Abstract
Purpose
Senescence related regulatory pathways serve as barriers to cancer immortalization and progression but they are currently not well defined. FILIP1L is a growth inhibitory gene with multiple isoforms whose expression is increased in senescent prostate and prostate cancer cells, and decreased in many cancers. We investigated whether DNA methylation regulates FILIP1L in senescence and in prostate cancer development.
Materials and Methods
FILIP1L mRNA expression was assessed in prostate cancer and associated normal prostate tissues using quantitative polymerase chain reaction. A tissue microarray was constructed using 95 prostate cancer specimens and 45 benign prostate specimens. Vectra™ imaging was used to quantitate nuclear and cytoplasmic FILIP1L protein expression. Bisulfite sequencing and Pyrosequencing® were used to assess methylation. Prostate cancer cell lines were treated with 2′-deoxy-5-azacytidine and mRNA expression was assessed.
Results
FILIP1L isoform 2 mRNA was increased in replicatively senescent human prostate epithelial cells and decreased in prostate cancer specimens. We verified a reduction in nuclear FILIP1L protein in prostate cancer using tissue microarrays (p = 0.006). A CpG island 5′ of the isoform 2 translational start site was identified that showed hypermethylation in prostate cancer cell lines and tumors compared to normal prostate cells and tissues. Pyrosequencing confirmed FILIP1L hypermethylation in all 14 tumors compared to paired normal tissues (p <0.0001). Isoform 2 expression was induced in prostate cancer cell lines using 2′-deoxy-5-azacytidine.
Conclusions
FILIP1L isoform 2 is one of the most commonly hypermethylated genes in prostate cancer. It may serve as an important marker of prostate cancer. Isoform 2 expression is associated with senescence and its down-regulation may represent an early important biological event in prostate cancer development.
Keywords: prostate, prostatic neoplasms, gene expression, methylation, cell aging
Some hallmarks of cancer are its extended life span and persistent cellular proliferation. Cellular senescence is a mechanism by which cells undergo terminal cell cycle arrest. It is characterized by a distinct phenotype, including absent proliferation, a flattened, enlarged morphology, senescence associated β-galactosidase staining and a unique gene expression profile.1 It was postulated that cellular senescence serves as a barrier against cancer growth and progression. Pathways involved in the senescence response, including Rb, p53, cyclin-dependent kinase inhibitors and others, are frequently inactivated during cancer development.1 Identifying additional genes associated with senescence is important because of the potential to identify new therapeutic targets that can slow cancer progression.
Hypermethylation of CpG dinucleotides in CGIs in or around the promoter region of endogenous genes can down-regulate gene expression.2 Aberrant DNA methylation, which is recognized as a major hallmark of cancer, includes global hypomethylation and regional CGI hypermethylation.3 PCa is characterized by a number of DNA methylation alterations that develop early in the disease.4 Hypermethylation of GSTpi, which occurs in approximately 90% of primary prostate tumors, is also frequently found in prostatic intraepithelial neoplasia.5
Currently, the role of the epigenetic changes that occur during senescence is not well understood. We hypothesized that alterations in DNA methylation in PCa can silence important genes in the senescence phenotype.
We previously identified by RNA microarray that FILIP1L was markedly induced during senescence and inactivated with immortalization in HPECs.6 FILIP1L, formerly known as down-regulated in ovarian cancer 1, was initially discovered by DNA fingerprinting to be down-regulated in ovarian cancer compared to normal ovarian tissue.7 Functionally, sequence analyses predict that FILIP1L is part of the filamin A/actin complex.
FILIP1L consists of 3 isoforms that are differentially expressed during development. In the current study we examined the isoform expression and regulation of FILIP1L during senescence in HPECs and in PCa. We identified that isoform 2 of FILIP1L was induced during senescence but down-regulated in PCa at the mRNA and protein levels. Furthermore, a CGI in the promoter of isoform 2 of FILIP1L is commonly hypermethylated in PCa and could serve as an epigenetic marker of the disease. We conclude that DNA hypermethylation may have an important role in silencing the expression of the known senescence marker FILIP1L, suggesting that loss of the senescence phenotype may favor PCa progression.
MATERIALS AND METHODS
Cell Culture
The metastasis derived PCa cell lines DU145, PC3, LNCaP and 22Rv1 were cultured and treated with 2′-deoxy-5-azacytidine, a methyltransferase inhibitor, as previously described.8,9 We used a culture system to generate and maintain normal HPECs.10 Cells were collected in accordance with University of Wisconsin institutional review board approval. DNA and RNA were harvested throughout serial passages using standard techniques since these cells typically undergo about 20 population doublings (4 or 5 passages). We monitored each culture passage for the signs of slowed replication and morphological differences seen in senescence. We then used positive SA-β-gal11 and measured the expression of genes previously found to be up-regulated in senescence, including FILIP1L, by quantitative PCR.6
Tissue Collection
We obtained 14 paired samples of tumor and benign adjacent tissues from radical prostatectomy or cystoprostatectomy samples using an approved institutional review board protocol. Tissues were snap frozen in liquid nitrogen. Histological evaluation ruled out the presence of cancer cells. Areas of tumor and benign adjacent tissues were collected for RNA and DNA using standard techniques.
