Abstract
Human ribonucleases A (hRNaseA) superfamily consists of thirteen members with high-structure similarities but exhibits divergent physiological functions other than RNase activity. Evolution of hRNaseA superfamily has gained novel functions which may be preserved in a unique region or domain to account for additional molecular interactions. hRNase3 has multiple functions including ribonucleolytic, heparan sulfate (HS) binding, cellular binding, endocytic, lipid destabilization, cytotoxic, and antimicrobial activities. In this study, three putative multifunctional regions, 34RWRCK38 (HBR1), 75RSRFR79 (HBR2), and 101RPGRR105 (HBR3), of hRNase3 have been identified employing in silico sequence analysis and validated employing in vitro activity assays. A heparin binding peptide containing HBR1 is characterized to act as a key element associated with HS binding, cellular binding, and lipid binding activities. In this study, we provide novel insights to identify functional regions of hRNase3 that may have implications for all hRNaseA superfamily members.
1. Introduction
Human ribonuclease A (hRNaseA) family members are encoded by unique genes located on human chromosome 14 [1]. The hRNaseA family is vertebrate cationic protein sharing conserved tertiary structure and specific enzymatic sites for RNase activity. It is in general considered to comprise eight members: RNase1 (pancreatic RNase), RNase2 (eosinophil derived neurotoxin/EDN), RNase3 (eosinophil cationic protein/ECP), RNase4, RNase5 (angiogenin), RNase6, RNase7 (skin-derived RNase), and RNase8 (divergent paralog of RNase7) [2]. Analysis of human genome sequence has revealed the existence of five additional RNases named as RNases9–13, although they appear to lose enzymatic activity [3]. All hRNaseA family members encode relatively small polypeptides of 14 to 16 kDa containing signal peptides of 20 to 28 amino acids for protein secretion. Mature hRNaseA members contain 6 to 8 cysteine residues that are crucial to hold the overall tertiary structure [4]. They possess an invariant catalytic triad including two histidines (one near the N terminus, and the other near the C terminus) and one lysine located within a conserved signature motif (CKXXNTF) [5]. These RNaseAs are catalytically active to various degrees against standard polymeric RNA substrates [6]. Interestingly, their host defense functions including cytotoxic [7, 8], helminthotoxic [9, 10], antibacterial [11, 12], and antiviral [5, 13] activities have also been reported. However, the mechanisms of noncatalytic functions of some hRNaseA members, especially the ones with low RNase activities, are poorly understood.
hRNase3 is found within the secondary granules of eosinophils and serves as a clinical asthma marker [14]. It is a multiple functional protein as the N-terminal domain1–45 possesses antipathogenic activities such as antibacterial, antihelminthic, and antiviral competencies [15–17]. In terms of key amino acids involving specific functions, Trp35 of hRNase3 interacts with cell membrane to form transmembrane pores in an artificial lipid bilayer, suggesting that lipid destruction is a crucial step in bactericidal activity [18]. In addition, Arg1, Trp10, Gln14, Lys38, and Gln40 located in antipathogenic domain are identified to bind to lipopolysaccharide (LPS) and peptidoglycan with high affinity, which may also be important for its bactericidal activity [19, 20]. Moreover, hRNase3 possesses cytotoxic activity against various mammalian cell lines including those derived from blood and epidermis [17], and Arg97 is the key residue for its cytotoxicity [21]. It is also highly associated with host inflammatory response and thus involved in tumor microenvironment to exercise its antitumor response [22–24]. Interestingly, hRNase3 reduces the infectivity of human respiratory syncytial virus in an RNase activity-dependent manner [25]. Treatment of bronchial epithelial cells with hRNase3 induces production of tumor necrosis factor alpha (TNF-α) and triggers apoptosis via a caspase-8-dependent pathway [26]. Furthermore, hRNase3 is reported to bind to a class of cell surface receptors termed as heparan sulfate proteoglycans [27] and thereby internalizes target cells through macropinocytosis [28]. Taken together, basic and aromatic residues, especially Arg and Trp, are considered to play important roles in enzymatic and other biological functions of hRNase3.
In this study, in combination with in silico analyses employing Reinforced Merging for Unique Segments (ReMUS) system, we have identified three heparin binding regions (HBRs) in hRNase3. We focused on their roles in heparin and cellular binding and endocytic and cytotoxic activities employing in vitro functional analyses. Our results showed that HBR1 (34RWRCK38) is crucial for enzymatic RNase function and serves as a major heparin binding site for endocytosis, HBR2 (73RSRFR77) contributes toward cell binding and endocytic activities, and HBR3 (101RPGRR105) plays a critical role in cytotoxicity. In addition, a noncytotoxic HBR1-derived peptide was characterized to bind to negatively charged molecules including glycosaminoglycans (GAGs) and lipids on cell surface. In summary, we have identified multifunctional regions of hRNase3, which may provide novel insights to implicate for all hRNaseA superfamily members.
2. Materials and Methods
2.1. In Silico Analysis
Unique peptides of query proteins, 13 hRNaseA family members, were identified employing Reinforced Merging for Unique Segments ReMUS system (ReMUS) (http://140.121.196.30/remus.asp) [42]. The system adopted a bottom-up strategy to extract unique patterns in each sequence at different unique levels. A fundamental unique peptide segment with previously defined pattern length, named as primary pattern was extracted at the first step. The rule of thumb for primary pattern lengths is that a shorter length setting for similar protein sequences and a longer length for dissimilar ones. The length of primary pattern in this study is set as 3 residues for hRNaseA protein family. After that Boyer Moore algorithm was performed to efficiently retrieve all primary patterns among all sequences. Each verified fundamental unique peptide segment was analyzed based on its frequencies of appearance, and its representation level of uniqueness was calculated for the merging processes in the next module. The last merging algorithm concatenated these extracted unique peptide segments through a bottom-up approach only if the primary unique peptide segments were overlapped within a sequence. The merged segments were guaranteed with unique features compared to all other protein sequences in the query dataset.
Clustal W2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) was used to align protein sequences of 13 hRNaseA family members on the basis of automatically progressive alignment mode. Protein sequences were retrieved from UniProtKB (http://www.uniprot.org/). To perform multiple-sequence alignment, gap open and extend penalties were set to 10 and 0.2, respectively. For secondary structure analysis in corresponding HBRs of hRNase1 to hRNase8, tertiary structures of hRNase1, 2, 3, 4, 5, and 7 were collected from protein data bank (PDB, http://www.rcsb.org/pdb/home/home.do), and those of hRNase6 and hRNase8 were simulated by Protein Structure Prediction Server (PS)2 (http://ps2.life.nctu.edu.tw/) using hRNase7 as a template. In addition, National Center for Biotechnology Information (NCBI) Blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was employed to compare sequence correspondent to HBPRNase3(32–41) among nonhuman primate hRNase2s and hRNase3s, as well as human RNaseA superfamily members.
2.2. Cell Line Strains
Beas-2B (ATCC number: CRL-9609) was a human lung bronchial epithelial cell line infected with an adenovirus 12-SV40 virus hybrid (Ad12SV40). Beas-2B cells were cultured in RPMI 1640 medium (Gibco, Invitrogen, USA) supplemented with 56°C heat-inactivated 10% (v/v) fetal bovine serum (FBS) (Gibco, Invitrogen, USA) and 1% (v/v) Glutamine-Penicillin-Streptomycin (Biosera). Wild-type Chinese Hamster Ovary- (CHO-) K1 and mutant cell lines CHO-pgsA-745 (lacking of all GAGs) and CHO-pgsD-677 (lacking of HS) were cultured in Vitacel Ham's F12 K medium (Sigma-Aldrich) supplemented with 10% FBS. Beas-2B and pgs D-677 cell lines were purchased from ATCC. CHO-K1 and CHO-pgsA745 cell lines were kindly provided by Dr. W.-G. Wu and Dr. C.-L. Yang, respectively (Department of Life Science, National Tsing Hua University, Taiwan). The cells were maintained at 37°C in a humidified atmosphere of 5% CO2.
2.3. Recombinant Protein Purification
Recombinant wild-type hRNase3 with a C-terminal His6 tag was expressed in Escherichia coli BL21(DE3) Codon Plus (Novagen), purified by affinity column chromatography, and refolded as previously described [27, 43]. The plasmids containing insert encoding mutant HBR1, HBR2, and HBR3 hRNase3 were generated by QuikChange site-directed mutagenesis with primer sets mtr1 forward: 5′-TATGCAGCGGCTTGCGCAAACCAAAAT-3′ and mtr1 reverse 5′-TTTGCGCAAGCCGCTGCATAATTGTTA-3′; mtr2 forward 5′-AGGCACGGCGGCGGCGGCGGCATGACAATTGTTGAG-3′ and mtr2 reverse 5′-TGTCATGCCGCCGCCGCCGCCGTGCCTTTACTCCAC-3′; mtr3 forward 5′-ATAGAAGGCGGCGGCGGCGGCGTCTGCATACCTGCA-3′ and mtr3 reverse 5′-GCAGACGCCGCCGCCGCCGCCTTCTATGTAGTTGCA-3′. For each preparation, 10 mL of overnight culture was subcultured into 1 L TB containing 100 μg/mL ampicillin, and grown at 37°C for 6 h. Isopropyl-β-D-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM. Wild-type and mutant hRNase3 were collected from inclusion bodies that were refolded by dialysis in refolding buffer (20 mM Tris, 0.5 M arginine, 0.2 mM GSSG, 2 mM EDTA, 10% glycerol, pH 8.5) at 4°C gently, concentrated by Amicon Ultra-15 (Millipore), and stored in phosphate-buffered saline (PBS).
