Abstract
The development of fleshy fruits involves complex physiological and biochemical changes. After fertilization, fruit growth usually begins with cell division, continues with both cell division and expansion, allowing fruit set to occur, and ends with cell expansion only. In spite of the economical importance of grapevine, the molecular mechanisms controlling berry growth are not fully understood. The present work identified and characterized Vitis vinifera cell elongation bHLH protein (VvCEB1), a basic helix–loop–helix (bHLH) transcription factor controlling cell expansion in grape. VvCEB1 was expressed specifically in berry-expanding tissues with a maximum around veraison. The study of VvCEB1 promoter activity in tomato confirmed its specific fruit expression during the expansion phase. Overexpression of VvCEB1 in grape embryos showed that this protein stimulates cell expansion and affects the expression of genes involved in cell expansion, including genes of auxin metabolism and signalling. Taken together, these data show that VvCEB1 is a fruit-specific bHLH transcription factor involved in grape berry development.
Key words: auxin, bHLH, cell expansion, fruit, grape, transcription factor.
Introduction
Determination of the final fruit size depends on both cell growth and proliferation. Cell division activity determines the final cell number, but cell expansion is critical, as it allows the increase in volume determining the final size of the fruit. Cell divisions occur immediately after fertilization and during the early phase of fruit development. This is followed by cell expansion involving cell-wall loosening and uptake of solutes and water (Coombe, 1992). Cell expansion requires primary cell-wall loosening and incorporation of newly synthesized cell-wall material. Cell-wall loosening results from the disruption of chemical bonds between the structural components of the cell wall through either acidification or the action of hydrolysing enzymes. All these modifications require a finely tuned and coordinated transcriptional regulation of genes involved in cell-wall biosynthesis and modification.
Fruit development is a complex process involving numerous physiological and biochemical changes that are initiated by hormonal signals generated after pollination (Coombe and McCarthy, 2000; Conde et al., 2007). Their abundance at specific stages of fruit development and ripening indicates their possible role during these developmental stages (Conde et al., 2007). In the berry of grapevine (Vitis vinifera L.), the first steps of development, from fertilization to nouaison (fruit set) are under the control of developmental hormones (auxins, cytokinins, and gibberellins) promoting cell division and expansion. Although part of these hormones can be imported into the berry, they are mostly produced by the seeds, or by the maternal tissues (unfertilized ovules) in case of seedless cultivars. The concentration of these hormones decreases from their maximum at flowering to a low level at veraison and throughout ripening (Coombe 1992; Blouin and Guimberteau, 2000; Böttcher et al., 2011).
Basic helix–loop–helix (bHLH) proteins are found throughout eukaryotic organisms (Pires and Dolan, 2010). Due to their propensity to form homodimers or heterodimers, bHLH proteins can participate in an extensive set of combinatorial interactions leading to the regulation of multiple transcriptional programmes. Some bHLH proteins only form homodimers or restrict their heterodimerization to closely related members (Toledo-Ortiz et al., 2003), while others can form heterodimers with one or several different partners (Littlewood and Evan, 1998). These properties allow bHLH proteins to participate in the regulation of a myriad of essential developmental and physiological processes. In plants, bHLH proteins function as transcriptional regulators modulating secondary metabolism pathways, fruit dehiscence, carpel and epidermal development, phytochrome signalling, and stress responses (Ramsay and Glover 2005; Castillon et al., 2007; Pires and Dolan, 2010; Feller et al., 2011). Recent studies have described the involvement of bHLH proteins in the determination of plant organ size. The SPATULA protein was shown to control cotyledon, leaf, and petal expansion by affecting cell proliferation in Arabidopsis thaliana (Ichihashi et al., 2010). The Capsicum annuum protein Upa20 (upregulated by AvrBs3) was described as a master regulator of cell enlargement stimulating cell growth (Kay et al., 2007), whereas BIGPETALp (BPEp) from A. thaliana limits petal growth by reducing cell size (Szécsi et al., 2006). In grapevine, few data are available about the role of bHLH proteins in reproductive development. Only two papers have described the identification of bHLH grapevine genes related to flavonoid synthesis (Hichri et al., 2010; Matus et al., 2010).
The present study identified and characterized Vitis vinifera cell elongation bHLH protein (VvCEB1), a novel bHLH-like protein from grapevine (cv. Cabernet Sauvignon). VvCEB1 transcripts accumulate predominantly in the berries, especially when auxin amounts are minimal. A time-course study of VvCEB1 expression showed that, among the many hormones tested, only auxin significantly affected VvCEB1 expression levels. Transformation experiments showed that VvCEB1 overexpression affected embryo development and increased cell size. Finally, a transcriptional analysis performed on 35S::VvCEB1 transgenic embryos confirmed that VvCEB1 overexpression stimulated cell expansion and suggested that its biological function is related to auxin responses.
Materials and methods
Isolation of VvCEB1 cDNA and construct production
VvCEB1 full-length clone was generated from a cDNA library of grape Cabernet Sauvignon berries (veraison stage) by PCR using synthetic oligonucleotide primers designed to begin and end at the start and stop codons of the open reading frame (forward primer, 5’-TAGAATTCCCCGGGATGGC AGCCTTTTCTCAGCAGTCTCACCAC-3’; reverse primer, 5’-AT GGATCCCCCGGGCTAGCGGCCGCAAAAAGAGTATCTG TTGCTGAAACCATA-3’).
This VvCEB1 complete open reading frame was amplified and cloned into the pGEM-T Easy vector (Promega) for DNA sequencing, prior to subcloning into a stable expression binary vector downstream of the 35S promoter of cauliflower mosaic virus (CaMV). pFB8 and Pk7m34GW binary vector (Gateway™; Karimi et al., 2002) were used to generate VvCEB1-overexpressing 41B cells and tomato plants, respectively.
Plant transformation and culture
Grapevine transformations were made with the 41B rootstock (V. vinifera ‘Chasselas’×Vitis berlandieri) according to Lecourieux et al. (2010). An embryogenic cell suspension culture was initiated as described previously (Coutos-Thévenot et al., 1992a ). This cell suspension was subcultured weekly in 25ml of glycerol/maltose culture medium (Coutos-Thévenot et al., 1992b ) supplemented with naphthoxyacetic acid at 1mg l–1 in the dark. Embryogenic cells were transformed using an Agrobacterium tumefaciens co-cultivation method (Mauro et al., 1995), and, after selection, the transgenic cells were subcultured in the same conditions in a medium supplemented with paromomycin at 2mg ml–1 and cefotaxime at 200mg ml–1 (Duchefa). Embryos were initiated from these 41B cells by removing auxin from the culture medium.
Transgenic tomato plants (Solanum lycopersicum L. cv. Wva106) were generated by A. tumefaciens-mediated transformation of tomato cotyledons as described by Gonzalez et al. (2007). Tomato plants were grown in a culture chamber with a 14h/10h day/night cycle. The temperature was 25 °C during the day and 20 °C during the night. Individual flowers were tagged on the day of anthesis (flower opening). Fruits were harvested at different developmental stages (expressed in days post-anthesis, d p.a.).
RNA and cDNA production
Roots, shoots, leaves, and inflorescences were collected from Cabernet Sauvignon fruit cuttings grown in a greenhouse. Berries from different varieties (Tite de crabe, Dodrelabi, Dabouki, Cardinal, Candicans 10089, Riparia 10202, Riparia 10525, Sylvestris 38, Rubra 10924, Cinerea 10137, and Cabernet Sauvignon) were harvested in Domaine du Grand Parc (INRA, Lastresne, France) or in Domaine de la Grande Ferrade (INRA, Villenave d’Ornon, France). In order to compare berries at the same level of maturity, Cabernet Sauvignon berries were sorted by weight before veraison, and on a NaCl density gradient after veraison.
All samples collected were quickly frozen in liquid nitrogen, ground to a fine powder, and stored at –80 °C until use. Total RNA from grape organs and berries was extracted according to the method of Lecourieux et al. (2010). Total RNA from grape embryos was extracted using a Spectrum™ Plant Total RNA kit (Sigma) following the manufacturer’s protocol. RNA isolation was followed by DNase I treatment. Reverse transcription was performed from 2 µg of purified RNA using Moloney murine leukemia virus reverse transcriptase (Promega) according to the manufacturer’s instructions. The cDNA obtained was diluted 1:20 in distilled water.
