Abstract
Psychiatric disorders are highly heritable, and in many individuals likely arise from the combined effects of genes and the environment. A substantial body of evidence points towards DISC1 being one of the genes that influence risk of schizophrenia, bipolar disorder and depression, and functional studies of DISC1 consequently have the potential to reveal much about the pathways that lead to major mental illness. Here, we review the evidence that DISC1 influences disease risk through effects upon multiple critical pathways in the developing and adult brain.
Keywords: DISC1, schizophrenia, depression, genetics, neural pathways
1 Introduction
Schizophrenia and bipolar disorder jointly affect approximately 2% of the world’s population and, together with depression, account for three of the top ten leading causes of disability (measured as missed days at work) (Lopez and Murray, 1998). These disorders are devastating for the affected individual and their family, and they impact upon society as a whole at many levels. Indeed, in terms of financial burden, the cost of schizophrenia alone was estimated to be >$60 billion in the United States in 2002 (McEvoy, 2007). Consequently, significant effort has been put into understanding the causes of major mental illness and finding effective treatments. Schizophrenia, bipolar disorder and depression are highly heritable, thus one approach towards understanding the underlying disease mechanisms is to identify the genes that influence disease risk. For psychiatric illness, this is proving extremely difficult, likely due to interplay between the effects of multiple genes plus environmental influences. In fact, psychiatric disorders are considered to be common complex genetic disorders, with polygenic or even oligogenic causation in some individuals (Porteous, 2008;Gershon et al., 2011;Sullivan et al., 2012). However, a small number of genes are emerging as convincing risk factors, at least in some patients, including Disrupted In Schizophrenia 1 (DISC1). In this review we will discuss the evidence supporting the role of DISC1 in risk of major mental illness. We will then go on to review what functional studies of DISC1 have revealed about its role in the brain, and hence the molecular events that may underlie psychiatric illness (Figure 1).
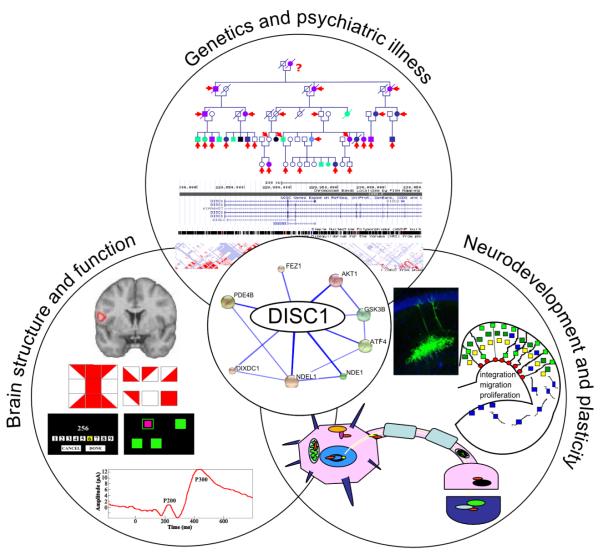
Figure 1.
DISC1 in neurodevelopment, brain functioning and psychiatric illness. DISC1 is a scaffold protein with many functional partners associated with brain development and disease. Genetics and psychiatric illness: The DISC1 protein network was modified from String DB (http://string-db.org/). The t(1;11) Scottish pedigree was modified, with permission, from Blackwood et al. (Blackwood et al., 2001) . A fragment of the DISC1 gene structure, SNPs and linkage disequilibrium blocks was generated using the UCSC genome browser (http://genome.ucsc.edu/). Brain structure and function: Examples of neurocognitive tests in which individuals carrying variants at the DISC1 locus show altered performance; differences in brain activation by DISC1 L607F genotype as measured by functional MRI (with thanks to Andrew McIntosh); the P300 event related potential, which is elicited in decision making, and shows reduced amplitude and latency in DISC1 translocation carriers. Neurodevelopment and plasticity: DISC1 is a multi-compartmentalised protein, present at the nucleus, centrosome, mitochondria, microtubules and synapse; DISC1 has roles in neurodevelopment and plasticity, in proliferation, migration and integration of neurons; neural stem cells are a promise in research and treatment of psychiatric and neurodegenerative disorders; GFP-labelled neural stem cells transplanted into the mouse brain can give rise to cells resembling normal hippocampal neurons (Image by Yirui Sun, Wellcome Imagescc).
2 The t(1;11)(q42.1,q14.3) translocation
In 1970 a survey of chromosomal rearrangements in samples referred for clinical cytogenetic analysis identified a reciprocal chromosomal t(1:11) translocation in an individual with adolescent conduct disorder. Investigation of the extended family generated a genome-wide significant linkage score for co-segregation of the translocation with multiple cases of schizophrenia and major depressive disorder and a single case of bipolar disorder (logarithm of the odds ratio (LOD) = 7.1). The linkage to schizophrenia alone also reached genome-wide significance (LOD= 3.6), as did linkage to affective disorders (LOD=4.5) (Blackwood et al., 2001). Inheritance of the translocation thus confers significantly increased risk of developing these disorders. However, it should be noted that penetrance is incomplete and there are family members with a normal karyotype who have been diagnosed with a psychiatric disorder, although none with major mental illness (Blackwood et al., 2001). This translocation is unique to this one Scottish family, and is one of the most convincing causative mutations identified for major mental illness to date. Studying the effects of this translocation therefore has the potential to reveal much about the pathways underlying psychiatric disorders.
The P300 event-related potential (ERP), a measure of the speed and efficiency of information processing, has been examined in multiple members of the t(1;11) family because it is believed to provide a measure of brain function that is independent of the environmental influences that affect disease penetrance and presentation. Translocation carriers were found to have significantly reduced P300 amplitude and latency, similar to subjects with schizophrenia, but different from karyotypically normal family members and unrelated controls (Blackwood et al., 2001). Furthermore, the translocation carriers in the study included individuals with no psychiatric diagnosis, indicating that inheritance of the translocation results in subtle disturbances of brain function in all individuals, even though it only leads to major psychiatric illness in a subgroup. A major unanswered question then, is how this mutation has such variable penetrance and presentation.
We demonstrated that on chromosome 1 the translocation directly disrupts the Disrupted In Schizophrenia 1 (DISC1) gene, and its non-coding antisense partner DISC2 (Millar et al., 2000b), and that DISC1 expression is reduced by approximately 50% at the transcript and protein level in lymphoblastoid cell lines derived from t(1;11) translocation carriers (Millar et al., 2005b). DISC1 haploinsufficiency is thus likely to be a major component of the mechanism by which the translocation increases disease risk. This is unlikely to be the only factor however, because DISC2, like DISC1, is brain-expressed (Millar et al., 2000b). Moreover, there is a third disrupted gene, DISC1 Fusion Partner 1 (DISC1FP1, also known as Boymaw) on chromosome 11 (Zhou et al., 2008b;Eykelenboom et al., 2012). Like DISC2, DISC1FP1 is non-coding and brain-expressed (Zhou et al., 2010). Loss of normal function of all three genes may thus contribute to the disease mechanism. However, unlike the DISC locus on chromosome 1 (see later), there is limited independent genetic evidence for involvement of DISC1FP1 in psychiatric disorders, although association with a nearby Single Nucleotide Polymorphism (SNP), rs2509382, has been reported in male patients diagnosed with schizophrenia (Debono et al., 2012). Analysis of the Psychiatric Genetics Consortium Genome-Wide Association Study (GWAS) data using Ricopili (www.broadinstitute.org/mpg/ricopili/) revealed that there is weak additional evidence for association of DISC1FP1 with schizophrenia (p=0.00096 with rs10501738), bipolar disorder (p=0.00052 with rs56163) and major depressive disorder (p=0.001 with rs4635043), although this did not reach genome-wide significance. This suggests that more associations may be found if this region is examined in detail.
The disease mechanism may be complicated still further by the fusion of the DISC1 and DISC1FP1 genes as a result of the translocation. Zhou et al speculated that this would result in production of chimeric transcripts (Zhou et al., 2008b), and indeed we demonstrated experimentally that this is the case (Eykelenboom et al., 2012). In lymphoblastoid cell lines derived from t(1;11) carriers at least four distinct chimeric transcripts are detectable. One, directed by the DISC1FP1 promoter on the derived 11 chromosome, encodes a C-terminal fragment of DISC1, but this is unlikely to be translated. Three, directed by the DISC1 promoter on the derived 1 chromosome, encode DISC1 amino acids 1-597 plus 1, 60 or 69 amino acids arising from translation of the normally non-coding DISC1FP1 sequence, and have been named CP1, CP60 and CP69, respectively (Eykelenboom et al., 2012). Notably, CP1 is almost equivalent to DISC11-597, a truncated species examined in many functional studies prior to discovery of the chimeric transcripts. The additional 69 novel amino acids in CP69 result in structural alterations and all three proteins behave aberrantly when exogenously overexpressed (Zhou et al., 2010;Eykelenboom et al., 2012). It is therefore likely that if expressed in the brains of t(1;11) translocation carriers the chimeric proteins would have multiple effects. However, while there is no obvious reason why the chimeric transcripts should not be brain-expressed (this cannot currently be tested due to lack of access to suitable tissue), we have been unable to demonstrate that the endogenous chimeric transcripts are translated in lymphoblastoid cell lines derived from translocation carriers (Eykelenboom et al., 2012). Thus it is not yet clear whether these chimeric transcripts and putative chimeric proteins contribute to the disease mechanism in translocation carriers. In the future it will be possible to address these issues using neural/glial material generated using induced pluripotent stem cells derived from t(1;11) translocation carriers.
3 Independent genetic evidence that the DISC locus is involved in risk of major mental illness
Evidence for independent linkage and association of the DISC locus to schizophrenia, schizoaffective disorder, schizophrenia spectrum, bipolar disorder, Asperger’s syndrome and autism spectrum disorder was summarised in a review by Chubb et al. in 2008 (Chubb et al., 2008). This review will cover the genetic studies published since Chubb et al., highlighting the evolving areas of interest. Following Chubb et al., recent candidate gene studies focussed on the DISC locus have reported significant findings for the psychiatric diagnoses schizophrenia (Saetre et al., 2008;Song et al., 2008;Rastogi et al., 2009;Hotta et al., 2011), schizoaffective spectrum (Green et al., 2011), bipolar disorder (Hennah et al., 2009;Schosser et al., 2010;Ram Murthy et al., 2012), bipolar spectrum disorder (Song et al., 2010), depression (Schosser et al., 2010;Carless et al., 2011;Thomson et al., 2012), autism (Zheng et al., 2011), and the related traits of age of onset for depression (Thomson et al., 2012) and depression, anxiety and emotional stability (Harris et al., 2010). However genome-wide approaches have identified no clear association between DISC locus variants and schizophrenia, bipolar disorder or recurrent major depression, indicating that no common variants at this locus act independently to substantially increase risk of developing these mental illnesses. Indeed, meta-analysis of all common variants within the DISC locus, from a total of 11626 cases and 15237 controls, and testing 1241 SNPs found no evidence that common variants at the DISC locus are significantly associated with schizophrenia (Mathieson et al., 2012). The lack of signal may result from the heterogeneity/oligogenic variation that is being revealed by the current genome-wide analyses, and the DISC locus continues to feature strongly in attempts to assess genome-wide association results in terms of networks (Jia et al., 2012) or in combination with known biology (Le-Niculescu et al., 2009;Ayalew et al., 2012;Tiwary, 2012).
Despite failing to show clear association of the DISC locus with psychiatric disorders, GWAS studies have found association between variation at the DISC locus and the neurodegenerative disorders late-onset Alzheimer’s disease (Beecham et al., 2009) and susceptibility to sporadic amyotrophic lateral sclerosis (OR = 0.499) (Landers et al., 2009). Although, neither of these results has been replicated, this further widens the potential effects of DISC1 variants. Intriguingly, there is biochemical evidence in support of the potential link between DISC1 and Alzheimers disease (Young-Pearse et al., 2010) which will be discussed later in this review.
For psychiatric illness, the focus has shifted to next generation sequencing approaches, with five studies reporting large-scale exon sequencing results. None of these studies identified major causative variants in DISC1, although there is evidence for rare sequence changes that influence disease risk (Song et al., 2008;Song et al., 2010;Green et al., 2011;Moens et al., 2011;Crowley et al., 2012). Song et al. reported increased burden of rare structural variants in both schizophrenia and bipolar disorder. Similarly, Green et al. reported an increase in exon 11 rare variants in a sample of schizoaffective disorder, bipolar type. Finally, Moens et al. used pooled sequencing to examine the burden of rare missense mutations in DISC1 and identified elevated burden, particularly in schizophrenic patients with a young onset age. By contrast, Crowley et al. reported no evidence for evidence for increased burden of rare variants in schizophrenia. It therefore remains to be seen whether surveying the regulatory/intronic regions of DISC1, or examining epistatic interactions within the gene, will uncover the variants underlying the previous reports of haplotypic associations derived from candidate gene studies.
GWAS and sequencing have highlighted the importance of rare genomic events such as the t(1;11) translocation and rare copy number variants (CNVs) in major mental illness, and a number of CNVs involving the DISC locus have now been identified. Subsequent to the identification of DISC1, a rare four base pair deletion at the 3′ end of DISC1 exon 12 was reported in a family with schizoaffective disorder (Sachs et al., 2005). This deletion exhibits incomplete segregation with psychiatric illness within this family and was subsequently identified in unaffected individuals (Green et al., 2006). Its relevance to psychiatric illness therefore remains equivocal, although it is likely to have functional consequences as it is predicted to result in expression of a C-terminally truncated protein (Sachs et al., 2005). In addition to this small deletion, a number of CNVs affecting the DISC locus have been reported. Duplications in the region of the DISC locus have been detected in three individuals, one affected by schizophrenia and two controls drawn from the same population (Buizer-Voskamp et al., 2011). A 2 Mb duplication encompassing seven genes, including DISC1 and DISC2, has also been described in two brothers with autism, ADHD and mild mental retardation (Crepel et al., 2010), although the clinical picture in this family is confused by the appearance of psychiatric symptoms in individuals not carrying the mutation. Additionally, a 2 Mb deletion, encompassing at least TSNAX (immediately upstream of DISC1), DISC1 and DISC2, was identified in a patient diagnosed with autism spectrum disorder (Williams et al., 2009). This deletion was inherited from the unaffected mother. An unenhanced head CT of the proband was normal at 3 years of age, but an MRI evaluation revealed multifocal scattered areas of gliosis and/or abnormal myelination in the bilateral cerebral white matter of unclear etiology. The carrier status of the child’s four unaffected siblings is unknown and there is no family history of consanguinity, psychiatric disorders, or autism. Sequencing of the paternal DISC1 coding regions identified two nonsynonymous polymorphisms, L607F and E661K. E661K has not been previously reported and, since amino acid 661 is the terminal amino acid in exon 9, it may have effects on splicing. However this amino acid is poorly conserved and the effect on the protein is likely to be benign. The presence of this paternal DISC1 allele may explain the lack of effect in the mother from whom the deletion was inherited. Altogether these studies indicate that copy number variants targeting the DISC locus may contribute to risk of mental illness, autism and other neurological disorders.
In addition to these behavioural effects, copy number variants affecting the DISC locus may also have gross effects upon brain structure, specifically, development of the corpus callosum, a fibre tract that connects the right and left hemispheres allowing communication between the two sides of the brain. Osbun et al. fine-mapped a 5.8 Mb deletion encompassing DISC1 in a mother and her two children (Osbun et al., 2011), all of whom have complete agenesis of the corpus callosum (Puthuran et al., 2005). In addition, they found a de novo 13.7 Mb deletion encompassing DISC1 in a second individual with agenesis of the corpus callosum, and four rare inherited, and potentially pathogenic, variants from a cohort of 144 well-characterised patients with this condition. One of these variants is a splice site mutation at the 5′ boundary of exon 11 that dramatically reduces expression of full-length DISC1 mRNA, but not of shorter forms. Consistent with these observations, transgenic mice expressing truncated DISC1 from a BAC exhibit partial agenesis of the corpus callosum (Shen et al., 2008). Indeed, DISC1 is highly expressed in the embryonic mouse corpus callosum at a critical time for callosal formation, suggesting that DISC1 may be critically involved in this process (Osbun et al., 2011). However other large deletions in the region that do not directly affect the DISC1 gene have also been identified in patients with this condition (Gentile et al., 2003;Filges et al., 2010), while another deletion that removes DISC1 was reported in a patient with a normal corpus callosum (Rice et al., 2006). Despite the inconsistencies, these observations are intriguing because agenesis of the corpus callosum is frequently associated with cognitive dysfunction and impaired social behaviour, both of which are present in mental illness and autism, as reviewed by Paul et al. (Paul et al., 2007).
In recent years, there has been an increase in association studies examining the effect of DISC sequence variation on brain structure, in particular grey matter and white matter volume, and cortical thickness. These studies have largely focussed on the nonsynonymous DISC1 exonic variants rs821616 (S704C) or rs6675281 (L607F), or other common variants.
Following the original observations that S704C is associated with altered grey matter volume in the hippocampus and prefrontal cortex (Callicott et al., 2005;Cannon et al., 2005), effects of variants upon grey matter volume have also been found in the prefrontal cortex with L607F (Szeszko et al., 2008), in the hippocampus with S704C (Di Giorgio et al., 2008), in the cingulate cortex with C704 (Hashimoto et al., 2006) and in the left parahippocampal gyrus and right orbitofrontal cortex with rs821597, located within intron 10 (Wei et al., 2012). In addition to the grey matter abnormalities in the prefrontal cortex, negative correlations between grey matter volume and the severity of hallucinations in patients carrying F607 compared to L/L homozygotes have been reported (Szeszko et al., 2008). Prefrontal white matter of individuals carrying the C704 allele exhibits decreased fractional anisotropy in comparison with S/S subjects (Hashimoto et al., 2006), and effects of this variant upon white matter integrity have subsequently been reported, although the allelic effects are somewhat inconsistent (Sprooten et al., 2011;Li et al., 2012).
Effects of L607F and S704C on cortical thickness have been studied and influences upon distinct cortical regions detected (Brauns et al., 2011;Raznahan et al., 2011;Chakravarty et al., 2012). In the right lateral temporal region there appears to be an additive effect of the two SNPs with L/L + C carriers exhibiting greater cortical thickness than F + C carriers (Raznahan et al., 2011). Cortical thickness in the temporal region is also reported to be affected by a SNP in intron 9, rs821589, and two SNPs in intron 1, rs11122319 and rs1417584, which also influence DISC1 expression, with differing effects in cases versus controls (Kahler et al., 2012). In the most comprehensive assessment of DISC1 variation and cortical thickness, Carless et al. identified a number of DISC1 variants, spanning an interval from intron 1 to the 3′ UTR, associated with cortical thickness in the temporal, parietal and frontal lobes (Carless et al., 2011). However they found no evidence of association with cortical thickness and S704C.
Lateral ventricular enlargement is a consistent characteristic of schizophrenia, and there is evidence that DISC1 influences this anatomical feature. Transgenic mice expressing C-terminally truncated forms of DISC1 exhibit lateral ventricular enlargement (Hikida et al., 2007;Pletnikov et al., 2008;Shen et al., 2008). Consistent with this, SNP rs2793092 within intron 3 is associated with lateral ventricle size in first episode schizophrenics (Mata et al., 2010). However, in this study rs2793092 genotype only predicted lateral ventricular enlargement among those patients also carrying the T allele at SNP rs6994992, located within the highly supported schizophrenia risk factor Neuregulin 1 (NRG1), suggesting that DISC1 and the risk factor NRG1 act in concert to regulate lateral ventricle size. There is further biological evidence that these two genes may act in common or related pathways (Wood et al., 2009;Seshadri et al., 2010) and this will be discussed later in this review.
A number of imaging studies have also examined the effects of variants on brain activation measured by functional MRI (fMRI) of the blood oxygen level-dependent (BOLD) contrast, as a measure of brain activation, during cognitive tasks. Association of the S704 allele has been reported with increased hippocampal activation during memory encoding and greater hippocampal formation–prefrontal cortex coupling in healthy individuals (Di Giorgio et al., 2008). Similarly, the F607 allele has been associated with increased activation of the dorsolateral prefrontal cortex in healthy individuals (Brauns et al.;2011). Prata et al. examined neural activity in healthy (Prata et al., 2008) and schizophrenic or bipolar patients (Prata et al., 2011), and detected increased activation of the prefrontal cortex, cingulate cortex, striatum and thalamus in healthy S704 homozygotes, but this was not replicated in patients with schizophrenia or bipolar disorder. Similarly, the F607 allele significantly affects brain activation in controls, but not in individuals at high risk of schizophrenia or bipolar disorder (Whalley et al., 2012). Prata et al proposed that the lack of effect in their patient population may reflect interaction with other genes associated with psychiatric illness, or indeed the effect of the disorders upon brain function.
Neurocognitive traits have also been studied both as endophenotypes for schizophrenia and to determine the functional consequences of variation at the DISC locus. Independent evidence for an effect of variation upon the P300 event related potential has been reported (Shaikh et al., 2011). This study detected significant associations between P300 amplitude and latency, and polymorphisms/haplotypes in a sample of patients with schizophrenia or psychotic bipolar disorder, their unaffected relatives, and unrelated healthy controls. These results are not robust to multiple-testing correction and will require replication, but they are consistent with the P300 data obtained from the t(1;11) family (Blackwood et al., 2001).
We have reported association been S704C and IQ at age 79, and cognitive ageing between age 11 and age 79 in the Lothian Birth Cohort 1921 (Thomson et al., 2005). However the association with IQ at the slightly younger age of 70 was not replicated in the Lothian Birth Cohort 1936, and in this study replication of the effect upon cognitive aging was not attempted (Houlihan et al., 2009). Positive associations of neurocognitive traits with multiple SNPs and domains have been reported, with the strongest association observed between rs751229, within intron 1, and perseverance (Palo et al., 2007). Carless et al. (Carless et al., 2011) reported association between rs2793094, within intron 3, and spatial working memory, a trait first linked to the DISC locus by Gasperoni et al. in 2003 (Gasperoni et al., 2003).
Genetic epistasis occurs when the genetic effect of one variant is modified by another variant, within either the same or different genes. Multiple studies point to epistatic effects involving DISC1. A joint association study using 67 SNPs tagging common haplotypes across a genomic region spanning DISC1/DISC2 and the upstream gene TSNAX, found no individually significant SNP in a combined European cohort of schizophrenia and bipolar disorder (Hennah et al., 2009). However, two individual SNPs, rs1538979, within intron 4, and rs821577, within intron 9, showed association in Finnish and London samples, respectively. Conditioning the analysis on rs1538979 identified a third SNP, rs821633, within intron 11. The minor allele at rs821633 increases risk dependent on the presence of the minor allele at rs1538979 or rs821577. It is protective if the major allele is present at both other SNPs. Interplay between rs1538979, within intron 4, and r821633, within intron 11, has also been detected in a European meta-analysis of schizophrenia (Schumacher et al., 2009). These studies indicate that interactions between variants in different regions of the DISC locus impact upon disease risk.
Numerous genetic association studies have also reported sex-specific association, suggesting an interaction between sex and genotype upon diagnosis. In contrast, there are no reported interactions between brain structure or function and sex, perhaps due to small sample sizes or the better proximity to the effects of DISC locus variation to these phenotypes compared to the phenotypic outcome as measured by the diagnoses. However interaction has been noted between genotype and diagnosis for fMRI activation (Prata et al., 2008;Chakirova et al., 2011;Prata et al., 2011;Whalley et al., 2012), cortical thickness (Kahler et al., 2012), grey matter density (Wei et al., 2012), and brain morphology (Takahashi et al., 2009). In these studies the effects are either detected in cases or controls or are in the opposite directions between cases and controls. It is possible that this is the result of interaction between risk genes for psychiatric illness. Consistent with this suggestion, as already mentioned, DISC1 and NRG1 may interact genetically to affect lateral ventricular volume (Mata et al., 2010), DISC1 genetic interactions with RELN and ERBB4, two molecules that are essential for many signalling pathways in the brain, also influence regional brain volume (Andreasen et al., 2011).
