Abstract
A single click ensemble segmentation (SCES) approach based on an existing “Click&Grow” algorithm is presented. The SCES approach requires only one operator selected seed point as compared with multiple operator inputs, which are typically needed. This facilitates processing large numbers of cases. Evaluation on a set of 129 CT lung tumor images using a similarity index (SI) was done. The average SI is above 93% using 20 different start seeds, showing stability. The average SI for 2 different readers was 79.53%. We then compared the SCES algorithm with the two readers, the level set algorithm and the skeleton graph cut algorithm obtaining an average SI of 78.29%, 77.72%, 63.77% and 63.76% respectively. We can conclude that the newly developed automatic lung lesion segmentation algorithm is stable, accurate and automated.
Keywords: Image Features, Delineation, Lung Tumor, Lesion, CT, Region growing, Ensemble Segmentation
1. Introduction
Lung cancer has become one of the most significant diseases in human history. The World Health Organization estimates the worldwide death toll from lung cancer will be 10,000,000 by 2030. The 5-year survival rate for advanced Non Small Cell Lung Cancer (NSCLC) [1] remains disappointingly low. It has been hypothesized that quantitative image feature analysis can improve diagnostic/prognostic or predictive accuracy, and therefore will have an impact on a significant number of patients [2]. In the current study, standard-of-care clinical computed tomography (CT) scans were used for image feature extraction. In order to reduce variability for feature extraction, the first and essential step is to accurately delineate the lung tumors. Accurate delineation of lung tumors is also crucial for optimal radiation oncology. A common approach to delineate tumor from CT scans involves radiologists or radiation oncologists manually drawing the boundary of the tumor. In the majority of cases, manual segmentation overestimates the lesion volume to ensure the entire lesion is identified [3] and the process is highly variable [4, 5]. A stable accurate segmentation is critical, as image features (such as texture and shape related features) are sensitive to small tumor boundary changes. Therefore, a highly automatic, accurate and reproducible lung tumor delineation algorithm would represent a significant advance.
Accurate extraction of soft tissue lesions from a given modality such as CT, PET or MRI is a topic of great interest for computer-aided diagnosis (CAD), computer-aided surgery, radiation treatment planning and medical research. However, segmentation of a lesion is typically a difficult task due to the large heterogeneity of cancer lesions (compared to normal tissues), noise that results from the image acquisition process and the characteristics of lesions often being very similar to those of the surrounding normal tissues. Traditional medical image segmentation techniques include intensity-based or morphological methods [6–9], yet these methods sometimes fail to provide accurate tumor segmentation.
A lung tumor analysis (LuTA) tool [10] within the Definiens Cognition Network Technology [11] was developed by Definiens AG [12] and Merck & Co., Inc. It is a prototype application that demonstrates the ability to automatically and semi-automatically identify and recognize organs and tumors in CT images. Its efficacy in automatic lung segmentation is described in [10] and we were able to obtain accurate lung segmentations from LUTA for all cases discussed here.
LuTA is designed to enable fast and easy annotation of lung tumors or other user-defined regions of interest. Flexible controls allow the annotation of structures of the user’s choice. Once a user has clicked on a region of interest, in a single two-dimensional (2D) slice, the application builds out the object three-dimensionally. The results [10] of the first prototype application built using the Definiens Cognition Network Technology for CT based scans provided a proof of concept enabling semi-automatic volumetric analysis of tumors. However, the current generation of the LuTA tool still has some drawbacks. First, although processing time is reduced compared to manual delineation, it still requires substantial operator input. For example, more than one user selected seed point may be required for tumor segmentation. Some tumor segmentations will not be completed in one step, thus additional operations are required (e.g. the radiologist must scroll over the CT slices and find out what part of the segmentation is missing). Additionally, lung tumor boundaries are often found to be incorrect during manual inspection, and thus require manual editing that takes additional time and creates additional sources of error. Finally, although segmentations can be performed in batch mode, which is appropriate for large studies, it is impractical if manual editing is required.
With the motivation of overcoming the above drawbacks of the “Click & grow” algorithm, we propose a new delineation algorithm based on using multiple seed points with region growing [13]. The new algorithm makes use of the original algorithm by using an original seed point to define an area, within which multiple seed points are automatically generated. An ensemble segmentation can be obtained from the multiple regions that were grown. Ensemble segmentation has played an important role in many medical image applications recently [14–17] and refers to a set of different input segmentations (multiple runs using the same segmentation technique with different initializations) that are combined in order to generate a consensus segmentation. In this paper, we demonstrate that such an approach reduces interobserver variability with significantly fewer operator interactions when compared to the original algorithm.
2. Related Work
More complex methods, such as energy minimization techniques, have been proposed and have been extensively applied in many studies within the last 10 years. Graph cut methods [18–24] and active contours (snake) [25–29] are two widely used methods that have been applied in many medical imaging applications. The graph cut method has been very popular in the area of image segmentation in recent years; it constructs an image-based graph and achieves a globally optimal solution of energy minimization functions. However, the biggest problem of the conventional graph cut algorithm is its computational cost, the running time and the memory consumption restricts its feasibility for many applications. In [24], a skeleton based-graph cut algorithm was introduced to more quickly classify volume data with high quality, extract important information about interesting structures, and decrease user interaction. A comparison with a skeleton based-graph cut algorithm is done in this paper. The active contours (snake) algorithm works similarly to a stretched elastic band being released. The initial points are defined around the object to be extracted. The points then move through an iterative process to a point with the lowest energy function value. The live wire or intelligent scissor [30–33] method is motivated by the general paradigm of the active contour algorithm, it changes the segmentation problem into an optimal graph search problem via local active contour analysis. Using dynamic programming the cost function is minimized. The live wire approach requires a user interactively define the boundary by moving a mouse along the region of interest, while the live wire process automatically computes a suggested boundary. Lu and Higgins [32, 33] recently proposed a single-section live wire based on a 2D section and a single-click live wire applied directly to 3D CT images to segment central-chest lymph nodes. The single-click live wire approach is similar to the single-section live wire, but is almost completely automatic. The single-click live wire idea is similar to the one we proposed here, only requiring one single seed in the desired location. Their method was applied to handle lymph node segmentation, which is quite different from lung tumor segmentation; it may be harder or easier than lung tumor segmentation depending on the lymph node location. Another extensively used method in recent years is the level set [34–39] algorithm. The level set method was first proposed by Osher and Sethian [39] in 1988 to track moving interfaces. The main idea behind the approach is to represent a contour as the zero level set of a higher dimensional function, called a level set function, and formulate the motion of the contour as the evolution of the level set function. A number of these approaches are finding commercial application. For example, the lesion sizing toolkit [38] is an open-source tool kit for CT lung lesions with integrated lesion sizing and level set algorithms. The CT lung lesion sizing tool is first used to detect four three-dimensional features corresponding to vasculature, the lung wall, lesion boundary edges and low density background lung parenchyma. Those features are the key to the segmentation process and they potentially prevent the segmentation from bleeding into non-lesion regions. Those features then were combined into a single feature using the feature aggregator method, the segmentation manager then applies a level-set region growing algorithm starting from a seed point and expanding until feature boundaries prohibit boundary advancement. This lesion sizing algorithm was added to ISP (Interactive Science Publishing) 2.3 [40], which is a 2D and 3D volume visualization application based on VolView [41]. ISP 2.3 is publicly available and was used in the current study to compare our results with the level set methods. In recently published works, statistical learning based approaches [42] [43] show us another way to handle segmentation problem. In Wu’s work [42], a system was created to mainly detect whether a lung nodule is attached to any of the major lung anatomies. The segmentation algorithm in their system played a very important role. It uses a conditional random field (CRF) model incorporating texture features, gray-level, shape, and edge cues to improve the segmentation of the nodule boundary. However, the purpose of their system at this segmentation stage is not to provide a perfect segmentation, but to apply a fast and robust method that can create a reasonable segmentation to serve as an input to a higher-level nodule connectivity classification system. Similarly, in Tao’s work [43], early detection of ground glass nodules (GGN) in lung CT images was presented, which is a multi-level statistical learning-based framework for automatic detection and segmentation of GGN. The system seems very promising. However, our work differs from above methods as we search for tumors.
