Abstract
Transcription activator-like effectors (TALEs) can be used as DNA-targeting modules by engineering their repeat domains to dictate user-selected sequence specificity. TALEs have been shown to function as site-specific transcriptional activators in a variety of cell types and organisms. TALE nucleases (TALENs), generated by fusing the FokI cleavage domain to TALE, have been used to create genomic double-strand breaks. The identity of the TALE repeat variable di-residues, their number, and their order dictate the DNA sequence specificity. Because TALE repeats are nearly identical, their assembly by cloning or even by synthesis is challenging and time consuming. Here, we report the development and use of a rapid and straightforward approach for the construction of designer TALE (dTALE) activators and nucleases with user-selected DNA target specificity. Using our plasmid set of 100 repeat modules, researchers can assemble repeat domains for any 14-nucleotide target sequence in one sequential restriction-ligation cloning step and in only 24 h. We generated several custom dTALEs and dTALENs with new target sequence specificities and validated their function by transient expression in tobacco leaves and in vitro DNA cleavage assays, respectively. Moreover, we developed a web tool, called idTALE, to facilitate the design of dTALENs and the identification of their genomic targets and potential off-targets in the genomes of several model species. Our dTALE repeat assembly approach along with the web tool idTALE will expedite genome-engineering applications in a variety of cell types and organisms including plants.
Keywords: Genome engineering, TALE-based activators and repressors, TALE nucleases (TALENs), Targeted mutagenesis, Targeted activation and repression, Genome modifications
Introduction
The interaction between Xanthomonas pathogenic bacteria and plants leads to the injection of bacterial proteins through the syringe-like type III secretion system. These injected bacterial proteins include transcription activator-like effectors (TALEs) that act as transcription factors to modulate the expression of disease resistance-related genes in the host nucleus (Boch and Bonas 2010; Romer et al. 2009, 2010). The existence of a large number of TALEs and information regarding their target genes have been essential for deciphering how these proteins bind to the promoter regions of their target genes (Boch et al. 2009; Moscou and Bogdanove 2009). TALEs possess specific structural features, including an N-terminal secretion signal, a DNA-binding domain (DBD) with a variable number of 34/35-amino acid repeats, and a nuclear localization signal and acidic activation domain at the C-terminus (Boch and Bonas 2010; Bonas et al. 1993; Bonas et al. 1989). Analysis of TALE structure, in particular their DBD repeats and the sequence of the corresponding DNA-binding box, led to the determination of the TALE protein DNA-binding code (Boch et al. 2009; Bogdanove et al. 2010; Moscou and Bogdanove 2009). The number of DBD repeats can range from 1.5 to 30. The repeat variable diresidue (RVD), at positions 12 and 13 of each repeat, dictates the specificity of the repeat binding to one nucleotide in the DNA target. Based on this code, the HD repeat specifies the C nucleotide, NI specifies the A nucleotide, NK specifies the G nucleotide, NG specifies the T nucleotide, NN specifies the A and G nucleotides, while NS can bind to A, C, G or T with equal affinity (Boch et al. 2009; Moscou and Bogdanove 2009; Scholze and Boch 2011). The DBD of TALE has been shown to be adaptable and can be engineered to bind to any user-selected DNA targets (Christian et al. 2010; Li et al. 2011a; Mahfouz et al. 2011b; Morbitzer et al. 2010; Mussolino et al. 2011; Sander et al. 2011; Zhang et al. 2011). Consequently, TALE proteins can be used as DNA-binding modules.
Adaptable DNA-binding modules are essential tools for genome-engineering applications. Different effector domains can be fused to these DNA-binding modules to produce a variety of sequence-specific genome modifications including epigenetic modifications (Mahfouz 2010; Mahfouz and Li 2011). Several DNA-binding modules have been used for genomic targeting, and these include zinc finger modules (Doyon et al. 2008; Durai et al. 2005; Kandavelou et al. 2009; Porteus 2008). Zinc fingers, however, suffer from low efficiency and reproducibility; moreover, their use is labor intensive and time consuming (Ramirez et al. 2008). Highly adaptable DNA-binding modules that can be easily engineered to dictate any user-selected sequence specificities are in high demand for a broad range of biotechnological applications in agriculture and medicine. TALE-based DNA-binding modules should be more efficient than zinc fingers because they confer high sequence specificity, exhibit low off-target binding and low cytotoxicity, and are highly adaptable to any user-selected sequence (Huang et al. 2011; Mahfouz et al. 2011b; Miller et al. 2011; Mussolino et al. 2011; Wood et al. 2011). By using the TALE DNA-binding code, researchers can develop a suite of precise and efficient molecular genome-modification tools that are applicable in many different organisms (Mahfouz and Li 2011). These genome-engineering tools are based on the design of chimeric proteins composed of TALE, which confers very high DNA sequence specificity when coupled to a variety of functional domains including nucleases, activation and repression domains, methylases, and integrases (Mahfouz and Li 2011; Mahfouz et al. 2011a). TALEs fused to the FokI cleavage domain can generate chimeric nucleases (TALENs) that bind to the DNA and create double-strand breaks (DSBs) in several systems and cell types (Cermak et al. 2011; Christian et al. 2010; Huang et al. 2011; Li et al. 2011a; Mahfouz et al. 2011b; Sander et al. 2011; Tesson et al. 2011; Wood et al. 2011). The DSBs, which are mainly repaired by the cellular non-homologous end-joining repair machinery, lead to small deletions or insertions (InDels) (Zhang et al. 2010). In the presence of a donor DNA, gene replacements or stacking can be achieved. In several systems, the DSBs stimulate the efficiency of gene targeting by several orders of magnitude, allowing highly efficient gene stacking and replacement (Cai et al. 2009; Weinthal et al. 2010).
