Abstract
TCRαβ thymocytes differentiate to either CD8αβ cytotoxic T lymphocytes or CD4+ T helper cells. This functional dichotomy is controlled by key transcription factors, including the T helper master regulator, ThPOK, which suppresses the cytolytic program in MHC class II-restricted CD4+ thymocytes. ThPOK continues to repress CD8-lineage genes in mature CD4+ T cells, even as they differentiate to T helper effector subsets. Here we show that the T helper-fate was not fixed and that mature antigen-stimulated CD4+ T cells could terminate Thpok expression and reactivate CD8-lineage genes. This unexpected plasticity resulted in the post-thymic termination of the T helper-program and the functional differentiation of distinct MHC class II-restricted CD4+ cytotoxic T lymphocytes.
CD4+ T cells are commonly classified as “helper” T cells based on their roles in providing “help” to promote or dampen cellular and humoral immune responses. In contrast CD8αβ expressing cytotoxic T lymphocytes (CTLs) provide direct protective immunity by killing infected or transformed cells. The T helper (TH)-program is initially induced during thymic development, where thymocytes expressing a major histocompatibility complex (MHC) class II-reactive T cell antigen receptor (TCR) develop into the CD4 TH-lineage, whereas thymocytes with MHC class I specificity differentiate to the CD8 CTL-lineage. The functional programming, which coincides with, but does not depend on, the MHC restriction and CD4 or CD8αβ co-receptor expression, is controlled by the action and counteraction of key transcription factors. Together with Tox and GATA3, the T helper transcription factor, ThPOK (also known as cKrox; encoded by the gene, Zbtb7b, hereafter referred to as Thpok) first induces the CD4 TH fate and prevents thymocytes from differentiating into CD8+ CTLs1–6. The Runx family member, Runx3, has the opposite effect and terminates CD4 expression while promoting CTL-lineage differentiation7,8. The CD8-CD4 lineage dichotomy persists in the periphery in mature T cells, where ThPOK continues to suppress the cytotoxic fate of MHC class II-restricted CD4+ T cells, even as they differentiate into TH effector subsets, controlled by additional transcription factors6.
This lineage separation, however, is not all encompassing, and repeated reports have indicated the presence of CD4+ T cells with cytolytic functions in various species, including humans and rodents9–12. At steady state, cytotoxic T cells, including CD4+ T cells, are enriched among the effector cells that reside as intraepithelial lymphocytes (IELs) in the intestine13–15, whereas under inflammatory conditions, including viral infections, autoimmune disorders and in response to tumor antigens, many cytolytic CD4+ T cells expand in the blood and peripheral tissues9–12,16,17. Although their widespread abundance and participation in various beneficial as well as pathogenic adaptive immune responses9–12,16,17, underscore the physiological relevance of cytolytic CD4+ effector cells, convincing evidence directly linking them to specific aspects of the adaptive immune response has been difficult since they have been merely viewed as functional variants of the well-defined TH1 subtype12,18. The lack of a defined gene signature has also greatly impaired progress on elucidating the biology of cytolytic CD4+ T cells and in designing clinical strategies that specifically target these cells in various diseases.
Here we provide compelling evidence indicating that cytotoxic CD4+ T cells represent a distinct subset of effector cells that could be defined by the absence of the master regulator, ThPOK, which maintains the TH fate in all classical CD4+ TH subsets by continuously suppressing the CD8 CTL-lineage program6. Nevertheless, we also show that the ThPOK-negative CD4+ CTLs originated from ThPOK-expressing progenitor cells that initially committed to the ThPOK-controlled TH-lineage during thymic selection. These findings therefore challenge the notion that the TH program in CD4+ T cells is fixed and show that mature CD4+ T cells can lose ThPOK expression post-thymically and functionally reprogram to become MHC class II-restricted CTLs. We also identify the Thpok silencer as the transcriptional switch that terminated Thpok transcription and by default drives the derepression of the CTL program in mature CD4+ effector cells. At steady state, CD4+ CTLs remained immune quiescent even in the continuous presence of their cognate antigens. However, in response to restimulation in the context of interleukin 15 (IL-15), CD4+ CTLs greatly increased their inflammatory and cytolytic functions and differentiated to potent killer effector cells. Overall the data demonstrate that CD4+ CTLs are not a simple variant of classical ThPOK-controlled TH1 cells, but that instead, they are distinct functional MHC class II-restricted effector cells that can be characterized by the loss of ThPOK expression and the derepression of aspects of the CD8-CTL lineage gene expression program.
RESULTS
Not all mature CD4 T cells express ThPOK
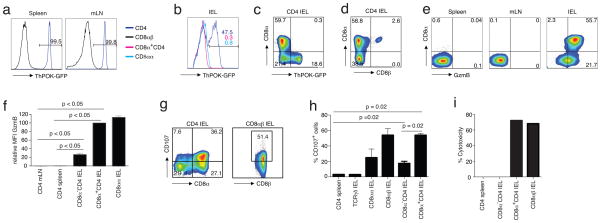
The reported cytolytic activity of mature CD4+ T cells is inconsistent with the notion that ThPOK continuously suppresses the CTL program in all mature MHC class II-restricted CD4+ T cells6 and suggests that these cells might not be under the negative control of ThPOK. To investigate this, we analyzed ThPOK expression in mature T cells isolated from ThPOK-green fluorescent protein (gfp)-knock-in (ThPOK-gfp) reporter mice19. As expected, CD4+ ThPOK-gfp lymphocytes isolated from the spleen or mesenteric lymph node (mLN), which are mostly naïve T cells, were GFP-positive (GFP+), indicating that they all expressed ThPOK as is typical of mature CD4+ TH-lineage cells (Fig. 1a). Conversely, all cells in the CD8+ fraction were GFP-negative (GFP−), consistent with the absence of ThPOK expression in CTL-lineage cells (Fig. 1a). Surprisingly, many of the CD4+ ThPOK-gfp effector T cells that at steady state accumulated in the intestine were GFP−, signifying that, like their CD8+ counterparts, they did not express ThPOK (Fig. 1b,c). Interestingly, the majority of the GFP−CD4+ cells resided in the subset of IELs that co-express CD8α (without CD8β)20 (Fig. 1b-d). Consistent with the lack of ThPOK-mediated suppression, these CD8α+CD4+ double-positive (DP) cells also displayed functional features that were remarkably similar to those of mature CD8+ CTLs, including abundant expression of granzyme (Fig. 1e,f) and substancial levels of the activation-induced degranulation marker, CD107a, also known as lysosome-associated membrane protein 1 (LAMP-1), a glycoprotein present in the membrane of cytotoxic granules and exposed on the cell surface of activated cytolytic cells21 (Fig. 1g,h). The induction of CD107a by the DP subset was comparable to that of typical CD8 TCRαβ CTLs, whereas activated SP CD4+ IELs or TH cells from the spleen did not induce this cytolytic marker (Fig. 1g,h). Moreover, activated DP CD4+ cells also effectively killed target cells in vitro as measured by the release of lactate dehydrogenase (LDH) upon target lysis (Fig. 1i and Supplementary Fig. 1a). In all, the data demonstrated that in normal mice, not all CD4+ effector cells expressed ThPOK and furthermore, that those CD4+ ThPOK-negative (ThPOK−) lymphocytes expressed CD8α and displayed cytolytic activity that closely resembled that of mature CD8+ CTLs.
Figure 1.
Some mature CD4 T cells do not maintain ThPOK expression in the periphery. (a) Frequency of GFP positive cells among gated CD45+TCRβ+ lymphocytes isolated from the spleen and mLN of naïve ThPOK-gfp reporter mice. (b) Frequency of GFP positive cells among gated CD45+TCRβ+ lymphocytes isolated from the small intestine epithelium of naïve ThPOK-gfp reporter mice. Data in a and b are representative of 2 independent experiments, n = 4 or 5 mice. (c) Cell surface staining for CD8α and GFP expression in gated CD45+TCRβ+CD8β−CD4+ IELs as in (b). (d) Cell surface staining for CD8α and CD8β expression in gated CD45+TCRβ+CD4+ IELs as in (b). (e) Cell surface staining for CD8α and intracellular for Granzyme B in gated CD45+TCRβ+CD8β−CD4+ lymphocytes isolated from spleen, mLN and sIELs of naïve WT B6 mice. Values in plots indicate percent positivity (f) MFI for Granzyme B expression by CD4+, CD8α+CD4+ or CD8αα+ T cells in gated CD45+TCRβ+CD8β− lymphocytes from different tissues of naïve wild type B6 mice. Data in c–f are representative of 3 independent experiments, n = 4 or 5 mice. ANOVA and Bonferroni as post test were used for statistics (g) Frequency of CD107 positive cells among total gated CD45+TCRβ+CD8β−CD4+- and CD45+TCRβ+CD8αβ+ IELs after anti-TCRβ stimulation. Representative data of 4 independent experiments. Values indicate percent positivity (h) Frequency of CD107 expressing CD4+ splenocytes or TCRγδ+-, CD8αα+-, CD8αβ+-, CD8α−- and CD8α+CD4+ IEL subsets after anti-TCRβ stimulation. Data are representative of 4 independent experiments. Each bar shows standard error of the mean (SEM; n=4). Nonparametric two tailed Mann-Whiteny test was used for statistics (i) % cytotoxicity of respective lymphocyte subsets as measured by LDH release assay. Representative data of 4 independent experiments.