In many medical image segmentation applications, one of the major drawbacks for most of the algorithms is that they require deliberate initialization, which becomes impractical when dealing with large numbers of images, this issue has been addressed in some of the existing works already. In Lu’s work [32, 33], a single click was required for live wire algorithm and Yan’s work [44] also shows it can find the minimal path for an object from a single starting point. One of our contributions in the paper is that the SCES requires only one initialization (one starting point).
3. Methods & Materials
3.1 LuTA Analysis Workflow
The overall goal for the LuTA implementation was to accurately, precisely and efficiently enable the analysis of lesions in the lung under the guidance of an operator. A standard analysis workflow was described in detail elsewhere [10]. The workflow is briefly described in the following:
A preprocessing step. This was designed to perform a segmentation of the lung as well as other off-line tasks, such as filtering, to improve the interactive performance of the analysis.
An optional step with semi-automated correction of the segmented lung. Since lesions are commonly found to be attached to the pleural surface, it was critical to enable efficient correction of the lung boundary in cases where the boundary between juxtapleural target lesions and the pleura had not been correctly determined during the automated preprocessing step.
A “Click&Grow” step with a user selected seed based segmentation of the lesions.
An optional manual refinement step of the semi-automated lesion segmentation to ensure medical expert agreement with any results that could influence patient management.
A reporting step generating volumes and statistics about other features, such as average density.
In this paper, we focused on how to accurately segment the lung lesion with minimum human interaction; the new algorithm is basically a substitution of steps 3 & 4 above and only requires one manual seed to be entered. The preprocessing and “Click&Grow” steps within the LuTA workflow are used by our new algorithm, as described below.
3.1.1 Preprocessing
The preprocessing step performs automated organ segmentation with the main goal of segmenting the aerated lung with correct identification of the pleural wall in order to facilitate the semi-automated segmentation of juxtapleural lesions. In Figure 1, a CT image of a representative patient is shown segmented after the preprocessing step. The tumor is located in the right lung field.
Figure 1.

Lung fields (left and right) were segmented after preprocessing.
3.1.2 Click & Grow
After the preprocessing step, the lung lesions must be located in one of the lung fields. In order to segment a target lesion the image analysts identified the lesion within the segmented lung and placed a seed point in its interior - typically at the perceived center of the lesion. Starting from the seed point, an initial seed object was automatically segmented using LuTa’s region growing based on similar intensities and proximity to areas with low intensity (“air”). This Definiens proprietary region growing process approximates the object’s surface tension T using an N3 voxels sized kernel locally by calculating the ratio of the object volume inside a kernel (Vi ) to the total kernel volume (Vk), T =Vi /Vk. With this approach a high relative kernel volume of the objects surface voxels corresponds to a high surface tension. The strength of the surface tension is mainly controlled by the volume of the grown object in order to impose a smoother surface for larger objects.
From the grown region, which consists of voxels with similar density, the intensity weighted center of gravity (IWCOG) was calculated. To decrease inter and intra-reader variability, the seed point was shifted closer to the IWCOG. Additionally, an approximation of the lesion radius and volume, as well as histogram based lower and upper bounds for the intensity were extracted.
These parameters were used to define an octahedron-shaped candidate region within the lung. The new seed object was then grown into the candidate region with adaptive surface tension and intensity constraints.
The intensity constraints restrict the growth into candidate regions defined by: 1) a pre-computed intensity range of the Gaussian smoothed CT image, where the intensity range was estimated from the intensity statistics of the seed region and 2) a bound on the distance to the seed region which was calculated using a distance map. The distance map was calculated solely for the candidate region within the CNL local processing framework, and provides the minimal distance for each voxel to the seed region as an intensity value. Using the distance map ensures an approximate convexity of the seed object when growing into regions with similar intensities.
3.2 Single click ensemble segmentation
The original “Click&Grow” algorithm is very useful for delineating the tumor from the lung field in the LuTA application. If the growing process does not sufficiently capture the target lesion, the operator can place additional seed points within the lesion and repeat the growing process outlined above. Upon completion of the segmentation, the individual image objects are merged to form a single image object representing the segmented target lesion. The algorithm also provides the capability for a user to manually edit the segmentation. However, it still has the following drawbacks:
The segmentation is not consistent, different readers may generate different tumor regions.
It can require many human interactions (multiple clicks) to delineate the tumor in the case where the growing process did not sufficiently capture the target lesion.
The tumor boundary is often not satisfactory upon visual examination; sometimes it obviously includes many areas that do not belong to the target tumor.
