Abstract
Advances in vaccine technology are occurring in the molecular techniques used to develop vaccines and in the assessment of vaccine efficacy, allowing more complete characterization of vaccine-induced immunity correlating to protection. FIV vaccine development has closely mirrored and occasionally surpassed the development of HIV-1 vaccine, leading to first licensed technology. This review will discuss technological advances in vaccine designs, challenge infection assessment, and characterization of vaccine immunity in the context of the protection detected with prototype and commercial dual-subtype FIV vaccines and in relation to HIV-1.
Keywords: feline immunodeficiency virus, vaccine, immune response
Introduction: FIV and HIV-1 classification and structural vaccine targets
Classification of virus isolates is essential to deriving lentiviral vaccines. Based on the genetic makeup of their full genome, isolates of both FIV and HIV-1 are classified as subtypes. FIV is currently classified into five subtypes, whereas HIV-1 is categorized into three groups (M, N, O) with the most common group M divided into at least nine subtypes (Yamamoto et al., 2007). Both FIV and HIV-1 subtypes are distributed throughout the world. There are generally 7-27% nucleic acid difference between subtypes (inter-subtype difference) and 2–17% nucleic acid difference within subtype (intra-subtype difference) for both FIV and HIV-1 (Table 1). FIV and HIV-1 vaccines are difficult to develop due to these large intra- and inter-subtype variations. Individual isolates of FIV and HIV-1 not only mutate within the infected host, but recombination can occur between isolates of either the same or different subtypes (Bachmann et al., 1997; Blackard et al., 2002). Major subtype recombination can be determined by sequencing the full viral genome (gag-pol-env) and analyzed using phylogenetic techniques. In comparison, small recombination within select regions of group associated antigen (gag), polymerase (pol), or envelope (env) genes is determined by comparative analysis of the select regions and is often undetectable by phylogenetic tree analysis. Conserved and variable regions exist within each viral protein. Moderately conserved sequence regions are generally retained amongst strains from both the same and different subtypes; while highly conserved regions are sometimes even retained between lentiviruses (Matsuo et al., 1992). If the goal is to make a single universal vaccine for all global isolates of either FIV or HIV-1, then such a vaccine needs to include protective conserved epitopes.
Table 1.
Intra-subtype and inter-subtype variations of HIV-1 and FIVa
| HIV-1 (Group M)b |
FIV (Felis catus)c |
|||
|---|---|---|---|---|
| Intra-subtype |
Inter-subtype |
Intra-subtype |
Inter-subtype |
|
| gag | 7.6 (2.3 – 13.2) | 14.2 (7.7 – 17.4) | 6.4 (3.7 – 8.4) | 18.6 (14.0 – 21.6) |
| env | 11.7 (4.3 – 16.6) | 19.7 (15.1 – 21.6) | 11.0 (6.7 – 14.1) | 21.9 (13.3 – 26.6) |
Average and range of % nucleic acid differences are shown for gag and env genes.
HIV-1 strains with subtype used for gag and env analyses are Q23-17 (A), 92UG037 (A), 97CDKTB48 (A), 94CY017.41 (A), HXB2CG (B), BK132 (B), 1058-11 (B), 92BR025 (C), 21068 (C), 04ZASK146B1 (C), ELI (D), 94UG114 (D), 93BR020 (F), 96FR-MP411 (F), 02CM.0016BBY (F), 95CM-MP255 (F), DRCBL (G), SE6165 (G), VI991 (H), VI997 (H), 90CR056 (H), SE9280 (J), SE9173 (J), 96CM-MP535 (K), and 97ZR-EQTB11 (K); and for env analysis alone is 01TZA280 (D).
FIV strains with subtype used for gag and env analyses are PPR (A), Petaluma (A), GL8 (A), MD (B), FC1 (B), BM3070 (C), and Shizuoka (D); and for env analysis alone are Yokohama (B), Aomori2 (B), and Fukuoka (D).
The most variable epitopes of FIV and HIV-1 are found in the surface envelope (SU Env) and transmembrane (TM) Env regions. The SU region contains important primary receptor binding sites, while the TM binds a secondary cellular receptor, allowing the virus to penetrate the cellular membrane (Zolla-Pazner, 2004; Miyazawa, 2005). Variations in these regions make it difficult for the host to produce virus neutralizing antibodies (VNAs) with broad neutralizing activities. Efforts have been made to identify regions conserved between different isolates that can induce broad VNAs. A region called membrane proximal external region of the HIV-1 transmembrane contains VNA epitopes generating the broadest VNAs (Zolla-Pazner, 2004). However, recent HIV-1 studies suggest this region has very poor immunogenicity and that antibodies to this region may function as autoimmune antibodies (Haynes et al., 2005). VNAs have also been induced to the binding regions of primary and secondary receptors (Zolla-Pazner, 2004, Pantophlet and Burton, 2006), but these antibodies are generally less cross-neutralizing and thus, lack the broad VN activities of those generated against membrane proximal external region (Zwick et al., 2001; Zolla-Pazner, 2004). Recent phase-III trials in humans using a recombinant surface SU Env (gp120) vaccine showed no protection against HIV-1 infection (Flynn et al., 2005). The gp120 vaccine induced type-specific VNAs but only few antibodies capable of neutralizing circulating primary isolates (Schultz and Bradac, 2001). Cytotoxic T lymphocyte (CTL) activities to gp120 were reported to be CD4+ (Stanhope et al., 1993; Gorse et al., 2000). Another phase-III trial using prime-boost with ALVAC-HIV (DNA vaccine with gp120/gp41/Gag/Protease) followed by AIDSVAX B/E (gp120 of subtype B and circulating Env E) is currently underway in Thailand (Girard et al., 2006). This vaccine combination induced type-specific VNAs in majority of vaccinates and HIV-specific CD8+ CTL in a small percentage of vaccinates in phase-I/II trials (Nitayaphan et al., 2004).
HIV-1 induced cellular immunity has broad multi-subtype activities (Cao et al., 1997; Norris et al., 2004). As a result, induction of virus-specific cellular immunity is thought to be critical for the efficacy of both HIV-1 and FIV vaccines (Mooij and Heeney, 2002; Yamamoto et. al., 2007). The two major cellular immune functions analyzed for vaccine protection are those produced by CD4+ T-helper (TH) cells and CD8+ CTLs (Mooij and Heeney, 2002; Nitayaphan et al., 2004). Epitopes for TH and CTL activities are located in most HIV-1 proteins (Los Alamos National Laboratory, 2006). T-cell immunity of TH and CTL is generally MHC-restricted; therefore, HIV-1 and FIV vaccines need to include TH and CTL epitopes presented in the context of a diverse set of MHC haplotypes. HIV-1 epitope mapping reveals Gag p24 and Nef have the most CTL epitopes that are recognized by many MHC-I alleles (Los Alamos National Laboratory, 2006). Of the known HIV-1 TH epitopes, Gag p24 and Env gp160 have the most TH epitopes recognized by the largest number of MHC-II alleles. To date, FIV Gag p24 and Env gp140 have been reported to have CTL epitopes (Flynn et al., 1995; Li et al., 1995). Little is known about TH and CTL epitopes for FIV. FIV has only few regulatory genes (vif, vpr, rev) (Myazawa et al., 1994; Gemeniano et al., 2004) whose gene products can serve as potential immunogens in addition to the viral structural proteins and enzymes. The majority of HIV-1 TH and CTL epitope mapping has been performed in HIV-1-infected humans (Addo et al., 2003; Cao et al., 2003; Kaufmann et al., 2004). Unfortunately, epitope mapping in infected individuals may not predict the epitopes that elicit protective immunity generated by vaccination. The FIV/cat model has a unique advantage in that vaccine epitopes can be tested in cats by viral challenge, thus the efficacy of the immunogen in eliciting sterilizing protection can be determined. In vivo challenge also allows for TH and CTL epitope mapping, enabling the FIV epitopes detected in infected cats to be compared with epitopes recognized in response to vaccination. Such comparisons provide information about epitopes important for lentiviral vaccine protection.
FIV vaccine approaches and challenge system
The majority of veterinary viral vaccines are modified live vaccines (MLV). MLV contain an inoculum of live avirulent virus, which is attenuated by molecular deletion or by modification induced from a natural event. This approach cannot be used to make an FIV vaccine, since recombination can occur between live wild-type FIV and MLV FIV, resulting in MLV reversion to wild type FIV. Further, such recombination may lead to recombinant viruses that can escape vaccine immunity (also called breakthrough) and induce immunodeficiency diseases. The same concern existed for vaccines against feline leukemia virus (FeLV), a feline retrovirus which causes immunodeficiency and hematopoietic cancer (Quackenbush et. al., 1990). FeLV vaccines are generally inactivated (also called killed) whole virus (IWV). Based on the success of FeLV-IWV vaccines, many of the experimental FIV vaccines were based on IWV or inactivated whole FIV-infected cells (IWC). MLV for FIV have been used to experimentally identify viral epitopes and immune mechanisms important for vaccine protection.
FIV vaccines based on IWV, IWC, or MLV have been evaluated as vaccine immunogens with varying success (Table 2–Table 3). Experimental MLV containing various molecular deletions of FIV enzymes or regulatory genes have been tested alone or as part of prime-boost approaches using an attenuated or vectored vaccine for priming and subunit or inactivated vaccine for boosting (Tellier et al., 1998; Dunham et al., 2006). The attenuated vaccines generally elicited moderate levels of cellular immunity and significant antibody responses but had minimal-to-no success against homologous FIV challenges (Table 2). Recombinant subunit vaccines have been delivered either in a vector as a genetic vaccine or in an adjuvant as a protein vaccine. Both vectored and protein subunit vaccines provided marginal-to-no success even against homologous FIV challenge (Table 2). The most successful experimental FIV vaccines against heterologous in vivo-derived strains were made using conventional vaccine approaches and were either IWV or IWC (Table 3) (Uhl et al., 2002; Yamamoto et al., 2007). Improvements in the production of IWC and IWV vaccines have been made through the use of bioreactors and synthetic media which facilitates mass production with minimal purification steps (Kallel et al., 2002, 2003; Yamamoto, 2002). The composition of adjuvants delivered concurrent with protein vaccines has also been improved in order to enhance immunity to the immunogen. Newer approaches currently being investigated are the use of toll-like receptor stimulants and T-helper-1 (TH1) cytokines as adjuvants (Naylor and Hadden, 2003; Seya et al., 2006). As more vaccines are being developed to induce antigen-specific CTL activities, TH1 cytokines are being used to polarize the immunity towards CTL functions. In addition, TH1 cytokine genes are being used as part of molecular vaccines to enhance CTL activity of the vaccine (Carlarota and Weiner, 2004).
