Abstract
We aimed to investigate whether the character of the immunodominant HIV-Gag peptide (variable or conserved) targeted by CD8+ T cells in early HIV infection would influence the quality and quantity of T cell responses, and whether this would affect the rate of disease progression. Treatment-naive HIV-infected study subjects within the OPTIONS cohort at the University of California, San Francisco, were monitored from an estimated 44 days postinfection for up to 6 years. CD8+ T cells responses targeting HLA-matched HIV-Gag-epitopes were identified and characterized by multicolor flow cytometry. The autologous HIV gag sequences were obtained. We demonstrate that patients targeting a conserved HIV-Gag-epitope in early infection maintained their epitope-specific CD8+ T cell response throughout the study period. Patients targeting a variable epitope showed decreased immune responses over time, although there was no limitation of the functional profile, and they were likely to target additional variable epitopes. Maintained immune responses to conserved epitopes were associated with no or limited sequence evolution within the targeted epitope. Patients with immune responses targeting conserved epitopes had a significantly lower median viral load over time compared to patients with responses targeting a variable epitope (0.63 log10 difference). Furthermore, the rate of CD4+ T cell decline was slower for subjects targeting a conserved epitope (0.85% per month) compared to subjects targeting a variable epitope (1.85% per month). Previous studies have shown that targeting of antigens based on specific HLA types is associated with a better disease course. In this study we show that categorizing epitopes based on their variability is associated with clinical outcome.
Introduction
CD8+ T cells play an important role in the control of human immunodeficiency virus type 1 (HIV) viremia1–4 and the immunological pressure from these cells is a major driving force of viral evolution.5–7 Genetic variability is a hallmark of HIV, giving the virus the capacity to rapidly escape the selective pressures of the immune system.8,9 Changes in the peptide sequence can abrogate binding to the HLA-molecule and/or inhibit recognition by the T cell receptor (TCR). It is well known that recognition of antigen by the TCR is extremely sensitive.10,11 However, there are regions in the HIV genome that are more conserved between the different strains and subtypes.12 Mutations in more conserved regions tend to have a higher impact on fitness and reduce the viral replication.13 Mutations that affect viral fitness may in turn influence disease progression.14,15
Several factors, both virological and immunological, are known to influence disease progression. The most prominent host factor associated with disease progression is the expression of certain HLA alleles.16–19 Recently, several studies revealed that one of the most important mechanisms behind the association between HLA alleles and disease outcome is the character of the peptide presented by these alleles. Alleles associated with a slower disease progression are more prone to bind conserved epitopes.15,20–22 Studies show that Env-specific T cell responses are more frequently observed in patients with faster disease progression, while patients with slower progression preferentially target epitopes in the more conserved Gag region.23–28 However, there are still gaps in our knowledge of how the level of conservation within targeted HLA class I-restricted epitopes influences clinical outcome over time. Most observations have been reported on cross-sectional studies, focusing on immune responses restricted by a single HLA allele or directed against whole regions using overlapping peptide sets. Usually these studies have not revealed when an epitope-specific response is initiated, or how the quality and quantity of responses over time are associated with the character of the targeted peptide. This is supported by a study indicating that measures of the breadth and magnitude of CD8+ T cell responses at 3 months postinfection cannot predict viral load and disease progression at 12 months postinfection.29 Answering these questions would be valuable for the characterization of effective CD8+ T cell responses and design of vaccine antigens.
We hypothesized that the character of the peptides targeted early in HIV infection influences the efficacy of T cell responses over time, where targeting of conserved epitopes would be associated with beneficial disease outcome. To test this hypothesis we conducted a longitudinal study of Gag-specific CD8+ T cell responses in HIV-infected study subjects monitored from primary infection.30 We found that the character of the HIV-Gag-peptide targeted in early infection was associated with viral load and the CD4+ T cell count over time. This study shows that the character of targeted antigens in early HIV infection is an important determinant for the efficacy of the immune responses that may influence disease outcome.
Materials and Methods
Study cohort
Thirteen study subjects were selected from HIV subtype B-infected patients enrolled and followed longitudinally from early infection within the OPTIONS cohort at the University of California, San Francisco30 based on identified HIV-Gag-p17 and/or Gag-p24 antigen-specific T cell responses in early infection.31 Each study subject was followed longitudinally for 3 to 6 years while remaining treatment naive. In addition, HLA-typing results were available for each subject. Thirteen patients fulfilled these criteria and were included in the study. The University of California, San Francisco (UCSF) Committee on Human Research approved this study and all patients provided written informed consent.
Peptides and level of variability
A total of 35 HLA class I-restricted peptides located within the HIV-Gag-p17 and HIV-Gag-p24 regions and matched to the HLA type of each subject were used to identify HIV-specific immune responses over time. Twenty of the HLA class I-restricted peptides had previously been verified to be targeted by the study subjects during early infection31 and 15 peptides were added from previously identified broadly immunogenic epitopes.32 All peptides consisted of eight to nine amino acid residues and were categorized as either conserved or variable. The level of variability within each epitope was based on the frequency of a particular amino acid among the HIV subtype B strains available at the HIV Molecular Immunology database (www.hiv.lanl.gov). The dataset contained 3,869 and 3,664 sequences for the Gag p17 and Gag p24 regions, respectively. After obtaining the genetic variability as a percentage of amino acid conservation of each epitope, the median and interquartile range were calculated for the set. An epitope was considered conserved if it belonged to the top 25 percentile (≥79.7% conservation), while the rest of the epitopes were defined as variable. Peptides synthesized by GeneScript (Piscataway, NJ) had a purity of at least 70%.
