Abstract
Chromatin modifications are sensitive to environmental and nutritional stimuli. Abnormalities in epigenetic regulation are associated with metabolic disorders such as obesity and diabetes that are often linked with defects in oxidative metabolism. Here, we evaluated the potential of class-specific synthetic inhibitors of histone deacetylases (HDACs), central chromatin-remodeling enzymes, to ameliorate metabolic dysfunction. Cultured myotubes and primary brown adipocytes treated with a class I–specific HDAC inhibitor showed higher expression of Pgc-1α, increased mitochondrial biogenesis, and augmented oxygen consumption. Treatment of obese diabetic mice with a class I– but not a class II–selective HDAC inhibitor enhanced oxidative metabolism in skeletal muscle and adipose tissue and promoted energy expenditure, thus reducing body weight and glucose and insulin levels. These effects can be ascribed to increased Pgc-1α action in skeletal muscle and enhanced PPARγ/PGC-1α signaling in adipose tissue. In vivo ChIP experiments indicated that inhibition of HDAC3 may account for the beneficial effect of the class I–selective HDAC inhibitor. These results suggest that class I HDAC inhibitors may provide a pharmacologic approach to treating type 2 diabetes.
Abnormalities in epigenetic regulation have been associated with multiple metabolic disorders, such as cardiovascular disease, obesity, and type 2 diabetes (1,2). Histone deacetylases (HDACs) regulate gene transcription by compacting chromatin and making it less accessible to transcriptional activators. Eighteen mammalian HDACs have been described, divided into four classes. While class I HDACs (HDACs 1, 2, 3, and 8) are broadly expressed and localize to the nucleus (3), class II HDACs (HDACs 4, 5, 6, 7, 9, and 10) can shuttle between cytoplasm and nucleus and exhibit minimal histone deacetylase activity (4,5). Class III HDACs (sirtuins) are good metabolic sensors (6); little is known about HDAC11, the single class IV HDAC in mammals (7).
Class II HDACs have been associated with the regulation of cardiac and skeletal muscle physiology (8,9). Genetic deletion of class II HDACs in skeletal muscle increases myocyte enhancer factor (MEF)2 activity and promotes the formation of slow-twitch type I fibers, rich in mitochondria and with high oxidative capacity. Less is known about the role of class I HDACs in skeletal muscle physiology, but cardiac-specific deletion of HDAC3 also results in increased expression of fatty acid oxidation and oxidative phosphorylation genes, though HDAC3 deletion is also associated with cardiac hypertrophy with deleterious consequences (10,11).
A recent report showed that sodium butyrate, an HDAC pan-inhibitor, has beneficial effects in mice with diet-induced obesity (12). To explore the promise of HDACs as targets in metabolic disorders, here we evaluated the therapeutic potential of selective class I and II HDAC synthetic inhibitors in obese diabetic mice. We found that class I, but not class II, HDAC inhibitors promote oxidative metabolism in db/db mice, reduce body weight, increase energy expenditure, and enhance insulin sensitivity, suggesting that class I HDAC inhibitors may be useful in conditions associated with suppressed oxidative metabolism, such as type 2 diabetes.
RESEARCH DESIGN AND METHODS
Reagents.
Suberoyl anilide hydroxamic acid (SAHA) was from Cayman. MS275 and MC1568 were synthesized in-house. Anti–acetyl-H3, anti-HDAC1, anti-cytochrome C (CytC), and anti–rabbit IgG (Cell Signaling); anti–acetyl-tubulin, anti–α-tubulin, anti–β-actin, anti-Tfam, and anti–mouse IgG (Sigma-Aldrich); anti-HDAC3 and anti–peroxisome proliferator–activated receptor (PPAR)γ (Santa Cruz); and anti-HDAC3 (ChIP), anti–uncoupling protein (UCP)1, and anti-LCAD (Abcam) antibodies were used. Anti-electron transfer chain complexes (MitoProfile Total OXPHOS Rodent WB Antibody Cocktail) were from Mitosciences.
Cell culture.
C2C12 cells were maintained in Dulbecco’s modified Eagle’s medium–10% FBS and differentiated in Dulbecco’s modified Eagle’s medium–2% horse serum. Cells were treated with SAHA (5 μmol/L), MS275 (5 μmol/L), MC1568 (5 μmol/L), or vehicle for 60 h. No toxicity was detected. Small interfering RNAs (Sigma-Aldrich) were transfected (30 nmol/L) into C2C12 myoblasts for 48 h prior to analysis. Adenoviruses expressing control or PPARγ coactivator (Pgc)-1α shRNAs were used to infect C2C12 myotubes at day 4 of differentiation. Myotubes were treated 24 h after infection and analyzed 16 h later. Primary brown preadipocytes were prepared from P0–P4 B6 mice as previously described (13).
Analysis of mitochondria.
C2C12 myotubes were stained with 200 nmol/L MitoTracker Green FM or 400 nmol/L MitoTracker Red CM-H2XRos (Invitrogen) for 30 min, 37°C, and then stained with Hoechst 33258. Fluorescence was measured with an EnVision (Perkin-Elmer). For electron microscopy, cells were processed as previously described (14). Ultrathin sections (200 nm) were evaluated using a JEM 1010 TEM (Jeol). Bioptic fragments from gastrocnemius (2 × 2 mm) were fixed in 3% glutaraldehyde in 0.1 mol/L Sorensen buffer, pH 7.4, overnight at 4°C and Araldite embedded. Two micron semithin sections were stained with toluidine blue. Ultrathin sections (60 nm) were stained with lead citrate and uranyl acetate and examined with a Jeol CX100 TEM (Jeol).
Gene expression and chromatin immunoprecipitation.
Real time quantitative PCR (qPCR) was performed as previously described (15). For measurement of mitochondrial DNA, genomic qPCR was performed on 12S mitochondrial DNA and normalized to a nuclear Cyp7a1 sequence. Primer sequences are available upon request. Microarray analysis was performed by Genopolis (Milan, Italy). Differentially expressed genes were identified using Linear Models for Microarray Data. Chromatin immunoprecipitations (ChIPs) were performed as previously described (16) on C2C12 myotubes treated for 60 h. For in vivo ChIP, tissues were minced and fixed in 0.5% paraformaldehyde for 10 min and processed as previously described (17).
Oxygen consumption.
Cells (5 × 105) were detached; resuspended in PBS containing 25 mmol/L glucose, 1 mmol/L sodium pyruvate, and 2% fatty acid-free BSA; and transferred to a Clark-type oxygen electrode chamber at 37°C. After recording of basal respiration, uncoupled respiration was determined with oligomycin (2.5 μg/mL), and maximal respiration was induced with 2.4 μmol/L carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone. Data were normalized to protein content.
Animal studies.
