22 °C for 5 d. Seedlings were then transferred to agar containing 0.5× MS salt with or without 10–7 mol l–1 1-naphthaleneacetic acid (NAA) or 2,4-dichlorophenoxyacetic acid (2,4-D), and grown horizontally (at 90° rotation) under the same growth conditions. The root gravity response was scored by measuring the angles formed 24h after the gravity change with use of ImageJ (NIH).
Auxin transport assay
Arabidopsis seedlings were grown for 6 d and a 10mm segment to the root tip was excised. [3H]-labelled IAA was applied to the cut and the root segments were incubated in the dark for 6h. After incubation, a 5mm segment to the tip was excised and washed with 0.5× MS salt. The washed root tips (15 tips for each measurement) were incubated in scintillation liquid and scintillation counting was carried out (PerkinElmer 1450 Microbeta scintillation counter) for [3H]IAA measurement.
Laser-scanning confocal microscopy
The subcellular localizations of yellow fluorescent protein (YFP)–AUX1 and green fluorescent protein (GFP)–aux1rcr1 were examined by laser scanning confocal microscopy with an Olympus FV1000 microscope.
β-Glucuronidase (GUS) staining
Histochemical staining for GUS activity in transgenic plants was performed as described previously (Jefferson et al., 1987). Seedlings were grown under light on MS salt-containing agar for 4 d and transferred to MS salt-containing agar with or without auxin (100nM NAA) in an air-tight chamber with or without ethylene treatment for 2 d. The seedlings were harvested, immersed in the reaction solution (1mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid, 100mM sodium phosphate, 0.1mM EDTA, 0.5mM ferricyanide, 0.5mM ferrocyanide, and 0.1% Triton X-100, pH 7.0) and incubated at 37 °C for 16h.
Results
The hypomorphic ctr1-10 mutation results in mild constitutive ethylene responses
Ethylene inhibits elongation of the hypocotyl and primary root of etiolated Arabidopsis seedlings. We sought to isolate the components involved in ethylene-induced root growth inhibition from a screen of suppressors in a mutation background that exhibited weak constitutive ethylene responses.
Currently known constitutive triple response1 (ctr1) loss-of-function mutants, except for ctr1-8 and ctr1btk, show strong constitutive ethylene responses, with a short seedling hypocotyl and root (Huang et al., 2003; Ikeda et al., 2009; Xie et al., 2012). Here, we found that ctr1-10 had a T-DNA insertion in the 5’-untranslated region (5’-UTR) and determined whether it was a weak allele that could be used for a suppressor screen (Fig. 1A).
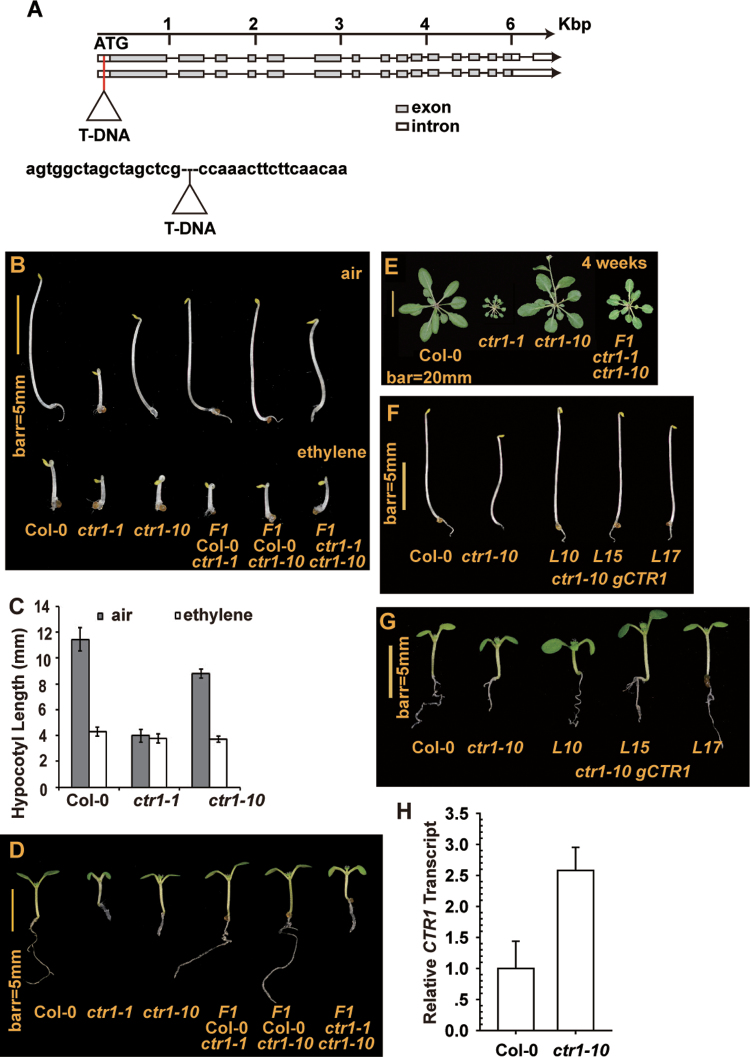
Fig. 1.
The ethylene response phenotype of ctr1-10. (A) The structure and T-DNA insertion position, with the flanking sequence shown, for ctr1-10. Two CTR1 alternative spliced isoforms are shown. (B, C) Phenotype (B) and hypocotyl measurement (C) for wild-type, ctr1-1, and ctr1-10 seedlings grown in the dark with or without ethylene (20 µl l–1). (D, E) Phenotype for light-grown seedlings (D) and rosettes (E) of the wild type (Col-0) and ctr1-1 and ctr1-10 mutants. (F, G) Expression of the genomic gCTR1 transgene rescued the constitutive ethylene-response phenotype of etiolated (F) and light-grown (G) ctr1-10 seedlings. (H) qRT-PCR analysis of CTR1 expression in the wild type (Col-0) and ctr1-10 mutant. Data are means ±SD or ±standard error (SE) for hypocotyl measurement and gene expression, respectively.
Under dark growth conditions, the seedling hypocotyl was shorter for ctr1-10 than for the wild type (Col-0) and was longer than ctr1-1 without ethylene treatment. F1 seedlings of ctr1-1 and ctr1-10 phenotypically resembled ctr1-10 seedlings. Hypocotyls were slightly shorter for F1 seedlings generated from the respective crosses of the wild type with ctr1-1 and ctr1-10 than for wild-type seedlings. Ethylene treatment inhibited seedling growth, and these genotypes showed a typical ethylene triple-response phenotype: shortening of the seedling hypocotyl and root, with an exaggerated apical hook (Fig. 1B). Hypocotyl measurement of seedlings gave the same results, with the hypocotyl shorter for ctr1-10 seedlings than for the wild type and longer than for ctr1-1 seedlings without ethylene treatment (Fig. 1C).
Under light growth conditions, the seedling growth inhibition phenotype was more severe for ctr1-10 than for the wild type (Col-0) and weaker than that of ctr1-1. The cotyledons were small and the hypocotyls and roots were shorter for ctr1-10 and ctr1-1 than for the wild-type seedlings, and ctr1-1 seedlings produced a shorter primary root and smaller cotyledons than ctr1-10 seedlings. The mutant phenotype was weaker for F1 ctr1-10/ctr1-1 than for ctr1-1 plants but was similar to that for ctr1-10 plants (Fig. 1D). At the adult stage, the ctr1-1 mutant produced a relatively small rosette, but wild-type and ctr1-10 plants did not differ in rosette size. The rosette was smaller for F1 ctr1-10/ctr1-1 than ctr1-10 plants but larger than for ctr1-1 plants (Fig. 1E). Thus, the constitutive ethylene response was weaker with the ctr1-10 than with the ctr1-1 mutation. Complementation tests showed that ectopic expression of the genomic CTR1 clone gCTR1 (driven by the native CTR1 promoter) rescued the ctr1-10 seedling growth inhibition (Fig. 1F, G).