Mature ThPOK− CD4 T cells derive from ThPOK+ thymocytes
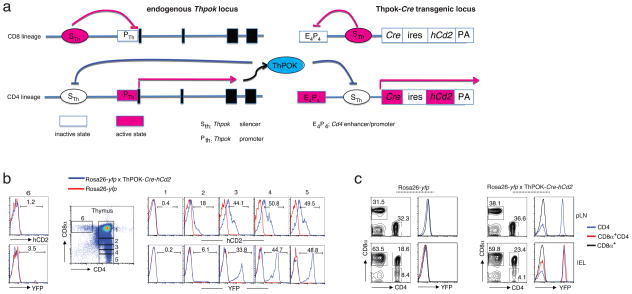
ThPOK is the master regulator of the TH-lineage and is first expressed in the thymus where it counteracts Runx3 and suppresses the CTL fate of MHC class II-restricted thymocytes4–6. The absence of ThPOK expression associated with cytotoxicity in mature CD4+ T cells could suggest that they might have originated from ThPOK− progenitors. To investigate this, we designed a fate-mapping mouse-model, in which we tracked previous ThPOK expression in mature T cell subsets (Fig. 2a). Inactivation of Thpok gene transcription in MHC class I-specific CD8+ CTL lineage thymocytes is mediated by repressive factors, such as Runx proteins, that bind to the Thpok silencer (Sth) DNA element22,23. In MHC class II-restricted cells however, ThPOK itself binds to the Thpok silencer and prevents Runx-mediated silencing, which results in a positive-feedback loop that warrants continuous expression of Thpok (Fig. 2a). A fate marker that reports on any previous activity of the Thpok silencer in thymocytes is therefore an accurate reporter for the lineage-origin and initial commitment of mature CD4+ T cells. Based on this rationale, we designed a new Cre-transgenic mouse strain, referred to here as ThPOK-Cre mice, using a Cre-transgene construct in which we inserted the Thpok silencer into a Cd4 enhancer/promoter locus that controls the Cre transgene expression (Fig. 2a). In contrast to the widely used CD4-Cre mice, which express Cre under the control of the Cd4 enhancer/promoter starting at the CD4+CD8αβ+DP thymocyte stage, in ThPOK-Cre mice, the Thpok silencer prevents Cd4 enhancer/promoter activity in DP and CD8-lineage cells, resulting in exclusive Cre expression in CD4+ SP thymocytes committed to the TH lineage (Fig. 2a). Therefore, when crossed to Rosa26-yellow fluorescent protein (YFP) reporter mice, only progeny derived from thymus-committed ThPOK-controlled CD4+ TH lineage-committed thymocytes are marked by the expression of the YFP reporter after Cre mediated recombination (Fig. 2b,c). In these gene-reporter mice, SP CD8+ thymocytes and mature peripheral CD8+ T cells, including CD4− CD8+ mucosal T cells, did not express YFP, whereas, as expected, the majority of SP CD4+ LN T cells and IELs were YFP+. Notably, also CD8α-expressing CD4+ IELs were YFP+, to a similar extent as the conventional CD4+ SP cells (Fig. 2b,c), indicating that they had induced ThPOK-dependent Cre expression, and therefore that ThPOK-negative mature CD4+ T cells must have derived from progenitor cells that expressed ThPOK at an earlier stage.
Figure 2.
ThPOK− mature CD4 T cells are progeny of ThPOK-expressing thymocytes. (a) A schematic presentation of the Thpok and the transgenic ThPOK-Cre locus in CD8- and CD4-lineage thymocytes. Red indicates “active” and white “inactive”. (b) Staining for CD4 and CD8α (dot plot), human CD2 and YFP (histograms) of thymocytes gated on CD4 and CD8α expression levels (gates 1–6) where 1 represents immature double-positive thymocytes and 5 and 6 represent mature, single-positive CD4 and CD8 thymocytes, respectively. Red histograms represent thymocytes from Rosa26-yfp mice and blue histograms from Rosa26-yfp x ThPOK-Cre-hCd2 mice. Values above histograms indicate percent positivity (c) Staining for CD4 and CD8α (contour plots) and YFP (histograms) of peripheral lymph node (pLN) and sIELs, gated on TCRβ+CD4+ lymphocytes isolated from the Rosa26-yfp (left)- or the Rosa26-yfp x ThPOK-Cre-hCd2 (right) mice. Values above histograms and in contour plots indicate percent positivity. Data in b and c are representative of at least 3 independent experiments.
Mature ThPOK− CD4 T cells lose ThPOK expression post-thymically
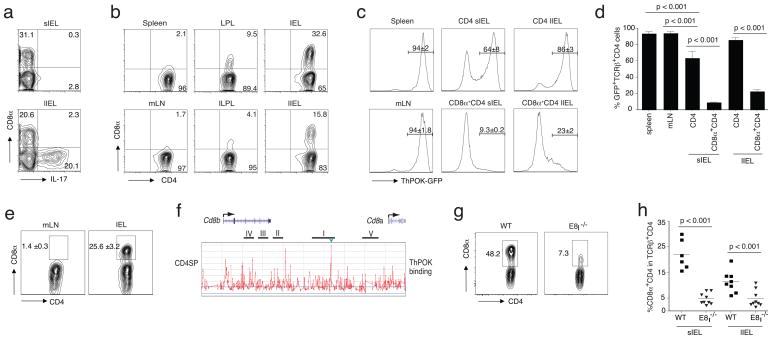
The notion that the mature ThPOK− CD4+ cells previously expressed ThPOK, together with the observation that these cells mainly accumulate among effector cells, suggests that the loss of ThPOK expression might be the result of a post-thymic activation or maturation process that mature T cells undergo in the periphery. To test this, we adoptively transferred highly purified ThPOK-expressing GFP+ naïve ThPOK-gfp CD4+ T cells to Rag1-deficient (Rag1−/−) recipient mice. The lymphopenic conditions in this model induce strong proliferation and differentiation of the donor cells, which accumulate mainly as TH17 cells (characterized by the expression of the transcription factor RORγt and the cytokine, IL-17) in the large intestine, and interestingly, also as DP effector cells especially in the small intestine of the Rag1−/− hosts (Fig. 3a,b). Donor cells in the spleen or mLNs were GFP+ implying that they continued to express ThPOK (Fig. 3c). In contrast, many of the ThPOK-gfp CD4+ T cells that accumulated as effector cells in the intestine did not express detectable levels of GFP, indicating that they strongly reduced or completely lost ThPOK expression (Fig. 3c). Consistent with the observations in immune-replete mice, the loss of GFP expression was again most pronounced in those CD4+ cells that also re-induced CD8α, although a small fraction of SP CD4+ IELs was also GFP−(Fig. 3c,d). Serial transfer of those SP donor cells to a second set of Rag1−/− recipients generated many more DP cells (Fig. 3e), indicating that the re-expression of CD8α on mature CD4+ T cells follows the loss of ThPOK expression and further implies that the Cd8a locus in conventional CD4+ TH cells might be constitutively suppressed by ThPOK. To analyze this, we performed chromatin immunoprecipitation (ChIP) combined with tiling arrays using cells from genetically engineered mice that express a Flag-hemagglutinin epitope-tagged ThPOK protein (FH-ThPOK) from the Thpok locus19. FH-ThPOK associated with the E8I enhancer element24 in the Cd8a locus in SP CD4+ thymocytes (Fig. 3f). This result was also confirmed in mature SP CD4+ T cells using a ChIP assay (Supplementary Fig. 2a), suggesting that in mature CD4+ thymocytes and lymphocytes, ThPOK prevents CD8α expression by direct suppression of the E8I enhancer element in the Cd8a locus, and also that the expression of CD8α without CD8β in mature ThPOK− CD4+ T cells is driven by the de-repression of the E8I enhancer. In support of this finding, the percent of CD8α-expressing CD4+ IELs and CD8 expression were both severely reduced in E8I-deficient mice (Fig. 3g) or in the progeny of E8I-deficient CD4 cells in Rag1−/− recipient mice that previously received an equal number of naïve wild-type and E8I-deficient CD4 donor cells (Fig. 3h and Supplementary Fig. 2b). Overall the data indicate that naive ThPOK+ CD4+ TH cells may lose expression of ThPOK in the periphery and consequently regain expression of ThPOK-suppressed genes such as Cd8a.
Figure 3.