Nine-week-old male C57BLKS/J-Leprdb/db mice (The Jackson Laboratory) were randomized into groups according to glucose levels and body weight and treated with 25 mg/kg i.p. SAHA, 10 mg/kg i.p. MS275, 6.5 mg/kg i.p. MC1568, or vehicle every other day. Compounds were dissolved in DMSO. Doses were based on pilot experiments (3–5 db/db mice per group). For blood chemistry analysis, animals were fasted for 16 h. Glycemia was determined using an Accu-Chek glucometer (Roche); commercial kits were used for other parameters (plasma triglyceride, Sentinel; NEFA-HR, Wako; ALT or AST Reagent, Teco Diagnostics; and Diagnostic Cholesterol, ABX Pentra). Insulin levels were determined with an AlphaLISA Immunoassay (Perkin-Elmer). Cholesterol distribution in lipoprotein fractions was determined by fast-protein liquid chromatography (FPLC). Lipids in vastus lateralis were extracted with the Folch method (18). Total triglycerides were quantified as described above using [3H]-triolein as a standard. In glucose tolerance tests, mice were fasted for 16 h, and glucose levels were determined at the indicated times after injection of 2 g/kg i.p. glucose. At day 15 of treatment, oxygen consumption, heat production, and activity were measured over 3 days using the Oxymax System (Columbus Instruments). Mice were individually housed and allowed to acclimate with free access to food and water. Data were analyzed using the OxymaxWin, version 3.32, software as previously described (19). At day 20 of treatment, mice underwent a cold challenge (4°C for 2 h); rectal body temperature was measured every 20 min. Studies were conducted in accordance with European Commission regulations (European Union Directive 63/2010) and Italian regulations (decree no. 116, 27 January 1992) and with approval of The Scripps Research Institute's Institutional Animal Care and Use Committee.
Magnetic resonance imaging.
At day 18 of treatment, mice were anesthetized and analyzed in a 4.7 Tesla Avance II magnetic resonance imaging (MRI) scanner (Bruker Corporation). After a gradient-echo scout, 16 axial 1-mm-thick T1-weighted slices were placed in the abdominal region spanning from kidneys to bladder inclusive. The field of view was 30 × 30 mm2 with a matrix of 128 × 128 pixels. Four averages of a spin echo sequence with time to echo 10 ms and time of repetition 400 ms were acquired in 3′25”. The slice immediately frontal with respect to the ilium bone was chosen for visceral fat estimation and was computed as follows: (fat area)/(slice area). Areas were measured with Photoshop (Adobe Systems).
Histology.
Skeletal muscle and adipose tissue were fixed with Carnoy solution/chloroform and embedded in paraffin, and 8-μm sections were stained with hematoxylin-eosin. For succinate dehydrogenase staining, slides were incubated for 30 min at 37°C in nitroblue tetrazolium–succinate solution. Quantification of dark fibers was performed counting multiple images (n = 14 for control, n = 6 for SAHA, and n = 16 for MS275) of nonconsecutive sections (four mice per group). Images were taken at ×20 magnification. For alkaline phosphatase staining, slides were incubated with borate buffer, pH 8.8, at 37°C for 1 h, fixed, and mounted. Esterase staining was performed by incubating slides at 37°C for 1 h in pararosaniline and sodium nitrite solution. For mitochondrial staining, white adipose tissue (WAT) sections were incubated with 200 nmol/L MitoTracker Green FM for 10 min and then washed and mounted. Fluorescence intensity was quantified using ImageJ.
Immunohistochemistry.
Sections of WAT (8 μm) were deparaffinized, and antigen retrieval was performed with 0.05 mol/L NH4Cl for 30 min, room temperature. Endogenous peroxidase activity was blocked with 1% H2O2 for 20 min. Blocking was performed in 1% BSA–0.1% Triton for 1 h. Anti-Mac1 (AbD Serotec) or anti-UCP1 (Abcam) was applied (1:300) overnight at 4°C. Incubation with biotinylated secondary antibodies (1:3000) followed. Histochemical reactions were performed using diaminobenzidine and sections counterstained with hematoxylin-eosin. Crown-like structures (Mac1 positive) were counted from multiple images (n = 14 for control, n = 9 for SAHA, and n = 16 for MS275) of nonconsecutive sections (four mice per group) taken at ×10 magnification.
Statistics.
Statistical analyses were performed with Student t test or one-way ANOVA followed by Dunnett posttest as indicated, using Prism 5.0b (GraphPad Software).
RESULTS
Inhibition of class I HDACs promotes mitochondrial biogenesis.
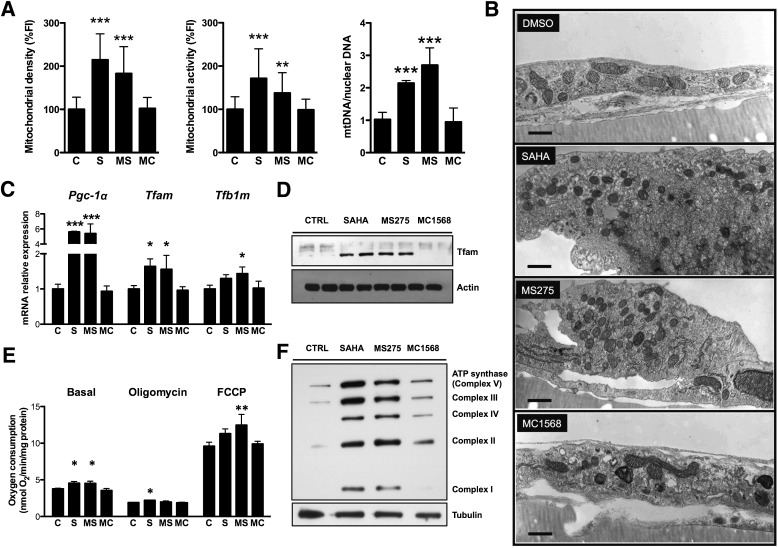
For evaluation of whether synthetic class I or class II HDAC inhibitors can enhance mitochondrial function, C2C12 myotubes were treated with the HDAC pan-inhibitor SAHA, a class I HDAC selective inhibitor (MS275), or a class II HDAC selective inhibitor (MC1568) (20–22). Concentrations were chosen based on dose-response curves (0.5–50 μmol/L) used to determine an effective, nontoxic, and selective concentration for each inhibitor based on the hyperacetylated state of histone H3 (a class I HDAC substrate) and α-tubulin (a class II HDAC substrate) (Supplementary Fig. 1A). After 60 h of treatment, global or class I HDAC inhibition resulted in increased mitochondrial density and activity, while inhibition of class II HDACs had no effect on these parameters (Fig. 1A and Supplementary Fig. 1B). These increases were accompanied by robust increases in mitochondrial DNA (Fig. 1A). Transmission electron microscopy confirmed that SAHA and MS275 induced mitochondrial biogenesis. Treatment with SAHA or MS275 resulted in an increase in mitochondrial density and greater electron opacity of the matrix typical of metabolically active cells. Cells treated with the class II HDAC inhibitor MC1568 showed mitochondria similar to those of controls (Fig. 1B).
FIG. 1.