If the contribution of DISC1 to mental illness does include epistatic interactions these will be difficult to unequivocally detect statistically even given the sample sizes of current meta-analyses (Zuk et al., 2012) and will likely rely on biological determination.
In addition to the accumulating genetic evidence for an involvement of the DISC locus in risk of major mental illness, autism spectrum disorders and others, through effects on brain structure and activation, some studies have found effects of DISC1 amino acid variants upon responses to drug treatment. It has been reported that the right medial superior frontal gyrus volume is significantly correlated with daily dosage of antipsychotic medication in S704 homozygote schizophrenia patients, suggesting this this variant may have some effect on medication induced-changes in brain morphology (Takahashi et al., 2009). Association between DISC1 and “ultra-treatment resistant schizophrenia” versus treatment-responsive schizophrenia has been reported in French Caucasian patients for missense variants Q264R and L607F, but not S704C (Mouaffak et al., 2011). However it is worth noting that a separate Japanese study failed to find evidence of association between S704C, and three other variants, and treatment-resistant schizophrenia versus treatment responsive cases (Hotta et al., 2011).
4 Regulation of DISC1 expression
DISC1 expression is extremely complex. A common alternative splicing event within exon 11 giving rise to transcripts encoding the long (L) and long variant (Lv) protein isoforms was initially identified (Millar et al., 2000b;Taylor et al., 2003), but since then more than 50 differentially spliced transcripts have been reported. It is not clear how many of the differentially spliced DISC1 transcripts identified from human brain tissue (Nakata et al., 2009) are translated, but western blot analyses indicate that there are multiple DISC1 protein species. The alternative transcripts arise principally from use of alternative terminal exons following exons 3 and 9, to encode extremely short (Es) or short (S) isoforms respectively, or from exon skipping, although there is also evidence for use of alternative exons (Taylor et al., 2003;Nakata et al., 2009). In addition we, and others, have identified several intergenic transcripts containing exons of the gene TSNAX, which has an evolutionarily conserved location upstream of the DISC1 gene (Millar et al., 2000a;Nakata et al., 2009). These intergenic transcripts lack the TSNAX terminal exon and the first exon of DISC1, and intriguingly, some contain an uninterrupted open reading frame fusing TSNAX amino acid sequence to that of DISC1. This raises the interesting possibility that TSNAX/DISC1 fusion proteins are biologically relevant, although the existence of such proteins has yet to be demonstrated.
In general, the abundance of human DISC1 transcripts in brain decreases with age, exhibiting greatest expression during development, decreasing at birth, and continuing to decline throughout adult life, with the Δ7Δ8, Δ3 and Esv1 forms found at particularly low abundance in adulthood (Nakata et al., 2009). This expression pattern is paralleled by DISC1 protein expression in rodents which peaks at early postnatal stages, followed by a gradual decline (Austin et al., 2004;Honda et al., 2004;Kuroda et al., 2011).
Notably, hippocampal expression of the Δ7Δ8, Δ3, Esv1 and Lv transcripts is reported to be elevated in patients diagnosed with schizophrenia (Nakata et al., 2009). Moreover, expression of Δ3 transcripts is associated with the intronic SNP rs821597 that influences grey matter volume (Wei et al., 2012), while expression of Δ7Δ8 transcripts is associated with the common disease-associated DISC1 variants L607F and S704C (Nakata et al., 2009). Additional evidence for disturbed DISC1 expression levels in psychiatric patients comes from a study that reported reduced DISC1 transcript levels in lymphoblastoid cell lines derived from patients diagnosed with bipolar disorder (Maeda et al., 2006), and our own demonstration that DISC1 expression, at the transcript and protein level, is reduced by approximately half in lymphoblastoid cell lines derived from carriers of the t(1;11) translocation (Millar et al., 2005b). Moreover, DISC1 transcript levels are increased by clinically relevant doses of the atypical antipsychotic drugs olanzapine and risperidone in mouse frontal cortex and hippocampus (Chiba et al., 2006) and there is preliminary evidence from peripheral blood mononuclear cells that downregulation of DISC1 isoforms may be correlated with treatment response (Olincy et al., 2011). This suggests that some of the drugs used to treat schizophrenia may exert their effects, in part, through modulation of DISC1 expression, and this may underlie the genetic effects described above.
A number of studies thus highlight altered DISC1 expression levels as a potential disease mechanism, indicating the importance of understanding the processes that govern DISC1 transcription and translation. Multiple SNPs across the 5′ region of DISC1 are associated with DISC1 expression levels (Hennah and Porteous, 2009;Carless et al., 2011). The pattern of linkage disequilibrium between these variants suggests that they identify a haplotype block encompassing a variant(s) that affects DISC1 expression level. Moreover, a region 300-177 nucleotides upstream of the transcription start site (TSS) has been identified as the core DISC1 promoter (Walker et al., 2012). However an additional region, 982-301 nucleotides upstream of the TSS, negatively regulates promoter activity (Walker et al., 2012). There are putative transcription factor FOXP2 binding sites within the core DISC1 promoter region (Spiteri et al., 2007;Rosenbloom et al., 2010) and, consistent with this, FOXP2 inhibits DISC1 promoter activity and protein expression (Walker et al., 2012). Intriguingly, this negative effect of FOXP2 is counteracted by two distinct FOXP2 mutations, R553H and R328X, that are associated with developmental verbal dyspraxia, a speech and language disorder (Vernes et al., 2006). It will be interesting to now discover whether FOXP2 regulates DISC1 expression in vivo, and whether the mutant FOXP2-induced clinical phenotype, which may include an element of cognitive deficit, is related to the predicted resultant increase in DISC1 expression.
There is also evidence that DISC1 expression is regulated by Neuregulins (Seshadri et al., 2010), which may provide a direct functional link between two of the best supported risk factors for major mental illness. Treatment with NRG1 or NRG2 increases expression of a novel 130 kDa DISC1 antibody-immunoreactive species in HEK293 cells, an effect that was observed using two independent antibodies. There is no effect, however, upon other DISC1 isoforms at 100-105 kDa. This effect is mediated by the neuregulin receptors ErbB2 and ErbB3 (but not ErbB4) and activation of PI3K/Akt. A corresponding decrease in the 130 kDa species, but not the 100-105 kDa species, was observed in NRG1 knockout mice, and in BACE1 knockout mice which exhibit dysregulated NRG1 signalling (Seshadri et al., 2010). Moreover, expression of human DISC11-597 in mouse forebrain influences expression of NRG1, and ErbB3 and ErbB4, and induces abnormalities in oligodendrocyte differentiation and function (Katsel et al., 2011). These observations are consistent with both the evidence for genetic epistasis between the DISC1 and NRG1 genes (Mata et al., 2010), and with the observation that DISC1 and NRG1 apparently function within a common pathway in developing neurons and oligodendrocytes in zebrafish (Wood et al., 2009).
5 DISC1 protein structure
Based upon bioinformatics analysis, the “long” full-length DISC1 sequence (854 amino acids) has been suggested to consist of two regions: (i) an N-terminal “head” region spanning amino acid residues ~1–325, that lacks secondary structure elements and is predicted to contain large stretches of disorder (Soares et al., 2011), but which contains two notable regions of sequence conservation corresponding to a nuclear localization signal motif (Ma et al., 2002) and a serine-phenylalanine-rich motif (Taylor et al., 2003); and (ii) an alpha-helix containing C-terminal coiled-coil domain (spanning ~326–854) that shows greater conservation among orthologues than the N-terminus (Millar et al., 2000b;Ma et al., 2002;Taylor et al., 2003;Chubb et al., 2008;Soares et al., 2011).
Initial insight into DISC1 oligomerisation was provided by Brandon et al. (Brandon et al., 2004a), who noted that DISC1 forms large (≥250 kDa) species that they believed to be DISC1 dimers, or higher order oligomers. A region spanning residues 403–504 of DISC1 was later shown to be a self-association domain by pull-down experiments with deletion mutants (Kamiya et al., 2005). Early attempts at biophysical characterisation focussed on truncated C-terminal constructs (Leliveld et al., 2008;Leliveld et al., 2009). This was in part due to an inherent difficulty in expressing and purifying folded full-length DISC1 protein. From these studies it emerged that the C-terminal regions of DISC1 harbour oligomerisation domains that facilitate formation of dimers, octamers, and multimeric species. Interestingly, the authors noted that only the octameric state is required for functional interaction with nuclear distribution protein E homologue like-1 (NDEL1) indicating that an oligomerisation optimum is necessary for interaction between the two proteins. Size-exclusion chromatography analysis of another C-terminal truncated fragment showed that the common disease-associated C704 variant form of DISC1 adopts a moderately higher fraction of oligomers than the S704 form.
The first biophysical characterisation of full-length native DISC1 was recently published (Narayanan et al., 2011), and confirmed the findings from the C-terminal fragment studies. Utilising size-exclusion chromatography and analytical ultracentrifugation, the authors observed that native DISC1 forms octamers via dimers. The authors confirmed that NDEL1 interacts with an octamer of DISC1. Narayanan et al. also evaluated the structural consequence of the DISC1 common variant S704C and noted that compared to wild-type DISC1, the DISC1-C704 protein exhibits a greater than 9mer-sized oligomeric state (Narayanan et al., 2011). Despite this improper oligomeric assembly, there is no influence upon the interaction of DISC1-C704 with NDEL1. Our laboratory (Eykelenboom et al., 2012) investigated a truncated central fragment in the C-terminus of DISC1 (amino acids 326-597), the region up to the translocation breakpoint. This fragment was compared with another fragment, DISC1 amino acids 326-597 plus the additional 69 amino acids in CP69, one of the putative aberrant chimeric proteins arising from the t(1;11) translocation (Eykelenboom et al., 2012). We found that on average, the fusion protein exhibited a higher oligomerisation propensity by dynamic light scattering experiments, with greater thermal stability, and that the additional 69 amino acids possess additional alpha-helical content by circular dichroism. Leliveld et al. demonstrated that detergent-insoluble DISC1 could be seen by immunoreactivity in ~20% of post-mortem brains of patients with chronic mental diseases, but not in healthy controls (Leliveld et al., 2008). These DISC1 aggregates did not bind NDEL1, whereas insoluble DISC1 (316–854) aggregates did co-purify with dysbindin, in brains of a subset of patients with mental illness, but not in healthy controls (Ottis et al., 2011). More recently, Atkin et al. (Atkin et al., 2012) showed that DISC1 forms large perinuclear aggregates and conclusively identified these structures as aggresomes. These findings highlight the potential of DISC1 protein aggregation and recruitment of binding partners as a biological mechanism in a subset of mental illness cases (Korth, 2009;2012).
A key remaining question is how common and rare amino acid sequence variants in DISC1 affect its structure, stability, oligomeric state and aggregation propensity, and ultimately how this might relate to protein interactions and phenotypic consequences. For further details on this see our earlier review (Soares et al., 2011). Additionally, while numerous DISC1 protein interaction partners have been confirmed, little is known about stoichiometry and thermodynamics of these interactions, precise interaction sites, or whether these interactions are competitive or co-operative. These, in addition to obtaining high resolution structural data on DISC1 (and of DISC1 in complex with its interaction partners) will be the next key goals from a biochemical and biophysical standpoint. This detailed knowledge of the structure-function relationships of DISC1 will be critical in understanding its role in disease and translating this knowledge for therapeutic benefit.
6 Mouse behavioural phenotypes
Several DISC1 mouse models have been generated. The first to be described carries a modified DISC1 allele from the mouse strain 129 that consists of a natural 25 nucleotide deletion in exon 6 resulting in a premature stop codon, plus targeted premature transcription termination and polyadenylation sites in intron 8 (Koike et al., 2006). This was followed by investigation of the effects of ethyl-nitrosourea (ENU)-generated point mutations, L100P or Q31L, within exon 2 of the endogenous gene (Clapcote et al., 2007). Three transgenic models express C-terminally truncated DISC1 species, truncated at the position of the t(1;11) translocation breakpoint, and thus referred to hereafter as DISC11-597. These mice constitutively (Hikida et al., 2007) or inducibly (Pletnikov et al., 2008) express truncated human cDNA under control of the CAMKII promoter, or express mouse transcripts from a truncated mouse BAC under control of the natural mouse DISC1 promoter (Shen et al., 2008). Expression of a C-terminal DISC1 fragment has also been described (Li et al., 2007). These models and their phenotypes were recently discussed in an excellent review by Brandon & Sawa (Brandon and Sawa, 2011) and will therefore not be reviewed in detail here. In general these mouse models exhibit overlapping phenotypes, including cognitive and behavioural abnormalities, and deficits in sensorimotor gating (PPI), working memory and latent inhibition, consistent with some characteristics of schizophrenia in humans (Brandon and Sawa, 2011), thus adding further weight to the evidence for involvement of DISC1 in mental illness. However it should be noted that a recent independent study (Shoji et al., 2012) failed to confirm the behavioural alterations reported in the missense mutant mice (Clapcote et al., 2007). This may be due to strain differences, backcrossing, or may even reflect differences in laboratory housing/testing conditions (Crabbe et al., 1999;Papaleo et al., 2012).
Following on from these models, a targeted mouse model lacking DISC1 exons 2 and 3 (Δ2-3) was recently reported (Kuroda et al., 2011). Removal of these exons was designed to abolish DISC1 expression, which was convincingly demonstrated using two new DISC1 antibodies directed against the N- or C-termini. However there is increasing evidence for expression of a myriad of additional isoforms (see section 4), some of which lack exons 2 and/or 3 (Nakata et al., 2009) and it thus remains to be seen whether this mouse is in fact a complete knockout. This deletion mouse model showed no overt brain developmental defect, but there were behavioural abnormalities, some of which could be rescued pharmacologically (Kuroda et al., 2011). Another recently generated mouse model allows inducible expression of human DISC11-597 in astrocytes (Ma et al., 2012). These mice exhibit heightened responses to the NMDA receptor antagonist MK-801 in open field and prepulse inhibition tests, consistent with NMDA receptor hypofunction, which is considered to be a major characteristic of schizophrenia (Kristiansen et al., 2007). Thus, these mice are useful additions to the growing number now available. Each of these models needs to be used and interpreted with caution as none precisely mimic the t(1;11) translocation event, nor any of the clinically associated DISC1 variants since reported.
A number of these mouse models have been used to study gene-environment interactions. Poly I:C is an immunostimulant that is frequently used to mimic the innate immune response to viral infection. When administered during pregnancy (gestation day 9) to mice inducibly expressing human DISC11-597, the resulting offspring exhibit increased anxiety and depressive-like symptoms, and decreased sociability, which may be related to effects of mutant DISC1 expression upon production of cytokines in response to Poly I:C (Abazyan et al., 2010). Postnatal Poly I:C treatment of mice constitutively expressing human DISC11-597 demonstrated combined action to produce cognitive impairment, and effects upon hippocampus-dependent fear memory, social interaction and hyperactivity in response to treatment with the NMDA antagonist MK-801 (Ibi et al., 2010). In addition, Poly I:C treatment of these mice reduces the number of parvalbumin-positive interneurons in the medial prefrontal cortex (Ibi et al., 2010). Consistent with the genetic data indicating a role in responses to drug treatment (see section 3), cognitive impairment in these mice is rescued by treatment with the antipsychotic clozapine, while their hyper-responsiveness to MK-801, but not their social interaction deficit, is reversed by clozapine or haloperidol (Nagai et al., 2011). Chronic social defeat, another environmental stressor, may also interact with DISC1 to influence behavior (Haque et al., 2012). When tested in several behavioural paradigms relevant to schizophrenia, the 31L mutation was found to interact with social defeat in hereozygous mice (Haque et al., 2012). These studies in mice indicate that environmental factors can interact with the effects of mutant DISC1 expression to induce or exacerbate behavioural deficits.
7 DISC1 brain function – neural precursor proliferation
A number of key studies demonstrate that DISC1 is critically involved in several stages of neurogenesis, the process by which newborn neurons are generated within the developing and adult brain. This process is widespread in the developing brain, but in adult brain is restricted essentially to the subventricular zone (SVZ) and subgranular zone of the hippocampal dentate gyrus.
In embryonic mouse brain DISC1 expression peaks at E14-E15, and then declines as development progresses (Mao et al., 2009). This peak of expression coincides with a period of active neurogenesis in the developing cortex, where DISC1 is prominent within neural progenitors in the ventricular zone (VZ) and SVZ at this time (Mao et al., 2009). DISC1 is also expressed in neural progenitors of the hippocampal dentate gyrus in adult mouse brain (Mao et al., 2009). Together these observations suggested a role for DISC1 in neurogenesis, and indeed, DISC1 RNAi in isolated adult hippocampal progenitor cells decreases their rate of proliferation, while DISC1 overexpression has the opposite effect (Mao et al., 2009). Consistent with these observations, DISC1 knock down in utero at E13 reduces proliferation within the SVZ, while overexpression increases proliferation (Mao et al., 2009). Moreover, reducing DISC1 expression leads to premature cell cycle exit and differentiation of neural progenitors within the VZ/SVZ (Mao et al., 2009). Altogether this results in depletion of the progenitor cell pool and, at least shortly after the treatment, overproduction of neurons (Mao et al., 2009). Neural progenitor proliferation and maintenance of the progenitor cell pool is controlled, in part, by the canonical Wnt signalling pathway, in which β-catenin modulates transcription of a set of genes that are critically required for cell proliferation (Wu and Pan, 2010). DISC1 regulates this process through direct binding to, and inhibition of, the kinase GSK3β (Mao et al., 2009), which in turn phosphorylates β-catenin to direct its degradation (Wu and Pan, 2010). Thus, DISC1 knockdown results in increased GSK3β activity, followed by increased β-catenin phosphorylation and degradation, which in turn inhibits cell proliferation while promoting differentiation, while DISC1 overexpression reduces GSK3β activity to stabilise β-catenin and stimulate gene transcription and cell proliferation (Mao et al., 2009). DIXDC1 is another DISC1 interactor involved in this process (Singh et al., 2010). Together with DISC1, DIXDC1 co-regulates Wnt-GSK3β/β-catenin signalling and cortical neural progenitor proliferation (Singh et al., 2010).
Consistent with the studies by Mao et al. and Singh et al., neurogenesis defects have also been observed in DISC1 mutant mice i) carrying the modified 129 allele (Koike et al., 2006;Kvajo et al., 2008), ii) expressing DISC1-597 fused to EGFP (Shen et al., 2008), and iii) carrying ENU-induced point mutations, 31L and 100P, in the endogenous gene (Clapcote et al., 2007;Lee et al., 2011).
The studies by Mao et al and Singh et al clearly demonstrate DISC1 involvement in neural progenitor proliferation and provide a mechanism for its regulatory action via GSK3β, β-catenin and DIXDC1 (Mao et al., 2009;Singh et al., 2010). Intriguingly, however, the DISC1 interactors LIS1 and NDE1 (Brandon et al., 2004b;Burdick et al., 2008;Bradshaw et al., 2009) are also directly involved in regulation of neurogenesis. LIS1 and NDE1 (or its paralogue NDEL1) complex together to regulate many cytoskeleton-related functions through their effects upon the motor protein dynein (Lam et al., 2010). LIS1 deficiency affects neural progenitor cell proliferation in the VZ through disruption of the orientation of cleavage (Yingling et al., 2008). This determines the symmetry of division, distribution of cell fate determinants, and thus the fate of the daughter cells, such that asymmetric division increases neuronal differentiation at the expense of the progenitor pool. This consequence of LIS1 deficiency is likely due to multiple deleterious effects on microtubule function which can be rescued by overexpression of NDEL1 (Yingling et al., 2008). NDE1 is enriched within progenitor cells of the VZ, where, like LIS1, it regulates the plane of progenitor cell division, cell fate and maintenance of the progenitor pool (Feng and Walsh, 2004). NDE1 mutant mice also display mitotic delay/arrest, chromosome mispositioning and other abnormalities that affect progenitor cell proliferation (Feng and Walsh, 2004). NDEL1 is however, largely absent from the embryonic cortical progenitors (Feng and Walsh, 2004), suggesting that it does not have a significant role in progenitor proliferation. It will be of interest to discover whether DISC1 modulates neural progenitor proliferation through interaction with LIS1 and NDE1, in addition to its regulatory role via GSK3β and β-catenin, and DIXDC1, and indeed whether these pathways are related.
The disease-associated variants R264Q, L607F and a novel rare variant, A83V, all impact upon DISC1 binding to GSK3β, and fail to activate canonical Wnt signalling (Singh et al., 2011). Intriguingly, these three variants also impact upon neural progenitor cell proliferation in vivo (Singh et al., 2011). Importantly, GSK3β is a major target of Lithium, in clinical use as a mood stabiliser for treatment of bipolar disorder patients (Serretti et al., 2009), which suggests that clinical symptoms in psychiatric patients may arise from aberrantly high GSK3β activity. Silencing DISC1 in the adult mouse dentate gyrus results in novelty-induced hyperlocomotion and increased immobility in the forced swim test, both of which are reversed through treatment with the specific GSK3β inhibitor SB-216763 (Mao et al., 2009). It is also reported that both the 31L and 100P mouse mutations reduce DISC1/GSK3β interaction, and that treatment with GSK3 inhibitors can rescue aspects of their behavioural phenotypes (Lipina et al., 2011;Lipina et al., 2012). Altogether these studies highlight reduced DISC1/GSK3β interaction, increased GSK3β activity, reduced canonical Wnt signalling and decreased neural progenitor proliferation as possible disease mechanisms in psychiatric illness. However it is worth pointing out that GSK3β performs multiple additional regulatory functions in vivo. Indeed, in zebrafish it has been demonstrated that DISC1, through interaction with GSK3β, has extensive effects upon brain development via canonical Wnt signalling, but also via a non-canonical Wnt pathway (De Rienzo et al., 2011). While these additional effects upon the brain could also influence disease risk if they occur in humans, it is notable that production of new neurons in the adult dentate gyrus is downregulated in response to stress. Since stress is a major trigger for psychiatric illness it is possible that reduced hippocampal neurogenesis is a contributory factor (Hanson et al., 2011;Lee et al., 2012). Moreover, there is evidence that antidepressant treatment may exert its effects, at least in some cases, through upregulation of neurogenesis in the subgranular zone. Hippocampal neurogenesis may thus be fundamentally involved in both the causation and treatment of mental illness, although this remains to be conclusively proven (Hanson et al., 2011).
8 DISC1 brain function - neuronal migration
In the developing cortex, pyramidal neurons produced in the VZ/SVZ migrate radially, along guide fibres provided by radial glia, to their final destination in an inside-out manner, where they become integrated into functional neuronal networks. Interneurons, however, migrate tangentially, independent of radial glia (Kriegstein and Noctor, 2004). Neuronal migration also proceeds in other developing brain regions including the hippocampus, where granule cells produced near the dentate notch migrate to the developing dentate gyrus, while pyramidal cells populate the hippocampus in an inside-out fashion. In the adult brain neuronal migration is restricted to neural precursors moving from the subgranular zone to the granular zone of the hippocampus, and from the SVZ to the olfactory bulbs via the rostral migratory stream. The migration process involves extension of the leading edge of the cell. This is followed by migration of the nucleus (nucleokinesis), and finally, retraction of the trailing edge of the cell.