Characterization of epitope-specific CD8+ T cell responses
Identification of HIV-specific cytokine-producing cells was made using an intracellular cytokine staining (ICS) assay.33 In brief, 5×105 peripheral blood mononuclear cells (PBMCs)/well were plated with HLA-matched HIV-Gag-peptides, positive control (staphylococcal enterotoxin B plus lipopolysaccharide), or medium alone (negative control). Brefeldin A (Sigma Aldrich) was added (5 μg/ml) after 1 h to each well and incubated at 37°C for an additional 5 h. Stimulated PBMCs were washed with FACS buffer [phosphate-buffered saline (PBS) with 2 mM ethylenediaminetetraacetic acid and 0.5% bovine serum albumin] and stained for surface markers with Pacific Blue-conjugated anti-CD4 antibody (clone RPA-T4), allophycocyanin (APC)-Cy7-conjugated CD8-APC-Cy7 (clone SK1), and aqua amine reactive dye to identify dead cells. The cells were permeabilized with FACS Permeabilizing Solution 2 (Becton Dickinson) followed by intracellular staining with phycoerythrin-Texas Red (ECD)-conjugated anti-CD3 (clone UCHT1 from Beckman Coulter), APC-conjugated anti-interferon (IFN)-γ (clone B27), phycoerythrin (PE)-conjugated anti-MIP-1β (clone D21-1351), Alexa700-conjugated anti-tumor necrosis factor (TNF)-α (clone MAb11), and fluorescein isothiocyanate (FITC)-conjugated anti-interleukin (IL)-2 (rat anti-human, clone MQ1-17H12). All antibodies were mouse anti-human from BD Biosciences unless otherwise stated. The sample analysis was performed within 12 h on a 4-laser LSR-II flow cytometer (Becton Dickinson Biosciences). Antimouse or antirat immunoglobulin G-coated beads were stained separately with each fluorochrome-conjugated mouse or rat antibody and used for software-based compensation. Final flow cytometric analysis was carried out using FlowJo (TreeStar). A positive response was defined as twice the negative background and at least 0.05% IFN-γ-producing CD8+ T cells. All data are presented after subtraction of the background.
HIV gag amplification and sequencing from viral RNA
Viral RNA from 1.0 ml of ACD or EDTA blood plasma was extracted using QIAamp Viral RNA kits (Qiagen). The HIV gag regions (HXB2 nucleotides 737 through 2096) were sequenced using a nested, two-step RT-PCR procedure. In brief, HIV cDNA was generated from 8 μl viral RNA using Thermoscript RT combined with Superscript II (Invitrogen Life Technologies, San Diego, CA) and primer RA01 (5′-CTGCTCCTGTATCTAATAGAGCTTC-3′; positions 2313–2337). A 1.6-kb amplicon was generated from 5.0 μl cDNA using the FastStart PCR kit (Applied Biosystems, Foster City CA), primers 737F (5′-GCGACTGGTGAGTACGCC-3′; positions 737–754) and RA01 under the following conditions: 94°C×2′, 35 (94°C×30”, 56°C×30′′, 68°C×2′), 68°C×4′, 10°C hold. Of the PCR product 1 μl was used for each of the three nested PCR using primer pairs 737F and JA155mod (5′-CTGATAATGCTGAAAACATGGGTA-3′; positions 1295–1318); 1232F (5′- ACCTAGAACTTTAAATGCATGGG-3′; positions 1233-1255) and 1754R (5′- CAACAAGGTTTCTGTCATCC-3′; positions 1736–1755); and 1503F (5′-GGAAGTGACATAGCAGGAA-3′; positions 1486–1504) and 2095R (5′-TTCCCTAAAAAATTAGCCTG-3′; positions 2077–2096) using the following conditions: 94°C×2′, 30(94°C×30”, 56°C×30”, 68°C×30′), 68°C×4′, 10°C hold. PCR products were treated with ExoSAP-IT (GE Healthcare Life Sciences) and quantified by PicoGreen (Invitrogen/Life Technologies). The nested PCR primers also served as the sequencing primers using BigDyev3 cycle sequencing chemistry (Applied Biosystems/Life Technologies). Amplicon sequences were determined using a 3730xl capillary array sequencer (Applied Biosystems/Life Technologies). Forward and reverse sequences were analyzed and edited using Sequencher (v. 4.6, Genecodes, Ann Arbor, MI). The sequences have been submitted to GenBank and received the following accession numbers: JF905572–JF905594.
Statistics
Statistical calculations and graphic presentations were performed using GraphPad Prism 5.0 statistical software and SAS version 9.2 (SAS Institute, Cary, NC). The cytokine production by CD8+ T cells was illustrated using SPICE 5.1 software.34 Differences in CD4+ T cells over time between groups were assessed using linear mixed models. CD4+ T cell count was log transformed in all models. The type of epitope targeted (variable or conserved) and follow-up time were considered as covariates. Random effects for time and intercept allowed for individual differences in CD4+ T cell slope and initial level. A term for covariance between the two random effects did not substantially improve fit to the data and so was omitted from the primary model. Comparing intercepts and slopes fitted by simple linear regression separately on each individual's data produced qualitatively similar results.
Results
The aim of this study was to investigate the association between genetic variability within targeted HLA class I-restricted HIV Gag epitopes, the quantity and quality of the specific CD8+ T cell response, and clinical predictors of disease outcome following early infection. The selection of patients, and their responding epitopes, was based on results from a previous study conducted by Streeck et al.,31 where the immunodominance pattern of the first virus-specific CD8+ T cell responses developed during primary HIV infection were obtained from 83 study subjects within the OPTIONS cohort at the University of California, San Francisco.30 From these 83 study subjects, only 13 fulfilled our inclusion criteria that consisted of the following: (1) identified HIV-Gag-p17 and/or Gag-p24 antigen-specific T cell responses in early infection, (2) longitudinal follow-up for at least 3 years, (3) remaining treatment naive during the study period, and (4) access to available plasma and PBMC samples from three to four time points collected approximately 1 year apart during the study period. However, the results from one subject were excluded due to low cell viability at the first time point (<60% live cells) and two subjects did not respond to any of the tested peptides and were thus excluded. Therefore, complete data sets from 10 subjects were used in this study (Table 1). The first blood sample was drawn during early infection (median: 101 days from infection; IQR: 76–120 days) with the following three to four PBMC samples collected approximately 1 year apart during the study period (median: 3.3; IQR: 2.8–5.2 years). The data for both CD4+ T cell count (median 20 times per patient) and plasma viral load (median 18 times per patient) were documented during the entire study period.
Table 1.