Inhibition of class I HDACs promotes mitochondrial biogenesis and oxidative metabolism in C2C12 myotubes. A: Quantification of mitochondrial density, activity, and mitochondrial DNA (mtDNA) in C2C12 myotubes after treatment with 5 μmol/L SAHA, 5 μmol/L MS275, 5 μmol/L MC1568, or vehicle. Fluorescence intensity (FI) of mitochondrial probes was normalized to a nuclear stain (Hoechst 33258). B: Representative electron microphotographs of ultrathin sections of C2C12 monolayers. The ultrastructural appearance of mitochondria in vehicle-treated cells was characterized by a dense matrix and well-organized cristae with dilated intracristae spaces in the typical condensed conformation of metabolically active cells (44). Treatment with SAHA or MS275 resulted in an increase in mitochondrial density and greater electron opacity of the matrix to the detriment of the development and organization of cristae. Cells treated with the class II HDAC inhibitor MC1568 showed rod-like mitochondria similar to those of controls (bars = 500 nm). C: Expression of genes associated with mitochondrial biogenesis (24 h) (C) and Western blot analysis of mitochondrial transcription factor A (Tfam) in C2C12 myotubes treated for 48 h with HDAC inhibitors (D). E: Measurement of oxygen consumption at the basal level and in the presence of oligomycin (2.5 μg/mL) or carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) (2.4 μmol/L) in C2C12 myotubes treated for 60 h with HDAC inhibitors. F: Western blot analysis of mitochondrial complexes I–V of the electron transfer chain in C2C12 myotubes treated with HDAC inhibitors for 48 h. Data are presented as means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control. C and CTRL, control; MC, MC1568; MS, MS275; S, SAHA.
Transcriptome analysis revealed that global or class I–selective HDAC inhibition increased expression of several key mitochondria-related transcription factors, such as Tfam, Tfb1m, and the coactivator Pgc-1α (Fig. 1C and D), as well as the levels of multiple genes involved in glucose and lipid metabolism (Supplementary Fig. 1D). These changes in gene expression and mitochondrial density translated to differences in oxidative metabolism, as global and class I–selective HDAC inhibitors induced a 20% increase in basal respiration (Fig. 1E). In the presence of oligomycin, only cells treated with SAHA showed a small but consistent increase (∼15%) in oxygen consumption, while MS275 treatment increased maximal respiratory capacity by ~30%. SAHA treatment showed a tendency to increase maximal respiratory capacity but to a lesser extent (∼20%). These changes were accompanied by corresponding increases in mitochondrial complex proteins (Fig. 1F). No differences were seen with the class II HDAC inhibitor. These results indicate that inhibition of class I HDACs reprograms myotubes toward a more oxidative state.
Class I–selective HDAC inhibitors ameliorate obesity and diabetes.
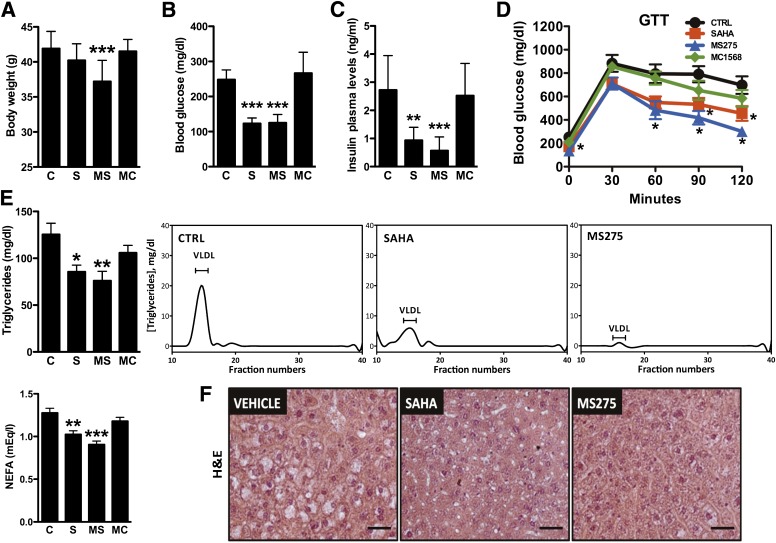
Next, we tested the physiologic relevance of HDAC inhibition in a model of obesity and diabetes, the db/db mouse. At the doses used (25 mg/kg SAHA, 10 mg/kg MS275, and 6.5 mg/kg MC1568 administered every other day for a 23-day period), the compounds reached skeletal muscle and retained their class-selective inhibitory activity (Supplementary Fig. 2A). Mice treated with MS275 showed a significant reduction of body weight (Fig. 2A and Supplementary Fig. 2B), in spite of similar food intake (Supplementary Fig. 2B). Interestingly, we observed a dramatic reduction of fasting glycemia, of circulating insulin, and of the homeostasis model assessment of insulin resistance index in animals treated with SAHA or MS275 but not in those treated with the class II HDAC inhibitor (Fig. 2B and C and Supplementary Fig. 2C). Moreover, global or class I–selective HDAC inhibition improved glucose clearance during glucose tolerance tests (Fig. 2D). Circulating triglycerides and nonesterified fatty acids were also decreased in SAHA and MS275 groups (Fig. 2E). MS275 completely cleared the lipids that accumulate in the liver of db/db mice, while SAHA had a significant but milder effect (Fig. 2F). The reduced hepatic steatosis was mirrored by decreased plasma transaminases, confirming that no toxic effects were observed with these compounds (Supplementary Fig. 2D and E). Interestingly, we did not observe any significant differences in hepatic gene expression or mitochondrial content, which suggests that the lack of hepatic steatosis is likely a reflection of the effect of MS275 in tissues other than liver (Supplementary Fig. 2F and G). MC1568 had no effect on any parameter, indicating that inhibition of class I HDACs underlies the observed improvements in metabolic profile.
FIG. 2.
MS275 ameliorates insulin resistance in db/db mice. A: Body weight of db/db mice treated with vehicle or HDAC inhibitors for 23 days (n = 10 per group). B and C: Fasting plasma glucose and insulin levels measured at day 23 of the treatment. D: Glucose tolerance test (GTT) performed on day 8 of treatment. E: Fasting serum triglycerides, nonesterified fatty acids (NEFA), and FPLC profile of plasma triglycerides of db/db mice treated with vehicle, SAHA, MS275, or MC1568. (FPLC samples are pools of 10 mice.) F: Hematoxylin-eosin (H&E) stain of liver sections from db/db mice treated with HDAC inhibitors. Data are presented as means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control. C and CTRL, control; MC, MC1568; MS, MS275; S, SAHA. (A high-quality color representation of this figure is available in the online issue.)
Class I HDAC inhibitors induce oxidative metabolism in skeletal muscle.