ThPOK− CD4 effector cells lose ThPOK as mature cells in the periphery. (a) CD8α and intracellular IL-17 staining of gated CD45+TCRβ+CD4+ small and large intestine IELs of Rag1−/−recipient mice 8 weeks after adoptive transfer of naïve TCRβ+CD8α−CD45RBhighCD25−CD4+ spleen T cells. Values in plots indicate percent positivity. (b) CD8α and CD4 staining of gated CD45+TCRβ+CD4+ T cells from various tissues of Rag1−/− recipient mice 8 weeks after adoptive transfer of naïve TCRβ+CD8α−CD45RBhighCD25−CD4 spleen T cells. Values in plots indicate percent positivity. (c) Thpok GFP expression in gated CD45+TCRβ+ -SP CD4+ or -DP CD8α+CD8β−CD4+ lymphocytes isolated from various tissues of Rag1−/− recipient mice 8 weeks after transfer of sorted TCRβ+CD8α−CD45RBhighCD25−CD4+ spleen T cells from Thpok GFP donor mice. Values above histograms indicate mean±SEM percent positivity. Data in a–c are representative of 3 independent experiments. (d) Frequency of GFP+ cells in gated CD45+TCRβ+ -SP CD4+ or -DP CD8α+CD8β−CD4+ lymphocytes from spleen, mLN or IELs of Rag1−/− recipients after transfer of sorted naïve TCRβ+CD8α−GFP+CD45RBhighCD25−CD4 spleen T cells from Thpok GFP donor mice. Data are representative of 3 independent experiments. ANOVA and Bonferroni as post test were used for statistics. Each bar shows the SEM (e) Surface Staining for CD8α on re-transferred sorted (> 99.7% purity) CD45+TCRβ+CD8α−CD4+ donor IELs (from 3 pooled recipient mice) isolated from mLN and sIELs of a second Rag1−/−recipient 8 weeks after the second transfer. Values in plots indicate mean±SEM percent positivity. Data are representative of 2 independent experiments. (f) Chromatin immuno-precipitation (ChIP) with tiling array using CD4 SP thymocytes from the FH-ThPOK knock-in mice expressing FLAG-hemagglutinin epitope tagged ThPOK protein. Binding of ThPOK to the E8I region was marked with a green arrowhead. Data are representative of 2 independent experiments. (g) Frequency of CD8α positive CD45+TCRβ+CD8β−CD4+ IELs isolated from wild type (left panel) and E8I-deficient (right panel) mice. Data are representative of 3 independent experiments, n = 4 or 5 mice. (h) Frequency of CD8α+CD4+ IELs from Rag1−/− recipient mice 8 weeks after transfer of wild type or E8I-deficient TCRβ+CD8α− CD45RBhighCD25−CD4+ spleen T cells.
ThPOK loss coincides with CTL differentiation
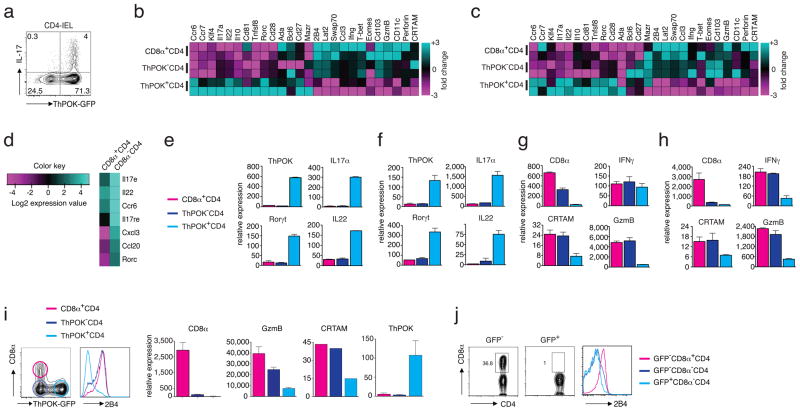
Although subsets of polarized CD4 effector cells are controlled by unique transcription factors, such as T-bet for TH1, GATA3 for TH2 and RORγt for TH17 cells, ThPOK continues to function as a master regulator of the TH-lineage and continues to suppress the CTL program. Consistent with this notion, in the adoptive transfer model, ThPOK-gfp donor cells that differentiated to TH17 effector cells in the intestine of the recipient mice remained GFP+, indicating that their gene expression profile was still regulated by the ThPOK repressor (Fig. 4a). To examine the impact of the loss of ThPOK expression on the gene expression pattern of CD4+ effector cells, we analyzed gene microarrays generated from RNA isolated from either sorted ThPOK+ or ThPOK− SP and DP effector cells from normal unmanipulated Thpok-GFP reporter mice19 (Fig. 4b) or from Rag1−/− recipient mice (Fig. 4c and Supplementary Fig. 3a). Interestingly, the DP and SP ThPOK− CD4+ T cells expressed a unique but similar gene expression pattern that clearly differed from that of the SP ThPOK+ CD4+ cells, although in the transfer experiments, the donor cells originated from the same pool of naive lymphocytes and differentiated in the same host environment (Fig. 4b,c and Supplementary Fig. 3a). As expected, many of the ThPOK+ SP CD4+ cells isolated from the intestine displayed a clear TH17 gene-expression pattern, whereas expression of these genes was barely detectable in the ThPOK− and DP subset (Fig. 4b–d). Furthermore, RT-PCR analysis of genes characteristic of TH17 cells, including those encoding the cytokines IL-17A, IL-17F and IL-22, the cytokine receptor IL-23R and the TH17 hallmark nuclear transcription factor, RORγt (Fig. 4e,f and Supplementary Fig. 3b) confirmed that ThPOK− CD4 effector cells were distinct from TH17 cells. Furthermore, analysis of genes characteristic of other CD4+ TH types, such as the TH1 and TH2 cells (Supplementary Fig. 3c), demonstrated that the gene expression pattern of SP and DP CD4+ lymphocytes that had lost ThPOK expression did not resemble the patterns expressed by the known TH effector subsets.
Figure 4.
Activated CD4 TH cells that lose ThPOK expression differentiate to CTL. (a) Intracellular IL-17 and GFP staining in gated CD45+TCRβ+CD4+ IELs from Rag1−/− recipient mice of Rag1−/− recipients that previously received sorted naïve TCRβ+CD8α−GFP+CD45RBhighCD25−CD4+ spleen T cells from ThPOK-gfp donor mice. Data are representative of 3 independent experiments. (b) Gene expression microarray analysis of mRNA from sorted CD45+TCRβ+-ThPOK+CD8α−CD4+, -ThPOK−CD8α−CD4+ and ThPOK−CD8α+CD4+ IELs of naïve ThPOK-gfp reporter mice. Each sample represents 2 independent experiments, sorting and duplicate microarray analyses (c) Gene expression microarray analysis of mRNA from sorted CD45+TCRβ+-ThPOK+CD8α−CD4+, -ThPOK−CD8α−CD4+ and ThPOK−CD8α+CD4+ IELs of Rag1−/− recipients that previously received sorted naïve TCRβ+CD8α− GFP+CD45RBhighCD25−CD4+ spleen T cells from ThPOK-gfp donor mice. Each sample represents 2 independent experiments, sorting and duplicate microarray analyses (d) Heat map of normalized gene expression levels of Th17 signature genes in donor CD4 T cells determined by microarray analysis as in (b). (e) Relative mRNA levels of Th17-associated genes from sorted T cell subsets as in (c). (f) Relative mRNA levels of Th17-associated genes from sorted T cell subsets as in (d). (g) Relative mRNA levels of CTL-associated genes from sorted T cell subsets as in (c). (h) Relative mRNA levels of CTL-associated genes from sorted T cell subsets as in (d). (i) ThPOK-GFP (left panel) and 2B4 (right panel) expression in gated CD45+TCRβ+CD4+ IELs as in (a). Relative mRNA expression of CD8α, Granzyme B, CRTAM and ThPOK in IELs sorted for (CD45+TCRβ+CD4+) GFP+, GFP−CD8α−, and GFP−CD8α+ subsets isolated from 4 individual Rag1−/− recipients as in (a). Data are representative of 3 independent experiments. Gene expression was analyzed by quantitative real-time PCR and expressed relative to L32 as an internal standard. (j) Staining for CD4, CD8α (left and middle panels) and 2B4 (right panel) of gated TCRβ+CD4+ IELs from Rag1−/− recipients that previously received ThPOK-transfected (GFP+) or untransfected (GFP−) TCRβ+CD8α−CD45RBhighCD25−CD4+ donor spleen T cells. Data are representative of 3 independent experiments, n = 3 mice.
In contrast, in addition to the re-expression of CD8α, ThPOK− CD4+ effector cells also expressed many other genes typically associated with the CD8 CTL-program, including the expression of various genes encoding cytolytic proteins, such as several granzymes and perforin, as well as interferon-γ (IFN-γ) and several receptors expressed by natural killer (NK) cells and mature CD8+ CTLs (Fig. 4b,c,g,h and Supplementary Fig. 3d,e). Of particular note was the expression of the cytotoxicity-related class I-restricted T cell-associated molecule, CRTAM, and the CD2 family member, CD244 (2B4), both known to promote the cytolytic function and IFN-γ production of CD8+ T cells25–27.