Silencing DISC1 in the developing cortex inhibits radial migration to the cortical plate, an effect that is recapitulated by overexpression of DISC11-597 (Kamiya et al., 2005;Duan et al., 2007;Kubo et al., 2010;Young-Pearse et al., 2010). DISC1 is also required for migration of cortical interneurons in the embryonic brain, with DISC1 knockdown inhibiting migration of neurons from the medial ganglionic eminence (Steinecke et al., 2012). Migrating interneurons extend a long branched leading process that detects guidance cues (Valiente and Martini, 2009), and this leading process is abnormal in cells where DISC1 expression has been silenced. Such cells exhibit abnormally extended processes with fewer branches (Steinecke et al., 2012). In the developing hippocampus DISC1 knockdown inhibits granule cell migration (Meyer and Morris, 2009), while there are conflicting reports about whether DISC1 is required for migration of CA1 pyramidal cells (Meyer and Morris, 2009;Tomita et al., 2011). In postnatal brain DISC1 is reported not to be involved in neuroblast migration via the rostral migratory stream (Wang et al., 2011b), while intriguingly, in the adult dentate gyrus, DISC1 RNA interference results in overextended migration, with newborn neurons from the subgranular zone migrating beyond their intended destination within the granular zone (Duan et al., 2007). The reason for this apparent discrepancy in the effect of DISC1 RNA interference in developing cortex and adult hippocampus is unknown, but it may be related to the different organisation of these two structures, with cortical layers laid down in ‘inside-out’ fashion while hippocampal layers are established in an ‘outside-in’ sequence (Duan et al., 2007). It has thus been suggested that DISC1 knockdown compromises the ability of the neuron to recognise the cues that guide its migration (Duan et al., 2007), and these cues may exhibit temporal and spatial specificity in cortex versus hippocampus. Intriguingly, in the adult hippocampus there is evidence that DISC1 acts in concert with NMDA receptors to modulate neuronal migration (Namba et al., 2011), Consistent with these studies revealing involvement of DISC1 in neuronal migration, evidence of disrupted radial migration was found in the neocortex of the 100P and the 31L DISC1 mutant mice (Lee et al., 2011).
The DISC1 interactors LIS1 and NDEL1 are known to be critically involved in neuronal migration (Sasaki et al., 2000), with mutations in LIS1 causing the neuronal migration disorder lissencephaly (Wynshaw-Boris, 2007). The DISC1 interactor DIXDC1 is also required for neuronal migration in the developing cortex, and intriguingly this requires both DISC1/DIXDC1 interaction, and CDK5-dependent recruitment of NDEL1 to DIXDC1, to which it also binds (Singh et al., 2010). Unlike their role in regulating neural progenitor proliferation, the role of DISC1 and DIXDC1 in neuronal migration is, however, independent of the Wnt-GSK3β/β-catenin pathway (Singh et al., 2010). Thus, in the absence of NDEL1 binding, DISC1/DIXDC1 direct neural progenitor proliferation via Wnt-GSK3β/β-catenin, while upon recruitment of NDEL1 the function of this protein complex shifts to neuronal migration.
There is an additional important mechanism that mediates the functional transition of DISC1 from activator of neural progenitor proliferation to promoter of newborn neuron migration in the developing cortex (Ishizuka et al., 2011). This switch is triggered by DISC1 phosphorylation at mouse S710 or its human equivalent S713, which abolishes DISC1 binding to, and inhibition of GSK3β, consequently inhibiting DISC1’s activity as a positive regulator of wnt/β-catenin signalling (Ishizuka et al., 2011). While losing interaction with GSK3β, DISC1 phosphorylated at S710 (DISC1 pS710) becomes enriched at the centrosome and displays markedly increased affinity for (Bardet-Biedl Syndrome) BBS1, leading to enhanced recruitment of this protein to the centrosome (Ishizuka et al., 2011). This is thought to be an important step in the transition from neural progenitor proliferation to migration, because the DISC1-mediated recruitment of BBS proteins to the centrosome is an established mechanism underlying neuronal migration in the developing cortex (Kamiya et al., 2008). Consistent with the proposed role of DISC1 S710 phosphorylation in the transition from neural progenitor proliferation to migration, DISC1 pS710 is predominantly enriched in the cortical plate and intermediate zone of the developing mouse cortex, where most migrating neurons are found, and is virtually absent in the ventricular and subventricular zones, sites of active cell proliferation (Ishizuka et al., 2011). Most strikingly, neuronal migration defects induced by DISC1 knock-down in the developing mouse cortex can be rescued by expression of wild-type DISC1 or phospho-mimic mutant DISC1 E710, but not by phospho-dead mutant DISC1 A710, whereas the opposite is true for progenitor proliferation deficits induced by DISC1 silencing (Ishizuka et al., 2011).
The common disease-associated S704C DISC1 variant influences the role of DISC1 in neuronal migration, with the C variant inhibiting migration in the developing cortex (Singh et al., 2011). Notably, the other variants tested, A83V, R264Q and L607F, do not affect cortical migration (Singh et al., 2011). Moreover, these variants, but not S704C, influence Wnt-GSK3β/β-catenin signalling, which again demonstrates that the role of DISC1 in cortical neuronal migration is independent of this pathway (Singh et al., 2011).
The Alzheimer’s disease risk factor amyloid precursor protein (APP) is also required for neuronal migration (Young-Pearse et al., 2007). Intriguingly, APP knockdown in mouse results in cortical migration defects that are rescued by overexpression of DISC1 (Young-Pearse et al., 2010). DISC1 and APP interact (Young-Pearse et al., 2010), therefore they likely function together during neuronal migration. This study thus provides a biochemical link between risk factors for psychiatric illness and Alzheimers disease, which suggests the possibility of overlapping pathological mechanisms.
DISC1’s role in migration has also been examined in zebrafish, where DISC1 knockdown dysregulates cranial neural crest cell migration (Drerup et al., 2009). The migration defect correlates with increased expression of the transcription factors FOXD3 and SOX10, leading to a suggested mechanism whereby DISC1 indirectly modulates migration through transcriptional repression of transcription factors with a known role in development (Stuhlmiller and Garcia-Castro, 2012).
9 DISC1 brain function - neuronal integration and maturation
As well as regulating production and migration of neurons, DISC1 is also required for neuronal integration (Duan et al., 2007). Silencing DISC1 in the adult mouse dentate gyrus affects the morphogenesis of newborn granule cells, with these cells exhibiting enlarged cell bodies, ectopic dendrites, increased dendritic length and branching, and accelerated acquisition of intrinsic excitability and synapse formation (Duan et al., 2007). Moreover, DISC1 knockdown results in aberrant axonal targeting to hippocampal subfield CA3 (Faulkner et al., 2008). DISC1 has thus been suggested to act as a ‘master regulator’, controlling the timing of neuronal integration through its many stages (Duan et al., 2007). Functional effects upon neurons in the adult mouse hippocampus have also been reported in mice carrying the endogenous modified 129 allele (Koike et al., 2006). Within the dentate gyrus of these mice there is neuronal mispositioning providing further evidence of overextended migration, and abnormal dendritic morphology. However, in contrast to the RNA interference experiments reported by Duan et al, these mice exhibit impaired dendritic growth, suggesting that newborn neurons may be unable to fully integrate into circuits within the dentate gyrus (Kvajo et al., 2008). Similar impairment of dendritic development has been reported in frontal cortical neurons of mice carrying 31L and 100P missense mutations (Lee et al., 2011). Mice carrying the modified 129 allele also exhibit altered axonal projections to CA3, and have short-term plasticity deficits at hippocampal CA3/CA1 synapses (Kvajo et al., 2011). Consistent with the observations in these mice, when heterologously expressed in C. elegans, mouse DISC1 induces axon guidance defects via activation of UNC-73/TRIO-RAC-PAK signalling (Chen et al., 2011). Finally, in addition to these effects upon neuronal positioning, morphology and axon pathfinding, when DISC1 is knocked down in pyramidal neurons of the prefrontal cortex early in development, maturation of dopaminergic neurons is defective, with consequent effects upon neural circuitry (Niwa et al., 2010).
DISC1 co-operates with its binding partners NDEL1 and FEZ1 in regulating many aspects of neuronal development in the adult hippocampus (Duan et al., 2007;Kang et al., 2011), but the most detailed analysis of underlying pathways involves Girdin, otherwise known as KIAA1212. Girdin activates the kinase AKT, however DISC1 interaction with Girdin blocks its ability to bind and activate AKT (Kim et al., 2009), thus DISC1 indirectly modulates activity of this important kinase. Overexpression of Girdin or a constitutively active form of AKT recapitulates many, but not all, of the effects of DISC1 knockdown upon granule cell development (Kim et al., 2009;Enomoto, 2011), indicating that these three proteins function in a common pathway to regulate maturation of newborn granule cells. Intriguingly, the effects of DISC1 knockdown or Girdin overexpression can be rescued by treatment with the mTOR inhibitor rapamycin, but not by inhibition of GSK3β (Kim et al., 2009). Moreover Girdin is weakly expressed in proliferating neural progenitors in the subgranular zone of the adult hippocampus, suggesting that it does not have a significant role in progenitor proliferation (Kim et al., 2009;Enomoto, 2011). Together these observations point towards a mechanism whereby DISC1 regulates the role of Girdin and AKT in neuronal maturation via the mTOR pathway (Kim et al., 2009). AKT also directly regulates GSK3β activity via inhibitory phosphorylation of S9 on GSK3β, but this kinase does not seem to be involved in the neuronal integration function of DISC1 (Kim et al., 2009).
The role of DISC1 in adult hippocampal neuronal maturation is also dependent upon the extrinsic factor GABA, via NKCC1, an ion transporter involved in GABA responses (Kim et al., 2012). In adult hippocampus NKCC1 knockdown rescues the dendritic growth defects in newborn neurons that result from DISC1 knockdown and knocking down the GABAA receptor γ2 subunit has a similar effect. Moreover, both treatments inhibit the effect of DISC1 knockdown upon the AKT/mTOR pathway. Thus this study provides evidence that, in adult hippocampal neuronal maturation/integration, DISC1 and GABA signalling synergise in regulating dendritic outgrowth via the AKT/mTOR pathway (Kim et al., 2012). DISC1 knockdown has no apparent effect upon early postnatal hippocampal neuronal maturation/integration, until mice are subjected to maternal deprivation stress, whereupon the same spectrum of deficits occurs as in adult hippocampus (Kim et al., 2012). Moreover, knockdown of NKCC1 suppresses the dendritic abnormalities and soma hypertrophy observed under these conditions at early postnatal stages (Kim et al., 2012). Similarly, treatment with rapamycin also reverses the defects induced by DISC1 knockdown plus stress (Kim et al., 2012). These intriguing observations indicate that early postnatal hippocampal neuronal maturation and integration may be particularly sensitive to the combined effects of aberrant DISC1 expression, as may occur in some psychiatric patients such as t(1;11) translocation carriers, and environmental stress, such as maternal deprivation.
There is evidence that cAMP signalling is another pathway that modulates neuronal maturation. DISC1 is known to function in the cAMP pathway through interaction with the phosphodiesterase 4 family of proteins (Millar et al., 2005b). PDE4s hydrolyse cAMP to terminate, and thus regulate, cAMP signalling in a compartmentalised manner (Houslay and Adams, 2003). Mammalian PDE4s are orthologues of Drosophila Dunce, so-called because of its role in cognition and synaptic plasticity (Houslay and Adams, 2003), and are specifically inhibited by drugs such as rolipram which have antidepressant and antipsychotic activity (Houslay and Adams, 2003;Siuciak et al., 2007). In a negative feedback loop, catalytic activity of certain PDE4 isoforms is induced by upregulation of cellular cAMP, resulting in cAMP hydrolysis and termination of the signalling cascade (Houslay and Adams, 2003). We have shown that DISC1 binds to PDE4s in a cAMP-dependent manner (Millar et al., 2005b;Murdoch et al., 2007), and have provided evidence that DISC1 inhibits the induction of PDE4 activity that normally occurs in response to elevated cAMP levels (Carlyle et al., 2011). Mice carrying the modified 129 DISC1 allele exhibit decreased PDE4 activity in the hippocampus, with expression of many PDE4B and PDE4D isoforms decreased at the protein level (Kvajo et al., 2011). Consistent with this, cAMP levels in the granular zone of the dentate gyrus are increased. This increase in cAMP leads to increased activation of the cAMP-dependent transcription factor CREB and dysregulated expression of molecules required for axon guidance and dendritic growth (Kvajo et al., 2011). In primary culture, mutant neurons exhibit altered axon pathfinding and dendritic branching defects, both of which are rescued by treatment with an adenylyl cyclase inhibitor to reduce cAMP levels (Kvajo et al., 2011). Consistent with these effects on PDE4 activity and cAMP signalling, we have observed decreased DISC1/PDE4 interaction in mice carrying 31L and 100P mutations, with the 31L mutation resulting in reduced total brain PDE4 activity (Clapcote et al., 2007), although it is not yet known how this relates to the anatomical defects that have been reported in these mice (Lee et al., 2011).
Altogether then, DISC1 regulates neurogenesis at many levels, including proliferation of neural progenitors via Wnt-GSK3β/β-catenin and DIXDC1, migration of neurons to their final destination via DIXDC1-NDEL1, BBS1 and APP, and possibly also through effects upon expression of transcription factors FOXD3 and SOX10, and maturation/integration into existing neural circuitry via AKT/mTOR/GABA, PDE4/cAMP, NDEL1 and FEZ1.
10 DISC1 brain function - neurosignalling
Several studies report that DISC1 is present at synapses. We (Bradshaw et al., 2008), and others (Hayashi-Takagi et al., 2010), have shown that, when overexpressed in primary cultured rodent neurons, DISC1 locates to dendritic spines where it co-localises with PSD-95 (Bradshaw et al., 2008), a marker of the post-synaptic density. At the ultrastructural level, endogenous DISC1 localises to the post-synaptic density in human frontal and parietal cortices (Kirkpatrick et al., 2006), rat hippocampus (Wang et al., 2011a), mouse striatum (Ramsey et al., 2011) and monkey prefrontal cortex (Paspalas et al., 2012), using three independent antibodies. This location is further confirmed by subcellular fractionation studies which show that DISC1 is loosely associated with the post-synaptic density (Hayashi-Takagi et al.;Clapcote et al., 2007;Wang et al., 2011a), again using three different antibodies. Finally, DISC1 co-immunoprecipitates from mature neurons with PSD-95, CIT and Kal-7, all core post-synaptic density components (Hayashi-Takagi et al.).
Intriguingly, DISC1 modulates the size and density of dendritic spines via interaction with Kal-7 (Hayashi-Takagi et al., 2010). Essentially, DISC1 dissociates Kal-7 interaction with RAC1, for which Kal-7 is a GDP/GTP exchange factor (GEF), and RAC1 activity is reduced as a result. DISC1 may thus inhibit activation of RAC1 by sequestering its GEF, Kal-7. This likely underlies DISC1’s effect upon spine morphology and density because RAC1 is an established modulator of dendritic spines (Hayashi-Takagi et al., 2010). These effects appear to be dependent upon NMDA receptor activity (Hayashi-Takagi et al., 2010), suggesting that DISC1 may play an important role in activity-dependent spine remodelling. There is also evidence that DISC1 modulates glutamatergic synapse composition via its interaction with the kinase TNIK at the post-synaptic density (Wang et al., 2011a). TNIK protein expression and phosphorylation are apparently glutamate receptor activity-dependent and TNIK may therefore be involved in synaptic signal transduction (Wang et al., 2011a). Inhibition of TNIK activity using a peptide derived from the TNIK binding site on DISC1 decreases synaptic expression of PSD-95, the AMPA receptor subunit GluR1 and the AMPA receptor regulator stargazin through protein degradation (Wang et al., 2011a). Altogether these data suggest that DISC1 mediates activity-dependent glutamatergic synapse remodelling. It is now apparent from these two studies that DISC1 may be a key modulator of some of the activity-dependent changes that underlie synaptic plasticity. In a separate study, it was demonstrated that mice carrying a DISC1 point mutation, 100P, display altered expression profiles of transcripts encoding the synaptic proteins NRXN1 and NRXN3 (Brown et al., 2011). How this occurs is not yet known, but it is likely to be independent of TNIK and synaptic proteolysis, and indicates that there must also be a transcriptional mechanism by which DISC1 regulates synapse composition.
We have demonstrated that a number of additional DISC1 interactors localise to the post-synaptic density, including the phosphodiesterase (PDE4) family of proteins (Bradshaw et al., 2008). While the role of synaptic PDE4 is currently unknown, its presence at the post-synaptic density suggests a role in modulation of cAMP signalling events downstream of receptor activation, a role that is likely to be modulated by DISC1.
In addition to its post-synaptic density localisation, within dendritic spines DISC1 is also present at perisynaptic and extrasynaptic sites in monkey prefrontal cortex, where it associates with Hyperpolarization-activated Cyclic Nucleotide-gated (HCN) channels (Paspalas et al., 2012). HCN channel activation is modulated by cAMP to regulate, for example, neuronal excitability, so it is notable that they also associate with the dopamine D1 receptor and with PDE4A within spines (Paspalas et al., 2012). Dopamine D1 receptor stimulation induces cAMP synthesis, thus within spines the HCN channels, PDE4A and DISC1 are strategically placed to respond to these cAMP signalling events. It has been suggested that the location of this ‘cAMP signalplex’ may render prefrontal neuronal networks particularly sensitive to dysregulated cAMP signalling (Paspalas et al., 2012).
There is also evidence that DISC1 functions presynaptically. Using optogenetics to stimulate presynaptic neurons, it has been demonstrated that DISC1 overexpression enhances the probability of glutamate release at the synapse, while DISC1 RNA interference has the opposite effect (Maher and LoTurco, 2012). Additionally, expression of DISC11-597 disrupts the synchronous nature of glutamate release (Maher and LoTurco, 2012). This is consistent with a report that DISC1 knockdown or expression of DISC11-597 reduces synaptic vesicle transport (Flores et al., 2011), while mice expressing the modified 129 allele have smaller synaptic vesicles (Kvajo et al., 2011). It is thus possible that the effect of DISC1 expression upon glutamate release at the synapse is related to its role in transporting vesicles to the synapse.
Because DISC1 has multiple roles at the synapse, and no doubt more have still to be uncovered, it is intriguing that synaptic expression of DISC1 is altered in mice mutant for synaptic proteins. The first study investigated Densin, a post-synaptic density scaffold molecule that regulates function of metabotropic glutamate receptors (mGluRs) and Calmodulin Kinase II (CAMKII) at the synapse (Carlisle et al., 2011). Knockout of Densin in mice leads to selective reduction of mGlur5 and DISC1 at the post-synaptic density (Carlisle et al., 2011). The second study used mice which express NMDA receptors at only 10% of the normal level. In these mice synaptic DISC1 and its binding partner 14-3-3Epsilon are reduced in striatum in an age-dependent manner (Ramsey et al., 2011). Moreover treatment of wild-type mice with NMDA receptor antagonists also reduces DISC1 levels (Namba et al., 2011;Ramsey et al., 2011). Together, these studies suggest that DISC1 expression is synaptic activity-dependent.
DISC1 may also modulate synaptic transmission via interaction with serine racemase (Ma et al., 2012). This enzyme generates D-serine, an NMDA receptor co-agonist. DISC11-597 cannot interact with serine racemase, and expression of this aberrant form of DISC1 decreases serine racemase expression via its ubiquitination and subsequent degradation (Ma et al., 2012). Expression of DISC11-597 may thus lead to impaired NMDA receptor transmission, consistent with the behavioural phenotype of mice expressing this form of DISC1, as discussed earlier. Aberrant DISC1 expression also influences dopamine D2 receptor expression (see section 13) (Lipina et al., 2010;Pogorelov et al., 2012). DISC1 thus modulates neurosignalling at many levels, including effects upon NMDA receptors and dopamine D2 receptors, which are involved in the actions of psychotropic drugs such as phencyclidine, and antipsychotic drugs respectively, and therefore strongly implicated in the pathophysiology of schizophrenia (Kristiansen et al., 2007;da Silva Alves et al., 2008).
11 DISC1 at the subcellular level - nucleus
DISC1 is involved in multiple functions at the subcellular level, via localisation to several compartments and interaction with an ever-increasing number of binding partners. Any/all of these locales and binding partners could contribute to its roles in neurogenesis and neurosignalling.
Multiple lines of evidence support a direct involvement of DISC1 in nuclear transcriptional regulation. DISC1 partially distributes to the nucleus (Sawamura et al., 2005;Malavasi et al., 2012), and here it can be prominently detected within promyelocytic leukaemia (PML) nuclear bodies, sites of active transcription (Sawamura et al., 2008). DISC1 nuclear targeting is mediated by at least two well conserved cis-acting elements in the protein: a classic tetra-arginine nuclear localisation motif at positions 35-RRRR-38 (NLS1) and a putative leucine zipper spanning residues 607-628, encoded by DISC1 exon 9 (LZ9) (Sawamura et al., 2008). In Drosophyla melanogaster the nucleus-restricted expression of full-length but not DISC11-597induces sleep homeostasis disturbances, supporting a potential role for nuclear DISC1 in the regulation of sleep/wake cycles (Sawamura et al., 2008). These findings must be cautiously interpreted, since it has not been established that invertebrates naturally express DISC1. However the possibility that nuclear DISC1 might be directly involved in regulation of sleep homeostasis is particularly intriguing because this process is often disturbed in psychiatric patients (Schulz and Steimer, 2009;Wulff et al., 2009;Wulff et al., 2012) and is partly regulated by cAMP signalling (Zimmerman et al., 2008), a pathway in which DISC1 is involved by virtue of its modulatory interaction with PDE4 (Millar et al., 2005b;Carlyle et al., 2011). A study conducted on human post-mortem brains also hinted at the potential clinical relevance of altered nuclear DISC1 function (Sawamura et al., 2005). In this study, increased nuclear expression of a 75-85kDa DISC1 protein species was detected in brains from patients diagnosed with schizophrenia or depression (Sawamura et al., 2005). While this effect was correlated with drug abuse in patients with major depression, none of the common brain-associated confounding factors explained the nuclear enrichment of DISC1 in brain tissue from schizophrenic patients, suggesting aberrant DISC1 nuclear targeting as a potential pathogenic mechanism (Sawamura et al., 2005). Additional evidence supporting the potential contribution of aberrant nuclear DISC1 expression to psychopathogenesis came from a recent study from our group, which revealed that the putatively causal amino acid substitution 37W (Song et al., 2008) located within DISC1 Nuclear Localisation Signal 1 (NLS1), and the disease-associated variant 607F located within DISC1 Leucine Zipper 9 (LZ9), significantly decrease DISC1 nuclear expression in a dominant-negative fashion (Malavasi et al., 2012). Interestingly, nuclear targeting of DISC11-597, which retains NLS1 but lacks LZ9, is also significantly decreased (Sawamura et al., 2008).