The T-DNA insertion occurs at the 5’-UTR and does not disrupt the CTR1 open reading frame. qRT-PCR revealed greater mRNA expression of CTR1 in ctr1-10 than in the wild type (Fig. 1H). Ethylene promotes CTR1 expression (Hall et al., 2012), and the increased CTR1 level was consistent with the elevated constitutive ethylene response in ctr1-10. Sequence analysis did not identify an alternative start codon in the 5’-UTR. The mutation nature that attenuates CTR1 functions in ctr1-10 is unclear; one possibility is that the corresponding CTR1 transcript may not be efficiently translated into protein. Our results indicated that ctr1-10 is a loss-of-function mutation and a hypomorph.
Isolation of REVERSING CTR1-10 ROOT1 (RCR1)
To isolate the components of ethylene-induced root growth inhibition, we mutagenized ctr1-10 with ethyl methanesulfonate and grew the resulting M2 seedlings under light on MS salt-containing agar. We identified a mutant that produced a longer primary root than ctr1-10 and named the mutation reversing ctr1-10 root1 (rcr1).
Etiolated seedlings of ctr1-10 rcr1 produced a longer primary root than ctr1-10, regardless of ethylene treatment, but the hypocotyls were similar in length. Ethylene treatment resulted in the formation of an exaggerated apical hook curvature in wild-type (Col-0) and ctr1-10 seedlings but not in ctr1-10 rcr1 seedlings (Fig. 2A). Consistently, light-grown crt1-10 and ctr1-10 rcr1 seedlings were phenotypically similar, except that ctr1-10 rcr1 produced a longer root, regardless of ethylene treatment (Fig. 2B). Of note, ctr1-10 rcr1 seedlings showed an agravitropic root growth phenotype (Fig. 2A, B).
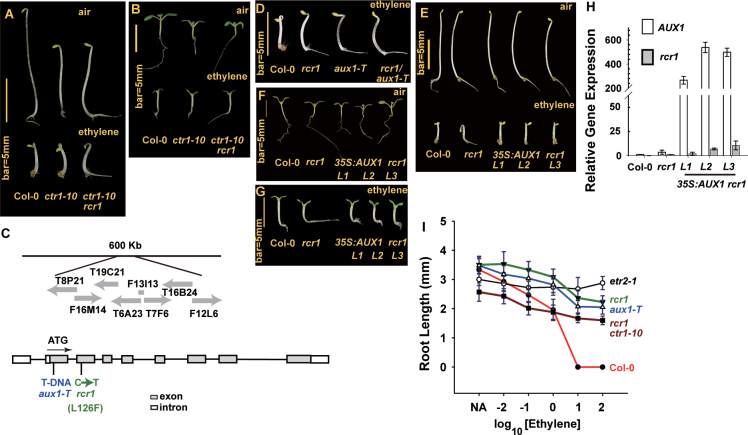
Fig. 2.
RCR1 is allelic to AUX1. (A, B) Phenotype for etiolated (A) and light-grown (B) seedlings of the wild type (Col-0) and ctr1-10 and ctr1-10 rcr1 mutants. (C) RCR1 maps to a 600kb region, and the rcr1 mutation results from a C→T mutation (L126F substitution); aux1-T is a T-DNA insertion allele for AUX1. Grey arrows indicate the positions of bacterial artificial clones in this region. The AUX1 gene structure and positions for the T-DNA insertion site and the rcr1 lesion are indicated. (D) Phenotype of etiolated seedlings of rcr1, aux1-T, and the F1 of rcr1 and aux1-T (rcr1/aux1-T) with ethylene (20 µl l–1) treatment. (E–G) Phenotype of rcr1 seedlings, grown in the dark (E) and light (F, G) expressing the 35S:AUX1 transgene without (F) or with (G) ethylene (20 µl l–1) treatment. (H) qRT-PCR of AUX1 expression in rcr1 expressing 35S:AUX1. (I) Ethylene dose-response curve for the root growth of light-grown seedlings as indicated. Data are means ±SD or ±SE for root length and gene expression, respectively. (This figure is available in colour at JXB online.)
To clone RCR1, crt1-10 rcr1 was crossed with the La-0 ecotype, and the resulting F2 seedlings exhibiting the ctr1-10 rcr1 root phenotype underwent map-based cloning. Using 472 individual F2 samples, RCR1 was mapped to a 600kb region on chromosome 2. Within this region, we identified a C→T transition mutation at the AUX1 locus, which resulted in the L126F substitution (Fig. 2C). Thus, RCR1 may be an AUX1 allele and the mutation may prevent the ethylene-induced root growth inhibition and apical hook formation. To support this suggestion, we crossed aux1-T (Fig. 2C; a T-DNA insertion mutation of aux1) with rcr1 for an allele test, and the root phenotype of the resulting F1 seedlings was similar to that of both parents. The ethylene-induced root growth inhibition and apical hook phenotype in wild-type (Col-0) seedlings was prevented in aux1-T, rcr1, and F1 seedlings (Fig. 2D). We performed a complementation test for rcr1 with expression of the 35S:AUX1 transgene. Following ethylene treatment, the roots were longer in etiolated rcr1 than in wild-type seedlings, with agravitropic root growth and no exaggerated apical hook curvature. As expected, seedlings of rcr1 lines expressing the 35S:AUX1 transgene were phenotypically similar to wild-type seedlings and showed root growth inhibition and an exaggerated apical hook with ethylene treatment (Fig. 2E). With light germination, the roots were longer for rcr1 than for wild-type seedlings and rcr1 lines expressing 35S:AUX1, regardless of ethylene treatment (Fig. 2F, G). qRT-PCR of AUX1 and rcr1 levels suggested that the 35S:AUX1 transgene was expressed (Fig. 2H). The aux1/rcr1 root ethylene-insensitive phenotype was consistent with the ethylene dose–response assay for root growth inhibition, which showed a much shorter seedling root for the wild type (Col-0) than for the aux1/rcr1 and the ethylene-insensitive etr2-1 mutant at elevated ethylene concentrations (Fig. 2I).
Genetic and complementation tests suggested that rcr1 is an AUX1 allele, and we designated rcr1 as aux1rcr1. The aux1rcr1 mutation, like the aux1-T mutation, largely attenuated but did not completely prevent the ethylene-induced root growth inhibition. The defect in apical hook formation with aux1rcr1 is consistent with the defective apical hook phenotype in aux1-21 (Vandenbussche et al., 2010).
aux1rcr1 is defective in auxin transport and has reduced sensitivity to auxin
AUX1 is an auxin influx carrier, and its loss-of-function mutation results in reduced auxin transport. To determine whether the aux1rcr1 mutation also attenuated auxin transport, we measured acropetal auxin transport (from the shoot towards the root apex) in aux1rcr1 seedling roots.
Measurement of the uptake of the tritiated auxin IAA showed an identical level of [3H]IAA in root apexes of aux1rcr1 and aux1-T (Fisher’s LSD, P=0.4) but lower than that in the wild type (Col-0) (Fisher’s LSD, P<0.003). With the level of [3H]IAA in the wild type set to 1, the level of [3H]IAA in aux1-T and aux1rcr1 was about 0.51 and 0.41, respectively, which suggested reduced acropetal auxin transport in the mutants (Fig. 3A) and was consistent with aux1-22 showing an approximate 50% reduction in auxin transport (Rahman et al., 2001).