Using TALENs, we and others have created site-specific DSBs in plant cells (Cermak et al. 2011; Mahfouz et al. 2011b). The ability to target TALENs to a specific genomic sequence permits researchers to generate a targeted mutation and to add, delete, or edit genes with extreme precision. The implications of TALEN technology for the modification of crops and other plant species are profound. For example, the ability to perform targeted genome modifications in plants can facilitate efficient and robust addition, deletion, activation, and inactivation of genes. The precise and efficient modification of the plant genome can expand the number of plant species amenable to genetic modification, accelerate beneficial trait development, and expand the range of traits that can be modified and developed (Mahfouz and Li 2011).
The development of an efficient and simple repeat assembly approach for the construction of designer TALEs (dTALEs) is needed to advance the use of TALE-based genome-engineering applications across different species and systems. Several repeat assembly approaches have recently been reported that include PCR-based methods coupled to Golden Gate cloning or Golden Gate cloning only (Cermak et al. 2011; Geissler et al. 2011; Huang et al. 2011; Morbitzer et al. 2011; Weber et al. 2011; Zhang et al. 2011). In general, these methods, apart from being labor-and resource- intensive, are sophisticated and require several restriction and ligation steps in different backbone vectors.
Here, we present a rapid and efficient approach for the modular assembly of TALE repeats using the dHax3 scaffold as backbone for TALE transcription factors (TALE-TFs) and TALENs. With this approach, a dTALE with 12.5 or more repeats can be assembled in just one sequential restriction-ligation step and in only 24 h from a set of 100 plasmids containing all possible combinations of repeat modules. In contrast to previously reported approaches, our approach involves just one sequential restriction-ligation step and results in a high efficiency and reproducibility of assembly, does not require highly skilled researchers, and can be performed in only 24 h. Furthermore, we developed a web-tool called idTALE that facilitates the design of dTALE-TFs and dTALENs and the identification of their targets in the genome of several model species including Arabidopsis thaliana. Using this approach, we generated several dTALEs with novel user-selected sequence specificities and tested their function in vivo. Because our assembly strategy facilitates the rapid construction of TALE-TFs and TALENs, it should expedite TALE-based genome-engineering applications in plants and other eukaryotes.
Results
Assembly of dTALE repeats in a one-step sequential restriction-ligation reaction
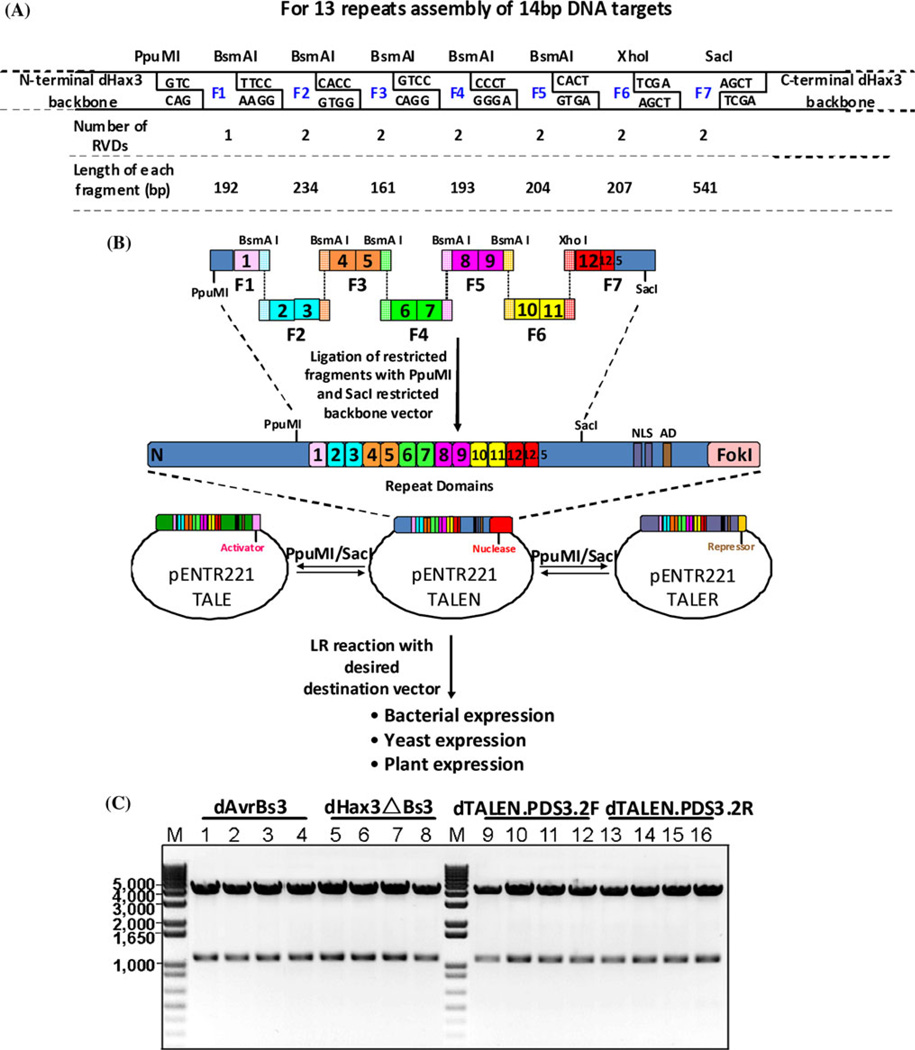
We selected the dHax3 TALE protein as a scaffold and backbone for the construction of dTALE-TFs and dTALENs. The dHax3 backbone has been successfully used in several systems as a transcriptional activator and chimeric nuclease. Our goal was to establish a tool kit that facilitates a rapid and ordered assembly and cloning of the repeats to any user-selected recognition sequence in dTALE-TF or dTALEN backbones. To achieve this goal, we designed, synthesized, and cloned a plasmid library of fragments containing all possible combinations of RVDs in the pUCMCS minus vector. The dHax3 repeat sequence was optimized to include a few restriction enzymes at specific positions so that their restriction produces several distinct fragments that can be correctly and orderly assembled by restriction-ligation cloning. These fragments can be assembled in a one-step sequential restriction-ligation cloning to produce a custom order of repeats for any user-selected target specificity. Our assembly approach is based on a 100-plasmid library of repeat fragments and type IIS restriction enzymes. We used PpuMI and SacI to clone the assembled repeats into the TALE backbone (Fig. 1 and Supplementary Information). To assemble the repeats, we used BsmAI and BsmBI that cleave outside their recognition sequence and produce 4-bp overhangs. Repeat fragments were designed to contain BsmAI and BsmBI enzymes so that ordered assembly of the fragments can be achieved after restriction with BsmAI and BsmBI by sequential restriction ligation cloning method (Fig. 1). We also used XhoI enzyme to assemble fragment 6–7. The introduction of this enzyme also serves another purpose—when confirming the clones by restriction digestion, we used PpuMI and XhoI because XhoI is absent in the original backbone.