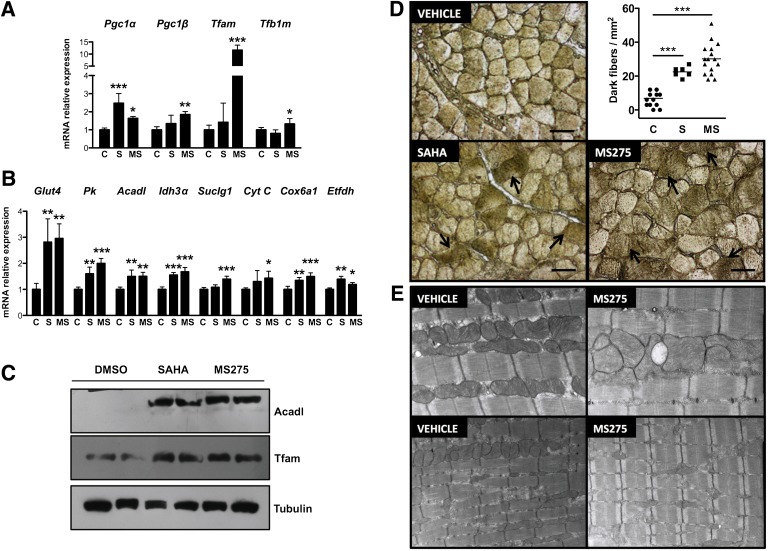
To explore the molecular basis of the beneficial effects of SAHA and MS275 on metabolic parameters, we measured the expression of metabolic genes in skeletal muscle. In gastrocnemius, SAHA and MS275 increased expression of transcription factors and cofactors that regulate mitochondrial function (e.g., Pgc-1α, Pgc-1β, Tfam, and Tfb1m) (Fig. 3A) and of genes involved in glucose (Glut4 and Pk) and lipid metabolism (Acadl), TCA cycle (Idh3α and Suclg1), and oxidative phosphorylation (CytC, Cox6a1, and Etfdh) (Fig. 3B). Similar effects were observed in the vastus lateralis and soleus (Supplementary Fig. 3A and D). Changes in gene expression translated to differences in protein levels (e.g., Tfam, Acadl [Fig. 3C]). Furthermore, succinate dehydrogenase staining demonstrated that mice treated with MS275, and to a lesser degree with SAHA, had an increased number of dark fibers, indicating greater oxidative capacity (Fig. 3D). Mitochondrial complex I and II proteins were also increased (Supplementary Fig. 3B). Electron microscopy provided suggestions of differences in mitochondrial content, but these were not conclusive (Fig. 3E). No changes were detected in ectopic lipid levels (Supplementary Fig. 3C), perhaps because the extreme obesity of db/db mice did not allow detection of modest changes. Absence of toxicity was confirmed by lack of increased alkaline phosphatase or esterase staining (Supplementary Fig. 4A). Notably, while Tnn1 mRNA levels increased in soleus, no concomitant changes in mRNA levels of contractile proteins characteristic of type I and II myofibers occurred in gastrocnemius or vastus lateralis (Supplementary Fig. 4B). These findings indicate that inhibition of class I HDACs in skeletal muscle contributes to ameliorating the phenotype of db/db mice at least in part by increasing expression of genes involved in fatty acid oxidation and glucose clearance.
FIG. 3.
Inhibition of class I HDACs promotes oxidative metabolism in skeletal muscle. mRNA expression levels of mitochondrial biogenesis–associated (A) and metabolic pathway (B) genes in skeletal muscle of db/db mice treated with HDAC inhibitors. C: Western blot analysis of Acadl and Tfam in skeletal muscle of db/db mice after treatment with HDAC inhibitors. D: Succinate dehydrogenase staining and quantification of dark fibers (arrows) in gastrocnemius sections (bars = 100 μm). E: Representative electron microphotographs of ultrathin sections of gastrocnemius from mice treated with vehicle or MS275 (magnification: upper panels, ×15,000; lower panels, ×8,000). Data are presented as means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control. C, control; MS, MS275; S, SAHA. (A high-quality color representation of this figure is available in the online issue.)
Inhibition of class I HDACs promotes energy expenditure in db/db mice.
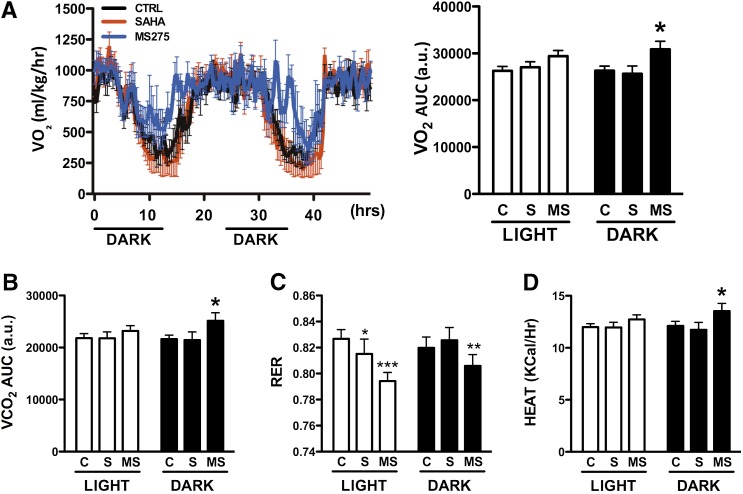
Since the oxidative pattern of gene expression induced by global and class I–selective HDAC inhibitors has been associated with increased energy expenditure, we assessed energy balance in db/db mice treated with SAHA or MS275. Animals treated for 15 days with the class I–selective HDAC inhibitor showed increased oxygen consumption and carbon dioxide release during the dark cycle, while only a trend for increased oxygen consumption was observed during the light cycle (Fig. 4A and B and Supplementary Fig. 5A). Nonetheless, these changes were sufficient to reduce the respiratory exchange ratio (RER) during both light and dark cycles, an indication that these mice use lipids preferentially as fuel (Fig. 4C). Heat production was also 12% greater in the MS275-treated group (Fig. 4D), while locomotor activity did not change (Supplementary Fig. 5B). SAHA had no significant effect on oxygen consumption, but treated mice showed a mild but significant reduction of RER during the day (Fig. 4A–D).
FIG. 4.
Effect of HDAC inhibitors on energy balance in db/db mice. Oxygen consumption (A and B), RER (C), and heat production (D) in db/db mice treated every other day with vehicle, 25 mg/kg SAHA, or 10 mg/kg MS275 for 2 weeks prior to analysis. All parameters were measured over three days using five individually housed db/db male mice per group. Statistical comparison of repeated measurements was made using ANOVA. Data are presented as means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control group. a.u., arbitrary units; AUC, area under the curve; C and CTRL, control; MS, MS275; S, SAHA.
HDAC3 regulates oxidative metabolism in a Pgc-1α–dependent manner.
To explore the mechanism whereby class I HDAC inhibition promotes mitochondrial biogenesis, oxidative metabolism, and increased energy expenditure, we evaluated the contribution of Pgc-1α to these effects. C2C12 myotubes infected with shRNA against Pgc-1α lost the ability to increase oxidative gene expression upon SAHA or MS275 exposure (Fig. 5A), indicating that Pgc-1α is a primary mediator of the effect of class I HDAC inhibition on oxidative gene expression. Absence of Rip140 did not abolish the response to treatment with SAHA or MS275 (Supplementary Fig. 6A and B), suggesting that Rip140 is not a central mediator of the effect of these compounds.