Clinical, Virological, and Immunological Characteristics
| Patient ID | HLA class I A | HLA class I B | CD4+T cell range (cells/mm3)a | CD4+ T cell slopeb | Viral load rangea(log10) | Viral load median (log10) | Estimated time since infection (month) | Study period (month) | Peptide characterc | Protein regiond | Peptide sequencee |
|---|---|---|---|---|---|---|---|---|---|---|---|
| OP562 | A1, A3 | B35, B51 | 378–1152 | −7,93 | 3.60–5.58 | 4.45 | 3 | 65 | Variable | Gag p17 | RLRPGGKKK |
| OP565 | A1, A24 | B8, B50 | 288–650 | −3,79 | 3.75–5.20 | 4.43 | 7 | 34 | Conserved | Gag p24 | EIYKRWII |
| OP584 | A3, A30 | B44, B51 | 328–760 | −5,80 | 2.74–4.97 | 4.25 | 4 | 56 | Conserved | Gag p17 | EEKAFSPEV |
| OP639 | A2, A3 | B7, B27 | 480–1155 | −4,66 | 3.39–5.30 | 4.39 | 5 | 73 | Variable | Gag p24 | GPGHKARVL |
| OP653 | A25, A68 | B7, B35 | 506–1404 | −20,91 | 3.96–4.88 | 4.65 | 15 | 34 | Variable | Gag p24 | GPGHKARVL |
| OP722 | A3, A11 | B7, B40/60 | 132–616 | −11,76 | 4.71–5.23 | 5.16 | 3 | 30 | Variable | Gag p17 | RLRPGGKKK |
| OP781 | A2, A25 | B44, B44 | 267–759 | −6,99 | 4.01–4.91 | 4.44 | 3 | 34 | Conserved | Gag p24 | ETINEEAAEW |
| OP835 | A1, A30 | B8, B39 | 420–1060 | 1,42 | 2.08–3.92 | 2.74 | 4 | 62 | Conserved | Gag p24 | TPQDLNTML |
| OP842 | A3, A25 | B40/60, 55 | 319–633 | −8,56 | 3.77–5.07 | 4.57 | 2 | 29 | Variable | Gag p17 | RLRPGGKKK |
| OP849 | A1, A2 | B7, B8 | 320–555 | −3,88 | 2.30–4.06 | 3.63 | 5 | 39 | Conserved | Gag p24 | EIYKRWII |
The highest and lowest CD4 or viral load measurements detected in the samples obtained during the study period, before initiation of treatment.
CD4+ T cell slope (cells/mm3) calculated from all available measurements during the study period, before initiation of treatment.
Peptide characteristics of immunodominant peptide identified in early infection.
Location of the peptide targeted by the initial immunodominant CD8+ T cell response in early HIV-1 infection.
Amino acid sequence of the peptide targeted by the initial immunodominant CD8+ T cell response in early HIV-1 infection.
HIV-specific T cell responses against the panel of 35 HLA-matched optimal HIV-Gag-p17 and HIV-Gag-p17-p24 peptides (mean eight peptides/patient; range 5–13), including the 20 previously verified Gag peptides targeted by the study subjects during early infection,31 were tested using the ICS assay. All time points of PBMC samples from a subject were analyzed simultaneously. The HIV-Gag-epitope inducing the strongest, immunodominant, CD8+ T cell response was confirmed at the first time point during early infection in each subject and longitudinally monitored. Patients were divided into two groups based on whether the targeted Gag peptide was considered conserved or variable (see Materials and Methods for details). The first identified HIV-Gag response in five study subjects was directed against a conserved peptide and the remaining five patients targeted a variable peptide. Samples were available and analyzed from at least three time points for all study subjects. At the fourth time point, samples were available from three patients targeting a variable epitope, and four of the patients targeting a conserved epitope (Table 1).
Quantitative but not qualitative differences of responses targeting conserved versus variable peptides over time
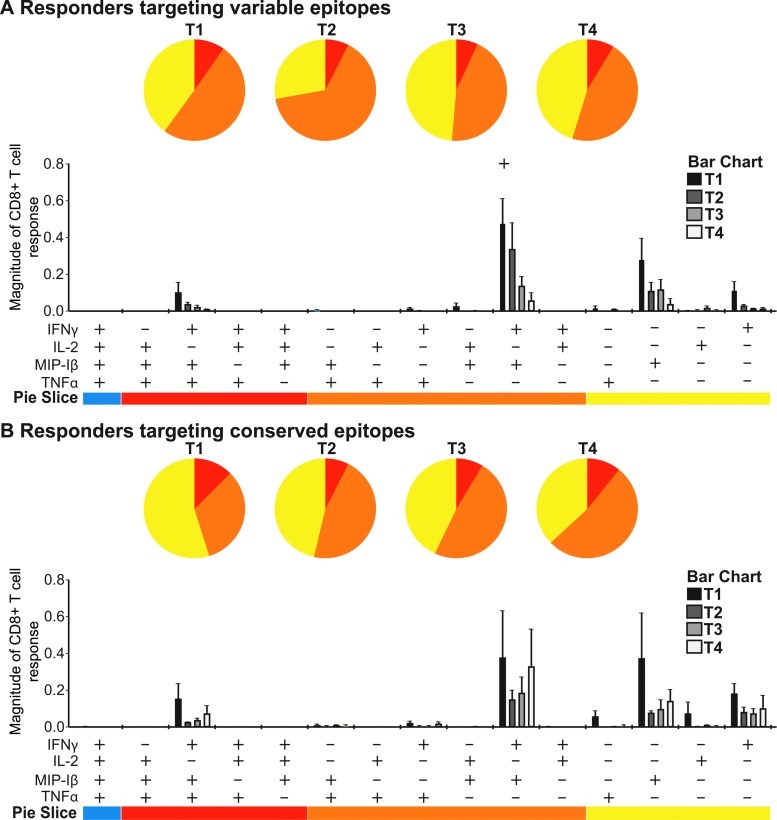
The quality of the responses was monitored over time as measured by the ability of the antigen-specific CD8+ T cells to produce IFN-γ, MIP-1β, TNF-α, and IL-2, i.e., if they were polyfunctional. Since the tested peptides were optimal for binding to HLA class I recognized by CD8+ T cells, we did not consider any CD4+ T cell responses. There was no clear difference in the percentage of polyfunctionality between the patients targeting a variable (Fig. 1A) versus conserved (Fig. 1B) epitope. Instead, the percentages of polyfunctional and monofunctional cells remained largely unchanged over time in both groups except for a significant decline in the magnitude of dually IFN-γ and MIP-1β-producing CD8+ T cells targeting a variable epitope between the first and fourth time point (p=0.046). Also, the magnitude of antigen-specific responses in patients targeting conserved peptides shows persistently higher values for all functional combinations at the third and fourth time point compared to patients targeting variable epitopes.
FIG. 1.