Table 2.
Efficacy of experiment FIV vaccines derived by molecular technologya
| Studyb | Vaccine(Route)cd | Challenge FIV CID50/Route)cd | Protection Rate | Comments (Reference) |
|---|---|---|---|---|
| Subunit Vaccine | ||||
| 1A | pDNA-Pet-env (ID/IM) | Pet | 1 / 4 | Enhanced plasma virus load in Group 1B (Richardson et al., 2002) |
| 1B | pDNA-Pet-env (IN) | (10/ IP) | 0 / 4 | |
| 1C | pDNA empty (ID/IM) | 1 / 8 | ||
| 2A | VRP-NCSU1-gag/env (SC) | NCSU1 | 0 / 4 | (Burkhard et al., 2002) |
| 2B | VRP-GFP (SC) | (cell / Vag) | 0 / 4 | |
| 3A | 19k1-Env protein (SC) | AM19 | 0 / 3 | Enhanced plasma virus load in vaccinates; 19k1 is molecular clone of AM19 (Huisman et al., 2004) |
| 3B | 19k1-Env(ΔV3–V5) protein (SC) | (20 / IM) | 0 / 4 | |
| 3C | PBS (SC) | 0 / 5 | ||
| 4A | LM-NCSU1-gag/pDNA-env (PO) | NCSU1 | 0 / 5 | Lower proviral load & higher CD4+ T cells (Stevens et al., 2004) |
| 4B | LM wt (PO) | (CF+cell /Vag) | 0 / 5 | |
| 4C | PBS (PO) | 0 / 5 | ||
| 5A | Pet-Orf-A protein (SC) | Plasma Pet | 0 / 5 | Early enhanced plasma virus load; later virus decrease & higher CD4+ T cells (Pistello et al., 2006) |
| 5B | pDNA-Pet-Orf-A (IM) | (10 / IV) | 0 / 5 | |
| 5C | pDNA-Orf-A (IM) + Orf-A protein (SC) | 0 / 5 | ||
| 5D | Alum (SC) or pDNA-empty (IM) | 0 / 8 | ||
| Attenuated Vaccine based on Deletion or Substitution | ||||
| 6A | GL8ΔIN+pDNA-IL18 (IM) | Pet | 1 / 6 | 4 protected cats became infected after GL8 challenge but has lower GL8 provirus & plasma virus loads than 4 controls (Dunham et al. 2002) |
| 6B | GL8ΔIN+pDNA-IL18+IL12(IM) | (25 / IP) | 2 / 6 | |
| 6C | GL8ΔRT+pDNA-IL18 (IM) | 2 / 6 | ||
| 6D | GL8ΔRT+pDNA-IL18+IL12 (IM) | 0 / 6 | ||
| 6E | pDNA + pDNA-IL18+IL12 (IM) | 0 / 6 | ||
| 7A | Pet-Δorf-A (SC) | Plasma Pet | 3 / 9 | (Pistello et al., 2005) |
| 7B | None | (10 / IV) | 0 / 6 | |
| 8A | Pet-env-TN14 (IP) | Wo | 0 / 5 | Pet and Wo from subtype A (Broche-Pierre et al., 2005) |
| 8B | Pet-env-TN92 (IP) | (10 / IP) | 1 / 5 | |
| 8C | Pet wt (IP) | 0 / 5 | ||
| 8D | None | 0 / 5 | ||
| 9A | GL8ΔIN+pDNA-IFNγ(IM), Pet-IWV (SC) | GL8 clone | 1 / 6 | Lower proviral PBMC & lymph node loads & higher CD4+ T cells in groups 9B & 9C; (Dunham et al., 2006) |
| 9B | GL8ΔIN+pDNA-IFNγ(IM) | (10 / IP) | 1 / 6 | |
| 9C | Pet-IWV (SC) | 0 / 6 | ||
| 9D | GL8ΔIN+pDNA-IFNγ(IM)+Pet-IWV (SC) | 0 / 6 | ||
| 9E | PBS (SC) | 0 / 6 | ||
| 10A | pDNA-PPRΔvifATG/IFNγ(IM) | PPR | 0 / 5 | (Gupta et al., 2006) |
| 10B | pDNA-PPRΔvif (IM) | (10 / IM) | 0 / 5 | |
| 10C | pDNA-PPRΔvif+pDNA-IFNγ (IM) | 0 / 5 | ||
| 10D | Saline (IM) | 0 / 5 | ||
Experimental vaccine trials reported in years 2002–2006 are shown for FIV vaccines derived by molecular technology. Earlier vaccine trials are summarized in previous publications (Elyar et al., 1997; Uhl et al., 2002).
FIV strains for vaccine and challenge are Petaluma (Pet), North Carolina State University-1 (NCSU1), Amsterdam-19k1 clone (19k1), Amsterdam-19 (AM19), Glasgow-8 (GL8), San Diego PPR (PPR), and France-Wo (Wo).
Subunit and attenuated vaccines are described with construct derivation, FIV strain of the vaccine, and FIV gene or protein of the vaccine. Vaccines consisted of plasmid DNA (pDNA), Venezuelan equine encephalitis virus-replicon particles (VRP), Listeria monocytogenes vector (LM), recombinant protein, inactivated whole virus (IWV), and deletion or substitution mutants. Adjuvants for protein vaccines were ISCOM (Study 3), alum (Study 5), and Quil A (Study 9). Attenuated vaccines consisted of substitution(s) in env gene (TN14, TN92) or deletions of integrase (ΔIN), reverse transcriptase (ΔRT), open-reading frame-A (Δorf-A), or vif (Δvif) gene. Controls were either non-immunized cats or cats immunized with wild type (wt) organism, saline, or PBS. Group 9A in Study 9 was primed IM with GL8ΔIN+pDNA-IFNγ and boosted SC with Pet-IWV.
Routes of immunization or challenge are intradermal (ID), intramuscular (IM), subcutaneous (SC), oral (PO), intraperitoneal (IP), vaginal (Vag), and intravenous (IV). The challenge inoculum for Study 4 was a combination of infected culture fluid and infected cells (CF+cell).
Table 3.
Summary of dual-subtype FIV vaccine studies
| FIV Challenge Inoculumc |
|||||||
|---|---|---|---|---|---|---|---|
| Studyab | Dual-subtype Vaccine | Strain (Subtype) | Source | Dose (Route) | Protection Rate of Vaccinates (%) | Protection Rate of Controls (%) | Preventable Fraction (P-value) |
| Short-duration Studies | |||||||
| 1 | IWV | Pet (A) | In vivo | 20,50 (IV) | 11 / 11 (100%) | 0 / 14 (0%) | 100% (<0.001) |
| Fel-O-Vax+IWV | |||||||
| 2 | IWV | Bang (A/B) | In vivo | 10,25,100 (IV) | 14 / 19 (74%) | 0 / 19 (0%) | 73.7% (<0.001) |
| 3 | Fel-O-Vax; IWV; | FC1 (B) | In vivo | 15 (IV) | 17 / 19 (89%) | 0 / 14 (0%) | 89.5% (<0.001) |
| Fel-O-Vax+IWV | |||||||
| 4 | Fel-O-Vax+IWV | FC1 (B) | In vitro | 100 (IV) | 1 / 4 (25%) | 0 / 4 (0%) | 25.0% (0.686) |
| 5 | Fel-O-Vax+IWV | Pet (A) | In vitro | 25 (Vaginal) | 5 / 6 (83%) | 0 / 7 (0%) | 83.3% (0.008) |
| Year-1 & Year-1.5 Contact Study (with 1-yr boost) | |||||||
| 6A/1yr | Fel-O-Vax | Ao2 (B) | In vivo | Contact | 6/ 6 (100%) | 5 / 8 (62%) | 100% (0.282) |
| 6B/1.5yr | Fel-O-Vax | Ao2 (B) | In vivo | Contact | 6 / 6 (100%) | 4 / 8 (50%) | 100% (0.142) |
| Year-1 Challenge Studies for USDA (no 1-yr boost) | |||||||
| 7 | Fel-O-Vax | ? | In vitro | ? (IM) | 18 / 27 (67%) | 9 / 34 (26%) | 54.7% (0.009) |
| 8 | Fel- O-Vax | ? | In vitro | ? (IM) | 21 / 25 (84%) | 2 / 19 (10%) | 82.1% (<0.001) |
In Study 1, 7 of 7 IWV(3X)- or combination(4X)-vaccinated cats and 4 of 4 IWV(4X)-vaccinated cats were protected against 20 and 50 CID50, respectively. In Study 2, 4 of 5 3X-vaccinated cats, 8 of 9 4X-vaccinated cats, and 2 of 5 3X-vaccinated cats were protected against 10, 25, and 100 CID50, respectively. In Study 3, 11 of 11 Fel-O-Vax-vaccinated cats, 3 of 4 IWV-vaccinated cats, and 3 of 4 combination-vaccinated cats were protected. Studies 3, 6A, 7, and 8 had three vaccinations and Studies 4 and 6B had four vaccinations consecutively (Study 4) or 4th vaccination as 1-year boost (Study 6B). Studies 5 had either three vaccinations or eight vaccinations with last three vaccinations as 1-year boosts. All vaccinations were given at 3–4 week intervals.
References for Studies 1–5 (Pu et al., 2001; Yamamoto et al., 2007); for Study 6 (Kusuhara et al., 2005); and Studies 7–8 (Uhl et al., 2002).
FIV challenge inocula were either in vivo-derived tissues directly from infected cats (pooled plasma or infected PBMC) or in vitro-derived infected tissue culture fluids from short-term PBMC cultures. Cats received intravenous (IV), intramuscular (IM), or intravaginal (Vaginal) inoculation with doses in CID50 or contact exposure with high-dose FIV-infected cats.