To overcome the drawbacks of the original method, we propose a new algorithm: the single click ensemble segmentation (SCES) algorithm, which is an advanced version of the previous algorithm. The SCES makes use of the original algorithm by choosing different seed points automatically within a specified area of the lesion and performing region growing with each generated seed point. Thus, an ensemble segmentation is obtained from the multiple regions that were grown, and the final segmentation is based on a voting strategy. In order to better describe the algorithm, we provide first several definitions:
Tumor Core: the area most likely belonging to the tumor.
Manual Seed input: the first seed point provided by the user.
Start Seed point: the seed point randomly selected from the initial tumor region after shrinking.
Parent Seed point: algorithm selected seed point from a specific location of the Tumor Core.
Child Seed point: algorithm selected seed point from the outside of the existing tumor.
The detailed algorithm is described below and the detailed algorithm work flow is shown in Figure 2:
Figure 2.
Schematic illustration of the flow of the single click ensemble segmentation method.
The user provides the manual seed point and calls the “Click&Grow” algorithm to create the tumor region.
The initial tumor region created was shrunk in the x, y and z direction. (This process is to ensure step c selects a good start seed point)
Find start seed point: randomly select seed from shrunken tumor region obtained from step b.
-
Find the Tumor Core process (Figure 3): take the center of the tumor region obtained and use it as a new seed point. Perform region growing using the new seed point, and then from the new tumor region, record the pixels which are assigned to the tumor class. Repeat the above step 10 times; we will have 10 tumor regions corresponding to 10 different seed points (tumor center). The intersection of the 10 tumor regions is defined as the Tumor Core.
The center of the tumor object is defined as the following: the image object can be treated as the voxel co-ordinates (x,y,z) of a set Pv, the center of gravity of the set Pv is
Locate 10 Parent Seed points from the Tumor Core in 3D: use 3 planes: xy, yz and xz that pass through the center of the Tumor Core to divide the Tumor Core into 8 regions, the center of the Tumor Core in each region is taken as a Parent Seed point. The 9th Parent Seed point is the center of the whole Tumor Core, and the 10th one is randomly selected from the Tumor Core region.
-
For each Parent Seed point,
Apply the “Click&Grow” algorithm and obtain the corresponding tumor region.
-
Based on the tumor region obtained from the Parent Seed point, the next step involves additional tumor growing as follows: select 3 slices from the existing tumor region: center slice, center−1 and center+1 slice (Figure 4). The Child Seed point (24 in total, 8 Child Seed points for each slice) is 3 pixels away from the existing tumor boundary. For the center slice, the Child Seed point is placed at a distance of 45° successively, starting from 0° to the x-axis. Similarly, the Child Seed points of center −1 and center+1 slices were placed in a similar fashion. Their starting point is at 15° and 30° respectively. Independently apply the “Click & Grow” algorithm to each child point.
The “Click&Grow” algorithm might not always return a tumor region. Or in the case of invalid seed points the “Click&Grow” algorithm does not return a tumor volume. Even if grown successfully, the region is kept only after several conditions are satisfied. The conditions we added for the rule-set are based on the intensity mean, standard deviation, shape and connection status with the main tumor region produced by the Parent Seed point.
The new rule-sets are listed as follows:
If mean (new region) < (mean (main tumor region) - 3 * SD (main tumor region))
Then Remove new region
Else If ConRborder > 0.2 Then keep new region
Else If 0.019 < ConRborder <= 0.2 And Roundness (new region) > 0.4
Then keep new region
Else Remove new region.
where ConRborder is the ratio of the area of the new region shared with the main tumor region to its total area. The roundness feature used here is mainly to remove the vessel area that happens to grow. All parameter values set here were from empirical tests.
Merge tumor regions: simply assigns the tumor regions created from 1 Parent Seed point and 24 Child Seed points to the same class. We now have a complete tumor segmentation.
Finally, we get an ensemble segmentation consisting of 10 (since we have 10 Parent Seed points that came from the Tumor Core) different/similar tumor segmentations. We use a voting strategy to determine the final tumor region. A voxel is assigned to the tumor class if in its 3x3x3 neighborhood window half or more of the voxels were labeled tumor voxels in at least half of the segmentations.
Figure 3.

Illustration of the workflow to find the tumor core process.
Figure 4.

Expand tumor region: locate 24 secondary seed points: child Seed point selection from the existing tumor region.
3.3 Lesion Dataset
The patient dataset (129 patients) from the H. Lee Moffitt Cancer Center and Research Institute in Tampa, FL, was used to evaluate the new segmentation algorithm. The dataset contains stage I-II NSCLC patients. The standard-of-care clinical CT scans are high resolution with contrast enhancement. The slice thicknesses for the CT scans vary among 3mm, 4mm, 5mm and 6mm. However, 5mm is the most common. The tumor types include both adeno- and squamous cell carcinoma. They were chosen because they cover the major types of lung tumor and were of reasonable resolution.
3.4 Metrics
The similarity index formula:
| (1) |
was used to evaluate the tumor segmentation, in which, VA and VB are two delineated tumor volumes (e.g. manual or automatic) respectively. The segmentation in the proposed single click ensemble segmentation algorithm was tested on lung tumors from the Moffitt dataset, which contained 129 patients in total. We first selected 15 cases out of 129 from the dataset, since most of these 15 cases required many human interactions (including multiple clicks and manual editing). Two expert readers used the “Click&Grow” method in addition to manual editing operations to delineate the lung tumor in those 15 cases. The segmentation results from the two readers were marked as R1 and R2 respectively. Reader 2 provided the manual seed input for the SCES algorithm whose result was labeled SCES. We also show results from the level set (LS) algorithm provided by ISP 2.3 [40] and skeleton graph cut (SGC) [24] by 3DMed [45]. The ISP provides a wizard to perform one-click segmentations of lesions. In this approach, the user identifies the region containing the lesion with a box, places the seed within the lesion and selects the lesion type (solid or part-solid). The software then segments the lesion. All the operations for the level set algorithm were performed by one of the readers. These include the selection of the seed point, the surrounding box for the tumor and the lesion type. The skeleton graph cut algorithm requires three initializations for most of the cases. In the segmentation process, we select seed points by painting the lesion and healthy parenchyma in the CT volume using a 3D brush. Each lesion region needs at least one seed and the healthy region needs at least two seeds.
We also tested the stability of the new algorithm for 129 cases from the Moffitt Cancer Center by providing 20 different start seed points (the 20 points were randomly generated inside the shrunken tumor region by the computer (detailed algorithm: step c)) for each case and then the similarity index for each case was calculated as in Equation (2):
| (2) |
Where i ∈[1,129] is the case index, SIim,in, is the similarity index from Equation (1) and measures the similarity between the two segmentation results from different start seed points for the same case i.