Fig. 1.
Construction of TALE-based transcription factors and nucleases by one sequential restriction ligation approach. a Schematic representation of the dHax3 repeat region showing the seven restriction fragments and the overhang sequence of each fragment produced by a restriction digestion. b Overall assembly strategy showing the steps required to produce the final expression clone of TALE-TF or TALEN. The repeat region can be transferred between different dHax3-based backbones, with different effectors, using the PpuMI and SacI restriction enzymes. c Confirmation of the assembly clones by restriction digestion analysis. The lower bands, indicated by the arrow, are the products of PpuMI and XhoI digestion and demonstrate the successful and correct assembly of the repeats. All restriction digestion positive clones showed the correct order and sequence of the repeats, indicating the extreme fidelity of this assembly approach. M 1-kb plus marker, lane 1–4, dAvrBs3, lane 5–8 dHax3deltaBs3, lane 9–12 dTALEN.PDS3.2F, and lanes 13–16 dTALEN.PDS3.2R
As indicated in Fig. 1, our plasmid library is based on seven fragments; the first fragment contains one RVD and hence has four plasmids. The second fragment, and also the third through seventh fragment, contains two RVDs, and hence each has 16 plasmids for all possible combinations of RVDs (Supplementary Information). The restriction of these fragments by the designated enzymes produces fragments that can be assembled in a desired order. The overlaps of the fragments were designed so that only an ordered and correct assembly is possible. For each fragment, we generated all possible variants that specify the RVDs, including HD that binds to the C nucleotide, NI that binds to the A nucleotide, NK that binds to the G nucleotide, NN that binds to the A or G nucleotides, and NG that binds to the T nucleotide.
We performed the assembly of the repeats using a one-step sequential restriction-ligation protocol and confirmed the correct assembly of the clones. More than 95% of the clones confirmed by restriction digestion followed by Sanger sequencing had the correct order and number of repeats, indicating that this assembly approach is highly efficient. Using this protocol, we can produce TALENs and TALE-TFs with 12 and 13 repeats in just 24 h with much less effort than required for other methods. The assembly of 13 repeats can be achieved if fragment 6 is restricted with BsmAI and XhoI enzymes. This number of repeats should be sufficient to produce dTALENs that are highly specific, with a single genomic target. Although a dTALE-TF with 13 or 14 repeats might have more than one target in the genome, it would still be sufficiently specific for targeting a group of genes.
Because the dTALEN and dTALE-TF backbone vectors are already in the Gateway-compatible vector pENTR221, an assembled dTALEN or dTALE-TF can be cloned in any Gateway-compatible destination vector for bacterial, yeast, plant, or mammalian expression. The LR Gateway reaction that moves the dTALEN or dTALE-TF clones from the pENTR221 vector into a plant expression vector can be avoided by the direct assembly of the repeats in the pK2GW7 expression vector. We performed the assembly of the dTALE repeats directly in the pK2GW7 plant expression vector. We first restricted the pK2GW7.dHax3 (for TALE-TF backbone) and pK2GW7.dHax3.N (for TALEN backbone) clones with the unique SacI and PpuMI and then assembled the repeats as described above. We generated correctly assembled TALE-TFs and TALENs in the destination expression vector in one step and with high efficiency. The ability to perform the assembly directly in the expression vector eliminates the need for a 2-day step and expensive Gateway reagents. Users could have the choice of assembly directly either in the expression vector or in an entry Gateway-compatible clone that can be transferred into a variety of expression destination vectors for different species and different purposes. This assembly approach is relatively simple, rapid, straightforward, and reproducible, and thus can be utilized by researchers with different levels of molecular biology skills. Hence, it can facilitate the generation of genome-engineering reagents and their application in many laboratories and for various systems and organisms. Using our assembly approach, we efficiently generated more than 20 TALEs for site-specific transcriptional activation and cleavage purposes.