Conventional IWC and IWV vaccines based on single FIV strains have been protective against low challenge doses of homologous or homologous-subtype in vitro-derived FIV challenge (Yamamoto et. al., 1991, 1993; Hosie et al., 2000; Pu et al., 2001). A conventional single-strain IWC vaccine was also successful against contact challenge with cats infected with homologous-subtype strains (Matteucci et al., 2000). However, IWC and IWV vaccine efficacy decreased significantly against moderate-to-high challenge doses even those composed of in vivo-derived homologous or same subtype strains (Pu et al., 2001; Yamamoto et al., 2007). Contact challenge is the best challenge system in terms of simulating the natural mode of transmission. However, this challenge system takes a very long time for 100% of the control cats to become infected. In fact, only 30–50% of the control cats were infected within 1.5 years (Matteucci et al., 2000; Kusuhara et al., 2005). The dose of virus transmitted by the contact challenge system is much lower than the challenge doses used in most FIV and SIV vaccine trials since majority of the studies use challenge dose of 10 ID50 or more. In order for FIV infection of domestic cats to serve as small animal model for human HIV/AIDS, the challenge doses need to be moderate to high, which means they are at least 25 times the natural transmission dose of cats (i.e., 25 CID50). Consequently, the best system for challenge is to inoculate with in vivo-derived inoculum and include both doses typical of natural transmission and moderate-to-high challenge doses (25–100 CID50) needed as AIDS model. Under such rigorous challenge conditions, to date only a dual-subtype FIV vaccine has successfully conferred protection against in vivo-derived inoculums of heterologous-subtype strain (Pu et al., 2005; Yamamoto et al., 2007). The dual-subtype FIV vaccine consisting of chemically inactivated subtype-A and -D viruses was commercially released in USA in 2002 (Uhl et al., 2002). Dual subtype (A + D) FIV vaccines have successfully conferred protection against low to high doses of homologous in vivo-derived inoculum as well as low to moderate doses of heterologous subtype-B viruses (Table 2) (Yamamoto et al., 2007).
Analysis of challenge infection has also improved with advances in molecular techniques. Polymerase chain reaction (PCR) technology for analysis of challenge-virus infection has improved sensitivity of detection. Hence, efficacy of FIV vaccine trials has been tested under increasingly stringent protocols using both conventional virus detection systems and highly sensitive PCR analysis for FIV infection in blood and tissues.
Characterization of protective immunity induced by dual-subtype FIV vaccine
Both humoral and cellular immunity induced by the prototype and commercial dual-subtype FIV vaccines are under investigation. Antibody immunity has been studied more extensively in vaccinated cats than cellular immunity. Two major antibody immunities thought to be important for HIV-1 and FIV vaccines are VNA and antibody-dependent cytotoxic cell (ADCC) activities (Karnasuta et al., 2005; Girard et al., 2006; Yamamoto et al., 2007). For both HIV-1 and FIV, the literature on VNAs is more extensive than that on ADCC. The key immunocyte involved in ADCC response is natural killer (NK) cell, which has antiviral activity with or without ADCC antibodies (Amed and Amad, 2003). Unlike T cells, NK cells are not MHC-restricted, although they respond to specific MHC ligands to direct their cytotoxic activity. The role of NK cells and innate immunity has become an important subject for HIV-1 vaccine development (Ahmed and Ahmad, 2003; Levy et al., 2003). However, little is known about the role of vaccination in inducing NK-cell and ADCC activities in dual-subtype vaccinated cats. As discussed earlier, CD4+ TH and CD8+ CTL are the two major cellular immunities most frequently investigated in both HIV-1-infected individuals and experimental HIV-1 vaccine immunized subjects. Analysis of cellular immune responses is much more difficult than antibody immunity due to MHC restriction of T-cell functions. VNA and T-cell immunity induced by dual-subtype vaccination are discussed below within the context of the technology available to assess these immunities.
Vaccine-induced VNA immunity
VNA titers induced by dual-subtype FIV vaccines have been the major humoral immunity so far evaluated. Specific pathogen free (SPF) cats vaccinated with commercial dual-subtype FIV vaccine had higher titers of VNAs compared to those given the prototype dual-subtype FIV vaccine (Pu et al., 2004). Commercial dual-subtype FIV vaccine is composed of IWC (1.5×107−2.5×107 cells/dose) with a small concentration of IWV (50 µg/dose) in Fort Dodge adjuvant (Pu et al., 2005); whereas, prototype vaccine is composed of a high concentration of IWV (250–500 µg/dose) in Fort Dodge adjuvant supplemented with IL-12. The IWV in prototype vaccine is produced by ultracentrifugation for a virus pellet without any gradient purification. Gradient purification is omitted to prevent the loss of the SU (gp95/100) attached to TM. Pooled virus pellets are then inactivated by paraformaldehyde (1.25–2.5 µg/ml) and dialyzed against PBS to remove paraformaldehyde. In general, IWC vaccines are known to induce higher VNA titers than IWV (Yamamoto et al., 1991). It is speculated that the higher VNA titers induced by commercial vaccine are due to the presence of various transitional stages of SU and TM interactions on the cell surface of IWC (Finnegan et al., 2001; Zolla-Pazner et al., 2004), which are not present on SU and TM of IWV. This is not caused by the antibody responses to MHC-II on the IWC, since IWV vaccine has as much or more MHC-II than the commercial vaccine (Pu et al., 2004). Furthermore, both sera from IWC-vaccinated and IWV-vaccinated cats preabsorbed extensively with uninfected vaccine cells retained the VNA titers even though ELISA anti-cell antibodies were significantly decreased by >70% (Hohdatsu et al., 1993).
The sera from commercial dual-subtype-vaccinated cats had much higher VNA titers to homologous vaccine strains (FIVPet and FIVShi) than to either heterologous subtype-B strains or homologous subtype-A FIVGL8 (Fig 1). In contrast, sera from prototype dual-subtype-vaccinated cats had high VNA titers to vaccine strain FIVPet, but had little-to-no VNA titers to vaccine strain FIVShi; recombinant subtype-A/B strain FIVBang; heterologous subtype-B FIVFC1 and FIVMD; and homologous subtype-A strain FIVGL8 (Fig 1). The majority of prototype-vaccinated cats developed ELISA antibodies to SU and TM, suggesting that VNA epitopes were not exposed even though SU and TM were present in the IWV. The inability of the dual-subtype vaccine to induce VNAs to homologous subtype-A FIVGL8 may result from the observation that FIVGL8 does not express high levels of Env (Hosie et al., 2005). Based on our immunoblot analysis, FIVGL8 virions do not retain high levels of SU when compared to FIVPet. In fact, FIVGL8-infected cats do not generally produce significant titers of VNAs to FIVGL8 (Pu R and Yamamoto JK, personal communiqué). These results suggest that the FIVGL8 isolate is indeed a VNA-resistant strain. Passive-transfer studies using inactivated sera from FIVPet-infected cats with high FIVPet–specific VNA titers protected passive-transfer recipients against FIVPet challenge, while SPF cats passively transferred with sera from uninfected/nonvaccinated cats were not protected against the same challenge (Hohdatsu et al., 1993). This finding demonstrates that FIVPet is a VNA-sensitive strain. Similar passive-transfer studies using inactivated sera from FIVUK8-infected cats should determine the importance of antibodies, especially the VNAs against VNA-resistant strains. Moreover, passive-transfer studies with vaccine-induced antibodies can determine the importance of antibody immunity in protection against homologous- and heterologous-subtype strains.
Fig. 1.
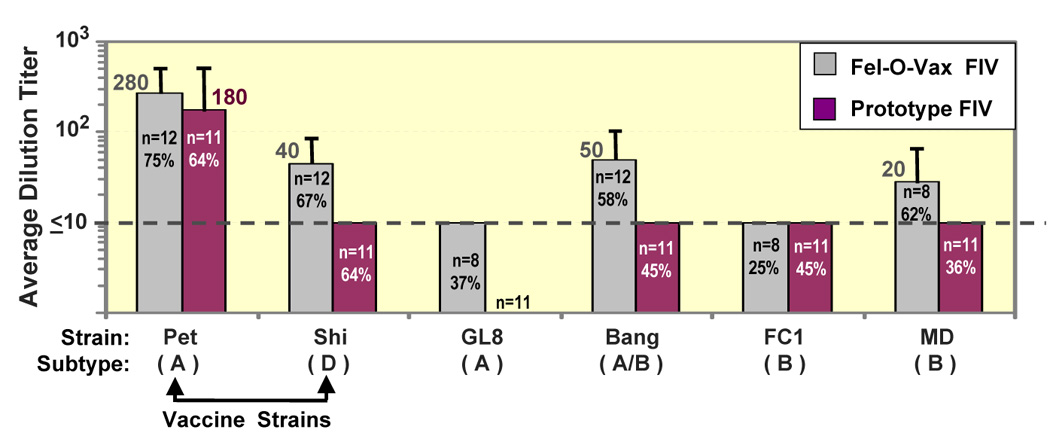
Vaccine-induced VNAs to heterologous and homologous subtype strains. SPF cats, immunized 3X at 3–4 week intervals with Fel-O-Vax FIV vaccine, induced the most VNAs to vaccine strains, recombinant subtype-A/B strain (FIVBang), and heterologous subtype-B strain (FIVMD). FIVBang is a recombinant of subtype-A gag/pol/envV1–V2 and subtype-B envV3–V9. VNA assay was performed with mitogen-stimulated PBMC and at 100 mean tissue culture infectious dose of low-passage FIV strains grown in primary PBMC (Pu et al., 2001). VNA titers are based on end-point dilution, which resulted in 50% inhibition of viral reverse transcriptase activity. The bar represents average VNA titers for each strain. The numbers of vaccinated cats tested are shown in the bar with percentage of cats that responded with ≥10 VNA titers.
Passive-transfer studies with sera from dual-subtype vaccinated cats
In order to analyze the protective efficacy of VNAs and antiviral antibodies, pooled sera from either commercial dual-subtype-vaccinated cats or nonvaccinated cats were passively transferred to naïve SPF cats (Table 4). Passive-transfer recipients were challenged IV 1-day post-first passive transfer with homologous FIVPet. All recipients of vaccine sera in this pilot study were protected, whereas all recipients of PBS or sera from nonvaccinated cats were not protected. The four vaccinated donors resisted challenge with heterologous subtype-B FIVFC1, while three PBS-immunized cats became infected with FIVFC1 (data not shown). Thus, FIVFC1 is resistant to vaccine-induced VNAs and yet dual-subtype-vaccinated cats are protected against FIVFC1 challenge.
Table 4.