4. Results
In Table 1, we show a comparison of the tumor segmentation results from people and algorithms; The average SI value over 15 cases for R1 vs. R2 is 79.53%, R1 vs. SCES is 78.29%, R2 vs. SCES is 77.72%, R1 vs. LS is 65.57%, R2 vs. LS is 64.47%, SCES vs. LS is 63.77%, SCES vs. SGC is 63.76%, SGC vs. R1 is 63.49%, SGC vs. R2 is 63.07% and SGC vs. LS is 61.15%. The number of human interactions for R1, R2, SCES, SGC and LS are shown in Figure 5. The average number of interactions from reader 1 was 6, from reader 2 was 3.87, SGC was 3.4, and SCES was always 1, which is the initial manual seed input, and for LS was 5.33. The level set algorithm in ISP requires the user to identify the box that contains the targeted lesion before doing the segmentation, which we counted as 4 operations to create the box, plus the initial seed point within the tumor region, we then get 5 user interactions for each case. However, for case P11, we did not get a segmentation result after the first trial, therefore we did the same procedure again with a different box and seed point. In total we counted 10 operations for this case. On the other hand, for one operator the LS approach only had to be re-done one time. The skeleton graph cut algorithm requires at least 3 initializations, however for cases P9 and P11, 2 seeds were required for the lesion object and 4 seeds for the healthy region. The SCES approach has the advantage in that it only required one simple input, as compared to multiple operator inputs used in other methods.
Table 1.
Pixel by pixel segmentation comparison (R1=reader1, R2=reader2, SCES=single click ensemble segmentation, LS=level set, SGC=skeleton graph cut, p-value from student t-test for each comparison was calculated based on R1 vs. R2). The fraction indicates the portion of pixels in agreement.
| Case | SI value % (R1 VS R2) | SI value % (R1 VS SCES) | SI value % (R2 VS SCES) | SI Value % (R1 VS LS) | SI Value % (R2 VS LS) | SI Value % (SCES VS LS) | SI Value % (SCES VS SGC) | SI Value % (SGC VS R1) | SI Value % (SGC VS R2) | SI Value % (SGC VS LS) |
|---|---|---|---|---|---|---|---|---|---|---|
| P1 | 93.14 | 77.95 | 77.67 | 65.5 | 62.73 | 54.19 | 80.41 | 70.08 | 69.75 | 52.16 |
| P2 | 83.43 | 81.86 | 85.83 | 71.91 | 77.07 | 77.60 | 73.99 | 63.72 | 73.66 | 70.45 |
| P3 | 64.23 | 82.13 | 72.37 | 60.19 | 64.36 | 65.64 | 62.59 | 55.27 | 63.62 | 62.71 |
| P4 | 63.77 | 63.52 | 75.11 | 62.26 | 66.49 | 58.72 | 26.63 | 35.9 | 32.28 | 40.37 |
| P5 | 77.57 | 67.00 | 73.40 | 65.61 | 63.48 | 62.22 | 45.77 | 51.75 | 52.17 | 44.84 |
| P6 | 75.79 | 78.06 | 81.37 | 64.56 | 54.34 | 55.37 | 57.54 | 69.26 | 58.19 | 62.28 |
| P7 | 94.90 | 97.13 | 93.61 | 81.48 | 79.18 | 82.36 | 79.36 | 77.94 | 78.01 | 77.66 |
| P8 | 84.11 | 69.70 | 71.51 | 63.86 | 53.73 | 54.01 | 68.6 | 72.11 | 61.29 | 59.13 |
| P9 | 78.65 | 80.49 | 72.57 | 55.53 | 54.18 | 48.11 | 39.06 | 44.59 | 43.06 | 40.58 |
| P10 | 90.20 | 81.36 | 84.27 | 60.59 | 66.66 | 69.25 | 79.91 | 70.48 | 75.13 | 65.94 |
| P11 | 78.23 | 76.34 | 71.23 | 54.71 | 55.55 | 59.97 | 66.47 | 53.77 | 53.93 | 55.52 |
| P12 | 73.52 | 88.85 | 68.93 | 67.04 | 65.51 | 62.24 | 63.95 | 67.27 | 70.11 | 75.32 |
| P13 | 76.30 | 84.48 | 88.04 | 78.96 | 80.84 | 84.10 | 80.22 | 73.58 | 77.79 | 76.58 |
| P14 | 82.30 | 78.41 | 81.67 | 56.12 | 52.99 | 60.69 | 75.68 | 74.81 | 73.15 | 62.05 |
| P15 | 76.80 | 67.02 | 68.25 | 75.18 | 69.89 | 62.04 | 56.19 | 71.88 | 63.9 | 71.63 |
| Average | 79.53 | 78.29 | 77.72 | 65.57 | 64.47 | 63.77 | 63.76 | 63.49 | 63.07 | 61.15 |
| p-value | - | 0.7062 | 0.5592 | 0.0002 | 0.0002 | 0.0002 | 0.0028 | 0.0004 | 0.0004 | <0.000001 |
Figure 5.
Number of interactions required for each method (R1=reader1, R2=reader2, SCES=single click ensemble segmentation, LS=level set algorithm, SGC=skeleton graph cut algorithm).
In Table 1, we calculated the P values of the student t-test for each comparison (with R1 vs. R2 as a reference). The P values for R1 vs. SCES and R2 vs. SCES are 0.7062 and 0.5592, other comparison’s P-values are very small and we can conclude that there are significant differences between the associated methods and the manual segmentations. For the SCES algorithm, the differences from manual segmentation are not statistically significant.
We further examined the stability of the level set algorithm and skeleton graph cut algorithm by providing initializations from two readers, to determine how much agreement there is when using the two algorithms twice. We only compared them on 15 cases for which multiple reader results exist. There was an average SI of 84.21% with 12.94% standard deviation for the level set algorithm and an average SI of 70.47% with 20.15% standard deviation for the skeleton graph cut. Our approach was more stable with an SI of 86.51% and 4.51% standard deviation (Table 2). Based on the average and standard deviation values, our approach seems better than level set, but the difference was not statistically significant (Wilcoxon signed-rank test shows we cannot reject the null hypothesis). However, there is a statistically significant difference between our approach and the skeleton graph cut according to the Wilcoxon signed-rank test at confidence level 0.01, which indicates our approach is more stable than the skeleton graph cut algorithm.