Quantitative PCR
Total RNA was isolated from drug treated cells using the RNeasy® RNA isolation kit and treated with DNase I (QIAGEN®). The Omniscript® Reverse Transcription Kit was used to synthesize cDNA using 200 ng RNA per sample. Primers were designed to amplify FILIP1L isoform 2 mRNA (RefSeq NM_014890.2). The conditions used for PCR amplification were 95C for 3 minutes, 40 cycles at 95C for 10 seconds and at 55C for 30 seconds, as previously described.12 All reactions were performed in duplicate. Quantification was done by monitoring the real-time fluorescence of SYTO® 9. PCR product amplification and detection were performed. Threshold cycles were measured using the CFX96™ Real-Time PCR Detection System. Target gene expression was normalized to 18S expression and calculated to generate the fold change. The t test was used to compare mean expression in the treatment vs the control group.
Tissue Microarray
Formalin fixed, paraffin embedded patient tissues were obtained from the Department of Pathology and Laboratory Medicine at our institution according to institutional review board approval and policies. A tissue microarray was constructed using tissues from 95 patients with a mean age of 62.8 years who had PCa. The archival prostate tissues from this cohort were collected from 1995 to 2006.
The tissue microarray consisted of 384 duplicate cores from different disease groups, including 43 localized (pT2), 30 aggressive (pT3 and 4) and 22 metastatic PCa, 25 HGPIN (from HGPIN tissue blocks of some patients with PCa in this cohort), 48 benign prostate (from the nontumor blocks of some patients with cancer in this cohort) and 24 benign prostatic hyperplasia. The tissue microarray slides were taken through routine deparaffinization and rehydration, and stained with antibodies against FILIP1L (Santa Cruz Biotechnology, Santa Cruz, California). E-cadherin antibodies (Cell Signaling Technology®) were used to define the epithelial compartment for better tissue segmentation.
For automated image acquisition and analysis the stained slides were loaded on the slide scanner. FILIP1L target signals per cell were quantitated for individual cores using the Vectra imaging system according to the manufacturer protocol. This system allows the automated selection of cellular subsets (epithelial vs stromal) and the quantitation of fluorescent staining. We used inForm1.2 software to segment tissue compartments (epithelium vs stroma) and subcellular compartments (nucleus vs cytoplasm). Nuclear and cytoplasmic expression was statistically compared using the t test. FILIP1L protein expression was compared against clinicopathological features. Statistical significance was considered at p <0.05.
Sodium Bisulfite DNA Treatment and Sequencing
After genomic DNA was extracted using standard methods, 1 µg DNA was treated with sodium bisulfite according to the EpiTect® Bisulfite kit protocol. Genomic DNA was isolated and treated with sodium bisulfite. Sodium bisulfite treatment changes unmethylated cytosine in the DNA sequence to a uracil (and subsequently to thymine), while methylated cytosines remain unchanged. The entire CGI of FILIP1L was PCR amplified and subcloned using the TOPO® TA vector system according to manufacturer instructions. After transformation using Escherichia coli, we selected 5 to 10 individual E. coli colonies for each cell line assessed. Plasmid DNA was isolated using a QIAPrep Miniprep Kit (QIAGEN). Plasmids were then sequenced using M13 forward and reverse primers maintained in the multiple cloning site of the vector.
Pyrosequencing FILIP1L CGI
Quantitative Pyrosequencing was done to assess methylation at 10 CpGs 700 bp upstream of the transcriptional start site, as previously described.13 Bisulfite converted DNA was amplified by PCR using forward or reverse biotinylated primers. PCR and sequence primers for Pyrosequencing were designed with PSQ™ Assay Design 1.0. Biotinylated PCR products were separated with streptavidin Sepharose® beads, denatured to single strands and annealed to sequencing primers for the Pyrosequencing assay. Each 25 µl PCR contained 1× HotStarTaq® buffer (including 1.5 mM MgCl2), 0.2 mM deoxynucleotide triphosphate, 5 pmol of each primer, 1.0 U HotStarTaq DNA Polymerase and 2 µl bisulfite converted DNA. Thermocycling conditions included initial denaturation at 95C for 10 minutes, 40 cycles at 95C for 40 seconds, 55C for 40 seconds and 72C for 40 seconds, and final extension at 72C for 7 minutes.
PCR products (10 µl) were sequenced using Pyro-Mark™ MD according to manufacturer directions. Pyro Q-CpG™, version 1.0.9 was used to analyze results. Methylation across 10 CpGs was statistically compared using 2-way ANOVA with the Bonferroni posttest for multiple comparisons. Statistical analyses were performed using GraphPad Prism®, version 5.03.