Two highly related members of the ATF/CREB family of transcription factors, ATF4 and ATF5, are DISC1 interactors (Millar et al., 2003;Morris et al., 2003;Ozeki et al., 2003;Bradshaw et al., 2008;Sawamura et al., 2008;Malavasi et al., 2012). ATF4 and ATF5 are preferentially translated in response to a variety of environmental insults, including nutrient deprivation, endoplasmic reticulum stress, viral infections and oxidative stress, and in turn govern the transcriptional responses to these damaging stimuli (Ameri and Harris, 2008;Watatani et al., 2008;Zhou et al., 2008a). ATF4 and ATF5 are also key regulators of the transition from neuroprogenitor cell proliferation to neuronal differentiation in the developing cortex (Greene et al., 2009;Frank et al., 2010). Moreover, ATF4, is a central controller of the transcriptional events governing establishment of long-term memory (Karpinski et al., 1992;Bartsch et al., 1995;Chen et al., 2003;Lee et al., 2003;Costa-Mattioli and Sonenberg, 2006;Costa-Mattioli et al., 2007), and is additionally involved in regulation of emotional behaviour (Green et al., 2008) and behavioural flexibility (Trinh et al., 2012) in rodent models. Collectively, these observations support the hypothesis that ATF4 dysregulation may contribute to the pathophysiology of schizophrenia, characteristics of which include impaired emotional behaviour and cognition, and for which various environmental stressors are believed to contribute significantly to risk, (Trinh et al., 2012). We, and others, have carried out in vitro reporter assays (Sawamura et al., 2008;Malavasi et al., 2012) which have established that DISC1 regulates the transcriptional activity of both exogenously expressed, and endogenous stress-induced, ATF4 on different promoter elements, and may thus be potentially involved in regulation of multiple ATF4-dependent transcriptional events. Both ATF4 and ATF5 expression is subject to circadian regulation (Igarashi et al., 2007;Lemos et al., 2007;Koyanagi et al., 2011), and a direct involvement in the control of circadian pathways has been demonstrated for ATF4 (Koyanagi et al., 2011;Ushijima et al., 2012), suggesting a potential mechanism linking nuclear DISC1 expression to altered sleep/wake cycles in the fruit fly (Sawamura et al., 2008). Since DISC1 is also capable of binding the nuclear receptor co-repressor NCoR, a transcriptional co-repressor that recruits histone deacetylase to the transcriptional machinery (Jepsen and Rosenfeld, 2002), it was hypothesised that DISC1 may act by recruiting this factor to the ATF4-containing complex on DNA (Sawamura et al., 2008). Intriguingly, and consistent with their detrimental effect on DISC1 nuclear expression, we recently demonstrated that amino acid substitutions 37W and 607F significantly impair DISC1’s ability to regulate ATF4-dependent transcription in vitro (Malavasi et al., 2012), suggesting that these DISC1 variants may contribute to susceptibility to psychiatric illness, at least in part, by affecting the DISC1-dependent regulation of ATF4 transcriptional activity.
12 DISC1 at the subcellular level - centrosome
In actively dividing animal cells, microtubules nucleate from the centrosome, which functions as the main microtubule-organising center (MTOC)(Nigg and Raff, 2009). The centrosome consists of a pair of microtubule-based structures, the centrioles, embedded in an amorphous protein matrix, the pericentriolar matrix (PCM) (Nigg and Raff, 2009). As well as being essential for the process of cell division, it is well established that the centrosome plays important roles throughout neurodevelopment. In proliferating neural progenitors, the centrosome regulates asymmetrical distribution of cell fate factors to daughter cells, thus determining whether they will continue dividing, or begin differentiating (Higginbotham and Gleeson, 2007). Moreover, correct positioning of the centrosome is necessary for establishment of cell polarity in newborn neurons, and for migration of developing neurons to their final destinations in the brain (Kuijpers and Hoogenraad, 2011).
Various centrosomal proteins, including NDE1, NDEL1, LIS1, PCM1, CAMDI, PCNT (also known as Kendrin) and multiple BBS proteins are DISC1 binding partners (Morris et al., 2003;Ozeki et al., 2003;Brandon et al., 2004a;Miyoshi et al., 2004;Camargo et al., 2007;Shinoda et al., 2007;Kamiya et al., 2008;Fukuda et al., 2010). All these proteins have well-established roles in centrosome-dependent aspects of neural development, and some of them cause severe brain development defects when mutated. For example, as previously mentioned, mutations affecting LIS1 are associated with lissencephaly, a condition arising from defective neuronal migration and characterised by lack of cortical folds, severe mental retardation and seizures (Reiner et al., 1993). On the other hand, mutations in PCNT result in primordial dwarfism and microcephaly (small brain size) (Rauch et al., 2008), mutations in NDE1 cause microcephaly and microlissencephaly (Alkuraya et al., 2011;Bakircioglu et al., 2011;Guven et al., 2012), whereas BBS4 mutations are associated with Bardet-Biedl syndrome, a pleiotropic disorder that is often accompanied by mental retardation (Mykytyn et al., 2001).
A number of studies independently reported localisation of DISC1 to the centrosome in cultured cell lines and primary neurons (Morris et al., 2003;Miyoshi et al., 2004;Kamiya et al., 2005;Bradshaw et al., 2008;Kamiya et al., 2008). DISC1’s centrosomal localisation requires interaction with Kendrin (Miyoshi et al., 2004;Shimizu et al., 2008). Disruption of DISC1-Kendrin binding prevents DISC1 recruitment to the centrosome and impedes microtubule aster formation, highlighting the importance of this interaction for correct centrosomal function (Shimizu et al., 2008). Further studies established that centrosomal DISC1 is involved in the recruitment and anchoring of several other essential centrosomal proteins to this organelle. For example, DISC1 cooperates with its interactor BBS4 to tether PCM1 to the centrosome (Kamiya et al., 2008). Here, PCM1 is in turn necessary for the correct localisation of other centrosomal proteins, including Kendrin (Kamiya et al., 2008), and for the correct organisation of microtubule arrays (Dammermann and Merdes, 2002). Interestingly, studies in cell models and post-mortem human brains have evidenced an influence of risk-conferring DISC1 amino acid substitutions L607F and S704C on PCM1 centrosomal recruitment (Eastwood et al., 2009;Eastwood et al., 2010). Exogenous expression of DISC1-607F in a neural cell line is associated with reduced centrosomal immunoreactivity of endogenous PCM1 compared to cells expressing DISC1-607L (Eastwood et al., 2009). Consistent with findings in cultured cells, analysis of brain tissue from DISC1 607F homozygotes revealed a trend towards reduced centrosomal PCM1 immunoreactivity in glial cells, and the same was observed in tissue from DISC1-704C carriers (Eastwood et al., 2010). Moreover, a potential cumulative effect between the two DISC1 variants was observed, whereby PCM1 centrosomal abundance was lowest in brain tissue from 607F homozygotes that were also 704C carriers (Eastwood et al., 2010). The molecular mechanism behind the effects of DISC1 amino acid substitutions L607F and S704C on PCM1 localisation is not known, but it is possible that these amino acid substitutions might directly disrupt DISC1 binding to PCM1 and/or reduce DISC1 localisation to the centrosome, perhaps by reducing its binding to Kendrin. Like PCM1, CAMDI is recruited to the centrosome in a DISC1-dependent manner (Fukuda et al., 2010).
Importantly, centrosomal DISC1 is also necessary for anchoring the dynein microtubule motor complex (Kamiya et al., 2005). Overexpression of DISC11-597 displaces endogenous wild-type DISC1 from the centrosome and prevents the DISC1-mediated recruitment of the dynein motor complex to the centrosome, which is consequently unable to organise the microtubule network (Kamiya et al., 2005). Failure of DISC1-dependent recruitment of the dynein motor complex to the centrosome has been suggested to underlie the neurite outgrowth and cortical migration defects observed in neurons after DISC1 knock-down or overexpression of DISC11-597(Kamiya et al., 2005). This hypothesis is supported by the observation that DISC1 knock-down in cortical neurons significantly increases the distance between the centrosome and the nucleus, which is one of the proposed mechanisms accounting for the migration defects (Kamiya et al., 2005).
The dynein motor complex also contains NDE1, NDEL1 and LIS1, which together regulate either cargo recruitment or dynein processivity (Lam et al., 2010). This complex is essential for the correct organisation and function of microtubule networks during neurite outgrowth and neuronal migration (Wynshaw-Boris and Gambello, 2001). Our work showing that the cAMP phosphodiesterase PDE4 co-localises with DISC1 at the centrosome, together with LIS1, NDEL1 and its paralogue NDE1, suggests a potential role for cAMP and DISC1/PDE4 in regulation of the NDE1/NDEL1/LIS1 complex at this location (Bradshaw et al., 2008). Indeed, we demonstrated that two NDE1 residues, S306 and T131, can be phosphorylated by PKA and that this is regulated by DISC1 and PDE4 (Bradshaw et al., 2011). NDE1 phosphorylation at T131 inhibits its binding to LIS1 while promoting its interaction with NDEL1 (Bradshaw et al., 2008;Bradshaw et al., 2011). Moreover PDE4 itself binds directly to NDEL1 to promote NDEL1/NDEL1 self-association, and this interaction is ablated by cAMP (Collins et al., 2008). PDE4 also binds directly to LIS1, an interaction that is augmented by cAMP (Murdoch et al., 2011). Cyclic AMP, via DISC1 and PDE4, thus substantially redistributes interactions within the LIS1/NDE1/NDEL1 complex. Critically, these effects of cAMP are accompanied by decreased LIS1 interaction with dynein and loss of dynein function (Murdoch et al., 2011).
13 DISC1 at the subcellular level – primary cilium
The observation that exogenous DISC1 localises at the base of primary cilia in NIH3T3 cells and primary striatal neurons, supports its potential involvement in the establishment and/or function of this structure (Marley and von Zastrow, 2010). Primary cilia are single immotile filamentous protrusions found on virtually all cell types, including neurons and glia (Louvi and Grove, 2011). They consist of a long membrane-enveloped axoneme projecting from a basal body or mother centriole, which is structurally indistinguishable from the one that originates the centrosome (Nigg and Raff, 2009). The basal body has the dual function of anchoring the cilium to the cell surface and organising the movement of cargo molecules along ciliary microtubules to and from the cytoplasm, in a process known as intraflagellar transport (IFT) (Nigg and Raff, 2009). The motor proteins Kinesin and Dynein are necessary for IFT which is, in turn, essential for the establishment and maintenance of primary cilia (Nigg and Raff, 2009), while NDE1 knockdown promotes ciliogenesis (Kim et al., 2011). Ciliogenesis additionally depends on the transport of membrane vesicles required for ciliary membrane assembly, a process mediated by a large complex of BBS proteins (the BBSome) located at the basal body (Nigg and Raff, 2009). Primary cilia are multifunctional organelles that act as sensors for both mechanical and extracellular stimuli, and they are additionally involved in the regulation of key developmental signalling pathways. Primary cilia also act as regulators of canonical and non-canonical wnt signalling, and mutations that prevent the development of primary cilia in mice result in markedly increased β-catenin levels, suggesting that primary cilia normally inhibit wnt signalling (Berbari et al., 2009). Loss-of-function mutations of proteins required for normal ciliogenesis, including BBS proteins, result in a broad spectrum of developmental and degenerative disorders collectively described as ciliopathies (Hildebrandt et al., 2011). Several ciliopathies, including Bardet-Biedl syndrome and Joubert syndrome, present neurodevelopmental and/or cognitive defects as part of their clinical spectrum (Louvi and Grove, 2011). Consistently, genetic ablation of ciliogenesis in mouse models causes brain development defects, and it also inhibits adult hippocampal neurogenesis by decreasing the formation, expansion and maintenance of the pool of proliferating neural progenitor cells (Breunig et al., 2008;Han et al., 2008;Amador-Arjona et al., 2011) while inhibiting the morphological and functional maturation of newborn neurons in the adult dentate gyrus (Kumamoto et al., 2012). Intriguingly, DISC1 knock-down in NIH3T3 cells or primary striatal neurons reduces the number of primary cilia to a similar extent as depletion of IFT88, a protein essential for ciliogenesis (Marley and von Zastrow, 2010). Moreover, the ciliogenesis defects induced by DISC1 knockdown in NIH3T3 cells and striatal neurons can be fully rescued by exogenous expression of GPF-DISC1, strongly supporting a central role of DISC1 in ciliogenesis in both neuronal and non-neuronal cells (Marley and von Zastrow, 2010). Interestingly, several neuroreceptors, including dopamine receptors 1, 2 and 5, serotonin receptor 6 and somatostatin receptor 3, are prominently or exclusively expressed in neuronal primary cilia, strongly implicating this organelle in neurotransmission (Handel et al., 1999;Marley and von Zastrow, 2010;Domire et al., 2011). In mice, the ENU-induced DISC1 single amino acid substitution L100P is associated with a behavioural phenotype related to characteristics of schizophrenia (Clapcote et al., 2007). DISC1 100P mice display hypersensitivity to the psychostimulant effects of amphetamine, which induces dopamine release, coupled to an increased proportion of high-affinity functional D2 receptors in the striatum (Lipina et al., 2010). Conversely, inducible expression of human DISC11-597 is linked to altered responses to the amphetamine analogue methamphetamine, and decreased D2 receptor expression (Pogorelov et al., 2012). Collectively, these findings raise the intriguing possibility that, besides being directly involved in the formation and maintenance of primary cilia, where D2 receptors are primarily expressed, DISC1 might also contribute to the trafficking and/or functional modulation of ciliary D2 receptors. It would thus be interesting to examine if and how the 100P mutation, as well as clinically relevant DISC1 mutations, affect the development and function of primary cilia in relevant neuronal populations.
14 DISC1 at the subcellular level - mitochondria
Mitochondria are the major source of ATP, through oxidative phosphorylation, but they also perform additional functions, including calcium homeostasis. Of all the organs in the body, the brain is particularly sensitive to mitochondrial dysfunction, in part due to its extremely high energy requirements, approximately 20% of total body energy consumption at rest. The majority of this energy consumption is due to neurotransmission. Neuronal action potential firing results in influx of sodium or calcium ions, among others, and neurons consequently must utilise the majority of their ATP for rebalancing the ionic gradients that are essential to maintain their excitability. Indeed, sodium and calcium transport, including mitochondrial calcium transport, are estimated to account for 40-50% or 3-7%, respectively, of ATP utilised by the brain (Ames, 2000). An estimated further 10-20% of ATP is used for additional aspects of neurotransmission, including neurotransmitter synthesis, packaging and transport to synapses (Ames, 2000).
Any factor affecting mitochondrial function and/or transport is likely to deleteriously affect neurotransmission and other cellular processes, ultimately influencing brain development/function at multiple levels. Not surprisingly then, mitochondrial dysfunction is believed to underpin a number of important neurological diseases, including Parkinson’s, Alzheimer’s and Huntington’s diseases (Han et al.) and there is now accumulating evidence pointing towards DISC1 being another factor that induces mitochondrial dysfunction to cause brain disease, indeed network analysis indicates enrichment of Huntingtin binding partners in networks of interactors built around DISC1, including a number of mitochondrial proteins (Boxall et al., 2011).
DISC1 associates with mitochondria, where it has been detected both at mitochondrial membranes in an unspecified location (Ramsey et al., 2011;Paspalas et al., 2012) and inside the outer mitochondrial membrane (James et al., 2004;Park et al., 2010), in association with mitofilin (Park et al., 2010), a core component of the Mitochondrial Inner Membrane Organising System (MINOS), the Mitochondrial Organising Structure (MitOS) or the Mitochondrial Intermembrane space Bridging complex (MIB)(Hoppins et al., 2011;Alkhaja et al., 2012;Ott et al., 2012). These multiprotein complexes, which likely represent the same entity, attach cristae membranes to the inner mitochondrial membrane and promote assembly of respiratory chain complexes (Hoppins et al., 2011;Alkhaja et al., 2012;Ott et al., 2012). Mitofilin knockdown induces mitochondrial morphological abnormalities visible at the ultrastructural level, including disorganisation of the mitochondrial inner membrane and loss of cristae junctions (John et al., 2005).DISC1 modulates mitofilin ubiquitination and stability (Park et al., 2010) and, intriguingly, DISC1 knockdown induces similar mitochondrial membrane architecture abnormalities to mitofilin knockdown (Park et al., 2010), suggesting that it may directly regulate mitochondrial inner membrane architecture via mitofilin.
In addition, knockdown of either DISC1 or mitofilin, or expression of DISC11-597, reduces the activity of mitochondrial NADH dehydrogenase, a component of the respiratory chain pathway, and consistent with this, cellular ATP levels are reduced (Park et al., 2010). Decreased mitochondrial monoamine oxidase activity and perturbed calcium dynamics also result from the same treatments (Park et al., 2010). Altogether these observations indicate that DISC1 and mitofilin influence a range of mitochondrial processes, and this may occur through effects on mitochondrial membrane organisation.In support of these observations, DISC1 and mitofilin both associate with CHCM1/CHCHD6, another protein of the inner mitochondrial membrane (An et al., 2012). Like mitofilin and DISC1, CHCM1/CHCHD6 regulates cristae morphology (An et al., 2012), and knockdown of this protein leads to reduced ATP synthesis, reduced oxygen consumption, and slowed cell growth (An et al., 2012).
As well as mitochondrial function per se, mitochondrial trafficking is also of critical importance to the brain because, for neuronal mitochondria to deliver their energy provision and calcium buffering to the synapse, they must be efficiently transported along axons and dendrites, which can be extremely lengthy in some cases. It is therefore notable that DISC1 expression levels influence the number of motile mitochondria within neurons, as assessed by live cell imaging (Atkin et al.;2011). The mechanism by which manipulation of DISC1 expression exerts these effects upon mitochondrial motility is not yet known, however it is possible that they are a consequence of effects on mitochondrial membrane architecture and general mitochondrial function: Mitochondrial transport is an active process that would be predicted to be affected by ATP production. Moreover, mitochondrial damage and intracellular calcium levels influence mitochondrial transport (Miller and Sheetz, 2004;Yi et al., 2004), thus the morphological and functional mitochondrial abnormalities caused by DISC1 knockdown (Park et al., 2010) could contribute to inhibition of axonal mitochondrial motility induced by the same treatment in neurons (Atkin et al.;2011). Alternatively, DISC1 may affect the mitochondrial transport process more directly (see section 15), although this possibility has yet to be investigated.
There is mounting evidence that DISC1-induced mitochondrial function and/or transport defects may contribute to disease risk. In addition to the observations discussed above, we have demonstrated that two of the aberrant DISC1 species putatively expressed by t(1;11) translocation carriers (Eykelenboom et al., 2012), CP60 and CP69, are almost exclusively mitochondrial and induce profound mitochondrial dysfunction, manifest as perinuclear mitochondrial clustering and loss of membrane potential (Eykelenboom et al., 2012). Notably, we have found that addition of GFP tags, or deletion of the C-terminal tail, also results in increased mitochondrial DISC1 expression and altered mitochondrial morphology (Millar et al., 2005a). While these latter two examples are not disease-related, they further illustrate the emerging impression that aberrant DISC1 expression frequently leads to increased mitochondrial targeting and mitochondrial dysfunction. Finally, the disease-associated DISC1 common sequence variant 607F fails to rescue the block on axonal mitochondrial transport induced by DISC1 knock down (Atkin et al., 2011). Altogether, these observations indicate that altered DISC1 expression and/or function compromises mitochondrial function and may contribute to disease risk. Consistent with this, there is evidence for mitochondrial dysfunction in schizophrenia and bipolar disorder (Rezin et al., 2009;Clay et al., 2011), and it is now essential to discover whether psychiatric patients carrying the t(1;11) translocation or the DISC1 607F variant, or otherwise exhibiting altered DISC1 expression, suffer mitochondrial dysfunction.
15 DISC1 functions at the subcellular level – motor proteins
DISC1 knockdown inhibits axonal transport of various cargoes, including mitochondria (see mitochondria section) and synaptic vesicles (Shinoda et al., 2007;Taya et al., 2007;Atkin et al., 2011;Flores et al., 2011), suggesting a general role in microtubule-based intracellular transport. This process is mediated by the co-ordinated actions of multiple molecular motors including kinesin and dynein which are respectively responsible for anterograde and retrograde microtubule-based transport. Anterograde transport is essential in all cells, but particularly so in neurons for trafficking of cargo, including newly synthesised proteins, receptors and synaptic vesicles or mitochondria, to distant regions such as axonal growth cones and synapses. Retrograde transport is required, for example, for synaptic vesicle recycling or transport of chemical messengers back to the soma, thus allowing the synapse to communicate with the cell body. Kinesin and dynein are also essential for additional functions including mitosis and nucleokinesis and thus, in the brain, are critical for processes such as neural precursor proliferation and neuronal migration.
DISC1 associates with kinesin1, and DISC1 knockdown reduces axonal transport of the proteins GRB2, LIS1, NDEL1 and 14-3-3Epsilon, which are all kinesin-associated (Shinoda et al., 2007;Taya et al., 2007), suggesting a direct role in anterograde axonal transport. Interestingly, the DISC1 interactor FEZ1 (Miyoshi et al., 2003) is also implicated in this transport process. FEZ1 complexes with kinesin 1 and tubulin to promote anterograde mitochondrial transport (Fujita et al., 2007;Ikuta et al., 2007). FEZ1 also promotes synaptic vesicle transport in association with kinesin 1, a process that is disrupted by expression of DISC1-597 (Flores et al., 2011). In Drosophila FEZ1 acts as an intermediary between kinesin 1 and the synaptic vesicle membrane protein Synaptotagmin 1 (SYT1) (Toda et al., 2008). DISC1, but not DISC11-597, promotes FEZ1 interaction with SYT1, suggesting that DISC1 may facilitate recruitment of synaptic vesicles to the transport complex for trafficking to the synapse (Flores et al., 2011). Intriguingly, lithium, in clinical use as a mood stabiliser, reverses the FEZ1/SYT1 association and synaptic vesicle trafficking defects induced by expression of DISC11-597 (Flores et al., 2011). It is tempting to speculate that DISC1 also participates in recruiting mitochondria to kinesin/FEZ1 complexes. It will be interesting to discover whether DISC1 has a general role in recruiting cargo to kinesin transport complexes, which could help to explain its very large number of interactors.
As already discussed (see section 12), DISC1 also complexes with Dynein Intermediate Chain (DIC) and the dynein adaptor protein Dynactin (p150glued) (Kamiya et al., 2005), and may thus be involved in retrograde intracellular transport, although this has yet to be demonstrated.
16 Involvement of DISC1-associated molecules in disease risk
There is evidence that a number of DISC1-associated proteins are themselves involved in conferring risk of psychiatric illness (summarised in Table 1 for schizophrenia) or are associated with brain-related traits (Table 2). For some of these the convergent evidence is becoming compelling. As for DISC1, we have demonstrated that the PDE4B gene is directly disrupted by a chromosomal translocation in psychiatric patients (Millar et al., 2005b) and a number of genetic studies have found evidence for involvement in schizophrenia (Table 1). The potential importance of AKT, GSK3β, NDEL1, LIS1 and 14-3-3Epsilon are all highlighted by several genetic association studies (Table 1), while NDE1 shows association with schizophrenia (Table 1) and is also targeted by a number of CNVs identified in psychiatric patients (Ingason et al., 2011). Moreover, there are recent reports of interplay/epistasis between DISC1 and its protein interactors from structural and fMRI studies (Mata et al., 2010;Nicodemus et al., 2010;Kang et al., 2011) adding to the initial reports suggesting interaction between candidate genes for schizophrenia (Hennah et al., 2007;Burdick et al., 2008), as proposed by Meyer-Lindenberg and Weinberger (Meyer-Lindenberg and Weinberger, 2006). These reports suggest that epistatic interactions affecting genetic risk may occur between DISC1 and NDEL1/NDE1 (Hennah et al., 2007), with Ser704Cys status altering both risk of schizophrenia detected using the NDEL1/NDE1 SNPs, and NDEL1/NDE1 binding to DISC1 (Burdick et al., 2008). An interaction between DISC1 3′ UTR SNP rs1411771 and an intronic SNP in the gene encoding another DISC1 interactor, CIT, has also been shown to increase the risk of developing schizophrenia, and this was validated by fMRI (Nicodemus et al., 2010). Genetic interaction between DISC1 and PDE4B influences regional brain volume (Andreasen et al., 2011), while epistasis between Ser704Cys and FEZ1 is reported to act on the risk of developing schizophrenia, and synergistic effects of the two proteins on dendritic growth of newborn neurons in the adult mouse hippocampus are reported (Kang et al., 2011). Finally, SNPs within DISC1 and the gene encoding NKCC1 also interact (Kim et al., 2012). Altogether, these studies indicate that pathways involving DISC1 and these associated molecules may be of particular relevance to the aetiology of mental illness.