2.4. Fluorescence-Assisted Carbohydrate Electrophoresis (FACE)
Carbohydrates were labeled with 2-aminoacridone (AMAC) according to previous study [44]. The AMAC-labeled carbohydrate and peptide were mixed and incubated at 25°C for 15 min. The complex was then loaded onto 1% agarose gels and electrophoresed in the buffer containing 40 mM Tris-acetic acid, 1 mM EDTA, pH 8.0 for 20 to 30 min. This experiment was performed in dark or under red light to prevent from light exposure. The AMAC labeled probe was observed under UV light (424 nm) and scanned by transilluminator.
2.5. RNase Activity Assay
RNase activity assay of recombinant wild-type and mutant hRNase3 were performed using yeast tRNA (Invitrogen), and bovine RNaseA (USB) was typically RNase and used as positive control. Three hundred microliters of 100 mM sodium phosphate (NaPO4) buffer, pH 7.4, and 500 μL diethylpyrocarbonate- (DEPC-) ddH2O were mixed including 50 μL of 0.05 μM RNaseA and 5 μM of wild-type and mutant hRNase3, separately. Ten microliters of 5 mg/mL yeast tRNA was added and incubated at 37°C for 0, 5, 10, and 15 min, respectively. Ice-cold 500 μL stop solution (1 : 1 (v/v) 40 mM lanthanum nitrate and 6% perchloric acid) was added and mixed for 10 min to stop reaction. Entire yeast tRNA was suspended by centrifugation at 16,100 ×g at 25°C for 5 min. One hundred microliters of supernatant in each tube was placed on to a 96-well plate. The amount of soluble tRNA in supernatant was determined by UV absorbance at 260 nm.
2.6. MTT Assay
Beas-2B cells (5 × 103cells/well) were plated in each well of a 96-well plate and allowed to incubate at 37°C for 24 h. Beas-2B cells were incubated with 0, 5, 10, 20, 40, 60, 80, and 100 μM HBPRNase3(32–41) and were incubated at 37°C for 24 h. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (10 μL of 10 mg/mL, Roche) was added to each well, and the cultures were incubated for an additional 4 h. After 24 h, the culture medium was replaced by 100 μL 5 mg/mL MTT for 3 h, then MTT solution was substituted by 100 μL 100% DMSO for 15 min. Finally, the absorbance of the sample was measured at 570 nm.
2.7. Uptake Assay and Western Blotting
Beas-2B cells were seeded into 100 mm dish at 37°C for 24 h and followed by washed with PBS and incubated with 1 μM wild type or mutant hRNase3 in serum-free RPMI 1640 medium at 37°C for 1 h. The cells were washed with PBS and trypsinized for at 37°C 15–20 min to remove surface-associated protein, and cells were lysed with Pro-prep (iNtRon). After electrophoresis, proteins were transferred to PVDF membrane (Pall) using a Bio-Rad Trans-Blot semidry transfer cell. Membranes were blocked with 3% BSA in 0.15 M NaCl, 0.5% (v/v) Tween-20, and 20 mM Tris, pH 7.4 (TBST) followed by primary antibody (anti-His antibody) diluted in 1% BSA/TBST. After washing with TBST, membranes were incubated with a secondary antibody (anti-mouse antibody) diluted in 1% BSA/TBST. Bound antibody was detected using the enhanced chemiluminescence detection system (Pierce).
2.8. Synthetic Peptides
HBPRNase3(32–41) (NYRWRCKNQNK), HBPRNase3(71–80) (NNCHRSRFRV) and HBPRNase3(96–105) (TYADRPGRRF) were synthesized by Genemed Synthesis Inc. (USA). All peptides were purified by analytical high-pressure liquid chromatography to a purity exceeding 90%. The identity of peptides was confirmed by matrix-assisted laser desorption ionization time of flight mass spectrometry.
2.9. Cell-Based Enzyme-Linked Immunosorbent Assay (cELISA)
Cells (2 × 104 cells/well) were seeded in a 96-well black plate and incubated with 5% CO2 at 37°C for 24 h. The fluorescein isothiocyanate- (FITC-) labeled HBPRNase3(32–41), HBPRNase3(71–80), and HBPRNase3(97–106) were diluted to 0, 1, 5, and 10 μM in medium for 1 h. After wash with 100 μL PBS, the plate was fixed by 2% (w/v) paraformaldehyde (PFA) in PBS for 15 min, and 100 μL 50 mM ammonium chloride in PBS was added to quench fluorescence. The plate was washed with 100 μL PBS, and 2% (w/v) BSA in PBS was added to block at room temperature for 1.5 h. The signals were measured using 488 nm laser with standard 530 nm ± 30 nm bandpass emission filter (Omega Optical, Brattleboro, VT, USA). In each set of the assays, analyses were carried out in duplicate or triplicate.
2.10. Lipid Overlay Blots
SphingoStrips (Invitrongen catalog no. S23753) and PIP Strips (Invitrongen catalog no. P23751) membranes were blocked at 37°C for 1 h with Tris-buffered Saline/0.05% Tween-20 (TBST) containing 3% (w/v) fatty acid free BSA. Membranes were incubated with 1 μg/mL hRNase3 or 0.5 μg/mL FITC-HBPRNase3(32–41) at 37°C for 2 h separately, the membranes were washed once using 0.1% BSA/TBST with a gentle shaking for 10 min. hRNase3 binding to lipids was probed by primary antibody (mouse anti-6His 1 : 5000) for 1 h. After wash, anti-mouse antibody conjugated with horseradish peroxidase (HRP) was used as secondary antibody (1 : 5000) in 1% BSA/TTBS. Finally, the immunoreactive bands were visualized by enhanced chemiluminescence (USA). For FITC-HBPRNase3(32–41) analysis, the membrane strips were initially incubated with FITC-HBPRNase3(32–41) followed by PBS wash and finally imaged using 488-nm laser with standard 530 nm ± 30 nm bandpass emission filter.
3. Result
3.1. In Silico Analysis of Unique Peptide Regions in hRNaseA Superfamily
To predict unique peptide regions possibly involved in multifunctions of hRNase3, ReMUS system was employed to analyze sequences of hRNase3 and the other 12 members of hRNaseA family. Eleven unique peptide motifs including HISLNPPR, RCTIAMRA, NYRWRC, SIRCPHNRTLNNC, RSRFRVP, PLLHCD, DLINP, PGAQN, NCTYADRPGRRFYV, DPRDSPRY, and LDTTI in hRNase3 were identified as shown in blue and light blue in Figure 1. Among which three unique segments rich in positively charged amino acids were denoted as putative HBRs including 34RWRCK38 (HBR1), 73RSRFR77 (HBR2), and 101RPGRRR105 (HBR3). Since heparin binding activity of HBR1 has been previously reported [27], the presence of 3 HBRs might possibly correlate with stronger heparin binding features of hRNase3 than other hRNaseA family members. Subsequently, Clustal W2 was applied to compare primary sequence of hRNase3 with the other hRNaseAs and alignment of putative HBRs of hRNase3 with correspondent segments of the other 12 hRNaseAs. Figure 2 revealed that HBR1 in hRNase3 was 60% identical to the corresponding segments of hRNase1, hRNase2, hRNase7, and hRNase8, but these HBRs were not conserved with any of the other hRNase family members, suggesting that these three HBRs might account for unique functions of hRNase3. Therefore, seven mutant hRNase3 constructs were generated by site-directed mutagenesis with selective alanine replacement in each HBR in order to investigate unique functions of HBR1, HBR2 and HBR3 in hRNase3 (Supplementary Figure 1 available online at http://dx.doi.org/10.1155/2013/170398).
Figure 1.

Unique peptide motifs in hRNase3 revealed from 13 hRNaseA members by Reinforced Merging for Unique Segments (ReMUS) system. Blue and light blue segments represent unique peptide motifs of hRNase3 compared to other 12 hRNaseA members. Three unique motifs 34RWRCK38, 75RSRFR79, and 101RPGRR105, representing, respectively, HBR1, HBR2, and HBR3 in hRNase3 are highlighted in yellow.
Figure 2.