RESULTS
FILIP1L Isoform 2
mRNA expression down-regulated in immortalized cells and cancer
We previously found that total FILIP1L expression was down-regulated in immortalized PC cell lines compared to normal HPECs.6,14 Furthermore, increased FILIP1L expression was observed when cells were passaged to senescence. To examine differential FILIP1L transcripts, we identified and designed primers to span the 3 known isoforms of FILIP1L (fig. 1, A). Primers in exon 5 are unique to isoform 2 and those in exon 7 are distinct for isoform 1. However, isoform 3 cannot be uniquely distinguished based on exon overlap with other isoforms.
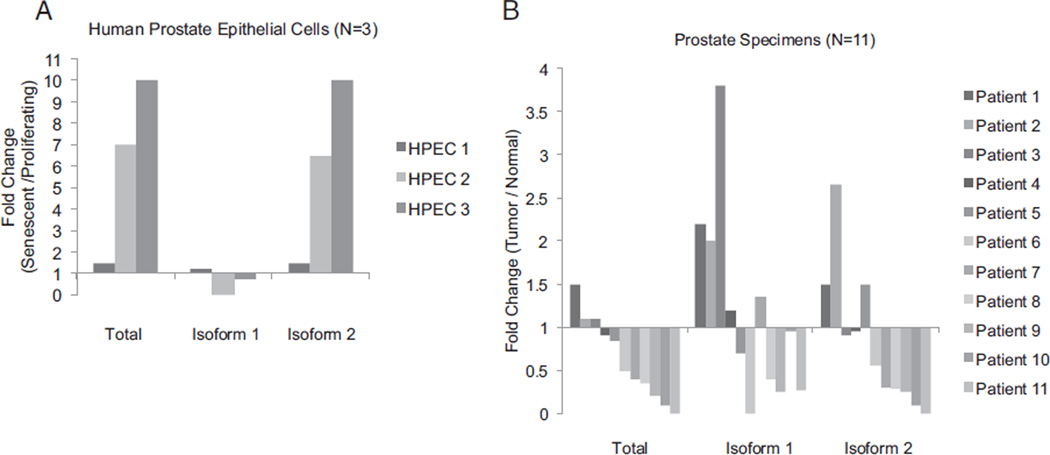
Figure 1.
Quantitative PCR was performed using isoform specific primers to determine FILIP1L RNA expression in senescence. Values were normalized to 18S. A, isoform specific mRNA levels in HPECs cultured to senescence. Fold change in expression for 3 cultures (HPEC1 to 3) was calculated by comparing mRNA expression of senescent late passage vs proliferating early passage cells. Isoform 2, only isoform up-regulated in senescence, reflected most of total FILIP1L expression. B, isoform specific mRNA expression in human PCa compared to areas of normal peripheral prostate tissue from same patient. Isoform 2 showed frequent down-regulation in tumor specimens.
Quantitative reverse transcriptase-PCR was performed on HPECs, comparing proliferating to senescent and immortalized cancer cell lines. Results demonstrated that isoform 2 and total FILIP1L were consistently induced during senescence (twofold to tenfold). Isoform 1 expression did not change (fig. 1, A). FILIP1L mRNA was not expressed in the DU145, PC3, LNCaP or 22Rv1 PCa cell lines.
We then analyzed FILIP1L isoforms from paired tumor and associated benign prostate tissues. Total FILIP1L mRNA expression was significantly decreased in 8 of 11 microdissected tumor samples (73%) compared to associated benign samples (fig. 1, B). The expression of isoforms 1 and 2 was down-regulated in 7 and 5 of 11 tumors, respectively. Thus, our expression analysis identified isoform 2 up-regulation in senescence and the frequent loss of its expression in tumors and PCa cell lines.