Gene expression analysis
Quantitative real-time RT-PCR (qRT-PCR) expression analysis was carried out using a CFX96 Real-Time PCR Detection system (Bio-Rad). Reaction mixes (10 µl) were prepared, which included 5 µl of iQ™ SYBR Green Supermix (Bio-Rad), 0.2 µM of each primer, and 2 µl of diluted cDNA. In this study, three V. vinifera reference genes were evaluated, elongation factor EF1 γ-chain (EF1γ; GenBank accession no. AF176496), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; XM_002263109) and actin (XM_002282480). GAPDH and actin were described by Reid et al. (2006) as relevant reference genes for normalization in grape berry development studies. We also included EF1γ as this gene displayed good expression stability according to the results of several unpublished microarrays produced in our laboratory and to data published previously by Deluc et al. (2007). The stability of all these genes was tested in our biological conditions (berry development, Cabernet Sauvignon berry cell suspensions, and 41B cells). VvEF1γ, whose stability was validated by the RefFinder program (http://www.leonxie.com/referencegene.php), was finally selected as the best reference gene. Thus, gene transcripts were quantified following normalization to VvEF1γ (GenBank accession no. AF176496) as an internal standard.
All biological samples were tested in triplicate, and means ±standard deviation (SD) values were calculated using standard statistical methods. Specific oligonucleotide primer pairs were designed with Beacon Designer 7 software (Premier Biosoft International). Specific annealing of the oligonucleotides was controlled by dissociation kinetics performed at the end of each PCR run. The efficiency of each primer pair was measured on a PCR product serial dilution. Primer sequences used in qRT-PCR experiments are listed in Supplementary Table S1 at JXB online.
In silico promoter region identification and analysis
Identification of the potential promoter regions and Myc-like bHLH binding factor (MYCL) binding sites was conducted using the Genomatix suite of programs (http://www.genomatix.de, Genomatix Software GmbH, Munich, Germany) (Quandt et al., 1995). The Gene2promotor program from the Genomatix software package was used to define 1000bp of the promoter regions (1000bp upstream of the transcription start site) for each gene. The 1000bp sequences obtained from the Gene2promotor program were then used as the target sequences for putative bHLH transcription factor recognition site identification using the MatInspector version 8.06 program (Cartharius et al., 2005). The parameters used were the Matrix Family Library version 8.4 (June 2011), the standard (0.75) core similarity, and the optimized matrix similarity.
Light microscopy and stereomicroscopy analyses
Grape embryos were placed in a quick-clearing solution of chloral hydrate:H2O:glycerol (8:2:1, w:v:v) on a microscope slide for 4–24h. The samples were then examined under differential interference contrast optics (Nomarski) using a light microscope (Axiophot, Zeiss). Photographs were taken using a Spot RTKE camera (Diagnostic Instruments).
β-Glucuronidase (GUS) histochemical staining
Tissues from stably transformed tomato plants were fixed in cold acetone, washed twice in sodium phosphate buffer (0.1M NaH2PO4, pH 7.2), soaked, vacuum infiltrated, and incubated overnight at 37 °C in GUS staining solution [1mM X-Gluc, 0.1M NaH2PO4 (pH 7), 1.5mM K3Fe(CN)6, 1.5mM K4Fe(CN)6, 0.05% Triton X-100]. GUS-stained tissues were cleared for 2 d in 96% ethanol and stored in 70% ethanol. For this experiment, five independent transgenic tomato lines were used, but only one representative staining pattern of each tissue or developing stage is shown.
Sequence analysis
Amino acid sequence alignments were performed using ClustalW (Thompson et al., 1994; http://www.ebi.ac.uk/Tools/msa/clustalw2/). The phylogenetic tree was constructed using MEGA version 4 (Tamura et al., 2007) with full-length protein sequences. The optimal tree with the sum of branch length (32.66568898) is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (2000 replicates) is shown next to the branches. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in the units of number of amino acid substitutions per site. All positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons (pairwise deletion option). There were a total of 1167 positions in the final dataset.
Results
Identification of a novel bHLH-like protein from grape
A transcriptomic analysis of developing grape berries was performed in order to identify new transcription factors that could affect berry development (D. Glissant and S. Delrot, unpublished data). Different genes encoding transcription factors whose expression was upregulated during berry development were identified. Among these, a bHLH-like gene was isolated and analysed further. The full-length cDNA was amplified by PCR using a grape berry cDNA library and was named VvCEB1 (GenBank accession no. JQ823168).
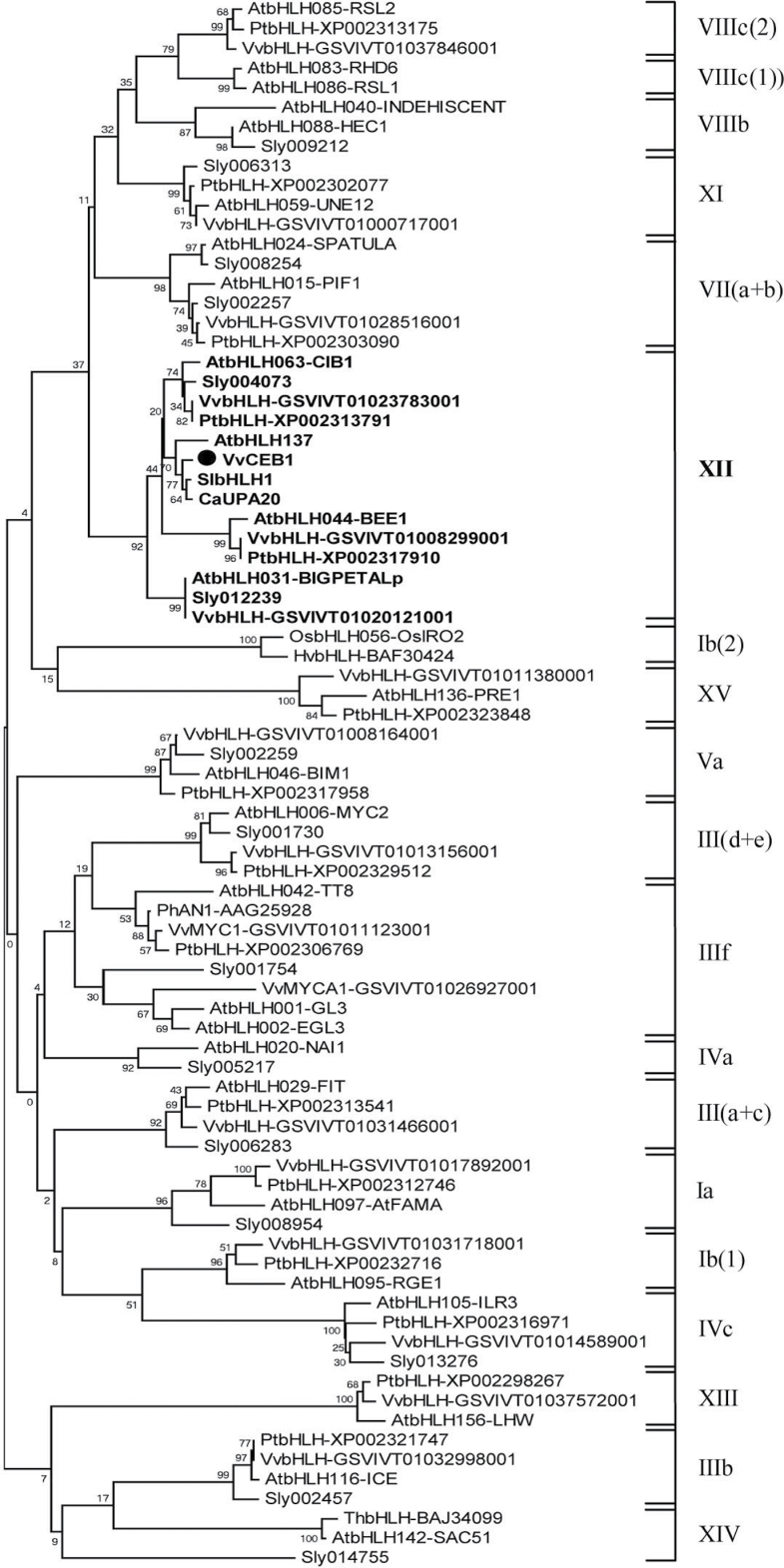
To identify the cluster to which VvCEB1 belongs, a rooted phylogenetic tree was constructed from the alignment of 67 full-length bHLH protein sequences of different plant species (Fig. 1). The grapevine sequences were identified by performing a BLASTP similarity search on the NCBI database using different Arabidopsis bHLH proteins as queries (Pires and Dolan, 2010). This phylogenetic analysis revealed that, among the 26 distinct plant bHLH protein subfamilies described by Pires and Dolan (2010), VvCEB1 belonged to subfamily XII and particularly to a cluster containing proteins involved in growth regulation such as UPA20 (Kay et al., 2007), BPEp (Szécsi et al., 2006) and bHLH137 (Zentalla et al., 2007).