Table 2.
Pixel by pixel segmentation comparison (SCES1=single click ensemble segmentation (manual seed input provided by 1st reader), SCES2=single click ensemble segmentation (manual seed input provided by 2nd reader), LS1=level set (1st reader), LS2=level set (2nd reader), SGC1=skeleton graph cut (1st reader), SGC2=skeleton graph cut (2nd reader)). The fraction indicates the portion of pixels in agreement.
| Case | SI Value % (SCES1 VS SCES2) | SI value % (LS1 VS LS2) | SI value % (SGC1 VS SGC2) |
|---|---|---|---|
|
| |||
| P1 | 89.72 | 83.48 | 88.67 |
| P2 | 92.48 | 80.46 | 52.34 |
| P3 | 84.71 | 96.74 | 62.62 |
| P4 | 81.04 | 85.26 | 53.83 |
| P5 | 85.20 | 88.39 | 63.72 |
| P6 | 80.18 | 71.53 | 67.97 |
| P7 | 86.31 | 98.36 | 83.45 |
| P8 | 92.22 | 97.54 | 91.12 |
| P9 | 81.59 | 51.36 | 23.13 |
| P10 | 89.36 | 97.75 | 86.39 |
| P11 | 94.92 | 75.19 | 50.33 |
| P12 | 82.57 | 88.95 | 89.85 |
| P13 | 85.59 | 82.51 | 94.1 |
| P14 | 83.50 | 71.82 | 83.97 |
| P15 | 88.27 | 93.91 | 65.58 |
| Average | 86.51 | 84.21 | 70.47 |
| SD | 4.51 | 12.94 | 20.15 |
Figure 6 shows the stability of the proposed algorithm when providing 20 different Start Seed points. The purpose of this experiment was to test the sensitivity of the segmentation algorithm. The average SI for 129 cases is above 93% and it reflects that the segmentation is very stable, no matter where the Start Seed points are located in the tumor region.
Figure 6.
Similarity Index of SCES algorithm for 129 cases (20 different start seed points) from the Moffitt Cancer Center.
Five segmentation results: Reader 1, Reader 2, single click ensemble segmentation, level set and skeleton graph cut are shown in Figures 7–9. The results of SCES seem to follow the tumor boundary and visually look better than the result from the readers and level set algorithm, the skeleton graph cut algorithm result looks very compact, however many vessel region have been segmented together with tumor region.
Figure 7.

Representative segmentation results (P13, slices 79–85) (a) Reader 1 (b) Reader 2 (c) SCES (d) Level Set (e) Skeleton Graph Cut.
Figure 9.
Representative segmentation results (P14, slices 44–54) (a) Reader 1 (b) Reader 2 (c) SCES (d) Level Set (e) Skeleton Graph Cut.
5. Discussion
The new algorithm has been tested on a large patient dataset. Figure 5 highlights perhaps the biggest advantage of the SCES algorithm, it only requires 1 human interaction, that is the manual seed input. The new algorithm saves a lot of human interaction while agreeing well with the results from a human expert. We compared the new segmentation result with the results from two expert readers. It is of note that the results from two readers agreed on 79.53% of the voxels. The average number of interactions involved for each reader was 6 and 3.87 respectively as shown in Figure 5. The SI results between SCES and R1, SCES and R2 are 78.29% and 77.72%, which implies that the new result is close to the reader’s 79.53%, SCES and the Level set method are not very close, the agreement is only 63.77%, the agreement between SCES and SGC is 63.76%. From the visual result of Figures 7–8, we can also conclude that the result of SCES algorithm seems to follow the tumor boundary on CT images, as well as the results of level set and graph cut. However, Figure 9 shows us a different scenario. Starting from slice 47 in Figure 9, the level set algorithm didn’t follow the boundary of the tumor and many tumor regions got cut off, which resulted in a bad segmentation. The skeleton graph cut algorithm seems to be performing well on this case, but the major issue for SGC again is getting too much vessel tissue. Our algorithm also started failing at slice 54, but it seems it performed better than the level set algorithm. Our new algorithm didn’t explore the search space entirely (Child Seed Point) in the Z-direction due to computational time limitations. As a result the top/bottom of a “big” tumor region will not be detected by the algorithm (in Figure 9, only the top of the tumor region was not correctly detected). We only performed the search on 3 slices in the new algorithm. Region growing helps find the additional tumor region in the Z-direction, but the growing in Z-direction stopped when it reached the criteria, which will result in incomplete tumor segmentation for some big tumor cases.
Figure 8.

Representative segmentation results (P10, slices 46–50) (a) Reader 1 (b) Reader 2 (c) SCES (d) Level Set (e) Skeleton Graph Cut.
For the stability test result shown in Figure 6, there are a few cases that have a similarity index lower than 60%. Further investigation found that most of them have a part-solid tumor [46] (Figure 10) with low density and their lesion boundary is not well defined. For those cases, SCES (and LS) was not stable, additional work needs to be done, one possible method “Shrink & Wrap” may be used instead of the “Click & Grow” method, which requires a pre-defined boundary surrounding the tumor area. Another solution is utilizing more than one modality while doing the segmentation (PET-CT based). Such a method has been reported which can reduce interobserver variability and increase delineation accuracy [47]. We also found many cases had 100% agreement in Figure 6, which indicates that they always found the same Tumor Core with a different Start Seed point, this usually took place for Solid tumor cases. Since the Parent Seed point and Child Seed points for SCES were kept unchanged when the Tumor Core was the same, this eventually resulted in same tumor segmentations.
Figure 10.
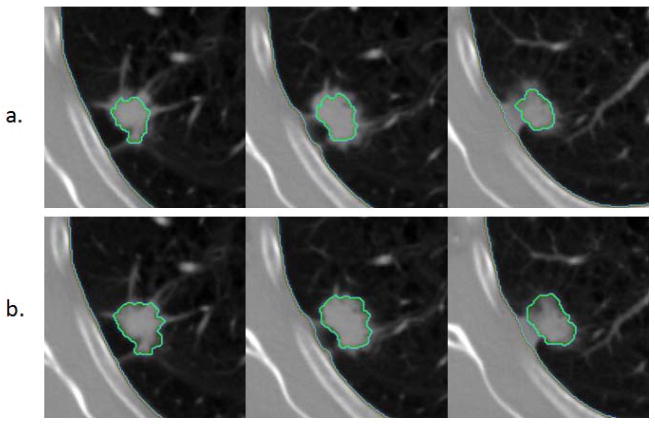
Single click ensemble segmentation result with a different start seed point (a and b) on part-solid tumors.