Nuclear protein expression down-regulated in cancer
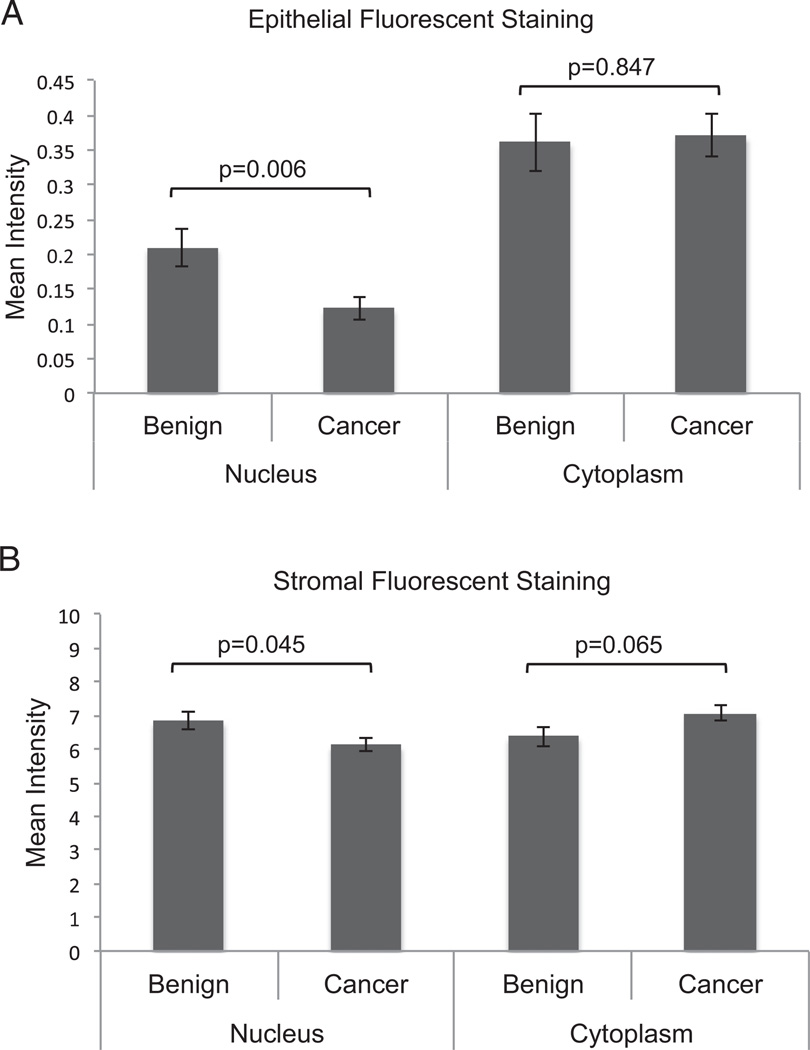
We then assessed FILIP1L protein expression using a tissue microarray and Vectra quantitative fluorescence analysis. Vectra is an automated quantitative imaging system that differentiates epithelial from stromal compartments. The FILIP1L antibody used recognizes multiple isoforms of FILIP1L protein. FILIP1L protein was expressed predominantly in the stroma of normal prostate tissues at a level approximately 15 to 30 times greater than in the epithelium. In the epithelium of benign and cancer specimens FILIP1L expression was greater in the cytoplasm than in the nucleus. Nuclear expression of FILIP1L was markedly decreased in cancer specimens compared to benign specimens (p = 0.006). However, no change was observed in the cytoplasm (p = 0.847, fig. 2, A). Stromal cells also showed decreased nuclear expression in cancer specimens (p = 0.045, fig. 2, B). Protein expression of FILIP1L was not associated with Gleason score, stage, seminal vesicle involvement, extracapsular extension or the presence of metastasis.
Figure 2.
FILIP1L protein expression in human PCa compared to normal prostate tissue. Microarrays containing 95 PCa samples and 48 normal peripheral prostate tissues were stained for FILIP1L using immunofluorescence. Automated image acquisition and analysis was performed using Vectra system according to manufacturer protocols. A, quantitation of nuclear and cytosplasmic FILIP1L protein expression in epithelial cells. Decreased nuclear expression was observed