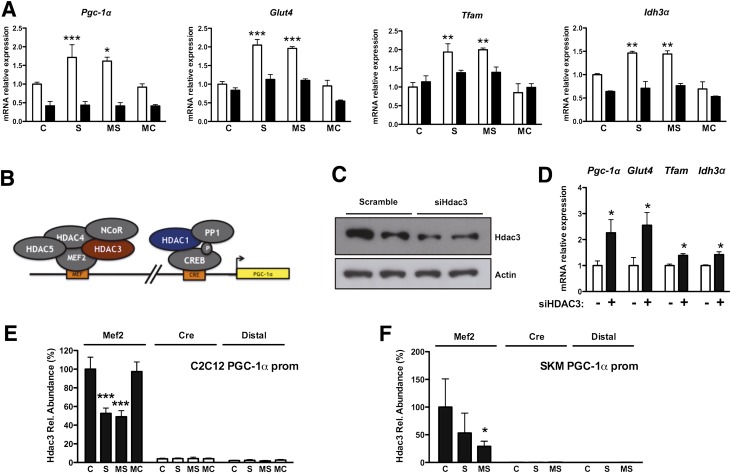
FIG. 5.
HDAC3 ablation mimics the effect of class I HDAC inhibitors in a PGC-1α–dependent manner. A: Pgc-1α, Glut4, Tfam, and Idh3α expression in C2C12 myotubes infected with adenoviruses expressing shRNA against Pgc-1α (■) or scramble control (□). Note that the effect of HDAC inhibitors is lost in the absence of Pgc-1α. B: Schematic representation of the HDACs known to be present on the Pgc-1α promoter (prom). C: Hdac3 protein levels in C2C12 myoblasts transfected with small interfering RNA against Hdac3 or control. D: Gene expression profile after silencing Hdac3 in C2C12 myoblasts. E and F: Hdac3 ChIP of C2C12 myotubes or skeletal muscle (SKM) of db/db mice treated with HDAC inhibitors. Bars represent presence of Hdac3 on the Pgc-1α promoter within the Mef2 or the cAMP-responsive element regions shown in B. A distal region was used as a negative control. Data are presented as means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control. C, control; MC, MC1568; MS, MS275; Rel., relative; S, SAHA. (A high-quality color representation of this figure is available in the online issue.)
Next, we sought to understand how class I HDAC inhibition enhances Pgc-1α expression. Two class I HDACs are recruited onto two different regions of the Pgc-1α promoter: Hdac1 represses Creb-mediated Pgc-1α transcription (23), while Hdac3, together with Hdac4, Hdac5, and the nuclear corepressor NCoR, is recruited onto members of the Mef2 family to repress transcriptional activation of Pgc-1α (24,25) (Fig. 5B). We found that a 70% reduction of Hdac3 (Fig. 5C) is sufficient to mimic the effect of class I HDAC inhibitors on the expression of Pgc-1α, Glut4, Tfam, and Idh3α (Fig. 5D). In contrast, similar silencing of Hdac1 had no effect on expression of these genes (Supplementary Fig. 6C and D). ChIP showed that treatment with SAHA or MS275 reduced Hdac3 recruitment onto the Mef-binding site in the Pgc-1α promoter (Fig. 5E). The presence of Hdac3 in the cAMP-responsive element region was barely detectable and was unaffected by compound treatment. Similar results were obtained when ChIP was performed in skeletal muscle extracts derived from db/db mice treated with SAHA or MS275: both compounds decreased Hdac3 recruitment on the Pgc-1α promoter in vivo, though MS275 had a greater effect (Fig. 5F). These results suggest that the beneficial effects of class I HDAC inhibition in muscle are primarily due to Hdac3 inhibition that results in increased Pgc-1α expression.
Inhibition of class I HDACs promotes uncoupled metabolism in brown adipose tissue.
As observed for skeletal muscle, the compounds retained their class-selective inhibitory activity in the brown adipose tissue (BAT) of treated animals (Fig. 6E). BAT of obese diabetic animals treated with SAHA or MS275 showed a significant reduction in cell size (Fig. 6A). MRI analysis showed a 13.6 and 15.7% increase of interscapular BAT in SAHA- and MS275-treated animals, respectively (Fig. 6B). db/db mice treated with MS275, but not SAHA, maintained body temperature better during an acute cold challenge, indicating that the increased BAT mass is functional (Fig. 6C). Expression of classical markers of brown fat (e.g., Ucp1, Elovl3, Dio2, Cidea, Prdm16, and the β3-adrenergic receptor Adrb3) was robustly increased (Fig. 6D and E). MS275 upregulated the expression of Pparγ, Pgc-1α, Pgc-1β, and mitochondrial biogenesis markers (e.g., Tfam, Tfb1m, and CytC) (Fig. 6F). The effects of SAHA were less pronounced, which may partially explain why animals treated with this compound did not show an increase in heat production (Fig. 4D). Similar to the in vivo scenario, primary brown adipocytes differentiated in vitro and treated with MS275 or SAHA showed higher levels of Pgc-1α, Ucp1, Adrb3, Tfam, and PPARγ mRNA (Fig. 6G) and increased mitochondrial DNA content (Fig. 6H). As a consequence, treated primary brown adipocytes showed higher basal, uncoupled, and maximal respiratory capacity (Fig. 6I).
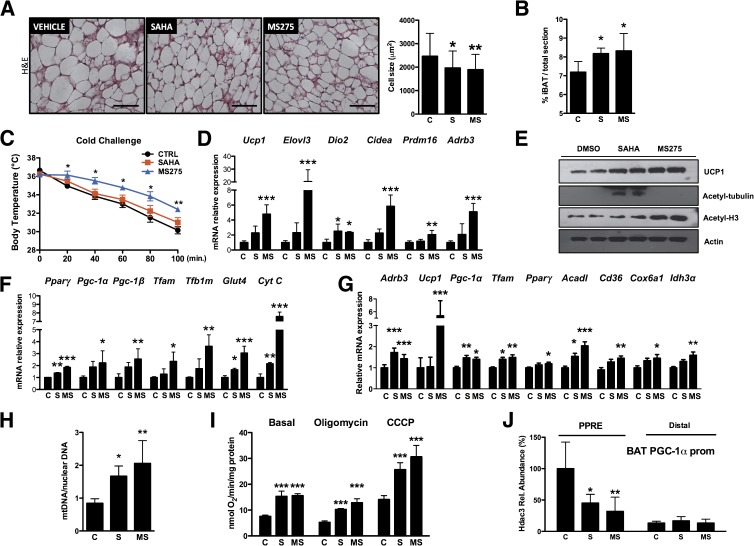
FIG. 6.