Sustained magnitude of CD8+ T cell responses against conserved epitopes. Polyfunctional responses were measured by detection of IFN-γ, IL-2, MIP-1β, and TNF-α production at T1 to T4. The distribution of polyfunctional CD8+ T cells is illustrated in the pie charts for variable (A) and conserved (B) responders, showing the ratio of T cells producing one, two, or three cytokines upon antigen stimulation in different colors. In the column-chart the magnitude of antigen-specific CD8+ T cells with a particular functionality is visualized at the four time points in different shades of gray. p-values were calculated in SPICE using the Student's t test and permutation tests. Samples were available and analyzed from three time points for all study subjects. At the fourth time point, samples were available from three patients targeting a variable epitope and four of the patients targeting a conserved epitope. Color images available online at www.liebertpub.com/aid
When investigating each cytokine separately, a decline in the epitope-specific IFN-γ production in subjects targeting a variable peptide in early infection was evident between the first and third time points (mean drop 0.53, 95% CI −0.033 to 1.095, p=0.059) and the first and fourth time points (mean drop 0.74, 95% CI −0.66 to 2.13, p=0.076), although the decrease was not statistically significant (data not shown). In contrast, patients targeting a conserved peptide maintained their responses throughout the study period after a nonsignificant initial drop in the magnitude of IFN-γ between the first and second time point (mean drop 0.48, 95% CI −0.65 to 1.61, p=0.30, data not shown).
Early variable epitope responders targeted a larger total number of variable compared to conserved Gag epitopes over time, and maintained the functional profile
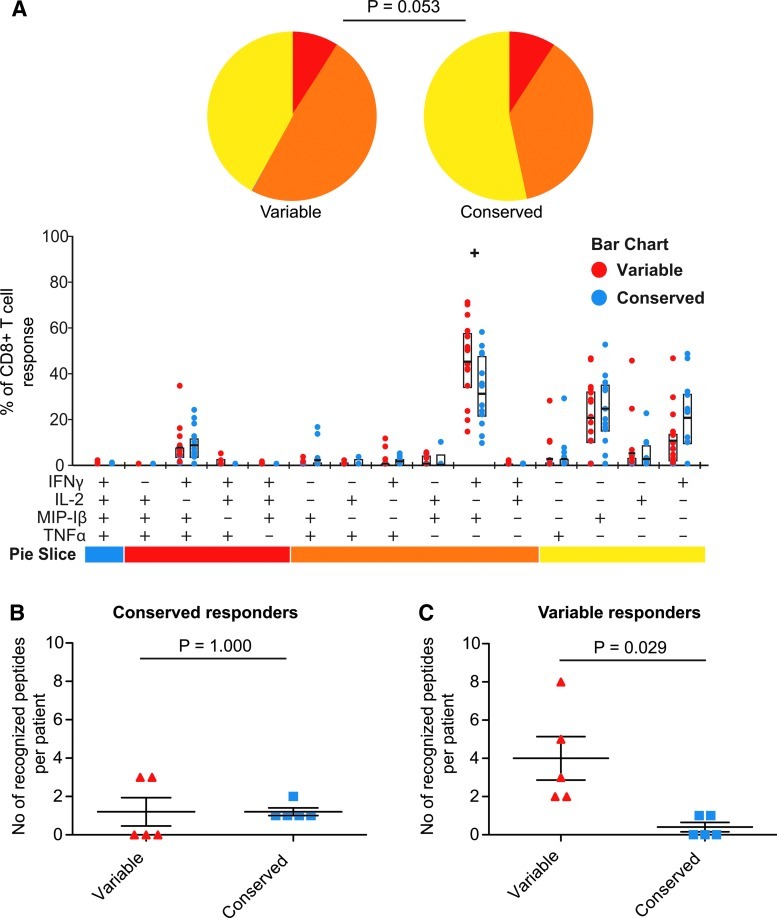
To further test whether the functional profiles differed between CD8+ T cell responses against variable and conserved epitopes we compiled the results from time point one to three to distinguish the overall polyfunctionality. We excluded time point four as immunological data were lacking from three individuals at this time point. We found that the individuals targeting variable epitopes generated a higher fraction of polyfunctional responses compared to the conserved epitopes (Fig. 2A; p=0.053), although the difference was not statistically significant. The detected difference was driven by a higher fraction of the variable-specific CD8+ T cells dually producing IFN-γ and MIP-1β (Fig. 2A; p=0.016). When we extended the analysis to include responses against all tested variable and conserved peptides, including subdominant epitope responses, we distinguished the same tendencies where CD8+ T cell responses in the variable group showed a higher degree of polyfunctionality (p=0.085, data not shown).
FIG. 2.
Early targeting of a variable Gag-epitope associated with IFN-γ and MIP-1β-producing CD8+ T cells. The functional distribution of all detected Gag-specific CD8+ T cell responses at T1 to T3 is illustrated in the pie chart and column chart in subjects with early targeting of a variable or conserved epitope (A). p-values were calculated in SPICE using the Student's t test and permutation tests. The total number of targeted epitopes characterized as variable and conserved at time point three in the subjects grouped as conserved (B) and variable (C) responders in early infection (mean and SEM indicated). p-values from pair t-test (two-tailed). Color images available online at www.liebertpub.com/aid
Next we investigated the character of all targeted epitopes in each subject at time point three. In the conserved group, a similar and low number of conserved and variable epitopes was targeted (Fig. 2B, p=1.000). Interestingly, subjects who had an early response against a variable epitope also targeted a larger total number of variable compared to conserved Gag epitopes at time point three (Fig. 2C, p=0.029). Taken together, our results show that although the magnitude of the responses decreases over time in patients targeting a variable epitope, there was no limitation of the functional profile and they were likely to target additional variable epitopes.