The reciprocal expression in mature CD4+ effector cells of either the transcription factor ThPOK or CTL signature genes (Fig. 4i), confirmed that ThPOK continuously suppresses the CTL program in conventional TH cells but also demonstrated that the differentiation of CD4+ CTL effector cells coincides with the post-thymic loss of the TH-lineage master regulator ThPOK. In agreement with this finding, enforced expression of ThPOK due to retroviral transduction, CD4+ donor cells no longer differentiated into CTLs in vivo (Fig. 4j), whereas transfection of a construct encoding a Thpok spontaneous mutation found in the helper-deficient mouse strain (HD-variant)4 did not prevent the CTL differentiation (Supplementary Fig. 3f). These data indicate that the loss of ThPOK coincided with the de-repression of the CTL phenotype in mature CD4+ effector cells.
Thpok silencer de-repression terminates Thpok expression
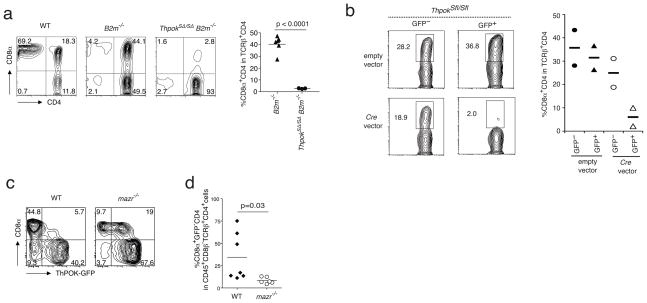
In CD8 CTL-lineage committed thymocytes ThPOK expression is switched off at the transcriptional level by the Thpok silencer19. Consequently, in mice, which harbor a germline deletion of this silencer (ThpokSΔ/SΔ), MHC class I-restricted thymocytes mature as CD4-expressing T cells in the periphery19. To investigate if the Thpok silencer engages in a similar role to terminate Thpok expression in mature MHC class II-restricted CD4+ CTL precursor cells, we analyzed peripheral CD4+ T cell maturation in mice in which the Thpok silencer had been deleted (ThpokSΔ/SΔ mice). To eliminate the MHC class I-restricted CD4-expressing cells in these mice, we crossed the ThpokSΔ/SΔ mice with MHC class I-deficient (B2m−/−) mice, to abrogate the development of MHC class I-restricted DP T cells. Interestingly, whereas in wild-type B2m −/−mice, in the absence of mature CD8+ T cells, all CD4 populations are greatly enhanced, including the DP CD4+ IELs (Fig. 5a), in ThpokSΔ/SΔ x B2m−/− mice however, mature MHC class II-restricted CD8-expressing CD4+ T cells were severely reduced (Fig. 5a). These observations indicate that the Thpok silencer is a critical genomic switch in the process of CD4 CTL differentiation, which turns off Thpok transcription in mature CD4+ T cells. This notion was further confirmed using CD4+ T cells, in which the floxed Thpok silencer (ThpokSfl/Sfl) was deleted at the mature stage by transfecting the cells with a Cre-gfp-containing retroviral construct. Transfer of sorted GFP+ ThpokSfl/Sfl CD4+ T cells, which expressed Cre (GFP+) and consequently deleted the Thpok silencer, showed impaired accumulation of CD8-expressing CD4 CTL in the intestine of Rag1−/− recipient mice, further underscoring the critical role of the Thpok silencer in terminating Thpok expression as part of the CTL differentiation process of CD4+ effector cells (Fig. 5b). In contrast to ThPOK, which suppresses the repressive activity of the Thpok silencer, the zinc finger transcription factor, MAZR, is known to activate the Thpok silencer28, resulting in MAZR-induced negative regulation of Thpok gene transcription. Consistent with the participation of MAZR in the reactivation of the Thpok silencer in mature CD4 T cells, Mazr−/− CD4+ donor cells formed fewer DP CTLs in the intestine of Rag1−/− hosts (Fig. 5c,d). These results indicate that the transcriptional regulation of Thpok is key for the control of the TH phenotype of mature CD4+ T cells, and that reactivation of its silencer leads to the termination of the TH program and conversely to the functional differentiation of MHC class II-restricted CD4+ CTLs.
Figure 5.
The Thpok silencer forms the switch that terminates Thpok expression in mature CD4 T cells. (a) Staining for CD4 and CD8α expression on gated TCRβ+CD4+ sIELs isolated from naïve wild type,B2m−/− and ThpokSΔ/SΔ (Thpok silencer deletion) x B2m−/− mice. Plots are representative of 3 independent experiments, n = 2 or 3 mice. (b) Staining as in (a) gated on (GFP+ or GFP−) sIELs isolated from Rag1−/−recipients 4–5 weeks after adoptive transfer of Cre-gfp-transfected TCRβ+CD4+ Thpok silencerflox/flox (ThpokSflox/Sflox) primary cells. Data are representative of 2 independent experiments. (c) Staining for GFP and CD8α expression on sIELs gated as in (a) and isolated from Rag1−/− recipients after transfer of sorted naïve TCRβ+CD8α−GFP+CD45RBhighCD25−CD4+ spleen T cells from wild type- and Mazr−/− ThPOK-gfp donor mice. Representative data of 3 independent experiments. (d) Frequency of CD8α+GFP−CD4 sIELs as in (c). Data are representative of 3 independent experiments.
The CD4 CTL differentiation in vivo is driven by antigen
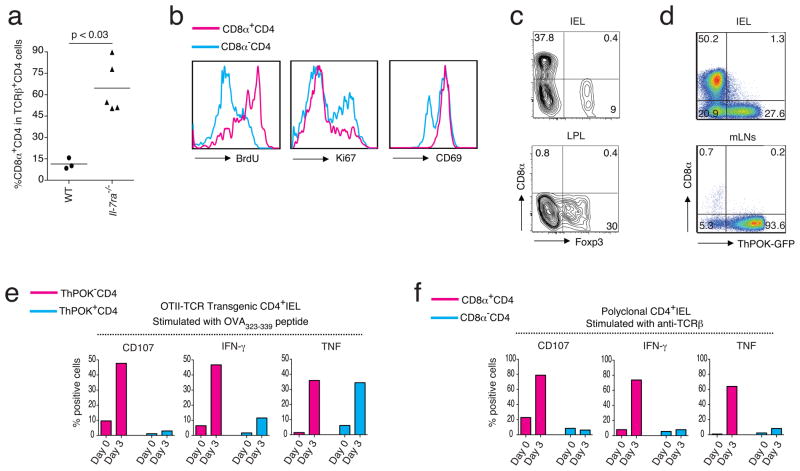
The loss of ThPOK observed in progeny of naïve ThPOK+ donor cells suggested that the induction of the CTL program in mature CD4+ T cells coincided with an activation or maturation process. In light of this, the re-expression of CD8α as well as the cytolytic functional differentiation of mature CD4+ T cells is reminiscent of the cytotoxic-lineage differentiation of positively selected CD8+ SP thymocytes mediated by IL-729. Despite this however, we found significant, and even increased numbers, of DP CD4+ T lymphocytes in Il7ra−/− animals (Fig. 6a), indicating that the in vivo differentiation of mature CD4+ CTLs is not an IL-7 cytokine-driven process. Similar to other effector cells in the intestine, DP cells are absent in germ-free (GF) mice (Supplementary Fig. 4a), and appear in normal numbers in germ-free animals reconstituted with specific pathogen free (SPF) microorganisms (Supplemental Fig. 4b), indicating that some microbial factors directly or indirectly promote the accumulation of CD4+ CTLs in the intestine. In contrast, unlike classical CD4+ TH17 effector cells, they did not expand in the intestine of germ-free mice mono-colonized with segmented filamentous bacteria (SFB) alone30 (Supplemental Fig. 4b) or in response to an infection with Citrobacter rodentium, a pathogen known to induce a strong TH17 response in the large intestine (Supplementary Fig. 4c), whereas they were present in normal numbers in the intestine of Myd88−/− animals. These observations indicate that the microbial conditions required for the steady state accumulation of CD4+ CTLs in the intestine are not likely established by a single microorganism and do not depend on IL-1 or TLR-induced signals (Supplementary Fig. 4d). Published reports have indicated that CD4+ CTLs are antigen-experienced cells that differentiated in response to repeated activation signals10,12. In agreement with this, in the adoptive transfer model, DP CD4+ donor cells, which displayed a CTL phenotype, also showed evidence of strong or repeated activation and marked BrdU uptake, but only weak staining for the active cell cycle marker, Ki67 (Fig. 6b), indicating that they were resting cells that had been activated and intensely proliferated previously. In addition, the CD4+ CTLs also showed enhanced expression of CD69 as compared to the SP CD4+ TH effector cells, suggesting that the activation process that coincided with the loss of ThPOK expression and de-repression of the CTL program also coincided with those CD4+ effector cells that received strong or repeated activation signals (Fig. 6b). To directly test this, we analyzed the effector differentiation of monoclonal OT-II TCR transgenic (OT-II) CD4+ T cells in response to a continuous activation in vivo with their cognate OVA peptide, (OVA323-339) presented by I-Ab. T cells only migrate as effector cells to peripheral tissues and in agreement with this, in the absence of OVA, very few OT-II CD4+ T cells accumulated in the intestine of OT-II TCR transgenic animals. Similarly, upon transfer of naïve OT-II CD4+ T cells to Rag1−/− recipients, only a limited number of these donor cells migrate to the intestine of the recipient mice in the absence of OVA antigen. In contrast, upon feeding an OVA-containing diet for at least 4 weeks, a large number of activated OT-II CD4+ cells accumulated in the intestine and interestingly, many re-expressed CD8α (Fig. 6c), whereas those that remained SP had a tendency to express Foxp3 (Fig. 6c). To investigate if the de-repression of CD8α on the responder OT-II CD4+ T cells coincided with the loss of ThPOK and the differentiation to CTLs, we transferred naïve donor cells isolated from OT-II ThPOK-gfp reporter mice to OVA-fed Rag1−/− recipient mice and analyzed the phenotype and function of the OVA-responding cells that accumulated as effector cells in the intestine of the hosts. As expected, many DP OT-II ThPOK-gfp cells accumulated in the small intestine but more importantly they were all GFP−, indicating that they had lost ThPOK expression (Fig. 6d). Furthermore, in addition to the re-expression of CD8α, OT-II GFP− effector cells also induced typical cytolytic markers, such as 2B4 and granzyme B (Supplementary Fig. 5a). More importantly, they displayed an antigen-specific cytolytic response when re-stimulated with the OT-II TCR specific OVA323-339 peptide (Fig. 6e) but not with the MHC class I restricted OVA peptide (SIINFEKL) (Supplementary Fig. 5b).