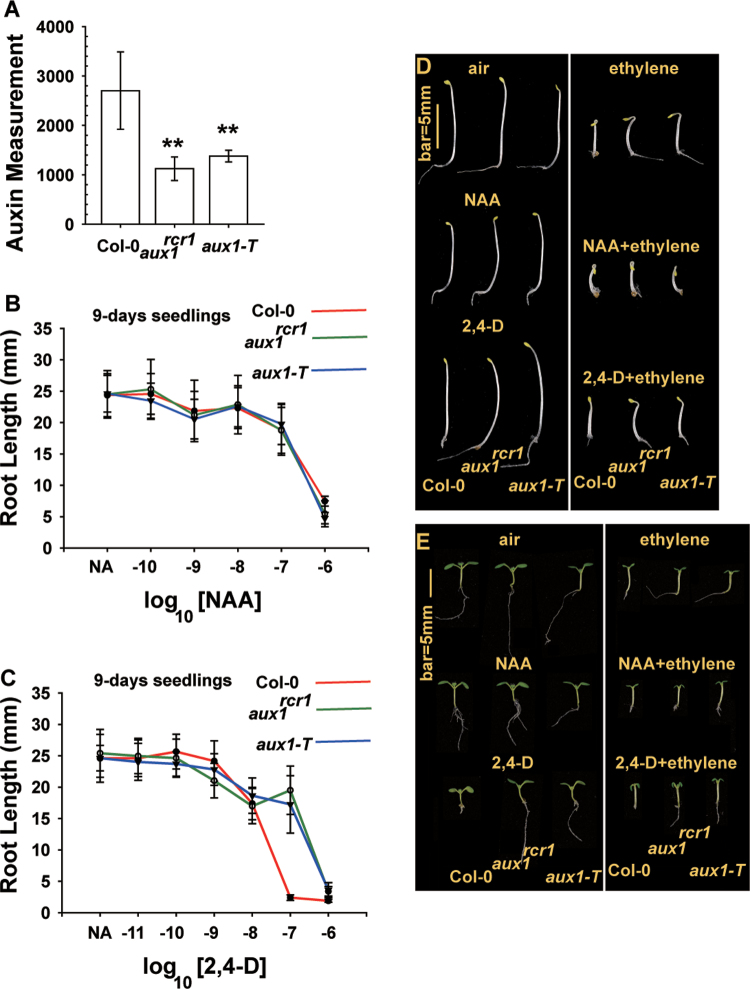
Fig. 3.
The aux1rcr1 mutant is defective in IAA transport. (A) Uptake of exogenously applied [3H]IAA in wild-type (Col-0), aux1rcr1, and aux1-T seedling roots. (B, C) Does–response curve for root growth with NAA (B) and 2,4-D (C) treatment. (D, E) Effects of ethylene and auxin on seedling growth of aux1rcr1 compared with wild-type (Col-0) and aux1-T seedlings. Data are means ±SD for root length. (This figure is available in colour at JXB online.)
Import of the synthetic auxins NAA and 2,4-D is in part independent of and dependent on, respectively, the auxin influx carrier AUX1. Conceivably, both aux1rcr1 and aux1-T seedlings are responsive to NAA but not to 2,4-D. We performed a dose–response assay to evaluate the effect of NAA and 2,4-D on the root growth of wild-type, aux1rcr1, and aux1-T seedlings. NAA inhibited the root elongation of light-grown aux1rcr1, aux1-T, and wild-type seedlings (9 d after germination) to a similar extent (Fig. 3B). Wild-type (Col-0) seedlings germinated under light conditions showed marked root growth inhibition with the auxin 2,4-D at a concentration of >10–8 mol l–1. Root growth was strongly inhibited in aux1rcr1 and aux1-T with 2,4-D at >10–7 mol l–1 (Fig. 3C).
Air-grown, etiolated aux1rcr1 and aux1-T seedlings were phenotypically identical. Following ethylene treatment, root growth was inhibited less in aux1rcr1 and aux1-T seedling than in the wild type, and neither mutant produced exaggerated apical hook curvature (Fig. 3D). Of note, NAA treatment (10–7 mol l–1) had minor effects on seedling root growth (Fig. 3B), and NAA but not 2,4-D (10–7 mol l–1) facilitated the ethylene-induced seedling triple-response phenotype in aux1rcr1 and aux1-T seedlings (Fig. 3D). 2,4-D treatment inhibited the root growth of wild-type but not aux1rcr1 and aux1-T seedlings. With 2,4-D and ethylene treatment, root growth was inhibited less in both mutants than in the wild type. Of note, the ethylene-induced apical hook formation in wild-type seedlings was prevented by 2,4-D and was not observed in aux1rcr1 and aux1-T seedlings (Fig. 3D).
Grown under light with ethylene treatment, root growth was inhibited less in the mutant than in the wild-type seedlings (Fig. 3E). With NAA treatment (10–7 mol l–1), the primary root of wild-type, aux1rcr1, and aux1-T seedlings was similar in length. In contrast, 2,4-D treatment (10–7 mol l–1) inhibited the root growth of the wild-type but not the aux1rcr1 and aux1-T seedlings (Fig. 3B, C, E). With ethylene treatment, NAA-treated wild-type, aux1rcr1, and aux1-T seedlings were phenotypically identical, whereas the root was shorter for 2,4-D-treated wild-type than aux1rcr1 and aux1-T seedlings (Fig. 3E).
Thus, the aux1rcr1 mutation attenuated auxin transport and reduced the sensitivity to auxin. Reduced auxin transport in aux1rcr1 and aux1-T root apexes was consistent with the ethylene-induced root growth inhibition and apical hook phenotype of aux1rcr1 and aux1-T rescued by NAA but not by 2,4-D treatment.
aux1rcr1 but not the aux1-T allele alleviates DR5:GUS expression in the root apex
The DR5:GUS construct comprises a synthetic auxin-responsive promoter (DR5) fused to the GUS-encoding reporter gene. The expression of DR5:GUS is thus auxin inducible and has been widely used as a reporter to indicate auxin responses (Ulmasov et al., 1995; Ivanchenko et al., 2008; Negi et al., 2008). With the DR5:GUS transgene, an auxin maximum in wild-type root apexes is associated with elevated GUS expression. Here, we evaluated whether the root apex auxin maximum would be altered by the aux1rcr1 allele by comparing DR5:GUS expression in the root apex of wild-type, aux1-T, and aux1rcr1 seedlings.
The DR5:GUS transgene was introduced into aux1-T and aux1rcr1 plants from a common wild-type line that carries the transgene (designated the DR5:GUS donor), and GUS expression was examined. With growth on MS medium, GUS staining was observed in wild-type (Col-0) and aux1-T root apexes (zone 1), as defined previously (Stepanova et al., 2007). Ethylene treatment promoted acropetal auxin transport and elevated GUS staining in the transition zone (zone 2) in wild-type but not in aux1-T root tips. The minor effects of ethylene treatment on the DR5:GUS maximum in aux1-T root apexes were consistent with DR5:GUS expression in aux1-7 root apexes unaffected by the ethylene biosynthesis precursor 1-aminocyclopropane-1-carboxylic acid (Stepanova et al., 2007). Unexpectedly, GUS staining was not observed or was extremely weak in aux1rcr1 root apexes, regardless of ethylene treatment (Fig. 4A).