Early targeting of conserved peptides was associated with lower plasma viral load and slower CD4+ T cell depletion
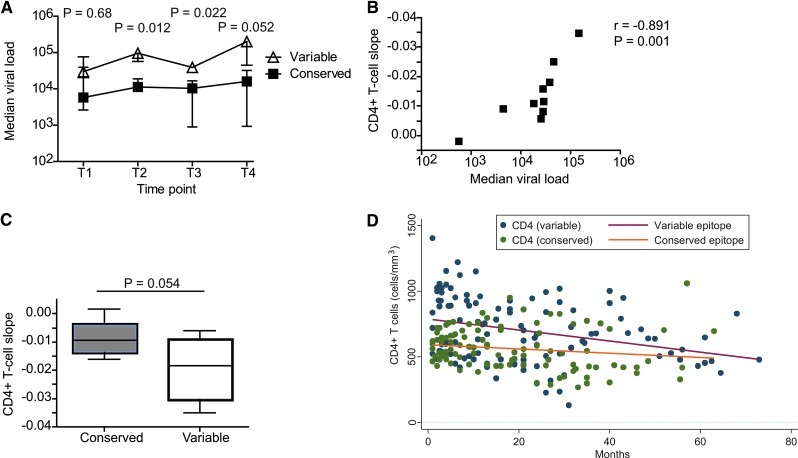
We next investigated whether the character of the peptide inducing the immunodominant Gag response during early infection had an effect on the clinical outcome. Table 2 summarizes the CD4+ T cell count and plasma viral loads at each selected time point for both the conserved and variable epitope-targeting groups. At the first sampling time point, which occurred during early infection, there was a nonsignificant viral load difference (0.76 log10 difference in medians, p=0.68) between the two groups (Fig. 3A). At the second time point the median viral load in patients targeting conserved epitopes was significantly lower (0.93 log10 difference, p=0.012), suggesting a better control of the viral replication in these individuals. The enhanced viral control in the group of patients targeting a conserved epitope was maintained at the third time point (0.58 log10 difference, p=0.022). At the fourth time point the viral load difference between the two groups was larger, but with greater uncertainty (1.55 log10 difference, p=0.052), which may relate to the smaller number of patients in the groups at this time point (conserved: n=3, and variable: n=4).
Table 2.
Summary Measures of CD4+ T Cell Counts and Plasma Viral Loads at Selected Time Points
| |
|
Median (IQR) |
Mean (SD) |
||
|---|---|---|---|---|---|
| Laboratory test | Time point | Variable epitope | Conserved epitope | Variable epitope | Conserved epitope |
| CD4 | Time 1 | 930 (546–990) | 532 (504–684) | 795 (258) | 574 (123) |
| Time 2 | 621 (445–756) | 486 (476–578) | 636 (205) | 586 (210) | |
| Time 3 | 637 (359–644) | 428 (420–546) | 491 (238) | 463 (101) | |
| Time 4 | 480 (378–607) | 362 (302–557) | 488 (115) | 429 (186) | |
| Viral load (log10) | Time 1 | 4.48 (4.31–4.88) | 3.76 (3.42–4.60) | 4.41 (0.63) | 3.94 (0.91) |
| Time 2 | 4.98 (4.76–5.07) | 4.05 (3.95–4.28) | 4.89 (0.29) | 3.89 (0.61) | |
| Time 3 | 4.59 (4.46–4.71) | 4.01 (2.95–4.22) | 4.55 (0.20) | 3.57 (0.91) | |
| Time 4 | 5.30 (4.65–5.31) | 3.75 (2.97–4.51) | 5.09 (0.38) | 3.74 (0.89) | |
FIG. 3.
Targeting of a conserved epitopes associated with lower viremia and slower CD4+ T cell depletion. The median and interquartile ranges of viral load (copies/ml) in patients with early targeting of conserved and variable epitopes are illustrated at time point one to four (A). p-values from a Mann–Whitney test of viral loads are given. From the last time point (T4) fewer samples were available, three from subjects targeting variable epitope and four from subjects in the conserved group. A clear correlation is seen between the median viral load and the CD4+ T cell slope over time, calculated using Spearman's rank test (two-tailed) (B). The median CD4+ T cell slope in patients targeting conserved epitopes compared to study subjects targeting a variable epitope (C). p-value from Mann–Whitney test is given. Scatter plot showing absolute CD4+ T cell numbers over time (months since first OPTIONS visit), with least squares fitted lines. Blue dots and red line indicate patients targeting variable epitopes, and the green dots and orange line indicate patients targeting conserved epitopes (D). Time point one to four: T1 to T4. Color images available online at www.liebertpub.com/aid
The strongest determinant of disease outcome is the rate of CD4+ T cell decline, i.e., a negative CD4+ T cell slope. We found a significant correlation (r=−0.89, p=0.001) between the CD4+ T cell slope and median viral load over the study period (Fig. 3B). Next we tested whether the character of the Gag peptide targeted in early infection was associated with the rate of CD4+ T cell decline. We found that patients targeting variable epitopes had an average CD4+ T cell decline of 1.85% per month (95% CI: 0.06% to 3.61% decline) while patients targeting conserved epitopes had a much slower average CD4+ T cell decline of 0.85% per month (95% CI: 0.11% to 1.59% decline). The difference in CD4+ T cell decline between the groups was substantial, though it was at the border of statistical significance (p=0.054) in this relatively small cohort of subjects (Fig. 3C). To visualize the change in CD4+ T cell counts in the two groups another way, all available CD4+ T cell counts for each subject in this study were graphed against time enrolled in the OPTIONS cohort (Fig. 3D). By fitting a least squares regression line, the rate of CD4+ T cell decline appears greater in subjects targeting variable epitopes. Although subjects targeting a variable epitope had an estimated 37.5% higher CD4+ T cell count (CI 5.2% to 79.9%, p=0.021) at the first time point, differences in fitted CD4+ T cell count were not statistically significant at any other time point. The more rapid loss of CD4+ T cells in the variable group appears to erode the initial advantage.
Comparison of the autologous viral sequences with the sequence of the tested peptides
To characterize how dependent the presence and preservation of the immune responses against conserved epitopes were to sequence identity with the tested epitope, we sequenced the autologous virus at time point one and three for all patients (Table 3). At time point one, all but one of the autologous conserved epitope sequences was identical to the tested peptide. A substantially larger number of amino acid substitutions was detected within the variable epitopes. Already at time point one, three out of five sequenced epitopes contained amino acid differences compared to the tested epitopes. Over time, changes were detected in four out of the five epitopes. Overall, these results are in agreement with the idea that the maintained HIV-specific CD8+ T cell responses against the conserved epitopes are due to the absence of, or limited and conserved, sequence changes within these epitopes.
Table 3.