Class I HDAC inhibition enhances oxidative metabolism in brown fat. A: Hematoxylin-eosin (H&E) stain of BAT from db/db mice treated with HDAC inhibitors (bar = 100 μm) and quantification of brown adipocyte cell size. B: Magnetic resonance analysis of interscapular BAT (iBAT) in db/db mice treated for 15 days with HDAC inhibitors. C: Body temperature of db/db mice treated with vehicle, SAHA, or MS275 and exposed to temperature of 4°C. Expression of classical markers of brown fat (D) and Western blot analysis of Ucp1 and HDAC targets, acetyl-tubulin, and acetyl-histone H3 (E). F: mRNA level of genes related to mitochondrial biogenesis or selected metabolic pathways in animals treated with vehicle, SAHA, or MS275. Note that MS275 has a more pronounced effect than SAHA on all genes examined. G: mRNA level of genes related to mitochondrial biogenesis or adrenergic response in primary brown adipocytes differentiated in vitro and treated with 1 μmol/L SAHA, 1 μmol/L MS275, or vehicle. H: Quantification of mitochondrial DNA (mtDNA) in primary brown adipocytes after treatment with 1 μmol/L SAHA, 1 μmol/L MS275, or vehicle. I: Measurement of oxygen consumption at the basal level and in the presence of oligomycin (2.5 μg/mL) or carbonyl cyanide 3-chlorophenylhydrazone (CCCP) (2.4 μmol/L) in primary brown adipocytes treated for 24 h with HDAC inhibitors. J: Hdac3 ChIP assay BAT of db/db mice treated with HDAC inhibitors. Bars represent presence of Hdac3 on the Pgc-1α promoter (prom) within the PPAR-responsive element (PPRE) region. A distal region was used as a negative control. Data are presented as means ± SD. C and CTRL, control; MS, MS275; Rel., relative; S, SAHA. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control. (A high-quality color representation of this figure is available in the online issue.)
In analogy to skeletal muscle, we performed ChIP in BAT and found significantly diminished amounts of Hdac3 associated with the region containing the PPAR-responsive element of the Pgc-1α promoter in animals treated with MS275 (Fig. 6J). This observation suggests that Hdac3 inhibition, and its consequent dissociation from the promoter, may be primarily responsible for Pgc-1α induction in mice treated with MS275.
WAT treated with a class I HDAC inhibitor acquires brown fat features.
White adipocyte size was reduced in db/db mice treated with the class I–selective HDAC inhibitor (Fig. 7A and Supplementary Fig. 7A), and MRI analysis also showed an 18% reduction of WAT in these animals (Supplementary Fig. 7B). These findings were associated with increased expression of genes that regulate lipid metabolism (Fig. 7B). Pparγ expression was also dramatically upregulated, as was that of its direct targets (Fig. 7C). Class I HDAC inhibition also resulted in changes of Pgc-1α, Tfam, and CytC transcripts (Fig. 7D) and increased mitochondrial content (Fig. 7E) and density (Supplementary Fig. 7C), indicating a greater oxidative state in WAT. In addition, as may be expected from increased Pparγ signaling, expression of inflammatory markers was robustly suppressed after SAHA or MS275 administration (Supplementary Fig. 7D). Immunohistochemical analysis confirmed reduced macrophage infiltration into WAT (Fig. 7A and Supplementary Fig. 7E).
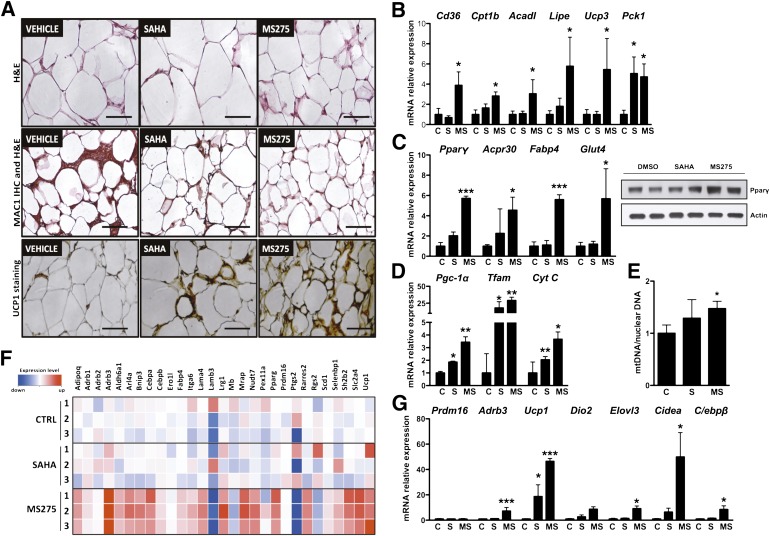
FIG. 7.
Inhibition of class I HDACs in WAT promotes oxidative metabolism, reduces inflammation, and induces the acquisition of brown fat features. A: top panel, hematoxylin-eosin (H&E) stain of WAT from db/db mice treated with HDAC inhibitors (bars = 100 μm); middle panel, Mac1 immunohistochemistry (IHC) confirms reduced macrophage infiltration (bars = 100 μm); and bottom panel, UCP1 staining in sections of WAT from treated db/db mice confirms the “browning” of WAT prompted by inhibition of class I HDACs (bars = 100 μm). B: Expression of genes involved in lipid metabolism and mitochondrial function. C: Increased expression of Pparγ and its direct targets in WAT of animals treated with MS275. D: Expression profile of genes involved in mitochondrial function after HDAC inhibitor treatment. E: Quantification of mitochondrial DNA in WAT after 23 days of treatment with SAHA, MS275, or vehicle. F: Heat map revealing the expression in MS275-treated WAT of multiple genes associated with BAT. G: Real time qPCR analysis showing that class I HDAC inhibition induces the expression of brown fat markers in white adipocytes. Data are presented as means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control. C and CTRL, control; MS, MS275; S, SAHA. (A high-quality digital representation of this figure is available in the online issue.)
Surprisingly, inhibition of class I HDACs also resulted in a dramatic increase in the expression of genes normally associated with brown fat (Fig. 7F and Supplementary Table 1). Ucp1 (∼50-fold increase), Cidea, Dio2, Adrb3, C/ebpβ, and other markers characteristic of brown adipocytes were robustly induced in WAT treated with MS275 and, to a lesser extent, with SAHA (Fig. 7A and G). As reported in other cases of “browning” of white adipose depots (26–29), the class I inhibitor appears to transcriptionally reprogram WAT toward a more oxidative phenotype characterized by a strong upregulation of Ucp1 expression.
DISCUSSION
In spite of provocative genetics data that demonstrate a role for class I and II HDACs in muscle physiology, significantly less is known about the ability of modulators of these HDAC classes to regulate systemic metabolism. Recent studies used the natural short-chain fatty acid sodium butyrate to show that this dual class I/II HDAC inhibitor increases insulin sensitivity and energy expenditure in high-fat–fed mice (12,30,31). However, sodium butyrate has a cornucopia of cellular effects, many of which are independent of its ability to block HDAC function (32). Conclusive association between chemical inhibition of specific HDAC classes and systemic energy metabolism has thus been lacking. In this study, we have used a pan-inhibitor and class I– or class II–selective synthetic HDAC inhibitors to establish the contribution of specific HDACs to whole-body metabolism.
In vitro, class I HDAC inhibition enhanced expression of critical mitochondrial regulators resulting in increased mitochondrial biogenesis and greater oxygen consumption in muscle cells and primary brown adipocytes. The lack of an effect of the class II HDAC inhibitor was surprising; however, class II HDACs are thought to have minimal deacetylase activity and to behave primarily as bridging molecules that recruit catalytically active HDAC complexes and other corepressors (33). Hence, it is possible that chemical inhibition of class II HDACs is not sufficient to interfere with assembly of silencing complexes.