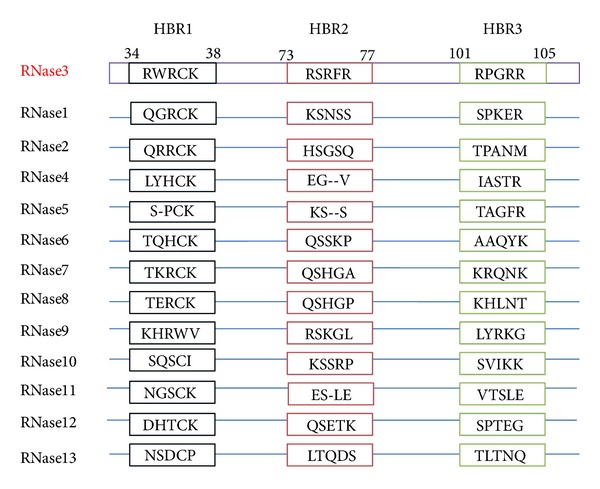
Sequences of HBR1, HBR2, and HBR3 in hRNase3 and corresponding regions of other 12 hRNaseA members. Sequences of hRNase3 and other hRNaseA superfamily members are aligned using ClustalW2 software. Putative HBR1, HBR2, and HBR3 separately located on residues 34–38, 73–77, and 101–105 are predicted from hRNase3. Residues in black, red, and green boxes indicate corresponding sequence motifs aligned with, respectively, HBR1, HBR2, and HBR3 in all hRNaseA superfamily members.
3.2. RNase Activity of Wild-Type and Mutant RNase3
After 0.05 μM bovine RNaseA, 5 μM wild-type or mutant hRNase3 was incubated with 50 μg tRNA at 37°C for 0, 5, 10, and 15 min, the amount of digested ribonucleotides was examined by UV absorbance at 260 nm. Figure 3(a) showed that RNase activity of HBR1-mt RNase3 was significantly reduced and that of HBR3-mt RNase3 was 15% less than wild type hRNase3. Besides, the RNase activity of HBR2-mt RNase3 was comparable to that of wild-type hRNase3. Moreover, double and triple mutation abolished hRNase activities of HBR12-, HBR13- and HBR123-mtRNase3 expect HBR23-mtRNase3 (Figure 3(b)). These results suggested that HBR1 of hRNase3 played a critical role in ribonucleolytic activity, mainly due to the presence of a catalytic residue Lys in the sequence.
Figure 3.
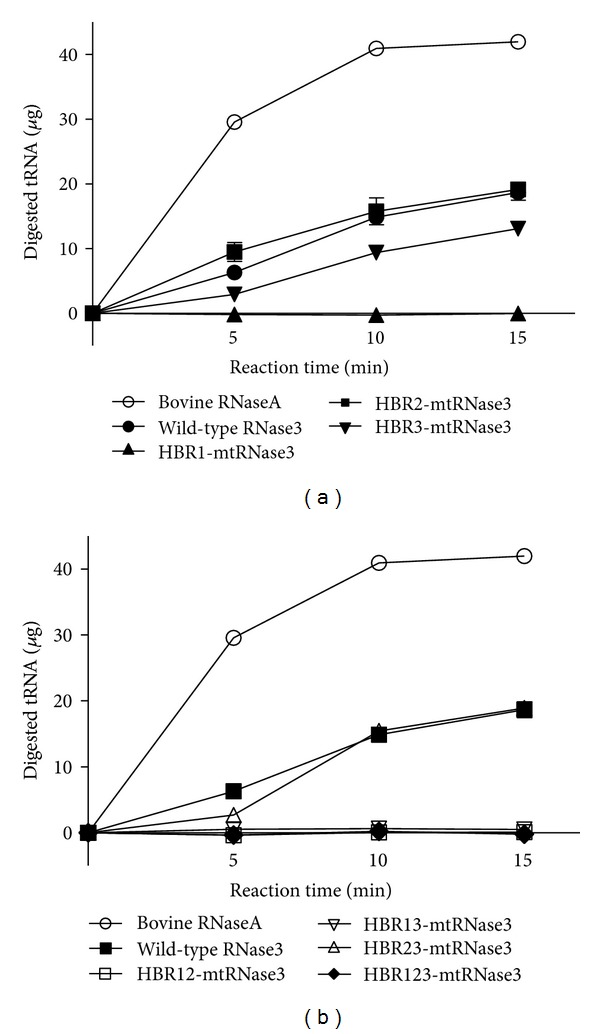
RNase activity of wild-type and mutant hRNase3. Five mg of yeast tRNA was added to 0.05 μM bovine RNaseA and 5 μM of (a) HBR1-mtRNase3, HBR2-mtRNase3, and HBR3-mtRNase3 and (b) HBR12-mtRNase, HBR13-mtRNase3, HBR23-mtRNase3, and HBR123-mtRNase3 separately to examine the RNase activity. The amount of digested ribonucleotides in supernatant was detected by monitoring OD260 and RNase activity of bovine RNaseA was set as a positive control.
3.3. Endocytosis Activity of Wild-Type and Mutant hRNase3 to Beas-2B Cells
To determine the influence of different HBRs on endocytosis activity of hRNase3, intracellular uptake assay was performed. Beas-2B cells were treated with wild type or mutant hRNase3 at 37°C for 1 h, followed by trypsin digestion for 15 min to remove surface-bound recombinant proteins before being analyzed by western blotting. When 40 μg of total cell lysates were examined with an exposure time of 1 min, only HBR2-mtRNase3 and HBR3-mtRNase3 could be detected in cytosol of Beas-2B cells (Figure 4, lanes 3 and 4). In addition, none of the double and triple HBR mutants of hRNase3 was able to enter Beas-2B cells (Figure 4, lanes 5, 6, 7, and 8). These results indicated that the importance of HBRs associated with endocytosisactivity of hRNase3 to Beas-2B cells in increasing order was HBR3, HBR2, and HBR1.
Figure 4.
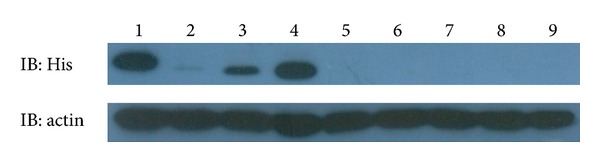
Endocytosis of wild-type and mutant hRNase3 to Beas-2B cells. Endocytosis activity of hRNase3 and its HBR mutants were assayed by western blotting employing 1 : 5000 dilution of anti-His-antibody. Actin was used to normalize the hRNase3-6His immunoblot signal. Forty micrograms of total protein in cell lysates were separated by 15% SDS-PAGE. IB: immunoblot; lane 1: wild-type hRNase3; lane 2-HBR1-mtRNase3; lane 3-HBR2-mtRNase3; lane 4-HBR3-mtRNase3; lane 5-HBR12-mtRNase3; lane 6-HBR13-mtRNase3; lane 7-HBR23-mtRNase3; lane 8-HBR123-mt RNase3; lane 9-cell only.
3.4. Cytotoxicity of Wild-Type and Mutant hRNase3 to Beas-2B Cells
Beas-2B cells were incubated individually with 15 μM of wild-type or mutant hRNase3 in serum-free medium at 37°C for 48 h followed by MTT assay. The cell viability of PBS treatment to Beas-2B cells was set as 100% to normalize that of different protein treatment. Figure 5 revealed that the viability of wild-type hRNase3-treated cells decreased to 50%, while that of HBR1-, HBR2-, HBR3-, HBR12-, HBR13-, HBR23-, and HBR123-mtRNase3-treated cells increased to 69%, 60%, 101%, 73%, 96%, 88%, and 97%, respectively. The cytotoxicity of HBR3-mtRNase3, HBR13-mtRNase3, HBR23-mtRNase3, and HBR123-mtRNase3 apparently diminished as compared to that of wild-type hRNase3, suggesting that HBR3 mutation played a major role in loss of cytotoxicity of hRNase3. It should be noted that HBR3 was located on β sheet 6 of hRNase3, hence mutation of HBR3 to alanine stretch might possibly lead to conformational change and subsequent of functional variation.
Figure 5.
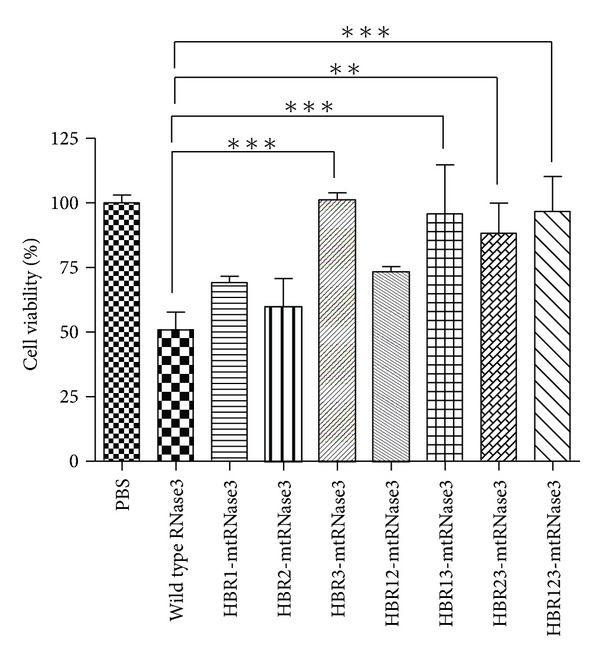
Cytotoxicity of wild-type and mutant hRNase3 to Beas-2B cells. Cytotoxicity of hRNase3 and its HBR mutants to Beas-2B cells were assessed by MTT assay. Cell viability of Beas-2B cells with PBS treatment was set as 100%. Results are presented as mean ± SD (n = 3). **P < 0.01, ***P < 0.005.