Figure 6.
The ThPOK loss and reprogramming of CD4 CTL is an antigen-driven process in vivo. (a) Frequency of CD8α+CD4+ T cells in gated CD45+TCRβ+CD8β−CD4+ IELs from wild type or Il-7ra−/− mice. Nonparametric two-tailed Mann-Whiteny test was used for statistics. (b) BrdU staining following an i.p. injection with 1 mg BrdU and 6 days of 0.8 mg/ml BrdU in the drinking water (left panel). Ki67 staining (middle panel) and CD69 staining (right panel) of CD8α+ or CD8α− gated CD45+TCRβ+CD8β−CD4+ IELs of naïve wild type mice. (c) Staining for CD8α and Foxp3 in gated CD45+TCRβ+CD8β−CD4 IELs (upper panel) and LPL (lower panel) of OVA-specific OTII TCR transgenic Rag1−/− mice after 4 weeks of OVA-diet feeding. Representative data of 2 independent experiments, n = 4 mice per group. (d) Frequency of CD8α and GFP positive cells among CD45+TCRβ+ OT-II TCR ThPOK-gfp CD4+ IELs isolated from Rag1−/− recipient mice after 4 weeks of OVA-diet feeding. Data represents 5 independent experiments. (e) Frequency of CD107, IFNγ and TNFα positive cells in ThPOK−(red) and ThPOK+ (blue) CD45+TCRβ+ OT-II TCR ThPOK-gfp CD4+ IELs as in (d) analyzed at day 0, without IL-15 exposure (red bars) or after 3 days in vitro culturing with rIL-15 (blue bars) prior to re-stimulation with OVA323-339 peptide. Data are representative of 3 independent experiments. (f) Frequency of CD107, IFNγ and TNF positive cells in CD8α+ (red) and CD8α− (green) CD45+TCRβ+CD8β−CD4+ polyclonal wild type IELs without or with IL-15 as in (e). Data are representative of 3 independent experiments.
Despite their functional potential ThPOK− OT-II CD4+ CTLs remained immune quiescent in vivo even in the continuous presence of their cognate antigen in the diet. This finding is similar to the MHC class I-restricted CD8αβ CTLs, which also appear inactive under steady state conditions31. Upon challenge in excess amounts of IL-15, however, as in active celiac disease18,32, CD8+ CTLs become pathogenic killer cells33,34 that also greatly upregulate secretion of the inflammatory cytokines, IFN-γ and tumor necrosis factor (TNF)18,35,36,. To investigate if CD4+ CTLs might be equally responsive to IL-15, we re-stimulated the diet-induced ThPOK− OT-II CD4+ CTLs in vitro in the context of IL-15. As expected, IL-15 increased CD8α expression37, however, IL-15 alone or together with the irrelevant MHC class I-restricted peptide, SIINFEKL, had no effect on the functional differentiation or maturation of the MHC class II-restricted OT-II CD4+ CTLs (Supplementary Fig. 5b). In contrast, IL-15 greatly increased the immune responses of OVA323-339-stimulated ThPOK− OT-II CD4+ CTLs, as measured by the marked upregulation of CD107a expression and the significant increase in production of IFN-γ and TNF (Fig. 6e). These data therefore demonstrate that antigen-induced CD4+ CTLs, generated in the absence of inflammation, are poised to exert potent effector functions when re-activated by their cognate antigen in the context of the inflammatory cytokine IL-15. We confirmed these data with polyclonal ThPOK− CD4+ CTLs isolated from the intestine of normal lympho-replete animals and showed that unmanipulated CD4+ CTLs analyzed ex vivo also harbor the potential to increase their cytolytic and inflammatory immune responses when exposed to IL-15. Although, IL-15 alone supported short-term survival of wild-type CD4+ and CD8αβ+ CTLs, this cytokine alone did not induce cytolytic effector functions in these polyclonal cells (Supplementary Fig. 5c). On the contrary, similar to the diet antigen-induced ThPOK− OT-II CD4+ CTLs, or normal CD8αβ+ CTLs (Supplementary Fig. 5d), IL-15 also strongly increased the cytolytic and inflammatory functions of wild-type TCR-stimulated CD4+ CTLs, whereas it had no effect on ThPOK+ CD4+ TH cells (Fig. 6f). These data underscore the likely pathogenic role of antigen-induced ThPOK− CD4+ CTLs, which are poised to kill under conditions where IL-15 is abundant.
DISCUSSION
The results reported here indicate an unexpected degree of plasticity for CD4+ TH cells, and demonstrate that in response to chronic or strong antigen stimulation, mature CD4+ cells can terminate the expression of the TH master regulator, ThPOK, and differentiate to functional CTLs. The identification of additional functional CD4+ T lymphocyte subsets defined by signature transcription factors, such as TH17 cells and Foxp3+ regulatory T cells, has been one of the most important recent advances in immunology research. Although cytotoxic activity carried out by CD4+ T cells has long been known, here we show that this is not simply a function of TH1-like cells. Instead, here we identify a new type of mature, antigen-experienced CD4+ cells characterized by the activation-induced loss of the CD4 lineage signature transcription factor, ThPOK. These cells display the concomitant appearance of a cell surface- and functional phenotype that is distinct from that of conventional TH subsets, but that strikingly resembles that of CD8+ CTL effector cells. The data therefore indicate that the ThPOK-driven thymic commitment to the CD4 TH lineage is not fixed, and that plasticity controlled at the level of ThPOK expression may lead to the functional differentiation of defined ThPOK-negative MHC class II-restricted CD4+ CTLs, despite their initial ThPOK expression and TH-lineage commitment or the MHC-class II restriction of their TCRs.
The differentiation of CD4+ T lymphocytes to CTL instead of suppressing them and prevent them from become inflammatory TH1 and TH17 cells, makes teleological sense at epithelial interfaces, where a rapid elimination of infected cells is important not only to resist the initial invasion of intracellular pathogens, but also to limit or prevent excessive infiltration of systemically activated cytokine secreting cells that could jeopardize the integrity of the mucosal barrier38. In addition, the cytotoxic function of the CD4+ CTLs may also contribute to reducing the infection and inflammation by eliminating infected migratory DCs that could further prime naïve cells or mediate systemic spreading of the pathogen.
The MHC class II restriction of CD4+ CTLs might render these lymphocytes capable of restraining viral infections tropic for MHC class II+ target cells, including infected MHC class II+ epithelial cells39, Epstein–Barr virus B cells40 or Human immunodeficiency virus ((HIV)-infected human CD4+ T cells41. These T cells might also be critical for protection against viruses, including HIV and Cytomegalovirus, that have developed mechanisms to escape surveillance by MHC class I-restricted CD8+ CTLs40,42 or under conditions where the CD8 CTL-control response becomes exhausted as is the case in many chronic infections. In addition, like CD8αβ CTLs, CD4+ CTL responsiveness to IL-15 might provide a means to promote their antigen-specific or bystander protective capacity against a variety of pathogens or for enhancing the efficacy of adoptive cellular anti-tumor therapies.
It is however also possible, in inflammatory disorders involving IL-15, such as celiac disease or virally induced inflammatory conditions32,43, that CD4+ CTLs may increasingly become pathogenic effector cells that harmfully destroy their target tissue and recruit other immune cell types, thereby contributing to inflammatory pathogenesis. In the context of celiac disease, it has been difficult to connect the known pathogenic factors, including dietary antigens, MHC class II association, IL-15, and TCR-dependent and -independent killer cells18,36. Our finding, that diet-antigen-induced MHC class II restricted CD4+ CTLs become potent cytotoxic effector cells, producing large amounts of IFN-γ and TNF, when stimulated by their cognate antigen in the presence of IL-15, may form the long sought “missing link” that ties together the genetic and environmental factors involved in the pathogenesis of celiac disease.