Functional testing of custom dTALE-TFs in tobacco leaf transient expression assays
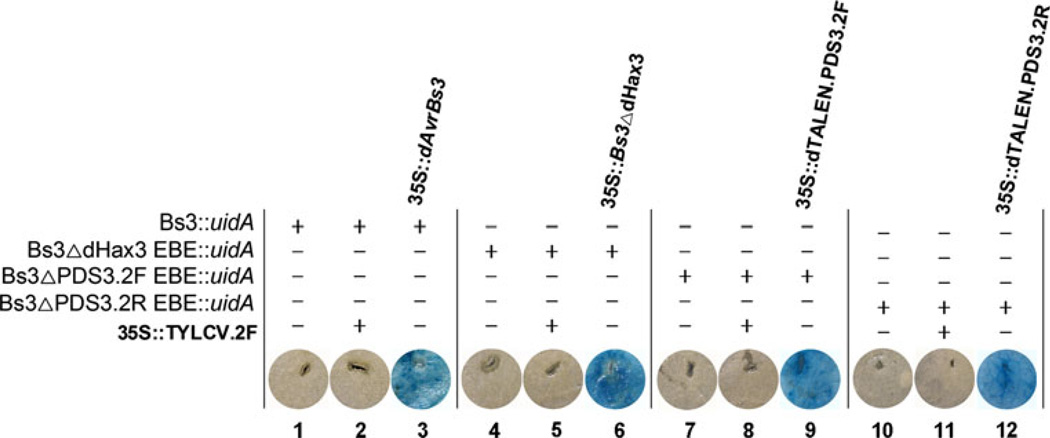
We tested the activity of the assembled dTALE-TFs in vivo in Nicotiana benthamiana leaves. We used our assembly strategy to assemble several dTALE-TFs in pENTR221. The pENTR221/dTALE-TF clones were then moved into the pK2GW7 plant expression vector by LR Gateway cloning. The pK2GW7/dTALE-TF clones were transformed into Agrobacterium tumefaciens, which was used to infiltrate N. benthamiana leaves. We used the WT Bs3 promoter and generated different versions of it by inserting different DNA-binding boxes. We assembled different dTALE-TFs that would recognize these DNA-binding boxes. Tobacco leaves agroinfiltrated with pK2GW7/dAvrBs3 and BS3::uidA vectors were collected 48 hpi and assayed for GUS (β-glucuronidase) activity. We tested several dTALE-TFs that bind to different DNA-binding boxes inserted in the Bs3 promoter, which is activated by AvrBs3 protein. To activate the WT Bs3 promoter, we assembled a dTALE-TF (dAvrBs3) with 12.5 repeats that specifically binds the 14-bp AvrBs3 target sequence. The assembled dAvrBs3-TFs activated the expression of the WT Bs3 promoter (Fig. 2). Transcriptional activation did not occur following agroinfiltration with the Bs3 promoter only or coinfiltration with other dTALE-TFs that do not contain a binding box in the Bs3 promoter. These data indicate that the assembled dAvrBs3-TF binds to the Bs3 promoter and activates transcription in vivo.
Fig. 2.
Validation of custom assembled dTALE-TFs in transcriptional activation of EBE containing promoters. Assembled dTALE-TFs, dAvrBs3, dHax3, PDS3.2F, and PDS3.2R EBE activate WT Bs3 promoter or modified Bs3 promoter containing their respective binding EBE boxes (lane 3, 6, 9, and 12). For a negative control, we used Bs3 promoter only (lane 1, 4, 7, and 10) or modified Bs3 promoter. The 35S::dTALE.TLYCV.2F, which bind to a different EBE was used as a negative control as well (lane 2, 5, 8, and 11)
Functional testing of custom dTALEN activity in DNA cleavage assays in vitro
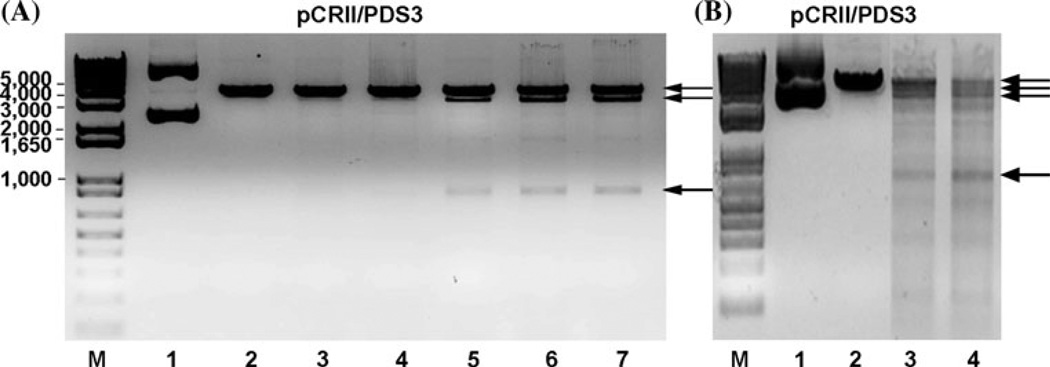
We tested the ability of the assembled dTALEN proteins to cleave their double-stranded DNA targets in vitro. Previous studies have shown that the nuclease activity of the FokI enzyme and the zinc finger hybrid nucleases require dimerization (Mani et al. 2005; Miller et al. 2007; Pruett-Miller et al. 2009). To test the activity of our dTALENs, we designed target sequences containing two effector-binding elements in a tail-to-tail orientation and separated by 16-bp spacers (Li et al. 2011a; Mahfouz et al. 2011b). We assembled the dTALEN heterodimers that bind to the phytoene desaturase cDNA from A. thaliana. The heterodimer pair was assembled in pENTR221 and then moved into Thioredoxin. His/pET32A by LR Gateway cloning. The linearized pCRII/PDS3 cDNA plasmid was gel-purified and used as substrate for the PDS3TALEN digestion reactions. The thiroedoxin-tagged PDS3 dTALEN heterodimer pair proteins were expressed in Escherichia coli and affinity purified using Ni-NTA. The DNA cleavage activity of the proteins was tested in vitro on the pCRII/PDS3 cDNA plasmid (Mahfouz et al. 2011b). The heterodimer pair was active in vitro and resulted in cleavage of the PDS3 target. Digestion reactions performed using only one monomer of the PDS3 dTALENs (either PDS3 TALEN-F or PDS3 TALEN-R) did not show any cleavage activity (Fig. 3).