Protection with vaccine-induced antibodies against homologous strain but not against heterologous-subtype strain based on passive-transfer studiesa
| Study | Pooled Sera or Antibodies | Challenge Strain (CID50) | Protection Rate (%) |
|---|---|---|---|
| 1A | Vaccinated cat sera | FIV-Pet (10) | 2 / 2 (100%) |
| 1B | Non-vaccinated cat sera | FIV-Pet (10) | 0 / 2 (0%) |
| 1C | PBS | FIV-Pet (10) | 0 / 2 (0%) |
| 2A | Vaccinated cat antibodies | FIV-Pet (15) | 4 / 4 (100%) |
| 2B | Non-vaccinated cat antibodies | FIV-Pet (15) | 0 / 2 (0%) |
| 2C | PBS | FIV-Pet (15) | 0 / 2 (0%) |
| 3A | Vaccinated cat antibodies | FIV-FC1 (15) | 0 / 3 (0%) |
| 3B | PBS or Non-vaccinated cat antibodies | FIV-FC1 (15) | 0 / 2 (0%) |
Passive transfer was administered by intravenous infusion and FIV challenge route was intravenous.
In the next passive-transfer study, partially purified antibodies from either vaccinated or nonvaccinated cats were passively transferred to naïve SPF cats. Partially purified antibodies were prepared by ammonium sulfate precipitation (Harlow and Lane, 1988). Four of seven recipients of vaccine antibodies, two of three recipients of antibodies from nonvaccinated cats, and two of three PBS-immunized cats were challenged with FIVPet, while remaining cats were challenged with FIVFC1. Cats that received vaccine antibodies were protected against FIVPet challenge but not against FIVFC1 challenge. These preliminary results suggest that vaccine protection was achieved with antibody immunity against a VNA-sensitive strain but not against a VNA-resistant strain. In this passive-transfer study, the partially purified vaccine antibodies had VNA titer of 750. Since the passive-transfer recipients received antibodies equivalent to 30% of their total blood volume, the vaccine-antibody recipients should have VNA titer of at least 100 immediately after the passive transfer. Based on our previous experience of 2–3 fold decreasing titer of transferred VNA per week (Hohdatsu et al., 1993), these cats at 1-week post transfer should have 37–56 VNA titers, which are similar to the protective VNA titers observed in passive-transfer study in SHIV/macaque model (Nishimura et al., 2002; 2003).
Vaccine-induced T-cell immunity
Initial cellular immunity induced by dual-subtype vaccine was determined by measuring mRNA levels and biological activities of TH cytokines and CTL mediators in response to FIV antigenic stimulation (Omori et al., 2004). TH1 cytokines are known to mediate cellular immunity, while TH2 cytokines mediate predominantly antibody immunity. Expression of the TH1 cytokines interferon-γ(IFNγ) and interleukin-2 (IL-2), and the TH2 cytokines IL-4 and IL-6 was analyzed. The CTL mediators monitored were perforin, tumor necrosis factor-α (TNFα), and IFNγ. The peripheral blood mononuclear cells (PBMC) of the vaccinated cats had FIV-specific IFNγ, IL-2, and perforin responses when compared to PBMC from placebo-immunized cats (Fig 2) (Omori et al., 2004). FIV-specific TH2 cytokine responses were not detected in the PBMC from vaccinated cats. These results suggested that TH1 cells and CTLs may be the basis for the vaccine immunity important in protection against FIV infection. In order to define the cell types involved in the vaccine protection, adoptive-transfer studies were performed using isolated cell populations of B cells, T cells, CD4+ T cells, and CD8+ T cells, described below.
Fig. 2.
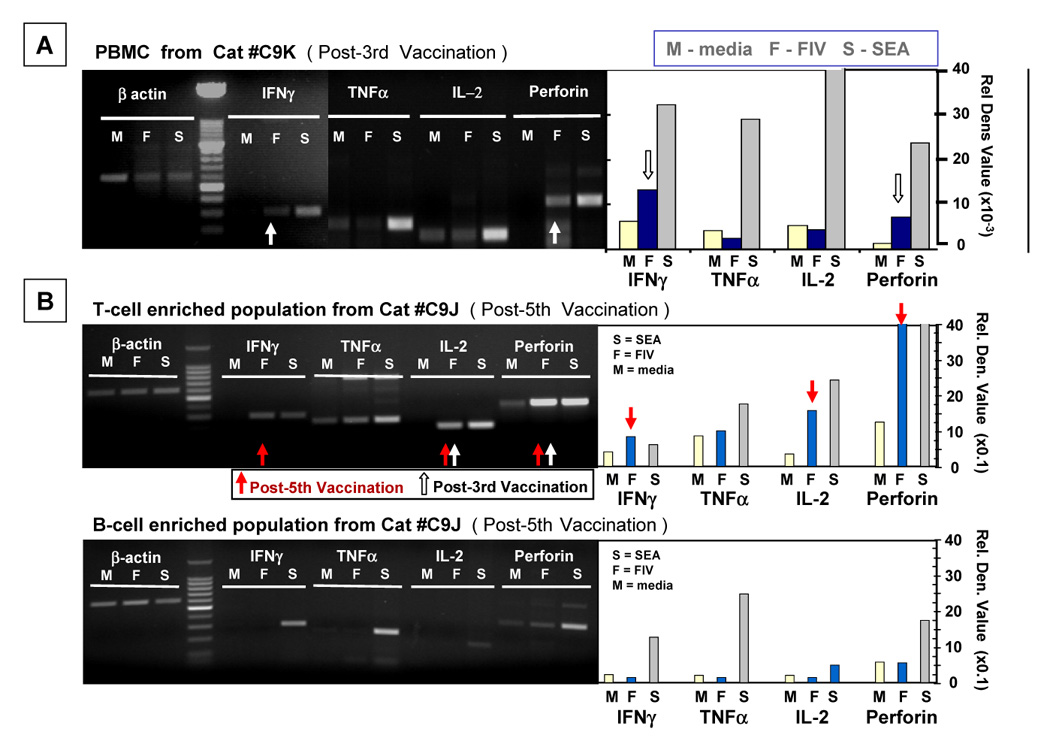
Monitoring vaccine-induced T-cell immunity by measuring mRNA for FIV-specific Th1 cytokines and CTL mediators. SPF cats were immunized 3X–5X with prototype dual-subtype FIV vaccine at 3–4 week intervals. PBMC from vaccinated cats at post-3rd vaccination (panel A) and post-5th vaccination (panel B) were cultured with inactivated FIV (F), T-cell mitogen staphylococcal enterotoxin A (S), or diluent media (M), and the cells were analyzed 18 hours later for mRNA. Th1 cytokine (IL-2 and IFNγ) mRNA, CTL mediator (TNFα, IFNγ, perforin) mRNA, and β-actin mRNA (housekeeping gene) were monitored. The mRNA was amplified by RT-PCR, and the amplified products were determined by agarose gel analysis. The intensity of the bands at predicted molecular weight sizes were determined by UV densitometry. The densitometric value representing each cytokine or CTL mediator mRNA was divided by the value for the β-actin mRNA to provide the relative densitometric value. The gel profiles and corresponding relative densitometirc value histograms of PBMC (panel A) from a cat post-3rd vaccination, and T-cell (panel B) and B-cell (panel C) enriched populations from a cat post-5th vaccination are shown with lanes for IFNγ, TNFα, IL-2, perforin, and β-actin. White and red arrows represent bands indicating high levels of cytokine or CTL mediator mRNAs present after 3rd vaccination and 5th vaccination, respectively.
Characterization of FIV-specific T-cell immunity by adoptive-transfer studies
The first adoptive-transfer studies were performed using MHC-half-matched parents and their progeny (Pu et al., 1999). These studies using single-strain FIVPet IWV vaccine suggested that recipients of adoptive transfer with peripheral blood cells from vaccinated/MHC-half-matched cats were more frequently protected against homologous FIVPet challenge. The adoptive-transfer protection was MHC-restricted based on the finding that recipients of cells from vaccinated/MHC-unmatched cats were not protected. In order increase the protection rate, improvements in generating MHC-matched cats were needed. To this end, SPF cats were inbred for MHC compatibility based on mixed leukocyte reaction (MLR) analysis. MLR analysis is frequently used as an additional analysis for matching MHC-II compatibility between human transplant donors and recipients (Jeras, 2002). The level of inbreeding was later determined by MHC-I and MHC-II sequence analyses using modified primers based on those described by Yuhki and O’Brien (Yuhki et al., 1989; Yuhki and O’Brien, 1990, 1997). The first generations (F1) were backcrosses between parent and progeny, and the subsequent generations were from breeding between MLR-matched siblings. Semi-inbred cats from F2–F4 generations were used in three adoptive-transfer studies against homologous FIVPet (Studies 1 and 2) or heterologous-subtype-B FIVFC1 (Study 3) at 25 CID50 given one day after the adoptive transfer (Table 5) (Pu et al., 2006). In Study 1, 3 of 4 adoptive-transfer recipients of T-cell-enriched population from vaccinated/MLR-matched siblings were protected, while all four recipients of either B cells from vaccinated/MLR-matched siblings or PBMC from unvaccinated/unrelated cats were infected. In Study 2, 4 of 4 recipients of T-cell-enriched population, 2 of 3 recipients of CD8+ T cells, and 2 of 3 recipients of CD4+ T cells from vaccinated/MLR-matched donors were protected. All four cats that received either PBS or T-cell-enriched populations from nonvaccinated/matched donors were infected. In Study 3, 2 of 4 recipients of T-cell-enriched population from vaccinated/MLR-matched siblings were protected against heterologous-subtype challenge, while both recipients of T-cell-enriched population from vaccinated/MLR-unmatched cats and the 2 recipients of PBS were not protected.
Table 5.