In summary, the results here identify CD4+ CTLs as distinct CD4+ effector cells, defined by their post-thymic loss of the TH master transcription factor ThPOK and the reciprocal functional differentiation of these MHC class II-restricted T lymphocytes to CTLs. The data also identify the Thpok-silencer as the molecular switch, which drives the unique post-thymic reprogramming of these CD4+ effector cells. The unexpected plasticity of mature CD4+ T cells to differentiate into CTLs expands the functional ability of MHC class II-restricted CD4+ effector cells to also include direct protective functions and provides the immune system with an alternative protective mechanism, although the findings here also demonstrate that these CD4+ CTLs may be central in driving the immune pathology associated with inflammatory MHC class II-restricted T cells.
CD4+ effector cells with cytolytic functions have been known for a long time and they have frequently been associated with beneficial as well as pathological immune responses. Nevertheless, progress in directly linking these cells to these events or targeting them as part of immune therapies, has been extremely difficult mainly because they could not be separated from classical TH effector cells. Therefore, the new insights reported here, defining CD4+ CTLs as a distinct subset of effector cells and identifying the mechanism that leads to the unique differentiation of these cells, provide much needed information to finally move forward the field of CD4+ CTLs and to specifically target these cells to enhance protective immunity or prevent or treat inflammatory diseases and immune pathology.
METHODS
Mice
C57BL/6 CD45.1 (Ly5.1) and CD45.2 (Ly5.2), OT-II TCR-transgenic, Rag1−/−, Il7ra−/− and Foxp3eGFP mice (all on a C57BL/6 background) were purchased from The Jackson Laboratories and CD4-Cre and C57BL/6 mice were obtained from Taconic Farms. Swiss-Webster and C57BL/6 GF and conventionally raised specific pathogen-free (SPF) mice were purchased from Taconic Farms or from University of North Carolina Gnotobiotic Core, respectively. GF IQI mice were purchased from Japan CLEA Inc. Mice monocolonized with SFB were previously described44. ThPOK-gfp, ThpokSΔ/SΔ and ThPOKFH were previously described19,22. OT-II ThPOK-gfp mice were generated by breeding OT-II TCR transgenic mice with ThPOK-gfp reporter mice. ThpokSΔ/SΔ mice were crossed with B2m−/− mice (The Jackson Laboratories) to obtain ThpokSΔ/SΔ B2m−/− mice. Generation of Thpok silencer flox (ThpokSfl) allele in ES cells and ThpokSfl/Sfl mice will be described elsewhere. In brief, 826 bp sequences including the core Thpok silencer were flanked by two loxP site in the ThpokSfl allele. E8I-deficient mice were provided by D. Littman (New York University, NY). Mazr−/− mice were generated as previously described28 and crossed with ThPOK-gfp mice. To generate CD4-lineage specific Cre-transgenic mice (ThPOK-Cre mice), we inserted a 562 bp ThPOK silencer fragment into a mini transgene harboring the minimal CD4 enhancer-promoter. To target transgene expression, we inserted the IRES-hCD2 sequence downstream of the Cre cDNA cassette. Mice were maintained at the La Jolla Institute for Allergy and Immunology vivarium under SPF conditions and sentinel mice from the Rag1−/− mice colony were tested to be negative for Helicobacter spp. and C. rodentium. Animal care and experimentation were consistent with the NIH guidelines and were approved by the institutional Animal Care and Use Committees of the La Jolla Institute for Allergy and Immunology, the RIKEN Research Center for Allergy and Immunology, the Department of Immunology, Graduate School of Medicine, University of Tokyo and the Medical University of Vienna.
Antibodies and flow cytometry analysis
Fluorescent-dye-conjugated antibodies, including conjugates; FITC, PE, PercPcy5.5, APC, APCef-780, AF700, PB, PECy7, PE-Texas Red, V500 and antigens; CD4 (RM4-5), CD8α (53-6.7), CD8β (YTS156.7.7), CD25 (PC61.5), CD44 (IM7), CD45.2 (104), CD45 (30-F11), CD107 (1D4B), CD107b (ABL-93), Vα2 (B20.1), Vβ5.1 and Vβ5.2 (MR9-4), CD244.2 (2B4), granzyme B (GB11) and TCRβ (H57-597) were from BD-PharMingen or eBioscience or BD-Biosciences or Biolegend or Invitrogen. Anti-mouse CD16/32 used for Fc receptor blocking was produced in our laboratory. In some experiments LIVE/DEAD Fixable Yellow dead cell stain kit (Invitogen) was used to analyze only LIVE cell events. Flow cytometry data were acquired on a LSR-II instrument (Becton Dickinson) and analysed using FlowJo software package (Tri-Star). Doublets were excluded by Fsc-area versus Fsc-width and Ssc-area versus Ssc-width. Intracellular staining of Foxp3 and Granzyme B was conducted using Foxp3 Mouse Regulatory T cell Staining Kit (eBioscience). For flow cytometric analysis of cytokine-secreting cells, cell populations were first stained with antibodies against the indicated cell surface markers, followed by permeabilization in Fix/Perm buffer, and intracellular staining in Perm/Wash buffer (BD PharMingen).
Preparation of intraepithelial and lamina propria lymphocytes
Intraepithelial and lamina propria lymphocytes were isolated as previously described45.
Culture of intraepithelial lymphocytes
Intraepithelial lymphocytes enriched by anti-CD45 microbeads (Miltenyi Biotec Inc.) or FACS Aria sorted, were cultured in RPMI medium in the presence of recombinant mouse IL-2 (BD Biosciences) and recombinant human IL-15 (R&D Systems Inc. at 37 °C with 5% CO2.
Cohousing and Microbiota Reconstitution
Cohousing and microbiota reconstitutions were performed as described previously30.
Bacterial infection of mice
Wild-type C. rodentium strain DBS100 rendered resistant to chloramphenicol was obtained and used in all studies46. Bacteria were grown overnight in 3 ml Luria-Bertani (LB) broth with shaking at 37 °C and bacterial titers were determined after each experiment by serial dilution. Mice were regularly infected by oral gavage with 0.5~1.5 × 109 C.F.U. of C. rodentium (depending on experiment settings) and were sacrificed at 14 days post infection.
Microarray and qRT–PCR
RNA was prepared from sorted IEL populations using a FACS Aria cell sorter flow cytometer (Becton Dickinson). For microarray analysis, RNA was labeled and hybridized to GeneChip Mouse Genome 430 2.0 arrays according to the Affymetrix protocols. RNA was prepared from sorted IEL populations using a FACS Aria cell sorter flow cytometer (Becton Dickinson). Data were analyzed in GeneSpring GX10. Complementary DNAs (cDNAs) were synthesized from RNA samples extrated with TRIZol (Invitrogen) using an iScript cDNA Synthesis kit (BIO-RAD). Real-time RT-PCR was performed with the Roche 480 real-time PCR system. Values were normalized by the amount of L32 in each sample. Primer sequences are reported under Primers for qPCR. For Illumina microarray analysis, RNA was labeled and hybridized to Illumina MouseRef-8 v2.0 Expression BeadChips array according to the Illumina protocols. Data was analyzed in GeneSpring GS v12.2. The heat-maps were generated with the heatmap.2 function of R language. The inputs are base 2 log transformed normalized microarray probe set intensities. For genes with multiple probe sets on the array, the average intensities were used to produce heat-map. Normalized values were used for unsupervised analysis using gene spring and gene ontology in IPA (ingenuity pathway analysis). This analysis showed that major changes were related to T cell differentiation and activation genes. Based on these data a supervised analysis of clusters of genes (or families) that most represented differences between the groups was performed and shown. The full data sets were submitted to the GEOArchives.
Adoptive transfer model
5 × 105 sorted naïve (CD4+CD45RBhi) T cells were transferred into Rag1−/−mice, as previously described45. Sorting purity of naive cells was higher than 99.7% and usually above 99.9%. 4 to 8 weeks after transfer, donor cells were isolated from various tissues of the Rag1−/− recipient mice and analyzed as described.
Generation of diet antigen induced OT-II TCR transgenic CD4+ IELs
For naïve OT-II Thpok-GFP+ CD4 isolation, splenocytes were enriched for CD4+ cells by positive selection using anti-CD4 microbeads according to the manufacturer’s instructions (Miltenyi Biotec) followed by sorting for CD44loCD62LhiVα2+Vβ5+Thpok-GFP+ CD4+ cells using a FACS Aria cell sorter. Naïve OT-II Thpok GFP+ CD4+ cells (1 × 106) were transferred into the Rag1−/− recipients via retro-orbital route. To activate the antigen-specific OT-II T cells, these Rag1−/− recipients were fed an ovalbumin-containing diet for 4 weeks.
ChIP and tiling array
Chip-on-chip assay was performed as previously described19. In brief, 10 × 106 CD4 SP thymocytes from FH-ThPOK transgenic mice were used to isolate chromatin DNA for chromatin immuno-precipitation with anti-Flag and amplified twice by ligation-mediated PCR according to the manufacture’s protocol (Agilent). Custom microarray generated by Agilent that tiled through the murine Cd8 locus by means of 60-nucleotide probes were used. Probe hybridization and scanning of oligonucleotide array data were done according to manufacturer’s protocol (Agilent). Feature Extraction software and ChIP Analytics software (Agilent) were used for data analysis.