Fig. 1.
Phylogenetic analysis of VvCEB1. VvCEB1 (black circle) and its homologues (bold) belong to a distinct bHLH gene subfamily. The different bHLH gene subfamily numbers and Arabidopsis thaliana (At), Thellungiella halophila (Th), Petunia hybrida (Ph), Oryza sativa (Os), and Hordeum vulgare (Hv) GenBank accession numbers reported in this figure are as described by Pires and Dolan (2010). The phylogenetic tree represents a non-exhaustive list of bHLH transcription factors; each bHLH subfamily is generally illustrated by a known A. thaliana bHLH and its closest homologues in V. vinifera (Vv), S. lycopersicum (Sl), and Populus trichocarpa (Pt). Sequence data were obtained from: http://www.genoscope.cns.fr/externe/GenomeBrowser/Vitis/ (V. vinifera), http://planttfdb.cbi.edu.cn/family.php?sp=Sly&fam=bHLH (S. lycopersicum), and http://www.ncbi.nlm.nih.gov/genbank/ (P. trichocarpa). The phylogenetic tree was constructed with MEGA version 4 (Tamura et al., 2007) using the neighbour-joining method with 2000 bootstrap replicates.
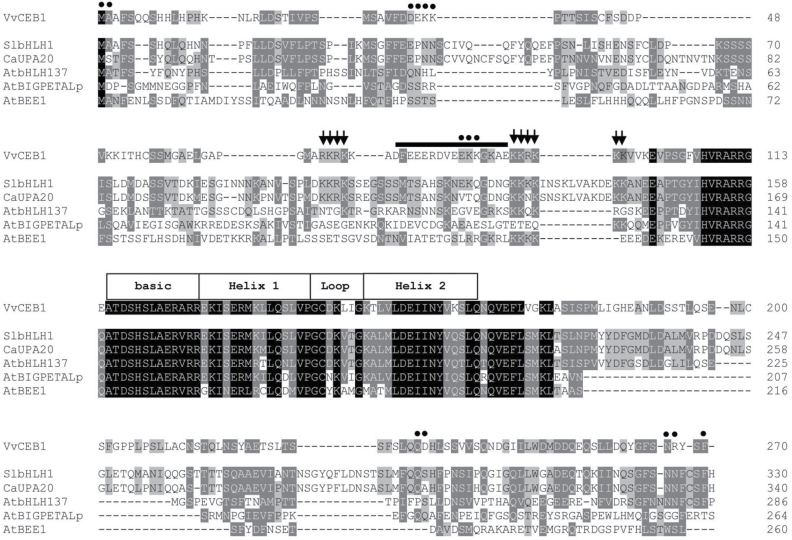
The full-length sequence alignment of VvCEB1 with its closest homologues from other plant species revealed that VvCEB1 exhibited 64 % amino acid sequence similarity with SlbHLH1 (Sly014317, S. lycopersicum), 59% with UPA20 (C. annum), and 46% with BPEp (A. thaliana).
The VvCEB1 transcript was 810bp and encoded a protein of 270 aa. VvCEB1 contained a nuclear localization site between aa 68 and 96 and a typical bHLH domain, which usually serves as a DNA binding and dimerization domain (Toledo-Ortiz et al., 2003), located in the region between aa 115 and 164 (Fig. 2).
Fig. 2.
Sequence analysis of VvCEB1. Full-length sequence comparison of VvCEB1 and its closest homologues SlbHLH1, CaUPA20, AtbHLH137, AtBIGPETAL and AtBEE1 using the ClustalW program with default parameters. Conserved residues are shaded in black; dark grey shading indicates conserved residues in at least four out of six of the sequences, and light grey shading indicates conserved residues in three out of six of the sequences. Basic residues that putatively function as a nuclear localization site for VvCEB1 are indicated by arrows above the alignment. Putative protein–protein binding domains and a bHLH domain for VvCEB1 are labelled above the alignment in black circles and open squares, respectively. Another conserved region with predicted secondary structure was identified for VvCEB1: an acidic α-helical domain, indicated by a black line.
Transient expression experiments using tobacco protoplasts expressing green fluorescent protein (GFP) fused in frame to the C terminus of VvCEB1 showed that, unlike the GFP control that was expressed throughout the cell, VvCEB1–GFP was only detected in the nucleus (Supplementary Fig. S1 at JXB online), in agreement with a putative role in the control of transcription.
Expression analysis of VvCEB1 in grapevine
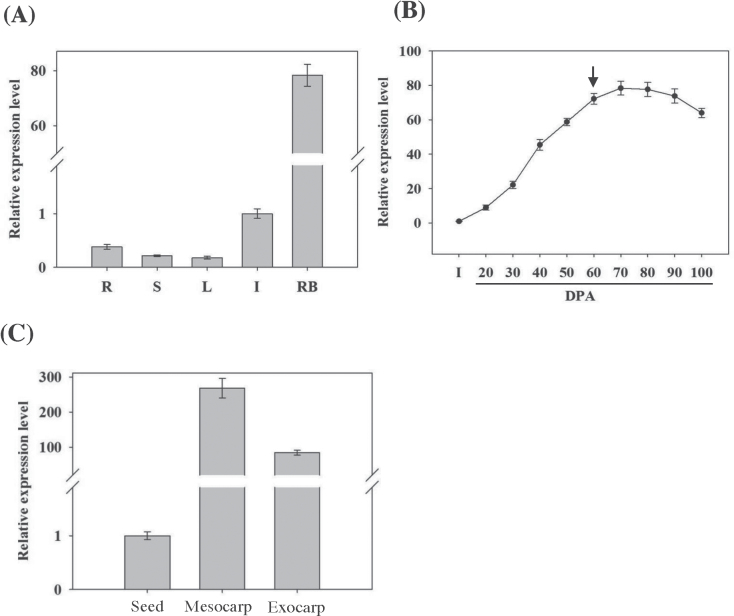
The expression profile of VvCEB1 was determined in different grapevine organs by qRT-PCR with RNA extracted from Cabernet Sauvignon roots, stems, leaves, flowers, and mature berries (80 d p.a.). VvCEB1 was expressed almost exclusively in berries (Fig. 3A). Indeed, in mature berries, VvCEB1 was highly expressed, whereas it was barely detectable in leaves, stems, roots, and inflorescences.
Fig. 3.
qRT-PCR analysis of VvCEB1 expression patterns in grapevine cv. Cabernet Sauvignon. (A) VvCEB1 expression in various grapevine organs: roots (R), stem (S), leaves (L), inflorescences (I), and ripening berries (RB) at 80 d p.a. Results are shown as means ±SD for three independent experiments. Gene expression was normalized against VvEF1γ expression. (B) VvCEB1 expression at different stages of berry development, from inflorescences (I) to mature berries at 100 d p.a. (DPA). The arrow indicates the veraison stage. Results are shown as means ±SD for four replicates from two independent experiments (carried out in the summer of 2006 and 2009). Gene expression was normalized against VvEF1γ. (C) VvCEB1 expression in different tissues from ripening berries at 80 d p.a. Results are shown as means ±SD for three independent experiments. Gene expression was normalized against VvEF1γ.
VvCEB1 transcript accumulation was also assessed during berry development (Fig. 3B). VvCEB1 expression showed a strong and gradual increase starting from nouaison (20 d p.a., berry set) and reached a maximum after veraison (60 d p.a., fruit ripening). Transcript levels were maintained maximal until 80 d p.a. and then slowly decreased until the mature stage (100 d p.a.).
VvCEB1 transcript amounts were also monitored in seeds and in different berry tissues (mesocarp and exocarp) after veraison. VvCEB1 transcripts were most abundant in the mesocarp, moderately abundant in the exocarp, and weakly detected in seeds (Fig. 3C).