in PCa specimens compared to benign prostate tissue (p = 0.006). No change in expression was observed in cytoplasmic compartment (p = 0.847). B, stromal cells showed decreased FILIP1L expression in nucleus in cores containing cancer (p = 0.045).
FILIP1L Hypermethylation
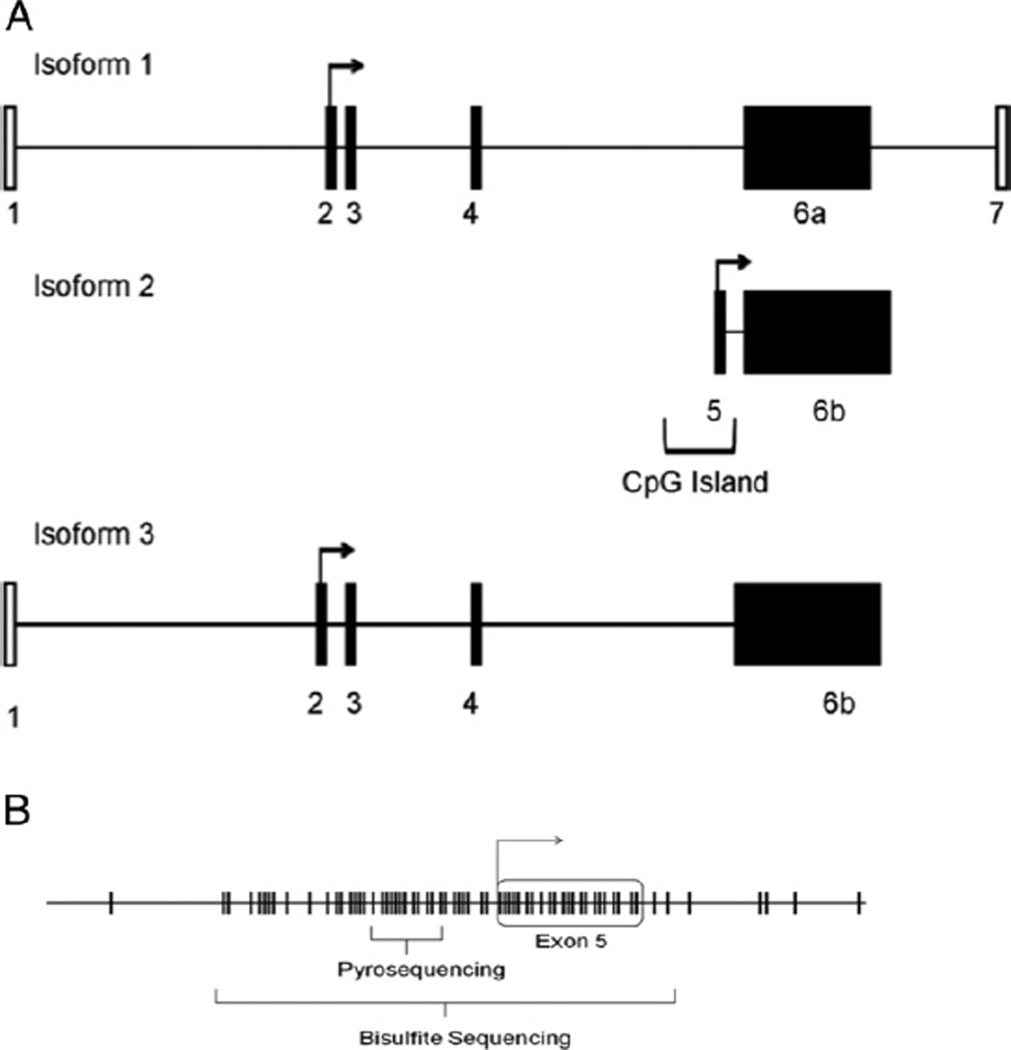
mrna expression regulation in pca cell lines
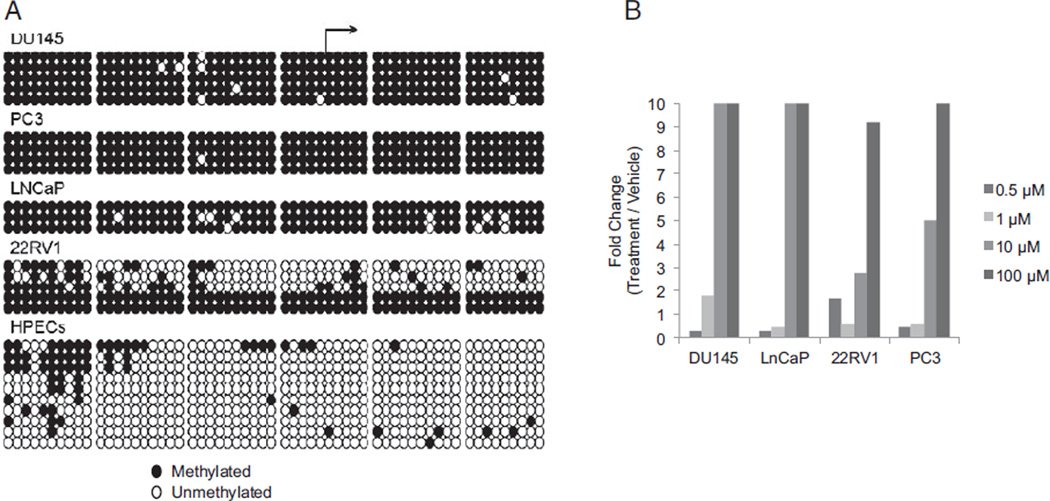
Using MethPrimer software,15 we identified a CGI in exon 5 that was immediately adjacent to the transcriptional start site of isoform 2 of FILIP1L (fig. 3, A). Previously described CGI criteria include a genomic region greater than 200 bp with an observed-to-expected CpG ratio of greater than 0.6 and a CG percent of greater than 50%.16 There are 59 individual CG sites in this region extending approximately 500 bp across the entire length of exon 5, which meets the criteria for a CGI. Methylation was assessed for isoform 2 using bisulfite sequencing and quantitative Pyrosequencing (fig. 2, B). Bisulfite sequencing revealed exon 5 hypermethylation of FILIP1L DU145, PC3, LNCaP and 22Rv1 in 85% of CG dinucleotides compared to HPECs (fig. 4, A). The p53 expressing cell line 22Rv117 was unique in demonstrating lower methylation in 3 of the 5 clones sequenced but overall hypermethylation occurred in 60% of the CG dinucleotides assessed. Bisulfite sequencing of proliferating and senescent cultured HPECs demonstrated significantly lower methylation compared with PCa cell lines (11% vs 85%, p <0.05), suggesting a role for hypermethylation in silencing FILIP1L.
Figure 3.