Table 1.
Evidence of association to schizophrenia from candidate gene studies for selected DISC1 interactors. Evidence for association was identified using SZGene (http://www.szgene.org/, updated 23 December 2011). The table shows those found in SZGene, the total number of studies (both case-control and familial) listed in SZGene, and whether any SNP remained significant after meta-analysis. Note: Only those SNPs with published genotype data in at least four independent case-control samples eligible for analysis were subjected to meta-analysis by SZGene (see http://www.szgene.org/methods.asp for details).
| gene | present in SZgene |
number of studies |
results | meta-analysis significant SNPs |
|---|---|---|---|---|
| DISC1 | Y | 45 studies | 31 positive/9 negative/ 3 trend |
rs3737597, rs999710 |
| AKT1 | Y | 17 studies | 9 positive/7 negative/1 trend | rs3803300 |
| ATF4 | Y | 1 study | negative | |
| ATF5 | Y | 1 study | negative | |
| BBS1 | ||||
| BBS4 | ||||
| CCDC141 | ||||
| CCDC88A (encodes KIAA 1212/Girdin) |
||||
| CIT | Y | 1 study | positive | |
| DIXDC1 | ||||
| FEZ1 | Y | 5 studies | negative | |
| GRB2 | ||||
| GSK3β | Y | 8 studies | 1 positive/6 negative/1 trend | |
| KALRN (encodes KAL-7) |
||||
| NDE1 | Y | 4 studies | negative | |
| NDEL1 | Y | 7 studies | 4 positive/3 negative | |
| PAFAH1B1 (encodes LIS1) |
Y | 5 studies | 1 positive/4 negative | |
| PCM1 | Y | 7 studies | 6 positive/1 negative | |
| PCNT | Y | 3 studies | 1 positive/1 negative/1 trend | |
| PDE4A | ||||
| PDE4B | Y | 12 studies | 9 positive/3 negative | rs910694 |
| PDE4C | ||||
| PDE4D | Y | 2 studies | positive | |
| SLC12A2 (encodes NKCC1) |
||||
| TNIK | ||||
| YWHAE (encodes 14-3-3s) |
Y | 5 studies | 2 positive/3 negative |
Table 2.
Evidence of association between DISC1 interactors and brain-related traits. Evidence from genome-wide studies was downloaded from the NCBI Phenotype-Genotype Integrator (http://www.ncbi.nlm.nih.gov/gap/PheGenI) for any category or trait with p-values p < 1 × 10−5. Brain-related traits were identified by inspection. The full list of genes examined is given in Table 1.
| trait | SNP | context | gene | p-value |
|---|---|---|---|---|
| neuroanatomy | rs12042938 | intron | DISC1 | 4.00E-36 |
| Alzheimer’s disease |
12044355 | intron | DISC1 | 9.00E-06 |
| schizophrenia | 2088885 | intron | TNIK | 6.00E-06 |
| neurotic disorders |
702543 | intron | PDE4D | 2.00E-06 |
| mental competency |
295973 | intron | PDE4D | 7.61E-07 |
| mental competency |
295973 | intron | PDE4D | 7.61E-07 |
| neuroblastoma | 9892996 | intergenic | SLC25A19/ GRB2 |
2.23E-07 |
| neuroblastoma | 16967789 | intron | GRB2 | 1.10E-06 |
17 Summary
Although the contribution of DISC1 to total risk of mental illness remains unknown, it is apparent that rare genetic events that target the DISC locus, such as the t(1;11) translocation and CNVs, can predispose individuals to developing a major psychiatric disorder. Indeed, the unequivocal segregation of the t(1;11) translocation with schizophrenia, bipolar disorder and major depressive disorder indicates that disruption of the DISC1 gene is likely to be a major predisposing factor in this family. Thus studying DISC1 function and the effect of the translocation will inform on the disease pathways operating in translocation carriers, and likely also in unrelated psychiatric patients as a result. Such studies have highlighted critical roles for DISC1 at many stages of neurogenesis (neural precursor proliferation, neuronal migration and neuronal integration/maturation) indicating that any factor that compromises normal DISC1 function is likely to impact upon brain development and also production of new neurons in the adult brain. Importantly, DISC1 also has multiple roles at the pre- and post-synapse, indicating that neurosignalling deficits are likely to occur, in addition to developmental defects. DISC1 regulates these pathways through interaction with an extensive array of protein interactors and most likely via a complex blend of alternative splicing, distribution to multiple subcellular compartments, oligomerisation and regulation of protein phosphorylation. Intriguingly, many DISC1 interactors are themselves implicated as risk factors for mental illness, and there is evidence for epistasis between DISC1 and its interactors in disease risk. Moreover, consistent with its multiple roles in the developing and adult brain, DISC1 is now also implicated in risk of autism spectrum disorders, and there is emerging genetic and molecular evidence of overlap with molecular pathways relating to proteins involved in Alzheimer’s disease, while protein network analysis indicates possible functional overlap with Huntingtin. It will therefore be interesting to find out if DISC1 dysfunction can result in the kind of neurodegeneration that characterises Alzheimer’s or Huntington’s diseases.
An increasing number of studies are addressing the effects of missense variants upon DISC1 function, and common variants are now known to influence DISC1 oligomerisation, and nuclear targeting, with effects upon signalling pathways, nuclear transcription, centrosome composition and mitochondrial trafficking. Normal variation thus influences DISC1 function, and consequently numerous brain processes, at many levels. Notably, some of the same processes are also affected by at least one rare, putatively causal variant. Consistent with these observations, imaging studies have demonstrated effects of common DISC1 variants upon brain structure in adults, either unaffected or diagnosed with a mental disorder. These effects upon DISC1 function and brain structure likely contribute to disease risk, as evidenced by the association of some of the common variants with psychiatric disorders. Moreover, the paradigm of interaction between genes and the environment in determining disease risk is well established for major mental illness (van Os et al., 2008), with maternal deprivation stress and maternal immune activation (for example due to viral infection) already believed to be critical environmental risk factors. It is thus intriguing that the genetic risk factor DISC1 apparently interacts with these environmental risk factors to adversely affect postnatal brain development or behavioural phenotypes in mice. DISC1 also interacts directly with transcription factors that regulate the pathways by which cells respond to a variety of environmental stressors, and may therefore directly mediate the effects of stress. Such DISC1-environment interactions may go some way towards explaining the incomplete penetrance of the t(1;11) translocation (Blackwood et al., 2001).
Studying DISC1 function has revealed much about the brain processes that are likely to be adversely affected by DISC1 dysfunction and one of the challenges now is to determine if/how these pathways involving DISC1 contribute to psychiatric disorders, and why the outcomes of DISC1 dysfunction are so variable - the t(1;11) translocation apparently causes brain dysfunction in all carriers, but some carriers are unaffected, while others suffer from schizophrenia, bipolar disorder or depression. Moreover, one of the major motivations for studying DISC1, or indeed any other risk factor for major mental illness, is ultimately to develop more effective therapeutic interventions than are currently available. DISC1 itself is not likely to be a viable drug target, but the ever-increasing information on DISC1 brain function is revealing pathways that are, or may be, druggable in patients. Another considerable challenge will therefore be to determine if/which pathways involving DISC1 can be targeted to effectively treat the symptoms of psychiatric illness. A number of DISC1 mutant mice with defined behavioural phenotypes have been generated that will aid in this process, and induced pluripotent stem cells will provide an essential complementary tool. IPSC can now be generated directly from patient-derived primary cells such as skin fibroblasts, and ultimately differentiated into neural progenitors, specified neuronal subtypes or even glial cells. Because these cells can be generated from patients and their unaffected family members it is possible to assay the effects of endogenous mutations, CNVs, common variants etc in human neural material, and importantly, against the relevant genetic background which may include genetic modifiers or other risk alleles. Such IPSC have already been generated for the four base pair deletion in exon 12 identified in one family (Chiang et al., 2011), and are in prospect for the t(1;11) translocation family.
Acknowledgements
We thank David Porteous for critical reading of this manuscript. DCS is funded by the Wellcome Trust (WT088179MA), MB is funded by an MRC studentship and EG is funded by the MRC (MR/J004367/1).
References
- Abazyan B, Nomura J, Kannan G, Ishizuka K, Tamashiro KL, Nucifora F, Pogorelov V, Ladenheim B, Yang C, Krasnova IN, Cadet JL, Pardo C, Mori S, Kamiya A, Vogel MW, Sawa A, Ross CA, Pletnikov MV. Prenatal interaction of mutant DISC1 and immune activation produces adult psychopathology. Biol Psychiatry. 2010;68:1172–1181. doi: 10.1016/j.biopsych.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhaja AK, Jans DC, Nikolov M, Vukotic M, Lytovchenko O, Ludewig F, Schliebs W, Riedel D, Urlaub H, Jakobs S, Deckers M. MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization. Mol Biol Cell. 2012;23:247–257. doi: 10.1091/mbc.E11-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkuraya FS, Cai X, Emery C, Mochida GH, Al-Dosari MS, Felie JM, Hill RS, Barry BJ, Partlow JN, Gascon GG, Kentab A, Jan M, Shaheen R, Feng Y, Walsh CA. Human mutations in NDE1 cause extreme microcephaly with lissencephaly [corrected] Am J Hum Genet. 2011;88:536–547. doi: 10.1016/j.ajhg.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador-Arjona A, Elliott J, Miller A, Ginbey A, Pazour GJ, Enikolopov G, Roberts AJ, Terskikh AV. Primary cilia regulate proliferation of amplifying progenitors in adult hippocampus: implications for learning and memory. J Neurosci. 2011;31:9933–9944. doi: 10.1523/JNEUROSCI.1062-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameri K, Harris AL. Activating transcription factor 4. Int J Biochem Cell Biol. 2008;40:14–21. doi: 10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- Ames A., 3rd CNS energy metabolism as related to function. Brain Res Brain Res Rev. 2000;34:42–68. doi: 10.1016/s0165-0173(00)00038-2. [DOI] [PubMed] [Google Scholar]
- An J, Shi J, He Q, Lui K, Liu Y, Huang Y, Sheikh MS. CHCM1/CHCHD6, novel mitochondrial protein linked to regulation of mitofilin and mitochondrial cristae morphology. J Biol Chem. 2012;287:7411–7426. doi: 10.1074/jbc.M111.277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen NC, Wilcox MA, Ho BC, Epping E, Ziebell S, Zeien E, Weiss B, Wassink T. Statistical epistasis and progressive brain change in schizophrenia: an approach for examining the relationships between multiple genes. Mol Psychiatry. 2011 doi: 10.1038/mp.2011.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin TA, Brandon NJ, Kittler JT. Disrupted in Schizophrenia 1 forms pathological aggresomes that disrupt its function in intracellular transport. Hum Mol Genet. 2012;21:2017–2028. doi: 10.1093/hmg/dds018. [DOI] [PubMed] [Google Scholar]
- Atkin TA, Macaskill AF, Brandon NJ, Kittler JT. Disrupted in Schizophrenia-1 regulates intracellular trafficking of mitochondria in neurons. Mol Psychiatry. 16:122–124. 121. doi: 10.1038/mp.2010.110. [DOI] [PubMed] [Google Scholar]
- Atkin TA, Macaskill AF, Brandon NJ, Kittler JT. Disrupted in Schizophrenia-1 regulates intracellular trafficking of mitochondria in neurons. Mol Psychiatry. 2011;16:122–124. 121. doi: 10.1038/mp.2010.110. [DOI] [PubMed] [Google Scholar]
- Austin CP, Ky B, Ma L, Morris JA, Shughrue PJ. Expression of Disrupted-In-Schizophrenia-1, a schizophrenia-associated gene, is prominent in the mouse hippocampus throughout brain development. Neuroscience. 2004;124:3–10. doi: 10.1016/j.neuroscience.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, Patel SD, Winiger E, Breier A, Shekhar A, Amdur R, Koller D, Nurnberger JI, Corvin A, Geyer M, Tsuang MT, Salomon D, Schork NJ, Fanous AH, O’donovan MC, Niculescu AB. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol Psychiatry. 2012;17:887–905. doi: 10.1038/mp.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakircioglu M, Carvalho OP, Khurshid M, Cox JJ, Tuysuz B, Barak T, Yilmaz S, Caglayan O, Dincer A, Nicholas AK, Quarrell O, Springell K, Karbani G, Malik S, Gannon C, Sheridan E, Crosier M, Lisgo SN, Lindsay S, Bilguvar K, Gergely F, Gunel M, Woods CG. The essential role of centrosomal NDE1 in human cerebral cortex neurogenesis. Am J Hum Genet. 2011;88:523–535. doi: 10.1016/j.ajhg.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch D, Ghirardi M, Skehel PA, Karl KA, Herder SP, Chen M, Bailey CH, Kandel ER. Aplysia CREB2 represses long-term facilitation: relief of repression converts transient facilitation into long-term functional and structural change. Cell. 1995;83:979–992. doi: 10.1016/0092-8674(95)90213-9. [DOI] [PubMed] [Google Scholar]
- Beecham GW, Martin ER, Li YJ, Slifer MA, Gilbert JR, Haines JL, Pericak-Vance MA. Genome-wide association study implicates a chromosome 12 risk locus for late-onset Alzheimer disease. Am J Hum Genet. 2009;84:35–43. doi: 10.1016/j.ajhg.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari NF, O’connor AK, Haycraft CJ, Yoder BK. The primary cilium as a complex signaling center. Curr Biol. 2009;19:R526–535. doi: 10.1016/j.cub.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood DH, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders--cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxall R, Porteous DJ, Thomson PA. DISC1 and Huntington’s disease-- overlapping pathways of vulnerability to neurological disorder? PLoS One. 2011;6:e16263. doi: 10.1371/journal.pone.0016263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw NJ, Christie S, Soares DC, Carlyle BC, Porteous DJ, Millar JK. NDE1 and NDEL1: multimerisation, alternate splicing and DISC1 interaction. Neurosci Lett. 2009;449:228–233. doi: 10.1016/j.neulet.2008.10.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw NJ, Ogawa F, Antolin-Fontes B, Chubb JE, Carlyle BC, Christie S, Claessens A, Porteous DJ, Millar JK. DISC1, PDE4B, and NDE1 at the centrosome and synapse. Biochem Biophys Res Commun. 2008;377:1091–1096. doi: 10.1016/j.bbrc.2008.10.120. [DOI] [PubMed] [Google Scholar]
- Bradshaw NJ, Soares DC, Carlyle BC, Ogawa F, Davidson-Smith H, Christie S, Mackie S, Thomson PA, Porteous DJ, Millar JK. PKA phosphorylation of NDE1 is DISC1/PDE4 dependent and modulates its interaction with LIS1 and NDEL1. J Neurosci. 2011;31:9043–9054. doi: 10.1523/JNEUROSCI.5410-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon NJ, Handford EJ, Schurov I, Rain JC, Pelling M, Duran-Jimeniz B, Camargo LM, Oliver KR, Beher D, Shearman MS, Whiting PJ. Disrupted in Schizophrenia 1 and Nudel form a neurodevelopmentally regulated protein complex: implications for schizophrenia and other major neurological disorders. Mol Cell Neurosci. 2004a;25:42–55. doi: 10.1016/j.mcn.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Brandon NJ, Handford EJ, Schurov I, Rain JC, Pelling M, Duran-Jimeniz B, Camargo LM, Oliver KR, Beher D, Shearman MS, Whiting PJ. Disrupted in Schizophrenia 1 and Nudel form a neurodevelopmentally regulated protein complex: implications for schizophrenia and other major neurological disorders. Molecular and Cellular Neuroscience. 2004b;25:42–55. doi: 10.1016/j.mcn.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Brandon NJ, Sawa A. Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nat Rev Neurosci. 2011;12:707–722. doi: 10.1038/nrn3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauns S, Gollub RL, Roffman JL, Yendiki A, Ho BC, Wassink TH, Heinz A, Ehrlich S. DISC1 is associated with cortical thickness and neural efficiency. Neuroimage. 57:1591–1600. doi: 10.1016/j.neuroimage.2011.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauns S, Gollub RL, Roffman JL, Yendiki A, Ho BC, Wassink TH, Heinz A, Ehrlich S. DISC1 is associated with cortical thickness and neural efficiency. Neuroimage. 2011;57:1591–1600. doi: 10.1016/j.neuroimage.2011.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig JJ, Sarkisian MR, Arellano JI, Morozov YM, Ayoub AE, Sojitra S, Wang B, Flavell RA, Rakic P, Town T. Primary cilia regulate hippocampal neurogenesis by mediating sonic hedgehog signaling. Proc Natl Acad Sci U S A. 2008;105:13127–13132. doi: 10.1073/pnas.0804558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SM, Clapcote SJ, Millar JK, Torrance HS, Anderson SM, Walker R, Rampino A, Roder JC, Thomson PA, Porteous DJ, Evans KL. Synaptic modulators Nrxn1 and Nrxn3 are disregulated in a Disc1 mouse model of schizophrenia. Mol Psychiatry. 2011;16:585–587. doi: 10.1038/mp.2010.134. [DOI] [PubMed] [Google Scholar]
- Buizer-Voskamp JE, Muntjewerff JW, Strengman E, Sabatti C, Stefansson H, Vorstman JA, Ophoff RA. Genome-wide analysis shows increased frequency of copy number variation deletions in Dutch schizophrenia patients. Biol Psychiatry. 2011;70:655–662. doi: 10.1016/j.biopsych.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick KE, Kamiya A, Hodgkinson CA, Lencz T, Derosse P, Ishizuka K, Elashvili S, Arai H, Goldman D, Sawa A, Malhotra AK. Elucidating the relationship between DISC1, NDEL1 and NDE1 and the risk for schizophrenia: evidence of epistasis and competitive binding. Hum Mol Genet. 2008;17:2462–2473. doi: 10.1093/hmg/ddn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callicott JH, Straub RE, Pezawas L, Egan MF, Mattay VS, Hariri AR, Verchinski BA, Meyer-Lindenberg A, Balkissoon R, Kolachana B, Goldberg TE, Weinberger DR. Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc Natl Acad Sci U S A. 2005;102:8627–8632. doi: 10.1073/pnas.0500515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo LM, Collura V, Rain JC, Mizuguchi K, Hermjakob H, Kerrien S, Bonnert TP, Whiting PJ, Brandon NJ. Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol Psychiatry. 2007;12:74–86. doi: 10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- Cannon TD, Hennah W, Van Erp TG, Thompson PM, Lonnqvist J, Huttunen M, Gasperoni T, Tuulio-Henriksson A, Pirkola T, Toga AW, Kaprio J, Mazziotta J, Peltonen L. Association of DISC1/TRAX haplotypes with schizophrenia, reduced prefrontal gray matter, and impaired short- and long-term memory. Arch Gen Psychiatry. 2005;62:1205–1213. doi: 10.1001/archpsyc.62.11.1205. [DOI] [PubMed] [Google Scholar]
- Carless MA, Glahn DC, Johnson MP, Curran JE, Bozaoglu K, Dyer TD, Winkler AM, Cole SA, Almasy L, Maccluer JW, Duggirala R, Moses EK, Goring HH, Blangero J. Impact of DISC1 variation on neuroanatomical and neurocognitive phenotypes. Mol Psychiatry. 2011;16:1096–1104. 1063. doi: 10.1038/mp.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle HJ, Luong TN, Medina-Marino A, Schenker L, Khorosheva E, Indersmitten T, Gunapala KM, Steele AD, O’dell TJ, Patterson PH, Kennedy MB. Deletion of densin-180 results in abnormal behaviors associated with mental illness and reduces mGluR5 and DISC1 in the postsynaptic density fraction. J Neurosci. 2011;31:16194–16207. doi: 10.1523/JNEUROSCI.5877-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlyle BC, Mackie S, Christie S, Millar JK, Porteous DJ. Co-ordinated action of DISC1, PDE4B and GSK3beta in modulation of cAMP signalling. Mol Psychiatry. 2011;16:693–694. doi: 10.1038/mp.2011.17. [DOI] [PubMed] [Google Scholar]
- Chakirova G, Whalley HC, Thomson PA, Hennah W, Moorhead TW, Welch KA, Giles S, Hall J, Johnstone EC, Lawrie SM, Porteous DJ, Brown VJ, Mcintosh AM. The effects of DISC1 risk variants on brain activation in controls, patients with bipolar disorder and patients with schizophrenia. Psychiatry Res. 2011;192:20–28. doi: 10.1016/j.pscychresns.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Chakravarty MM, Felsky D, Tampakeras M, Lerch JP, Mulsant BH, Kennedy JL, Voineskos AN. DISC1 and Striatal Volume: A Potential Risk Phenotype For mental Illness. Front Psychiatry. 2012;3:57. doi: 10.3389/fpsyt.2012.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Muzzio IA, Malleret G, Bartsch D, Verbitsky M, Pavlidis P, Yonan AL, Vronskaya S, Grody MB, Cepeda I, Gilliam TC, Kandel ER. Inducible enhancement of memory storage and synaptic plasticity in transgenic mice expressing an inhibitor of ATF4 (CREB-2) and C/EBP proteins. Neuron. 2003;39:655–669. doi: 10.1016/s0896-6273(03)00501-4. [DOI] [PubMed] [Google Scholar]
- Chen SY, Huang PH, Cheng HJ. Disrupted-in-Schizophrenia 1-mediated axon guidance involves TRIO-RAC-PAK small GTPase pathway signaling. Proc Natl Acad Sci U S A. 2011;108:5861–5866. doi: 10.1073/pnas.1018128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang CH, Su Y, Wen Z, Yoritomo N, Ross CA, Margolis RL, Song H, Ming GL. Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Mol Psychiatry. 2011;16:358–360. doi: 10.1038/mp.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S, Hashimoto R, Hattori S, Yohda M, Lipska B, Weinberger DR, Kunugi H. Effect of antipsychotic drugs on DISC1 and dysbindin expression in mouse frontal cortex and hippocampus. J Neural Transm. 2006;113:1337–1346. doi: 10.1007/s00702-005-0414-1. [DOI] [PubMed] [Google Scholar]