Finally, a time-course study of VvCEB1 expression in response to 20 µM treatment with different hormones revealed that, at this concentration, only auxin significantly affected VvCEB1 expression and decreased the abundance of its transcripts (Supplementary Fig. S3 at JXB online).
Relationship between VvCEB1 expression and grape berry size
To investigate further the relationship between VvCEB1 expression and grape fruit size, VvCEB1 transcript abundance was determined in grapevine varieties exhibiting differences in berry size. Five varieties producing small berries (Vitis rubra cv. 10924, Vitis cinerea cv. 10137, Vitis riparia cv. 10525, V. riparia cv. 10202, and V. vinifera cv. Sylvestris 38) and five varieties producing large berries (Vitis candicans cv. 10089, V. vinifera cv. Tite de crabe, V. vinifera cv. Cardinal, V. vinifera cv. Dodrelabi, and V. vinifera cv. Dabouki) were analysed. This study was performed with berries collected 3 weeks after veraison had occurred, when VvCEB1 expression peaks. The results are summarized in Fig. 4. In small berries, VvCEB1 expression varied from a relative expression level of 1 to 1.2, whereas it was almost tripled in large berries (relative expression level from 2.8 to 3.2). These data suggested a relationship between VvCEB1 expression level and berry size.
Fig. 4.
Correlation between VvCEB1 expression and grape berry size. qRT-PCR analysis showing VvCEB1 transcript accumulation (black squares) in different grape varieties exhibiting different berry weight (grey bars). Berries were harvested 3 weeks after veraison and seeds were removed for this analysis. Gene expression was normalized against VvEF1γ. All data are means ±SD of four replicates from two independent experiments (summer 2009 and 2010).
VvCEB1 overexpression affects grapevine embryo development
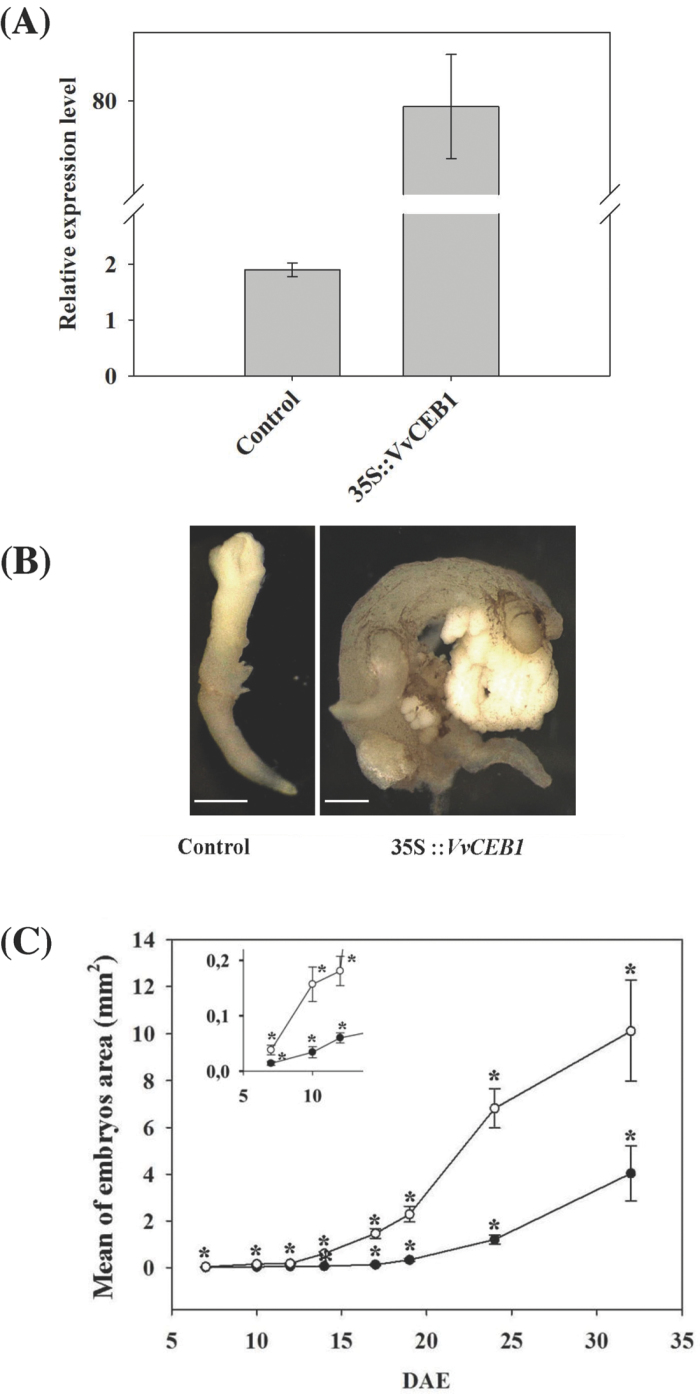
To investigate the function of VvCEB1 in grape, transgenic cells overexpressing VvCEB1 were produced using 35S promoter-driven VvCEB1 constructs. After stabilization of the cell suspension, expression of VvCEB1 was tested by qRT-PCR using VvCEB1-specific primers. In cells expressing 35S::VvCEB1, VvCEB1 transcripts accumulated 75-fold more than in cells expressing the empty vector (Fig. 5A).
Fig. 5.
Overexpression of VvCEB1 strongly affects grape embryo development. (A) Relative expression level of VvCEB1 transcript accumulation in transgenic grape 41B embryos. The VvCEB1 transcript level was quantified by qRT-PCR in control (pFB8 empty vector) and VvCEB1-overexpressing (35S::VvCEB1) lines. These embryos were collected 7 d after initiation of embryogenesis. Gene expression was normalized against VvEF1γ. Data are means ±SD of three independent experiments. (B) Control (pFB8 empty vector) and VvCEB1-overexpressing (35S::VvCEB1) embryos lines were observed with a stereomicroscope 32 d after the initiation of embryogenesis. Bars, 1mm. (C) Embryos sizes were measured at different stages of 41B embryo development, from 7 to 32 d after initiation of embryogenesis (DAE), both for control (pFB8 empty vector, black circles) and VvCEB1-overexpressing (35S::VvCEB1, open circles) lines. Thirty embryos were measured for each developmental stage in both lines. Results are shown as means ±SD. Kruskal-Wallis one way analysis of variance on ranks was performed and asterisks indicate statistically significant differences between lines at the same time point (P<0.001).