Vaccine-induced T-cell immunity important in protection observed in adoptive-transfer (A-T) studies
| Study-Group | A-T Donor Cats |
A-T Donor-Recipient Matching |
Number of Recipients | FIV Statusc | Protection Rate (%) | |||
|---|---|---|---|---|---|---|---|---|
| Immune Status | Cell Typea | MLR | MHC-I b | MHC-II b | ||||
| F2 generation against homologous FIVPet challenge | ||||||||
| 1A | Vaccinate | T cell | Matched | ND | ND | 3 | − | 3 / 4 (75%) |
| Vaccinate | T cell | Matched | ND | ND | 1 | + | ||
| 1B | Vaccinate | B cell | Matched | ND | ND | 2 | + | 0 / 2 ( 0% ) |
| 1C | Non-vaccinate | PBMC | Unmatched | ND | ND | 2 | + | 0 / 2 ( 0% ) |
| F3/F4 generations against homologous FIVPet challenge | ||||||||
| 2A | Vaccinate | T cell | Matched | Identical | ND | 4 | − | 4 / 4 (100%) |
| 2B | Vaccinate | CD8+ T cell | Matched | Identical | ND | 2 | − | 2 / 3 (67%) |
| Vaccinate | CD8+ T cell | Matched | None | ND | 1 | + | ||
| 2C | Vaccinate | CD4+ T cell | Matched | Identicald | ND | 2 | − | 2 / 3 (67%) |
| Vaccinate | CD4+ T cell | Matched | Identical | None | 1 | + | ||
| 2D | Non-vaccinate | T cell | Matched | ND | ND | 1 | + | 0 / 2 ( 0% ) |
| Non-vaccinate | PBMC | Unmatched | ND | ND | 1 | + | ||
| 2E | None | PBS | None | NA | NA | 2 | + | 0 / 2 ( 0% ) |
| F3/F4 generations against heterologous-subtype-B FIVFC1 challenge | ||||||||
| 3A | Vaccinate | T cell | Matched | ND | ND | 2 | − | 2 / 4 (50%) |
| Vaccinate | T cell | Matched | ND | ND | 2 | + | ||
| 3B | Vaccinate | T cell | Unmatched | ND | ND | 2 | + | 0 / 2 ( 0% ) |
| 3C | None | PBS | None | NA | NA | 4 | + | 0 / 4 ( 0% ) |
Cell type from donor cat adoptively transferred to recipient cat. PBMC were purified by magnetic cell separation systems of Miltenyi Biotec, Inc. (Auburn, CA) in Study 1 and R&D Systems (Minneapolis, MN) in Studies 2 and 3.
Not done (ND); not applicable (NA); >37% of the MHC sequences were identical (Identical); none or <10% of the MHC sequences were identical (none).
FIV status (+ or −) after challenge with either homologous strain (Studies 1 & 2) or heterologous-subtype-B strain (Study 3).
One pair was not tested but the other pair had >50% of identical MHC-I sequences.
These cats were matched by MLR. Since this does not assess MHC-I homology, the cats in Study 2 were extensively evaluated by MHC-I sequencing. Thirty-seven percent of the MHC-I sequences tested were identical at the peptide binding region between the pairs of protected recipient and vaccinated/MLR-matched donor. The unprotected recipient of CD8+ T-cell population was not significantly matched with the vaccinated donor at MHC-I, since 90% of MHC-I sequences were unmatched at the peptide binding region. These observations suggest that the lack of protection by this pair was most likely caused by the lack of MHC-I compatibility. In contrast, the unprotected recipient of CD4+ T-cell population was identical in at least 80% of MHC-I sequences with vaccinated/MLR-matched donor. However, the adoptively transferred CD4+ T cells require MHC-II compatibility for their effector function. Hence, a careful analysis of MHC-II may be needed between this pair. Preliminary MHC-II sequence analysis of this pair indicates identical sequences at the MHC-II DRA peptide binding region, but only 50% of the MHC-II DRB sequences are identical at the peptide binding region. Since the MHC-II molecule consists of one molecule each of DRA and DRB, this lack of MHC-II sequence identity at DRB may result in differences in peptide recognition and enhanced donor cell rejection. Therefore, the lack of protection observed in this recipient of CD4+ T cells can be explained by the lack of MHC-II compatibility between the donor and recipient cells. Similar MHC sequence analysis is currently underway for Study 3.
Vaccine epitope mapping for FIV-specific TH and CTL functions
Several technical advances have improved evaluation of antigen-specific responses and have enabled T-cell epitopes to be both identified and functionally characterized. These include: IFN-γ enzyme-linked immunospot (IFN-γ ELISPOT), intracellular staining (ICS), Microbeta Trilux-based proliferation assays, and immunomic microarrays. The use of overlapping peptides provides a rapid method for screening large numbers of sequences, and T-cell epitopes can be mapped using these assays by assessing the cellular responses to target peptides.
Feline IFN-γELISPOT
IFN-γ ELISPOT assays are used to quantify antigen-specific T cells and have generally replaced the chromium release CTL assay, as they are easier and faster to perform. In addition, IFN-γ production generally occurs before T cells proliferate, and therefore is an early event in T-cell activation. A feline IFN-γ ELISPOT assay has been developed and the ability to use it on both PBMC and targeted subsets of T cells (i.e., CD4+ or CD8+) has allowed for more complete characterizations of the feline immune response (Dean et al., 2004, Sirriyah et al., 2004). For example, the immunogenicity of Gag-p24 has been analyzed using feline IFN-γ ELISPOT assays to measure the responses of PBMC isolated from both FIV-infected and dual-subtype vaccinated cats following exposure to overlapping p24 peptide pools (15mers with 11 aa overlap; 3–4 peptides/pool). PBMC from both vaccinated cats and cats infected with a highly pathogenic strain of FIV (FIVFC1) had high IFNγ responses to multiple p24 peptide pools (Fig 3). In contrast, PBMC isolated from cats infected with low pathogenic FIVPet generally had minimal IFNγ responses. The p24 peptide pools (epitopes) recognized most frequently by PBMC from vaccinated cats were completely different from those recognized by PBMC from either FIVFC1 or FIV Pet infected cats (Fig 3). Interestingly, the pattern of p24 epitope recognition by vaccinated cats was also significantly different from that recognized by cats infected with the live vaccine virus (FIV Pet). In addition, PBMC from vaccinated cats recognized recombinant p24 proteins and to some extent whole-FIV immunogen (IWV: inactivated FIV Pet plus FIV shi) more efficiently than the PBMC from FIV-infected cats. These results suggest that the dual-subtype FIV vaccinated cats have a different recognition pattern for the whole FIV-immunogen and p24 proteins. Further characterization of this pattern of epitope recognition will provide information critical to the understanding of vaccine protection and lead to the development of new vaccines.
Fig. 3.
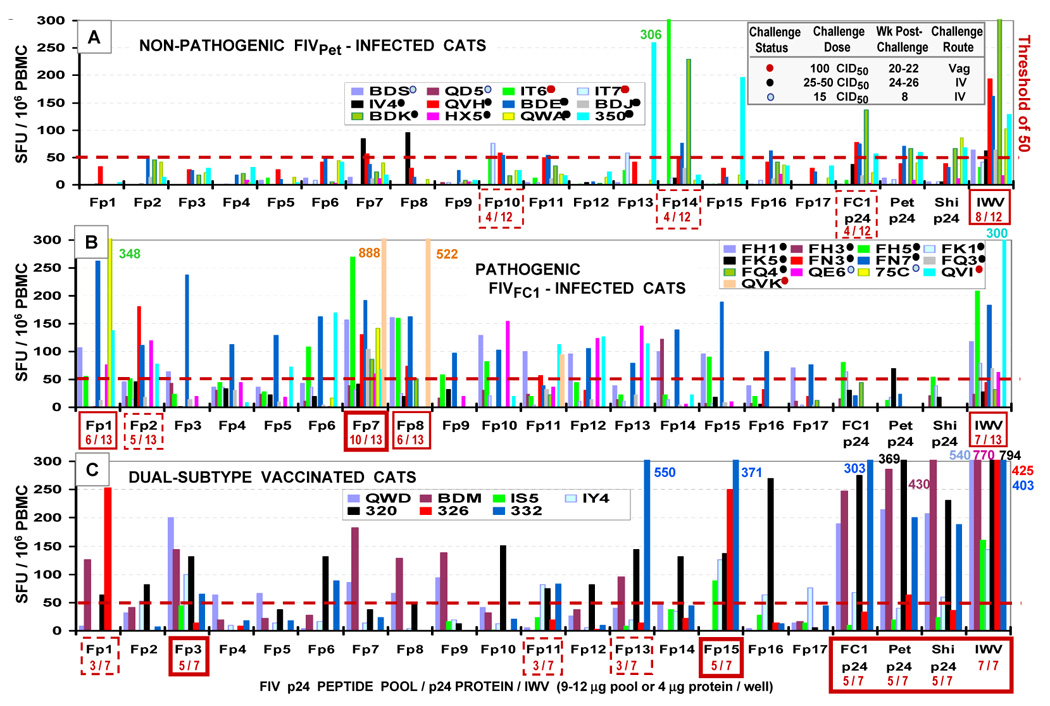
FIV p24-specific IFNγELISpot responses of PBMC from FIV-infected cats and dual-subtype vaccinated cats. Twelve and 13 SPF cats were inoculated with non-pathogenic FIVPet (panel A) and pathogenic FIVFC1 (panel B), respectively. The challenge dose, challenge route, and the time of blood collection for ELISpot analysis are shown in panel A (right top). All of these cats were positive for FIV by virus isolation and proviral PCR and by the development of FIV antibodies (Pu et al., 2001). These cats were divided into three groups according to the following challenge doses: 15 CID50 (light-blue dot next to the cat identification number), 25–50 CID50 (black dot), and 100 CID50 (red dot). Seven SPF cats were immunized 3X–4X at 3–4 week intervals with prototype dual-subtype FIV vaccine (panel C). PBMC from FIVPet-infected cats, FIVFC1-infected cats, and dual-subtype vaccinated cats were incubated with FIV p24 peptide pools, FIV p24 proteins, or IWV for 18 hour in ELISpot plates. Overlapping 15mer peptides with 11 aa overlap were synthesized based on subtype-A FIV p24 sequence and the 3–4 consecutive peptides were pooled to derive 17 FIV p24 peptide pools (Fp1–Fp17). Recombinant FIV p24 proteins were produced using E. coli expression system (Coleman et al., 2005). Feline IFNγELISpot development was performed according to manufacturer’s protocol (R&D Systems). ELISpot analyses were performed on PBMC from vaccinated cats at post-3rd (cats QWD, BDM, IS5) and 5th (cats IY4, 320, 326, 332) vaccinations and from infected cats at 8–26 weeks post-challenge. Those peptide pools, which induced responses in 33–43%, 46–67%, and ≥71% of the cats, are shown with dotted-line, solid-line, and bolded-line boxes, respectively. The ratio within the box is the number of responding cats over total number of cats tested. In general, PBMC from nonpathogenic FIVPet-infected cats had lower IFNγresponses to overlapping peptide pools than PBMC from pathogenic FIVFC1-infected cats. Furthermore, the peptide pools most frequently recognized above threshold were different between the PBMC from FIVPet-infected cats and those from FIVFC1-infected cats. The FIV doses or routes used for infection were most likely not the cause of the difference since the cats with different inoculation doses and routes were evenly distributed between the two strains. The PBMC from vaccinated cats had robust IFNγresponses to FIV p24 proteins, IWV, and to multiple peptide pools. Furthermore, the vaccinated cats recognized p24 peptide pools, which were generally different from the pools recognized by the infected cats.