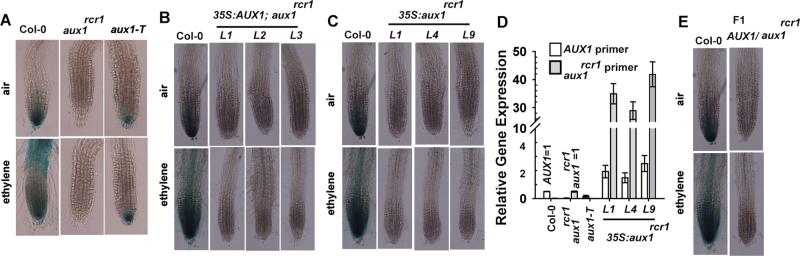
Fig. 4.
DR5:GUS expression in the root tip is alleviated by the aux1rcr1 allele. (A) Expression of DR5:GUS in root tips of wild-type (Col-0), aux1rcr1, and aux1-T seedlings. (B, C) Expression of DR5:GUS in wild-type and aux1rcr1 lines expressing 35S:AUX1 (B) and in wild-type lines expressing 35S:aux1rcr1 (C). (D) qRT-PCR of AUX1 and aux1rcr1 expression in wild-type, aux1rcr1, and wild-type (Col-0) DR5:GUS lines expressing 35S:aux1rcr1. Data are means ±SE of three measurements from three independent biological samples. (E) Expression of DR5:GUS in root tips of the wild type (Col-0) and F1 aux1rcr1 seedlings. The ethylene concentration is 20 µl l–1. L, transformation line. (This figure is available in colour at JXB online.)
Therefore, the aux1rcr1 allele prevented DR5:GUS expression. Given that expression of the 35S:AUX1 transgene complemented the aux1rcr1 mutation (Fig. 2), we examined whether the transgene rescued DR5:GUS expression in aux1rcr1. The 35S:AUX1 transgene was transformed into aux1rcr1 plants expressing DR5:GUS (Fig. 4A), but expression of DR5:GUS was not rescued by the 35S:AUX1 transgene, regardless of ethylene treatment (Fig. 4B). AUX1 levels in the transformed lines were highly elevated (Supplementary Fig. S2 at JXB online); hence, aux1rcr1 could prevent DR5:GUS expression, even in the presence of the wild-type AUX1. We examined the reciprocal negative effects of aux1rcr1 on DR5:GUS expression in the DR5:GUS donor expressing 35S:aux1rcr1. The 35S:aux1rcr1 transgene was introduced into the DR5:GUS donor (Fig. 4A) by transformation, and DR5:GUS was expressed at the same locus in the donor and transformed lines. In 27 independent lines that we examined, expression of 35S:aux1rcr1 prevented DR5:GUS expression in the root apex; Fig. 4C shows DR5:GUS expression in three representative lines. Expression of the 35S:aux1rcr1 transgene was confirmed by qRT-PCR in these three lines with reference to AUX1 and aux1rcr1 levels in the wild-type (Col-0) and aux1rcr1, which were each given a value of 1, respectively (Fig. 4D; i.e. AUX1=1 in the wild type and aux1rcr1=1 in aux1rcr1).
Thus, aux1rcr1 may have dominant-negative effects on DR5:GUS expression in the root apex. To support this scenario, we examined DR5:GUS expression in the heterozygous AUX1/aux1rcr1 line expressing the DR5:GUS transgene. The wild-type DR5:GUS donor and aux1rcr1 that expressed the DR5:GUS from the donor were genetically crossed to produce the heterozygous F1 AUX1/aux1rcr1; DR5:GUS, in which the DR5:GUS transgene was from a common donor. As expected, DR5:GUS expression was prevented (Fig. 4E).
DR5:GUS expression is NAA inducible in genotypes with aux1rcr1
We showed that DR5:GUS expression was largely prevented by the aux1rcr1 allele, even in the presence of the wild-type AUX1. The mutant aux1rcr1 could have a dominant-negative effect on DR5:GUS expression. Alternatively, DR5:GUS was not expressed (or silenced) for unknown reasons. The intercellular transport of NAA is independent of AUX1. If DR5:GUS expression is NAA inducible in genotypes with aux1rcr1, prevention of DR5:GUS expression by aux1rcr1 was probably not due to the silencing of DR5:GUS.
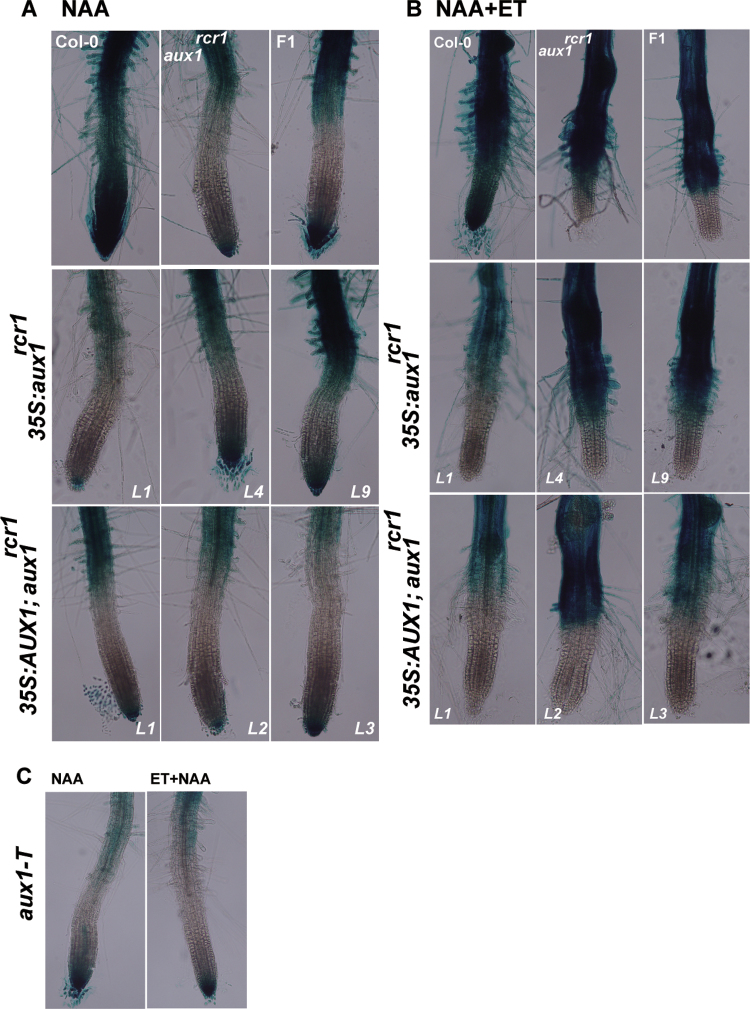
The DR5:GUS donor (the wild type expressing DR5:GUS) showed strong GUS staining in the root apex, zone 2, and mature zone with NAA treatment. Expression of DR5:GUS was induced by NAA in the root apex and mature zone in aux1rcr1, the wild type (Col-0) expressing 35S:aux1rcr1, aux1rcr 1 lines expressing 35S:AUX1, and the F1 aux1rcr1/AUX1 expressing the DR5:GUS transgene; however, GUS staining in the elongation zone was barely detectable (Fig. 5A). These results did not favour the DR5:GUS transgene being silenced in these genotypes. The aux1rcr1 mutation had dominant-negative effects on DR5:GUS expression in a domain-specific manner.
Fig. 5.