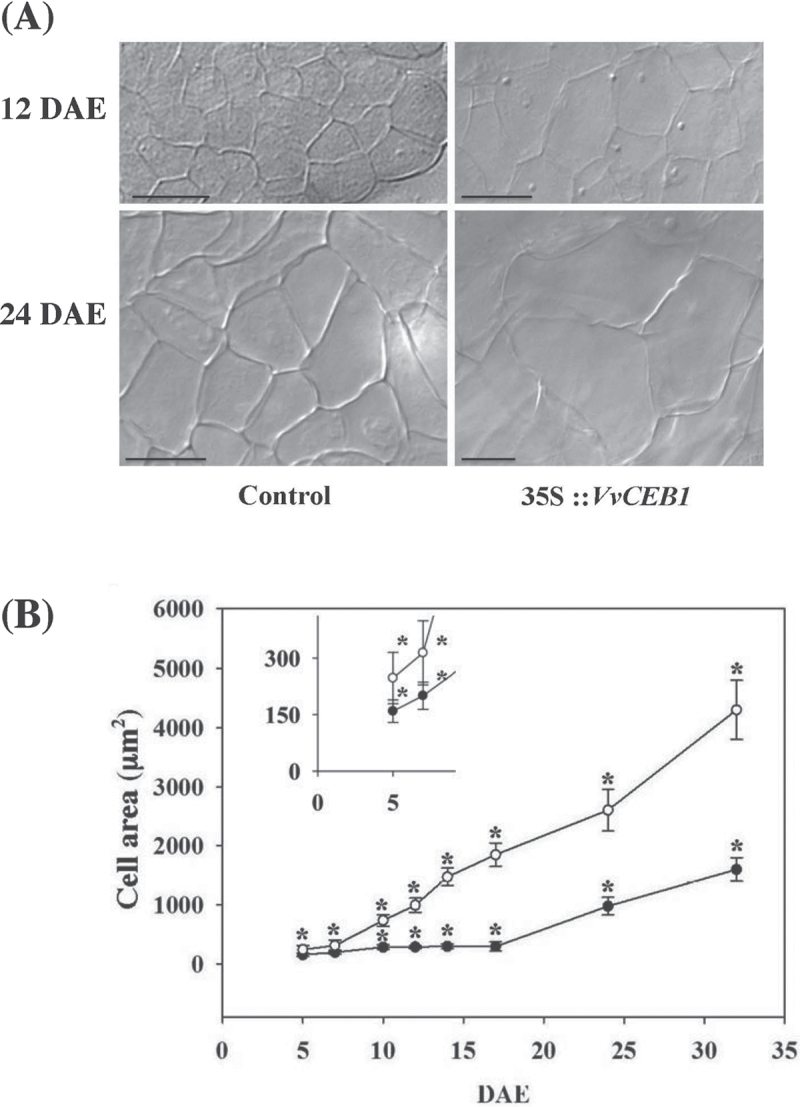
To study the effect of VvCEB1 on grape development, regeneration was initiated from VvCEB1 overexpressing cells. The development of control somatic embryos and of embryos expressing 35S::VvCEB1 was observed under a light microscope (Supplementary Fig. S2 at JXB online). 35S::VvCEB1 lines displayed strong phenotypical defects when compared with control embryos. Up to 7 d, embryos of similar appearance were formed in both control and 35S::VvCEB1 embryos. Between 7 and 14 d, control embryos switched from the heart to the torpedo stage (d 14) and to mature embryos, with fully formed cotyledons; the hypocotyls–root axis was visible at d 19. During the same period, the transgenic embryos rapidly elongated and still failed to develop cotyledons and to acquire bilateral symmetry (Supplementary Fig. S2). Moreover, secondary embryos started to develop from the primary transgenic ones and led to abnormal somatic embryos that could never develop into grape plantlets (Fig. 5B). Detailed measurements revealed that 35S::VvCEB1 embryos grew faster than the control and could reach, during the growth period, a size exceeding 11-fold the size of the control embryos (i.e. 17 d after initiation of embryogenesis) (Fig. 5C). In addition, bright-field microscopy observations after chloral hydrate treatment showed that the increased size of the transgenic embryos was associated with the presence of larger cells than in the control embryos (Fig. 6A). The difference in cell size increased during embryo development (Fig. 6B).
Fig. 6.
Overexpression of VvCEB1 increases cell size in grape 41B embryos. (A) Difference in cell size between control (PFB8 empty vector) and VvCEB1-overexpressing (35S::VvCEB1) embryos lines. Observations were assessed at 12 and 24 d after initiation of embryogenesis (DAE). Bars, 25 µm. (B) The cell area of control (pFB8 empty vector, black circles) and VvCEB1-overexpressing (35S::VvCEB1, open circles) lines was assessed at different stages of 41B embryo development, from 5 to 32 d after initiation of embryogenesis (DAE). Data are means ±SD of the ten largest cells measured in this area for each embryo and 20 embryos for each line were used to collect these data. Kruskal–Wallis one way analysis of variance on ranks was performed and asterisks indicate statistically significant differences between lines at the same time point (P<0.001).
These phenotypic changes suggested that VvCEB1 might be involved in cell and organ growth, and also that its ectopic overexpression might affect organogenesis.
Specific activity of the VvCEB1 promoter in tomato fruits
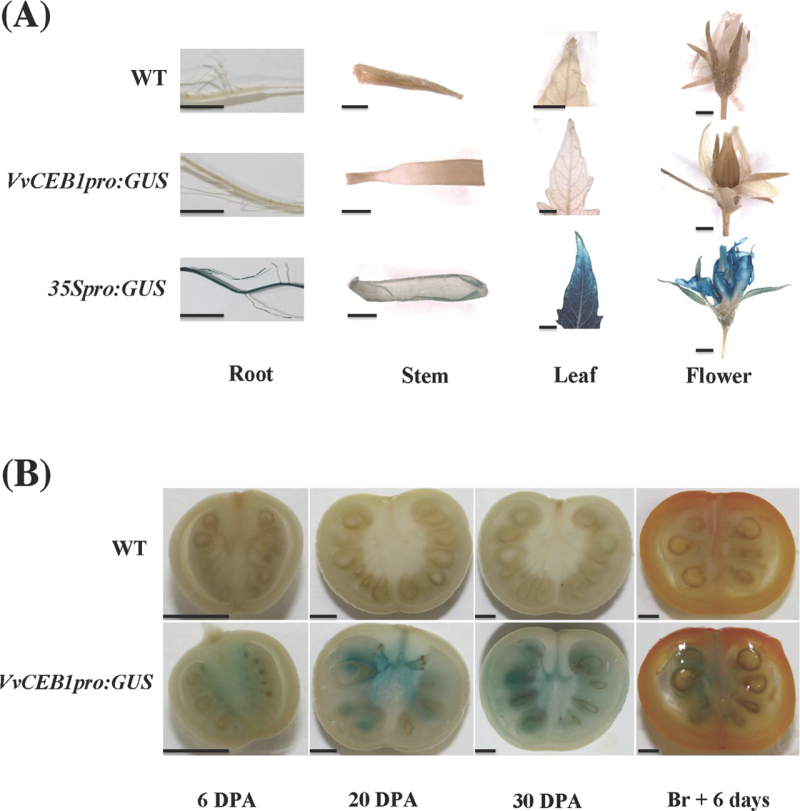
VvCEB1 transcripts accumulated predominantly in the berry, indicating a fruit-specific expression of this gene (Fig. 3A). To investigate further the expression of VvCEB1 in planta and to confirm this observation, the activity the VvCEB1 promoter fused to the GUS reporter gene was examined in transgenic tomatoes. Strong GUS activity was observed in all 35Spro:GUS organs, while no staining was detected in wild-type controls (Fig. 7A). The VvCEB1 promoter only resulted in fruit-specific expression, with no GUS staining in vegetative organs (Fig. 7), confirming that, even when expressed in tomato, VvCEB1 keeps its fruit specificity. Analysis of VvCEB1pro:GUS during fruit development showed GUS staining from early stages until the mature green stage (30 d p.a.), after which the activity of the VvCEB1 promoter decreased (Fig. 7B). These data were consistent with a specific expression of VvCEB1 in the fruit during the expansion phase spanning 10–40 d p.a. (Lemaire-Chamley et al., 2005).
Fig. 7.
Expression of VvCEB1 promoter in tomato. (A) GUS activity in various organs of tomato plants stably transformed with VvCEB1 promoter: GUS (VvCEB1pro:GUS) transgene. CaMV 35S promoter: GUS (35Spro:GUS) and Wild-Type (WT) plant lines were used as controls. Bars = 2.5mm. (B) GUS activity in tomato fruits stably transformed with VvCEB1promoter: GUS (VvCEB1pro:GUS) transgene at different development stages: 6 d post anthesis (DPA), 20 DPA, 30 DPA, and Breaker + 6 d (Br + 6 d). Wild-Type (WT) line was used as control. Bars = 2.5mm.
Modulation of cell expansion- and auxin-related gene expression in grape embryos overexpressing VvCEB1
To understand how VvCEB1 can affect cell expansion and/or auxin responses, the expression of several genes involved in these processes was tested in transgenic embryos overexpressing VvCEB1. These included some members of early auxin-responsive genes [auxin/indole-3-acetic acid (AUX/IAA), small auxin upregulated (SAUR) and Gretchen Hagen3 (GH3)], cell-wall metabolism (XET, PECL, AMY, AGP, and EXP) and aquaporin (AQP).
This expression analysis, assessed by qRT-PCR, showed that all the auxin signalling components tested were affected (Table 1). Indeed, VvIAA9 and VvIAA17 genes were downregulated in the transgenic embryos overexpressing VvCEB1, whereas VvIAA14, VvIAA16, VvIAA19, VvSAUR1, and VvSAUR5 were strongly upregulated compared with the control lines. The genes VvGH3-2 and VvGH3-6, encoding auxin-conjugating enzymes, were also dramatically upregulated in the transgenic line.
Table 1.