3.5. Binding Activity of Wild Type and Mutant hRNase3 to LMWH
The influence of wild type and mutant hRNase3 on LMWH binding activity was illustrated in Figure 6. Initially, AMAC-labeled LMWH was coincubated with wild-type or mutant hRNase3 individually at a molar ratio of protein to LMWH of 0.3. Free probe was separated by 1% gel electrophoresis. Relative intensities of free probes of wild type, HBR1-, HBR2-, HBR3-, HBR12-, HBR13-, HBR23-, and HBR123-mt RNase3 apparently decreased to, respectively, 17%, 31%, 15%, 9%, 76%, 59%, 76%, and 81%. The binding activity of HBR1-mtRNase3 to LMWH decreased 14% as compared to that of wild type hRNase3 (Figure 6, lanes 2 and 3), while HBR2- and HBR3-mt RNase3 did not show much difference in LMWH binding activity (Figure 6, lanes 4 and 5). However, the binding activity of double- and triple-mutant hRNase3 to LMWH decreased more significantly, especially when HBR1 was mutated (Figure 6, lanes 6, 7, 8, and 9). Therefore, in terms of facilitating hRNase3 binding to LMWH, the importance of three HBRs in decreasing order was HBR1, HBR2, and HBR3.
Figure 6.
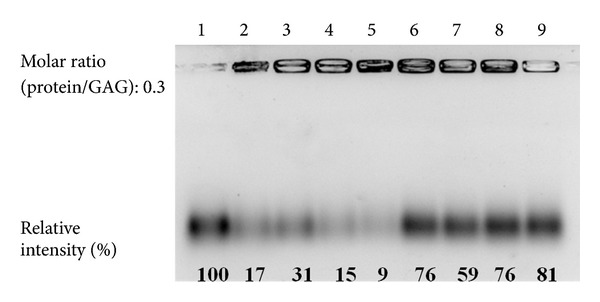
Binding activity of wild-type and mutant hRNase3 to LMWH. Heparin binding activity of hRNase3 and its HBR mutants were assessed by FACE. The value of labeled LMWH in the absence of protein was measured and set as 100%. Relative intensity of LMWH signal of each protein was normalized to LMWH only signal and was marked at the bottom; lane 1-LMWH only; Lane 2-hRNase3; lane 3-HBR1-mtRNase3; lane 4-HBR2-mtRNase3; lane 5-HBR3-mtRNase3; lane 6-HBR12-mtRNase3; lane 7-HBR13-mtRNase3; lane 8-HBR23-mtRNase3; lane 9-HBR123-mt RNase3.
3.6. Binding Activity of Synthetic Peptides FITC-HBPRNase3(32–41), FITC-HBPRNase3(71–80), and HBPRNase3(97–106) to Beas-2B Cells
FITC-labeled HBPRNase3(32–41) (NYRWRCKNQNK), HBPRNase3(71–80) (NNCHRSRFRV) and HBPRNase3(97–106) (TYADRPGRRF), peptides containing, respectively, HBR1 (34RWRCK38), HBR2 (73RSRFR77) and HBR3 (101RPGRR105) sequences were synthesized to investigate the cellular binding activity by cELISA. Figure 7 indicated that the binding activity of FITC-HBPRNase3(32–41), FITC-HBPRNase3(71–80), and HBPRNase3(97–106) to Beas-2B cells increased with elevated peptide concentration ranging from 1 μM to 10 μM in a dose-dependent manner, the binding activity of FITC-HBPRNase3(32–41) was 2 times stronger than that of FITC-HBPRNase3(71–80) and HBPRNase3(97–106). Since HBR1 was a stronger heparin binding site than HBR2 and HBR3, its multiple functions were further investigated.
Figure 7.
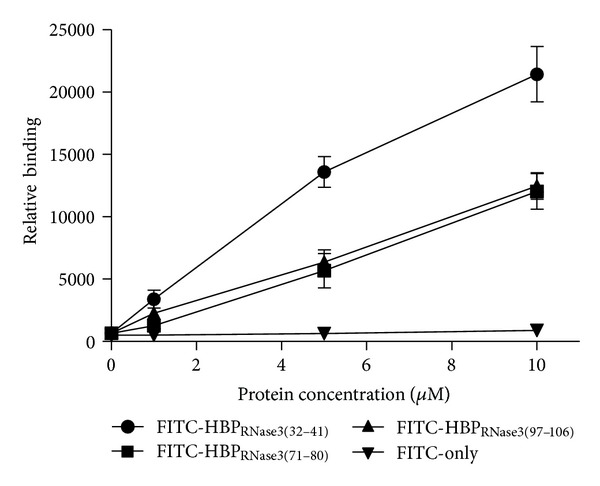
Binding activity of FITC-HBPRNase3 peptides to Beas-2B cells. Cellular binding activity of HBPs derived from hRNase3 including FITC-HBPRNase3(32–41), FITC-HBPRNase3(71–80), and FITC-HBPRNase3(97–106) were assayed using cELISA. The level of bound FITC signal was set as negative control. Data represented the means of triplicate incubations. Error bars showed standard deviations (SD) among triplicate experiments.
3.7. Cellular Binding Activity of hRNase3 and HBPRNase3(32–41)
A series of wild-type and mutant CHO cell lines with specific defect in HS or GAG biosynthesis were employed to study the binding target ofhRNase3 and HBPRNase3(32–41) on plasma membrane. Figure 8(a) showed that 1000 nM hRNase3 binding to CHO-pgsD677 and CHO-pgsA745 cells was 50% lower than that of wild-type CHO-K1 cells. Similarly, 5 μM FITC-HBPRNase3(32–41) binding to CHO-pgsD677 and CHO-pgsA745 cells significantly decreased, respectively, 40% and 50% as compared to that of wild-type CHO-K1cells (Figure 8(b)), and FITC-only was used a negative control and its relative binding intensities to each cell line as only 10%. While lacking of HS or GAGs on cell surface, relative hRNase3 and HBPRNase3(32–41) binding intensities to both CHO-pgsD677 and CHO-pgsA745 cells dropped about 50%. These results indicated that the presence of HS on the cell surface was most essential for molecular interaction with hRNase3 and HBPRNase3(32–41). In addition, cell surface components, possibly lipid moiety on A745 cells, might be involved in residual hRNase3 and HBPRNase3(32–41) binding activities.
Figure 8.
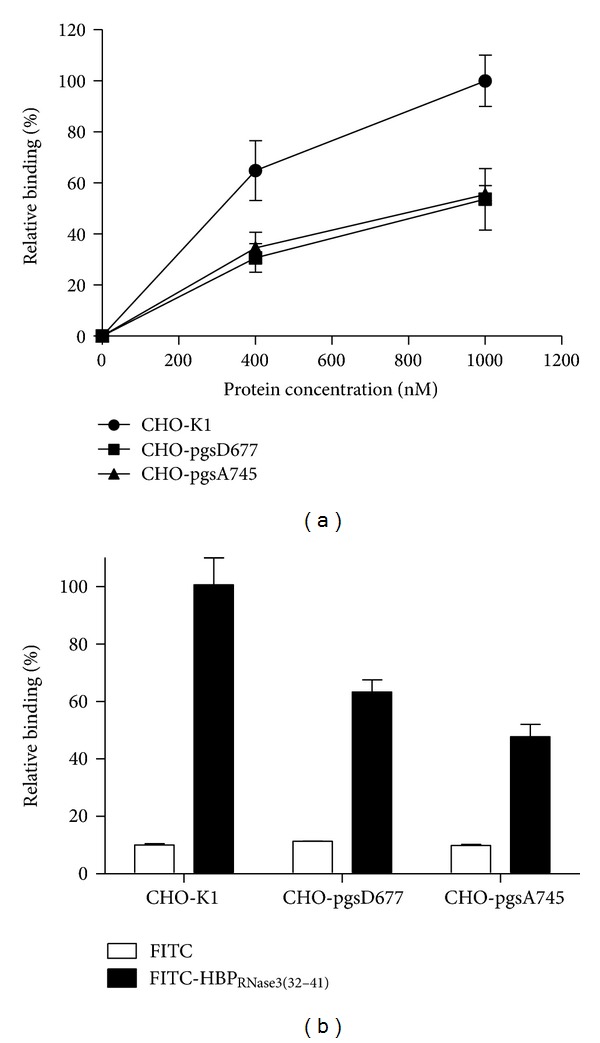
Binding activity of hRNase3 and FITC-HBPRNase3(32–41) to wild-type and mutant CHO cell lines. Cellular binding activity of hRNase3 (a) and FITC-HBPRNase3(32–41) (b) to CHO-K1, CHO-pgsD677, and CHO-pgsA745 cells were assayed using cELISA. The amount of 1000 nM hRNase3 and 5 μM FITC-HBPRNase3(32–41) bound to wild-type CHO-K1cell was set as 100% binding, respectively. The data represented the mean value of triple independent experiments and the error bar was shown as standard deviation (SD).