Future enhancements will focus on part-solid tumor types, which result in unstable tumor boundaries when given different start seed points. The problem of incomplete tumor segmentation with big tumors is also a remaining challenge.
The lung tumor segmentation problem in our study can mainly be divided into two parts: lung segmentation and lung lesion segmentation. These are two separate algorithms but the 2nd part relies on the quality of the lung segmentation. In our experiments, we sometimes found the lung segmentation failed. In other words, the tumor in which we are interested may be excluded by the lung segmentation process. However one of the important assumptions of our lung tumor segmentation is that the lung tumor must be inside of the lung field. Our newly designed algorithm cannot intelligently correct for any tumor that was excluded from the lung field. If such a case occurs, an optional step with semi-automated correction of the segmented lung must be done before using our lung lesion segmentation algorithm.
6. Conclusions
We proposed a stable, accurate and automatic single click ensemble segmentation algorithm in this paper. The important component of this work is to reduce the human interactions while lesion delineation remains accurate and consistent as a result of ensemble segmentation. Though the computation time was increased for each case since multiple “Click&Grow” algorithms were applied, we saved a lot of manpower (average (6+3.87)/2=4.94 clicks from two readers for the 15 cases). With the automated batch mode method built into the Definiens’ software platform, the lung lesion segmentation workload has been tremendously reduced (in other words, we can save a significant amount of valuable time for readers/oncologist/radiologists in the segmentation of lung tumors), the only thing requiring human interaction is the choice of the manual seed input. The aim of the algorithm is to provide a delineation tool that can be used by many readers to obtain the same segmentation results. The tumor segmentation should not differ much with different manual seeds provided by different readers. The single click ensemble segmentation algorithm we proposed here is a nice upgrade. As the new algorithm evolves to better address some issues we discussed above, it will become an even more powerful tool that can be further tested in clinical environment and it will be also very useful in future multi-center clinical trials.
Highlights.
We proposed an automatic, stable and accurate segmentation algorithm for lung tumor CT scans.
The approach requires just 1 seed point to obtain a good segmentation.
High agreement between new algorithm and two reader’s results.
It is consistent, the average SI is above 93% using 20 different start seeds.
Acknowledgments
This work is supported by grant U01CA143062-01, Radiomics of NSCLC from the National Institutes of Health.
Biographies
Yuhua Gu
He received M.S. in Department of Mathematics in 2003 and the Ph.D. degree in Department of CSE in 2009 from University of South Florida. He is currently working as Postdoctoral Fellow at Moffitt Cancer Center, FL, USA. His research interests include image processing, data mining and machine learning.
Virendra Kumar
He received the M.Sc in biotechnology from IIT - Roorkee, India, in 2000 and the Ph.D. from All India Institute of Medical Sciences, India in 2007. He is currently working as Postdoctoral Fellow at Moffitt Cancer Center, Tampa, FL, USA. His research interests are currently focused on medical image analysis.
Lawrence O. Hall
Lawrence O. Hall is a Distinguished University Professor and Chair of the Department of CSE at University of South Florida. He is a fellow of the IEEE and IAPR. His research interests lie in distributed machine learning, extreme data mining, bioinformatics, pattern recognition and integrating AI into image processing.
Dmitry B. Goldgof
Professor Dmitry B. Goldgof (Fellow IEEE, IAPR) is Professor and Associate Chair, Department of Computer Science & Engineering, University of South Florida. His research interests include Image and Video Analysis, Pattern Recognition and Bioengineering. Dr. Goldgof has graduated 19 Ph.D. students, has published over 65 journal and 160 conference papers.
Ching-Yen Lee
He is a PH.D student of Ultrasound Image Laboratory, Department of Biomedical Engineering and Bioinformatics, National Taiwan University. His research interests include image Processing and digital signal processing.
René Korn
René Korn has a PhD equivalent in Bioinformatics and a diploma in Mathematics. He is Product Manager Clinical Applications at Definiens AG, Germany. In his role he is responsible for the radiology product portfolio, containing image analysis applications for volumetric quantification. He has (co-)authored about 20 peer-reviewed scientific publications.
Claus Bendtsen
Claus Bendtsen holds a Ph.D. in applied mathematics from the Danish Technical University (DK). After spending several years in academia he joined the pharmaceutical industry: first Merck & Co., then Novartis and he is currently section head for computational biology at AstraZeneca.
Emmanuel Rios Velazquez
He received his BSc and MSc in Biomedical Engineering from the National Polytechnic Institute (Mexico, 2007) and the Eindhoven University of Technology (The Netherlands, 2009), respectively. Currently he is a PhD candidate in department of radiation oncology, Maastricht University. His research interests include intelligent decision support systems and medical imaging.
Andre Dekker
Andre Dekker is a medical physicist specialized in information technology and head of the division of knowledge engineering at MAASTRO Clinic, The Netherlands. He is current projects aim to develop a data pooling and machine learning network between radiotherapy institutions. He has (co-)authored more than 50 peer-reviewed articles.
Hugo Aerts
Hugo Aerts is a scientist and project-leader of the Radiomics research line at MAASTRO Clinic, The Netherlands. His scientific interests include advanced image analysis, system-biology, bioinformatics and artificial intelligence. He has (co-)authored more than 25 peer-reviewed journal articles and currently supervises three PhD students.
Philippe Lambin
Philippe Lambin is a world renowned radiation oncologist who is a thought leader in the application of evidence based medicine to clinical practice at MAASTRO Clinic, The Netherlands. His expertise is in the incorporation of medical and imaging data into predictive models of patient response to radiation therapy.
Xiuli Li
Xiuli Li ia a Ph.D. candidate in Computer Science and Technology, Intelligent Medical Research Center, Institute of Automation, Chinese Academy of Sciences, Beijing, China. Her research interests are mainly focused on medical image processing, including medical image segmentation, statistical shape modeling.
Jie Tian
Jie Tian received the Ph.D degree from the Institute of Automation, Chinese Academy of Sciences, China, in 1992. His research interests include the medical image process and analysis and pattern recognition. Dr. Tian is the Beijing Chapter Chair of The Engineering in Medicine and Biology Society of the IEEE.
Robert A. Gatenby
Robert Gatenby is chairman of the Radiology and Director of the Integrated Mathematical Oncology program at the Moffitt Cancer Center. He has published numerous articles in Nature and is on the editorial board of Molecular Cancer Therapeutics. His research interests include quantitative imaging and mathematical modeling of tumor size and growth.