Ratios of transcript levels of selected auxin-related genes in transgenic grape embryos compared with the control (C) and determination of the presence of bHLH-binding sites in the corresponding promoter regions.
| Description | Name | Accession no. | Ratio (35S/C)a | bHLH binding sitesb |
|---|---|---|---|---|
| AUX/IAA transcription factor | VvIAA9 | HQ337788.1 | 0.4 | + |
| VvIAA14 | XM_002284097.1 | 508 | +++ | |
| VvIAA16 | HQ337789.1 | 11 | +++++ | |
| VvIAA17 | XM_002280488.1 | 0.2 | + | |
| VvIAA19 | HQ337790.1 | 12.2 | + | |
| Auxin response transcription factor | VvARF6 | XM_002282794.1 | 1.3 | + |
| VvARF9 | XM_002265126.1 | 0.8 | + | |
| VvARF17 | XM_002284292.1 | 0.6 | ++ | |
| Small auxin upregulated protein | VvSAUR1 | XM_002271526.1 | 6 | +++ |
| VvSAUR5 | XM_002279234.1 | 17 | + | |
| IAA-amido synthetase | VvGH3-2 | XM_002283850.1 | 168 | +++++++++ |
| VvGH3-6 | XM_002268242.1 | 79 | – |
a Gene transcript levels were quantified by qRT-PCR in control (C, pFB8 empty vector) and VvCEB1-overexpressing (35S) grape 41B embryo lines. The embryos were collected 7 d after initiation of embryogenesis. Gene expression was normalized against VvEF1γ Data are means of three independent experiments.
b The promoter regions (1000bp upstream of the transcription start site) for each gene were used as the target sequences for putative bHLH transcription factor recognition site identification using the MatInspector program (Genomatix software package). Each ‘+’ indicates the presence of one MYCL family binding site whereas ‘–’ indicates the absence of a MYCL binding site.
qRT-PCR experiments confirmed that most of the cell expansion genes tested were strongly upregulated in 35S::VvCEB1 transgenic embryos compared with the control lines (Table 2). These include the cell-wall modification genes encoding expansins (VvEXP1, VvEXP8, VvEXP11, VvEXP12 and VvEXP17), α-amylase (VvAMY1), xyloglucan endotransglucosylase (VvXET2), pectate lyase (VvPECL1 and VvPECL8), arabinogalactan protein (VvAGP20) and three out of four aquaporin genes tested (VvAQP1, VvAQP3, and VvAQP4).
Table 2.
Ratios of transcript levels of selected cell expansion-related genes in transgenic grape embryos compared with the control (C) and determination of the presence of bHLH-binding sites in the corresponding promoter regions.
| Description | Name | Accession no. | Ratio (35S/C)a | bHLH-binding sitesb |
|---|---|---|---|---|
| Expansin | VvEXPA1 | XM_002269481.1 | 7.7 | – |
| VvEXPA8 | XM_002280264.1 | 38 | + | |
| VvEXPA11 | XM_002285855.1 | 1154 | + | |
| VvEXPA12 | XM_002284822.1 | 52 | + | |
| VvEXPA17 | XM_002273247.1 | 252 | ++ | |
| α-Amylase | VvAMY1 | XM_002285177.1 | 4.3 | – |
| Xyloglucan endotransglycosylase | VvXET2 | XM_002274484.1 | 4 | ++ |
| Pectate lyase | VvPECL1 | XM_002285603.1 | 4.8 | ++++++++ |
| VvPECL8 | XM_002275745.1 | 82 | ++++++ | |
| Arabinogalactan protein | VvAGP20 | XM_002280458.1 | 5 | +++ |
| Aquaporin (TIP) | VvAQP1 | XM_002274502.1 | 8.3 | +++++ |
| VvAQP2 | XM_002262942.1 | 0.2 | + | |
| VvAQP3 | XM_002274691.1 | 4.1 | ++ | |
| VvAQP4 | XM_002274519.1 | 2.4 | + |
a Gene transcript levels were quantified by qRT-PCR in control (C, pFB8 empty vector) and VvCEB1-overexpressing (35S) grape 41B embryo lines. The embryos were collected 7 d after initiation of embryogenesis. Gene expression was normalized against VvEF1γ. Data are means of three independent experiments.
b The promoter regions (1000bp upstream of the transcription start site) for each gene were used as the target sequences for putative bHLH transcription factor recognition site identification using the MatInspector program (Genomatix software package). Each ‘+’ indicates the presence of one MYCL family binding site whereas ‘–’ indicates the absence of MYCL-binding site.
Some of the genes that were significantly affected in the grape transgenic lines were also analysed throughout berry development. In this context, expression of the auxin-responsive transcription factor genes VvIAA9 and VvIAA17 that were negatively regulated by VvCEB1 overexpression was significant at the green stage and dropped during berry development. By contrast, the VvIAA19, VvIAA16, VvEXP11, VvAQP4, VvXET2, VvAMY1, VvPECL8, and VvSAUR genes that strongly accumulated in 35S::VvCEB1 lines were shown to be upregulated during the time course of VvCEB1 expression in the developing berry (Supplementary Fig. S4 at JXB online).
A region of 1000bp of upstream sequence for all of auxin and expansion-related genes was analysed for putative bHLH regulatory elements (MYCL) using the MatInspector version 8.06 program (Cartharius et al., 2005). The results are presented in Tables 1 and 2. The promoter regions of three genes analysed (VvGH3-6, VvEXPA1, and VvAMY1) did not show any MYCL binding sites, whereas the 23 others exhibited one or more (up to nine) MYCL binding sites, suggesting that they might be direct targets for VvCEB1.
Taken together, these data suggested a coordinated regulation of several genes encoding proteins affecting cell expansion and VvCEB1 expression, and reinforce the conclusion that the berry-specific transcription factor VvCEB1 may affect cell expansion processes during grape berry development.
Discussion
This work identified and characterized VvCEB1, a new bHLH transcription factor from grape that is preferentially expressed in the berry and particularly in the expanding tissues. Its expression starts at fruit set, is maximal at veraison, and is maintained until harvest. VvCEB1 overexpression strongly affected cell size in grape embryos. Together, these results suggest a key role for VvCEB1 in cell expansion during fruit development.
VvCEB1 is a nuclear bHLH transcription factor clustering with genes controlling cell growth and organ size
Phylogenetic analysis revealed that VvCEB1 belongs to subfamily XII of the bHLH transcription factors (Pires and Dolan, 2010), which contains proteins previously described as being involved in growth regulation. This cluster includes, among others, UPA20 and BPEp, both known to regulate cell growth and organ size. BPEp affects cell expansion and petal growth in an auxin-dependent manner (Szécsi et al., 2006; Varaud et al., 2011), whereas UPA20 is involved in cell elongation in C. annum (Kay et al., 2007). The A. thaliana bHLH137 was reported as a DELLA-responsive gene that may repress gibberellic acid signalling (Zentalla et al., 2007). The other known proteins of this subfamily are involved in brassinosteroid (BR) signalling (BEE) (Friedrichsen et al., 2002) and in cryptochrome interaction (CIB) (Liu et al., 2008). Like auxins, BRs promote plant growth and participate in a wide array of plant developmental processes (Yang et al., 2011). Recently, Chung et al. (2011) made a direct link between auxin and BRs. They showed that auxin regulates BR biosynthesis in Arabidopsis, and suggested that some of the growth-promoting effects of auxin are mediated through BR biosynthesis. Taken together, these data suggest similarities in the mode of action of proteins belonging to clade XII on the regulation of cell elongation. This hypothesis is further supported by data from the literature indicating that members of the same bHLH subfamily are frequently involved in the same biological process and present partially or totally redundant functions (Pires and Dolan, 2010).
The predominant expression of VvCEB1 in the fruit suggests a key role in berry development
VvCEB1 was expressed almost exclusively in berries and more particularly in the expanding tissues (mesocarp and exocarp) (Fig. 3). Its expression increased gradually throughout berry development, without following the bimodal expression pattern usually attributed to the two phases of berry enlargement (stages I and III). This maintained accumulation of VvCEB1 transcripts during the lag phase (stage II) may suggest that its role in cell expansion is linked to its interaction with other proteins and/or that VvCEB1 is involved in additional aspects of berry development when combined with other proteins.