Pgc-1α mediates the effects of class I HDAC inhibition, as silencing of Pgc-1α abolished the effect of these compounds on oxidative gene expression and genetic knockdown of Hdac3 recapitulated the effects seen with the chemical inhibitor of class I HDACs on Pgc-1α expression. We have shown in vitro and in vivo that the class I–selective inhibitor induces Pgc-1α transcription by blunting Hdac3 recruitment onto the Pgc-1α promoter, thus driving oxidative gene expression in skeletal muscle and BAT and browning of WAT.
Treatment of db/db mice with SAHA or MS275 resulted in enhanced glucose tolerance and insulin sensitivity, clearance of liver lipids, and decreased plasma triglycerides and free fatty acids. Given the large mass of skeletal muscle, its contribution to glucose clearance is likely to be comparable in mice treated with either SAHA or MS275, perhaps explaining why glucose and insulin levels were similarly decreased in these two groups. The reduction in hepatic steatosis contrasts with the accumulation of liver lipids seen in liver-specific Hdac3-null mice (34). This difference may be ascribed to the effect of global versus local HDAC3 inhibition: in diabetic mice, systemic HDAC3 inhibition (i.e., MS275 treatment) increases peripheral oxidative metabolism and energy expenditure, thus preventing hepatic lipid buildup. The increased PPARγ activity that we observed in WAT of MS275-treated mice also likely means that the capacity of this tissue to remove from the circulation and store excess fatty acids is enhanced, further preventing ectopic lipid deposition. The greater effect of the class I HDAC inhibitor relative to SAHA may be due to the fact that MS275 treatment resulted in significantly more dramatic changes in gene expression in adipose tissue. This could be a reflection of lower adipose tissue exposure to SAHA, perhaps a result of its pharmacokinetic profile (the in vivo half-life of SAHA is significantly shorter than that of MS275: 2 vs. 80 h) (35). This result suggests that robust suppression of class I HDACs in both skeletal muscle and adipose tissue is necessary to obtain the full metabolic benefit of inhibition of class I HDACs.
MS275 potentiates BAT function by increasing expression of markers of oxidative and uncoupled metabolism. These changes underlie the increased heat production and may contribute to the improvement of circulating lipid levels, as BAT plays a major role in triglyceride clearance (36,37). In vivo ChIP experiments indicate that inhibition of Hdac3 may be responsible for the beneficial effects of MS275 treatment in BAT.
MS275 treatment suppressed inflammatory markers and macrophage infiltration into white adipose, yet its most dramatic effect in WAT was its ability to induce robust expression of markers of BAT. This browning occurred in the absence of a change in expression of Prdm16, a major determinant of interscapular brown fat development (38). Consistent with work showing that chronic treatment of primary white adipocytes with Pparγ ligands results in the acquisition of brown adipocyte features in the absence of Prdm16 expression (26), we found that treatment with the class I HDAC inhibitor increased Pparγ expression and that of its targets in WAT. Increased Pparγ signaling in WAT could be a major determinant of the improvement of metabolic parameters seen in MS275-treated mice, for increased expression or enhanced activity of Pparγ exclusively in WAT has profound effects on systemic insulin sensitivity and lipid homeostasis (19,39). In analogy with our results in muscle and BAT, it is likely that enhanced Pparγ activity in WAT upon MS275 treatment is due to inhibition of Hdac3, as this HDAC has been shown to associate with Pparγ to block its function, though we cannot exclude the possibility of contribution of other class I HDACs to this effect (40).
Mice lacking the nuclear corepressor NCoR1 in either skeletal muscle or adipose tissue exhibit a phenotype similar to animals treated with MS275 (41,42). NCoR1 participates in transcriptional repression together with silencing mediator for retinoid and thyroid hormone receptors (SMRTs) and Hdac3 (43); thus, these observations are consistent with the notion that interfering with Hdac3 activity improves skeletal muscle and adipose tissue function.
Our results highlight the pivotal role of class I HDAC activity in the regulation of energy homeostasis. We have shown that pharmacologic inhibition of class I HDACs in the context of obesity and diabetes potentiates mitochondrial function and oxidative capacity in skeletal muscle and adipose tissue. These observations suggest that synthetic class I HDAC inhibitors may have promise in the treatment of obesity and associated disorders.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the European Union (FP6 LSHM-CT2006-037498 to M.C.), Fondazione Cariplo (2008.2511 to M.C.), the Giovanni Armenise-Harvard Foundation (to N.M.), the Italian Ministry of University (PRIN 2008 ZTN724 to E.D.F.), the American Diabetes Association (to E.S.), and the National Institutes of Health (DK-081003 to E.S.).
No potential conflicts of interest relevant to this article were reported.
A.Ga. and N.M. conceived the study, designed the experimental plan, performed most of the experiments, analyzed data, and wrote and edited the manuscript. A.F. isolated primary brown adipocytes, measured oxygen consumption, performed immunoblots and some molecular experiments, and read and edited the manuscript. E.G, F.G., and C.G. participated in the initial elaboration of the project and read and edited the manuscript. G.C. performed some biological and biochemical experiments and read and edited the manuscript. A.Gu. and E.D. performed electron microscopy analysis, contributed to image interpretation, and read and edited the manuscript. D.R. and S.V. synthesized MS275 and MC1568 and read and edited the manuscript. U.G. provided expertise for MRI experiments and read and edited the manuscript. D.C. provided suggestions for some biological experiments and read and edited the manuscript. A.M. synthesized MS275 and MC1568 and read and edited the manuscript. E.S. conceived the study, designed the experimental plan, analyzed data, wrote the manuscript, and edited the manuscript. E.D.F. and M.C. conceived the study, designed the experimental plan, analyzed data, wrote the manuscript, supervised the entire work, and edited the manuscript. E.D.F. and M.C. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
The authors thank Dr. Anastasia Kralli (Scripps Research Institute) and Dr. Mark Montminy (Salk Institute, La Jolla, CA) for adenoviruses expressing shRNA against Pgc-1α, Dr. Malcolm Parker (Imperial College of London) and Dr. Asha Seth (Imperial College of London) for providing RIP140-null cells, Dr. Franco Salerno (Istituto Neurologico “Carlo Besta”) for help in histological analysis of muscle sections, Marianna Gaman (Università degli Studi di Milano) and Paolo Monti (Università degli Studi di Milano) for electron microscopy, Dr. Anastasia Kralli (Scripps Research Institute) and Mari Gantner (Scripps Research Institute) for help with oxygen consumption measurements, and Erika Fiorino (Università degli Studi di Milano) for help in the setup of primary brown adipocytes. The authors also thank Dr. Anastasia Kralli (Scripps Research Institute) and Dr. Malcolm Parker (Imperial College of London) for discussion and comments on the manuscript. The authors thank Elda Desiderio Pinto (Università degli Studi di Milano) for administrative management.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db12-0548/-/DC1.
A.Ga., N.M., E.D.F., and M.C. contributed equally to this work.