Expression of DR5:GUS expression with NAA treatment and NAA plus ethylene treatment. DR5:GUS expression in the wild type, aux1rcr1, F1 aux1rcr1 and the wild type, wild-type lines expressing 35S:aux1rcr1, and aux1rcr1 lines expressing 35S:AUX1 following NAA (A) and NAA plus ethylene treatment (B). (C) DR5:GUS expression in aux1-T in response to NAA and NAA plus ethylene treatment. L, transformation line. NAA was used 10–7 mol l–1, and ethylene (ET) at 20 µl l–1. (This figure is available in colour at JXB online.)
Ethylene promotes auxin biosynthesis and acropetal transport, and DR5:GUS expression is elevated in the root tip. We examined whether ethylene treatment could synergistically elevate NAA-induced DR5:GUS levels in the root tip of genotypes with aux1rcr1. Of note, ethylene inhibited root elongation, and the region below the mature zone was largely shortened compared with no-ethylene treatment (Fig. 5B, C). In wild-type root tips, GUS staining was strong in the root apex, zone 2, and the mature zone with ethylene and NAA treatment. Unexpectedly, DR5:GUS expression was strong in the mature zone but nearly abolished in the root apex and elongation zone in genotypes with aux1rcr1 (Fig. 5B). Thus, the DR5:GUS transgene was probably not silenced; rather, its expression was affected by the aux1rcr1 allele in a domain-specific manner. Measurement of GUS staining intensity supported the association of DR5:GUS expression inhibition with aux1rcr1 in the root apex (Supplementary Fig. S3 at JXB online). In contrast to the aux1rcr1 allele, which prevented DR5:GUS expression, DR5:GUS expression was not prevented in the root apex of aux1-T. NAA treatment induced DR5:GUS levels in the root apex and mature zone but in not the region in between, and the induction was not prevented by ethylene treatment (Fig. 5C).
AUX1 is expressed mainly in the root apex (Péret et al., 2012). Our results suggested that aux1rcr1 affected DR5:GUS expression in AUX1-expressing domains.
aux1rcr1 and aux1-T mutations but not aux1rcr1 overexpression impair root gravitropism
In response to gravity changes, AUX1 and the auxin efflux carrier PIN2 protein mediate differential, basipetal auxin transport from the columella via the lateral root cap (LRC) cells to the expanding epidermis (Swarup et al., 2005; Rahman et al., 2010). As a result, root cells grow faster with lower than with higher auxin concentrations, and differential cell growth is facilitated. The differential root cell growth facilitates a curvature formation that re-orients the root growth towards gravity (Luschnig et al., 1998; Marchant et al., 1999; Rashotte et al., 2000; Ottenschläger et al., 2003).
Both aux1rcr1 and aux1-T seedling roots showed an agravitropic growth phenotype (Fig. 2) and showed distinct DR5:GUS expression patterns in air and ethylene. We evaluated the association of the root gravity response with root DR5:GUS expression in aux1-T, aux1rcr1, and aux1rcr1ox lines (wild-type lines expressing 35S:aux1rcr1).
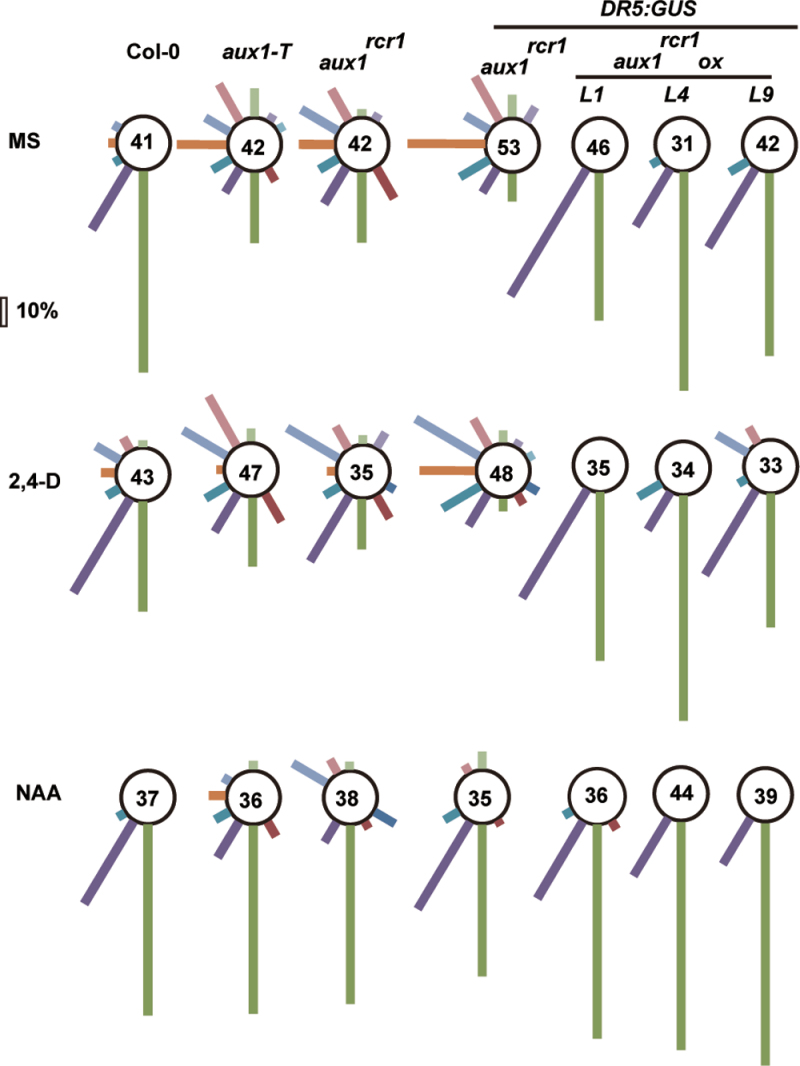
We quantified the extent of altered root gravity by measuring the root angles formed after a gravity change of 90° for vertically grown seedlings. The root growth angles were grouped in 12 classes of 30°, and we have presented the gravity response for the wild type (Col-0), aux1-T, aux1rcr1, and aux1rcr1ox lines diagrammatically (Fig. 6). Both aux1-T and aux1rcr1 seedlings showed a root-growth lack of gravitropism after the gravistimulation, whereas the gravity response of wild-type seedlings and aux1rcr1ox lines was similar. 2,4-D treatment did not rescue the agravitropic phenotype in aux1-T and aux1rcr1 seedlings, and its effects on the gravity response in wild-type seedlings and aux1rcr1ox lines were minor. As expected, NAA treatment rescued the agravitropic phenotype in aux1-T and aux1rcr1 seedlings to a similar degree as in the wild type and aux1-T seedlings.
Fig. 6.
Root gravity response assay. Illustration of the root response to gravistimulation with 12 classes of 30°, 24h after a 90° rotation on agar medium containing MS salt. The bar indicates 10% of the seedlings. The 2,4-D and NAA concentrations were both 10–7 mol l–1. Numbers indicate the population size for each scoring.
Roots of aux1-T and aux1rcr1 but not aux1rcr1ox seedlings showed defects in response to a 90° gravity stimulus, and the aux1rcr1 allele and aux1rcr1 overexpression impaired DR5:GUS expression in the root apex. The aux1rcr1 isoform could actively affect auxin distribution or concentration in the root apex but was insufficient to affect the root gravitropism in the presence of the wild-type AUX1. The wild-type AUX1 had a role in the root gravity response, even in the presence of the aux1rcr1 allele that has dominant-negative effects on DR5:GUS expression maximum.