Fig. 3.
Validation of custom assembled TALEN activity using the DNA cleavage assays. TALEN heterodimer dTALEN.PDS3-1F/1R a and dTALEN.homodimer b digested target DNA of PDS3 cDNA fragment in the pCRII vector (pCRII/PDS3). Arrows indicate the digestion products of the TALEN proteins. M 1-kb plus marker. Lanes in a are 1 undigested pCRII/PDS3 plasmid, 2 SacI-digested pCRII/PDS3, 3 pCRII/PDS3 digested with dTALEN.PDS3-1F, 4 pCRII/PDS3 digested with dTALEN.PDS31R, 5–7, pCRII/PDS3 digested with dTALEN.PDS3-1F/1R heterodimer. Lanes in b are 1 plasmid of pCRII/PDS3, 2 SacI-digested pCRII/PDS3, 3 and 4 pCRII/PDS3 digested with dTALEN.PDS3 homodimer
idTALE web-based tool: a designer and target finder of dTALE-TFs and dTALENs
To expedite the in silico design of TALE-TFs and TALENs and to find their potential genomic targets and off-targets (Fu et al. 2009; Sander et al. 2007), we developed a web-based tool called idTALE. idTALE, available at (http://idtale.kaust.edu.sa/), provides users with options to either design a dTALE and/or to check for potential dTALE targets and off-targets within the genome of choice among several model species. To design a dTALE, users can submit either the Ensembl Gene ID or the gene sequence. These two options require information regarding the spacer length and the TALE repeat number as input. Based on this input, the program outputs the result back to the web browser. The idTALE web tool offers searching options for dTALE and dTALEN target sites within the genome of choice. Providing the dTALEN repeat sequence in either homodimer or heterodimers format, the idTALE web tool can identify the potential target and off-target loci (Supplementary Information). In either case, users must provide a spacer length range and the distance upstream of the gene within which they would like to search for binding sites (Sander et al. 2010). When the search for TALE design is performed, possible TALENs as homodimers and heterodimers are returned in a tabulated form. A simple scoring algorithm is applied to filter out the common results, which are listed as distinct tabs for clarity. Because naturally occurring TALEs bind only to targets with a preceding thymine, this nucleotide is included in the calculation but not in the output. As a result of searching for dTALE target sites in the genome of choice, coordinates meeting predefined criteria are returned to the user together with other potential binding loci within the genome. The coordinates link to publicly available genome browsers such as Ensembl and NCBI.
Discussion
TALEs provide powerful and adaptable DNA-binding modules with great potential for a variety of biotechnological applications in agriculture and medicine. TALE RVDs dictate the DNA sequence specificity. The engineering of the RVDs of TALE repeats to bind to a user-selected DNA specificity is a challenging task. Because of their repetitive nature, engineering by PCR approaches is complex and not reproducible. Moreover, commercial synthesis is feasible but relatively slow (8–12 weeks for a single TALE) and expensive (around 3,500 USD per TALE). To fully realize the potential of TALEs in different applications and organisms, a robust, efficient and inexpensive custom construction is required.
Here, we report the development of a rapid, robust, and highly efficient approach for dTALE repeat assembly in dTALE-TF and dTALEN backbones. To our knowledge, this approach is the simplest, quickest, and most efficient among the approaches reported in the literature. A library of 100 plasmids can be used to assemble 12 and 13 TALE repeats, thereby dictating DNA-binding specificity to any user-selected 13- or 14-bp targets, respectively. Our approach can also be used to construct dTALEs with a greater number of repeats—a library of fragment 5.1 (Supplementary Information) would be used to construct TALEs with two additional repeats. Because nine DNA fragments can be efficiently assembled using sequential restriction ligation cloning, our assembly approach can be used to assemble dTALEs with 18 repeats. Because the method uses sequence-verified clones, the integrity of the assembled clones is guaranteed.
Our dTALE assembly approach relies on the use of the type II restriction enzymes PpuMI, BsmAI, BsmBI, XhoI, and SacI. These restriction enzymes are used in one sequential restriction-ligation reaction as indicated in Fig. 1. A further advantage of our approach is that the assembly can be done in a variety of backbones, including activator, nuclease, repressor, etc. Additionally, the assembled repeats can be transferred from one backbone to another by PpuMI and SacI restriction ligation. This is particularly advantageous for the simultaneous study of transcriptional activation and repression of a specific gene target (Mahfouz et al. 2011a). Our modular assembly tool kit is composed of HD, NG, NI, and NK repeat modules, which have been shown to be highly specific in binding the C, T, A, and G nucleotides, respectively. This allows the design of highly specific dTALENs and dTALE activators and dTALEs fused with any functional domains. It was recently reported that the efficiency of dTALEs with NN repeat modules is much higher than those with NK repeat modules (Huang et al. 2011). Accordingly, we also generated a library containing NN instead of NK so that users would have the choice of using either one or both. The number and identity of the repeats influence TALE target specificity (Boch et al. 2009; Cermak et al. 2011; Morbitzer et al. 2011). Other factors, however, may also influence the binding in vivo. These factors include, but are not limited to, the chromatin status and the neighboring effects from the DNA sequence.
Because dTALENs require a dimerization step for their cleavage activity, two monomers in a homo- or heterodimers form are needed. Several studies have shown that dTALENs based on the dHax3 backbone are highly efficient in generating DSBs in mammalian and plant cells. Given that two monomers (two heterodimers that bind 13 or 14 bp in a tail-to-tail orientation and that are separated by a 12–30-bp spacer region) are required, dTALENs with 12 or 13 repeats should be sufficient for the design of effective and highly specific TALE-based nucleases.