Robert J. Gillies
Robert Gillies is vice-chair of Radiology and Director of the Imaging Research program at the Moffitt Cancer Center. He is President of the Society of Molecular Imaging and serve as a member in NIH grant review sections. His research interests include quantitative imaging biomarkers, tumor microenvironment and targeting ligands.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Johnson DH, Blot WJ, Carbone DP. Cancer of the lung: Non-small cell lung cancer and small cell lung cancer. In: Abeloff MD, Armitage JO, Niederbuber JE, Kastan MB, McKenna WG, editors. Abeloff’s clinical oncology. Churchill Livingstone/Elsevier; Philadelphia: 2008. [Google Scholar]
- 2.Gillies RJ, Anderson AR, Gatenby RA, Morse DL. The biology underlying molecular imaging in oncology: from genome to anatome and back again. Clin Radiol. 2010;65:517–521. doi: 10.1016/j.crad.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rexilius J, Hahn HK, Schluter M, Bourquain H, Peitgen HO. Evaluation of accuracy in MS lesion volumetry using realistic lesion phantoms. Acad Radiol. 2005;12:17–24. doi: 10.1016/j.acra.2004.10.059. [DOI] [PubMed] [Google Scholar]
- 4.Tai P, Van Dyk J, Yu E, Battista J, Stitt L, Coad T. Variability of target volume delineation in cervical esophageal cancer. Int J Radiat Oncol Biol Phys. 1998;42:277–288. doi: 10.1016/s0360-3016(98)00216-8. [DOI] [PubMed] [Google Scholar]
- 5.Cooper JS, Mukherji SK, Toledano AY, Beldon C, Schmalfuss IM, Amdur R, Sailer S, Loevner LA, Kousouboris P, Ang KK, Cormack J, Sicks J. An evaluation of the variability of tumor-shape definition derived by experienced observers from CT images of supraglottic carcinomas (ACRIN protocol 6658) Int J Radiat Oncol Biol Phys. 2007;67:972–975. doi: 10.1016/j.ijrobp.2006.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hojjatoleslami S, Kittler J. Region growing: A new approach, Image Processing. IEEE Transactions on. 1998;7:1079–1084. doi: 10.1109/83.701170. [DOI] [PubMed] [Google Scholar]
- 7.Dehmeshki J, Amin H, Valdivieso M, Ye X. Segmentation of pulmonary nodules in thoracic CT scans: a region growing approach, Medical Imaging. IEEE Transactions on. 2008;27:467–480. doi: 10.1109/TMI.2007.907555. [DOI] [PubMed] [Google Scholar]
- 8.Dijkers J, Van Wijk C, Vos F, Florie J, Nio Y, Venema H, Truyen R, van Vliet L. Segmentation and size measurement of polyps in CT colonography. Medical Image Computing and Computer-Assisted Intervention–MICCAI. 2005;2005:712–719. doi: 10.1007/11566465_88. [DOI] [PubMed] [Google Scholar]
- 9.Le Lu AB, Wolf M, Liang J, Salganicoff M, Comaniciu D. Accurate polyp segmentation for 3D CT colongraphy using multi-staged probabilistic binary learning and compositional model. IEEE Conference on Computer Vision and Pattern Recognition; 2008. pp. 1–8. [Google Scholar]
- 10.Bendtsen C, Kietzmann M, Korn R, Mozley P, Schmidt G, Binnig G. X-Ray Computed Tomography: Semiautomated Volumetric Analysis of Late-Stage Lung Tumors as a Basis for Response Assessments. International Journal of Biomedical Imaging. 2011;2011:Article ID 361589. doi: 10.1155/2011/361589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Athelogou M, Schmidt G, Schäpe A, Baatz M, Binnig G. Cognition Network Technology–A Novel Multimodal Image Analysis Technique for Automatic Identification and Quantification of Biological Image Contents. Imaging cellular and molecular biological functions. 2007:407–422. [Google Scholar]
- 12.Definiens AG. www.definiens.com.
- 13.Gu Y, Kumar V, Hall LO, Goldgof DB, Korn R, Bendtsen C, Gatenby RA, Gillies RJ. Automated Delineation of Lung Tumors from CT Images: Method and Evaluation. World Molecular Imaging Congress; San Diego, CA, USA. 2011. p. 373. [Google Scholar]
- 14.Huo J, Okada K, Pope W, Brown M. Sampling-based ensemble segmentation against inter-operator variability. Proc. SPIE; 2011. p. 796315. [Google Scholar]
- 15.Huo J, van Rikxoort EM, Okada K, Kim HJ, Pope W, Goldin J, Brown M. Confidence-based ensemble for GBM brain tumor segmentation. Proc. SPIE; 2011. p. 79622P. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Allende PB, Conde OM, Krishnaswamy V, Hoopes PJ, Pogue BW, Mirapeix J, Lopez-Higuera JM. Automated ensemble segmentation of epithelial proliferation, necrosis, and fibrosis using scatter tumor imaging. Proc. SPIE; 2010. p. 77151B. [Google Scholar]
- 17.Oost E, Akatsuka Y, Shimizu A, Kobatake H, Furukawa D, Katayama A. Vessel segmentation in eye fundus images using ensemble learning and curve fitting. IEEE International symposium on Biomedical Imaging: From Nano to Macro; 2010. pp. 676–679. [Google Scholar]
- 18.Boykov Y, Veksler O, Zabih R. Fast approximate energy minimization via graph cuts. IEEE Transactions on pattern analysis and machine intelligence. 2001:1222–1239. [Google Scholar]
- 19.So RWK, Tang TWH, Chung A. Non-rigid image registration of brain magnetic resonance images using graph-cuts. Pattern Recognition. 2011;44:2450–2467. [Google Scholar]
- 20.Xu N, Bansal R, Ahuja N. Object segmentation using graph cuts based active contours. IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR ‘03); 2003. p. 46. [Google Scholar]
- 21.Slabaugh G, Unal G. Graph cuts segmentation using an elliptical shape prior. IEEE International Conference on Image Processing; 2005. pp. II-1222–1225. [Google Scholar]
- 22.Liu X, Veksler O, Samarabandu J. Graph cut with ordering constraints on labels and its applications. IEEE Conference on Computer Vision and Pattern Recognition; 2008. pp. 1–8. [Google Scholar]