The Autologous Sequences of the Identified Immunodominant Peptide in Early Infection at Time Point One and Three
| Patient ID | Tested peptide sequencea | Level of variability | T1 date | T1 sequenceb | T3 date | T3 sequencec | Peptide characterd |
|---|---|---|---|---|---|---|---|
| OP565 | EIYKRWII | 82.8 | Dec 2000 | --------- | Oct 2003 | --------- | Conserved |
| OP584 | EEKAFSPEV | 89.5 | Apr 2001 | --------- | Sept 2005 | --------- | Conserved |
| OP849 | EIYKRWII | 82.8 | Apr 2004 | ----K---- | Jul 2006 | -V--K---- | Conserved |
| OP781 | ETINEEAAEW | 87.7 | Aug 2002 | --------- | Aug 2005 | --------- | Conserved |
| OP835 | TPQDLNTML | 94.4 | Apr 2003 | --------- | Nov 2007 | NA | Conserved |
| OP562 | RLRPGGKKK | 57.8 | Feb 2002 | --------- | May 2006 | ------X-X | Variable |
| - - | |||||||
| N R | |||||||
| R | |||||||
| S | |||||||
| OP639 | GPGHKARVL | 68.0 | Jul 2001 | --S------ | Aug 2007 | --------- | Variable |
| OP653 | GPGHKARVL | 68.0 | Oct 2001 | --X---K-- | Jun 2004 | --------- | Variable |
| - | |||||||
| S | |||||||
| OP722 | RLRPGGKKK | 57.8 | Mar 2003 | --------X | Aug 2004 | ------X-R | Variable |
| - | - | ||||||
| R | N | ||||||
| R | |||||||
| S | |||||||
| OP842 | RLRPGGKKK | 57.8 | Apr 2003 | --------- | Aug 2005 | --------- | Variable |
Sequence of the tested peptide that induced the immunodominant response identified in early infection by population-based sequencing.
Sequence of the autologous epitope at time point one. A dash indicates identity with the tested peptide and an X indicates a polymorphic position that could encode more than one amino acid (identical with the tested peptide or any of the other amino acids given for the autologous sequence).
Sequence of the autologous epitope at time point three. A dash indicates identity with the tested peptide and an X indicates a polymorphic position that could encode more than one amino acid (identical with the tested peptide or any of the other amino acids given for the autologous sequence at time point three).
Peptide characteristics of immunodominant peptide identified in early infection.
T1, time point one; T3, time point three.
Discussion
We investigated how the genetic variability within the targeted Gag peptides in early HIV infection influenced the quality and quantity of the antigen-specific CD8+ T cell responses over time. We hypothesized that early targeting of conserved epitopes would be beneficial, as responses against such epitopes would maintain their antiviral effect for a longer time period. The study subjects were untreated and followed longitudinally for at least 3 years from early HIV infection. The analyses were restricted to responses against Gag-peptides as we hypothesized that the putative beneficial role of immune responses against this region31 is dependent on the level of conservation of the targeted epitopes rather than the location of the epitope. We showed that patients targeting a variable Gag epitope in early infection continued to preferentially target variable epitopes over conserved epitopes, while the subjects targeting conserved epitopes maintained their response against the conserved epitopes throughout the study period. Maintenance of immune responses was associated with no or limited sequence evolution within the conserved epitope. Importantly, we found that patients targeting a conserved epitope had a slower CD4+ T cell decline and significantly lower viral load during the study period compared to patients targeting variable epitopes. While restricting our analyses to Gag epitopes likely underestimates the contribution of other antigen variable and conserved epitopes to mediating viral load and disease progression, our results are in agreement with a larger study that performed comprehensive analyses of T cell responses across the HIV proteome.28 In that study, T cell responses to conserved epitopes remained detectable out to 12 months postinfection and were enriched in individuals maintaining lower viral loads.28 Our study suggests that the type of epitope targeted in Gag during acute infection, conserved versus variable, sets the stage for the type of epitopes targeted during chronic infection.
HIV is a chronic infection with great variation in disease outcome among infected individuals.35 The strongest host factor associated with a slower disease progression has been the expression of certain HLA-alleles in the host.36 Recent studies have shown that HLA-alleles associated with better disease outcome frequently bind conserved peptides.15,20–22 Binding of conserved peptides is likely to be beneficial due to the slower rate of mutations and because mutations, especially within structural proteins, can result in a virus with reduced replicative capacity (i.e., lower viral fitness).13,26,27,37 Troyer et al. showed that escape mutations in the conserved Gag-p24 capsid protein lead to changes in the secondary structure and a reduced viral fitness, while no effect or even enhanced viral fitness was seen when escape mutations occurred in the Env region.13 Hence, several factors associated with better disease progression are based on the character of the targeted peptide. However, many of these studies have been cross-sectional.21–24,38,39 It is therefore difficult to know which factors are causing the enhanced control and which are the result of a more intact immune system and lower antigen exposure in the slow progressor.40 Furthermore, it is not clear when these beneficial responses develop, since most studies have been performed during chronic infection. In this study we could monitor variable and conserved epitope responses identified in early infection and longitudinally follow them for up to 4 years of untreated infection. In agreement with a previous longitudinal study,28 we show that the character of peptides targeted early in infection affected the magnitude of the antigen-specific responses as well as several important clinical markers of disease outcome over time.
The fact that the peptide character influenced persistence or loss in cytokine production by antigen-specific T cells was expected. Responses targeting conserved peptides are likely to be maintained longer than responses against variable peptides due to the lower rates of mutations that permit immunological escape. Escape mutations occur in early infection as well as continuously during chronic infection.8,41–43 In our study we sequenced the autologous virus at the first and third time point and identified amino acid substitutions within the targeted Gag peptide. Interestingly, several amino acid substitutions were already present at the initial sequencing time point in variable epitopes. While we cannot determine whether these mutations represent transmitted mutations or early escape, it is interesting to postulate that such early mutations may further drive the propensity of variable epitope responders to continue to target variable epitopes. Indeed, we found that subjects targeting a variable peptide had mutations accruing over time within this epitope, while most of the subjects targeting a conserved peptide had an identical peptide sequence at time point one and three. Thus, we could confirm that the loss of response in patients targeting a variable peptide was likely a result of an abrogated HLA-peptide-TCR interaction due to mutations within the epitope. In addition, the loss of responses was not associated with limitations in the functional profile. Instead, the fraction of CD8+ T cells producing IFN-γ and MIP-1β was higher than in the conserved group despite the fact that the majority of their detected responses were directed against Gag epitopes that were characterized as variable. Unexpectedly, and despite the small number of participants in this study, we saw an effect on the CD4+ T cell slope and viral load depending on the peptide character initially targeted by the first Gag-specific CD8+ T cells response in the subjects. Even though patients targeting a variable epitope had a higher CD4+ T cell count at the initiation of this study, they also had a faster depletion of the CD4+ T cells in circulation. The higher CD4+ T cell count in subjects targeting a variable epitope at time point one could not be explained by the estimated duration of infection in months at the time for the first specimen, based on our estimated infection date. Thus, there must be another explanation for the difference in initial CD4+ T cell levels or it could be a random result due to the limited numbers of subjects studied. Our results indicate an important role for the initial immune response to the partial control of viral replication during the first years of infection. Two of the study subjects targeted epitopes that were HLA-B7 restricted, which is an allele that has not been shown to be associated with slow progression.44 The targeted epitope in these two study subjects was considered variable and the subjects belonged to the group that had a faster CD4+ T cell decline and higher viral load over time. Our findings are also in agreement with our previous study showing that early viral escape is associated with the development of new epitope-specific T cell responses and loss of viral control in HLA-A2-positive subjects.9 In addition, we showed that the majority of the epitope-specific responses were against other variable epitopes in the variable responders during the study period. We found limited associations between the functional profile of the peptide responses and the differences in viral load and CD4+ T cell decline. Instead, our results support the theory that the character of the peptide bound and presented on the HLA-alleles influences disease progression.