Several strategies have been previously described for dTALE assembly and for construction of chimeric activators and nucleases (Cermak et al. 2011; Geissler et al. 2011; Huang et al. 2011; Li et al. 2011b; Morbitzer et al. 2011; Weber et al. 2011; Zhang et al. 2011). Some of these previously reported protocols rely on PCR amplification of repeat modules followed by subsequent restriction ligations and transfer between more than one backbone vectors to achieve the final assembled clone.Zhang et al. (2011), for example, have reported a Golden Gate-based assembly approach. Their strategy involves two rounds of PCR amplification, restriction digestions and ligations of the assembled repeats in the final backbone (http://www.taleffectors.com). Although this approach has allowed the assembly and functional testing of several dTALE-TFs (Zhang et al. 2011), it depends on PCR amplification and is therefore prone to sequence mutations and recombination between the repeats. Using a different approach,Li et al. (2011b) have reported a modular construction of dTALENs by synthesis of eight sets of four RVD modules; these eight sets are used to assemble eight repeat subarrays, and the confirmed sequence of the eight repeat subarrays (for 16- or 24-bp targets) can be ligated together in a final dTALE backbone. In addition, Weber et al. (2011) have used the native AvrBs3 scaffold to assemble dTALEs in two successive cloning steps; in the first step, five or six repeats are assembled in three preassembly vectors for positions 1–6, 7–12, and 13–17. Then, these three preassembled repeats are ligated together in a second cloning step to produce dTALEs with 17.5 repeats. Similarly, Morbitzer et al. (2011) and Geissler et al. (2011) have reported dTALE assembly by two successive cut-ligation cloning steps that produced 17.5 repeats sufficient to confer high DNA-binding specificity.
Compared to these previously reported protocols, our tool kit represents a major improvement because it depends on a plasmid library of repeat modules to ensure the reliability of the sequence and the efficiency and reproducibility of a robust assembly. Moreover, our assembly strategy offers the following technical advantages: it does not require PCR amplification, which has the risk of repeat recombination; it requires only one sequential restriction-ligation cloning step, which can be performed in only 1 day (including confirmation of the clones by restriction digestion analysis); and the assembled repeats can be transferred between different backbones with different functional domains, which facilitates the study of different genomic modifications at a particular locus.
Our approach will be extremely useful when a library of dTALE activators or repressors and dTALENs is needed for genome-wide applications. All the restricted fragments of all possible RVDs can be ligated, and the library of pENTR221/dTALE activator, repressor, or nuclease can be transferred to a destination expression vector specific to a certain organism, including yeast, plant, mammalian cells, etc. The library of dTALE in a plant expression vector can be transformed into A. tumefaciens and then into the plant species of interest. The generation of an overexpression TALE library in plants can be used, for example, to screen for phenotypes with useful agronomic traits. Following phenotypic selection and heritability analysis, the TALE repeat sequences can be determined and their targets identified.
Our entire plasmid library for dTALE construction is available from King Abdullah University of Science and Technology (KAUST) for non-profit use. Because a user-friendly software tool for the in silico design of dTALEs and the identification of their potential targets and off-targets was lacking, we developed a web interface, idTALE, that allows the design and interrogation of the genomes of several model species for dTALE targets.
Because the currently available tool, TALE-NT, provides very limited design and search options, idTALE can significantly facilitate the design of dTALEs for genome-wide applications and thus should be extremely useful to researchers (Cermak et al. 2011). The currently available TALE-NT software (http://boglabx.plp.iastate.edu/TALENT) exhibits several limitations: it allows users to generate dTALENs based on an input sequence with a maximal length of only 5,000 bp; it does not allow the user to specify a desired dTALE RVD number, thereby creating by default all possible RVDs between 15–30 bp long and making the output enormously confusing and unreadable unless other software is used; and it is not possible to check whether individual dTALEN monomers would form a heterodimer in the genome of interest. idTALE, in contrast, allows researchers to design dTALE RVDs by submitting the Ensembl Gene ID or the gene sequence in FASTA format. Moreover, an option to design RVDs around the input sequence means that idTALE has no limitations for input length or model organism. Providing idTALE with the Ensembl Gene ID ensures that the output of RVDs will generate a heterodimer or homodimer for the coding sequence of the gene of interest. Furthermore, idTALE allows users to choose and check for the same dTALEN targets within multiple species. In conclusion, our assembly approach and web interface should facilitate the application of TALE-based genome modifications in plants and other eukaryotes.
Note: Our assembly plasmid library set is available for non-profit use, and the idTALE web tool is freely available online.
Materials and methods
Design and efficient construction of TALE-TFs and TALENs using the dHax3 and dHax3.N scaffolds
We previously reported the optimization of dHax3 cDNA to reduce the homology between the 102-bp repeat units and the codon optimization for in planta expression (Mahfouz et al. 2011b). The dHax3 and dHax3.N cDNAs, in the pENTR221 vector, were used as backbones to construct the TALE-TFs and TALENs, respectively. The PpuMI/SacI fragment containing the repeat regions was divided into seven different fragments. Each fragment had two repeats and therefore two RVDs, except fragment 1, which had only one repeat. The sequence of each fragment and the primers used for amplification and cloning in pUC-minusMCS are provided in SI. These fragment clones were confirmed by sequencing, and their repeat RVDs were mutagenized to produce different RVDs. Moreover, because these fragments were short, we generated a whole library of all possible RVDs in each fragment by commercial synthesis (Blue Heron Bothell, WA, USA). As a result, we produced and amplified a library of fragments 1–7 containing all possible RVDs in every repeat in every fragment. We performed in silico assembly of the seven fragments, and the produced clones were identical to our dHax3 and dHax3.N except for the user-selected RVDs (Supplementary Information). pUC-minusMCS was chosen to clone the library of all repeat fragments because it confers ampicillin resistance while the pENTR221 backbone assembly vector confers kanamycin resistance; these antibiotic markers facilitate the screening of the correctly assembled dTALE-TFs or dTALENs. The correctly assembled dTALE-TF- or dTALEN-confirmed clones could be sub-cloned in any Gateway-based destination vector by LR recombination reactions for bacterial, yeast, plant, and mammalian expression.