Chromatin immunoprecipitation (ChIP)
Cells were fixed with a 10% formaldehyde solution for 10 min, followed by cell lysis and sonication to solubilize and shear cross-linked chromatin DNA. The resulting whole cell extract was incubated with anti-mouse Ig or anti-rabbit Ig beads (Invitrogen) and the appropriate antibody, washed and bound complexes were eluted from the beads. Cells were fixed with a 10% formaldehyde solution for 10 min, followed by cell lysis and sonication to solubilize and shear cross-linked chromatin DNA. The resulting whole cell extract was incubated overnight at 4 °C with anti-mouse Ig or anti-rabbit Ig beads (Invitrogen) and the appropriate antibody. Beads were then washed 5 times with RIPA buffer and once with TBS. Bound complexes were eluted from the beads and cross-linking was reversed by overnight incubation at 65 °C. The primers used are as follows: E8I-forward 5′-CAA TGC GAA TGT GAC TCA AG-3′, E8I-reverse 5′-TAA TGC GGT GTG ATC AGT ATG-3′, Thpok silencer-forward 5′-TGG TTT CGA GAC TGG CTG GT-3′ and Thpok silencer -reverse 5′-GAC CGA GGA GCT GCT TTC AG-3′.
Retroviral transduction of CD4 T cells
Transduction of Thpok, Thpok-HD or Cre retroviral vectors into primary CD4 T cells was performed as previously described47. Thpok vectors were a gift from D. Kappes (Fox Chase Cancer Center, PA). The transfected cells were purified using a FACS Aria flow cytometer, and 1–2 × 106 cells were transferred into Rag1−/− mice. Four to five weeks after transfer, tissues were recovered from recipient mice for flow cytometery analyses.
Primers for qPCR
GATA-3-forward 5′-AGG ATG TCC CTG CTC TCC TT-3′, GATA-3-reverse 5′-GCC TGC GGA CTC TAC CAT AA-3′, ThPOK-forward 5′-ATG GGA TTC CAA TCA GGT CA-3′, ThPOK-Reverse 5′-TTC TTC CTA CAC CCT GTG CC-3′, T-bet-forward 5′-ATC CTG TAA TGG CTT GTG GG-3′, T-bet-Reverse 5′-TCA ACC AGC ACC AGA CAG AG-3′, CRTAM-forward 5′-TTA GAG TGA GCG TTT GGC CT-3′, CRTAM-Reverse 5′-GGG AGT CCT CAG TTT GCT GT-3′, RORγT-forward 5′-CCG CTG AGA GGG CTT CAC-3′, RORγT-Reverse 5′-TGC AGG AGT AGG CCA CAT TAC A-3′, L32 -forward 5′-GAA ACT GGC GGA AAC CCA-3′, L32 -Reverse 5′-GGA TCT GGC CCT TGA ACC TT-3′, IL23R-forward 5′-TTC AGA TGG GCA TGA ATG TTT CT-3′, IL23R--Reverse 5′-CCA AAT CCG AGC TGT TGT TCT AT-3′, IL17A-forward 5′-TGA GAG CTG CCC CTT CAC TT-3′, IL17A-Reverse 5′-ACG CAG GTG CAG CCC A-3′, IL-22-forward 5′-CAG GAG GTG GTA CCT TTC CTG A-3′, IL-22-Reverse 5′- TCT GGT CGT CAC CGC TGA T-3′, IL-2-forward 5′-CCT GAG CAG GAT GGA GAA TTA CA-3′, IL-2-Reverse 5′-TCC AGA ACA TGC CGC AGA G-3′, IL-4 -forward 5′-ACA GGA GAA GGG ACG CCA T-3′, IL-4-Reverse 5′- GAA GCC CTA CAG ACG AGC TCA-3′, IL-13-forward 5′-AGA CCA GAC TCC CCT GTG CA-3′, IL-13-Reverse 5′-TGG GTC CTG TAG ATG GCA TTG-3′, GRANZYME A-forward 5′-ATT CCT GAA GGA GGC TGT GAA-3′, GRANZYME A-Reverse 5′-GCA GGA GTC CTT TCC ACC AC-3′, GRANZYME B-forward 5′-GCC CAC AAC ATC AAA GAA CAG-3′, GRANZYME B-Reverse 5′-AAC CAG CCA CAT AGC ACA CAT-3′, CD8α-forward 5′-ACT GCA AGG AAG CAA GTG GT-3′, CD8α-Reverse 5′-CAC CGC TAA AGG CAG TTC TC-3′, IFNγ- forward 5′- ATG AAC GCT ACA CAC TGC ATC -3′, IFNγ-reverse 5′- CCA TCC TTT TGC CAG TTC CTC -3′.
CD107 mobilization assay
The degranulation activity of cytolytic cells was measured as described48. Briefly, enriched polyclonal IELs were activated with plate-bound anti-TCRβ (2 μg/ml) for 4 h in the presence of CD107 antibody and Brefeldin-A in 96-well U bottom plates at 37 °C and 5% CO2. OT-II TCR transgenic IELs were co-cultured with LPS overnight-treated LB27.4 (Mouse B cell lymphoma) APCs in 5:1 ratio and MHC-II class II specific OVA323-339 peptide or MHC-I specific SIINFEKL peptide for 6 h in the presence of CD107 antibody and Brefeldin-A in 96-well V bottom plates at 37 °C and 5% CO2. In some cases, sorted IEL were cultured for 3 days in the presence of recombinant mouse IL2 (BD Biosciences, USA) and recombinant human IL-15 (R&D systems Inc.). Following stimulation, CD107 and cytokine expression was assessed by flow cytometry.
In vitro cytotoxicity assay
Cytotoxicity by anti-TCRβ cross-linked T cells was measured 6 h after coculturing with P815 target cells at 37 °C and 5% CO2 using the CytoTox 96 Non-Radioactive cytotoxicity assay kit (Promega). The % cytotoxicity was calculated based on the release of lactate dehydrogenase. In some cases, cultured IELs were used.
Statistical Analysis
Data are presented as mean ± SEM. Comparisons between groups used the Student’s t test assuming two-tailed distributions unless otherwise stated, with an alpha level of 0.01–0.05. In some samples Nonparametric two tailed Mann-Whitney test or ANOVA and Bonferroni as post-test were used.
Supplementary Material
Acknowledgments
We thank A. Larange and I. Vicente-Suarez for helpful discussions and critical reading of the manuscript. M. Cheroutre for her contribution in conceiving the project. Y. Wang-Zhu for preparing TL-tetramers and breeding Rag1−/− mice, C. Kim and K. Van Gunst for cell sorting. D. Littman ((New York University, NY) for the E8I-deficient mice and D. Kappes (Fox Chase Cancer Center, PA) for the Thpok vectors. This work was supported by NIH grants RO1 AI050265 and DP1 OD006433 (H.C.), RCAI international collaboration awards (H.C. and I.T.), NIH fellowship F32 DK082249 (J.-W.S.), Ter Meulen fund KNAW fellowship (F.v W.), CCFA Career Development Award (D.M.) and PO1 DK46763 (M.K. and H. C.).
Footnotes
Author contributions
H. C., I. T., M.K., D. M. and M. M. H. conceived the project. M.M.H., D.M. and F. v W generated the phenotypic and functional data. I. T., S. M, Y. N. and C. M. generated the fate mapping- and Thpok silencer deletion data and performed the ChIP assays. R.S. performed the transfections. Y.H. provided the IL-7R KO data. B. S. R., M. D. and A. A. generated the gene arrays. G. K, F. L. and C. L. performed transfers and analyzed mice. J-W. S and D.M. performed the Citrobacter infections. K. A. and K. H. reconstituted germ-free mice. S. S. generated the data related to the role of MAZR. Y. P. analyzed the Myd88−/− mice. P. W., D. M., F. v W., B. S. R. and H.C. analyzed the gene array data. T. N. and W. E. provided expertise. M. K. provided conceptual advice and helped with data analysis and the writing of the manuscript. I. T. and H. C. generated the concepts, designed experiments, analyzed data and wrote the manuscript with all authors contributing to the writing and providing advice.
Competing Interests statement.
The authors declare no competing financial interests.