- Chubb JE, Bradshaw NJ, Soares DC, Porteous DJ, Millar JK. The DISC locus in psychiatric illness. Mol Psychiatry. 2008;13:36–64. doi: 10.1038/sj.mp.4002106. [DOI] [PubMed] [Google Scholar]
- Clapcote SJ, Lipina TV, Millar JK, Mackie S, Christie S, Ogawa F, Lerch JP, Trimble K, Uchiyama M, Sakuraba Y, Kaneda H, Shiroishi T, Houslay MD, Henkelman RM, Sled JG, Gondo Y, Porteous DJ, Roder JC. Behavioral phenotypes of Disc1 missense mutations in mice. Neuron. 2007;54:387–402. doi: 10.1016/j.neuron.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Clay HB, Sillivan S, Konradi C. Mitochondrial dysfunction and pathology in bipolar disorder and schizophrenia. Int J Dev Neurosci. 2011;29:311–324. doi: 10.1016/j.ijdevneu.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins DM, Murdoch H, Dunlop AJ, Charych E, Baillie GS, Wang Q, Herberg FW, Brandon N, Prinz A, Houslay MD. Ndel1 alters its conformation by sequestering cAMP-specific phosphodiesterase-4D3 (PDE4D3) in a manner that is dynamically regulated through Protein Kinase A (PKA) Cell Signal. 2008;20:2356–2369. doi: 10.1016/j.cellsig.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M, Gobert D, Stern E, Gamache K, Colina R, Cuello C, Sossin W, Kaufman R, Pelletier J, Rosenblum K, Krnjevic K, Lacaille JC, Nader K, Sonenberg N. eIF2alpha phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell. 2007;129:195–206. doi: 10.1016/j.cell.2007.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sonenberg N. Translational control of long-term synaptic plasticity and memory storage by eIF2alpha. Crit Rev Neurobiol. 2006;18:187–195. doi: 10.1615/critrevneurobiol.v18.i1-2.190. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Wahlsten D, Dudek BC. Genetics of mouse behavior: interactions with laboratory environment. Science. 1999;284:1670–1672. doi: 10.1126/science.284.5420.1670. [DOI] [PubMed] [Google Scholar]
- Crepel A, Breckpot J, Fryns JP, De La Marche W, Steyaert J, Devriendt K, Peeters H. DISC1 duplication in two brothers with autism and mild mental retardation. Clin Genet. 2010;77:389–394. doi: 10.1111/j.1399-0004.2009.01318.x. [DOI] [PubMed] [Google Scholar]
- Crowley JJ, Hilliard CE, Kim Y, Morgan MB, Lewis LR, Muzny DM, Hawes AC, Sabo A, Wheeler DA, Lieberman JA, Sullivan PF, Gibbs RA. Deep resequencing and association analysis of schizophrenia candidate genes. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva Alves F, Figee M, Van Avamelsvoort T, Veltman D, De Haan L. The revised dopamine hypothesis of schizophrenia: evidence from pharmacological MRI studies with atypical antipsychotic medication. Psychopharmacol Bull. 2008;41:121–132. [PubMed] [Google Scholar]
- Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rienzo G, Bishop JA, Mao Y, Pan L, Ma TP, Moens CB, Tsai LH, Sive H. Disc1 regulates both beta-catenin-mediated and noncanonical Wnt signaling during vertebrate embryogenesis. FASEB J. 2011;25:4184–4197. doi: 10.1096/fj.11-186239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debono R, Topless R, Markie D, Black MA, Merriman TR. Analysis of the DISC1 translocation partner (11q14.3) in genetic risk of schizophrenia. Genes Brain Behav. 2012 doi: 10.1111/j.1601-183X.2012.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio A, Blasi G, Sambataro F, Rampino A, Papazacharias A, Gambi F, Romano R, Caforio G, Rizzo M, Latorre V, Popolizio T, Kolachana B, Callicott JH, Nardini M, Weinberger DR, Bertolino A. Association of the SerCys DISC1 polymorphism with human hippocampal formation gray matter and function during memory encoding. Eur J Neurosci. 2008;28:2129–2136. doi: 10.1111/j.1460-9568.2008.06482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domire JS, Green JA, Lee KG, Johnson AD, Askwith CC, Mykytyn K. Dopamine receptor 1 localizes to neuronal cilia in a dynamic process that requires the Bardet-Biedl syndrome proteins. Cell Mol Life Sci. 2011;68:2951–2960. doi: 10.1007/s00018-010-0603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drerup CM, Wiora HM, Topczewski J, Morris JA. Disc1 regulates foxd3 and sox10 expression, affecting neural crest migration and differentiation. Development. 2009;136:2623–2632. doi: 10.1242/dev.030577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Chang JH, Ge S, Faulkner RL, Kim JY, Kitabatake Y, Liu XB, Yang CH, Jordan JD, Ma DK, Liu CY, Ganesan S, Cheng HJ, Ming GL, Lu B, Song H. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130:1146–1158. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood SL, Hodgkinson CA, Harrison PJ. DISC-1 Leu607Phe alleles differentially affect centrosomal PCM1 localization and neurotransmitter release. Mol Psychiatry. 2009;14:556–557. doi: 10.1038/mp.2009.13. [DOI] [PubMed] [Google Scholar]
- Eastwood SL, Walker M, Hyde TM, Kleinman JE, Harrison PJ. The DISC1 Ser704Cys substitution affects centrosomal localization of its binding partner PCM1 in glia in human brain. Hum Mol Genet. 2010;19:2487–2496. doi: 10.1093/hmg/ddq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto A. [Roles of DISC1-interacting protein Girdin in postnatal development and adult neurogenesis in the dentate gyrus] Nihon Shinkei Seishin Yakurigaku Zasshi. 2011;31:23–28. [PubMed] [Google Scholar]
- Eykelenboom JE, Briggs GJ, Bradshaw NJ, Soares DC, Ogawa F, Christie S, Malavasi EL, Makedonopoulou P, Mackie S, Malloy MP, Wear MA, Blackburn EA, Bramham J, Mcintosh AM, Blackwood DH, Muir WJ, Porteous DJ, Millar JK. A t(1;11) translocation linked to schizophrenia and affective disorders gives rise to aberrant chimeric DISC1 transcripts that encode structurally altered, deleterious mitochondrial proteins. Hum Mol Genet. 2012;21:3374–3386. doi: 10.1093/hmg/dds169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner RL, Jang MH, Liu XB, Duan X, Sailor KA, Kim JY, Ge S, Jones EG, Ming GL, Song H, Cheng HJ. Development of hippocampal mossy fiber synaptic outputs by new neurons in the adult brain. Proc Natl Acad Sci U S A. 2008;105:14157–14162. doi: 10.1073/pnas.0806658105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Walsh CA. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44:279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Filges I, Rothlisberger B, Boesch N, Weber P, Wenzel F, Huber AR, Heinimann K, Miny P. Interstitial deletion 1q42 in a patient with agenesis of corpus callosum: Phenotype-genotype comparison to the 1q41q42 microdeletion suggests a contiguous 1q4 syndrome. Am J Med Genet A. 2010;152A:987–993. doi: 10.1002/ajmg.a.33330. [DOI] [PubMed] [Google Scholar]
- Flores R, 3rd, Hirota Y, Armstrong B, Sawa A, Tomoda T. DISC1 regulates synaptic vesicle transport via a lithium-sensitive pathway. Neurosci Res. 2011;71:71–77. doi: 10.1016/j.neures.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank CL, Ge X, Xie Z, Zhou Y, Tsai LH. Control of activating transcription factor 4 (ATF4) persistence by multisite phosphorylation impacts cell cycle progression and neurogenesis. J Biol Chem. 2010;285:33324–33337. doi: 10.1074/jbc.M110.140699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Maturana AD, Ikuta J, Hamada J, Walchli S, Suzuki T, Sawa H, Wooten MW, Okajima T, Tatematsu K, Tanizawa K, Kuroda S. Axonal guidance protein FEZ1 associates with tubulin and kinesin motor protein to transport mitochondria in neurites of NGF-stimulated PC12 cells. Biochem Biophys Res Commun. 2007;361:605–610. doi: 10.1016/j.bbrc.2007.07.050. [DOI] [PubMed] [Google Scholar]
- Fukuda T, Sugita S, Inatome R, Yanagi S. CAMDI, a novel disrupted in schizophrenia 1 (DISC1)-binding protein, is required for radial migration. J Biol Chem. 2010;285:40554–40561. doi: 10.1074/jbc.M110.179481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperoni TL, Ekelund J, Huttunen M, Palmer CG, Tuulio-Henriksson A, Lonnqvist J, Kaprio J, Peltonen L, Cannon TD. Genetic linkage and association between chromosome 1q and working memory function in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2003;116B:8–16. doi: 10.1002/ajmg.b.10757. [DOI] [PubMed] [Google Scholar]
- Gentile M, Di Carlo A, Volpe P, Pansini A, Nanna P, Valenzano MC, Buonadonna AL. FISH and cytogenetic characterization of a terminal chromosome 1q deletion: clinical case report and phenotypic implications. Am J Med Genet A. 2003;117A:251–254. doi: 10.1002/ajmg.a.10018. [DOI] [PubMed] [Google Scholar]
- Gershon ES, Alliey-Rodriguez N, Liu C. After GWAS: searching for genetic risk for schizophrenia and bipolar disorder. Am J Psychiatry. 2011;168:253–256. doi: 10.1176/appi.ajp.2010.10091340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EK, Grozeva D, Sims R, Raybould R, Forty L, Gordon-Smith K, Russell E, St Clair D, Young AH, Ferrier IN, Kirov G, Jones I, Jones L, Owen MJ, O’donovan MC, Craddock N. DISC1 exon 11 rare variants found more commonly in schizoaffective spectrum cases than controls. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:490–492. doi: 10.1002/ajmg.b.31187. [DOI] [PubMed] [Google Scholar]
- Green EK, Norton N, Peirce T, Grozeva D, Kirov G, Owen MJ, O’donovan MC, Craddock N. Evidence that a DISC1 frame-shift deletion associated with psychosis in a single family may not be a pathogenic mutation. Mol Psychiatry. 2006;11:798–799. doi: 10.1038/sj.mp.4001853. [DOI] [PubMed] [Google Scholar]
- Green TA, Alibhai IN, Unterberg S, Neve RL, Ghose S, Tamminga CA, Nestler EJ. Induction of activating transcription factors (ATFs) ATF2, ATF3, and ATF4 in the nucleus accumbens and their regulation of emotional behavior. J Neurosci. 2008;28:2025–2032. doi: 10.1523/JNEUROSCI.5273-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene LA, Lee HY, Angelastro JM. The transcription factor ATF5: role in neurodevelopment and neural tumors. J Neurochem. 2009;108:11–22. doi: 10.1111/j.1471-4159.2008.05749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guven A, Gunduz A, Bozoglu TM, Yalcinkaya C, Tolun A. Novel NDE1 homozygous mutation resulting in microhydranencephaly and not microlyssencephaly. Neurogenetics. 2012;13:189–194. doi: 10.1007/s10048-012-0326-9. [DOI] [PubMed] [Google Scholar]
- Han XJ, Tomizawa K, Fujimura A, Ohmori I, Nishiki T, Matsushita M, Matsui H. Regulation of mitochondrial dynamics and neurodegenerative diseases. Acta Med Okayama. 65:1–10. doi: 10.18926/AMO/43824. [DOI] [PubMed] [Google Scholar]
- Han YG, Spassky N, Romaguera-Ros M, Garcia-Verdugo JM, Aguilar A, Schneider-Maunoury S, Alvarez-Buylla A. Hedgehog signaling and primary cilia are required for the formation of adult neural stem cells. Nat Neurosci. 2008;11:277–284. doi: 10.1038/nn2059. [DOI] [PubMed] [Google Scholar]
- Handel M, Schulz S, Stanarius A, Schreff M, Erdtmann-Vourliotis M, Schmidt H, Wolf G, Hollt V. Selective targeting of somatostatin receptor 3 to neuronal cilia. Neuroscience. 1999;89:909–926. doi: 10.1016/s0306-4522(98)00354-6. [DOI] [PubMed] [Google Scholar]
- Hanson ND, Owens MJ, Nemeroff CB. Depression, antidepressants, and neurogenesis: a critical reappraisal. Neuropsychopharmacology. 2011;36:2589–2602. doi: 10.1038/npp.2011.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque FN, Lipina TV, Roder JC, Wong AH. Social defeat interacts with Disc1 mutations in the mouse to affect behavior. Behav Brain Res. 2012;233:337–344. doi: 10.1016/j.bbr.2012.05.037. [DOI] [PubMed] [Google Scholar]
- Harris SE, Hennah W, Thomson PA, Luciano M, Starr JM, Porteous DJ, Deary IJ. Variation in DISC1 is associated with anxiety, depression and emotional stability in elderly women. Mol Psychiatry. 2010;15:232–234. doi: 10.1038/mp.2009.88. [DOI] [PubMed] [Google Scholar]
- Hashimoto R, Numakawa T, Ohnishi T, Kumamaru E, Yagasaki Y, Ishimoto T, Mori T, Nemoto K, Adachi N, Izumi A, Chiba S, Noguchi H, Suzuki T, Iwata N, Ozaki N, Taguchi T, Kamiya A, Kosuga A, Tatsumi M, Kamijima K, Weinberger DR, Sawa A, Kunugi H. Impact of the DISC1 Ser704Cys polymorphism on risk for major depression, brain morphology and ERK signaling. Hum Mol Genet. 2006;15:3024–3033. doi: 10.1093/hmg/ddl244. [DOI] [PubMed] [Google Scholar]
- Hayashi-Takagi A, Takaki M, Graziane N, Seshadri S, Murdoch H, Dunlop AJ, Makino Y, Seshadri AJ, Ishizuka K, Srivastava DP, Xie Z, Baraban JM, Houslay MD, Tomoda T, Brandon NJ, Kamiya A, Yan Z, Penzes P, Sawa A. Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat Neurosci. 13:327–332. doi: 10.1038/nn.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Takagi A, Takaki M, Graziane N, Seshadri S, Murdoch H, Dunlop AJ, Makino Y, Seshadri AJ, Ishizuka K, Srivastava DP, Xie Z, Baraban JM, Houslay MD, Tomoda T, Brandon NJ, Kamiya A, Yan Z, Penzes P, Sawa A. Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat Neurosci. 2010;13:327–332. doi: 10.1038/nn.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennah W, Porteous D. The DISC1 pathway modulates expression of neurodevelopmental, synaptogenic and sensory perception genes. PLoS One. 2009;4:e4906. doi: 10.1371/journal.pone.0004906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennah W, Thomson P, Mcquillin A, Bass N, Loukola A, Anjorin A, Blackwood D, Curtis D, Deary IJ, Harris SE, Isometsa ET, Lawrence J, Lonnqvist J, Muir W, Palotie A, Partonen T, Paunio T, Pylkko E, Robinson M, Soronen P, Suominen K, Suvisaari J, Thirumalai S, St Clair D, Gurling H, Peltonen L, Porteous D. DISC1 association, heterogeneity and interplay in schizophrenia and bipolar disorder. Mol Psychiatry. 2009;14:865–873. doi: 10.1038/mp.2008.22. [DOI] [PubMed] [Google Scholar]
- Hennah W, Tomppo L, Hiekkalinna T, Palo OM, Kilpinen H, Ekelund J, Tuulio-Henriksson A, Silander K, Partonen T, Paunio T, Terwilliger JD, Lonnqvist J, Peltonen L. Families with the risk allele of DISC1 reveal a link between schizophrenia and another component of the same molecular pathway, NDE1. Hum Mol Genet. 2007;16:453–462. doi: 10.1093/hmg/ddl462. [DOI] [PubMed] [Google Scholar]
- Higginbotham HR, Gleeson JG. The centrosome in neuronal development. Trends Neurosci. 2007;30:276–283. doi: 10.1016/j.tins.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Hikida T, Jaaro-Peled H, Seshadri S, Oishi K, Hookway C, Kong S, Wu D, Xue R, Andrade M, Tankou S, Mori S, Gallagher M, Ishizuka K, Pletnikov M, Kida S, Sawa A. Dominant-negative DISC1 transgenic mice display schizophrenia-associated phenotypes detected by measures translatable to humans. Proc Natl Acad Sci U S A. 2007;104:14501–14506. doi: 10.1073/pnas.0704774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda A, Miyoshi K, Baba K, Taniguchi M, Koyama Y, Kuroda S, Katayama T, Tohyama M. Expression of fasciculation and elongation protein zeta-1 (FEZ1) in the developing rat brain. Brain Res Mol Brain Res. 2004;122:89–92. doi: 10.1016/j.molbrainres.2003.11.020. [DOI] [PubMed] [Google Scholar]
- Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, Devay RM, Lackner LL, Westermann B, Schuldiner M, Weissman JS, Nunnari J. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J Cell Biol. 2011;195:323–340. doi: 10.1083/jcb.201107053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta Y, Ohnuma T, Hanzawa R, Shibata N, Maeshima H, Baba H, Hatano T, Takebayashi Y, Kitazawa M, Higa M, Suzuki T, Arai H. Association study between Disrupted-in-Schizophrenia-1 (DISC1) and Japanese patients with treatment-resistant schizophrenia (TRS) Prog Neuropsychopharmacol Biol Psychiatry. 2011;35:636–639. doi: 10.1016/j.pnpbp.2011.01.011. [DOI] [PubMed] [Google Scholar]
- Houlihan LM, Harris SE, Luciano M, Gow AJ, Starr JM, Visscher PM, Deary IJ. Replication study of candidate genes for cognitive abilities: the Lothian Birth Cohort 1936. Genes Brain Behav. 2009;8:238–247. doi: 10.1111/j.1601-183X.2008.00470.x. [DOI] [PubMed] [Google Scholar]
- Houslay MD, Adams DR. PDE4 cAMP phosphodiesterases: modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem J. 2003;370:1–18. doi: 10.1042/BJ20021698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibi D, Nagai T, Koike H, Kitahara Y, Mizoguchi H, Niwa M, Jaaro-Peled H, Nitta A, Yoneda Y, Nabeshima T, Sawa A, Yamada K. Combined effect of neonatal immune activation and mutant DISC1 on phenotypic changes in adulthood. Behav Brain Res. 2010;206:32–37. doi: 10.1016/j.bbr.2009.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi T, Izumi H, Uchiumi T, Nishio K, Arao T, Tanabe M, Uramoto H, Sugio K, Yasumoto K, Sasaguri Y, Wang KY, Otsuji Y, Kohno K. Clock and ATF4 transcription system regulates drug resistance in human cancer cell lines. Oncogene. 2007;26:4749–4760. doi: 10.1038/sj.onc.1210289. [DOI] [PubMed] [Google Scholar]
- Ikuta J, Maturana A, Fujita T, Okajima T, Tatematsu K, Tanizawa K, Kuroda S. Fasciculation and elongation protein zeta-1 (FEZ1) participates in the polarization of hippocampal neuron by controlling the mitochondrial motility. Biochem Biophys Res Commun. 2007;353:127–132. doi: 10.1016/j.bbrc.2006.11.142. [DOI] [PubMed] [Google Scholar]
- Ingason A, Rujescu D, Cichon S, Sigurdsson E, Sigmundsson T, Pietilainen OP, Buizer-Voskamp JE, Strengman E, Francks C, Muglia P, Gylfason A, Gustafsson O, Olason PI, Steinberg S, Hansen T, Jakobsen KD, Rasmussen HB, Giegling I, Moller HJ, Hartmann A, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Bramon E, Kiemeney LA, Franke B, Murray R, Vassos E, Toulopoulou T, Muhleisen TW, Tosato S, Ruggeri M, Djurovic S, Andreassen OA, Zhang Z, Werge T, Ophoff RA, Rietschel M, Nothen MM, Petursson H, Stefansson H, Peltonen L, Collier D, Stefansson K, St Clair DM. Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol Psychiatry. 2011;16:17–25. doi: 10.1038/mp.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka K, Kamiya A, Oh EC, Kanki H, Seshadri S, Robinson JF, Murdoch H, Dunlop AJ, Kubo K, Furukori K, Huang B, Zeledon M, Hayashi-Takagi A, Okano H, Nakajima K, Houslay MD, Katsanis N, Sawa A. DISC1-dependent switch from progenitor proliferation to migration in the developing cortex. Nature. 2011;473:92–96. doi: 10.1038/nature09859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James R, Adams RR, Christie S, Buchanan SR, Porteous DJ, Millar JK. Disrupted in Schizophrenia 1 (DISC1) is a multicompartmentalized protein that predominantly localizes to mitochondria. Mol Cell Neurosci. 2004;26:112–122. doi: 10.1016/j.mcn.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Jepsen K, Rosenfeld MG. Biological roles and mechanistic actions of co-repressor complexes. J Cell Sci. 2002;115:689–698. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- Jia P, Wang L, Fanous AH, Pato CN, Edwards TL, Zhao Z. Network-Assisted Investigation of Combined Causal Signals from Genome-Wide Association Studies in Schizophrenia. PLoS Comput Biol. 2012;8:e1002587. doi: 10.1371/journal.pcbi.1002587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John GB, Shang Y, Li L, Renken C, Mannella CA, Selker JM, Rangell L, Bennett MJ, Zha J. The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol Biol Cell. 2005;16:1543–1554. doi: 10.1091/mbc.E04-08-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahler AK, Rimol LM, Brown AA, Djurovic S, Hartberg CB, Melle I, Dale AM, Andreassen OA, Agartz I. Effect of DISC1 SNPs on brain structure in healthy controls and patients with a history of psychosis. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:722–730. doi: 10.1002/ajmg.b.32076. [DOI] [PubMed] [Google Scholar]
- Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, Ozeki Y, Sawamura N, Park U, Kudo C, Okawa M, Ross CA, Hatten ME, Nakajima K, Sawa A. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- Kamiya A, Tan PL, Kubo K, Engelhard C, Ishizuka K, Kubo A, Tsukita S, Pulver AE, Nakajima K, Cascella NG, Katsanis N, Sawa A. Recruitment of PCM1 to the centrosome by the cooperative action of DISC1 and BBS4: a candidate for psychiatric illnesses. Arch Gen Psychiatry. 2008;65:996–1006. doi: 10.1001/archpsyc.65.9.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang E, Burdick KE, Kim JY, Duan X, Guo JU, Sailor KA, Jung DE, Ganesan S, Choi S, Pradhan D, Lu B, Avramopoulos D, Christian K, Malhotra AK, Song H, Ming GL. Interaction between FEZ1 and DISC1 in regulation of neuronal development and risk for schizophrenia. Neuron. 2011;72:559–571. doi: 10.1016/j.neuron.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski BA, Morle GD, Huggenvik J, Uhler MD, Leiden JM. Molecular cloning of human CREB-2: an ATF/CREB transcription factor that can negatively regulate transcription from the cAMP response element. Proc Natl Acad Sci U S A. 1992;89:4820–4824. doi: 10.1073/pnas.89.11.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsel P, Tan W, Abazyan B, Davis KL, Ross C, Pletnikov MV, Haroutunian V. Expression of mutant human DISC1 in mice supports abnormalities in differentiation of oligodendrocytes. Schizophr Res. 2011;130:238–249. doi: 10.1016/j.schres.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Duan X, Liu CY, Jang MH, Guo JU, Pow-Anpongkul N, Kang E, Song H, Ming GL. DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron. 2009;63:761–773. doi: 10.1016/j.neuron.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Liu CY, Zhang F, Duan X, Wen Z, Song J, Feighery E, Lu B, Rujescu D, St Clair D, Christian K, Callicott JH, Weinberger DR, Song H, Ming GL. Interplay between DISC1 and GABA signaling regulates neurogenesis in mice and risk for schizophrenia. Cell. 2012;148:1051–1064. doi: 10.1016/j.cell.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Zaghloul NA, Bubenshchikova E, Oh EC, Rankin S, Katsanis N, Obara T, Tsiokas L. Nde1-mediated inhibition of ciliogenesis affects cell cycle reentry. Nat Cell Biol. 2011;13:351–360. doi: 10.1038/ncb2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick B, Xu L, Cascella N, Ozeki Y, Sawa A, Roberts RC. DISC1 immunoreactivity at the light and ultrastructural level in the human neocortex. J Comp Neurol. 2006;497:436–450. doi: 10.1002/cne.21007. [DOI] [PubMed] [Google Scholar]