3.8. Identification of Specific Lipids Interacting with RNase3 and HBPRNase3(32–41)
hRNase3 has been associated with lipid membrane destabilization [45]. To investigate whether any other membrane-associated negatively charged moieties other than HS was involved in hRNase3 and HBPRNase3(32–41) binding, overlay blots (SphingoStrips and PIPStrips) containing 100 pmoles of 26 different biologically active sphingolipids and glycerophospholipids were examined [46]. Using SphingoStrips membrane, a specific interaction between sulfatide, a sulfated glycosphingolipid expressed on the cell surface [47], and refold hRNase3 (Figure 9(a), left panel) as well as FITC- HBPRNase3(32–41) (Figure 9(a), right panel) was observed. For the PIP Strips membrane, hRNase3 interacted with PI3P, PI4P, PI5P, PI (3,4) P2, PI (3,5) P2, PI (3,4,5) P3, PA, and PS (Figure 9(b), left panel). FITC- HBPRNase3(32–41) interacted with PI3P, PI4P, PI5P, PE, PA, and PS (Figure 9(b), right panel). These results indicated that FITC-HBPRNase3(32–41) possessed quite similar lipid binding pattern to hRNase3, and the HBPRNase3(32–41) involved in hRNase3 binding to plasma membrane not only through HS binding but also through direct molecular interaction with phospholipids. Our data suggested that hRNase3 interacted with more lipids than HBPRNase3(32–41), presumably due to more diverse lipid binding regions on hRNase3. Besides, variation in the exposed level and conformation of 32NYRWRCKNQN41 sequence between refold hRNase3 and HBPRNase3(32–41) might also account for the difference in lipid recognition.
Figure 9.
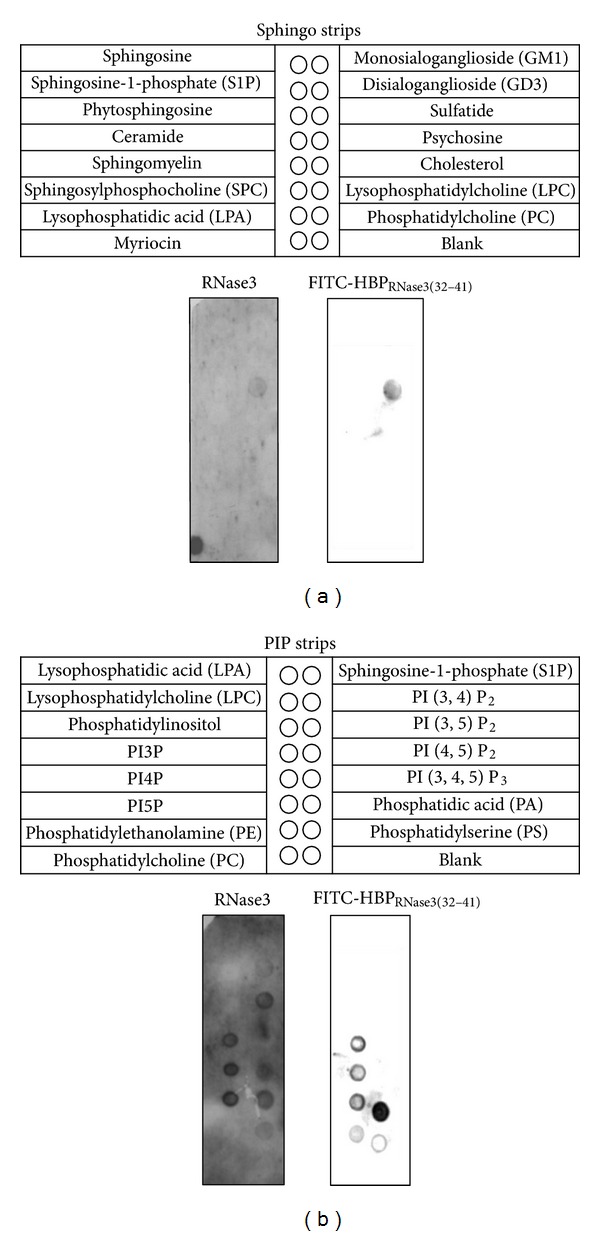
Interaction of hRNase3 and FITC-HBPRNase3(32–41) with membrane lipid fractions. Interaction of hRNase3 and FITC-HBPRNase3(32–41) on lipid overlay blot of SpingoStrips membranes (a) and PIP Strips membranes (b). Membranes were incubated with 1 μg/mL hRNase3 or 0.5 μg/mL FITC-HBPRNase3(32–41) at 37°C for 2 h, separately. The immunoreactive blot of hRNase3 treatment was visualized by enhanced chemiluminescence, and the FITC-HBPRNase3(32–41) treatment was using 488 nm laser with standard 530 nm ± 30 nm bandpass emission filter.
3.9. Cytotoxicity of hRNase3 and HBPRNase3(32–41) to Beas-2B Cells
Cytotoxicity of hRNase3 towards Beas-2B cells has been reported in previous study [27]. To examine the cytotoxic effect of HBPRNase3(32–41), Beas-2B cells were incubated with 0, 5, 10, 20, 40, 60, 80, and 100 μM HBPRNase3(32–41) in serum-free medium at 37°C for 24 h and cell viability was measured by MTT assay. Figure 10 revealed that no cytotoxicity of HBPRNase3(32–41) was detected, even if high concentration of 100 μM was applied. Thus, although HBPRNase3(32–41) conserved key heparin binding activity of hRNase3, it possessed no cytotoxic effect to mammal cell.
Figure 10.
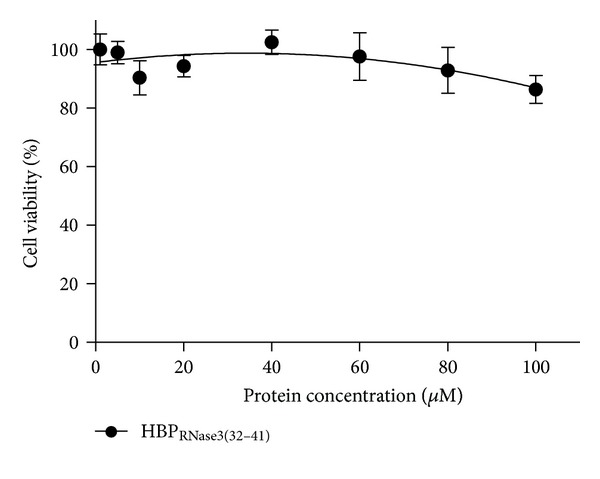
Cell viability of Beas-2B cells upon treatment with HBPRNase3(32–41). Cytotoxicity of HBPRNase3(32–41) to Beas-2B cell was assessed by MTT assay. Beas-2B cells were treated with various concentrations of HBPRNase3(32–41). The amount of cell viability of PBS treated Beas-2B cells was set as 100%.
4. Discussion
hRNaseA superfamily members share diverse protein identities to hRNase3 even though they contain conserved 3-dimensional structures and enzymatic functions. In addition to RNase activity, a variety of biological features including immune-regulatory, cytotoxic, antimicrobial, antitumor, and heparin/HS binding activities have been reported. In terms of RNase activities, hRNases1-to-8 have obvious catalytic activities; however, such function is unidentified in hRNases9-to-13 and remains to be elucidated. As for cytotoxicity, hRNases1-to-5 are harmful to various cells or organisms, but hRNases6-to-13 are indistinct. In terms of lipid binding activity, hRNase2, hRNase3, and hRNase7 have been reported to interact with lipids [30–32], while that of the other hRNaseA family members are not well studied. As for antimicrobial activity, hRNase2, hRNase3, hRNase5, hRNase7, and hRNase8 are harmful to microorganisms (Table 1). In addition, hRNase2 and hRNase3 have been elucidated to mediate immune responses [31, 32]. Taken together, comparison of hRNaseA family members indicates that only hRNase2 has similar sequences and functions to hRNase3 (Table 1).
Table 1.
Specific functions of hRNaseA superfamily.
| Function | RNase | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| Protein identity | 30% | 67% | 100% | 28% | 32% | 43% | 39% | 39% |
| RNase activity | O | O | O | O | O | O | O | O |
| Cytotoxicity | O | O | O | O | O | ND | ND | ND |
| Heparin binding motif | X | O | O | X | O | O | O | X |
| Lipid binding activity | ND | O | O | ND | ND | ND | O | ND |
| Antimicrobial activity | ND | O | O | ND | O | ND | O | O |
| Inflammatory mediators | ND | O | O | ND | ND | ND | ND | ND |
| Reference | [29] | [30] | [31, 32] | [33, 34] | [35, 36] | [37] | [38, 39] | [40, 41] |
Note: O, X, and ND, respectively, represent active, inactive, and not determined.