It is still not clear whether a T cell-based vaccine should aim for induction of broad responses targeting several peptides or induce a strong and narrow response.9,24 Our study does not answer these questions, but supports the importance of immune responses against conserved epitopes in early infection, even if the numbers of targeted epitopes are limited. However, CD8+ T cell responses against a conserved Gag epitope might not be more beneficial than a response against conserved epitopes in other regions or broader responses targeting multiple epitopes. Also, our study was restricted to a limited number of study subjects, which is why the lack of significance for some of the comparisons might be due to lack of statistical power. Despite these limitations our results show that the level of conservation of epitopes targeted by the initial HIV-Gag-specific CD8+ T cell responses is a factor that by itself is a predictor for disease outcome (i.e., viral load and CD4+ T cell count) that should be carefully considered in T cell-based vaccine development.
Sequence Data
The sequences generated in the gag region of HIV have been submitted to GenBank and received the following accession numbers: JF905572–JF905594.
Acknowledgments
We thank the study subjects for their participation. We thank Drs. Tim Hayes, Bill Critchfield,and Barbara Shacklett (University of California, Davis) for assistance with SPICE and FlowJo data analysis, and Gordon Bentley (University of California, San Francisco) for help with the sequence analysis. This work was supported by grants from the Swedish Agency for International Development Cooperation-SIDA (2005-001756), the Swedish Research Council (K2007-56X-20345-01-3 and K2010-56X-20345-04-3), the Swedish Physicians Against AIDS Research Foundation (FO2010-0019), Karolinska Institutet, the Swedish Society of Medicine (2009-22091 and SLS-101021), the Clas Groschinskys Memorial fund (M9 21 and M10 33), Åke Wibergs Foundation (40418186), and P01 AI071713 from the National Institute of Allergy and Infectious Diseases of the United States.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Borrow P. Lewicki H. Hahn BH. Shaw GM. Oldstone MB. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goonetilleke N. Liu MK. Salazar-Gonzalez JF. Ferrari G. Giorgi E. Ganusov VV, et al. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med. 2009;206:1253–1272. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koup RA. Safrit JT. Cao Y. Andrews CA. McLeod G. Borkowsky W, et al. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Norstrom MM. Buggert M. Tauriainen J. Hartogensis W. Prosperi MC. Wallet MA, et al. Combination of immune and viral factors distinguishes low-risk versus high-risk HIV-1 disease progression in HLA-B*5701 subjects. J Virol. 2012;86:9802–9816. doi: 10.1128/JVI.01165-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen TM. Altfeld M. Geer SC. Kalife ET. Moore C. O'Sullivan K M, et al. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J Virol. 2005;79:13239–13249. doi: 10.1128/JVI.79.21.13239-13249.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trachtenberg E. Korber B. Sollars C. Kepler TB. Hraber PT. Hayes E, et al. Advantage of rare HLA supertype in HIV disease progression. Nat Med. 2003;9:928–935. doi: 10.1038/nm893. [DOI] [PubMed] [Google Scholar]
- 7.Wolinsky SM. Korber BT. Neumann AU. Daniels M. Kunstman KJ. Whetsell AJ, et al. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science. 1996;272:537–542. doi: 10.1126/science.272.5261.537. [DOI] [PubMed] [Google Scholar]
- 8.Price DA. Goulder PJ. Klenerman P. Sewell AK. Easterbrook PJ. Troop M, et al. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc Natl Acad Sci USA. 1997;94:1890–1895. doi: 10.1073/pnas.94.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlsson AC. Iversen AK. Chapman JM. de Oliviera T. Spotts G. McMichael AJ, et al. Sequential broadening of CTL responses in early HIV-1 infection is associated with viral escape. PLoS One. 2007;2:e225. doi: 10.1371/journal.pone.0000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoof I. Perez CL. Buggert M. Gustafsson RK. Nielsen M. Lund O, et al. Interdisciplinary analysis of HIV-specific CD8+ T cell responses against variant epitopes reveals restricted TCR promiscuity. J Immunol. 2010;184:5383–5391. doi: 10.4049/jimmunol.0903516. [DOI] [PubMed] [Google Scholar]
- 11.Malhotra U. Nolin J. Horton H. Li F. Corey L. Mullins JI, et al. Functional properties and epitope characteristics of T-cells recognizing natural HIV-1 variants. Vaccine. 2009;27:6678–6687. doi: 10.1016/j.vaccine.2009.08.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goulder PJ. Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2008;8:619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Troyer RM. McNevin J. Liu Y. Zhang SC. Krizan RW. Abraha A, et al. Variable fitness impact of HIV-1 escape mutations to cytotoxic T lymphocyte (CTL) response. PLoS Pathog. 2009;5:e1000365. doi: 10.1371/journal.ppat.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miura T. Brumme ZL. Brockman MA. Rosato P. Sela J. Brumme CJ, et al. Impaired replication capacity of acute/early viruses in persons who become HIV controllers. J Virol. 2010;84:7581–7591. doi: 10.1128/JVI.00286-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brockman MA. Brumme ZL. Brumme CJ. Miura T. Sela J. Rosato PC, et al. Early selection in Gag by protective HLA alleles contributes to reduced HIV-1 replication capacity that may be largely compensated in chronic infection. J Virol. 2010;84:11937–11949. doi: 10.1128/JVI.01086-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrington M. Nelson GW. Martin MP. Kissner T. Vlahov D. Goedert JJ, et al. HLA and HIV-1: Heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 17.Migueles SA. Sabbaghian MS. Shupert WL. Bettinotti MP. Marincola FM. Martino L, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci USA. 2000;97:2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fellay J. Shianna KV. Ge D. Colombo S. Ledergerber B. Weale M, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altfeld M. Kalife ET. Qi Y. Streeck H. Lichterfeld M. Johnston MN, et al. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8(+) T cell response against HIV-1. PLoS Med. 2006;3:e403. doi: 10.1371/journal.pmed.0030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borghans JA. Molgaard A. de Boer RJ. Kesmir C. HLA alleles associated with slow progression to AIDS truly prefer to present HIV-1 p24. PLoS One. 2007;2:e920. doi: 10.1371/journal.pone.0000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dinges WL. Richardt J. Friedrich D. Jalbert E. Liu Y. Stevens CE, et al. Virus-specific CD8+ T-cell responses better define HIV disease progression than HLA genotype. J Virol. 2010;84:4461–4468. doi: 10.1128/JVI.02438-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang YE. Li B. Carlson JM. Streeck H. Gladden AD. Goodman R, et al. Protective HLA class I alleles that restrict acute-phase CD8+ T-cell responses are associated with viral escape mutations located in highly conserved regions of human immunodeficiency virus type 1. J Virol. 2009;83:1845–1855. doi: 10.1128/JVI.01061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Julg B. Williams KL. Reddy S. Bishop K. Qi Y. Carrington M, et al. Enhanced anti-HIV functional activity associated with Gag-specific CD8 T-cell responses. J Virol. 2010;84:5540–5549. doi: 10.1128/JVI.02031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiepiela P. Ngumbela K. Thobakgale C. Ramduth D. Honeyborne I. Moodley E, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 25.Rolland M. Heckerman D. Deng W. Rousseau CM. Coovadia H. Bishop K, et al. Broad and Gag-biased HIV-1 epitope repertoires are associated with lower viral loads. PLoS One. 2008;3:e1424. doi: 10.1371/journal.pone.0001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Picado J. Prado JG. Fry EE. Pfafferott K. Leslie A. Chetty S, et al. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol. 2006;80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peyerl FW. Bazick HS. Newberg MH. Barouch DH. Sodroski J. Letvin NL. Fitness costs limit viral escape from cytotoxic T lymphocytes at a structurally constrained epitope. J Virol. 2004;78:13901–13910. doi: 10.1128/JVI.78.24.13901-13910.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mlotshwa M. Riou C. Chopera D. de Assis Rosa D. Ntale R. Treunicht F, et al. Fluidity of HIV-1-specific T-cell responses during acute and early subtype C HIV-1 infection and associations with early disease progression. J Virol. 2010;84:12018–12029. doi: 10.1128/JVI.01472-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gray CM. Mlotshwa M. Riou C. Mathebula T. de Assis Rosa D. Mashishi T, et al. Human immunodeficiency virus-specific gamma interferon enzyme-linked immunospot assay responses targeting specific regions of the proteome during primary subtype C infection are poor predictors of the course of viremia and set point. J Virol. 2009;83:470–478. doi: 10.1128/JVI.01678-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hecht FM. Busch MP. Rawal B. Webb M. Rosenberg E. Swanson M, et al. Use of laboratory tests and clinical symptoms for identification of primary HIV infection. AIDS. 2002;16:1119–1129. doi: 10.1097/00002030-200205240-00005. [DOI] [PubMed] [Google Scholar]
- 31.Streeck H. Jolin JS. Qi Y. Yassine-Diab B. Johnson RC. Kwon DS, et al. Human immunodeficiency virus type 1-specific CD8+ T-cell responses during primary infection are major determinants of the viral set point and loss of CD4+ T cells. J Virol. 2009;83:7641–7648. doi: 10.1128/JVI.00182-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez CL. Larsen MV. Gustafsson R. Norstrom MM. Atlas A. Nixon DF, et al. Broadly immunogenic HLA class I supertype-restricted elite CTL epitopes recognized in a diverse population infected with different HIV-1 subtypes. J Immunol. 2008;180:5092–5100. doi: 10.4049/jimmunol.180.7.5092. [DOI] [PubMed] [Google Scholar]
- 33.Karlsson AC. Martin JN. Younger SR. Bredt BM. Epling L. Ronquillo R, et al. Comparison of the ELISPOT and cytokine flow cytometry assays for the enumeration of antigen-specific T cells. J Immunol Methods. 2003;283:141–153. doi: 10.1016/j.jim.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Roederer M. Nozzi JL. Nason MX. SPICE: Exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A. 2011;79:167–174. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kahn JO. Walker BD. Acute human immunodeficiency virus type 1 infection. N Engl J Med. 1998;339:33–39. doi: 10.1056/NEJM199807023390107. [DOI] [PubMed] [Google Scholar]
- 36.The_International_HIV_Controllers_Study: The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore CB. John M. James IR. Christiansen FT. Witt CS. Mallal SA. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 38.Betts MR. Nason MC. West SM. De Rosa SC. Migueles SA. Abraham J, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Owen RE. Heitman JW. Hirschkorn DF. Lanteri MC. Biswas HH. Martin JN, et al. HIV+ elite controllers have low HIV-specific T-cell activation yet maintain strong, polyfunctional T-cell responses. AIDS. 2010;24:1095–1105. doi: 10.1097/QAD.0b013e3283377a1e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Streeck H. Brumme ZL. Anastario M. Cohen KW. Jolin JS. Meier A, et al. Antigen load and viral sequence diversification determine the functional profile of HIV-1-specific CD8+ T cells. PLoS Med. 2008;5:e100. doi: 10.1371/journal.pmed.0050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Couillin I. Culmann-Penciolelli B. Gomard E. Choppin J. Levy JP. Guillet JG, et al. Impaired cytotoxic T lymphocyte recognition due to genetic variations in the main immunogenic region of the human immunodeficiency virus 1 NEF protein. J Exp Med. 1994;180:1129–1134. doi: 10.1084/jem.180.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goulder PJ. Phillips RE. Colbert RA. McAdam S. Ogg G. Nowak MA, et al. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 43.Phillips RE. Rowland-Jones S. Nixon DF. Gotch FM. Edwards JP. Ogunlesi AO, et al. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature. 1991;354:453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- 44.Kiepiela P. Leslie AJ. Honeyborne I. Ramduth D. Thobakgale C. Chetty S, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]