- Koike H, Arguello PA, Kvajo M, Karayiorgou M, Gogos JA. Disc1 is mutated in the 129S6/SvEv strain and modulates working memory in mice. Proc Natl Acad Sci U S A. 2006;103:3693–3697. doi: 10.1073/pnas.0511189103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korth C. DISCopathies: brain disorders related to DISC1 dysfunction. Rev Neurosci. 2009;20:321–330. doi: 10.1515/revneuro.2009.20.5-6.321. [DOI] [PubMed] [Google Scholar]
- Korth C. Aggregated proteins in schizophrenia and other chronic mental diseases: DISC1opathies. Prion. 2012;6:134–141. doi: 10.4161/pri.18989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi S, Hamdan AM, Horiguchi M, Kusunose N, Okamoto A, Matsunaga N, Ohdo S. cAMP-response element (CRE)-mediated transcription by activating transcription factor-4 (ATF4) is essential for circadian expression of the Period2 gene. J Biol Chem. 2011;286:32416–32423. doi: 10.1074/jbc.M111.258970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein AR, Noctor SC. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 2004;27:392–399. doi: 10.1016/j.tins.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Kristiansen LV, Huerta I, Beneyto M, Meador-Woodruff JH. NMDA receptors and schizophrenia. Curr Opin Pharmacol. 2007;7:48–55. doi: 10.1016/j.coph.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Kubo K, Tomita K, Uto A, Kuroda K, Seshadri S, Cohen J, Kaibuchi K, Kamiya A, Nakajima K. Migration defects by DISC1 knockdown in C57BL/6, 129X1/SvJ, and ICR strains via in utero gene transfer and virus-mediated RNAi. Biochem Biophys Res Commun. 2010;400:631–637. doi: 10.1016/j.bbrc.2010.08.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijpers M, Hoogenraad CC. Centrosomes, microtubules and neuronal development. Mol Cell Neurosci. 2011;48:349–358. doi: 10.1016/j.mcn.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Kumamoto N, Gu Y, Wang J, Janoschka S, Takemaru K, Levine J, Ge S. A role for primary cilia in glutamatergic synaptic integration of adult-born neurons. Nat Neurosci. 2012;15:399–405. S391. doi: 10.1038/nn.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda K, Yamada S, Tanaka M, Iizuka M, Yano H, Mori D, Tsuboi D, Nishioka T, Namba T, Iizuka Y, Kubota S, Nagai T, Ibi D, Wang R, Enomoto A, Isotani-Sakakibara M, Asai N, Kimura K, Kiyonari H, Abe T, Mizoguchi A, Sokabe M, Takahashi M, Yamada K, Kaibuchi K. Behavioral alterations associated with targeted disruption of exons 2 and 3 of the Disc1 gene in the mouse. Hum Mol Genet. 2011;20:4666–4683. doi: 10.1093/hmg/ddr400. [DOI] [PubMed] [Google Scholar]
- Kvajo M, Mckellar H, Arguello PA, Drew LJ, Moore H, Macdermott AB, Karayiorgou M, Gogos JA. A mutation in mouse Disc1 that models a schizophrenia risk allele leads to specific alterations in neuronal architecture and cognition. Proc Natl Acad Sci U S A. 2008;105:7076–7081. doi: 10.1073/pnas.0802615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvajo M, Mckellar H, Drew LJ, Lepagnol-Bestel AM, Xiao L, Levy RJ, Blazeski R, Arguello PA, Lacefield CO, Mason CA, Simonneau M, O’donnell JM, Macdermott AB, Karayiorgou M, Gogos JA. Altered axonal targeting and short-term plasticity in the hippocampus of Disc1 mutant mice. Proc Natl Acad Sci U S A. 2011;108:E1349–1358. doi: 10.1073/pnas.1114113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam C, Vergnolle MA, Thorpe L, Woodman PG, Allan VJ. Functional interplay between LIS1, NDE1 and NDEL1 in dynein-dependent organelle positioning. J Cell Sci. 2010;123:202–212. doi: 10.1242/jcs.059337. [DOI] [PubMed] [Google Scholar]
- Landers JE, Melki J, Meininger V, Glass JD, Van Den Berg LH, Van Es MA, Sapp PC, Van Vught PW, Mckenna-Yasek DM, Blauw HM, Cho TJ, Polak M, Shi L, Wills AM, Broom WJ, Ticozzi N, Silani V, Ozoguz A, Rodriguez-Leyva I, Veldink JH, Ivinson AJ, Saris CG, Hosler BA, Barnes-Nessa A, Couture N, Wokke JH, Kwiatkowski TJ, Jr., Ophoff RA, Cronin S, Hardiman O, Diekstra FP, Leigh PN, Shaw CE, Simpson CL, Hansen VK, Powell JF, Corcia P, Salachas F, Heath S, Galan P, Georges F, Horvitz HR, Lathrop M, Purcell S, Al-Chalabi A, Brown RH., Jr. Reduced expression of the Kinesin-Associated Protein 3 (KIFAP3) gene increases survival in sporadic amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2009;106:9004–9009. doi: 10.1073/pnas.0812937106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le-Niculescu H, Patel SD, Bhat M, Kuczenski R, Faraone SV, Tsuang MT, Mcmahon FJ, Schork NJ, Nurnberger JI, Jr., Niculescu AB., 3rd Convergent functional genomics of genome-wide association data for bipolar disorder: comprehensive identification of candidate genes, pathways and mechanisms. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:155–181. doi: 10.1002/ajmg.b.30887. [DOI] [PubMed] [Google Scholar]
- Lee FH, Fadel MP, Preston-Maher K, Cordes SP, Clapcote SJ, Price DJ, Roder JC, Wong AH. Disc1 point mutations in mice affect development of the cerebral cortex. J Neurosci. 2011;31:3197–3206. doi: 10.1523/JNEUROSCI.4219-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Kim H, Lee YS, Kaang BK. Overexpression and RNA interference of Ap-cyclic AMP-response element binding protein-2, a repressor of long-term facilitation, in Aplysia kurodai sensory-to-motor synapses. Neurosci Lett. 2003;337:9–12. doi: 10.1016/s0304-3940(02)01285-5. [DOI] [PubMed] [Google Scholar]
- Lee MM, Reif A, Schmitt AG. Major Depression: A Role for Hippocampal Neurogenesis? Curr Top Behav Neurosci. 2012 doi: 10.1007/7854_2012_226. [DOI] [PubMed] [Google Scholar]
- Leliveld SR, Bader V, Hendriks P, Prikulis I, Sajnani G, Requena JR, Korth C. Insolubility of disrupted-in-schizophrenia 1 disrupts oligomer-dependent interactions with nuclear distribution element 1 and is associated with sporadic mental disease. J Neurosci. 2008;28:3839–3845. doi: 10.1523/JNEUROSCI.5389-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leliveld SR, Hendriks P, Michel M, Sajnani G, Bader V, Trossbach S, Prikulis I, Hartmann R, Jonas E, Willbold D, Requena JR, Korth C. Oligomer assembly of the C-terminal DISC1 domain (640-854) is controlled by self-association motifs and disease-associated polymorphism S704C. Biochemistry. 2009;48:7746–7755. doi: 10.1021/bi900901e. [DOI] [PubMed] [Google Scholar]
- Lemos DR, Goodspeed L, Tonelli L, Antoch MP, Ojeda SR, Urbanski HF. Evidence for circadian regulation of activating transcription factor 5 but not tyrosine hydroxylase by the chromaffin cell clock. Endocrinology. 2007;148:5811–5821. doi: 10.1210/en.2007-0610. [DOI] [PubMed] [Google Scholar]
- Li W, Zhou Y, Jentsch JD, Brown RA, Tian X, Ehninger D, Hennah W, Peltonen L, Lonnqvist J, Huttunen MO, Kaprio J, Trachtenberg JT, Silva AJ, Cannon TD. Specific developmental disruption of disrupted-in-schizophrenia-1 function results in schizophrenia-related phenotypes in mice. Proc Natl Acad Sci U S A. 2007;104:18280–18285. doi: 10.1073/pnas.0706900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu B, Hou B, Qin W, Wang D, Yu C, Jiang T. Less Efficient Information Transfer in Cys-Allele Carriers of DISC1: A Brain Network Study Based on Diffusion MRI. Cereb Cortex. 2012 doi: 10.1093/cercor/bhs167. [DOI] [PubMed] [Google Scholar]
- Lipina TV, Kaidanovich-Beilin O, Patel S, Wang M, Clapcote SJ, Liu F, Woodgett JR, Roder JC. Genetic and pharmacological evidence for schizophrenia-related Disc1 interaction with GSK-3. Synapse. 2011;65:234–248. doi: 10.1002/syn.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipina TV, Niwa M, Jaaro-Peled H, Fletcher PJ, Seeman P, Sawa A, Roder JC. Enhanced dopamine function in DISC1-L100P mutant mice: implications for schizophrenia. Genes Brain Behav. 2010;9:777–789. doi: 10.1111/j.1601-183X.2010.00615.x. [DOI] [PubMed] [Google Scholar]
- Lipina TV, Wang M, Liu F, Roder JC. Synergistic interactions between PDE4B and GSK-3: DISC1 mutant mice. Neuropharmacology. 2012;62:1252–1262. doi: 10.1016/j.neuropharm.2011.02.020. [DOI] [PubMed] [Google Scholar]
- Lopez AD, Murray CC. The global burden of disease, 1990-2020. Nat Med. 1998;4:1241–1243. doi: 10.1038/3218. [DOI] [PubMed] [Google Scholar]
- Louvi A, Grove EA. Cilia in the CNS: the quiet organelle claims center stage. Neuron. 2011;69:1046–1060. doi: 10.1016/j.neuron.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Liu Y, Ky B, Shughrue PJ, Austin CP, Morris JA. Cloning and characterization of Disc1, the mouse ortholog of DISC1 (Disrupted-in-Schizophrenia 1) Genomics. 2002;80:662–672. doi: 10.1006/geno.2002.7012. [DOI] [PubMed] [Google Scholar]
- Ma TM, Abazyan S, Abazyan B, Nomura J, Yang C, Seshadri S, Sawa A, Snyder SH, Pletnikov MV. Pathogenic disruption of DISC1-serine racemase binding elicits schizophrenia-like behavior via D-serine depletion. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Nwulia E, Chang J, Balkissoon R, Ishizuka K, Chen H, Zandi P, Mcinnis MG, Sawa A. Differential expression of disrupted-in-schizophrenia (DISC1) in bipolar disorder. Biol Psychiatry. 2006;60:929–935. doi: 10.1016/j.biopsych.2006.03.032. [DOI] [PubMed] [Google Scholar]
- Maher BJ, Loturco JJ. Disrupted-in-schizophrenia (DISC1) functions presynaptically at glutamatergic synapses. PLoS One. 2012;7:e34053. doi: 10.1371/journal.pone.0034053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malavasi EL, Ogawa F, Porteous DJ, Millar JK. DISC1 variants 37W and 607F disrupt its nuclear targeting and regulatory role in ATF4-mediated transcription. Hum Mol Genet. 2012;21:2779–2792. doi: 10.1093/hmg/dds106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Ge X, Frank CL, Madison JM, Koehler AN, Doud MK, Tassa C, Berry EM, Soda T, Singh KK, Biechele T, Petryshen TL, Moon RT, Haggarty SJ, Tsai LH. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell. 2009;136:1017–1031. doi: 10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marley A, Von Zastrow M. DISC1 regulates primary cilia that display specific dopamine receptors. PLoS One. 2010;5:e10902. doi: 10.1371/journal.pone.0010902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata I, Perez-Iglesias R, Roiz-Santianez R, Tordesillas-Gutierrez D, Gonzalez-Mandly A, Berja A, Vazquez-Barquero JL, Crespo-Facorro B. Additive effect of NRG1 and DISC1 genes on lateral ventricle enlargement in first episode schizophrenia. Neuroimage. 2010;53:1016–1022. doi: 10.1016/j.neuroimage.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Mathieson I, Munafo MR, Flint J. Meta-analysis indicates that common variants at the DISC1 locus are not associated with schizophrenia. Mol Psychiatry. 2012;17:634–641. doi: 10.1038/mp.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcevoy JP. The costs of schizophrenia. J Clin Psychiatry. 2007;68(Suppl 14):4–7. [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Weinberger DR. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci. 2006;7:818–827. doi: 10.1038/nrn1993. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Morris JA. Disc1 regulates granule cell migration in the developing hippocampus. Hum Mol Genet. 2009;18:3286–3297. doi: 10.1093/hmg/ddp266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JK, Christie S, Porteous DJ. Yeast two-hybrid screens implicate DISC1 in brain development and function. Biochem Biophys Res Commun. 2003;311:1019–1025. doi: 10.1016/j.bbrc.2003.10.101. [DOI] [PubMed] [Google Scholar]
- Millar JK, Christie S, Semple CA, Porteous DJ. Chromosomal location and genomic structure of the human translin-associated factor X gene (TRAX; TSNAX) revealed by intergenic splicing to DISC1, a gene disrupted by a translocation segregating with schizophrenia. Genomics. 2000a;67:69–77. doi: 10.1006/geno.2000.6239. [DOI] [PubMed] [Google Scholar]
- Millar JK, James R, Christie S, Porteous DJ. Disrupted in schizophrenia 1 (DISC1): subcellular targeting and induction of ring mitochondria. Mol Cell Neurosci. 2005a;30:477–484. doi: 10.1016/j.mcn.2005.08.021. [DOI] [PubMed] [Google Scholar]
- Millar JK, Pickard BS, Mackie S, James R, Christie S, Buchanan SR, Malloy MP, Chubb JE, Huston E, Baillie GS, Thomson PA, Hill EV, Brandon NJ, Rain JC, Camargo LM, Whiting PJ, Houslay MD, Blackwood DH, Muir WJ, Porteous DJ. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005b;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, St Clair DM, Muir WJ, Blackwood DH, Porteous DJ. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000b;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- Miller KE, Sheetz MP. Axonal mitochondrial transport and potential are correlated. J Cell Sci. 2004;117:2791–2804. doi: 10.1242/jcs.01130. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Asanuma M, Miyazaki I, Diaz-Corrales FJ, Katayama T, Tohyama M, Ogawa N. DISC1 localizes to the centrosome by binding to kendrin. Biochem Biophys Res Commun. 2004;317:1195–1199. doi: 10.1016/j.bbrc.2004.03.163. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Honda A, Baba K, Taniguchi M, Oono K, Fujita T, Kuroda S, Katayama T, Tohyama M. Disrupted-In-Schizophrenia 1, a candidate gene for schizophrenia, participates in neurite outgrowth. Mol Psychiatry. 2003;8:685–694. doi: 10.1038/sj.mp.4001352. [DOI] [PubMed] [Google Scholar]
- Moens LN, De Rijk P, Reumers J, Van Den Bossche MJ, Glassee W, De Zutter S, Lenaerts AS, Nordin A, Nilsson LG, Medina Castello I, Norrback KF, Goossens D, Van Steen K, Adolfsson R, Del-Favero J. Sequencing of DISC1 pathway genes reveals increased burden of rare missense variants in schizophrenia patients from a northern Swedish population. PLoS One. 2011;6:e23450. doi: 10.1371/journal.pone.0023450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JA, Kandpal G, Ma L, Austin CP. DISC1 (Disrupted-In-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Hum Mol Genet. 2003;12:1591–1608. doi: 10.1093/hmg/ddg162. [DOI] [PubMed] [Google Scholar]
- Mouaffak F, Kebir O, Chayet M, Tordjman S, Vacheron MN, Millet B, Jaafari N, Bellon A, Olie JP, Krebs MO. Association of Disrupted in Schizophrenia 1 (DISC1) missense variants with ultra-resistant schizophrenia. Pharmacogenomics J. 2011;11:267–273. doi: 10.1038/tpj.2010.40. [DOI] [PubMed] [Google Scholar]
- Murdoch H, Mackie S, Collins DM, Hill EV, Bolger GB, Klussmann E, Porteous DJ, Millar JK, Houslay MD. Isoform-selective susceptibility of DISC1/phosphodiesterase-4 complexes to dissociation by elevated intracellular cAMP levels. J Neurosci. 2007;27:9513–9524. doi: 10.1523/JNEUROSCI.1493-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch H, Vadrevu S, Prinz A, Dunlop AJ, Klussmann E, Bolger GB, Norman JC, Houslay MD. Interaction between LIS1 and PDE4, and its role in cytoplasmic dynein function. J Cell Sci. 2011;124:2253–2266. doi: 10.1242/jcs.082982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mykytyn K, Braun T, Carmi R, Haider NB, Searby CC, Shastri M, Beck G, Wright AF, Iannaccone A, Elbedour K, Riise R, Baldi A, Raas-Rothschild A, Gorman SW, Duhl DM, Jacobson SG, Casavant T, Stone EM, Sheffield VC. Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nat Genet. 2001;28:188–191. doi: 10.1038/88925. [DOI] [PubMed] [Google Scholar]
- Nagai T, Kitahara Y, Ibi D, Nabeshima T, Sawa A, Yamada K. Effects of antipsychotics on the behavioral deficits in human dominant-negative DISC1 transgenic mice with neonatal polyI:C treatment. Behav Brain Res. 2011;225:305–310. doi: 10.1016/j.bbr.2011.07.049. [DOI] [PubMed] [Google Scholar]
- Nakata K, Lipska BK, Hyde TM, Ye T, Newburn EN, Morita Y, Vakkalanka R, Barenboim M, Sei Y, Weinberger DR, Kleinman JE. DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc Natl Acad Sci U S A. 2009;106:15873–15878. doi: 10.1073/pnas.0903413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba T, Ming GL, Song H, Waga C, Enomoto A, Kaibuchi K, Kohsaka S, Uchino S. NMDA receptor regulates migration of newly generated neurons in the adult hippocampus via Disrupted-In-Schizophrenia 1 (DISC1) J Neurochem. 2011;118:34–44. doi: 10.1111/j.1471-4159.2011.07282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan S, Arthanari H, Wolfe MS, Wagner G. Molecular characterization of disrupted in schizophrenia-1 risk variant S704C reveals the formation of altered oligomeric assembly. J Biol Chem. 2011;286:44266–44276. doi: 10.1074/jbc.M111.271593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodemus KK, Callicott JH, Higier RG, Luna A, Nixon DC, Lipska BK, Vakkalanka R, Giegling I, Rujescu D, St Clair D, Muglia P, Shugart YY, Weinberger DR. Evidence of statistical epistasis between DISC1, CIT and NDEL1 impacting risk for schizophrenia: biological validation with functional neuroimaging. Hum Genet. 2010;127:441–452. doi: 10.1007/s00439-009-0782-y. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Niwa M, Kamiya A, Murai R, Kubo K, Gruber AJ, Tomita K, Lu L, Tomisato S, Jaaro-Peled H, Seshadri S, Hiyama H, Huang B, Kohda K, Noda Y, O’donnell P, Nakajima K, Sawa A, Nabeshima T. Knockdown of DISC1 by in utero gene transfer disturbs postnatal dopaminergic maturation in the frontal cortex and leads to adult behavioral deficits. Neuron. 2010;65:480–489. doi: 10.1016/j.neuron.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olincy A, House R, Gao B, Recksiek P, Phang TL, Sullivan B, Hollis JP, Hopkins J, Shade T, Edwards MG, Vianzon R, Griffiths C, Ceilley J, Helfrich RW, Ritvo J, Weis E, Weiss D, Gault J. Elevated DISC1 transcript levels in PBMCs during acute psychosis in patients with schizophrenia. Transl Biomed. 2011;2 [PMC free article] [PubMed] [Google Scholar]
- Osbun N, Li J, O’driscoll MC, Strominger Z, Wakahiro M, Rider E, Bukshpun P, Boland E, Spurrell CH, Schackwitz W, Pennacchio LA, Dobyns WB, Black GC, Sherr EH. Genetic and functional analyses identify DISC1 as a novel callosal agenesis candidate gene. Am J Med Genet A. 2011;155A:1865–1876. doi: 10.1002/ajmg.a.34081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott C, Ross K, Straub S, Thiede B, Gotz M, Goosmann C, Krischke M, Mueller MJ, Krohne G, Rudel T, Kozjak-Pavlovic V. Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol Cell Biol. 2012;32:1173–1188. doi: 10.1128/MCB.06388-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottis P, Bader V, Trossbach SV, Kretzschmar H, Michel M, Leliveld SR, Korth C. Convergence of two independent mental disease genes on the protein level: recruitment of dysbindin to cell-invasive disrupted-in-schizophrenia 1 aggresomes. Biol Psychiatry. 2011;70:604–610. doi: 10.1016/j.biopsych.2011.03.027. [DOI] [PubMed] [Google Scholar]
- Ozeki Y, Tomoda T, Kleiderlein J, Kamiya A, Bord L, Fujii K, Okawa M, Yamada N, Hatten ME, Snyder SH, Ross CA, Sawa A. Disrupted-in-Schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc Natl Acad Sci U S A. 2003;100:289–294. doi: 10.1073/pnas.0136913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palo OM, Antila M, Silander K, Hennah W, Kilpinen H, Soronen P, Tuulio-Henriksson A, Kieseppa T, Partonen T, Lonnqvist J, Peltonen L, Paunio T. Association of distinct allelic haplotypes of DISC1 with psychotic and bipolar spectrum disorders and with underlying cognitive impairments. Hum Mol Genet. 2007;16:2517–2528. doi: 10.1093/hmg/ddm207. [DOI] [PubMed] [Google Scholar]
- Papaleo F, Lipska BK, Weinberger DR. Mouse models of genetic effects on cognition: relevance to schizophrenia. Neuropharmacology. 2012;62:1204–1220. doi: 10.1016/j.neuropharm.2011.04.025. [DOI] [PubMed] [Google Scholar]
- Park YU, Jeong J, Lee H, Mun JY, Kim JH, Lee JS, Nguyen MD, Han SS, Suh PG, Park SK. Disrupted-in-schizophrenia 1 (DISC1) plays essential roles in mitochondria in collaboration with Mitofilin. Proc Natl Acad Sci U S A. 2010;107:17785–17790. doi: 10.1073/pnas.1004361107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paspalas CD, Wang M, Arnsten AF. Constellation of HCN Channels and cAMP Regulating Proteins in Dendritic Spines of the Primate Prefrontal Cortex: Potential Substrate for Working Memory Deficits in Schizophrenia. Cereb Cortex. 2012 doi: 10.1093/cercor/bhs152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul LK, Brown WS, Adolphs R, Tyszka JM, Richards LJ, Mukherjee P, Sherr EH. Agenesis of the corpus callosum: genetic, developmental and functional aspects of connectivity. Nat Rev Neurosci. 2007;8:287–299. doi: 10.1038/nrn2107. [DOI] [PubMed] [Google Scholar]
- Pletnikov MV, Ayhan Y, Nikolskaia O, Xu Y, Ovanesov MV, Huang H, Mori S, Moran TH, Ross CA. Inducible expression of mutant human DISC1 in mice is associated with brain and behavioral abnormalities reminiscent of schizophrenia. Mol Psychiatry. 2008;13:173–186. 115. doi: 10.1038/sj.mp.4002079. [DOI] [PubMed] [Google Scholar]