Structural analysis reveals that all hRNaseA family members share very similar secondary structures in three putative HBRs (Table 2). Since hRNase7 shares high primary sequence identity to hRNase6 and hRNase8, 58% and 75%, respectively, its structure was used as a template for structure simulation of hRNase6 and hRNase8 which have no resolved 3D structures yet. The HBR1 and HBR3 in all hRNaseA family members are present, respectively, in loop and β-strand conformation. The secondary structure for HBR2 of hRNase4 is β-strand, and that of the others is loop conformation. Hence, these results suggested that sequence composition, rather than secondary structure contents of each HBR-like segment in hRNaseA family members, is crucial for differential heparin binding activities. Among all hRNaseA superfamily members, hRNase2, hRNase3, and hRNase7 possess conventional heparin binding motifs in several HBRs. However, only hRNase2, hRNase3, and hRNase5 have been reported to demonstrate heparin binding activities [48, 49]. Here, three unique functional peptides encoded HBRs have been predicted in hRNase3 by ReMUS and demonstrated heparin binding properties at both molecular and cellular levels, and the correspondent HBR1 of hRNase2 has been identified with heparin binding features too. Interestingly, in the primary sequence hRNase5 positively charged residue-rich regions, that is, 31RRR33 and R70 have been reported to involve heparin binding by site-directed mutagenesis [49], and they are located pretty close to putative HBR1 and HBR2 of hRNase5 in this study. Finally, hRNase7 has been reported to be purified through a heparin affinity column despite unclear heparin binding mechanism, indicating that hRNase7 also possesses heparin binding potency. This finding will contribute to further understanding of protein-ligand interaction in hRNaseA members.
Table 2.
Secondary structures of correspondent HBRs in hRNaseA members.
| HBR1 | HBR2 | HBR3 | PDB number | |
|---|---|---|---|---|
| hRNase1 | Loop | Loop | β-strand | 2K11 |
| hRNase2 | Loop | Loop | β-strand | 2C01 |
| hRNase3 | Loop | Loop | β-strand | 2KB5 |
| hRNase4 | Loop | β-strand | β-strand | 1RNF |
| hRNase5 | Loop | Loop | β-strand | 1H53 |
| hRNase6 | Loop | Loop | β-strand | 2HKY* |
| hRNase7 | Loop | Loop | β-strand | 2HKY |
| hRNase8 | Loop | Loop | β-strand | 2HKY* |
The structures of hRNase1, 2, 3, 4, 5, and 7 are collected from protein data bank (PDB), and those of RNase6, 8 were simulated by database (PS)2—Protein Structure Prediction Server (http://ps2.life.nctu.edu.tw/). *Denotes simulated structures using hRNase7 as template.
hRNase3 is a ribonuclease and its antimicrobial function has been shown to be dependent on its enzymatic activity [5, 19, 25]. Our study has identified three HBRs including 34RWRCK38, 73RSRFR77, and 101RPGRRR105, respectively, located on loop3, loop5, and strand β4 of hRNase3 to interact with heparin. Key roles including heparin binding, cytotoxic, endocytic, and lipid binding activities were contributed by these major HBRs and have been demonstrated. hRNase3 can also modulate Beas-2B cells to release TNF-α leading to apoptosis facilitated by first step attachment on cell surface GAGs especially HS [26]. In 2010, Garcia-Mayoral group reported that the major HS binding site on hRNase3 was located on a cavity composed by A8-Q14 in helix α1, Y33-R36 in loop3, Q40-L44 in strand β1, and H128-D130 in strand β6 [18]. Here HBR1 mutants significantly diminished their RNase activity due to replacement of the crucial catalytic residue Lys38 to Ala, in addition, it also decreased heparin binding and subsequent endocytosis by replacing three basic residues, Arg34, Arg36, and Lys38 and aromatic residue Trp35 responsible for binding and disrupting microbial membrane [50]. Mutation in HBR3 (101RPGRR105) significantly decreased the cytotoxicity of hRNase3 to Beas-2B cells, revealing that the three cationic residues Arg101, Arg104, and Arg105in HBR3 might play a crucial role in exterminate Beas-2B cells.
HBPRNase3(31–41) segment was demonstrated to be involved in multiple functions of hRNase3 (Table 3). Alignment with primate RNase3 sequences has revealed that the sequence of HBPRNase3(32–41) is conserved only in RNaseA family members of higher primates such as G. Gorilla and P. troglodytes, the closest living relatives of human. Interestingly, higher primates and the closest living relatives of humans, P. troglodytes and G. Gorilla [51] showed 100% sequence identity in corresponding regions to the HBPRNase3(32–41) motif of H. sapiens RNase3; while M. fascicularis and M. nemestrina showed 80% sequence identity with HBPRNase3(32–41) motif of H. sapiens RNase3 but 100% identity with H. sapiens RNase2. In addition, the motif sequence of P. pygmaeus was 70% and 90% identical to that of hRNase3 and hRNase2, respectively (Table 4). Our results strongly supported the notion that hRNase3 was generated from an RNase2/RNase3 precursor gene about 30 million years ago, at an evolutionary rate among the highest primate genes [52].
Table 3.
Correlation of HBR motifs and characteristic functions of hRNase3.
| Location | Function | |||||
|---|---|---|---|---|---|---|
| RNase | Cytotoxicity | Cell binding activity | HS binding activity | Endocytic activity | Lipid binding activity | |
| RNase3 | O | O | O | O | O | O |
| HBR1-mt RNase3 | No activity | Decrease 50% | Increase 50% | Decrease 40% | No activity | |
| HBR2-mt RNase3 | Similar | Similar | Decrease 20% | Similar | Decrease 50% | |
| HBR3-mt RNase3 | Decrease 30% | No activity | Decrease 20% | Similar | Similar | |
| HBPRNase3(32–41) | X | X | O | O | ND | O |
Note: O, X, and ND, respectively, represent active, inactive, and not determined.
Table 4.
Comparison of HBPRNase3(32–41) motif among hRNase3 and other primate RNase3s and RNase2s.
| Protein | Organism | Protein identity | HBPRNase3(32–41) identity | Sequence |
|---|---|---|---|---|
| RNase3 | Homo sapiens | 100% | 100% | N YRWRCKNQN |
| Pan troglodytes | 97% | 100% | NYRWRCKNQN | |
| Gorilla gorilla | 97% | 100% | NYRWRCKNQN | |
| Macaca fascicularis | 88% | 80% | NYQRRCKNQN | |
| Macaca nemestrina | 88% | 80% | NYQRRCKNQN | |
| Pongo pygmaeus | 88% | 70% | NYQRRCKDQN | |
|
| ||||
| RNase2 | Homo sapiens | 67% | 80% | NYQRQCKNQN |
| Pan troglodytes | 67% | 80% | NYQRQCKNQN | |
| Gorilla gorilla | 69% | 80% | NYQRQCKNQN | |
| Macaca fascicularis | 67% | 80% | NYQRQCKNQN | |
| Macaca nemestrina | 66% | 80% | NYQRQCKNQN | |
| Pongo pygmaeus | 68% | 70% | NFQRRCKNQN | |
Sequence identity was calculated employing National Center for Biotechnology Information Blast (NCBI Blast: http://blast.ncbi.nlm.nih.gov/Blast.cgi).
To date, 13 members of the RNaseA superfamily have been identified in humans; however, the functions of newly identified human hRNases9–13 remain unclear [53]. Blast analysis of HBPRNase3(32–41) motif among hRNase3 and other hRNaseA members was shown in Table 5, in which only hRNase2 and hRNase8 showed, respectively, 80% and 50% sequence identity, while the others showed lower than 50% identity with the HBPRNase3(32–41) of hRNase3. In summary, HBPRNase3 motif was not a conserved motif among hRNaseA superfamily, but a specific motif being present only in higher primates.
Table 5.
Comparison of HBPRNase3(32–41) motifs among human RNaseA superfamily members.
| Name | Accession number | Protein identity | HBPRNase3(32–41) identity | Sequence |
|---|---|---|---|---|
| RNase1 | P07998 | 30% | 40% | M TQGRCKPVN |
| RNase2 | P10153 | 67% | 80% | NYQRRCKNQN |
| RNase3 | P12724 | 100% | 100% | NYRWRCKNQN |
| RNase4 | P34096 | 28% | 30% | MTLYHCKRFN |
| RNase5 | P03950 | 32% | 30% | LTSP - CKDIN |
| RNase6 | Q93061 | 43% | 20% | KYFGRSLELY |
| RNase7 | Q9H1E1 | 39% | 40% | KHTKRCKDLN |
| RNase8 | Q8TDE3 | 39% | 50% | KYTERCKDLN |
| RNase9 | P60153 | 23% | 20% | YYKHRWVAEH |
| RNase10 | Q5GAN6 | 26% | 10% | EPSQSCIAQY |
| RNase11 | Q5GAN5 | 30% | 30% | EANGSCKWSN |
| RNase12 | Q5GAN4 | 23% | 20% | EPDHTCKKEH |
| RNase13 | Q5GAN3 | 25% | 10% | MQNSDCPKIH |
Sequence identity was calculated employing National Center for Biotechnology Information Blast (NCBI Blast: http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Herein we reported that HBPRNase3(32–41) accounted for major cellular binding activity than HBPRNase3(71–80) and HBPRNase3(97–106). Interestingly, although binding of HBPRNase3(32–41) was severely impaired in CHO-pgsD677 cells which had no HS but expressed 3-fold more CS than wild-type CHO-K1 cells, residual cellular binding activities of HBPRNase3(32–41) to CHO-pgsD677 and CHO-pgsA745 cells strongly implied that HBPRNase3(32–41) possessed certain interaction to cell surface CS. In addition, both hRNase3 and HBPRNase3(32–41) showed quite similar binding activities to membrane lipids.