Hypermethylation silenced isoform 2 FILIP1L CGI in PCa cell lines. A, schematic map of FILIP1L gene shows exons that make up isoforms 1 to 3. CGI that was identified and investigated encompasses isoform 2 transcription start site. B, identification of individual CG dinucleotides (tick marks) in CGI encompassing exon 5 and transcriptional start site. Bisulfite sequencing was performed at 59 sites across CGI. Quantitative methylation Pyrosequencing was also done to validate findings in separate tissue set.
Figure 4.
Hypermethylation silenced isoform 2 FILIP1L CGI in PCa cell lines. A, methylation status of cancer cell lines DU145, PC3, LNCaP and 22RV1, and HPECs was determined using bisulfite treatment of DNA, PCR amplification, cloning and sequencing. HPECs represent 5 pooled samples. HPECs were hypomethylated vs PCa cell lines (11% vs 85%, p <0.05). Each row represents individual DNA strand. B, PCa cell lines were treated with increasing concentrations of 5-aza-2′-deoxycytidine or dimethyl sulfoxide only. Seven-day 5-aza-2′-deoxycytidine treatment resulted in dose dependent increase in FILIP1L mRNA expression. Experiment was done 3 independent times and results of 1 experiment are shown. Expression is shown as fold change vs vehicle only.
To determine whether CGI methylation was primarily responsible for FILIP1L silencing, we treated PCa cell lines with a single treatment of 5-aza-2-deoxycytidine. Dose ranging studies were initially performed and concentrations were chosen to avoid apoptosis and cell death. It was previously reported that 5-aza-2′-deoxycytidine is irreversibly incorporated into DNA and results in DNA methylation loss.18 Exposure to 10 to 100 µM restored FILIP1L isoform 2 expression in all PCa cell lines (fig. 4, B). Cells also developed senescent morphology at this dose and expressed SA-β-gal staining (data not shown). These data indicate that the expression of FILIP1L isoform 2 is regulated by hypermethylation of the exon 5 CGI.
PCa specimens
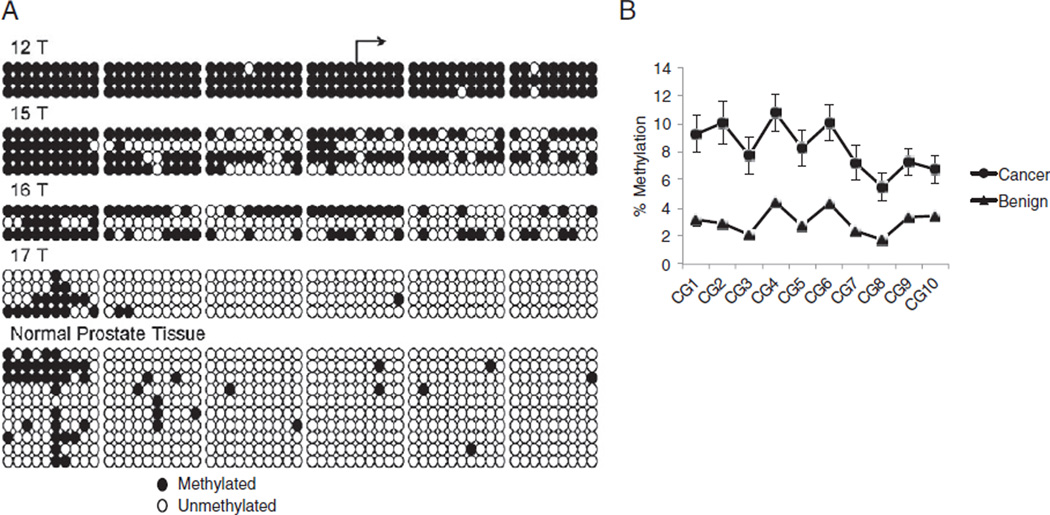
We examined exon 5 hypermethylation in PCa samples. Bisulfite sequencing of normal prostate tissues revealed minimal methylation of this region except the 5= edge of the CGI (fig. 5, A). Analysis of tumor samples revealed significantly greater methylation in 3 of 4 samples. To validate our findings, we used quantitative Pyrosequencing to assess methylation in normal and PCa cancer specimens. Based on the restraints of the Pyrosequencing reaction, the entire CGI was not evaluated. However, the region 150 bp upstream from the transcriptional start site was assessed covering CGs 17 to 27 in the CGI (fig. 3, B). Pyrosequencing revealed that tumor tissues were significantly more methylated than associated benign tissues across CpGs (2-way ANOVA p <0.0001, fig. 5, B). The Bonferroni posttest demonstrated significant hyper-methylation in tumor specimens at all 10 individual CpG sites (p <0.05). In all 14 paired specimens (100%) the tumor specimen showed higher methylation than the paired benign tissue.
Figure 5.
Hypermethylation of FILIP1L isoform 2 in PCa samples. A, bisulfite treatment of DNA, cloning and sequencing. Analysis of individual tumors revealed increased methylation in 3 of 4. Normal prostate tissue represents 6 samples. B, quantitative Pyrosequencing methylation changes across 10 CpGs were measured quantitatively in 14 paired normal and tumor samples run in triplicate. Tumor tissues were significantly more methylated than associated benign tissues across all CpGs tested (2-way ANOVA with Bonferroni post test p <0.0001).