Subcellular localization of GFP-fused aux1rcr1
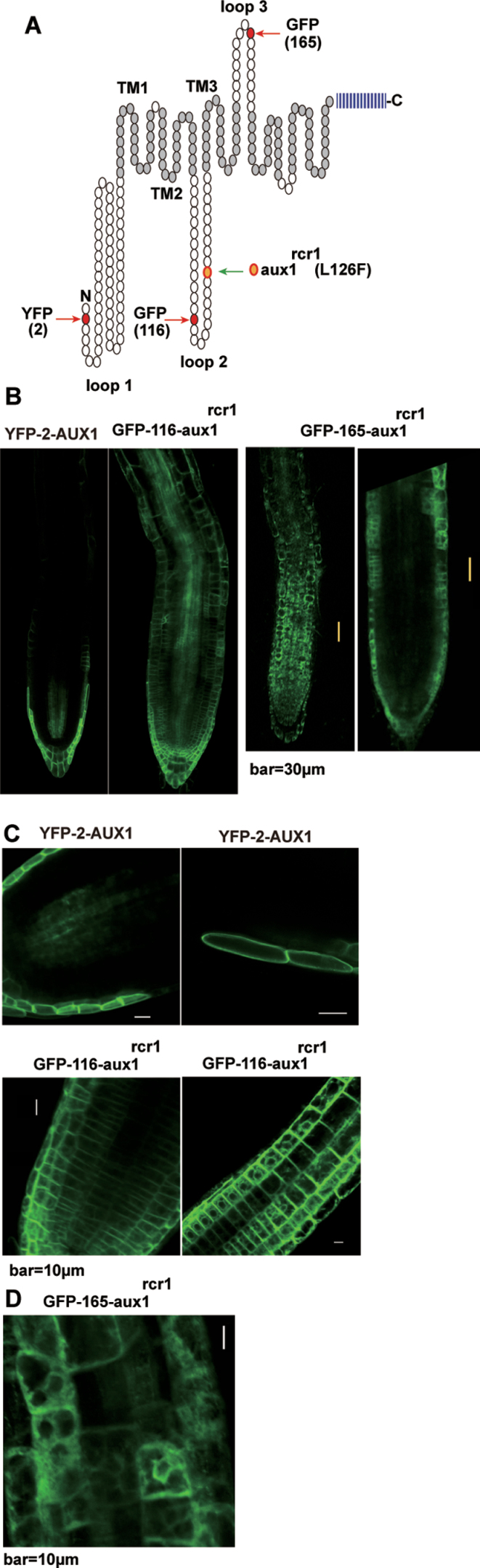
AUX1 is predicted to have ten transmembrane helixes. YFP fused to AUX1 at position 116 (loop 2) has revealed the fused YFP portion localized within the cytoplasm. However, YFP does not generate fluorescence when fused at position 165 (loop 3), and the YFP portion is extracytoplasmic (Swarup et al., 2004) (Fig. 7A). We examined the fluorescence of GFP fused to aux1rcr1 at positions 116 (designated GFP-116-aux1rcr1) and 165 (GFP-165-aux1rcr1) to evaluate aux1rcr1 targeting (Fig. 7A).
Fig. 7.
Subcellular localization of GFP–aux1rcr1. (A) Schematic illustration of the N-terminal structure of AUX1 and residues in which YFP or GFP were inserted. Red and green arrowheads indicate YFP/GFP insertion sites and the aux1rcr1 mutation, respectively. Blue vertical bars indicate the C-terminal portion of AUX1 not graphically shown. (B) Expression patterns of AUX1p:YFP-AUX1, 35S:GFP-116-aux1rcr1, and 35S:GFP-165-aux1rcr1 in the root tip. The fluorescence of GFP-165-aux1rcr1 was observed at different focal planes for cells on the surface (left panel) and in the middle (right panel) of a root. (C) Subcellular localization of YFP-AUX1 in LRC cells and GFP-116-aux1rcr1 in cells of the root tip (left panel) and elongation zone (right panel). (D) Subcellular localization of GFP-165-aux1rcr1 in root tip cells. (This figure is available in colour at JXB online.)
YFP fused with the wild-type AUX1 at position 2 (loop 1) (Fig. 7A), designated YFP-2-AUX1, was expressed (driven by the native AUX1 promoter) in columella, LRC, epidermis, and protophloem cells, as described previously, and localized to the PM (Péret et al., 2012) (Fig. 7B, C). Driven by the constitutive 35S promoter, GFP-116-aux1rcr1 was expressed over nearly all the root tip cells (Fig. 7B). GFP fluorescence with GFP-165-aux1rcr1 (driven by the 35S promoter) was observed mainly in cells of the outer layers (Fig. 7B). Unexpectedly, the fluorescence of GFP-116-aux1rcr1 and GFP-165-aux1rcr1 showed a pattern characteristic of the endoplasmic reticulum (ER) structure and in part of the PM (Fig. 7C, D). Subcellular compartments that GFP-aux1rcr1 could be associated with need to be investigated. AUX1 recycles between the PM and cytoplasm (Kleine-Vehn et al., 2006; Spitzer et al., 2009); the L126F substitution by the aux1rcr1 mutation could impact on AUX1 trafficking. AUXIN RESISTANT4 (AXR4) is an ER protein important to AUX1 targeting (Dharmasiri et al., 2006; Hobbie, 2006). The L126F mutation could impair aux1rcr1 targeting mediated by AXR4.
Of note, the YFP-165-AUX1 fusion did not produce fluorescence, possibly because the YFP portion may face the acidic apolastic space (Swarup et al., 2004). The fused GFP portion of GFP-165-aux1rcr1 was expected to be endocytoplasmic because the fusion produced fluorescence. GFP-116-aux1rcr1 and GFP-165-aux1rcr1 are probably not associated with the PM; rather, both could aggregate in part near by the PM and localize to the ER.
Discussion
The plant hormone ethylene inhibits many aspects of Arabidopsis seedling growth and development that depend in part on or are coordinated with auxin actions. AUX1 binds and transports auxin; the association of AUX1 structure and domain functions needs to be fully addressed. The isolation of aux1rcr1, which suppressed ctr1-10 root growth inhibition, is consistent with the central role of AUX1 in root tip auxin transport, which involves the synergy of auxin and ethylene regulating root growth and development. aux1-22 is not serologically detectable and the mutant is probably a null mutant (Swarup et al., 2004). The mutants aux1rcr1, aux1-T, and aux1-22, but not the hypomorphic mutant aux1-7, have a similar effect on auxin transport in the root apex (Fig. 3) (Rahman et al., 2001; Swarup et al., 2004; Negi et al., 2008) and a similar effect on many aspects of the auxin response, which suggests that aux1rcr1 is a strong allele.
AUX1 is a PM protein, with loops 1 and 2 being intracytoplasmic and loop 3 extracytoplasmic (Fig. 7A) (Swarup et al., 2004). Both GFP-116-aux1rcr1 and GFP-165-aux1rcr1 appeared in the cytoplasm and possibly in part at the PM, which suggests that aux1rcr1 alters AUX1 localization. Given that the AUX1 loop 3 locates to the acidic apoplastic space, GFP-165-aux1rcr1 was probably not able to produce fluorescence if located at the PM. Therefore, GFP-116-aux1rcr1 and GFP-165-aux1rcr1 were probably not localized in part at the PM. The L126F substitution may not alter AUX1 topology for the loop 3 to face the cytoplasm. The exact subcellular localizations of these GFP–aux1rcr1 fusions remain for further investigation. The ER protein AXR4 is essential for AUX1 targeting to the PM (Dharmasiri et al., 2006); the association of aux1rcr1 with the ER could be possible. Alternatively, aux1rcr1 may not have been recycled effectively to the PM. Our results imply an involvement of Lys126 of the AUX1 loop 2 in correct AUX1 targeting.