One-step sequential restriction-ligation assembly protocol
Plasmids that contained the seven fragments were selected based on the desired TALE repeat sequence. Each fragment was restricted as follows: fragment 1 (F1) with PpuMI and BsmAI, purify target band of 192 bp; fragments 2, 3, 4, and 5 (F2, F3, F4, and F5) with BsmAI, purify target bands of 234, 161, 193, and 204 bp, respectively; fragment 6 (F6) first with XhoI and then with BsmAI, purify target band of 207 bp; fragment 7 (F7) with XhoI and SacI, purify target band of 541 bp. Equi-molar amounts of fragments were ligated with the restricted backbone using T7 ligase at 23°C for 2 h or T4 ligase at 16°C for 6 h. A 2-µL volume of the ligation product was used to transform to TOP10 competent cells. Positive colonies were selected, and recombinant plasmid was purified and checked by restriction digestion with XhoI and PpuMI enzymes—the product band was expected to be about 1,190 bp. Sanger sequencing was used to further confirm the positive clones.
Agrobacterium tumefaciens-mediated transient expression and dTALE-TF activity assays
To test the activity of dTALE-TFs, we performed LR reactions between pENTR221/dTALE-TF entry clones and the pK2GW7 Gateway-compatible binary vector and generated overexpression clones pK2GW7/dTALE-TFs. These expression clones were transformed into A. tumefaciens GV3101 and used for transient expression in tobacco leaves. To assay the activity of dTALE-TFs in planta, we coinfiltrated or separately infiltrated A. tumefaciens GV3101 harboring BS3::uidA and its dTALE activator into tobacco leaves. Infiltrated discs were collected 48 hpi. The GUS qualitative assay was performed by immersing leaf discs in GUS-staining solution (10 mM sodium phosphate, pH 7, 10 mM EDTA, 0.1% Triton X-100, 0.1% 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, 1 mM potassium ferricyanide, 1 mM potassium ferrocyanide) at 37°C for 24 h. The discs were cleared in ethanol.
Bacterial expression and purification of dTALENs
The assembled dTALEN clones were confirmed by restriction digestions and sequencing. LR reactions were performed between the entry clones, pENTR221/dTALENs, and the Gateway-compatible pET32a expression vector (following the manufacturer’s instructions) to generate pET32a.dTALENs. The expression clones were transformed into E. coli BL21, and protein expression was induced at 25°C for 5 h with 1 mM isopropyl β-d-1-thiogalactopyranoside. The TRX.6His.dHax3.N proteins were purified using Qiagen Ni-NTA agarose resin according to the manufacturer’s instructions. The purified proteins, as homodimers or heterodimers for one specific target, were used for DNA cleavage assays as previously described (Mahfouz et al. 2011b).
Supplementary Material
Acknowledgments
We thank Thomas Lahaye for providing the Bs3::uidA binary construct. We also thank the members of the Genome Engineering Group at KAUST for their technical assistance.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11103-012-9875-4) contains supplementary material, which is available to authorized users.
Contributor Information
Lixin Li, Division of Chemical and Life Sciences and Engineering, King Abdullah University of Science and Technology, Thuwal 23955-6900, Kingdom of Saudi Arabia.
Marek J. Piatek, Division of Chemical and Life Sciences and Engineering, King Abdullah University of Science and Technology, Thuwal 23955-6900, Kingdom of Saudi Arabia
Ahmed Atef, Department of Biological Sciences, Faculty of Sciences, King Abdulaziz University, P.O. Box 80141, Jeddah 21589, Kingdom of Saudi Arabia.
Agnieszka Piatek, Division of Chemical and Life Sciences and Engineering, King Abdullah University of Science and Technology, Thuwal 23955-6900, Kingdom of Saudi Arabia.
Anjar Wibowo, Division of Chemical and Life Sciences and Engineering, King Abdullah University of Science and Technology, Thuwal 23955-6900, Kingdom of Saudi Arabia.
Xiaoyun Fang, Division of Chemical and Life Sciences and Engineering, King Abdullah University of Science and Technology, Thuwal 23955-6900, Kingdom of Saudi Arabia.
J. S. M. Sabir, Department of Biological Sciences, Faculty of Sciences, King Abdulaziz University, P.O. Box 80141, Jeddah 21589, Kingdom of Saudi Arabia
Jian-Kang Zhu, Email: jkzhu@purdue.edu, Department of Horticulture and Landscape Architecture, Purdue University, West Lafayette, IN 47907, USA.
Magdy M. Mahfouz, Email: magdy.mahfouz@kaust.edu.sa, Division of Chemical and Life Sciences and Engineering, King Abdullah University of Science and Technology, Thuwal 23955-6900, Kingdom of Saudi Arabia.