- Pogorelov VM, Nomura J, Kim J, Kannan G, Ayhan Y, Yang C, Taniguchi Y, Abazyan B, Valentine H, Krasnova IN, Kamiya A, Cadet JL, Wong DF, Pletnikov MV. Mutant DISC1 affects methamphetamine-induced sensitization and conditioned place preference: a comorbidity model. Neuropharmacology. 2012;62:1242–1251. doi: 10.1016/j.neuropharm.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteous D. Genetic causality in schizophrenia and bipolar disorder: out with the old and in with the new. Curr Opin Genet Dev. 2008;18:229–234. doi: 10.1016/j.gde.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Prata DP, Mechelli A, Fu CH, Picchioni M, Kane F, Kalidindi S, Mcdonald C, Kravariti E, Toulopoulou T, Miorelli A, Murray R, Collier DA, Mcguire PK. Effect of disrupted-in-schizophrenia-1 on pre-frontal cortical function. Mol Psychiatry. 2008;13:915–917. 909. doi: 10.1038/mp.2008.76. [DOI] [PubMed] [Google Scholar]
- Prata DP, Mechelli A, Picchioni M, Fu CH, Kane F, Kalidindi S, Mcdonald C, Kravariti E, Toulopoulou T, Bramon E, Walshe M, Murray R, Collier DA, Mcguire PK. No association of Disrupted-in-Schizophrenia-1 variation with prefrontal function in patients with schizophrenia and bipolar disorder. Genes Brain Behav. 2011;10:276–285. doi: 10.1111/j.1601-183X.2010.00665.x. [DOI] [PubMed] [Google Scholar]
- Puthuran MJ, Rowland-Hill CA, Simpson J, Pairaudeau PW, Mabbott JL, Morris SM, Crow YJ. Chromosome 1q42 deletion and agenesis of the corpus callosum. Am J Med Genet A. 2005;138:68–69. doi: 10.1002/ajmg.a.30888. [DOI] [PubMed] [Google Scholar]
- Ram Murthy A, Purushottam M, Kiran Kumar HB, Vallikiran M, Krishna N, Jayramu Sriharsha K, Janardhan Reddy YC, Ghosh S, Jain S. Gender-specific association of TSNAX/DISC1 locus for schizophrenia and bipolar affective disorder in South Indian population. J Hum Genet. 2012 doi: 10.1038/jhg.2012.62. [DOI] [PubMed] [Google Scholar]
- Ramsey AJ, Milenkovic M, Oliveira AF, Escobedo-Lozoya Y, Seshadri S, Salahpour A, Sawa A, Yasuda R, Caron MG. Impaired NMDA receptor transmission alters striatal synapses and DISC1 protein in an age-dependent manner. Proc Natl Acad Sci U S A. 2011;108:5795–5800. doi: 10.1073/pnas.1012621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi A, Zai C, Likhodi O, Kennedy JL, Wong AH. Genetic association and post-mortem brain mRNA analysis of DISC1 and related genes in schizophrenia. Schizophr Res. 2009;114:39–49. doi: 10.1016/j.schres.2009.06.019. [DOI] [PubMed] [Google Scholar]
- Rauch A, Thiel CT, Schindler D, Wick U, Crow YJ, Ekici AB, Van Essen AJ, Goecke TO, Al-Gazali L, Chrzanowska KH, Zweier C, Brunner HG, Becker K, Curry CJ, Dallapiccola B, Devriendt K, Dorfler A, Kinning E, Megarbane A, Meinecke P, Semple RK, Spranger S, Toutain A, Trembath RC, Voss E, Wilson L, Hennekam R, De Zegher F, Dorr HG, Reis A. Mutations in the pericentrin (PCNT) gene cause primordial dwarfism. Science. 2008;319:816–819. doi: 10.1126/science.1151174. [DOI] [PubMed] [Google Scholar]
- Raznahan A, Lee Y, Long R, Greenstein D, Clasen L, Addington A, Rapoport JL, Giedd JN. Common functional polymorphisms of DISC1 and cortical maturation in typically developing children and adolescents. Mol Psychiatry. 2011;16:917–926. doi: 10.1038/mp.2010.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- Rezin GT, Amboni G, Zugno AI, Quevedo J, Streck EL. Mitochondrial dysfunction and psychiatric disorders. Neurochem Res. 2009;34:1021–1029. doi: 10.1007/s11064-008-9865-8. [DOI] [PubMed] [Google Scholar]
- Rice GM, Qi Z, Selzer R, Richmond T, Thompson K, Pauli RM, Yu J. Microdissection-based high-resolution genomic array analysis of two patients with cytogenetically identical interstitial deletions of chromosome 1q but distinct clinical phenotypes. Am J Med Genet A. 2006;140:1637–1643. doi: 10.1002/ajmg.a.31349. [DOI] [PubMed] [Google Scholar]
- Rosenbloom KR, Dreszer TR, Pheasant M, Barber GP, Meyer LR, Pohl A, Raney BJ, Wang T, Hinrichs AS, Zweig AS, Fujita PA, Learned K, Rhead B, Smith KE, Kuhn RM, Karolchik D, Haussler D, Kent WJ. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 2010;38:D620–625. doi: 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs NA, Sawa A, Holmes SE, Ross CA, Delisi LE, Margolis RL. A frameshift mutation in Disrupted in Schizophrenia 1 in an American family with schizophrenia and schizoaffective disorder. Mol Psychiatry. 2005;10:758–764. doi: 10.1038/sj.mp.4001667. [DOI] [PubMed] [Google Scholar]
- Saetre P, Agartz I, De Franciscis A, Lundmark P, Djurovic S, Kahler A, Andreassen OA, Jakobsen KD, Rasmussen HB, Werge T, Hall H, Terenius L, Jonsson EG. Association between a disrupted-in-schizophrenia 1 (DISC1) single nucleotide polymorphism and schizophrenia in a combined Scandinavian case-control sample. Schizophr Res. 2008;106:237–241. doi: 10.1016/j.schres.2008.08.024. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Shionoya A, Ishida M, Gambello MJ, Yingling J, Wynshaw-Boris A, Hirotsune S. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- Sawamura N, Ando T, Maruyama Y, Fujimuro M, Mochizuki H, Honjo K, Shimoda M, Toda H, Sawamura-Yamamoto T, Makuch LA, Hayashi A, Ishizuka K, Cascella NG, Kamiya A, Ishida N, Tomoda T, Hai T, Furukubo-Tokunaga K, Sawa A. Nuclear DISC1 regulates CRE-mediated gene transcription and sleep homeostasis in the fruit fly. Mol Psychiatry. 2008;13:1138–1148. doi: 10.1038/mp.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamura N, Sawamura-Yamamoto T, Ozeki Y, Ross CA, Sawa A. A form of DISC1 enriched in nucleus: altered subcellular distribution in orbitofrontal cortex in psychosis and substance/alcohol abuse. Proc Natl Acad Sci U S A. 2005;102:1187–1192. doi: 10.1073/pnas.0406543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schosser A, Gaysina D, Cohen-Woods S, Chow PC, Martucci L, Craddock N, Farmer A, Korszun A, Gunasinghe C, Gray J, Jones L, Tozzi F, Perry J, Muglia P, Owen MJ, Craig IW, Mcguffin P. Association of DISC1 and TSNAX genes and affective disorders in the depression case-control (DeCC) and bipolar affective case-control (BACCS) studies. Mol Psychiatry. 2010;15:844–849. doi: 10.1038/mp.2009.21. [DOI] [PubMed] [Google Scholar]
- Schulz P, Steimer T. Neurobiology of circadian systems. CNS Drugs. 2009;23(Suppl 2):3–13. doi: 10.2165/11318620-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Schumacher J, Laje G, Abou Jamra R, Becker T, Muhleisen TW, Vasilescu C, Mattheisen M, Herms S, Hoffmann P, Hillmer AM, Georgi A, Herold C, Schulze TG, Propping P, Rietschel M, Mcmahon FJ, Nothen MM, Cichon S. The DISC locus and schizophrenia: evidence from an association study in a central European sample and from a meta-analysis across different European populations. Hum Mol Genet. 2009;18:2719–2727. doi: 10.1093/hmg/ddp204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serretti A, Drago A, De Ronchi D. Lithium pharmacodynamics and pharmacogenetics: focus on inositol mono phosphatase (IMPase), inositol poliphosphatase (IPPase) and glycogen sinthase kinase 3 beta (GSK-3 beta) Curr Med Chem. 2009;16:1917–1948. doi: 10.2174/092986709788186101. [DOI] [PubMed] [Google Scholar]
- Seshadri S, Kamiya A, Yokota Y, Prikulis I, Kano S, Hayashi-Takagi A, Stanco A, Eom TY, Rao S, Ishizuka K, Wong P, Korth C, Anton ES, Sawa A. Disrupted-in-Schizophrenia-1 expression is regulated by beta-site amyloid precursor protein cleaving enzyme-1-neuregulin cascade. Proc Natl Acad Sci U S A. 2010;107:5622–5627. doi: 10.1073/pnas.0909284107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh M, Hall MH, Schulze K, Dutt A, Li K, Williams I, Walshe M, Constante M, Broome M, Picchioni M, Toulopoulou T, Collier D, Stahl D, Rijsdijk F, Powell J, Murray RM, Arranz M, Bramon E. Effect of DISC1 on the P300 Waveform in Psychosis. Schizophr Bull. 2011 doi: 10.1093/schbul/sbr101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Lang B, Nakamoto C, Zhang F, Pu J, Kuan SL, Chatzi C, He S, Mackie I, Brandon NJ, Marquis KL, Day M, Hurko O, Mccaig CD, Riedel G, St Clair D. Schizophrenia-related neural and behavioral phenotypes in transgenic mice expressing truncated Disc1. J Neurosci. 2008;28:10893–10904. doi: 10.1523/JNEUROSCI.3299-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Matsuzaki S, Hattori T, Kumamoto N, Miyoshi K, Katayama T, Tohyama M. DISC1-kendrin interaction is involved in centrosomal microtubule network formation. Biochem Biophys Res Commun. 2008;377:1051–1056. doi: 10.1016/j.bbrc.2008.10.100. [DOI] [PubMed] [Google Scholar]
- Shinoda T, Taya S, Tsuboi D, Hikita T, Matsuzawa R, Kuroda S, Iwamatsu A, Kaibuchi K. DISC1 regulates neurotrophin-induced axon elongation via interaction with Grb2. J Neurosci. 2007;27:4–14. doi: 10.1523/JNEUROSCI.3825-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji H, Toyama K, Takamiya Y, Wakana S, Gondo Y, Miyakawa T. Comprehensive behavioral analysis of ENU-induced Disc1-Q31L and -L100P mutant mice. BMC Res Notes. 2012;5:108. doi: 10.1186/1756-0500-5-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, De Rienzo G, Drane L, Mao Y, Flood Z, Madison J, Ferreira M, Bergen S, King C, Sklar P, Sive H, Tsai LH. Common DISC1 polymorphisms disrupt Wnt/GSK3beta signaling and brain development. Neuron. 2011;72:545–558. doi: 10.1016/j.neuron.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, Ge X, Mao Y, Drane L, Meletis K, Samuels BA, Tsai LH. Dixdc1 is a critical regulator of DISC1 and embryonic cortical development. Neuron. 2010;67:33–48. doi: 10.1016/j.neuron.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siuciak JA, Chapin DS, Mccarthy SA, Martin AN. Antipsychotic profile of rolipram: efficacy in rats and reduced sensitivity in mice deficient in the phosphodiesterase-4B (PDE4B) enzyme. Psychopharmacology (Berl) 2007;192:415–424. doi: 10.1007/s00213-007-0727-x. [DOI] [PubMed] [Google Scholar]
- Soares DC, Carlyle BC, Bradshaw NJ, Porteous DJ. DISC1: Structure, Function, and Therapeutic Potential for Major Mental Illness. ACS Chem Neurosci. 2011;2:609–632. doi: 10.1021/cn200062k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Li W, Feng J, Heston LL, Scaringe WA, Sommer SS. Identification of high risk DISC1 structural variants with a 2% attributable risk for schizophrenia. Biochem Biophys Res Commun. 2008;367:700–706. doi: 10.1016/j.bbrc.2007.12.117. [DOI] [PubMed] [Google Scholar]
- Song W, Li W, Noltner K, Yan J, Green E, Grozeva D, Jones IR, Craddock N, Longmate J, Feng J, Sommer SS. Identification of high risk DISC1 protein structural variants in patients with bipolar spectrum disorder. Neurosci Lett. 2010;486:136–140. doi: 10.1016/j.neulet.2010.09.027. [DOI] [PubMed] [Google Scholar]
- Spiteri E, Konopka G, Coppola G, Bomar J, Oldham M, Ou J, Vernes SC, Fisher SE, Ren B, Geschwind DH. Identification of the transcriptional targets of FOXP2, a gene linked to speech and language, in developing human brain. Am J Hum Genet. 2007;81:1144–1157. doi: 10.1086/522237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprooten E, Sussmann JE, Moorhead TW, Whalley HC, Ffrench-Constant C, Blumberg HP, Bastin ME, Hall J, Lawrie SM, Mcintosh AM. Association of white matter integrity with genetic variation in an exonic DISC1 SNP. Mol Psychiatry. 2011;16:685, 688–689. doi: 10.1038/mp.2011.15. [DOI] [PubMed] [Google Scholar]
- Steinecke A, Gampe C, Valkova C, Kaether C, Bolz J. Disrupted-in-Schizophrenia 1 (DISC1) is necessary for the correct migration of cortical interneurons. J Neurosci. 2012;32:738–745. doi: 10.1523/JNEUROSCI.5036-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhlmiller TJ, Garcia-Castro MI. Current perspectives of the signaling pathways directing neural crest induction. Cell Mol Life Sci. 2012 doi: 10.1007/s00018-012-0991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ, O’donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeszko PR, Hodgkinson CA, Robinson DG, Derosse P, Bilder RM, Lencz T, Burdick KE, Napolitano B, Betensky JD, Kane JM, Goldman D, Malhotra AK. DISC1 is associated with prefrontal cortical gray matter and positive symptoms in schizophrenia. Biol Psychol. 2008;79:103–110. doi: 10.1016/j.biopsycho.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Suzuki M, Tsunoda M, Maeno N, Kawasaki Y, Zhou SY, Hagino H, Niu L, Tsuneki H, Kobayashi S, Sasaoka T, Seto H, Kurachi M, Ozaki N. The Disrupted-in-Schizophrenia-1 Ser704Cys polymorphism and brain morphology in schizophrenia. Psychiatry Res. 2009;172:128–135. doi: 10.1016/j.pscychresns.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Taya S, Shinoda T, Tsuboi D, Asaki J, Nagai K, Hikita T, Kuroda S, Kuroda K, Shimizu M, Hirotsune S, Iwamatsu A, Kaibuchi K. DISC1 regulates the transport of the NUDEL/LIS1/14-3-3epsilon complex through kinesin-1. J Neurosci. 2007;27:15–26. doi: 10.1523/JNEUROSCI.3826-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MS, Devon RS, Millar JK, Porteous DJ. Evolutionary constraints on the Disrupted in Schizophrenia locus. Genomics. 2003;81:67–77. doi: 10.1016/s0888-7543(02)00026-5. [DOI] [PubMed] [Google Scholar]
- Thomson PA, Harris SE, Starr JM, Whalley LJ, Porteous DJ, Deary IJ. Association between genotype at an exonic SNP in DISC1 and normal cognitive aging. Neurosci Lett. 2005;389:41–45. doi: 10.1016/j.neulet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Thomson PA, Macintyre DJ, Hamilton G, Dominiczak A, Smith BH, Morris A, Evans KL, Porteous DJ. Association of DISC1 variants with age of onset in a population-based sample of recurrent major depression. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.117. [DOI] [PubMed] [Google Scholar]
- Tiwary BK. The severity of mental disorders is linked to interaction among candidate genes. Integr Biol (Camb) 2012;4:1096–1101. doi: 10.1039/c2ib20066j. [DOI] [PubMed] [Google Scholar]
- Toda H, Mochizuki H, Flores R, 3rd, Josowitz R, Krasieva TB, Lamorte VJ, Suzuki E, Gindhart JG, Furukubo-Tokunaga K, Tomoda T. UNC-51/ATG1 kinase regulates axonal transport by mediating motor-cargo assembly. Genes Dev. 2008;22:3292–3307. doi: 10.1101/gad.1734608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Kubo K, Ishii K, Nakajima K. Disrupted-in-Schizophrenia-1 (Disc1) is necessary for migration of the pyramidal neurons during mouse hippocampal development. Hum Mol Genet. 2011;20:2834–2845. doi: 10.1093/hmg/ddr194. [DOI] [PubMed] [Google Scholar]
- Trinh MA, Kaphzan H, Wek RC, Pierre P, Cavener DR, Klann E. Brain-Specific Disruption of the eIF2alpha Kinase PERK Decreases ATF4 Expression and Impairs Behavioral Flexibility. Cell Rep. 2012;1:676–688. doi: 10.1016/j.celrep.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima K, Koyanagi S, Sato Y, Ogata T, Matsunaga N, Fujimura A, Ohdo S. Role of activating transcription factor-4 in 24-hour rhythm of serotonin transporter expression in the mouse midbrain. Mol Pharmacol. 2012;82:264–270. doi: 10.1124/mol.112.079079. [DOI] [PubMed] [Google Scholar]
- Valiente M, Martini FJ. Migration of cortical interneurons relies on branched leading process dynamics. Cell Adh Migr. 2009;3:278–280. doi: 10.4161/cam.3.3.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Os J, Rutten BP, Poulton R. Gene-environment interactions in schizophrenia: review of epidemiological findings and future directions. Schizophr Bull. 2008;34:1066–1082. doi: 10.1093/schbul/sbn117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernes SC, Nicod J, Elahi FM, Coventry JA, Kenny N, Coupe AM, Bird LE, Davies KE, Fisher SE. Functional genetic analysis of mutations implicated in a human speech and language disorder. Hum Mol Genet. 2006;15:3154–3167. doi: 10.1093/hmg/ddl392. [DOI] [PubMed] [Google Scholar]
- Walker RM, Hill AE, Newman AC, Hamilton G, Torrance HS, Anderson SM, Ogawa F, Derizioti P, Nicod J, Vernes SC, Fisher SE, Thomson PA, Porteous DJ, Evans KL. The DISC1 promoter: characterization and regulation by FOXP2. Hum Mol Genet. 2012;21:2862–2872. doi: 10.1093/hmg/dds111. [DOI] [PubMed] [Google Scholar]
- Wang Q, Charych EI, Pulito VL, Lee JB, Graziane NM, Crozier RA, Revilla-Sanchez R, Kelly MP, Dunlop AJ, Murdoch H, Taylor N, Xie Y, Pausch M, Hayashi-Takagi A, Ishizuka K, Seshadri S, Bates B, Kariya K, Sawa A, Weinberg RJ, Moss SJ, Houslay MD, Yan Z, Brandon NJ. The psychiatric disease risk factors DISC1 and TNIK interact to regulate synapse composition and function. Mol Psychiatry. 2011a;16:1006–1023. doi: 10.1038/mp.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kaneko N, Asai N, Enomoto A, Isotani-Sakakibara M, Kato T, Asai M, Murakumo Y, Ota H, Hikita T, Namba T, Kuroda K, Kaibuchi K, Ming GL, Song H, Sawamoto K, Takahashi M. Girdin is an intrinsic regulator of neuroblast chain migration in the rostral migratory stream of the postnatal brain. J Neurosci. 2011b;31:8109–8122. doi: 10.1523/JNEUROSCI.1130-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watatani Y, Ichikawa K, Nakanishi N, Fujimoto M, Takeda H, Kimura N, Hirose H, Takahashi S, Takahashi Y. Stress-induced translation of ATF5 mRNA is regulated by the 5′-untranslated region. J Biol Chem. 2008;283:2543–2553. doi: 10.1074/jbc.M707781200. [DOI] [PubMed] [Google Scholar]
- Wei Q, Diao F, Kang Z, Gan Z, Han Z, Zheng L, Li L, Guo X, Shan B, Liu C, Zhao J, Zhang J. The effect of DISC1 on regional gray matter density of schizophrenia in Han Chinese population. Neurosci Lett. 2012;517:21–24. doi: 10.1016/j.neulet.2012.03.098. [DOI] [PubMed] [Google Scholar]
- Whalley HC, Sussmann JE, Johnstone M, Romaniuk L, Redpath H, Chakirova G, Mukherjee P, Hall J, Johnstone EC, Lawrie SM, Mcintosh AM. Effects of a mis-sense DISC1 variant on brain activation in two cohorts at high risk of bipolar disorder or schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:343–353. doi: 10.1002/ajmg.b.32035. [DOI] [PubMed] [Google Scholar]
- Williams JM, Beck TF, Pearson DM, Proud MB, Cheung SW, Scott DA. A 1q42 deletion involving DISC1, DISC2, and TSNAX in an autism spectrum disorder. Am J Med Genet A. 2009;149A:1758–1762. doi: 10.1002/ajmg.a.32941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JD, Bonath F, Kumar S, Ross CA, Cunliffe VT. Disrupted-in-schizophrenia 1 and neuregulin 1 are required for the specification of oligodendrocytes and neurones in the zebrafish brain. Hum Mol Genet. 2009;18:391–404. doi: 10.1093/hmg/ddn361. [DOI] [PubMed] [Google Scholar]
- Wu D, Pan W. GSK3: a multifaceted kinase in Wnt signaling. Trends Biochem Sci. 2010;35:161–168. doi: 10.1016/j.tibs.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff K, Dijk DJ, Middleton B, Foster RG, Joyce EM. Sleep and circadian rhythm disruption in schizophrenia. Br J Psychiatry. 2012;200:308–316. doi: 10.1192/bjp.bp.111.096321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff K, Porcheret K, Cussans E, Foster RG. Sleep and circadian rhythm disturbances: multiple genes and multiple phenotypes. Curr Opin Genet Dev. 2009;19:237–246. doi: 10.1016/j.gde.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Wynshaw-Boris A. Lissencephaly and LIS1: insights into the molecular mechanisms of neuronal migration and development. Clin Genet. 2007;72:296–304. doi: 10.1111/j.1399-0004.2007.00888.x. [DOI] [PubMed] [Google Scholar]
- Wynshaw-Boris A, Gambello MJ. LIS1 and dynein motor function in neuronal migration and development. Genes Dev. 2001;15:639–651. doi: 10.1101/gad.886801. [DOI] [PubMed] [Google Scholar]
- Yi M, Weaver D, Hajnoczky G. Control of mitochondrial motility and distribution by the calcium signal: a homeostatic circuit. J Cell Biol. 2004;167:661–672. doi: 10.1083/jcb.200406038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingling J, Youn YH, Darling D, Toyo-Oka K, Pramparo T, Hirotsune S, Wynshaw-Boris A. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell. 2008;132:474–486. doi: 10.1016/j.cell.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young-Pearse TL, Bai J, Chang R, Zheng JB, Loturco JJ, Selkoe DJ. A critical function for beta-amyloid precursor protein in neuronal migration revealed by in utero RNA interference. J Neurosci. 2007;27:14459–14469. doi: 10.1523/JNEUROSCI.4701-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young-Pearse TL, Suth S, Luth ES, Sawa A, Selkoe DJ. Biochemical and functional interaction of disrupted-in-schizophrenia 1 and amyloid precursor protein regulates neuronal migration during mammalian cortical development. J Neurosci. 2010;30:10431–10440. doi: 10.1523/JNEUROSCI.1445-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng F, Wang L, Jia M, Yue W, Ruan Y, Lu T, Liu J, Li J, Zhang D. Evidence for association between Disrupted-in-Schizophrenia 1 (DISC1) gene polymorphisms and autism in Chinese Han population: a family-based association study. Behav Brain Funct. 2011;7:14. doi: 10.1186/1744-9081-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Palam LR, Jiang L, Narasimhan J, Staschke KA, Wek RC. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J Biol Chem. 2008a;283:7064–7073. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]
- Zhou X, Chen Q, Schaukowitch K, Kelsoe JR, Geyer MA. Insoluble DISC1-Boymaw fusion proteins generated by DISC1 translocation. Mol Psychiatry. 2010;15:669–672. doi: 10.1038/mp.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Geyer MA, Kelsoe JR. Does disrupted-in-schizophrenia (DISC1) generate fusion transcripts? Mol Psychiatry. 2008b;13:361–363. doi: 10.1038/sj.mp.4002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman JE, Naidoo N, Raizen DM, Pack AI. Conservation of sleep: insights from non-mammalian model systems. Trends Neurosci. 2008;31:371–376. doi: 10.1016/j.tins.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci U S A. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]