Intracellular staining (ICS), Microbeta Trilux-based proliferation assays and Immunomics
Other techniques used to assess T-cell functions include ICS, Microbeta Trilux-based proliferation assays and immunomic microarrays, and the feline reagents for use in these assays are available or are being developed. Perforin is a cytotoxic effector molecule found in CTL and natural killer cells. It is upregulated in PBMC from vaccinated cats following exposure to FIV (Omori et al., 2004). ICS for perforin can be used to compare the cytotoxic capacity of CD8+ T cells following vaccination/challenge versus FIV infection. The lytic activity of CTL or NK cells, and cellular proliferation in response to feline IL-2 can be measured using Benchtop Microbeta Trilux-based proliferation assays, which offer the advantages of smaller sample size and reduction of radioactive waste over the traditional chromium releases assays (Wallace et al., 2004). Advances in microarray technology are allowing cellular activation profiles to be increasingly characterized. For example, protein/DNA arrays, which simultaneously screen over 50 transcription factors, can be used on nuclear extracts of feline cells to determine the transcription factor activation profiles associated with FIV infection and potentially protective immunity (Uhl, personal communiqué). Finally, T-cell epitope mapping is only part of the new field of immunomics, which characterizes the interface between host and pathogen and bridges informatics, genomics, proteinomics, immunology and clinical medicine (De Groot A, 2006). Such an integrative approach has the potential to make dramatic advances in vaccine technology.
Computational modeling for T-cell immunity-based FIV vaccines
The adoptive-transfer studies with semi-inbred cats suggest that protection induced by the dual-subtype vaccine is mediated by MHC-restricted T cells and is effective against both homologous- and heterologous-subtype challenges. Both vaccine-induced CD4+ T cells and CD8+ T cells are important for protection. Studies are in progress to identify the vaccine epitopes recognized by these T cells using the epitope mapping methods described above. Developing databases for viral peptides and the binding pockets of feline MHC-I and -II in the context of efficacy trials should provide a computational approach for the development of T-cell based vaccines as well as T-helper based antibody vaccines (Fig 4). Computerized assessment of human MHC-I and –II binding sites on sequences of interest is available through website databases (De Groot et al., 2002; De Groot and Berzofsky, 2004). However, such databases for feline MHC-peptide binding sites have not yet been developed. Such MHC-based computerized modeling is still in the early stages of development for use in designing vaccine immunogens for human diseases (De Groot et al., 2005A, 2005B). Once feline MHC-peptide binding patterns are determined the integration of efficacy results to such a database can follow. Since unlike with human vaccines, feline vaccine efficacies can be directly determined in cats, the establishment of a feline MHC database would be expected to more quickly contribute to the development of new vaccines.
Fig. 4.
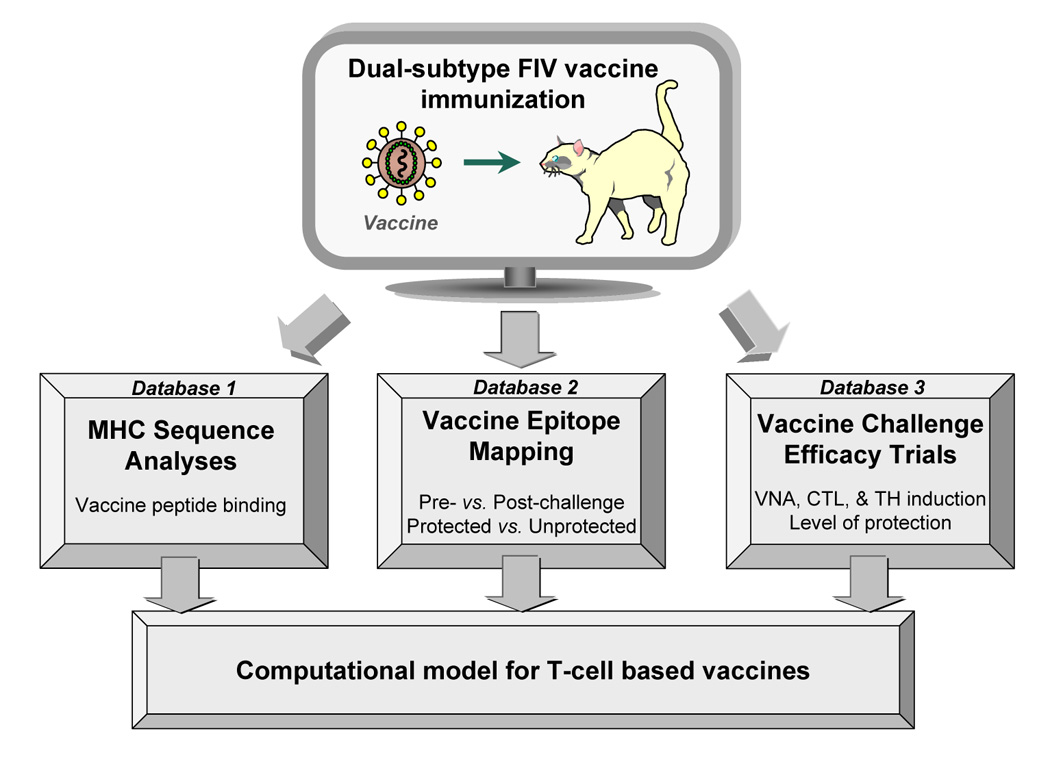
Computational modeling for T-cell immunity-based FIV vaccines. Propred-I and Propred-II epitope prediction tools are few of the known databases for identifying peptides binding to HLA class-I and class-II, respectively (De Groot et al., 2002). Los Alamos National Laboratory (LANL) database provides antibody, TH, and CTL epitope mapping for HIV-1 mainly derived from infected humans or vaccinated humans or animals (LANL, 2006). Similar epitope prediction tools based on feline MHC-I and –II can be produced concurrent to the sequencing of MHC alleles in domestic cat population (database 1) and to the epitope mapping generated by T-cell assays using semi-inbred cats (database 2). Vaccine epitope analysis can be performed directly in the natural host by immunizing potential protective peptides and evaluating the protective efficacy of these peptides against FIV challenge (database 3).
Acknowledgements
This work was funded by NIH R01AI30904, NIH R01AI065276, and JKY Miscellaneous Donors Fund. J. Yamamoto is the inventor of record on a University of Florida held patent and may be entitled to royalties from companies that are developing commercial products that are related to the research described in this paper.
Footnotes
Conflict of Interest Statement None of the authors has a financial or personal relationship with other people or organizations that could inappropriately influence or bias the paper entitled “Advances in FIV vaccine technology”.
Conflict of interest statement required by NIH to be placed in acknowledgment: Dr. Yamamoto is the inventor of record on a University of Florida patent and may be entitled to royalties from companies that are developing commercial products that are related to the research described in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Addo MM, Yu XG, Rathod A, Cohen D, Eldridge RL, Strick D, Johnston MN, Corcoran C, Wurcel AG, Fitzpatrick CA, Feeney ME, Rodriguez WR, Basgoz N, Draenert R, Stone DR, Brander C, Goulder PJ, Rosenberg ES, Altfeld M, Walker BD. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 2003;77:2081–2092. doi: 10.1128/JVI.77.3.2081-2092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed A, Ahmad R. HIV’s evasion of host’s NK cell response and novel ways of its countering and boosting anti-HIV immunity. Curr. HIV Res. 2003;1:295–307. doi: 10.2174/1570162033485267. [DOI] [PubMed] [Google Scholar]
- Bachmann MH, Mathiason-Dubard C, Learn GH, Rodrigo AG, Sodora DL, Mazzetti P, Hoover EA, Mullins JI. Genetic diversity of feline immunodeficiency virus: Dual infection, recombination, and distinct evolutionary rates among envelope sequence clades. J. Virol. 1997;71:4241–4253. doi: 10.1128/jvi.71.6.4241-4253.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackard JT, Cohen DE, Mayer KH. Human immunodeficiency virus superinfection and recombination: current state of knowledge and potential clinical consequences. Clin. Infect. Dis. 2002;34:1108–1114. doi: 10.1086/339547. [DOI] [PubMed] [Google Scholar]
- Broche-Pierre S, Richardson J, Moraillon A, Sonigo P. Evaluation of live feline immunodeficiency virus vaccines with modified antigenic properties. J. Gen. Virol. 2005;86:2495–2506. doi: 10.1099/vir.0.80469-0. [DOI] [PubMed] [Google Scholar]
- Burkhard MJ, Valenski L, Leavell S, Dean GA, Tompkins WA. Evaluation of FIV protein-expressing VEE-replicon vaccine vectors in cats. Vaccine. 2002;21:258–268. doi: 10.1016/s0264-410x(02)00455-3. [DOI] [PubMed] [Google Scholar]
- Capmanella JJ, Bitincka L, Smalley J. MatGAT: An application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics. 2003;4:29–32. doi: 10.1186/1471-2105-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Kank P, Sankale J-L, Dieng-Sarr A, Mazzara GP, Kalams SA, Korber B, Mboup S, Walker BD. Cytotoxic T-lymphocyte cross-reactivity among different human immunodeficiency virus type 1 clades: implications for vaccine development. J. Virol. 1997;71:8615–8623. doi: 10.1128/jvi.71.11.8615-8623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, McNevin J, Holte S, Fink L, Corey L, McElrath MJ. Comprehensive analysis of human immunodeficiency virus type 1 (HIV-1)-specific gamma interferon-secreting CD8+ T cells in primary HIV-1 infection. J. Virol. 2003;77:6867–6878. doi: 10.1128/JVI.77.12.6867-6878.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarota SA, Weiner DB. Enhancement of human immunodeficiency virus type 1-DNA vaccine potency through incorporation of T-helper 1 molecular adjuvants. Immunol. Rev. 2004;199:87–99. doi: 10.1111/j.0105-2896.2004.00150.x. [DOI] [PubMed] [Google Scholar]
- Dean GA, LaVoy A, Burkhard MJ. Peptide mapping of feline immunodeficiency virus by IFN-γ ELISPOT. Vet. Immunol. Immunopathol. 2004;100:49–59. doi: 10.1016/j.vetimm.2004.03.001. [DOI] [PubMed] [Google Scholar]
- De Groot AS. Immunomics: discovering new targets for vaccines and therapeutics. DDT. 2006;11:203–209. doi: 10.1016/S1359-6446(05)03720-7. [DOI] [PubMed] [Google Scholar]
- De Groot AS, Sbai H, Saint Aubin C, Mcmurry J, Martin W. Immuno-informatics: mining genomes for vaccine components. Immunol. Cell Biol. 2002;80:255–269. doi: 10.1046/j.1440-1711.2002.01092.x. [DOI] [PubMed] [Google Scholar]
- De Groot AS, Bersofsky JA. From genome to vaccine – new immunoinformatics tools for vaccine design. Methods. 2004;34:425–428. doi: 10.1016/j.ymeth.2004.06.004. [DOI] [PubMed] [Google Scholar]