The spatial expression of both YFP-2-AUX1 (Fig. 7) and YFP-116-AUX1 (Swarup et al., 2004) was consistent with AUX1 being predominant in columella, stele (protophloem), epidermis, and LRC cells in the root tip region (Swarup et al., 2001; Péret et al., 2012). Interestingly, a recent study showed that expression of the chimaeric protein consisting of the AUX1 N terminus and the LAX2 C terminus (DS2), but not the LAX2 N terminus and AUX1 C terminus (DS1), rescued aux1-22 gravity responses. Driven by the native AUX1 promoter, DS2 but not DS1 was expressed in AUX1-expressing LRC and epidermis cells, and DS1 was not targeted to the PM. Thus, the AUX1 N terminus may be involved in cell type-specific AUX1 expression and PM targeting (Péret et al., 2012). Given that the AUX1 N terminus is required for correct AUX1 expression in certain cell types, cell-specific AUX1 expression could be affected by the aux1rcr1 mutation and could involve the AUX1 loop 2. Our argument for roles of Lys126 at the AUX loop 2 in coupling cell type-specific expression and PM targeting is consistent with DS1 not being expressed in epidermis and LRC cells, or targeted to the PM.
The aux1-7 isoform, and possibly aux1-T, fails to mediate auxin transport across the PM (Yang et al., 2006). We showed that acropetal auxin transport in aux1rcr1 and aux1-T was prevented to a similar degree to that in aux1-22 (Rahman et al., 2001); polar auxin transport in the aux1rcr1 root tip was prevented, probably because of altered aux1rcr1 targeting. Whether aux1rcr1 can bind and transport auxin across membranes remains to be investigated. The DR5:GUS maximum was probably independent of acropetal auxin transport because it was affected in aux1rcr1 but not in aux1-T and aux1-7, with aux1-T affecting polar auxin transport and aux1-7 not (Stepanova et al., 2007; Negi et al., 2008) (Fig. 3). Basipetal transport for auxin, which is de novo biosynthesized in the root apex, was probably affected in aux1-T, which facilitated auxin accumulation in the root apex, so that root gravitropism but not DR5:GUS expression was impaired. This argument, however, does not explain the dominant-negative effect of aux1rcr1 on maximal DR5:GUS expression. We do not favour a second mutation in the aux1rcr1 mutant preventing DR5:GUS expression, because 35S:aux1rcr1 expression in the wild type also prevented DR5:GUS expression. DR5:GUS expression was probably not silenced after genetic crossing or transformation in the genotypes we studied, as the aux1rcr1-containing genotypes showed a similar DR5:GUS induction pattern to that of aux1-T following NAA treatment.
The effect of aux1rcr1 on DR5:GUS expression was associated with sites expressing AUX1. The PM-localized PINs are auxin-efflux carrier proteins that transport intracellular auxin to the apoplast. Interestingly, ER-localized PIN5 and PIN-LIKEs (PILs) facilitate intracellular auxin transport to the ER lumen, where auxin metabolism occurs to reduce auxin availability for nuclear auxin signalling (Mravec et al., 2009; Ganguly et al., 2010; Barbez et al., 2012; Feraru et al., 2012; Swarup and Péret, 2012). AUX1 and PIN5/PILs have 10 or 11 transmembrane helixes and transport auxin across membranes, which suggests similarity in protein structure and function. These features prompted us to hypothesize that aux1rcr1 could localize at the ER and gain a new function to transport the intracellular auxin to the ER lumen in AUX1-expressing domains. Alternatively, aux1rcr1 could transport intracellular auxin to other subcellular compartments. Either scenario would suggest a mechanism by which the nuclear auxin is reduced to a level that is insufficient for DR5:GUS expression.
The polar auxin transport that facilitates auxin redistribution plays important roles in root growth and gravitropism (Marchant et al., 1999; Swarup et al., 2005; Swarup and Péret, 2012). With disturbed polar auxin transport, aux1rcr1 and other aux1 alleles show the same root growth defect phenotypes. For aux1rcr1 with the 35S:AUX1 transgene and for the wild type with the 35S:aux1rcr1 transgene, the wild-type AUX1 restored polar auxin transport in the presence of aux1rcr1 and thus these genotypes showed a normal root growth phenotype. In contrast, the dominant-negative effects of auxrcr1 prevented DR5:GUS expression, even with the wild-type AUX1, in AUX1-expressing domains. The hypothesis that aux1rcr1 could promote auxin transport to the ER lumen to affect auxin homeostasis needs to be demonstrated, and this scenario would suggest a higher auxin concentration required for the maximal DR5:GUS expression than for gravitropic root growth. Our findings could lead to further studies of AUX1 domain functions and structure.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Data S1. Primer sequences and cloning of transgenes.
Supplementary Fig. S2. qRT-PCR of AUX1 in aux1rcr1 35S:AUX1 DR5:GUS lines.
Supplementary Fig. S3. DR5:GUS expression in root apexes for genotypes with aux1rcr1.
Acknowledgements
We thank Dr S. Z. Men for the seed stock carrying AUX1p:YFP-AUX1, our colleague Dr H. X. Lin for sharing [3H]-labelled IAA, and Mr Y. J. Chu for instructions for [3H]IAA measurement. This work was supported by the National Natural Sciences Foundation of China (grants 31123006, 31070249, and 30770199), the Chinese Ministry of Science and Technology (grants 2011CB100700 and 2012AA10A302-2), and the Shanghai Institutes for Biological Sciences (grant SIBS2008004).
Glossary
Abbreviations:
- 2,4-D
2,4-dichlorophenoxyacetic acid
- ARF
auxin response factor
- AUX
auxin
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- IAA
indole acetic acid
- LRC
lateral root cap
- MS
Murashige and Skoog
- NAA
1-naphthaleneacetic acid
- PM
plasma membrane
- qRT-PCR
quantitative RT-PCR
- SD
standard deviation
- SE
standard error
- UTR
untranslated region
- YFP
yellow fluorescent protein.