The heterologous analysis of the activity of the VvCEB1 promoter in tomato also showed that VvCEB1 is expressed specifically in the fruit. Interestingly, this fruit-specific expression was maintained during the fruit expansion period of phase III, again underlining a link between VvCEB1 expression and cell expansion. Finally, VvCEB1 expression studies in grape varieties exhibiting differences in berry size revealed a relationship between VvCEB1 transcript accumulation and fruit size.
Together, these data strengthen evidence for the role of VvCEB1 in regulating berry size during development.
Overexpression of VvCEB1 affects embryo development
The production of grape embryos overexpressing VvCEB1 also showed that its overexpression affects cell growth. Indeed, the transgenic lines ectopically overexpressing VvCEB1 exhibited drastic phenotypic differences. The 35S::VvCEB1 embryos rapidly elongated and failed to develop cotyledons and to acquire bilateral symmetry, thus leading to abnormal somatic embryos that could never develop into grape plantlets. Compared with the controls, these embryos grew faster and reached a more important size because of the presence of larger cells. These differences may result from a stimulation of cell-expansion processes and from an alteration of hormone fluxes, especially of auxin fluxes that are necessary for the establishment of bilateral symmetry, growth, and organogenesis (Liu et al., 1993; Jenik and Barton, 2005; Moller and Weijers, 2009; Vanneste and Friml, 2009; de Smet et al., 2010).
VvCEB1 strongly affects the expression of genes involved in early auxin response and cell expansion
In order to understand better how VvCEB1 might affect cell size and interact with auxin, expression analyses were performed on 35S::VvCEB1 embryos. The data showed that cell-expansion gene expression was affected, and indicated that auxin could be involved through some signalling components related to fruit growth and development. Additionally, a time-course study of VvCEB1 expression in response to different hormones revealed that, at the concentration used, only auxin significantly affected VvCEB1 expression and decreased the abundance of its transcripts (Supplementary Fig. S3).
In this context, the early auxin-responsive genes AUX/IAA, SAUR, and GH3 were tested. These genes encode very low-abundance nuclear proteins with short half-lives that control secondary downstream genes (Hagen and Guilfoyle, 2002; Knauss et al., 2003). In the VvCEB1 overexpressing line, the VvIAA genes tested were either induced or repressed, whereas the VvSAUR and VvGH3 genes tested were only upregulated. These disparities in the transcript levels of auxin-regulated genes may reflect their complex regulation, which involves various combinations of transcriptional regulators, some of which are modulated by cell specificity, developmental stage, or abiotic signals (Paponov et al., 2008). Among the AUX/IAA genes tested in this study, VvIAA19 caught our attention because, in addition to its high expression in the 35S:VvCEB1 line, this gene was strongly expressed during berry development. Furthermore, a recent study (Kohno et al., 2012) showed that, although no morphological change was observed, transgenic Arabidopsis plants overexpressing VvIAA19 grew faster and flowered earlier than control plants, which indicates that the constitutive expression of VvIAA19 promotes growth. SAUR genes that are upregulated in the 35S::VvCEB1 line are abundant in the elongation zone of soybean hypocotyls and are expressed most strongly in epidermal and cortical cells (Gee et al., 1991). Very recently, Spartz et al. (2012) showed that the SAUR19 subfamily function as positive effectors of cell expansion. Together, these data suggest that VvCEB1 might contribute to the stimulation of cell expansion by regulating the expression of these genes.
To coordinate auxin-mediated processes, plants need to maintain the endogenous pool of auxins at an appropriate level. This can be achieved by regulating auxin biosynthesis and distribution among different organs and by conjugation (Berleth et al., 2004; Woodward and Bartel, 2005). Although the physiological importance of conjugates in auxin homeostasis is not yet fully understood, it is generally accepted that conjugate formation plays a critical role in auxin action. In grape berries, Böttcher et al. (2011) recently identified a GH3-1 gene that displays a developmental expression pattern correlated with the third phase of berry development and suggested its involvement in ripening processes. In longan, another non-climacteric fruit, the role of GH3 genes was also investigated (Kuang et al., 2011). The authors suggested that DlGH3.1 and DlGH3.2 are involved in fruit growth and particularly in pericarp growth and fruit ripening. Our data showed a high transcript accumulation of two VvGH3 genes in the 35S::VvCEB1 lines, suggesting the presence of a high amount of conjugated auxin and possible control of GH3 expression by VvCEB1. Together, this transcriptional analysis suggested that VvCEB1 could participate in berry growth, and reinforces the hypothesis that auxin is involved in the mode of action of VvCEB1.
Cell expansion involves changes in cell-wall composition and the accumulation of different compounds maintaining both osmotic pressure and water flow in the expanding cells (Carrari and Fernie, 2006). This process implies, among other aspects, cell-wall modification proteins (e.g. expansin, arabinogalactan proteins, xyloglucan endotransglycosylases), starch degradation enzymes (α-amylase and pectate lyase), and water channels (AQPs). In agreement with these data, the present study showed that VvCEB1 overexpression strongly upregulated expansion-related genes (Table 2) and therefore stimulated cell-expansion processes. These results also agree with the observations showing that overexpression of VvCEB1 in grape embryos enhanced cell size (Fig. 5). In addition, the effect of VvCEB1 on auxin-associated genes also fits with its putative role in cell expansion. Indeed, the classical effect of auxin is described as a very rapid stimulation of cell expansion by modifying the cell-wall network, followed by sustained growth over a longer time period, although this hormone is also important in other responses, such as cell division and differentiation (Schenck et al., 2010).
In conclusion, this work showed that VvCEB1 is a bHLH transcription factor expressed specifically in the expanding tissues of the fruit and is probably involved in the regulation of cell size. The present work also suggests that genes involved in cell expansion and auxin responses are potential direct and/or indirect targets of VvCEB1. These data linking a fruit-specific bHLH transcription factor to cell expansion and auxin responses open new perspectives for the understanding of fleshy fruit development. However, additional work is needed to strengthen the hypothesis of VvCEB1 function in planta.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Nuclear localization of GFP-VvCEB1 fusion protein in tobacco protoplasts.
Fig. S2. Kinetics of development of control (pFB8 empty vector) and VvCEB1-overexpressing (35S::VvCEB1) 41B grape embryos.
Fig. S3. Study of the hormonal regulation of VvCEB1 expression in grape cell suspension.
Fig. S4. Time-course study of the expression of VvCEB1 putative target genes during grape berry development.
Fig. S5. List of GenBank accession numbers used for qRT-PCR analysis.
Table S1. PCR primers used to amplify gene-specific regions for expression analyses.
Acknowledgements
We thank G. Cramer and C. Chevalier for critical reading of the manuscript, M. Hernould for scientific discussions, L. Bordenave for help in the choice of the different grape varieties, and C. Cheniclet for microscopy assistance. P. N. was supported by a grant from Conseil Régional d’Aquitaine.
Glossary
Abbreviations:
- bHLH
basic helix–loop–helix
- BR
brassinosteroid
- CaMV
cauliflower mosaic virus
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- MYCL
Myc-like bHLH binding factor
- p.a.
post-anthesis
- qRT-PCR
quantitative real-time RT-PCR
- SD
standard deviation.