- De Groot AS, Marcon L, Bishop EA, Rivera D, Kutzler M, Weiner DB, Martin W. HIV vaccine development by computer assisted design: the GAIA vaccine. Vaccine. 2005A;23:2136–2148. doi: 10.1016/j.vaccine.2005.01.097. [DOI] [PubMed] [Google Scholar]
- De Groot AS, McMurry J, Marcon L, Franco J, Rivera D, Kutzler M, Weiner D, Martin W. Developing an epitope-driven tuberculosis (TB) vaccine. Vaccine. 2005B;23:2121–2131. doi: 10.1016/j.vaccine.2005.01.059. [DOI] [PubMed] [Google Scholar]
- Dunham SP, Flynn JN, Rigby MA, Macdonald J, Bruce J, Cannon C, Golder MC, Hanlon L, Harbour DA, Mackay NA, Spibey N, Jarrett O, Neil JC. Protection against feline immunodeficiency virus using replication defective proviral DNA vaccines with feline interleukin-12 and -18. Vaccine. 2002;20:1483–1496. doi: 10.1016/s0264-410x(01)00507-2. [DOI] [PubMed] [Google Scholar]
- Dunham SP, Bruce J, Klein D, Flynn JN, Golder MC, MacDonald S, Jarrett O, Neil JC. Prime-boost vaccination using DNA and whole inactivated virus vaccines provides limited protection against virulent feline immunodeficiency virus. Vaccine. 2006;24:7095–7108. doi: 10.1016/j.vaccine.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Elyar JS, Tellier MC, Soos JM, Yamamoto JK. Perspectives on FIV vaccine development. Vaccine. 1997;15:1437–1444. doi: 10.1016/s0264-410x(97)00056-x. [DOI] [PubMed] [Google Scholar]
- Finnegan CM, Berg W, Lewis GK, DeVico AL. Antigenic properties of the human immunodeficiency virus envelope during cell-cell fusion. J. Virol. 2001;75:11096–11105. doi: 10.1128/JVI.75.22.11096-11105.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JN, Beatty JA, Cannon CA, Stephens EB, Hosie MJ, Neil JC, Jarrett O. Involvement of gag- and env-specific cytotoxic T lymphocytes in protective immunity to feline immunodeficiency virus. AIDS Res. Hum. Retrovir. 1995;11:1107–1113. doi: 10.1089/aid.1995.11.1107. [DOI] [PubMed] [Google Scholar]
- Flynn NM, Forthal DN, Harro CD, Judson FN, Mayer KH, Para MF The rgp120 HIV Vaccine Study Group. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J. Infect. Dis. 2005;191:654–664. doi: 10.1086/428404. [DOI] [PubMed] [Google Scholar]
- Gemeniano MC, Sawai ET, Sparger EE. Feline immunodeficiency virus Orf-A localizes to the nucleus and induces cell cycle arrest. Virology. 2004;325:167–174. doi: 10.1016/j.virol.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Girard MP, Osmanov SK, Kieny MP. A review of vaccine research and development: the human immunodeficiency virus (HIV) Vaccine. 2006;24:4062–4081. doi: 10.1016/j.vaccine.2006.02.031. [DOI] [PubMed] [Google Scholar]
- Gorse GJ, Patel GB, Mandava MD, Belshe RB National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. Vaccine-induced cytotoxic T lymphocytes against human immunodeficiency virus type 1 using two complementary in vitro stimulation strategies. Vaccine. 2000;18:835–849. doi: 10.1016/s0264-410x(99)00323-0. [DOI] [PubMed] [Google Scholar]
- Gupta S, Leutenegger CM, Dean GA, Steckbeck JD, Cole KS, Sparger EE. Vaccination with attenuated feline immunodeficiency virus proviral DNA vaccine expressing interferon gamma. J. Virol. 2006 Nov 1; doi: 10.1128/JVI.00815-06. [Epub] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, Robinson J, Scearce RM, Plonk K, Staats HF, Ortel TL, Liao HX, Alam SM. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D, editors. Antibodies: A Laboratory Manual. New York: Cold Spring Harbor Laboratory; 1988. Storing and purifying antibodies; pp. 282–317. [Google Scholar]
- Hohdatsu T, Pu R, Torres BA, Trujillo S, Gardner MB, Yamamoto JK. Passive protection of cats against feline immunodeficiency virus infection. J. Virol. 1993;67:2344–2348. doi: 10.1128/jvi.67.4.2344-2348.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover EA, Mullins JI, Chu H-J, Wasmoen TL. Efficacy of an inactivated leukemia virus vaccine. AIDS Res. Hum. Retrovir. 1996;12:379–383. doi: 10.1089/aid.1996.12.379. [DOI] [PubMed] [Google Scholar]
- Hosie MJ, Dunsford T, Klein D, Willett BJ, Cannon C, Osborne R, Macdonald J, Spibey N, Mackay N, Jarrett O, Neil JC. Vaccination with inactivated virus but not viral DNA reduces virus load following challenge with a heterologous and virulent isolate of feline immunodeficiency virus. J. Virol. 2000;74:9403–9411. doi: 10.1128/jvi.74.20.9403-9411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosie MJ, Klein D, Binley JM, Dunsford TH, Jarrett O, Neil JC, Knapp E, Giannecchini S, Matteucci D, Bendinelli M, Hoxie JA, Willett BJ. Vaccination with an inactivated virulent feline immunodeficiency virus engineered to express high levels of Env. J. Virol. 2005;79:1954–1957. doi: 10.1128/JVI.79.3.1954-1957.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman W, Schrauwen EJ, Pas SD, Karlas JA, Rimmelzwaan GF, Osterhaus AD. Antibodies specific for hypervariable regions 3 to 5 of the feline immunodeficiency virus envelope glycoprotein are not solely responsible for vaccine-induced acceleration of challenge infection in cats. J. Gen. Virol. 2004;85:1833–1841. doi: 10.1099/vir.0.79949-0. [DOI] [PubMed] [Google Scholar]
- Jeras M. The role of in vitro alloreactive T-cell functional tests in the selection of HLA matched and mismatched haematopoietic stem cell donors. Transpl. Immunol. 2002;10:205–214. doi: 10.1016/s0966-3274(02)00067-9. [DOI] [PubMed] [Google Scholar]
- Kaufmann DE, Bailey PM, Sidney J, Wagner B, Norris PJ, Johnston MN, Cosimi LA, Addo MM, Lichterfeld M, Altfeld M, Frahm N, Brander C, Sette A, Walker BD, Rosenberg ES. Comprehensive analysis of human immunodeficiency virus type 1- specific CD4 responses reveals marked immunodominance of gag and nef and the presence of broadly recognized peptides. J. Virol. 2004;78:4463–4477. doi: 10.1128/JVI.78.9.4463-4477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallel H, Jouini A, Majoul S, Rourou S. Evaluation of various serum and animal protein free media for the production of a veterinary rabies vaccine in BHK-21 cells. J. Biotechnol. 2002;95:195–204. doi: 10.1016/s0168-1656(02)00009-3. [DOI] [PubMed] [Google Scholar]
- Kallel H, Rourou S, Majoul S, Loukil H. A novel process for the production of a veterinary rabies vaccine in BHK-21 cells grown on microcarriers in a 20-l bioreactor. Appl. Microbiol. Biotechnol. 2003;61:441–446. doi: 10.1007/s00253-003-1245-3. [DOI] [PubMed] [Google Scholar]
- Karnasuta C, Paris RM, Cox JH, Nitayaphan S, Pitisuttithum P, Thongcharoen P, Brown AE, Gurunathan S, Tartaglia J, Heyward WL, McNeil JG, Birx DL, de Souza MS Thai AIDS Vaccine Evaluation Group, Thailand. Antibody-dependent cell-mediated cytotoxic responses in participants enrolled in a phase I/II ALVAC-HIV/AIDSVAX B/E prime-boost HIV-1 vaccine trial in Thailand. Vaccine. 2005;23:2522–2529. doi: 10.1016/j.vaccine.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Kusuhara H, Hohdatsu T, Okumura M, Kayoko S, Suzuki Y, Motokawa K, Gemma T, Watanabe R, Huang C, Arai S, Koyama H. Dual-subtype vaccine (Fel-O-Vax FIV) protects cats against contact challenge with heterologous subtype B FIV infected cats. Vet. Microbiol. 2005;108:155–165. doi: 10.1016/j.vetmic.2005.02.014. [DOI] [PubMed] [Google Scholar]
- Levy JA, Scott I, Mackewicz C. Protection from HIV/AIDS: the importance of innate immunity. Clin. Immunol. 2003;108:167–174. doi: 10.1016/s1521-6616(03)00178-5. [DOI] [PubMed] [Google Scholar]
- Los Alamos National Laboratory. Epitope maps. [(Accessed 25 October 2006)]. //hiv-web.lanl.gov/content/immunology/maps/maps.html/
- Li J, Brown WC, Song W, Carpino MR, Wolf AM, Grant CK, Elder JH, Collisson EW. Retroviral vector-transduced cells expressing the core polyprotein induce feline immunodeficiency virus-specific cytotoxic T-lymphocytes from infected cats. Virus Res. 1995;38:93–109. doi: 10.1016/0168-1702(95)00050-z. [DOI] [PubMed] [Google Scholar]
- Matteucci D, Poli A, Mazzetti P, Sozzi S, Bonci F, Isola P, Zaccaro L, Giannecchini S, Calandrella M, Pistello M, Specter S, Bendinelli M. Immunogenicity of an anti-clade B feline immunodeficiency fixed-cell virus vaccine in field cats. J. Virol. 2000;74:10911–10919. doi: 10.1128/jvi.74.23.10911-10919.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo K, Nishino Y, Kimura T, Yamaguchi R, Yamazaki A, Mikami T, Ikuta K. Highly conserved epitope domain in major core protein p24 is structurally similar among human, simian and feline immunodeficiency viruses. J. Gen. Virol. 1992;73:2445–2450. doi: 10.1099/0022-1317-73-9-2445. [DOI] [PubMed] [Google Scholar]
- Miyazawa T. Evolution of lentiviruses and receptor specificity. Uirusu. 2005;55:27–34. doi: 10.2222/jsv.55.27. [DOI] [PubMed] [Google Scholar]