References
- Alonso JM, Stepanova AN, Solano R, Wisman E, Ferrari S, Ausubel FM, Ecker JR. 2003. Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 100, 2992–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbez E, Kubes M, Rolcik J, et al. 2012. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature. 485, 119–122 [DOI] [PubMed] [Google Scholar]
- Blakeslee JJ, Peer WA, Murphy AS. 2005. Auxin transport. Current Opinion in Plant Biology. 8, 494–500 [DOI] [PubMed] [Google Scholar]
- Carrier DJ, Bakar NTA, Swarup R, Callaghan R, Napier RM, Bennett MJ, Kerr ID. 2008. The binding of auxin to the Arabidopsis auxin influx transporter AUX1. Plant Physiology. 148, 529–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Gao Z, Kerris RJ, III, Wang W, Binder BM, Schaller GE. 2010. Ethylene receptors function as components of high-molecular-mass protein complexes in Arabidopsis . PLoS ONE. 5, e8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C. 1998. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proceedings of the National Academy of Sciences, USA. 95, 5401–5406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. 2005. The F-box protein TIR1 is an auxin receptor. Nature. 435, 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri S, Swarup R, Mockaitis K, et al. 2006. AXR4 is required for localization of the auxin influx facilitator AUX1. Science. 312, 1218–1220 [DOI] [PubMed] [Google Scholar]
- Dos Santos Maraschin F, Memelink J, Offringa R. 2009. Auxin-induced, SCFTIR1-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. The Plant Journal. 59, 100–109 [DOI] [PubMed] [Google Scholar]
- Feraru E, Vosolsobe S, Feraru MI, Petrášek J, Kleine-Vehn J. 2012. Evolution and structural diversification of PILS putative auxin carriers in plants. Frontiers in Plant Science. 3, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A, Lee SH, Cho M, Lee OR, Yoo H, Cho HT. 2010. Differential auxin-transporting activities of PIN-FORMED proteins in Arabidopsis root hair cells. Plant Physiology. 153, 1046–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Wen CK, Binder BM, Chen YF, Chang J, Chiang YH, Kerris RJ, III, Chang C, Schaller GE. 2008. Heteromeric interactions among ethylene receptors mediate signaling in Arabidopsis . Journal of Biological Chemistry. 283, 23801–23810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman P, Ecker J. 1990. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell. 2, 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BP, Shakeel SN, Amir M, Haq NU, Qu X, Schaller GE. 2012. Histidine kinase activity of the ethylene receptor ETR1 facilitates the ethylene response in Arabidopsis. Plant Physiology. 159, 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie LJ. 2006. Auxin and cell polarity: the emergence of AXR4. Trends in Plant Science. 11, 517–518 [DOI] [PubMed] [Google Scholar]
- Huang Y, Li H, Hutchison CE, Laskey J, Kieber JJ. 2003. Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. The Plant Journal. 33, 221–233 [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Men S, Fischer U, Stepanova AN, Alonso JM, Ljung K, Grebe M. 2009. Local auxin biosynthesis modulates gradient-directed planar polarity in Arabidopsis . Nature Cell Biology. 11, 731–738 [DOI] [PubMed] [Google Scholar]
- Ivanchenko MG, Muday GK, Dubrovsky JG. 2008. Ethylene–auxin interactions regulate lateral root initiation and emergence in Arabidopsis thaliana . The Plant Journal. 55, 335–347 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusion: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal. 6, 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr ID, Bennett MJ. 2007. New insight into the biochemical mechanisms regulating auxin transport in plants. Biochemical Journal. 401, 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Dhonukshe P, Swarup R, Bennett M, Friml J. 2006. Subcellular trafficking of the Arabidopsis auxin influx carrier AUX1 uses a novel pathway distinct from PIN1. Plant Cell. 18, 3171–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman A, Black R, Ecker JR. 1996. HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell. 85, 183–194 [DOI] [PubMed] [Google Scholar]
- Li H, Johnson P, Stepanova A, Alonso JM, Ecker JR. 2004. Convergence of signaling pathways in the control of differential cell growth in Arabidopsis . Developmental Cell. 7, 193–204 [DOI] [PubMed] [Google Scholar]
- Liu Q, Wen CK. 2012. Cooperative ethylene receptor signaling. Plant Signaling & Behavior. 7, 1042–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR. 1998. EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana . Genes & Development. 12, 2175–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ. 1999. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO Journal. 18, 2066–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec J, Skupa P, Bailly A, et al. 2009. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature. 459, 1136–1140 [DOI] [PubMed] [Google Scholar]
- Muller A, Guan C, Galweiler L, Tanzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K. 1998. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO Journal. 17, 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi S, Ivanchenko MG, Muday GK. 2008. Ethylene regulates lateral root formation and auxin transport in Arabidopsis thaliana . The Plant Journal. 55, 175–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenschläger I, Wolff P, Wolverton C, Bhalerao RP, Sandberg G, Ishikawa H, Evans M, Palme K. 2003. Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proceedings of the National Academy of Sciences, USA. 100, 2987–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, Estelle M. 2009. Complex regulation of the TIR1/AFB family of auxin receptors. Proceedings of the National Academy of Sciences, USA. 106, 22540–22545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Swarup K, Ferguson A, et al. 2012. AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell. 24, 2874–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L, Xie F, Yu J, Wen CK. 2012. Arabidopsis RTE1 is essential to ethylene receptor ETR1 amino-terminal signaling independent of CTR1. Plant Physiology. 159, 1263–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Ahamed A, Amakawa T, Goto N, Tsurumi S. 2001. Chromosaponin I specifically interacts with AUX1 protein in regulating the gravitropic response of Arabidopsis roots. Plant Physiology. 125, 990–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Takahashi M, Shibasaki K, Wu S, Inaba T, Tsurumi S, Baskin TI. 2010. Gravitropism of Arabidopsis thaliana roots requires the polarization of PIN2 toward the root tip in meristematic cortical cells. Plant Cell. 22, 1762–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Brady SR, Reed RC, Ante SJ, Muday GK. 2000. Basipetal auxin transport is required for gravitropism in roots of Arabidopsis. Plant Physiology. 122, 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert HS, Friml J. 2009. Auxin and other signals on the move in plants. Nature Chemical Biology. 5, 325–332 [DOI] [PubMed] [Google Scholar]
- Roman G, Lubarsky B, Kieber JJ, Rothenberg M, Ecker JR. 1995. Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: five novel mutant loci integrated into a stress response pathway. Genetics. 139, 1393–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer C, Reyes FC, Buono R, Sliwinski MK, Haas TJ, Otegui MS. 2009. The ESCRT-related CHMP1A and B proteins mediate multivesicular body sorting of auxin carriers in Arabidopsis and are required for plant development. Plant Cell. 21, 749–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM. 2005. A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis . Plant Cell. 17, 2230–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Doležal K, Schlereth A, Jürgens G, Alonso JM. 2008. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 133, 177–191 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Likhacheva AV, Alonso JM. 2007. Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell. 19, 2169–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Friml J, Marchant A, Ljung K, Sandberg G, Palme K, Bennett M. 2001. Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes & Development. 15, 2648–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Kargul J, Marchant A, et al. 2004. Structure–function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell. 16, 3069–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Kramer EM, Perry P, Knox K, Leyser HMO, Haseloff J, Beemster GTS, Bhalerao R, Bennett MJ. 2005. Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nature Cell Biology. 7, 1057–1065 [DOI] [PubMed] [Google Scholar]
- Swarup R, Péret B. 2012. AUX/LAX family of auxin influx carriers—an overview. Frontiers in Plant Science. 3, 225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LIA, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. 2007. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 446, 640–645 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ. 1995. Composite structure of auxin response elements. Plant Cell. 7, 1611–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Petrášek J, Žádníková P, et al. 2010. The auxin influx carriers AUX1 and LAX3 are involved in auxin–ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development. 137, 597–606 [DOI] [PubMed] [Google Scholar]
- Xie F, Liu Q, Wen CK. 2006. Receptor signal output mediated by the ETR1 N terminus is primarily subfamily I receptor dependent. Plant Physiology. 142, 492–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Qiu L, Wen CK. 2012. Possible modulation of Arabidopsis ETR1 N-terminal signaling by CTR1. Plant Signaling & Behavior. 7, 1243–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E. 2006. High-affinity auxin transport by the AUX1 influx carrier protein. Current Biology. 16, 1123–1127 [DOI] [PubMed] [Google Scholar]
- Zažímalová E, Murphy AS, Yang H, Hoyerová K, Hošek P. 2010. Auxin transporters—why so many?. Cold Spring Harbor Perspectives in Biology. 2, a001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Liu Q, Xie F, Wen C-K. 2007. RTE1 is a Golgi-associated and ETR1-dependent negative regulator of ethylene responses. Plant Physiology. 145, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.