References
- Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol. 2010;48:419–436. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Bogdanove AJ, Schornack S, Lahaye T. TAL effectors: finding plant genes for disease and defense. Curr Opin Plant Biol. 2010;13:394–401. doi: 10.1016/j.pbi.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Bonas U, Stall RE, Staskawicz B. Genetic and structural characterization of the avirulence gene AvrBs3 from Xanthomonas campestris pv. vesicatoria. Mol Gen Genet. 1989;218:127–136. doi: 10.1007/BF00330575. [DOI] [PubMed] [Google Scholar]
- Bonas U, Conrads-Strauch J, Balbo I. Resistance in tomato to Xanthomonas campestris pv vesicatoria is determined by alleles of the pepper-specific avirulence gene avrBs3. Mol Gen Genet. 1993;238:261–269. doi: 10.1007/BF00279555. [DOI] [PubMed] [Google Scholar]
- Cai CQ, Doyon Y, Ainley WM, Miller JC, Dekelver RC, Moehle EA, Rock JM, Lee YL, Garrison R, Schulenberg L, et al. Targeted transgene integration in plant cells using designed zinc finger nucleases. Plant Mol Biol. 2009;69:699–709. doi: 10.1007/s11103-008-9449-7. [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33:5978–5990. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu F, Sander JD, Maeder M, Thibodeau-Beganny S, Joung JK, Dobbs D, Miller L, Voytas DF. Zinc finger database (ZiFDB): a repository for information on C2H2 zinc fingers and engineered zinc-finger arrays. Nucleic Acids Res. 2009;37:D279–D283. doi: 10.1093/nar/gkn606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler R, Scholze H, Hahn S, Streubel J, Bonas U, Behrens SE, Boch J. Transcriptional activators of human genes with programmable DNA-specificity. PLoS One. 2011;6:e19509. doi: 10.1371/journal.pone.0019509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- Kandavelou K, Ramalingam S, London V, Mani M, Wu J, Alexeev V, Civin CI, Chandrasegaran S. Targeted manipulation of mammalian genomes using designed zinc finger nucleases. Biochem Biophys Res Commun. 2009;388:56–61. doi: 10.1016/j.bbrc.2009.07.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011a;39:359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Huang S, Zhao X, Wright DA, Carpenter S, Spalding MH, Weeks DP, Yang B. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011b;39:6315–6325. doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz MM. RNA-directed DNA methylation: mechanisms and functions. Plant Signal Behav. 2010;5:806–816. doi: 10.4161/psb.5.7.11695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz MM, Li L. TALE nucleases and next generation GM crops. GM Crop. 2011;2:1236–1256. doi: 10.4161/gmcr.2.2.17254. [DOI] [PubMed] [Google Scholar]
- Mahfouz MM, Li L, Piatek M, Fang X, Mansour H, Bangarusamy D, Zhu J-K. Targeted transcriptional repression using a chimeric TALE-SRDX repressor protein. Plant Mol Biol. 2011a doi: 10.1007/s11103-011-9866-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz MM, Li L, Shamimuzzaman M, Wibowo A, Fang X, Zhu JK. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc Natl Acad Sci USA. 2011b;108:2623–2628. doi: 10.1073/pnas.1019533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani M, Smith J, Kandavelou K, Berg JM, Chandrasegaran S. Binding of two zinc finger nuclease monomers to two specific sites is required for effective double-strand DNA cleavage. Biochem Biophys Res Commun. 2005;334:1191–1197. doi: 10.1016/j.bbrc.2005.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Morbitzer R, Romer P, Boch J, Lahaye T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc Natl Acad Sci USA. 2010;107:21617–21622. doi: 10.1073/pnas.1013133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morbitzer R, Elsaesser J, Hausner J, Lahaye T. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res. 2011;39:5790–5799. doi: 10.1093/nar/gkr151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Mussolino C, Morbitzer R, Lutge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39(21):9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus M. Design and testing of zinc finger nucleases for use in mammalian cells. Methods Mol Biol. 2008;435:47–61. doi: 10.1007/978-1-59745-232-8_4. [DOI] [PubMed] [Google Scholar]
- Pruett-Miller SM, Reading DW, Porter SN, Porteus MH. Attenuation of zinc finger nuclease toxicity by small-molecule regulation of protein levels. PLoS Genet. 2009;5:e1000376. doi: 10.1371/journal.pgen.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer P, Recht S, Lahaye T. A single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens. Proc Natl Acad Sci USA. 2009;106:20526–20531. doi: 10.1073/pnas.0908812106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer P, Recht S, Strauss T, Elsaesser J, Schornack S, Boch J, Wang S, Lahaye T. Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae. New Phytol. 2010;187:1048–1057. doi: 10.1111/j.1469-8137.2010.03217.x. [DOI] [PubMed] [Google Scholar]
- Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. Zinc finger targeter (ZiFiT): an engineered zinc finger/target site design tool. Nucleic Acids Res. 2007;35:W599–W605. doi: 10.1093/nar/gkm349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Maeder ML, Reyon D, Voytas DF, Joung JK, Dobbs D. ZiFiT (zinc finger targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010;38:W462–W468. doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholze H, Boch J. TAL effectors are remote controls for gene activation. Curr Opin Microbiol. 2011;14:47–53. doi: 10.1016/j.mib.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Tesson L, Usal C, Menoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L, et al. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29:695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- Weber E, Gruetzner R, Werner S, Engler C, Marillonnet S. Assembly of designer TAL effectors by golden gate cloning. PLoS One. 2011;6:e19722. doi: 10.1371/journal.pone.0019722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinthal D, Tovkach A, Zeevi V, Tzfira T. Genome editing in plant cells by zinc finger nucleases. Trends Plant Sci. 2010;15(6):308–321. doi: 10.1016/j.tplants.2010.03.001. [DOI] [PubMed] [Google Scholar]
- Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Maeder ML, Unger-Wallace E, Hoshaw JP, Reyon D, Christian M, Li X, Pierick CJ, Dobbs D, Peterson T, et al. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc Natl Acad Sci USA. 2010;107:12028–12033. doi: 10.1073/pnas.0914991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.