DISCUSSION
Bypassing the senescence barrier has an important role in tumor development and progression.19–22 Several senescence pathways are inactivated during tumorigenesis, including p53 and pRb, but many underlying mechanisms remain unclear.
FILIP1L is a novel protein predicted to bind to actin that was originally identified as an area of common deletion in ovarian cancer.7 Examination of the Oncomine™ RNA array database (http://www.oncomine.com) reveals frequent loss of FILIP1L expression in multiple tumor types, including prostate, lung, bladder, breast and liver cancer, and melanoma. To date little data are available on FILIP1L regulation in cancer. We initially identified FILIP1L on an array analysis of genes up-regulated during senescence and down-regulated during immortalization.6,14 These data suggested that FILIP1L may have a role in senescence. Furthermore, it appeared commonly down-regulated during tumor formation. In the current study FILIP1L mRNA expression was commonly down-regulated in PCa. Using a tissue microarray and quantitative immunofluorescence, we characterized FILIP1L protein expression and found that FILIP1L silencing occurs primarily in the nuclear compartment of epithelial cancer cells.
Little is known regarding alterations in FILIP1L expression during biological processes. FILIP1L has 3 known isoforms generated from 7 exons. None of these isoforms uses all exons for transcription (fig. 1). We applied this unique arrangement to distinguish between isoforms 1 and 2 during senescence and tumor formation. Our first finding was the consistent induction of isoform 2 during senescence in HPECs. This increase in expression mimics the increase seen in total FILIP1L. No change in isoform 1 was noted. Senescence is associated with inhibition of cell proliferation as well as characteristic phenotypic changes. Recently, Kwon et al transiently over expressed isoform 2 in endothelial cells and found inhibited cell proliferation and migration.23 These data suggest that isoform 2 may have a role in inhibiting cancer cell progression. We also found FILIP1L down-regulation in immortalized PCa cell lines as well as decreased mRNA and protein expression in the majority of tumors. These findings are consistent with studies of other cancers.24 Future studies will evaluate the biological role of FILIP1L expression changes in PCa.
Hypermethylation of promoter CGIs frequently leads to silencing of the associated gene.2,9 In the current study we identified a region in exon 2 and the isoform 2 transcription start site that meets the criteria for a CGI.16 We then identified an association between hypermethylation of this FILIP1L CGI and decreased mRNA expression. Treatment with 5-aza-2′-deoxycytidine led to mRNA re-expression in PCa cell lines (fig. 4, A). Thus, isoform 2 silencing is likely mediated by CGI hypermethylation in PCa. Hypermethylation in the PCa cell line 22Rv1 revealed less methylation in approximately 50% of alleles, although it was responsive to demethylation. Involvement of complex regulatory mechanisms in drug induced DNA demethylation may also explain the observed low methylation efficiency at many hypermethylated loci during epigenetic cancer therapy.25,26
Evaluation of a series of primary cancers using Pyrosequencing revealed hypermethylation in all 14 tumors (100%) compared to benign adjacent tissue. A study was recently published that used genome-wide methylation profiling in tumor and benign adjacent tissue from patients with radical prostatectomy.27 We also examined this data set and found that 55 of 59 tumors (93%) showed FILIP1L hypermethylation. FILIP1L is a highly specific methylation marker for PCa. Given the remarkably high frequency of methylation, we could not correlate methylation with Gleason score and other pathological features. To date FILIP1L has been one of the most frequently hypermethylated genes in PCa. The majority of tumors show a lower percent of methylation (less than 30%) for other genes examined, including p16, pRb and MTP1.28 Most but not all tumors showed CGI hypermethylation. This suggests that alternate pathways, such as mutation or deletion, may contribute to the silencing of this gene.
CONCLUSIONS
Little is known about the intrinsic details of senescence pathways in cancer cells. The isoform specific silencing occurring with FILIP1L suggests that FILIP1L isoforms may have separate functional roles. The ultimate function of FILIP1L is currently unclear. Given its predicted relationship to filamin A/actin, it may signal through this pathway. Furthermore, FILIP1L hypermethylation appears to be a frequent alteration in PCa, occurring in approximately 90% of prostate tumors.
Acknowledgments
Supported by 5R01CA097131.
Abbreviations and Acronyms
- CGI
CpG Island
- FILIP1L
filamin A interacting protein 1-like
- HPEC
human prostate epithelial cell
- PCa
prostate cancer
- PCR
polymerase chain reaction
- SA-β-gal
senescence associated-β-galactosidase
Footnotes
Study received University of Wisconsin institutional review board approval.
Financial interest and/or other relationship with Johnson & Johnson.