References
- 1.Aliahmad P, Kadavallore A, de la Torre B, Kappes D, Kaye J. TOX is required for development of the CD4 T cell lineage gene program. J Immunol. 2011;187:5931–5940. doi: 10.4049/jimmunol.1101474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hernandez-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 2003;19:83–94. doi: 10.1016/s1074-7613(03)00176-6. [DOI] [PubMed] [Google Scholar]
- 3.Pai SY, et al. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- 4.He X, et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 5.Sun G, et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 6.Wang L, et al. The zinc finger transcription factor Zbtb7b represses CD8-lineage gene expression in peripheral CD4+ T cells. Immunity. 2008;29:876–887. doi: 10.1016/j.immuni.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taniuchi I, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 8.Woolf E, et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci U S A. 2003;100:7731–7736. doi: 10.1073/pnas.1232420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Appay V, et al. Characterization of CD4(+) CTLs ex vivo. J Immunol. 2002;168:5954–5958. doi: 10.4049/jimmunol.168.11.5954. [DOI] [PubMed] [Google Scholar]
- 10.Appay V. The physiological role of cytotoxic CD4(+) T-cells: the holy grail? Clin Exp Immunol. 2004;138:10–13. doi: 10.1111/j.1365-2249.2004.02605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown DM. Cytolytic CD4 cells: Direct mediators in infectious disease and malignancy. Cell Immunol. 2010;262:89–95. doi: 10.1016/j.cellimm.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marshall NB, Swain SL. Cytotoxic CD4 T cells in antiviral immunity. J Biomed Biotechnol. 2011 doi: 10.1155/2011/954602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guy-Grand D, Malassis-Seris M, Briottet C, Vassalli P. Cytotoxic differentiation of mouse gut thymodependent and independent intraepithelial T lymphocytes is induced locally. Correlation between functional assays, presence of perforin and granzyme transcripts, and cytoplasmic granules. J Exp Med. 1991;173:1549–1552. doi: 10.1084/jem.173.6.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sydora BC, et al. Intestinal intraepithelial lymphocytes are activated and cytolytic but do not proliferate as well as other T cells in response to mitogenic signals. J Immunol. 1993;150:2179–2191. [PubMed] [Google Scholar]
- 15.Sasahara T, Tamauchi H, Ikewaki N, Kubota K. Unique properties of a cytotoxic CD4+CD8+ intraepithelial T-cell line established from the mouse intestinal epithelium. Microbiol Immunol. 1994;38:191–199. doi: 10.1111/j.1348-0421.1994.tb01764.x. [DOI] [PubMed] [Google Scholar]
- 16.Nascimbeni M, Shin EC, Chiriboga L, Kleiner DE, Rehermann B. Peripheral CD4(+)CD8(+) T cells are differentiated effector memory cells with antiviral functions. Blood. 2004;104:478–486. doi: 10.1182/blood-2003-12-4395. [DOI] [PubMed] [Google Scholar]
- 17.Strutt TM, McKinstry KK, Swain SL. Functionally diverse subsets in CD4 T cell responses against influenza. J Clin Immunol. 2009;29:145–150. doi: 10.1007/s10875-008-9266-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DePaolo RW, et al. Co-adjuvant effects of retinoic acid and IL-15 induce inflammatory immunity to dietary antigens. Nature. 2011;471:220–224. doi: 10.1038/nature09849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muroi S, et al. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat Immunol. 2008;9:1113–1121. doi: 10.1038/ni.1650. [DOI] [PubMed] [Google Scholar]
- 20.Cheroutre H. Starting at the beginning: new perspectives on the biology of mucosal T cells. Annu Rev Immunol. 2004;22:217–246. doi: 10.1146/annurev.immunol.22.012703.104522. [DOI] [PubMed] [Google Scholar]
- 21.Burkett MW, Shafer-Weaver KA, Strobl S, Baseler M, Malyguine A. A novel flow cytometric assay for evaluating cell-mediated cytotoxicity. J Immunother. 2005;28:396–402. doi: 10.1097/01.cji.0000165357.11548.6d. [DOI] [PubMed] [Google Scholar]
- 22.Setoguchi R, et al. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 2008;319:822–825. doi: 10.1126/science.1151844. [DOI] [PubMed] [Google Scholar]
- 23.He X, Park K, Kappes DJ. The role of ThPOK in control of CD4/CD8 lineage commitment. Annu Rev Immunol. 2010;28:295–320. doi: 10.1146/annurev.immunol.25.022106.141715. [DOI] [PubMed] [Google Scholar]
- 24.Ellmeier W, Sunshine MJ, Losos K, Hatam F, Littman DR. An enhancer that directs lineage-specific expression of CD8 in positively selected thymocytes and mature T cells. Immunity. 1997;7:537–547. doi: 10.1016/s1074-7613(00)80375-1. [DOI] [PubMed] [Google Scholar]
- 25.Kennedy J, et al. A molecular analysis of NKT cells: identification of a class-I restricted T cell-associated molecule (CRTAM) J Leukoc Biol. 2000;67:725–734. doi: 10.1002/jlb.67.5.725. [DOI] [PubMed] [Google Scholar]
- 26.Boles KS, Barchet W, Diacovo T, Cella M, Colonna M. The tumor suppressor TSLC1/NECL-2 triggers NK-cell and CD8+ T-cell responses through the cell-surface receptor CRTAM. Blood. 2005;106:779–786. doi: 10.1182/blood-2005-02-0817. [DOI] [PubMed] [Google Scholar]
- 27.Boles KS, Stepp SE, Bennett M, Kumar V, Mathew PA. 2B4 (CD244) and CS1: novel members of the CD2 subset of the immunoglobulin superfamily molecules expressed on natural killer cells and other leukocytes. Immunol Rev. 2001;181:234–249. doi: 10.1034/j.1600-065x.2001.1810120.x. [DOI] [PubMed] [Google Scholar]
- 28.Sakaguchi S, et al. The zinc-finger protein MAZR is part of the transcription factor network that controls the CD4 versus CD8 lineage fate of double-positive thymocytes. Nat Immunol. 2010;11:442–448. doi: 10.1038/ni.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park JH, et al. Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat Immunol. 2010;11:257–264. doi: 10.1038/ni.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivanov II, et al. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheroutre H, Lambolez F. Doubting the TCR coreceptor function of CD8alphaalpha. Immunity. 2008;28:149–159. doi: 10.1016/j.immuni.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 32.Mention JJ, et al. Interleukin 15: a key to disrupted intraepithelial lymphocyte homeostasis and lymphomagenesis in celiac disease. Gastroenterology. 2003;125:730–745. doi: 10.1016/s0016-5085(03)01047-3. [DOI] [PubMed] [Google Scholar]
- 33.Ebert EC. Interleukin 15 is a potent stimulant of intraepithelial lymphocytes. Gastroenterology. 1998;115:1439–1445. doi: 10.1016/s0016-5085(98)70022-8. [DOI] [PubMed] [Google Scholar]
- 34.Hirose K, et al. Interleukin-15 may be responsible for early activation of intestinal intraepithelial lymphocytes after oral infection with Listeria monocytogenes in rats. Infect Immun. 1998;66:5677–5683. doi: 10.1128/iai.66.12.5677-5683.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye W, Young JD, Liu CC. Interleukin-15 induces the expression of mRNAs of cytolytic mediators and augments cytotoxic activities in primary murine lymphocytes. Cell Immunol. 1996;174:54–62. doi: 10.1006/cimm.1996.0293. [DOI] [PubMed] [Google Scholar]
- 36.Abadie V, Discepolo V, Jabri B. Intraepithelial lymphocytes in celiac disease immunopathology. Semin Immunopathol. 2012;34:551–566. doi: 10.1007/s00281-012-0316-x. [DOI] [PubMed] [Google Scholar]
- 37.Gangadharan D, et al. Identification of pre- and postselection TCRalphabeta+ intraepithelial lymphocyte precursors in the thymus. Immunity. 2006;25:631–641. doi: 10.1016/j.immuni.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 38.Cheroutre H, Lambolez F, Mucida D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat Rev Immunol. 2011;11:445–456. doi: 10.1038/nri3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hershberg RM, et al. Highly polarized HLA class II antigen processing and presentation by human intestinal epithelial cells. J Clin Invest. 1998;102:792–803. doi: 10.1172/JCI3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khanna R, et al. Class I processing-defective Burkitt’s lymphoma cells are recognized efficiently by CD4+ EBV-specific CTLs. J Immunol. 1997;158:3619–3625. [PubMed] [Google Scholar]
- 41.Ko HS, Fu SM, Winchester RJ, Yu DT, Kunkel HG. Ia determinants on stimulated human T lymphocytes. Occurrence on mitogen- and antigen-activated T cells. J Exp Med. 1979;150:246–255. doi: 10.1084/jem.150.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alcami A, Koszinowski UH. Viral mechanisms of immune evasion. Immunol Today. 2000;21:447–455. doi: 10.1016/S0167-5699(00)01699-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McInnes IB, Gracie JA. Interleukin-15: a new cytokine target for the treatment of inflammatory diseases. Curr Opin Pharmacol. 2004;4:392–397. doi: 10.1016/j.coph.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 44.Ivanov II, et al. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 45.Mucida D, et al. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317:256–260. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 46.LeBlanc PM, et al. Caspase-12 modulates NOD signaling and regulates antimicrobial peptide production and mucosal immunity. Cell Host Microbe. 2008;3:146–157. doi: 10.1016/j.chom.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 47.Kimura M, et al. Regulation of Th2 cell differentiation by mel-18, a mammalian polycomb group gene. Immunity. 2001;15:275–287. doi: 10.1016/s1074-7613(01)00182-0. [DOI] [PubMed] [Google Scholar]
- 48.Betts MR, et al. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J Immunol Methods. 2003;281:65–78. doi: 10.1016/s0022-1759(03)00265-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.