5. Conclusion
In this study, we identify three functionally important HBRs in hRNase3, including 34RWRCK38 (HBR1), 75RSRFR79 (HBR2), and 101RPGRR105 (HBR3). HBR1 (34RWRCK38) in hRNase3 is required for RNase activity and serves as a major heparin binding site. HBR2 (73RSRFR77) contributes to both cell binding and endocytic activities, and HBR3 (101RPGRR105) plays an important role in cytotoxicity. Moreover, a noncytotoxic HBR1-derived peptide prefers to interact with negatively charged molecules including heparan sulfate, chondroitin sulfate and lipids present on cell surface. Understanding of the roles of key functional residues of hRNase3 in ribonucleolytic, heparin binding, cellular binding, endocytic, cytotoxic, and lipid binding activities provides informative correlation among sequence, structure, and functional features of hRNaseA family members. This finding will contribute to further investigation of molecular mechanisms and multiple functions of hRNaseA family in general.
Supplementary Material
Supplementary Figure 1: showed that mutant hRNase3 constructs containing alanine replacement in HBRs were generated by site-directed mutagenesis. Replacement with alanine stretch in HBR1, HBR2 and HBR3 were illustrated in black, red, and green boxes, respectively.
Authors' Contribution
P.-C. Lien and P.-H. Kuo contributed equally to this work.
Acknowledgments
The authors thank Mr. Ta-Jen Hung, Dr. Chao-Sheng Cheng, and Dr. Wen-Chi Cheng for critical comments and proofreading. No other potential conflict of interests relevant to this paper was reported. This work was supported by National Tsing Hua University (NTHU100N7051E1 and NTHU101N2051E1), National Science Counsil (NSC101-2622-B-007-001-CC1 and NSC101-2325-B-007-002), and Chang-Gung Memorial Hospital-National Tsing Hua University Joint Research Program (NTHU100N2710E1 and NTHU100N2711E1) to M. D.-T. Chang, and National Science Council (NSC98-2320-B-007-003-MY3) to Y.-K. Lai. P.-H. Kuo and C.-J. Chen were awarded a scholarship sponsored by Shen's Culture and Education Foundation.
References
- 1.Beintema JJ, Kleineidam RG. The ribonuclease A superfamily: general discussion. Cellular and Molecular Life Sciences. 1998;54(8):825–832. doi: 10.1007/s000180050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dyer KD, Rosenberg HF. The RNase a superfamily: generation of diversity and innate host defense. Molecular Diversity. 2006;10(4):585–597. doi: 10.1007/s11030-006-9028-2. [DOI] [PubMed] [Google Scholar]
- 3.Cho S, Beintema JJ, Zhang J. The ribonuclease A superfamily of mammals and birds: identifying new members and tracing evolutionary histories. Genomics. 2005;85(2):208–220. doi: 10.1016/j.ygeno.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Rosenberg HF. The eosinophil ribonucleases. Cellular and Molecular Life Sciences. 1998;54(8):795–803. doi: 10.1007/s000180050208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rugeles MT, Trubey CM, Bedoya VI, et al. Ribonuclease is partly responsible for the HIV-1 inhibitory effect activated by HLA alloantigen recognition. AIDS. 2003;17(4):481–486. doi: 10.1097/00002030-200303070-00002. [DOI] [PubMed] [Google Scholar]
- 6.Sorrentino S. The eight human “canonical” ribonucleases: molecular diversity, catalytic properties, and special biological actions of the enzyme proteins. FEBS Letters. 2010;584(11):2194–2200. doi: 10.1016/j.febslet.2010.04.018. [DOI] [PubMed] [Google Scholar]
- 7.Maeda T, Kitazoe M, Tada H, et al. Growth inhibition of mammalian cells by eosinophil cationic protein. European Journal of Biochemistry. 2002;269(1):307–316. doi: 10.1046/j.0014-2956.2001.02653.x. [DOI] [PubMed] [Google Scholar]
- 8.Lee JE, Raines RT. Cytotoxicity of bovine seminal ribonuclease: monomer versus dimer. Biochemistry. 2005;44(48):15760–15767. doi: 10.1021/bi051668z. [DOI] [PubMed] [Google Scholar]
- 9.Hamann KJ, Gleich GJ, Checkel JL, Loegering DA, McCall JW, Barker RL. In vitro killing of microfilariae of Brugia pahangi and Brugia malayi by eosinophil granule proteins. Journal of Immunology. 1990;144(8):3166–3173. [PubMed] [Google Scholar]
- 10.Hamann KJ, Barker RL, Loegering DA, Gleich GJ. Comparative toxicity of purified human eosinophil granule proteins for newborn larvae of Trichinella spiralis . Journal of Parasitology. 1987;73(3):523–529. [PubMed] [Google Scholar]
- 11.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nature Immunology. 2003;4(3):269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 12.Rudolph B, Podschun R, Sahly H, Schubert S, Schröder JM, Harder J. Identification of RNase 8 as a novel human antimicrobial protein. Antimicrobial Agents and Chemotherapy. 2006;50(9):3194–3196. doi: 10.1128/AAC.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee-Huang S, Huang PL, Sun Y, et al. Lysozyme and RNases as anti-HIV components in β-core preparations of human chorionic gonadotropin. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(6):2678–2681. doi: 10.1073/pnas.96.6.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sorkness C, McGill K, Busse WW. Evaluation of serum eosinophil cationic protein as a predictive marker for asthma exacerbation in patients with persistent disease. Clinical and Experimental Allergy. 2002;32(9):1355–1359. doi: 10.1046/j.1365-2222.2002.01471.x. [DOI] [PubMed] [Google Scholar]
- 15.Torrent M, de la Torre BG, Nogués VM, Andreu D, Boix E. Bactericidal and membrane disruption activities of the eosinophil cationic protein are largely retained in an N-terminal fragment. Biochemical Journal. 2009;421(3):425–434. doi: 10.1042/BJ20082330. [DOI] [PubMed] [Google Scholar]
- 16.Torrent M, Badia M, Moussaoui M, Sanchez D, Nogués MV, Boix E. Comparison of human RNase 3 and RNase 7 bactericidal action at the gram-negative and gram-positive bacterial cell wall. FEBS Journal. 2010;277(7):1713–1725. doi: 10.1111/j.1742-4658.2010.07595.x. [DOI] [PubMed] [Google Scholar]
- 17.Navarro S, Aleu J, Jiménez M, Boix E, Cuchillo CM, Nogués MV. The cytotoxicity of eosinophil cationic protein/ribonuclease 3 on eukaryotic cell lines takes place through its aggregation on the cell membrane. Cellular and Molecular Life Sciences. 2008;65(2):324–337. doi: 10.1007/s00018-007-7499-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Mayoral MF, Moussaoui M, de la Torre BG, et al. NMR structural determinants of eosinophil cationic protein binding to membrane and heparin mimetics. Biophysical Journal. 2010;98(11):2702–2711. doi: 10.1016/j.bpj.2010.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torrent M, Navarro S, Moussaoui M, Nogués MV, Boix E. Eosinophil cationic protein high-affinity binding to bacteria-wall lipopolysaccharides and peptidoglycans. Biochemistry. 2008;47(11):3544–3555. doi: 10.1021/bi702065b. [DOI] [PubMed] [Google Scholar]
- 20.Pulido D, Moussaoui M, Andreu D, Nogues MV, Torrent M, Boix E. Antimicrobial action and cell agglutination by the eosinophil cationic protein are modulated by the cell wall lipopolysaccharide structure. Antimicrobial Agents and Chemotherapy. 2012;56(5):2378–2385. doi: 10.1128/AAC.06107-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trulson A, Byström J, Engström A, Larsson R, Venge P. The functional heterogeneity of eosinophil cationic protein is determined by a gene polymorphism and post-translational modifications. Clinical and Experimental Allergy. 2007;37(2):208–218. doi: 10.1111/j.1365-2222.2007.02644.x. [DOI] [PubMed] [Google Scholar]
- 22.Sugihara R, Kumamoto T, Ito T, Ueyama H, Toyoshima I, Tsuda T. Human muscle protein degradation in vitro by eosinophil cationic protein (ECP) Muscle and Nerve. 2001;24(12):1627–1634. doi: 10.1002/mus.1198. [DOI] [PubMed] [Google Scholar]
- 23.Pereira MC, Oliveira DT, Kowalski LP. The role of eosinophils and eosinophil cationic protein in oral cancer: a review. Archives of Oral Biology. 2011;56(4):353–358. doi: 10.1016/j.archoralbio.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 24.Zheutlin LM, Ackerman SJ, Gleich GJ, Thomas LL. Stimulation of basophil and rat mast cell histamine release by eosinophil granule-derived cationic protein. Journal of Immunology. 1984;133(4):2180–2185. [PubMed] [Google Scholar]