References
- Berleth T, Krogan NT, Scarpella E. 2004. Auxin signals—turning genes on and turning cells around. Current Opinion in Plant Biology 7, 553–563 [DOI] [PubMed] [Google Scholar]
- Blouin J, Guimberteau G. 2000. Maturation et maturité des raisins. France:Féret; [Google Scholar]
- Böttcher C, Boss PK, Davies C. 2011. Acyl substrate preferences of an IAA-amido synthetase account for variations in grape (Vitis vinifera L.) berry ripening caused by different auxinic compounds indicating the importance of auxin conjugation in plant development. Journal of Experimental Botany 62, 4267–4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Fernie AR. 2006. Metabolic regulation underlying tomato fruit development. Journal of Experimental Botany 57, 1883–1897 [DOI] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. 2005. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21, 2933–2942 [DOI] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E. 2007. Phytochrome interacting factors, central players in phytochrome-mediated light signaling networks. Trends Plant Science 12, 514–521 [DOI] [PubMed] [Google Scholar]
- Chervin C, Tira-umphon A, Terrier N, Zouine M, Severac D, Roustan JP. 2008. Stimulation of the grape berry expansion by ethylene and effects on related gene transcripts, over the ripening phase. Physiology Plantarum 134, 534–546 [DOI] [PubMed] [Google Scholar]
- Chung Y, Maharjan PM, Lee O, et al. 2011. Auxin stimulates DWARF4 expression and brassinosteroid biosynthesis in Arabidopsis. The Plant Journal 66, 564–578 [DOI] [PubMed] [Google Scholar]
- Conde C, Silva P, Fontes N, Dias ACP, Tavares RM, Sousa MJ, Agasse A, Delrot S, Gerós H. 2007. Biochemical changes throughout grape berry development and fruit and wine quality. Food 1, 1–22 [Google Scholar]
- Coombe BG, McCarthy MG. 2000. Dynamics of grape berry growth and physiology of ripening. Australian Journal of Grape Wine Research 6, 131–135 [Google Scholar]
- Coombe BG. 1992. Research on development and ripening of the grape berry. American Journal of Enology and Viticulture 43, 101–110 [Google Scholar]
- Coutos-Thévenot P, Goebel-Tourand I, Mauro MC, Jouanneau JP, Boulay M, Deloire A, Guern J. 1992a. Somatic embryogenesis from grapevine cells. I. Improvement of embryo development by changes in culture conditions. Plant Cell Tissue Organ Culture 29, 125–133 [Google Scholar]
- Coutos-Thévenot P, Maës O, Jouenne T, Mauro MC, Boulay M, Deloire A, Guern J. 1992b. Extracellular protein patterns of grapevine cell suspensions in embryogenic and non-embryogenic situations. Plant Science 86, 137–145 [Google Scholar]
- de Smet I, Lau S, Voß U, et al. 2010. Bimodular auxin response controls organogenesis in Arabidopsis. Proceedings of the National Academy of Sciences, USA 107, 2705–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc LG, Grimplet J, Wheatley MD, Tillett RL, Quilici DR, Osborne C, Schooley DA, Schlauch KA, Cushman JC, Cramer GR. 2007. Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development. BMC Genomics 8, 429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E. 2011. Evolutionary and comparative genomics of MYB and bHLH plant transcription factors. The Plant Journal 66, 94–116 [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Nemhauser J, Muramitsu T, Maloof JN, Alonso J, Ecker JR, Furuya M, Chory J. 2002. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162, 144–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee MA, Hagen G, Guilfoyle TJ. 1991. Tissue-specific and organ-specific expression of soybean auxin-responsive transcripts GH3 and SAURs. Plant Cell 3, 419–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, Gévaudant F, Hernould M, Chevalier C, Mouras A. 2007. The cell cycle-associated protein kinase WEE1 regulates cell size in relation to endoreduplication in developing tomato fruit. The Plant Journal 51, 642–655 [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T. 2002. Auxin-responsive gene expression, genes, promoters and regulatory factors. Plant Molecular Biology 49, 373–385 [PubMed] [Google Scholar]
- Hichri I, Heppel SC, Pillet J, Léon C, Czemmel S, Delrot S, Lauvergeat V, Bogs J. 2010. The basic helix–loop–helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Molecular Plant 3, 509–523 [DOI] [PubMed] [Google Scholar]
- Hosy E, Duby G, Véry AA, Costa A, Sentenac H, Thibaud JB. 2005. A procedure for localisation and electrophysiological characterisation of ion channels heterologously expressed in a plant context. Plant Methods 1, 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y, Horiguchi G, Gleissberg S, Tsukaya H. 2010. The bHLH transcription factor SPATULA controls final leaf size in Arabidopsis thaliana. Plant Cell Physiology 51, 252–261 [DOI] [PubMed] [Google Scholar]
- Jenik PD, Barton MK. 2005. Surge and destroy, the role of auxin in plant embryogenesis. Development 132, 3577–3585 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195 [DOI] [PubMed] [Google Scholar]
- Kay S, Hahn S, Marois E, Hause G, Bonas U. 2007. A bacteria effector acts as a plant transcription factor and induces a cell size regulator. Science 318, 648–651 [DOI] [PubMed] [Google Scholar]
- Kiegerl S, Cardinale F, Siligan C, et al. 2000. SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress-induced MAPK, SIMK. Plant Cell 12, 2247–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauss S, Rohrmeier T, Lehle L. 2003. The auxin-induced maize gene ZmSAUR2 encodes a short-lived nuclear protein expressed in elongating tissues. Journal of Biological Chemistry 278, 23936–23943 [DOI] [PubMed] [Google Scholar]
- Kohno M, Takato H, Horiuchi H, Fujita K, Suzuki S. 2012. Auxin-nonresponsive grape Aux/IAA19 is a positive regulator of plant growth. Molecular Biology 39, 911–917 [DOI] [PubMed] [Google Scholar]
- Kuang JF, Zhang Y, Chen JY, Chen QJ, Jiang YM, Lin HT, Xu SJ, Lu WJ. 2011. Two GH3 genes from longan are differentially regulated during fruit growth and development. Gene 485, 1–6 [DOI] [PubMed] [Google Scholar]
- Lecourieux F, Lecourieux D, Vignault C, Delrot S. 2010. A sugar inducible protein kinase, VvSK1, regulates hexose transport and sugar accumulation in grape cells. Plant Physiology 152, 1096–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire-Chamley M, Petit J, Garcia V, Just D, Baldet P, Germain V, Fagard M, Mouassite M, Cheniclet C, Rothan C. 2005. Changes in transcriptional profiles are associated with early fruit tissue specialization in tomato. Plant Physiology 139, 750–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood T, Evan GI. 1998. Helix–loop–helix transcription factors, 3rd edn New York: Oxford University Press; [Google Scholar]
- Liu C, Xu Z, Chua NH. 1993. Auxin polar transport is essential for the establishment of bilateral symmetry during early plant embryogenesis. Plant Cell 5, 621–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C. 2008. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322, 1535–1539 [DOI] [PubMed] [Google Scholar]
- Matus JT, Poupin MJ, Cañón P, Bordeu E, Alcalde JA, Arce-Johnson P. 2010. Isolation of WDR and bHLH genes related to flavonoid synthesis in grapevine (Vitis vinifera L.). Plant Molecular Biology 72, 607–620 [DOI] [PubMed] [Google Scholar]
- Mauro MC, Toutain S, Walter B, Pinck L, Otten L, Coutos-Thévenot P, Deloire A, Barbier P. 1995. High efficiency regeneration of grapevine plants transformed with the GFLV coat protein gene. Plant Science 112, 97–106 [Google Scholar]
- Moller B Weijers D 2009. Auxin control of embryo patterning. Cold Spring Harbor Perspectives in Biology 1, a001545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov IA, Paponov M, Teale WD, Menges M, Chakrabortee S, Murray JAH, Palme K. 2008. Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Molecular Plant 1, 321–337 [DOI] [PubMed] [Google Scholar]
- Pires N, Dolan L. 2010. Origin and diversification of basic-helix-loop-helix proteins in plants. Molecular Biology and Evolution 27, 862–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Research 23, 4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay NA, Glover BJ. 2005. MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends in Plant Science 10, 63–70 [DOI] [PubMed] [Google Scholar]
- Reid KE, Olsson N, Schlosser J, Peng F, Lund ST. 2006. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biology 6, 27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck D, Christian M, Jones A, Lüthen H. 2010. Rapid auxin-induced cell expansion and gene expression: a four-decade-old question revisited. Plant Physiology 152, 1183–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inzé D, Peer WA, Murphy AS, Overvoorde PJ, Gray WM. 2012. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. The Plant Journal 70, 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szécsi J, Joly C, Bordji K, Varaud E, Cock JM, Dumas C, Bendahmane M. 2006. BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO Journal 25, 3912–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4, Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W, improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Research 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. 2003. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15, 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J. 2009. Auxin, a trigger for change in plant development. Cell 136, 1005–1016 [DOI] [PubMed] [Google Scholar]
- Varaud E, Brioudes F, Szécsi J, Leroux J, Brown S, Perrot-Rechenmann C, Bendahmane M. 2011. AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell 23, 973–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. 2005. Auxin: regulation, action, and interaction. Annals of Botany 95, 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CJ, Zhang C, Lu YN, Jin JQ, Wang XL. 2011. The mechanisms of brassinosteroids’ action: from signal transduction to plant development. Molecular Plantarum 4, 588–600 [DOI] [PubMed] [Google Scholar]
- Zentella R, Zhang ZL, Park M, et al. 2007. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19, 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.