- 23.Ye X, Beddoe G, Slabaugh G. Automatic graph cut segmentation of lesions in CT using mean shift superpixels. Journal of Biomedical Imaging. 2010;2010:19. doi: 10.1155/2010/983963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dehui X, Jie T, Fei Y, Qi Y, Xing Z, Qingde L, Xin L. Skeleton Cuts-An Efficient Segmentation Method for Volume Rendering, Visualization and Computer Graphics. IEEE Transactions on. 2011;17:1295–1306. doi: 10.1109/TVCG.2010.239. [DOI] [PubMed] [Google Scholar]
- 25.Liu W, Zagzebski JA, Varghese T, Dyer CR, Techavipoo U, Hall TJ. Segmentation of elastographic images using a coarse-to-fine active contour model. Ultrasound in medicine & biology. 2006;32:397–408. doi: 10.1016/j.ultrasmedbio.2005.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He Q, Duan Y, Miles J, Takahashi N. A context-sensitive active contour for 2D corpus callosum segmentation. International Journal of Biomedical Imaging. 2007;2007:24826. doi: 10.1155/2007/24826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C, Li H, Zhou X, Wong S. Constraint factor graph cut–based active contour method for automated cellular image segmentation in RNAi screening. Journal of microscopy. 2008;230:177–191. doi: 10.1111/j.1365-2818.2008.01974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki K, Kohlbrenner R, Epstein ML, Obajuluwa AM, Xu J, Hori M. Computer-aided measurement of liver volumes in CT by means of geodesic active contour segmentation coupled with level-set algorithms. Medical Physics. 2010;37:2159. doi: 10.1118/1.3395579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Li C, Sun Q, Xia D, Kao CY. Active contours driven by local and global intensity fitting energy with application to brain MR image segmentation. Computerized Medical Imaging and Graphics. 2009;33:520–531. doi: 10.1016/j.compmedimag.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 30.Mortensen EN, Barrett WA. Interactive segmentation with intelligent scissors. Graphical Models and Image Processing. 1998;60:349–384. [Google Scholar]
- 31.Souza A, Udupa JK, Grevera G, Sun Y, Odhner D, Suri N, Schnall MD. Iterative live wire and live snake: new user-steered 3D image segmentation paradigms. Proceedings of SPIE Medical Imaging: Physiology, Function, and Structure from Medical Images; March 2006; pp. 1159–1165. [Google Scholar]
- 32.Lu K, Higgins WE. Interactive segmentation based on the live wire for 3D CT chest image analysis. International Journal of Computer Assisted Radiology and Surgery. 2007;2:151–167. [Google Scholar]
- 33.Lu K, Higgins WE. Segmentation of the central-chest lymph nodes in 3D MDCT images. Comput Biol Med. 2011;41:780–789. doi: 10.1016/j.compbiomed.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sethian JA. Level set methods and fast marching methods : evolving interfaces in computational geometry, fluid mechanics, computer vision, and materials science. 2. Cambridge University Press; Cambridge, U.K.; New York: 1999. [Google Scholar]
- 35.Malladi R, Sethian JA, Vemuri BC. Shape modeling with front propagation: A level set approach, Pattern Analysis and Machine Intelligence. IEEE Transactions on. 1995;17:158–175. [Google Scholar]
- 36.Gao H, Chae O. Individual tooth segmentation from CT images using level set method with shape and intensity prior. Pattern Recognition. 2010;43:2406–2417. [Google Scholar]
- 37.Chen YT. A level set method based on the Bayesian risk for medical image segmentation. Pattern Recognition. 2010;43:3699–3711. [Google Scholar]
- 38.Krishnan K, Ibanez L, Turner WD, Jomier J, Avila RS. An open-source toolkit for the volumetric measurement of CT lung lesions. Optics Express. 2010;18:15256–15266. doi: 10.1364/OE.18.015256. [DOI] [PubMed] [Google Scholar]
- 39.Osher S, Sethian JA. Fronts propagating with curvature-dependent speed: algorithms based on Hamilton-Jacobi formulations. Journal of computational physics. 1988;79:12–49. [Google Scholar]
- 40.Interactive Scientific Publications (ISP 2.3) software. http://www.opticsinfobase.org/isp.cfm.
- 41.Volview software. http://www.kitware.com/products/volview.html.
- 42.Wu D, Lu L, Bi J, Shinagawa Y, Boyer K, Krishnan A, Salganicoff M. Stratified learning of local anatomical context for lung nodules in CT images. IEEE; 2010. pp. 2791–2798. [Google Scholar]
- 43.Tao Y, Lu L, Dewan M, Chen A, Corso J, Xuan J, Salganicoff M, Krishnan A. In: Multilevel Ground Glass Nodule Detection and Segmentation in CT Lung Images Medical Image Computing and Computer-Assisted Intervention – MICCAI 2009. Yang G-Z, Hawkes D, Rueckert D, Noble A, Taylor C, editors. Springer; Berlin / Heidelberg: 2009. pp. 715–723. [DOI] [PubMed] [Google Scholar]
- 44.Pingkun Y, Kassim AA. Medical Image Segmentation Using Minimal Path Deformable Models With Implicit Shape Priors, Information Technology in Biomedicine. IEEE Transactions on. 2006;10:677–684. doi: 10.1109/titb.2006.874199. [DOI] [PubMed] [Google Scholar]
- 45.Tian J, Xue J, Dai Y, Chen J, Zheng J. A novel software platform for medical image processing and analyzing. IEEE Trans Inf Technol Biomed. 2008;12:800–812. doi: 10.1109/TITB.2008.926395. [DOI] [PubMed] [Google Scholar]
- 46.Henschke CI, Yankelevitz DF, Mirtcheva R, McGuinness G, McCauley D, Miettinen OS. CT screening for lung cancer: frequency and significance of part-solid and nonsolid nodules. American Journal of Roentgenology. 2002;178:1053. doi: 10.2214/ajr.178.5.1781053. [DOI] [PubMed] [Google Scholar]
- 47.van Baardwijk A, Bosmans G, Boersma L, Buijsen J, Wanders S, Hochstenbag M, van Suylen RJ, Dekker A, Dehing-Oberije C, Houben R. PET-CT-based auto-contouring in non-small-cell lung cancer correlates with pathology and reduces interobserver variability in the delineation of the primary tumor and involved nodal volumes. International Journal of Radiation Oncology*Biology*Physics. 2007;68:771–778. doi: 10.1016/j.ijrobp.2006.12.067. [DOI] [PubMed] [Google Scholar]