- 25.Domachowske JB, Dyer KD, Adams AG, Leto TL, Rosenberg HF. Eosinophil cationic protein/RNase 3 is another RNase A-family ribonuclease with direct antiviral activity. Nucleic Acids Research. 1998;26(14):3358–3363. doi: 10.1093/nar/26.14.3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang KC, Lo CW, Fan TC, et al. TNF-α mediates eosinophil cationic protein-induced apoptosis in BEAS-2B cells. BMC Cell Biology. 2010;11, article 6 doi: 10.1186/1471-2121-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan TC, Fang SL, Hwang CS, et al. Characterization of molecular interactions between eosinophil cationic protein and heparin. The Journal of Biological Chemistry. 2008;283(37):25468–25474. doi: 10.1074/jbc.M803516200. [DOI] [PubMed] [Google Scholar]
- 28.Fan TC, Chang HT, Chen IW, Wang HY, Chang MDT. A heparan sulfate-facilitated and raft-dependent macropinocytosis of eosinophil cationic protein. Traffic. 2007;8(12):1778–1795. doi: 10.1111/j.1600-0854.2007.00650.x. [DOI] [PubMed] [Google Scholar]
- 29.Johnson RJ, McCoy JG, Bingman CA, Phillips GN, Jr., Raines RT. Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein. Journal of Molecular Biology. 2007;368(2):434–449. doi: 10.1016/j.jmb.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gleich GJ, Loegering DA, Bell MP, Checkel JL, Ackerman SJ, McKean DJ. Biochemical and functional similarities between human eosinophil-derived neurotoxin and eosinophil cationic protein: homology with ribonuclease. Proceedings of the National Academy of Sciences of the United States of America. 1986;83(10):3146–3150. doi: 10.1073/pnas.83.10.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gabay JE, Scott RW, Campanelli D, et al. Antibiotic proteins of human polymorphonuclear leukocytes. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(14):5610–5614. doi: 10.1073/pnas.86.14.5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang D, Chen Q, Rosenberg HF, et al. Human ribonuclease A superfamily members, eosinophil-derived neurotoxin and pancreatic ribonuclease, induce dendritic cell maturation and activation. Journal of Immunology. 2004;173(10):6134–6142. doi: 10.4049/jimmunol.173.10.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seno M, Futami JI, Tsushima Y, et al. Molecular cloning and expression of human ribonuclease 4 cDNA. Biochimica et Biophysica Acta. 1995;1261(3):424–426. doi: 10.1016/0167-4781(95)00040-n. [DOI] [PubMed] [Google Scholar]
- 34.Liddo RDI, Dalzoppo D, Baiguera S, et al. In vitro biological activity of bovine milk ribonuclease-4. Molecular Medicine Reports. 2010;3(1):127–132. doi: 10.3892/mmr_00000229. [DOI] [PubMed] [Google Scholar]
- 35.Saxena SK, Rybak SM, Davey RT, Jr., Youle RJ, Ackerman EJ. Angiogenin is a cytotoxic, tRNA-specific ribonuclease in the RNase A superfamily. The Journal of Biological Chemistry. 1992;267(30):21982–21986. [PubMed] [Google Scholar]
- 36.Tsirakis G, Pappa CA, Kanellou P, et al. Role of platelet-derived growth factor-AB in tumour growth and angiogenesis in relation with other angiogenic cytokines in multiple myeloma. Hematological Oncology. 2012;30(3):131–136. doi: 10.1002/hon.1014. [DOI] [PubMed] [Google Scholar]
- 37.Rosenberg HF, Dyer KD. Molecular cloning and characterization of a novel human ribonuclease (RNase k6): increasing diversity in the enlarging ribonuclease gene family. Nucleic Acids Research. 1996;24(18):3507–3513. doi: 10.1093/nar/24.18.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torrent M, Sánchez D, Buzón V, Nogués MV, Cladera J, Boix E. Comparison of the membrane interaction mechanism of two antimicrobial RNases: RNase 3/ECP and RNase 7. Biochimica et Biophysica Acta. 2009;1788(5):1116–1125. doi: 10.1016/j.bbamem.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Harder J, Schröder JM. RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. The Journal of Biological Chemistry. 2002;277(48):46779–46784. doi: 10.1074/jbc.M207587200. [DOI] [PubMed] [Google Scholar]
- 40.Huang YC, Lin YM, Chang TW, et al. The flexible and clustered lysine residues of human ribonuclease 7 are critical for membrane permeability and antimicrobial activity. The Journal of Biological Chemistry. 2007;282(7):4626–4633. doi: 10.1074/jbc.M607321200. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, Dyer KD, Rosenberg HF. RNase 8, a novel RNase A superfamily ribonuclease expressed uniquely in placenta. Nucleic Acids Research. 2002;30(5):1169–1175. doi: 10.1093/nar/30.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pai TW, Chang MDT, Tzou WS, et al. REMUS: a tool for identification of unique peptide segments as epitopes. Nucleic Acids Research. 2006;34:W198–W201. doi: 10.1093/nar/gkl188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boix E, Nikolovski Z, Moiseyev GP, Rosenberg HF, Cuchillo CM, Nogués MV. Kinetic and product distribution analysis of human eosinophil cationic protein indicates a subsite arrangement that favors exonuclease-type activity. The Journal of Biological Chemistry. 1999;274(22):15605–15614. doi: 10.1074/jbc.274.22.15605. [DOI] [PubMed] [Google Scholar]
- 44.Calabro A, Benavides M, Tammi M, Hascall VC, Midura RJ. Microanalysis of enzyme digests of hyaluronan and chondroitin/dermatan sulfate by fluorophore-assisted carbohydrate electrophoresis (FACE) Glycobiology. 2000;10(3):273–281. doi: 10.1093/glycob/10.3.273. [DOI] [PubMed] [Google Scholar]
- 45.Young JDE, Peterson CGB, Venge P, Cohn ZA. Mechanims of membrane damage mediated by human eosinophil cationic protein. Nature. 1986;321(6070):613–616. doi: 10.1038/321613a0. [DOI] [PubMed] [Google Scholar]
- 46.D’Angelo I, Welti S, Bonneau F, Scheffzek K. A novel bipartite phospholipid-binding module in the neurofibromatosis type 1 protein. EMBO Reports. 2006;7(2):174–179. doi: 10.1038/sj.embor.7400602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishizuka I. Chemistry and functional distribution of sulfoglycolipids. Progress in Lipid Research. 1997;36(4):245–319. doi: 10.1016/s0163-7827(97)00011-8. [DOI] [PubMed] [Google Scholar]
- 48.Hung SC, Lu XA, Lee JC, et al. Synthesis of heparin oligosaccharides and their interaction with eosinophil-derived neurotoxin. Organic and Biomolecular Chemistry. 2012;10(4):760–772. doi: 10.1039/c1ob06415k. [DOI] [PubMed] [Google Scholar]
- 49.Soncin F, Strydom DJ, Shapiro R. Interaction of heparin with human angiogenin. The Journal of Biological Chemistry. 1997;272(15):9818–9824. doi: 10.1074/jbc.272.15.9818. [DOI] [PubMed] [Google Scholar]
- 50.Carreras E, Boix E, Rosenberg HF, Cuchillo CM, Nogués MV. Both aromatic and cationic residues contribute to the membrane-lytic and bactericidal activity of eosinophil cationic protein. Biochemistry. 2003;42(22):6636–6644. doi: 10.1021/bi0273011. [DOI] [PubMed] [Google Scholar]
- 51.Miyamoto MM, Koop BF, Slightom JL, Goodman M, Tennant MR. Molecular systematics of higher primates: genealogical relations and classification. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(20):7627–7631. doi: 10.1073/pnas.85.20.7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenberg HF, Dyer KD, Tiffany HL, Gonzalez M. Rapid evolution of a unique family of primate ribonuclease genes. Nature Genetics. 1995;10(2):219–223. doi: 10.1038/ng0695-219. [DOI] [PubMed] [Google Scholar]
- 53.Cheng GZ, Li JY, Li F, Wang HY, Shi GX. Human ribonuclease 9, a member of ribonuclease A superfamily, specifically expressed in epididymis, is a novel sperm-binding protein. Asian Journal of Andrology. 2009;11(2):240–251. doi: 10.1038/aja.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: showed that mutant hRNase3 constructs containing alanine replacement in HBRs were generated by site-directed mutagenesis. Replacement with alanine stretch in HBR1, HBR2 and HBR3 were illustrated in black, red, and green boxes, respectively.