REFERENCES
- 1.Ewald JA, Desotelle JA, Wilding G, et al. Therapy-induced senescence in cancer. J Natl Cancer Inst. 2010;102:1536. doi: 10.1093/jnci/djq364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baylin SB, Herman JG, Graff JR, et al. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141. [PubMed] [Google Scholar]
- 3.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herman JG, Baylin SB. Mechanisms of disease: Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 5.Lee WH, Morton RA, Epstein JI, et al. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc Natl Acad Sci U S A. 1994;91:11733. doi: 10.1073/pnas.91.24.11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwarze SR, Fu VX, Desotelle JA, et al. The identification of senescence-specific genes during the induction of senescence in prostate cancer cells. Neoplasia. 2005;7:816. doi: 10.1593/neo.05250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mok SC, Wong KK, Chan RK, et al. Molecular cloning of differentially expressed genes in human epithelial ovarian cancer. Gynecol Oncol. 1994;52:247. doi: 10.1006/gyno.1994.1040. [DOI] [PubMed] [Google Scholar]
- 8.Kinoshita H, Shi Y, Sandefur C, et al. Methylation of the androgen receptor minimal promoter silences transcription in human prostate cancer. Cancer Res. 2000;60:3623. [PubMed] [Google Scholar]
- 9.Jarrard DF, Kinoshita H, Shi Y, et al. Methylation of the androgen receptor promoter CpG island is associated with loss of androgen receptor expression in prostate cancer cells. Cancer Res. 1998;58:5310. [PubMed] [Google Scholar]
- 10.Jarrard DF, Sarkar S, Shi Y, et al. p16/pRb pathway alterations are required for bypassing senescence in human prostate epithelial cells. Cancer Res. 1999;59:2957. [PubMed] [Google Scholar]
- 11.Dimri GP, Lee X, Basile G, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhusari S, Yang B, Kueck J, et al. Insulin-like growth factor-2 (IGF2) loss of imprinting marks a field defect within human prostates containing cancer. Prostate. 2011;71:1621. doi: 10.1002/pros.21379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bourque DK, Avila L, Peñaherrera M, et al. Decreased placental methylation at the H19/IGF2 imprinting control region is associated with normotensive intrauterine growth restriction but not preeclampsia. Placenta. 2012;31:197. doi: 10.1016/j.placenta.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Schwarze SR, DePrimo SE, Grabert LM, et al. Novel pathways associated with bypassing cellular senescence in human prostate epithelial cells. J Biol Chem. 2002;277:14877. doi: 10.1074/jbc.M200373200. [DOI] [PubMed] [Google Scholar]
- 15.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 16.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 17.Sramkoski RM, Pretlow TG, 2nd, Giaconia JM, et al. A new human prostate carcinoma cell line, 22Rv1. In Vitro Cell Dev Biol Anim. 1999;35:403. doi: 10.1007/s11626-999-0115-4. [DOI] [PubMed] [Google Scholar]
- 18.Christman JK. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 19.Schmitt CA, Fridman JS, Yang M, et al. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell. 2002;109:335. doi: 10.1016/s0092-8674(02)00734-1. [DOI] [PubMed] [Google Scholar]
- 20.Petti C, Molla A, Vegetti C, et al. Coexpression of NRASQ61R and BRAFV600E in human melanoma cells activates senescence and increases susceptibility to cell-mediated cytotoxicity. Cancer Res. 2006;66:6503. doi: 10.1158/0008-5472.CAN-05-4671. [DOI] [PubMed] [Google Scholar]
- 21.Xue W, Zender L, Miething C, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roninson IB. Tumor cell senescence in cancer treatment. Cancer Res. 2003;63:2705. [PubMed] [Google Scholar]
- 23.Kwon M, Hanna E, Lorang D, et al. Functional characterization of filamin a interacting protein 1-like, a novel candidate for antivascular cancer therapy. Cancer Res. 2008;68:7332. doi: 10.1158/0008-5472.CAN-08-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burton ER, Gaffar A, Lee SJ, et al. Downregulation of Filamin A interacting protein 1-like is associated with promoter methylation and induces an invasive phenotype in ovarian cancer. Mol Cancer Res. 2011;9:1126. doi: 10.1158/1541-7786.MCR-11-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagemann S, Heil O, Lyko F, et al. Azacytidine and decitabine induce gene-specific and non-random DNA demethylation in human cancer cell lines. PLoS One. 6:e17388. doi: 10.1371/journal.pone.0017388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stresemann C, Bokelmann I, Mahlknecht U, et al. Azacytidine causes complex DNA methylation responses in myeloid leukemia. Mol Cancer Ther. 2008;7:2998. doi: 10.1158/1535-7163.MCT-08-0411. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi Y, Absher DM, Gulzar ZG, et al. DNA methylation profiling reveals novel biomarkers and important roles for DNA methyltransferases in prostate cancer. Genome Res. 2011;21:1017. doi: 10.1101/gr.119487.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baylin SB. The cancer epigenome as a prevention/therapy target. Medicina (Buenos Aires) 2007;67:11. [Google Scholar]