- Miyazawa T, Tomonaga K, Kawaguchi Y, Mikami T. The genome of feline immunodeficiency virus. Arch. Virol. 1994;134:221–234. doi: 10.1007/BF01310563. [DOI] [PubMed] [Google Scholar]
- Naylor PH, Hadden JW. T cell targeted immune enhancement yields effective T cell adjuvants. Int. Immunopharmacol. 2003;3:1205–1215. doi: 10.1016/S1567-5769(03)00025-0. [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Igarashi T, Haigwood N, Sadjadpour R, Plishka RJ, Buckler-White A, Shibata R, Martin MA. Determination of a statistically valid neutralization titer in plasma that confers protection against simian-human immunodeficiency virus challenge following passive transfer of high-titered neutralizing antibodies. J. Virol. 2002;76:2123–2130. doi: 10.1128/jvi.76.5.2123-2130.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Igarashi T, Haigwood NL, Sadjadpour R, Donau OK, Buckler C, Plishka RJ, Buckler-White A, Martin MA. Transfer of neutralizing IgG to macaques 6 h but not 24 h after SHIV infection confers sterilizing protection: implications for HIV-1 vaccine development. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15131–15136. doi: 10.1073/pnas.2436476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitayaphan S, Pitisuttithum P, Karnasuta C, Eamsila C, de Souza M, Morgan P, Polonis V, Benenson M, VanCott T, Ratto-Kim S, Kim J, Thapinta D, Garner R, Bussaratid V, Singharaj P, el-Habib R, Gurunathan S, Heyward W, Birx D, McNeil J, Brown AE Thai AIDS Vaccine Evaluation Group. Safety and immunogenicity of an HIV subtype B and E prime-boost vaccine combination in HIV-negative Thai adults. J. Infect. Dis. 2004;90:702–706. doi: 10.1086/422258. [DOI] [PubMed] [Google Scholar]
- Norris PJ, Moffett HF, Brander C, Allen TM, O’Sullivan KM, Cosimi LA, Kaufmann DE, Walker BD, Rosenberg ES. Fine specificity and cross-clade reactivity of HIV type I Gag-specific CD4+ T cells. AIDS Res. Hum. Retrovir. 2004;20:315–325. doi: 10.1089/088922204322996554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori M, Pu R, Tanabe T, Hou W, Coleman J, Arai M, Yamamoto JK. Cellular immune responses to feline immunodeficiency virus (FIV) induced by dual-subtype FIV vaccine. Vaccine. 2004;23:386–398. doi: 10.1016/j.vaccine.2004.05.032. [DOI] [PubMed] [Google Scholar]
- Pantophlet R, Burton DR. Gp120: target for neutralizing HIV-1 antibodies. Annu. Rev. Immunol. 2006;24:739–769. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- Pistello M, Bonci F, Flynn JN, Mazzetti P, Isola P, Zabogli E, Camerini V, Matteucci D, Freer G, Pelosi P, Bendinelli M. AIDS vaccination studies with an ex vivo feline immunodeficiency virus model: analysis of the accessory ORF-A protein and DNA as protective immunogens. J. Virol. 2006;80:8856–8868. doi: 10.1128/JVI.00397-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistello M, Bonci F, Isola P, Mazzetti P, Merico A, Zaccaro L, Matteucci D, Bendinelli M. Evaluation of feline immunodeficiency virus ORF-A mutants as candidate attenuated vaccine. Virology. 2005;332:676–690. doi: 10.1016/j.virol.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Pu R, Okada S, Omori M, Rine SL, Lewis BA, Lipton E, Nemcik L, Glass P, Yamamoto JK. MHC-restricted protection of cats against FIV infection with adoptive transfer of immunocytes from FIV-vaccinated cats. Cell. Immunol. 1999;198:30–43. doi: 10.1006/cimm.1999.1574. [DOI] [PubMed] [Google Scholar]
- Pu R, Coleman J, Omori M, Mison M, Huang C, Arai M, Tanabe T, Yamamoto JK. Dual-subtype FIV vaccine protects cats against in vivo swarms of both homologous and heterologous subtype FIV isolates. AIDS. 2001;15:1–13. doi: 10.1097/00002030-200107060-00004. [DOI] [PubMed] [Google Scholar]
- Pu R, Paredes G, Yamamoto JK. FIV vaccine update [Proceeding] Am. Assoc. Feline Practitioners. 2004:103–107. [Google Scholar]
- Pu R, Coisman J, Coleman J, Sato E, Tanabe T, Arai M, Yamamoto JK. Dual-subtype FIV vaccine (Fel-O-Vax® FIV) protection against heterologous subtype B FIV isolate. Feline Med. Surg. 2005;7:65–70. doi: 10.1016/j.jfms.2004.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu R, Sato E, Yamamoto JK. Feline immunodeficiency virus (FIV)-cat model for AIDS: T-cell immunity important for prophylactic vaccine protection. XVI Intl. Confer. AIDS. 2006:290. [Abstract THP0016] [Google Scholar]
- Quackenbush SL, Donahue PR, Dean GA, Myles MH, Ackley CD, Cooper MD, Mullins JI, Hoover EA. Lymphocyte subset alterations and viral determinants of immunodeficiency disease induction by the feline leukemia virus FeLV-FAIDS. J. Virol. 1990;64:5465–5474. doi: 10.1128/jvi.64.11.5465-5474.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J, Broche S, Baud S, Leste-Lasserre T, Femenia F, Levy D, Moraillon A, Pancino G, Sonigo P. Lymphoid activation: a confounding factor in AIDS vaccine development? J. Gen. Virol. 2002;83:2515–2521. doi: 10.1099/0022-1317-83-10-2515. [DOI] [PubMed] [Google Scholar]
- Schutz AM, Bradac JA. The HIV vaccine pipeline, from preclinical to phase III. AIDS. 2001;15:S147–S158. doi: 10.1097/00002030-200100005-00018. [DOI] [PubMed] [Google Scholar]
- Seya T, Akazawa T, Tsujita T, Matsumoto M. Role of Toll-like receptors in adjuvant-augmented immune therapies. Evid. Based Complement Alternat. Med. 2006;3:31–38. doi: 10.1093/ecam/nek010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojima M, Miyazawa T, Ikeda Y, McMonagle EL, Haining H, Akashi H, Takeuchi Y, Hosie MJ, Willett BJ. Use of CD134 as a primary receptor by the feline immunodeficiency virus. Science. 2004;303:1192–1195. doi: 10.1126/science.1092124. [DOI] [PubMed] [Google Scholar]
- Sirriyah J, Dean GA, LaVoy A, Burkhard MJ. Assessment of CD4+ and CD8+ IFN-gamma producing cells by ELISPOT in naïve and FIV-infected cats. Vet. Immunol. Immunopathol. 2004;102:77–84. doi: 10.1016/j.vetimm.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Stanhope PE, Liu AY, Pavlat W, Pitha PM, Clements ML, Siliciano RF. An HIV-1 envelope protein vaccine elicits a functionally complex human CD4+ T cell response that includes cytolytic T lymphocytes. J. Immunol. 1993;150:4672–4686. [PubMed] [Google Scholar]
- Stevens R, Howard KE, Nordone S, Burkhard M, Dean GA. Oral immunization with recombinant listeria monocytogenes controls virus load after vaginal challenge with feline immunodeficiency virus. J. Virol. 2004;78:8210–8218. doi: 10.1128/JVI.78.15.8210-8218.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl EW, Heaton-Jones T, Pu R, Yamamoto JK. FIV vaccine development and its importance to veterinary and human medicine: a review. Vet. Immunol. Immunopathol. 2002;90:113–132. doi: 10.1016/S0165-2427(02)00227-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D, Hildesheim A, Pinto LA. Comparison of benchtop microplate beta counters with the traditional gamma counting method for measurement of chromium-51 release in cytotoxic assays. Clin. Diagn. Lab Immunol. 2004;11:255–260. doi: 10.1128/CDLI.11.2.255-260.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto JK, Okuda T, Ackley CD, Louie H, Zochlinski H, Pembroke E, Gardner MB. Experimental vaccine protection against feline immunodeficiency virus. AIDS Res. Hum. Retrovir. 1991;7:911–922. doi: 10.1089/aid.1991.7.911. [DOI] [PubMed] [Google Scholar]
- Yamamoto JK, Hohdatsu T, Olmsted RA, Pu R, Louie H, Zochlinski H, Acevedo V, Johnson HM, Soulds GA, Gardner MB. Experimental vaccine protection against homologous and heterologous strains of feline immunodeficiency virus. J. Virol. 1993;67:601–605. doi: 10.1128/jvi.67.1.601-605.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto JK. Vaccine for Prophylaxis and Therapeutics; Wild West Veterinary Conference; October 9–16, 2002; Reno, NV. 2002. [Google Scholar]
- Yamamoto JK, Pu R, Sato E, Hohdatsu T. FIV pathogenesis and development of a dual-subtype FIV vaccine. AIDS. 2007;21 doi: 10.1097/QAD.0b013e328013d88a. (in press) [DOI] [PubMed] [Google Scholar]
- Yuhki N, Heidecker GF, O'Brien SJ. Characterization of MHC cDNA clones in the domestic cat. Diversity and evolution of class I genes. J. Immunol. 1989;142:3676–3682. [PubMed] [Google Scholar]
- Yuhki N, O'Brien SJ. DNA recombination and natural selection pressure sustain genetic sequence diversity of the feline MHC class 1 genes. J. Exp. Med. 1990;172:621–630. doi: 10.1084/jem.172.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuhki N, O'Brien SJ. Nature and origin of polymorphism in feline MHC class II DRA and DRB genes. J. Immunol. 1997;158:2822–2833. [PubMed] [Google Scholar]
- Zolla-Pazner S. Identifying epitopes of HIV-1 that induce protective antibodies. Nat. Rev. Immunol. 2004;4:199–210. doi: 10.1038/nri1307. [DOI] [PMC free article] [PubMed] [Google Scholar]