A.Ga. and C.G. are currently affiliated with the Department of Chemical Physiology, The Scripps Research Institute, La Jolla, California.
F.G. is currently affiliated with the Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland.
See accompanying commentary, p. 685.
REFERENCES
- 1.Franks PW, Ling C. Epigenetics and obesity: the devil is in the details. BMC Med 2010;8:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ling C, Groop L. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes 2009;58:2718–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J 2003;370:737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lahm A, Paolini C, Pallaoro M, et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci USA 2007;104:17335–17340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet 2009;10:32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guarente L. Sirtuins as potential targets for metabolic syndrome. Nature 2006;444:868–874 [DOI] [PubMed] [Google Scholar]
- 7.Gao L, Cueto MA, Asselbergs F, Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol Chem 2002;277:25748–25755 [DOI] [PubMed] [Google Scholar]
- 8.McKinsey TA, Zhang C-L, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 2000;408:106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Potthoff MJ, Wu H, Arnold MA, et al. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J Clin Invest 2007;117:2459–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montgomery RL, Potthoff MJ, Haberland M, et al. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest 2008;118:3588–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Z, Singh N, Mullican SE, et al. Diet-induced lethality due to deletion of the Hdac3 gene in heart and skeletal muscle. J Biol Chem 2011;286:33301–33309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Z, Yin J, Zhang J, et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 2009;58:1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein J, Fasshauer M, Ito M, Lowell BB, Benito M, Kahn CR. β(3)-adrenergic stimulation differentially inhibits insulin signaling and decreases insulin-induced glucose uptake in brown adipocytes. J Biol Chem 1999;274:34795–34802 [DOI] [PubMed] [Google Scholar]
- 14.Guareschi S, Cova E, Cereda C, et al. An over-oxidized form of superoxide dismutase found in sporadic amyotrophic lateral sclerosis with bulbar onset shares a toxic mechanism with mutant SOD1. Proc Natl Acad Sci USA 2012;109:5074–5079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Fabiani E, Mitro N, Gilardi F, Caruso D, Galli G, Crestani M. Coordinated control of cholesterol catabolism to bile acids and of gluconeogenesis via a novel mechanism of transcription regulation linked to the fasted-to-fed cycle. J Biol Chem 2003;278:39124–39132 [DOI] [PubMed] [Google Scholar]
- 16.Mitro N, Godio C, De Fabiani E, et al. Insights in the regulation of cholesterol 7alpha-hydroxylase gene reveal a target for modulating bile acid synthesis. Hepatology 2007;46:885–897 [DOI] [PubMed] [Google Scholar]
- 17.Villena JA, Hock MB, Chang WY, Barcas JE, Giguère V, Kralli A. Orphan nuclear receptor estrogen-related receptor alpha is essential for adaptive thermogenesis. Proc Natl Acad Sci USA 2007;104:1418–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 1957;226:497–509 [PubMed] [Google Scholar]
- 19.Waki H, Park KW, Mitro N, et al. The small molecule harmine is an antidiabetic cell-type-specific regulator of PPARgamma expression. Cell Metab 2007;5:357–370 [DOI] [PubMed] [Google Scholar]
- 20.Hess-Stumpp H, Bracker TU, Henderson D, Politz O. MS-275, a potent orally available inhibitor of histone deacetylases—the development of an anticancer agent. Int J Biochem Cell Biol 2007;39:1388–1405 [DOI] [PubMed] [Google Scholar]
- 21.Mai A, Massa S, Rotili D, et al. Synthesis and biological properties of novel, uracil-containing histone deacetylase inhibitors. J Med Chem 2006;49:6046–6056 [DOI] [PubMed] [Google Scholar]
- 22.Nebbioso A, Manzo F, Miceli M, et al. Selective class II HDAC inhibitors impair myogenesis by modulating the stability and activity of HDAC-MEF2 complexes. EMBO Rep 2009;10:776–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Canettieri G, Morantte I, Guzmán E, et al. Attenuation of a phosphorylation-dependent activator by an HDAC-PP1 complex. Nat Struct Biol 2003;10:175–181 [DOI] [PubMed] [Google Scholar]
- 24.Grégoire S, Xiao L, Nie J, et al. Histone deacetylase 3 interacts with and deacetylates myocyte enhancer factor 2. Mol Cell Biol 2007;27:1280–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc Natl Acad Sci USA 2003;100:7111–7116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrovic N, Walden TB, Shabalina IG, Timmons JA, Cannon B, Nedergaard J. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J Biol Chem 2010;285:7153–7164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao L, Choi EY, Liu X, et al. White to brown fat phenotypic switch induced by genetic and environmental activation of a hypothalamic-adipocyte axis. Cell Metab 2011;14:324–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yadav H, Quijano C, Kamaraju AK, et al. Protection from obesity and diabetes by blockade of TGF-β/Smad3 signaling. Cell Metab 2011;14:67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seale P, Conroe HM, Estall J, et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest 2011;121:96–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr 2003;133(Suppl):2485S–2493S [DOI] [PubMed] [Google Scholar]
- 31.Li H, Gao Z, Zhang J, et al. Sodium butyrate stimulates expression of fibroblast growth factor 21 in liver by inhibition of histone deacetylase 3. Diabetes 2012;61:797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haberland M, Carrer M, Mokalled MH, Montgomery RL, Olson EN. Redundant control of adipogenesis by histone deacetylases 1 and 2. J Biol Chem 2010;285:14663–14670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischle W, Dequiedt F, Hendzel MJ, et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell 2002;9:45–57 [DOI] [PubMed] [Google Scholar]
- 34.Knutson SK, Chyla BJ, Amann JM, Bhaskara S, Huppert SS, Hiebert SW. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J 2008;27:1017–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer 2006;6:38–51 [DOI] [PubMed] [Google Scholar]
- 36.Bartelt A, Bruns OT, Reimer R, et al. Brown adipose tissue activity controls triglyceride clearance. Nat Med 2011;17:200–205 [DOI] [PubMed] [Google Scholar]
- 37.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev 2004;84:277–359 [DOI] [PubMed] [Google Scholar]
- 38.Seale P, Bjork B, Yang W, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature 2008;454:961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugii S, Olson P, Sears DD, et al. PPARgamma activation in adipocytes is sufficient for systemic insulin sensitization. Proc Natl Acad Sci USA 2009;106:22504–22509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fajas L, Egler V, Reiter R, et al. The retinoblastoma-histone deacetylase 3 complex inhibits PPARgamma and adipocyte differentiation. Dev Cell 2002;3:903–910 [DOI] [PubMed] [Google Scholar]
- 41.Li P, Fan W, Xu J, et al. Adipocyte NCoR knockout decreases PPARγ phosphorylation and enhances PPARγ activity and insulin sensitivity. Cell 2011;147:815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto H, Williams EG, Mouchiroud L, et al. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell 2011;147:827–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol 2001;21:6091–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hackenbrock CR, Rehn TG, Weinbach EC, Lemasters JJ. Oxidative phosphorylation and ultrastructural transformation in mitochondria in the intact ascites tumor cell. J Cell Biol 1971;51:123–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.